Abstract
Monitoring wastewater for the traces of viruses allows effective surveillance of entire communities, including symptomatic and asymptomatic infected individuals, providing information on whether a specific pathogen is circulating in a population. In the context of the COVID-19 pandemic, 261 wastewater samples from six communities of the province of Córdoba, Argentina were analyzed. From mid-May 2020 to the end of August 2021, raw sewage samples were collected from the central network pipe that enters into the Wastewater Treatment Plants (WWTP) in Córdoba city and five communities in the Punilla Valley. SARS-CoV-2 was concentrated by using the polyethylene glycol-6000 precipitation method. Viral genomes were extracted from concentrated samples, and N- and E-SARS-CoV-2 genes were detected by using real time RT-PCR. Wastewater samples that resulted positive for SARS-CoV-2 genome detection were subjected to viral variants of concern (VOCs) identification by real time RT-PCR. Overall, just by using the identification of the N gene or E gene, the rates of viral genome detection were 43.4% (86/198) and 51.5% (102/198) respectively, and by using both methodologies (positivity criterion: detection of N and / or E gene), the detection rate was 71.2% (141/198). Thereby, the optimal strategy to study the SARS-CoV-2 genome in wastewater would be the use of the combined detection of both genes. Detection of SARS-CoV-2 variants in wastewater reflected their circulation in the community, showing no VOCs detection in the first COVID-19 wave and their co-circulation with Gamma, Alpha and Delta VOCs during 2021. Therefore, SARS-CoV-2 Wastewater Based Epidemiology (WBE) described the introduction, permanence and/or the co-circulation of viral variants in the community. In geographical areas with a stable population, SARS-CoV-2 WBE could be used as an early warning sign of new COVID-19 cases, whereas in localities with a low number of inhabitants and high tourist influx, WBE may only be useful to reflect the circulation of the virus in the community. Overall, the monitoring of SARS-CoV-2 in wastewater can become a silent sentinel of the trend of viral circulation in the community, providing supplementary information for clinical surveillance to support public health measures.
Keywords: SARS-CoV-2, COVID-19, Wastewater-based epidemiology, SARS-COV-2 variants in wastewater, Pandemic
Abbreviations: ACE2, angiotensin converting enzyme-2; COVID-19, coronavirus disease 19; E, envelope E; GI, gastrointestinal; N, nucleocapsid; Orf1ab, open reading fragment 1ab; PEG-6000, polyethylene glycol-6000; PV, polioviruses; RT-PCR, retrotranscriptase polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; VOCs, viral variants of concern; VOI, variants of interest; WBE, wastewater-based epidemiology; WHO, The World Health Organization; WWTF, wastewater treatment facilities; WWTP, wastewater treatment plants
Graphical Abstract
1. Introduction
Since the coronavirus disease 19 (COVID-19) pandemic first broke out in December 2019, it has disrupted lives around the world for more than a year. The number of infected people until November 15th, 2021 (the time this paper was written) was approximately 250 million worldwide and the death toll reached more than 5 million people. Whereas fever and respiratory tract manifestations such as cough are the most common reported symptoms in patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), some patients present gastrointestinal (GI) symptoms like diarrhea, loss of appetite, nausea/vomiting and abdominal pain (Su S, 2020) (Megyeri K, 2021), (Ramachandran P, 2020), (Redd WD, 2020), (Calitri, 2021). This is because SARS-CoV-2 has been shown to enter the cell by the Angiotensin Converting Enzyme-2 (ACE2) receptor (Hikmet, 2020), (Zou X, 2020) which is widely expressed in many types of cells and tissues of the GI tract. Specifically, SARS-CoV-2 can infect and replicate in esophageal cells and enterocytes, leading to direct damage to the intestinal epithelium. Different studies report that individuals infected with SARS-CoV-2 can excrete the virus by feces (Tian Y, 2020), (Wu Y, 2020), (Wang W, 2020). It is estimated that feces of people infected with SARS-CoV-2 contain 104-108 RNA copies per gram up to approximately three weeks after the onset of symptoms, with a mean excretion time of 17 days, regardless of the presence or absence of the GI and other COVID-19 symptoms (Sender R, 2021), (Pan Y, 2020), (Xu Z, 2021), (Akiyama Y, 2021) (Yuan C, 2021), (Guo M, 2021).
Monitoring wastewater for traces of enteric viruses allows effective surveillance of entire communities (including symptomatic and asymptomatic infected individuals), providing information on whether the pathogen is circulating in the population. This kind of surveillance, named wastewater-based epidemiology (WBE), has a long tradition in public health, especially in the case of polioviruses (PV) (WHO, Guidelines for environmental surveillance of poliovirus circulation, 2003).
The recently published European Union report highlights that surveillance of SARS-CoV-2 and its variants in wastewater can be a fast and reliable source of information about the spread of SARS-CoV-2 in the population and can be a valuable part of an enhanced genomic and epidemiological surveillance (Official Journal of the European Union, 2021). As a consequence, wastewater monitoring should be included in national SARS-CoV-2 detection strategies. In the context of the COVID-19 pandemic, different researchers have suggested that a temporal correlation between SARS-CoV-2 RNA detection in sewage and the number of reported cases in a city may exist. In addition, it is suggested that WBE could provide an early warning sign of possible disease outbreaks in a community (Giraud-Billoud M, 2021), (La Rosa G, 2020), (Randazzo W, 2020), (Ahmed W A. N., 2020), (Medema G, 2020), (Izquierdo-Lara R E. G., 2021), (Peccia J, 2020), (Wurtz N, 2021), (Sharif S, 2021), (Haramoto E, 2020), (Ai Y, 2021), (Fongaro G, 2021), (Chavarria-Miró G, 2021). In this way, wastewater data could supplement current clinical measures of the spread of the virus at a community level.
It is known that SARS-CoV-2 variants with multiple amino acid changes at the spike protein are emerging in different parts of the world, raising concerns regarding their possible impact on human health and vaccine immune response efficacy against the virus (Wilton T, 2021), (Tegally H, 2021), (Faria NR, 2021). The World Health Organization (WHO) has classified SARS-CoV-2 variants into variants of concern (VOC) and variants of interest (VOI), according to the virus's features given by the mutations (transmissibility, virulence, clinical disease presentation, response to the vaccine) (WHO, https://www.who.int/activities/tracking-SARS-CoV-2-variants/tracking-SARS-CoV-2-variants., 2021). In this sense, environmental surveillance of SARS-CoV-2 could constitute a useful tool for viral surveillance in a community, providing an early warning of the introduction of variants into the population (Wilton T, 2021).
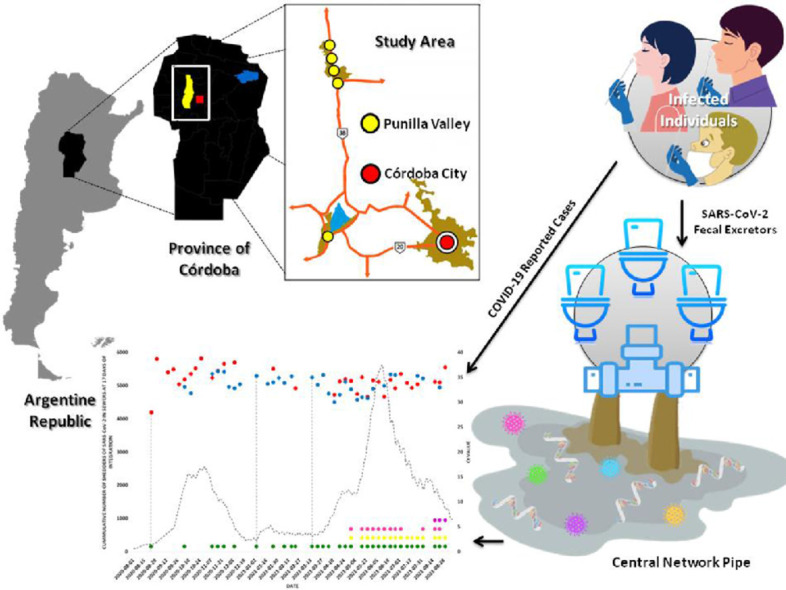
In the present study, the wastewater samples of six communities in the province of Córdoba, Argentina, were analyzed. The goal of this work was to obtain information about the presence of SARS-CoV-2 in the community and the description of the circulation dynamics of SARS-CoV-2 variants in the population through the analysis of SARS-CoV-2 in wastewater. The monitoring of SARS-CoV-2 in wastewater provides different information depending on the size and tourist influx of the community to be studied. The use of the point mutation detection strategy for the identification of SARS-COV-2 variants with particular attention on VOCs in wastewater samples expands the range of application of this methodology to samples other than clinical ones.
2. Study methodology
2.1. Study Area
2.1.1. Córdoba city
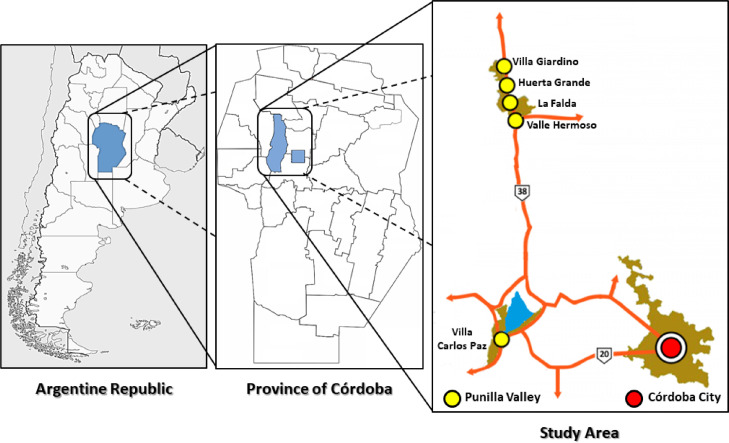
Cordoba city is the capital of the province of Córdoba, located in the central region of Argentina (31°25′00″S 64°11′00″O) and has 1,317,298 inhabitants with a population, density of 2308 habitants/km2 (INDEC, 2010) (Fig. 1 ). The sewerage system covers 50% of the population and no industrial wastewater is treated in this facility. The main wastewater treatment plant (WWTP) is named “Bajo Grande” and receives approximately the discharge of 50% of the sewage network coverage.
Fig. 1.
Geographical representation of the area under study: Argentina and the province of Córdoba, Argentina. The sampling points are indicated in colors. Córdoba City in red and Punilla Valley (Villa Giardino, Huerta Grande, La Falda, Valle Hermoso and Villa Carlos Paz) in yellow.
2.1.2. Punilla valley
Punilla Valley is a quasi-urbanized valley in the metropolitan area, which is located in the center-northwest of the province of Córdoba (31°02′S 64°30′O). At present, it is one of the main tourist centers of Argentina (Fig. 1). In this region, there are a lot of lakes-reservoirs, such as San Roque dam, on whose shores is located the city of Villa Carlos Paz, the largest city in Punilla Valley. According to the most recent National Census, Villa Carlos Paz has a population of 62,423 inhabitants and 37% of the population is connected to the sewage network. In summer, this city triples its population due to the large influx of tourists. La Falda additionally resides in this valley and maintains a population of 16,335 inhabitants. There are also smaller towns, that stand out as tourist centers, as Villa Giardino (6702 inhabitants), Huerta Grande (5925 inhabitants), and Valle Hermoso (6187 inhabitants). La Falda has a sewage effluent plant that treats its own effluents and wastewater from Villa Giardino and Huerta Grande towns. In total, the sewage network coverage of these three localities is 51%. Valle Hermoso, whose main economic activity is tourism due to its spas and rivers with crystal clear waters, has a sewer network coverage of 20%.
2.2. Wastewater sampling
From mid-May 2020 to the end of August 2021, raw sewage samples were collected from central network pipes from four wastewater treatment plants (WWTPs). WWTPs included in the study as well as the period and frequency of wastewater samples collected are depicted in Table 1 .
Table 1.
Wastewater treatment plants (WWTP) included in the study, sampling frequency and number of samples collected.
| City | WWTP Name | Period of Samples Collection | Sampling Frequency | Number of Samples Collected |
|---|---|---|---|---|
| CAPITAL DEPARTMENT | ||||
| Córdoba | Bajo Grande | mid-May 2020 - August 2021 | Once a week | 69 |
| PUNILLA DEPARTMENT (Punilla Valley) | ||||
| Carlos Paz | Costa Azul | May 2020 - October 2020 November 2020 - August 2021 | Biweekly Once a week | 63 |
| Valle Hermoso | Valle Hermoso | 63 | ||
| La Falda | La Falda* | 63 | ||
The WWTP La Falda, treats the effluents of La Falda, Villa Giardino and Huerta Grande localities
For each sample, 0.5 l of wastewater were taken on weekday mornings by the grab collection method described in the WHO Guidelines for Environmental Surveillance of Poliovirus Circulation (WHO, Guidelines for environmental surveillance of poliovirus circulation, 2003). Each sample was transported within 6 h at 4 °C to 8 °C to the Institute of Virology, National University of Córdoba, Córdoba City, for further processing and analysis.
2.3. Concentration of SARS-CoV-2 in wastewater samples
SARS-CoV-2 was concentrated in sewage specimens by using the polyethylene glycol-6000 (PEG-6000) precipitation method, according to the WHO guideline for environmental surveillance of poliovirus circulation (WHO, Guidelines for environmental surveillance of poliovirus circulation, 2003) using the method described previously by Lewis and Metcalf (1988). Briefly, sewage was centrifuged at 4750 x g for 20 min at 4 °C. Supernatant (S1) was maintained at 4 °C to be used later, and the sediment was mixed with 3% Beef extract (Lab-Lemco’ Poweder, OXOID LTD, Basingstoke, Hammpshire, England) /2 M NaNO3 Titolo min 99.5% (acidimetrico) (Analyticals, Farmitalia Carlo Erba S.p.A, Milano, Italy) eluant (pH 5.5) and stirred for 1 h at 4 °C. Solids were then removed by centrifugation at 10,000 x g for 20 min, and the eluate was mixed with the first supernatant obtained (S1) and adjusted to pH 7.2. PEG-6000 (Parafarm, Saporiti S.A.C.I.F.I.A, Buenos Aires, Argentina) was added to a final concentration of 10% (w/v) and NaCl pro-analysis A.C.S (Cicarelli laboratories, reagents S.A. Santa Fe, Argentina) to 2% (w/v). The resulting suspension was stirred overnight at 4 °C and centrifuged at 10,000 x g for 25 min. The PEG-containing supernatant was discarded, and the pellet was suspended in 5 mL PBS (Gibco, Invitrogen Corporation, New York, USA) (pH 7.2), adjusted to pH 8.0, incubated for 1 h with occasional vortex, and centrifuged at 10,000 x g for 20 min. The supernatant was stored at −70 °C.
2.4. Genomic RNA extraction from viral concentrates
Each concentrated sample (1 mL) was subjected to RNA automated extraction using MagNA Pure 96 DNA and Viral NA Large Volume Kit in MagNA Pure 96 system Roche (Roche Diagnostics GmbH – Mannheim, Germany), according to the manufacturer's instructions. The extracted RNA was eluted in a final volume of 50 µL of a 60 mM Tris-HCl buffer. Blank control was included during the RNA extraction to monitor any possible cross contamination during samples processing.
2.5. SARS-CoV-2 genome detection
2.5.1. Amplification reactions
Molecular detection of SARS-CoV-2 RNA was performed by real time RT-PCR using the DisCoVery SARS-CoV-2 RT-PCR Detection Kit (Safecare Biotech Hangzhou Co. Ltd., China), that identifies the nucleocapsid (N) and open reading fragment 1ab (Orf1ab) genes. In this study, only results derived from N gene detection were analyzed since it has a higher analytical sensitivity for detection than the Orf1ab gene (i.e., from the total samples that resulted positive for N gene detection (n=88), only 54.5% (n=48) were also positive for ORF 1ab gene. None of the samples resulted negative for N gene and positive for Orf1ab gene detection).
As of September 2020, N gene detection was coupled to envelope (E) detection, using the LightMix® Modular SARS and Wuhan CoV E-gene kit (TIB MOLBIOL GmbH, Berlin, Germany, distributed by Roche), to increase the SARS-CoV-2 genome detection sensitivity.
In all cases, assays were run according to the manufacturer's instructions. Real time RT-PCR reactions were carried out using a Cobas Z 480 equipment (Roche, Germany). The cycle conditions and the temperature ramps used for each of the amplification kits used are described in Tables 2 and 3 .
Table 2.
Cycling conditions for the DisCoVery SARS-CoV-2 RT-PCR Detection Kit, carried out in a Z480 Equipment (ROCHE, Germany).
| Program Step: | RT Step | Denaturation | Cycling | Cooling | |
|---|---|---|---|---|---|
| Analysis Mode | None | None | Quantification mode | None | |
| Cycles | 1 | 1 | 45 | 1 | |
| Target [°C] | 50 | 95 | 95 | 60 | 40 |
| Hold [hh:mm:ss] | 00:05:00 | 00:00:30 | 00:00:05 | 00:00:35 | 00:00:30 |
| Ramp Rate [°C/s] | 4.4 | 4.4 | 4.4 | 2.2 | 1.5 |
| Acquisition Mode | None | None | None | Single | None |
Table 3.
Cycling conditions for the LightMix® Modular SARS and Wuhan CoV E-gene kit carried out in a Z480 Equipment (ROCHE, Germany).
| Program Step: | RT Step | Denaturation | Cycling | Cooling | ||
|---|---|---|---|---|---|---|
| Analysis Mode | None | None | Quantification mode | None | ||
| Cycles | 1 | 1 | 45 | 1 | ||
| Target [°C] | 55 | 95 | 95 | 60 | 72 | 40 |
| Hold [hh:mm:ss] | 0:05:00 | 0:05:00 | 0:00:05 | 0:00:15 | 0:00:15 | 0:00:30 |
| Ramp Rate [°C/s] | 4.4 | 4.4 | 4.4 | 2.2 | 4.4 | 1.5 |
| Acquisition Mode | None | None | None | Single | None | None |
2.5.2. Controls used
Before nucleic acid extraction, each sample was contaminated with an exogenous internal control (RNA Process Control - LightCycler® Multiplex RNA Virus Master-Roche) which was detected in parallel with the specific viral targets. This control allowed monitoring the extraction process and the presence of inhibitors. In those negative real time -RT-PCR wastewater samples, serial dilutions (1:5 v/v and 1:10 v/v) were performed to rule out the presence of inhibitors.
In addition, the SARS-CoV-2 RNA detection kit includes RNaseP as an endogenous internal control. This control allows evaluating the quality of the samples. Finally, a positive control, included in the detection kit, was added to each amplification batch to monitor the amplification and detection of specific SARS-CoV-2 RNA viral genes.
To validate the results, the software associated with the Cobas Z 480 (Roche) analyzer was used, which applies an advanced result algorithm that interprets 3 key aspects of the PCR data: growth curve shape, growth curve height (curve amplitude) and Cycle threshold (Ct value). The validation criteria of the run were the following: No Template Control (NTC) (reagents blank): no Ct value in all targets evaluated (Gen N, Gen Orf-1ab, RNAseP and Gen E), extraction control: no Ct value in all specific targets evaluated (Gen N, Gen Orf-1ab and Gen E), Positive Template Control (PTC): for the kit DisCoVery Ct≤34 for Gen N, Gen Orf-1ab and RNAseP and for the kit LightMix-Roche Ct≤32 for Gen E. For the analysis of the samples: they were considered positive if typical S-shaped curve was observed in both the RNA Process Control and RNAseP specific detection channels of each specimen and Ct<40 for viral specific targets.
A NTC blank was included. It consisted of the PCR mix with all necessary reagents for virus detection, but instead of an eluate, an equivalent volume of molecular biology quality water was used for viral amplification and detection.
2.6. SARS-CoV-2 recovery rate from wastewater
Serial dilutions (from 10−1-10−7) of a SARS-CoV-2 live wild type strain B.1 linage (hCoV-19/Argentina/PAIS-6001/2020, GISAID ID: EPI-ISL-499083) (viral titer 2.106 plaque forming units (PFU)/mL) were analyzed by real time -RT-PCR as it is described in items 2.4-2.5. The Ct values obtained for each viral dilution was plotted against the corresponding viral titer, expressed in PFU/mL. The relationship between the Ct values versus the titer of each serial dilution showed a curve with a value of R2 = 0.96.
To estimate the SARS-CoV-2 recovery rate from wastewater by the PEG method, sewage subsamples from a sewage matrix previously determined as SARS-CoV-2 negative were seeded with a SARS-CoV-2 viral suspension to a final concentration of 300 PFU/mL. As negative control sewage did not seed with the SARS-CoV-2 viral dilution was tested in parallel. Samples were then concentrated and analyzed by real time -RT-PCR following the methodology described above (2.3, 2.4 and 2.5 sections). Ct values obtained were extrapolated in the curve to obtain the corresponding viral PFU/mL.
SARS-CoV-2 recovery rate was calculated based on the PFU/mL as follows: Recovery rate (%) = (viral PFU/mL recovered / viral PFU/mL seeded) × 100. The recovery rate mean value was 15.75%; with a range of 12.65–22.85%.
2.7. SARS-CoV-2 VOC detection
VOC identification was performed by consecutive real time -RT-PCRs to detect relevant mutations/deletions present in the Spike protein using the TaqMan™ SARS-CoV-2 Mutation Panel reagent (Life Technologies Corporation-Pleasanton CA) in SARS-CoV-2 positive concentrated wastewater samples.
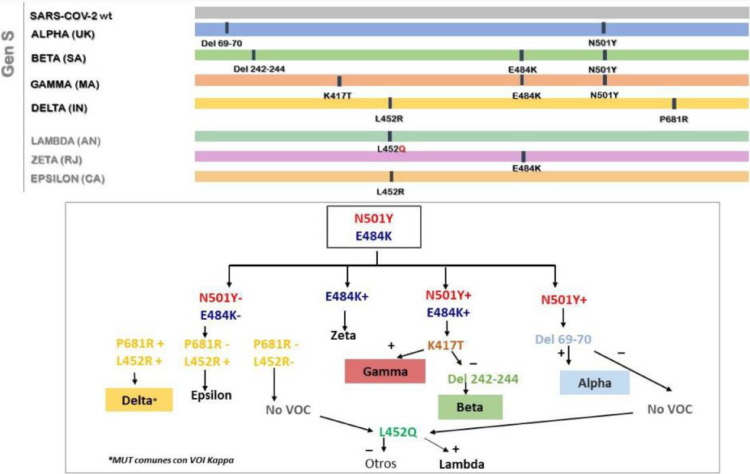
With this methodology, the identification of mutations N501Y, E484K, delH69V70, K417T, delL242_244L, L452R and P681R was performed. Combined mutation analysis allowed the detection of a mutation profile compatible with VOCs Alpha, Beta, Gamma and Delta. Fig. 2 shows the strategy used for variant detection.
Fig. 2.
Real-time RT-PCR variant detection strategy for the detection of relevant VOC/VOI mutations/deletions present in the spike protein (mutation/deletions identification: N501Y, E484K, delH69V70, K417T, delL242_244L, L452R and P681R).
Assays were run according to the manufacturer's instructions. Real time RT-PCR reactions were carried out using an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, USA). The cycle conditions and the temperature ramps used for each of the amplification kits used are described in Table 4 .
Table 4.
The conditions for the TaqMan™ SARS-CoV-2 Mutation Panel reagent (Applied Biosystems) on the Applied Biosystem 7500 Equipment.
| Step | Temperature | Time | Number of Cycles |
|---|---|---|---|
| Reverse Transcription[1] | 50 °C | 10 s | 1 |
| Fast DNA polymerase activation[2] | 90 °C | 2 s | 1 |
| Denaturation | 90 °C | 3 s | 45 |
| Anneal / extend | 60 °C | 30 s | |
| Post-read | 60 °C | 30 s | 1 |
2.8. Estimation of SARS-CoV-2 excretors in sewers
The number of individuals excreting the virus by faecal matter at the time the wastewater sample was obtained was estimated based on the following: 1) the 35% of individuals infected with SARS-CoV-2 excrete the virus via faecal matter (Wu Y, 2020), (Wang W, 2020); 2) the average SARS-CoV-2 genome excretion time is 17 days (Tian Y, 2020); 3) the sewage network coverage of the communities studied (official data available from the study communities) (WHO, Guidelines for environmental surveillance of poliovirus circulation, 2003); and 4) the daily report of SARS-CoV-2 cases in each studied locality studied.
3. Results
From the total samples collected (n = 261), 24.1% (63/261) were only assayed for the detection of SARS-CoV-2 RNA N gene, and the 198 remaining samples (75.9%) were analyzed for the detection of both SARS-CoV-2 RNA N and E genes. Fifty-eight samples from the total analyzed for both genes (29.3%) resulted negative; 47 (23.7%) were positive for both genes; 39 samples (19.7%) were positive only for the N gene and 55 samples (27.8%) were positive only for the E gene. Overall, by using the DisCoVery SARS-CoV-2RT-PCR Detection Kit, which identifies the N gene; the detection rate of the viral genome was 43.4% (86/198); by using the LightMix® Modular SARS and Wuhan CoV E-gene kit, the detection rate was 51.5% (102/198); by and using both methodologies (positivity criterion: detection of N and / or E gene), the detection rate was 71.2% (141/198).
3.1. SARS-CoV-2 genome and VOC detection in Córdoba city
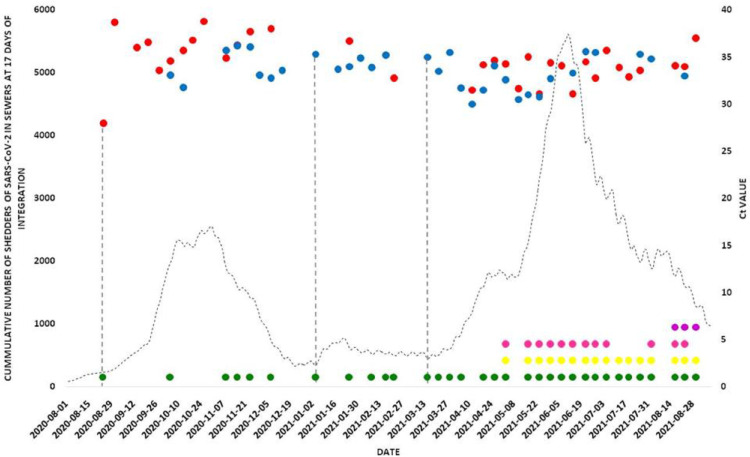
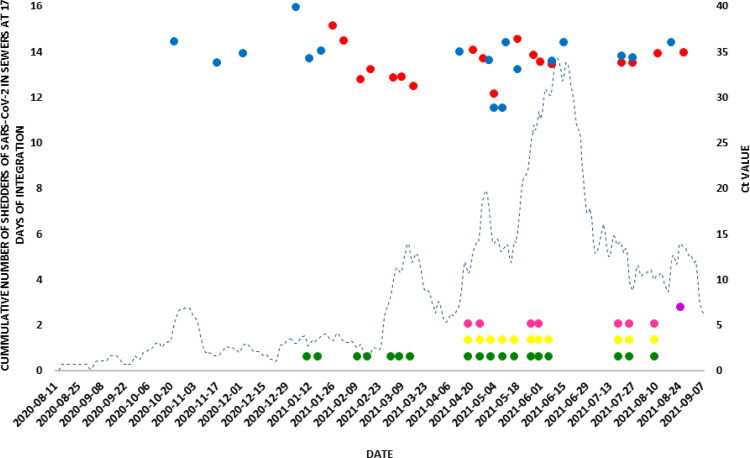
The sampling of wastewater from the main pipeline of Bajo Grande WWTP started on 11th May 2020, when clinical cases continued to increase gradually in Córdoba city. The first wastewater sample in which the virus genome was detected corresponded to 24th August 2020 (Fig. 3 ). From that moment onwards, the detection of SARS-CoV-2 RNA in sewage reflected the viral circulation in the community. The detection of the viral genome in wastewater can be divided into three periods: the first comprises the period 24th August-14th December 2020 and reflected the first epidemic wave in the city of Cordoba. The first detection of viral genome in sewage was an early warning sign of the subsequent increase in cases of COVID-19 in Córdoba city. Since mid to end of December 2020, no additional SARS-CoV-2 genome was detected (Fig. 3).
Fig. 3.
Detection of SARS-CoV-2 and VOCs in wastewater from Córdoba city. The curve with dotted line shows the number of cases of reported COVID 19 accumulated in 17 days in Córdoba city (Ministry of Health of Cordoba Province).Bullets  and
and  : detection of the SARS-CoV-2 genome by identifying the N and E genes respectively. Pointed Wheels: Identification of SARS-CoV-2 and VOCs by Typing:
: detection of the SARS-CoV-2 genome by identifying the N and E genes respectively. Pointed Wheels: Identification of SARS-CoV-2 and VOCs by Typing:  No VOCs;
No VOCs;  Alpha,
Alpha,  Gamma and
Gamma and  Delta.Vertical line: represents the first SARS-CoV-2 genome detection in wastewater on each period.
Delta.Vertical line: represents the first SARS-CoV-2 genome detection in wastewater on each period.
A second period of SARS-CoV-2 genome detection in sewage began in January 2021 to mid-February 2021, matching with the report of a summer COVID-19 epidemic outbreak. Genome detection in early January was predictive of the summer outbreak. From mid-February to mid-March 2021, no additional SARS-CoV-2 genome was detected.
The third period was registered from the middle of March 2021 (15th March 2021), displaying that the detection of SARS-CoV-2 RNA in sewage samples had a predictive value two weeks in advance of the beginning of the second epidemic wave in Córdoba city. In this second epidemic wave, the virus genome was detected continuously, accompanied by an increase in COVID-19 cases. The lowest Ct values (between 30 and 32) for gene E in this period were detected in two occasions with a similar pattern (Fig. 3). One pattern spans from 5th to 19th April 2021. In the sample collected on April 5th, a Ct value of 31.7 was obtained, coinciding with the beginning of an increase in reported COVID-19 cases. In the following two weeks, Ct values for the E gene of 30 and 31.5 were obtained, accompanying the increase in reported COVID-19 cases. The second moment spans from 3rd to 17th May 2021. In the sample collected on May 3rd, a Ct value of 30.5 for the E gene was obtained, matching with the beginning of an exponential increase in reported COVID-19 cases. In the samples collected in the following weeks, Ct values of 30.5 and 31 were detected.
From the SARS-CoV-2 variant analysis, in the first epidemic wave in Córdoba city, no VOC circulation was identified. At the beginning of March 2021, VOCs Gamma and Alpha were first identified in wastewaters. In August 2021, VOC Delta was first detected in wastewater samples, in co-circulation with Gamma, Alpha and no VOC variants (Fig. 3).
3.2. SARS-CoV-2 genome and VOC detection in Punilla Valley
The SARS-CoV-2 detection in wastewater collected in WWTP from the localities of Valle Hermoso and La Falda (which includes sewage from La Falda, Huerta Grande and Villa Giardino) reflected a common viral pattern, which was characterized by a continued SARS-CoV-2 genome detection. This pattern reflected the circulation of the virus in the studied localities. Besides, the epidemic curves in all the studied localities were defined by peaks of increasing and decreasing incidence of clinical COVID-19 cases over time (Figs. 5 and 6).
Fig. 5.
Number of daily cases of COVID-19, detection of SARS-CoV-2 in wastewater from La Falda, Villa Giardino and Huerta Grande, and circulation dynamics of VOCs. Number of COVID-19 cases accumulated at 17 days of integration from La Falda, Villa Giardino and Huerta Grande. Bullet  and
and  : detection of the SARS-CoV-2 genome by identifying the N and E gene respectively. Pointed Wheels: Identification of SARS-CoV-2 and VOCs by Typing:
: detection of the SARS-CoV-2 genome by identifying the N and E gene respectively. Pointed Wheels: Identification of SARS-CoV-2 and VOCs by Typing:  No VOCs;
No VOCs;  Alpha,
Alpha,  Gamma and
Gamma and  Delta.
Delta.
Fig. 6.
Number of daily cases of COVID-19, detection of SARS-CoV-2 in wastewater from Valle Hermoso city and circulation dynamics of VOCs. A. Number of daily COVID-19 cases reported. B. Number of COVID-19 cases accumulated at 17 days of integration from Valle Hermoso. Bullet  and
and  : detection of the SARS-CoV-2 genome by identifying the N and E gene respectively. Pointed Wheels: Identification of SARS-CoV-2 and VOCs by Typing:
: detection of the SARS-CoV-2 genome by identifying the N and E gene respectively. Pointed Wheels: Identification of SARS-CoV-2 and VOCs by Typing:  No VOCs;
No VOCs;  Alpha,
Alpha,  Gamma and
Gamma and  Delta.
Delta.
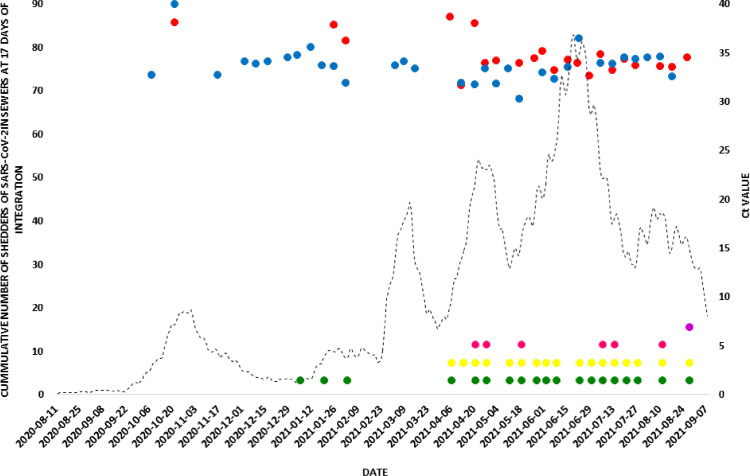
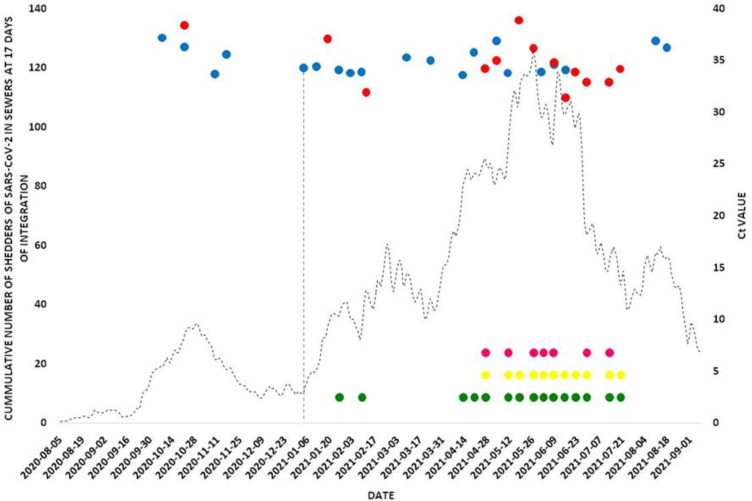
By contrast, in Carlos Paz city, the detection of the viral genome in wastewater can be divided into two periods: the first one, from October 2020 to the end of November 2020, corresponded to the first epidemic wave, as occurred in Córdoba city. Then, during the period November 25 - December 31, SARS-CoV-2 genome was not detected in sewage samples. From the beginning of January 2021, a second period of viral detection in sewage could be defined, matching with the second epidemic wave. This period was between the first week of January 2021 and the end of August 2021, when the present study was completed. The presence of SARS-CoV-2 genomes in sewage on January 4th (just before the tourist season started) was an early warning signal of the following increase of COVID-19 clinical cases in Carlos Paz city.
From the SARS-CoV-2 analysis variants, no VOCs circulated during the whole period studied in all the localities of Punilla Valley. In April 2021, VOC variants Gamma and Alpha were detected for the first time in wastewaters in co-circulation with no VOCs. In the month of August 2021, VOC Delta was registered in co-circulation with Gamma and no VOCs (Figs. 4 , Fig. 5, Fig. 6 ).
Fig. 4.
Number of daily cases of COVID-19, detection of SARS-CoV-2 in wastewater from Carlos Paz city and circulation dynamics of VOCs. A. Number of COVID-19 cases accumulated at 17 days of integration in Carlos Paz city. Bullets  and
and  : detection of the SARS-CoV-2 genome by identifying the N and E genes respectively. Pointed Wheels: Identification of SARS-CoV-2 and VOCs by Typing: No VOCs;
: detection of the SARS-CoV-2 genome by identifying the N and E genes respectively. Pointed Wheels: Identification of SARS-CoV-2 and VOCs by Typing: No VOCs;  Alpha and
Alpha and  Gamma.
Gamma.
4. Discussion
The study of SARS-CoV-2 epidemiology based on wastewater from Córdoba and Carlos Paz cities reflected the epidemic curve based on COVID-19 cases, with the detection of the viral genome in sewers predictive of epidemic waves and isolated outbreaks. Our finding is in concordance with the result obtained by Giraud-Billoud et al. (2021) which indicates that WBE is a useful epidemiological indicator to anticipate the increase in COVID-19 cases. The predictive value of the presence of SARS-CoV-2 genome in wastewater could be explained by a significant number of asymptomatic, pre-symptomatic and slightly symptomatic excretors. Data published by Pan et al. (2020) and Wu et al. (2020) indicate that SARS-CoV-2 genome shedding may occur soon after infection. Therefore, different authors highlight the utility of SARS-CoV-2 monitoring in wastewater as a non-invasive early warning tool to support health surveillance (Fongaro G, 2021), (Larsen DA, 2020), (Mousazadeh M, 2021).
However, in La Falda, Villa Giardino, Huerta Grande and Valle Hermoso, the SARS-CoV-2 genome in sewage was continuously detected throughout the studied period, accompanying a pattern characterized by abrupt increases and decreases in the reported incidence of COVID-19 cases. This could be explained because the studied localities have a low number of inhabitants and a high influx of tourists. During summer and winter holidays and special holiday periods such as Easter and carnival, their populations can even be four times higher with tourists entering and leaving the place week after week. It is likely that infected individuals remain a few days in the locality, being registered as cases in the health system but no longer contributing to the excretion of SARS-CoV-2 in the sewage system. Therefore, WBE is a population-based study tool, which necessarily requires a stable population.
The sensitive detection of SARS-CoV-2 RNA in wastewater depends in part on the molecular-based methods employed, which remain diverse and unstandardized (Hamouda M, 2021). Different authors have reported discrepancies among CDC N1 trials with CDC N2, CDC N3, and E_Sarbeco for wastewater samples in the emerging epidemic in different parts of the world (Medema G, 2020), (Wu F, 2020), (Ahmed W B. A., 2020), (Giraud-Billoud M, 2021). In our work, commercial kits were used to detect N and E SARS-CoV-2 genes. The results obtained showed discrepancies in the detection of the SARS-CoV-2 genome using one or another, indicating that the optimal strategy to study the SARS-CoV-2 genome in wastewater would be the use of the combined detection of both genes.
One of the limitations of this work is that the genomic load was not quantified in the positive samples. Different studies have performed the quantification of the SARS-CoV-2 genome in wastewater, reporting a general trend between the genomic load detected and the number of COVID-19 cases registered. However, other works have shown low genomic loads in wastewater samples obtained during a period with high number of reported COVID-19 cases (Barrios ME, 2021), (Giraud-Billoud M, 2021). Similarly, high genomic loads detected in wastewater are observed with a low number of reported COVID-19 cases (Giraud-Billoud M, 2021), (Barrios ME, 2021), (Ahmed W B. A., 2020), (Randazzo W, 2020). Multiple variables involved in the epidemiological scenario could explain these discrepancies. Regarding the number of reported cases of COVID-19 in a community, the most important variables involved are as follows: a) the strength of the health system for diagnosis, b) the capacity of diagnostic tests, c) the criteria to define and register COVID-19 cases and d) the ability to access the health system. However, genomic load detected in wastewater depends on the following: a) the percentage of individuals connected to the sewage system in a community, b) the methodology used for sample concentration and detection of the genome in wastewater, and c) virus dilution events in wastewater due to weather conditions (I.e. storm water runoff), among others (Hamouda M, 2021). Moreover, Barrios et al. (2021) noted that changes in the SARS-CoV-2 genome concentration in sewage samples collected from small communities do not correlate with the number of COVID-19 cases in the same time period. However, Melvin et al. (2021) proposed the Melvin Index, that can be applied both to wastewater treatment facilities (WWTF) that serve a wide range of population sizes and to large regions that are served by multiple WWTFs. The authors proposed that the Melvin index could avoid the influence of the population served and the rate of wastewater flow. Therefore, the relationship between genomic load and COVID-19 cases in a community must be interpreted in the context of the characteristics and size of the analyzed population and the health system in that community.
Detection of SARS-CoV-2 variants in wastewater reflected their circulation in the community (Córdoba, 2021), showing no VOCs detection in the first COVID-19 wave and its co-circulation with Gamma, Alpha and Delta VOCs during 2021, as they were introduced into the community. Coinciding with what was recorded in clinical surveillance, the implemented strategy resulted in a useful tool that allowed the characterization of circulation dynamics of variants in the studied localities (Castro G. M., 2021). To our knowledge, this is the first research work that implements this point mutation detection strategy for the identification of SARS-COV-2 variants in wastewater samples. In this sense, in addition to all the advantages previously described for this strategy (cost-effective, short time-consuming, no extra equipment required or specialized personnel and massive rapid typification) (Castro G. M., 2021), it was possible to typify samples with Cts>30, which is often difficult in traditional sequencing typing techniques (Crits-Christoph A, 2021) (Izquierdo-Lara R E. G., 2021). Additionally, this methodology allowed detection of more than one VOC simultaneously in the same sample. In Córdoba province, as in some countries around the world, like Australia (Medema G, 2020), New Zealand (Ahmed W B. A., 2020), the Netherlands (Bhattacharya P, 2021) and parts of Brazil (Claro ICM, 2021), environmental surveillance was adopted by the Ministry of Health as a tool to track the circulation dynamics of SARS-CoV-2 in the community.
5. Conclusion
-
•
The monitoring of SARS-CoV-2 in wastewater can become a silent sentinel of the trend of viral circulation in the community, providing different information according to the size and dynamics of the population.
-
•
In geographical areas with a stable population, the first detection of the SARS-CoV-2 viral genome in wastewater after a period of non-detection, is an early warning sign of the subsequent increase in COVID-19 cases.
-
•
In localities with a low number of inhabitants (lower than 16,000 inhabitants) and high tourist influx, in which a continuous detection of SARS-CoV-2 genome in wastewater was detected through the studied period, this tool may be useful only to reflect the circulation of the virus in the community.
-
•
The profile of variants detected in wastewater showed that WBE of SARS-CoV-2 allows to know the dynamics of circulation of the viral variants in the studied communities.
-
•
WBE of SARS-CoV-2 could provide supplementary information for clinical surveillance to support public health measures.
Funding
This work was supported by the Ministry of Science, Technology and Innovation of the Argentine Nation through the Federal Program for Coordination and Strengthening in Science and Technology COVID-19 (COR-28), by the National Agency for the Promotion of Research, Technological Development and Innovation - IP -COVID (IP-558 to G.M.), by the Program of Accreditation and Financing of Research and Development Projects of the National Defense University and by the Ministry of Health of Córdoba. G.M., M.B.P and V.R. are members of the researcher career program of CONICET, Argentina.
Declaration of Competing Interest
The authors declare that they do not have competing financial interests or personal relationships that could have influenced the work reported in this paper.
Acknowledgements
This work was possible thanks to the collaboration and coordination of different government agencies and institutions: The Municipality of the City of Córdoba and the Cooperativa Integral Regional de Provisión de Servicios Públicos, Vivienda y Consumo Limitada (COOPI) of Villa Carlos Paz, which managed the permits to perform the sampling of wastewater as well as assuming the responsibility for each sampling; and the Ministry of Health of the Province of Córdoba, which declared this investigation of public interest and provided financial support. We also thank the Ministry of Science, Technology and Innovation of Argentina (MinCyT) for its collaboration in coordinating institutional interactions. We express our gratitude to the entire team of the Central Laboratory of the Province for the support and responsibility in processing the detection of the SARS-CoV-2 genome in each of the stages. We also want to thank MinCyT, the National Agency for the Promotion of Research, Technological Development and Innovation and the National Defense University for subsidizing this research work.
Referencies
- Ahmed W A.N. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 1 de Aug de 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W B.A. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 30 de Sep de 2020 doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai Y D.A. SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States. Sci. Total Environ. 19 de Aug de 2021;801 doi: 10.1016/j.scitotenv.2021.149757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama Y K.N. A pilot study of viral load in stools of patients with COVID-19 and diarrhea. Jpn. J. Infect. Dis. 31 de May de 2021 doi: 10.7883/yoken.JJID.2021.018. [DOI] [PubMed] [Google Scholar]
- Barrios ME D.S. Dynamics of SARS-CoV-2 in wastewater in three districts of the Buenos Aires metropolitan region, Argentina, throughout nine months of surveillance: a pilot study. Sci. Total Environ. 15 de Dec de 2021;800 doi: 10.1016/j.scitotenv.2021.149578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya P K.M. Prevalence of SARS-CoV-2 in communities through wastewater surveillance-a potential approach for estimation of disease burden. Curr. Pollut. Rep. 6 de Apr de 2021:1–7. doi: 10.1007/s40726-021-00178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calitri C.F. Gastrointestinal involvement in paediatric COVID-19 - from pathogenesis to clinical management: a comprehensive review. World J. Gastroenterol. 2021;27(23):3303–3316. doi: 10.3748/wjg.v27.i23.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro G.M., S P. Strategy for a rapid screening and surveillance of SARS-CoV-2 variants by real time RT-PCR: a key tool that allowed control and delay in Delta spread in Cordoba, Argentina. COVID-19 SARS-CoV-2 preprints from medRxiv and bioRxiv. 2021 doi: 10.1101/2021.11.16.21266265. [DOI] [Google Scholar]
- Chavarria-Miró G A.-E.E.-V.-P. Time evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in wastewater during the first pandemic wave of COVID-19 in the metropolitan area of Barcelona, Spain. Appl. Environ. Microbiol. 11 de Mar de 2021;87(7):e02750. doi: 10.1128/AEM.02750-20. -20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claro ICM C.A. Long-term monitoring of SARS-COV-2 RNA in wastewater in Brazil: a more responsive and economical approach. Water Res. 15 de Sep de 2021;203 doi: 10.1016/j.watres.2021.117534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Córdoba G.d. 2021. Gobierno de la provincia de Córdoba, Vigilancia de variantes de preocupación.https://www.cba.gov.ar/coronavirus/vigilancia-de-variantes/?csrt=10691462293584337513 Obtenido de. [Google Scholar]
- Crits-Christoph A K.R.-S. Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. mBio. 19 de Jan de 2021;12(1):e02703–e02720. doi: 10.1128/mBio.02703-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Official Journal of the European Union, 2021.https://eur-lex.europa.eu/legal-content/ES/TXT/HTML/?uri=CELEX:32021H0472&from=EN.
- Faria NR C.I. 2021. Genomic Characterization of an Emergent SARS-CoV-2 Lineage in Manaus: Preliminary Findings.https://virological.org/t/genomic-characterization-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 Obtenido de. [Google Scholar]
- Fongaro G R.P.-L. ARS-CoV-2 in human sewage and river water from a remote and vulnerable area as a surveillance tool in Brazil. Food Environ. Virol. 8 de Jul de 2021:1–4. doi: 10.1007/s12560-021-09487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud-Billoud M C.P. Monitoring of SARS-CoV-2 RNA in wastewater as an epidemiological surveillance tool in Mendoza, Argentina. Sci. Total Environ. 2021, Aug doi: 10.1016/j.scitotenv.2021.148887. Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M T.W. Potential intestinal infection and faecal-oral transmission of SARS-CoV-2. Nat. Rev. Gastroenterol. Hepatol. Apr de 2021;18(4):269–283. doi: 10.1038/s41575-021-00416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamouda M M.F. Wastewater surveillance for SARS-CoV-2: lessons learnt from recent studies to define future applications. Sci. Total Environ. 10 de Mar de 2021;759 doi: 10.1016/j.scitotenv.2020.143493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E M.B. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 1 de Oct de 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikmet F.M. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020;16(7):e9610. doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Instituto Nacional de Estadísticas y Censos (INDEC) de Argentina. 2010. 2010;2:41–135. https://www.indec.gob.ar/indec/web/Nivel4-Tema-2-41-135. [Google Scholar]
- Izquierdo-Lara R E.G. Monitoring SARS-CoV-2 circulation and diversity through community wastewater sequencing, the Netherlands and Belgium. Emerg. Infect. Dis. 27 de May de 2021;(5):1405–1415. doi: 10.3201/eid2705.204410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G I.M. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 20 de Sep de 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen DA W.K. Tracking COVID-19 with wastewater. Nat. Biotechnol. 2020;38:1151–1153. doi: 10.1038/s41587-020-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis G.D. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 1988;54(8):1983–1988. doi: 10.1128/aem.54.8.1983-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G H.L. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Megyeri K D.Á.-L. COVID-19-associated diarrhea. World J. Gastroenterol. 21 de Jun de 2021;27(23):3208–3222. doi: 10.3748/wjg.v27.i23.3208. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melvin RG H.E. A novel wastewater-based epidemiology indexing method predicts SARS-CoV-2 disease prevalence across treatment facilities in metropolitan and regional populations. Sci. Rep. 1 de Nov de 2021;11(1):21368. doi: 10.1038/s41598-021-00853-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousazadeh M A.R. Wastewater based epidemiology perspective as a faster protocol for detecting coronavirus RNA in human populations: a review with specific reference to SARS-CoV-2 virus. Pathogens. Aug de 2021;10(8):1008. doi: 10.3390/pathogens10081008. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y Z.D. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. Apr de 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J Z.A.-M. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. Oct de 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P O.I. Gastrointestinal symptoms and outcomes in hospitalized coronavirus disease 2019 patients. Dig. Dis. 2020;38:373–379. doi: 10.1159/000509774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W T.P.-F. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 15 de Aug de 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd WD Z.J. Prevalence and characteristics of gastrointestinal symptoms in patients with severe acute respiratory syndrome coronavirus 2 infection in the United States: a multicenter cohort study. Gastroenterology. Aug de 2020;159(2):765–767. doi: 10.1053/j.gastro.2020.04.045. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R B.-O.Y. The total number and mass of SARS-CoV-2 virions. Proc. Natl. Acad. Sci. USA. 22` de Jun de 2021;118(25) doi: 10.1073/pnas.2024815118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif S I.A. Detection of SARs-CoV-2 in wastewater using the existing environmental surveillance network: a potential supplementary system for monitoring COVID-19 transmission. PLoS One. 29 de Jun de 2021;16(6) doi: 10.1371/journal.pone.0249568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S S.J. Involvement of digestive system in COVID-19: manifestations, pathology, management and challenges. Therap. Adv. Gastroenterol. 18 de Jun de 2020;13 doi: 10.1177/1756284820934626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H W.E. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. Apr de 2021;592(7854):438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Tian Y R.L.-1. Review article: gastrointestinal features in COVID-19 and the possibility of faecal transmission. Aliment. Pharmacol. Ther. May de 2020;51(9):843–851. doi: 10.1111/apt.15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W X.Y. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 12 de May de 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; 2003. Guidelines for Environmental Surveillance of Poliovirus Circulation.https://apps.who.int/iris/handle/10665/67854 Obtenido de. [Google Scholar]
- Wilton T B.E. Rapid increase of SARS-CoV-2 variant B.1.1.7 detected in sewage samples from England between October 2020 and January 2021. mSystems. 29 de Jun de 2021;6(3) doi: 10.1128/mSystems.00353-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F Z.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 21 de Jul de 2020;5(4):e00614–e00620. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y G.C. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. May de 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz N L.A.-G. Viral RNA in city wastewater as a key indicator of COVID-19 recrudescence and containment measures effectiveness. Front. Microbiol. 17 de May de 2021;12 doi: 10.3389/fmicb.2021.664477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z T.M. SARS-CoV-2 gastrointestinal infection prolongs the time to recover from COVID-19. Front. Med. (Lausanne) 4 de Jun de 2021;8 doi: 10.3389/fmed.2021.683551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C W.H. SARS-CoV-2 viral shedding characteristics and potential evidence for the priority for faecal specimen testing in diagnosis. PLoS One. 22 de Feb de 2021;16(2) doi: 10.1371/journal.pone.0247367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X C.K. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. Apr de 2020;14(2):185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]