Abstract
We report that alteration in MurM, an enzyme involved in the biosynthesis of branched-stem cell wall muropeptides, is required for maximal expression of penicillin and cefotaxime resistance in the pneumococcus. Hungarian isolate 3191 (penicillin MIC, 16 μg/ml; cefotaxime MIC, 4 μg/ml) was a source of donor DNA in transformation experiments. Penicillin-binding protein DNA was insufficient to transform recipient strain R6 to full resistance. Further transformation with altered murM DNA was required for full expression of donor penicillin and cefotaxime resistance.
Beta-lactam antibiotics inhibit the growth of pneumococci by the inactivation of penicillin-binding proteins (PBPs). PBPs are serine peptidases which catalyze polymerization of peptidoglycan precursors during cell wall synthesis (15). Pneumococcal resistance to β-lactams is the result of altered PBPs with decreased antibiotic affinities (8, 9, 16). Pneumococci contain a set of six PBPs (7). High-level penicillin resistance can be established by alteration in only three of these PBPs, that is, PBPs 2X, 2B, and 1A (2), while only altered PBPs 2X and 1A are required for high-level cefotaxime resistance (10). The role of PBPs in mediating β-lactam resistance in pneumococci was first described in the early 1980s (8, 16). More recently, further mechanisms for β-lactam resistance in pneumococci have been described, i.e., mutations in the histidine protein kinase CiaH (6) and mutations in the glycosyltransferase CpoA (5). These non-PBP mechanisms have been identified only in laboratory mutants and account for very low-level resistance.
In this paper we report a non-PBP resistance determinant that is essential for the complete development of high-level penicillin and cephalosporin resistance in pneumococcal isolates. This resistance mechanism involves alteration in MurM, an enzyme involved in the biosynthesis of branched-stem cell wall muropeptides. The major peptide species in susceptible cell walls are of a linear-stem structure, compared to an abnormal branched-stem structure found in resistant cell walls (4). Branched-stem peptides presumably have superior binding to structurally altered PBPs and therefore become the preferred substrate for cell wall synthesis in resistant bacteria. Filipe and Tomasz (3) recently described the murMN operon in the pneumococcus that codes for the MurM and MurN proteins, which control the biosynthesis of branched-stem-structured cell wall muropeptides. They showed that a functional murMN operon is critical for the expression of penicillin resistance. We extend their findings by showing that alterations in MurM contribute to development of high-level penicillin and cephalosporin resistance in the pneumococcus.
Properties of the pneumococcal strains studied are shown in Table 1. Chromosomal DNAs were extracted from bacterial cells, and genes were amplified from the chromosomal DNAs by PCR using methods that have been described previously (13). For pbp gene PCR, primers have been described previously (11, 12). For murMN gene PCR, the following primer pairs were used: (i) murMN-up (TTCAAACGAAAGTAGTAGAATAG) and murMN-down3 (CCTATCAAACGAAAAAGCCAGCGCA) and (ii) murMN-up2 (TTTATAAATGAACCACTATTTATAG) and MurMN-down (GCATGTCTCTCCACCTTTCTAGC). PCR products were sequenced using the BigDye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, Calif.) and an Applied Biosystems model 310 automated DNA sequencer. Pneumococcal strain R6 was used as the recipient in transformation studies.
TABLE 1.
Properties of pneumococci
| Strain | MIC (μg/ml)
|
Origin | |
|---|---|---|---|
| Penicillin | Cefotaxime | ||
| R6 | 0.015 | 0.015 | United States (1930s) |
| 3191 | 16 | 4 | Hungary (1997–1998) |
| 149193 | 2 | 2 | South Africa (1982) |
| 50012 | 2 | 2 | South Africa (1982) |
| 8303 | 4 | 1 | South Africa (1995) |
| 20475 | 1 | 1 | South Africa (1996) |
| 806 | 1 | 1 | South Africa (1998) |
Chromosomal DNA and cloned genes were used as transforming DNA. Pneumococcal strains were made competent as follows. Bacteria were cultured in C medium (14) until the mid-exponential phase (optical density at 620 nm, 0.15) and, after the addition of glycerol to 10%, were frozen at −70°C in 500-μl aliquots. For transformation, 1 μg of DNA was added to 500 μl of competent cells, which were then incubated at 30°C for 45 min and at 37°C for 90 min. Eighty-microliter amounts were then plated onto Mueller-Hinton blood agar containing increasing concentrations of antibiotic, and the plates were incubated at 37°C for 48 h. Transformants were picked from the plates containing the highest antibiotic concentration. Transformation frequencies were on the order of 10−4 to 10−5.
Our study is based on isolate 3191, a representative of a Hungarian pneumococcal clone, isolated during the period 1997 to 1998, that was found to have notably high levels of penicillin (MIC, 16 μg/ml) and cefotaxime (MIC, 4 μg/ml) resistance (12). In that study, transformation of susceptible strain R6 with combinations of all six pbp genes from isolate 3191 resulted in transformants for which the maximum MICs were 4 μg of penicillin per ml and 2 μg of cefotaxime per ml. Resistance in these R63191/2X/2B/1A transformants was due to altered PBPs 2X, 2B, and 1A. The full MICs for the donor (penicillin MIC, 16 μg/ml; cefotaxime MIC, 4 μg/ml) could be reached only by further transformation of R63191/2X/2B/1A strains with chromosomal 3191 DNA, demonstrating the involvement of a non-PBP resistance determinant.
Our present study has now identified this resistance determinant. Experiments were initiated along the lines of the methods described by Adrian and coworkers (1). The following steps were taken. The chromosomal DNA was digested, and fragments of DNA with transforming ability were identified. In the process of identifying open reading frames with transforming ability, Filipe and Tomasz (3) described the murMN operon in the pneumococcus and proved that a functional murMN operon was critical for the expression of penicillin resistance. We therefore decided to investigate whether the product of this murMN operon was our non-PBP resistance determinant. PCR primers were designed, and the murMN operon was amplified from isolate 3191. The murMN DNA was shown to successfully transform R63191/2X/2B/1A to requiring the full MICs of the donor (penicillin MIC, 16 μg/ml; cefotaxime MIC, 4 μg/ml). R63191/2X/2B/1A/mur transformants could be selected with either penicillin or cefotaxime.
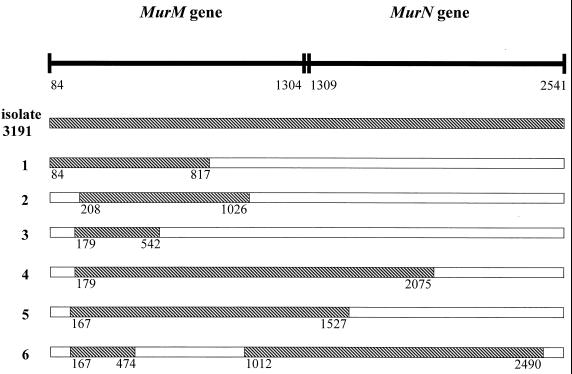
The murMN genes from isolate 3191 and from R63191/2X/2B/1A/mur transformants were then sequenced. The nucleotide sequence of the genes from susceptible strain R6 was used as the basis for comparison with resistant strains. The murMN genes from isolate 3191 displayed a mosaic pattern with 9.5% nucleotide sequence divergence from the genes of strain R6. The murMN operon is divided into the murM and murN genes, with the major sequence divergence occurring in the murM gene. The murM gene revealed 16.2% nucleotide sequence divergence, resulting in 74 amino acid mutations in the 406-amino-acid MurM protein, while the murN gene revealed a nucleotide sequence diversity of only 2.9%, which resulted in only 6 mutations in the 410-amino-acid MurN protein. Sequence analysis of the murMN genes from R63191/2X/2B/1A/mur transformants showed that altered MurM was the resistance determinant. Figure 1 schematically illustrates murMN genes from six R63191/2X/2B/1A/mur transformants, compared to the genes from donor isolate 3191, and indicates the regions of the genes where altered DNA from isolate 3191 has been introduced. A common area of alteration (nucleotides 208 to 474) could be identified among all transformants, which indicated that this area (located in the murM gene) housed the alterations leading to the development of resistance. This area corresponds to amino acid positions 42 to 131 of the MurM protein (Fig. 2), which account for 20 of the 74 mutations in the altered protein.
FIG. 1.
Schematic representation of murMN genes from six R63191/2X/2B/1A/mur transformants, compared to the genes from donor isolate 3191. The hatched areas in the transformants indicate the regions of the genes where altered DNA from isolate 3191 has been introduced. The nucleotide positions are numbered according to the sequence published by Filipe and Tomasz (3).
FIG. 2.
Amino acid sequences of MurM proteins from resistant pneumococcal isolates (lines b to d) compared to that from susceptible strain R6 (line a). Line b, isolate 3191; line c, isolate 149193; line d, isolate 50012. For resistant isolates, amino acid positions are shown only where a mutation has occurred compared to strain R6.
We then investigated whether the role played by altered MurM in the development of high-level penicillin and cefotaxime resistance was a unique characteristic of MurM from the Hungarian clone or whether murM genes from other resistant strains were also able to increase β-lactam MICs in pneumococci. For this analysis, we selected two South African isolates (149193 and 50012) with high-level resistance to both penicillin and cefotaxime (MICs, 2 μg/ml). Transformation experiments with murMN DNAs from these South African strains successfully transformed R63191/2X/2B/1A to full levels of resistance, in the same manner as did murMN DNA from Hungarian isolate 3191. This indicated that these South African strains may also have the requirement of altered MurM for high-level resistance development and that this resistance determinant may be widespread among pneumococci. Comparison of MurM sequences from isolates 3191, 149193, and 50012 revealed very similar patterns of alteration, and many common amino acid mutations were identified (Fig. 2). murM genes from three other isolates (8303, 20475, and 806) with lower levels of penicillin and cefotaxime resistance were then analyzed, all of which revealed MurM proteins with only a single V101A substitution. The murM genes from these isolates showed no resistance-transforming ability; therefore, in effect these isolates had unaltered MurM proteins. This surprisingly also included isolate 8303, for which the penicillin MIC was 4 μg/ml.
Our data show that for the pneumococcus to resist β-lactam antibiotics at extreme concentrations, a functional but altered MurM is required. The branched-stem peptide precursors produced by an unaltered MurM may be adequate for cells with the capacity to resist penicillin at concentrations of up to 4 μg/ml and cefotaxime at concentrations of up to 1 μg/ml, but at higher levels of resistance, an alteration in MurM is required. Altered MurM presumably results in a new species of branched-stem peptides, with superior binding to restructured PBPs in resistant bacteria, compared to the binding of the normal branched-stem peptides. Previous studies (2, 10) have shown that high-level penicillin and cefotaxime resistance can be transferred to susceptible strains using only altered pbp DNA; therefore, it is clear that high-level resistance is obtainable in the absence of altered MurM. Because our data have shown a requirement for altered MurM in the development of resistance, we have revealed an alternative pathway in the development of high-level penicillin and cefotaxime resistance.
In conclusion, we have shown that in conjunction with altered PBPs, a non-PBP resistance determinant (altered MurM) can be used as an alternative pathway in the development of high-level penicillin and cephalosporin resistance in the pneumococcus. Alteration of MurM is a mechanism particularly essential for full resistance development in a Hungarian clone.
Nucleotide sequence accession numbers.
murMN sequence data appear in the EMBL, GenBank, and DDBJ nucleotide sequence data libraries under the following accession numbers: AJ250764 (strain R6), AF319941 (strain 3191), and AF319942 (strains 149193 and 50012).
Acknowledgments
This work was supported by grants from the Medical Research Council, the South African Institute for Medical Research, and the University of the Witwatersrand.
REFERENCES
- 1.Adrian P V, Zhao W, Black T O, Shaw K J, Hare R S, Klugman K P. Mutations in ribosomal protein L16 conferring reduced susceptibility to evernimicin ( SCH27899): implications for mechanism of action. Antimicrob Agents Chemother. 2000;44:732–738. doi: 10.1128/aac.44.3.732-738.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barcus V A, Ghanckar K, Yeo M, Coffey T J, Dowson C G. Genetics of high level penicillin resistance in clinical isolates of Streptococcus pneumoniae. FEMS Microbiol Lett. 1995;126:299–303. doi: 10.1111/j.1574-6968.1995.tb07433.x. [DOI] [PubMed] [Google Scholar]
- 3.Filipe S R, Tomasz A. Inhibition of the expression of penicillin resistance in Streptococcus pneumoniae by inactivation of cell wall muropeptide branching genes. Proc Natl Acad Sci USA. 2000;97:4891–4896. doi: 10.1073/pnas.080067697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Bustos J, Tomasz A. A biological price of antibiotic resistance: major changes in the peptidoglycan structure of penicillin-resistant pneumococci. Proc Natl Acad Sci USA. 1990;87:5415–5419. doi: 10.1073/pnas.87.14.5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grebe T, Paik J, Hakenbeck R. A novel resistance mechanism against β-lactams in Streptococcus pneumoniae involves CpoA, a putative glycosyltransferase. J Bacteriol. 1997;179:3342–3349. doi: 10.1128/jb.179.10.3342-3349.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guenzi E, Gasc A-M, Sicard M A, Hakenbeck R. A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol Microbiol. 1994;12:505–515. doi: 10.1111/j.1365-2958.1994.tb01038.x. [DOI] [PubMed] [Google Scholar]
- 7.Hakenbeck R, Kaminski K, König A, van der Linden M, Paik J, Reichmann P, Zähner D. Penicillin-binding proteins in β-lactam resistant Streptococcus pneumoniae. Microb Drug Resist. 1999;5:91–99. doi: 10.1089/mdr.1999.5.91. [DOI] [PubMed] [Google Scholar]
- 8.Hakenbeck R, Tarpay M, Tomasz A. Multiple changes of penicillin-binding proteins in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1980;17:364–371. doi: 10.1128/aac.17.3.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laible G, Spratt B G, Hakenbeck R. Interspecies recombinational events during the evolution of altered PBP 2X genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1991;5:1993–2002. doi: 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 10.Muñoz R, Dowson C G, Daniels M, Coffey T J, Martin C, Hakenbeck R, Spratt B G. Genetics of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1992;6:2461–2465. doi: 10.1111/j.1365-2958.1992.tb01422.x. [DOI] [PubMed] [Google Scholar]
- 11.Smith A M, Klugman K P. Alterations in PBP 1A essential for high-level penicillin resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:1329–1333. doi: 10.1128/aac.42.6.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith A M, Klugman K P. Non-penicillin-binding protein mediated high-level penicillin and cephalosporin resistance in a Hungarian clone of Streptococcus pneumoniae. Microb Drug Resist. 2000;6:105–110. doi: 10.1089/107662900419401. [DOI] [PubMed] [Google Scholar]
- 13.Smith A M, Klugman K P, Coffey T J, Spratt B G. Genetic diversity of penicillin-binding protein 2B and 2X genes from Streptococcus pneumoniae in South Africa. Antimicrob Agents Chemother. 1993;37:1938–1944. doi: 10.1128/aac.37.9.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomasz A, Hotchkiss R D. Regulation of the transformability of pneumococcal cultures by macromolecular cell products. Proc Natl Acad Sci USA. 1964;51:480–487. doi: 10.1073/pnas.51.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waxman D J, Strominger J L. Penicillin-binding proteins and mechanism of action of β-lactam antibiotics. Annu Rev Biochem. 1983;52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 16.Zighelboim S, Tomasz A. Penicillin-binding proteins of multiply antibiotic-resistant South African strains of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1980;17:434–442. doi: 10.1128/aac.17.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]