Abstract
Introduction: Circular RNAs (circRNAs) are important regulators in various cancers, especially hepatocellular carcinoma. However, the role of circ RNA PTPRM (circPTPRM) in the development of non-small-cell lung cancer (NSCLC) remains unclear. Methods: We collected 26 clinical specimens (corresponding to 26 normal lung tissues) of lung adenocarcinoma and the expression of mir-139-5p and circPTPRM were first detected. Cell proliferation was detected by EdU method, invasion/migration ability of cells was evaluated by transwell method. And the correlation between circPTPRM and mir-139-5p was detected by luciferase reporter gene and RNA pull-down assay. Finally, we verified our hypothesis with BALB/c nude mice. Results: Through bioinformatics software, we found that circPTPRM was negatively correlated with mir-139-5p, and then we used human adenocarcinoma tissue samples to further verify their relationship and get the same result. EdU method, transwell method, and luciferase assay, RNA pull-down assay were applied, and the results show that the knockdown of circPTPRM inhibit proliferation, migration, and invasion of cells can be reversed by mir-139-5p inhibitor. Next, we used Starbase v2.0 to identify the target site of miR-139-5p and focused on SET domain containing 5 (SETD5). We derive the hypothesis by verifying the relationship between miR-139-5p and SETD5 that circPTPRM may interact with miR-139-5p/SETD5 axis. At last, we evaluated the effects of circPTPRM, SETD5, and miR-139-5p on tumor growth in vivo using BALB/c nude mice to prove the hypothesis. Conclusion: We thus conclude that circPTPRM promotes the progression of NSCLC by regulating the miR-139-5p/SETD5 axis.
Keywords: NSCLC, circPTPRM, miR-139-5p, SETD5, microRNA sponge
Introduction
Lung cancer has the highest morbidity and mortality of all cancers and is the leading cause of cancer death worldwide.1,2 According to global data from the International Agency for Research on Cancer (IARC), there were 2,093,876 new lung cancer patients and 176,007 deaths in 2018, about 85% of all cases are non-small cell lung cancer. 3 In addition to the traditional treatment methods of surgery, chemotherapy, and radiotherapy, NSCLC is now commonly targeted via biological immunotherapy. 4 Despite generally improved outcomes, the current situation is still poor for advanced NSCLC patients, whose median overall survival times are less than 1 year. 5 Recent guidelines from the National Comprehensive Cancer Network show that patients with specific predictive biomarkers who receive targeted therapy have a better quality of life and are more likely to achieve extended progression-free survival than those who receive conventional chemotherapy. 6 This emphasizes the importance of studying the mechanisms of NSCLC for diagnosis and treatment.
As an emerging and important biological regulatory factor, circular RNAs (circRNAs) are active in many physiological and pathological processes, and may become biomarker for tumor treatment and prognosis.7,8 CircRNA PTPRM (circPTPRM; ID: Hsa_circ_0007144) is located on CHR18:8076452-8143777. Its parent gene is the Protein Tyrosine Phosphatase receptor type M (PTPRM), whose members are affiliated with the Protein Tyrosine Phosphatase (PTP) family. PTP can regulate several cell biological processes including cell growth, cell connection, gene transcription, immune response, and oncogenic transformation.9,10 PTPRM is similar in structure to cell–cell adhesion molecules, and shows homophilic binding. It is involved in cell–cell adhesion for epithelial and cancer cells, and may be a key factor in the maintenance of tumor genesis and progression. 11 Recent studies have shown that circPTPRM is significantly upregulated in Hepatocellular Carcinoma (HCC) and promotes the proliferation, invasion, and migration of hepatocellular carcinoma cells. 12 However, how circPTPRM affects NSCLC progression remains unclear, and there is insufficient evidence on its underlying mechanisms. CircRNAs interact with mRNA splicing factors and 3′ untranslated region (3′-UTR) to degrade or process mRNA, thereby regulating gene expression. This is one of the mechanisms of epigenetics.13–15
Recently, circRNAs have been identified as sponges of microRNAs, known as competitive endogenous RNAs (ceRNAs).16,17 These ceRNAs share sequences recognized by miRNAs known as microRNA recognition elements (MREs).18–20 MiRNAs are short (20-25 nucleotides) non-coding RNAs (ncRNAs) that play important functions in many biological processes; 21 they pair with the target mRNA in the 3′-UTR to regulate gene expression at the posttranscriptional level 22 and regulate different targets with important functions across a wide range of biological and medical processes. 23 Studies have also shown that miRNAs are involved in the progression of NSCLC. MiR-340-5p inhibits the growth and metastasis of NSCLC cells by regulating ZNF503, 24 while miR-1269a acts as a carcinogenic miRNA in NSCLC by inhibiting the expression of SOX6. 25 miR-139-5p also plays an important regulatory role in tumorigenesis by interacting with circRNA and the target gene. 26 However, there is still a lack of evidence about the specific role of miR-139-5p in NSCLC development.
Bioinformatics approaches have shown that the SET domain containing 5 (SETD5) is the target gene of miR-139-5p. SETD5 plays an important role in the co-transcriptional regulation of mammalian development and histone acetylation.27,28 However, there is a research gap concerning the expression pattern and biological role of SETD5 in human malignant tumors. In another research, Kuechler et al demonstrated that SETD5 dysfunction is related to intellectual disability, and is a key factor for the 3p25.3 microdeletion syndrome phenotype.29,30 Poissonnier et al showed that miR-126-5p inhibits SETD5 to eliminate leukocyte migration across the endothelium, thus indicating possible involvement in tumor migration and invasion. 31 Further, a microarray analysis showed that the SETD5 locus is associated with prostate cancer aggressiveness, 32 while a transcriptomics study showed that SETD5 is associated with the treatment response of metastatic prostate tumors. 33 Particularly, high levels of SETD5 mRNA levels are also associated with poor prognoses among NSCLC patients. 34
However, we are less known about the possible relationships among SETD5, circPTPRM, and miR-139-5p in the progression of NSCLC. This study explored the role of circPTPRM in the occurrence and development of NSCLC, thus revealing a previously unknown function of circPTPRM in NSCLC promotion, specifically by adjusting the miR-139-5p/SETD5 axis.
Materials and Methods
The reporting of this study conforms to ARRIVE 2.0 guidelines. 35
NSCLC (Adenocarcinoma) Tissue Samples
We collected 26 clinical specimens (corresponding to 26 normal lung tissues) of NSCLC between September 2019 and June 2020. All patients were histopathologically diagnosed as lung adenocarcinoma. All cases were independently diagnosed and reviewed by 2 clinicians. No patients received systemic or local treatments prior to surgery. The tissues and corresponding adjacent tissues were immediately frozen in liquid nitrogen and stored at −80°C. All patients provided written informed consent prior to enrollment in the study.
Cell Culture Treatment and Cell Transfection
The normal human bronchial epithelial 16HE cells and 4 lung adenocarcinoma cell lines (eg A549, H1299, PC-9, and H1975) were obtained from the American Type Tissue Culture Collection (ATCC) and Procell Life Science & Technology Co. Ltd. They all derivate from human and authenticate by Short Tandem Repeat (STR). They were cultured in an Roswell Park Memorial Institute (RPMI) 1640 culture medium (Gibco™ catalog number: 31870082) containing 10% fetal bovine serum, then stored in a 37°C 5% CO2 environment. MiR-139-5p mimics or inhibitors, PTPRM siRNA or vectors, and their corresponding controls were provided by GenePharma (Shanghai, China). Via the Lipofectamine 2000 reagent (Invitrogen 11668-027), these were sent to A549 and PC-9 cells. We collected cells for further analysis 48 h after transfection.
CCK8 Assays
We used the CCK8 method (CCK-8 kit Sigma-Aldrich 96992) to detect the survival and proliferation of NSCLC cells. After seeding A549 and PC-9 cells (4 × 104 cells/cm2) in 96-well plates, we added 10 μL/ of CCK8 solution to each well, followed by incubation at 37°C for 1 h. We dissolved the supernatant in 150 μL dimethylsulfoxide (DMSO), then measured the optical density of each well via absorbance at 450 nm with a microplate reader.
EdU Assay
We used an EdU detection kit (Invitrogen™ C10644) to detect cell proliferation. The NSCLC cells were briefly cultured with EdU for 2 h, then fixed with paraformaldehyde (4%, room temperature, 30 min). Triton X-100 was then used to permeabilize the cells (0.4%, 10 min), which were treated with an EdU staining cocktail in the dark (room temperature, 30 min). Next, we stained the cell nucleus with Hoechst (room temperature, 30 min) and analyzed the image through a fluorescence microscope.
Transwell Assay
The cells and matrix gel were incubated overnight at 4°C. We diluted the cells with a serum-free medium at a ratio of 1:4 and added the cells to the top transwell chamber (Corning 3412). The room was kept at equilibrium while these remained in the incubator for 30 min. We then separated the cells, washed them with a serum-free medium, counted them, and created a cell suspension. We washed the matrix gel once with a serum-free medium, then inoculated a cell suspension of about 1 to 3 × 105 cells/mL in the top chamber. We added cells containing 10% fetal bovine serum medium to the basolateral compartment and incubated them for 24 h at 37°C. Next, we washed the transwell chamber twice with phosphate buffer saline (PBS) for 5 min, then fixed it with 5% pentanediol at 4°C, stained it with 0.1% crystal violet for 30 min, washed it twice with PBS, and observed the results under a microscope. We used the number of cells that passed through the matrix gel as an index to assess their invasion ability.
Flow Cytometry Assay
We used an Annexin V-FITC/propidium iodide (PI) Apoptosis Kit (Sigma-Aldrich 08168) to detect cell apoptosis according to the manufacturer’s instructions. 36 We collected the cells, washed them twice with frozen PBS, then stained them with 5 μL FluoresceinIsothiocyanate (FITC)-labeled Annexin-V and 10 μL PI at room temperature for 20 min. We used flow cytometry to detect stained cells, and calculated the apoptosis rate as the percentage of early/primary apoptotic cells (Annexin V + /PI−) and late/secondary apoptotic cells (Annexin V + /PI + ).
Luciferase Activity Assay
According to the bioinformatics website Starbase v2.0, the sequence of circPTPRM and 3′-UTR of SETD5 containing the potential binding sites of miR-139-5p were synthesized and purchased from RiboBio. We cloned or mutated the combined sequences, then inserted them into the luciferase reporter gene plasmid pmirGLO (Promega E1330) to construct wild-type reporter vectors PTPRM (PTPRM⁃WT) and SETD5 (SETD5-WT), mutant-type reporter vectors PTPRM (PTPRM⁃MUT), and SETD5 (SETD5-MUT). The vectors of PTPRM-WT or PTPRM-MUT were transfected into cell together with NC mimics, miR-139-5p mimics, and an NC or miR-139-5p inhibitor. Similarly, the SETD5-WT or SETD5-MUT were transfected into cell with NC or miR-139-5p mimics. We evaluated relative luciferase activity using a dual-luciferase reporter assay system (Dual-Luciferase Reporter Assay Kit Vazyme DL101-01) 48 h after transfection, according to the manufacturer’s instructions.
RNA Pull-Down
We transduced the cells with 50 nM biotin-labeled wild-type (WT) bio-miR-139-5p and mutant (MUT)-bio-miR-139-5p for 48 h. We collected and washed the cells with PBS, then incubated them in a specific incubation lysis buffer for 10 minutes. The lysate was incubated with M-280 magnetic beads streptavidin pre-coated with BSA. The magnetic beads were incubated at 4°C for 3 h, 1X combined with washing buffer and cell lysis buffer. Finally, we purified the bound RNA with Trizol (Generay GK3004) and determined the concentration.
Analysis of Tumorigenicity in Nude Mice
We followed the Guide for the Care and Use of Laboratory Animals, 8th Edition. 37
The following is a list of species, sex, strain, age, source, husbandry conditions and other information.
Species: Nude mice
Sex: Male and female
Strain: BALB/c
Age: 12 to 13 weeks
Source: Abcam
Husbandry conditions: Same-sex littermates were housed together in individual ventilated cages with 2 or 4 mice per cage. All mice were maintained on a regular diurnal lighting cycle (12:12 light–dark) with ad libitum access to food and water. Chopped corn cob was used as bedding. Environmental enrichment included nesting material, Poly Vinyl Chloride (PVC) pipe, and shelter. Mice were housed under broken barrier-specific pathogen-free conditions.
We analyzed the effects of circPTPRM and miR-139-5p on tumor growth in BALB/c nude mice, which we randomly divided into 3 groups (n = 9 small sample not power calculation). To establish an in vivo tumor model, we treated the A549 cells with control shRNA (Ctrl) and PTPRM shRNA (sh-PTPRM), or co-treated them with sh-PTPRM and miR-139-5p inhibitors. We subcutaneously injected approximately 1 × 107 cells into the treated mice, then measured tumor growth every 7 days following an initial 7-day buffer period. The mice were sacrificed 28 days post-injection, at which point we graded the tumors. We estimated tumor volume (V) based on length and width measurements taken with calipers, then used SETD5 (Abcam ab204363) antibody immunohistochemical staining to detect the expression level of SETD5 in tumor tissues. Next, we subjected the tumor tissues to the western blot method to detect the protein expression levels of STED5 (1:100) and reduced glyceraldehyde-phosphate dehydrogenase (GAPDH) (1:100) (Beyotime Biotechnology AF1186). Finally, we used quantitative real-time polymerase chain reaction (qRT-PCR) to detect the expression of miR-139-5p.
RNA Extraction and qRT-PCR
We extracted total RNA from cell and lung cancer tissues using the Trizol reagent, then synthesized 400 ng of RNA into cDNA via reverse transcription. The following procedure was used: transcribe at 16°C for 30 minutes, then incubate at 42°C for 30 min, and inactivate the enzyme at 85°C for 5 min. We used the SYBRH Select Master Mix (Applied Biosystems™ 4472908) for rapid quantitative PCR. The transcription reaction used the following parameters: 16°C, 30 min, 42°C, 30 min, 84°C, 5 min. The qRT-PCR reaction used the following parameters: 95°C, 2 min, then 95°C, 10 s, 60°C, 20 s; 40 cycles. We normalized all results to GAPDH expressions and adopted the 2−ΔΔct method for qualitative analysis. The primer sequence was as follows: miR-139-5p, 5′-TCTACAGTGCACGTGTCTCCAG-3′ and 5′-ACCTGCGTAGGTAGTTTCATGT-3′; U6, 5′-CTCGCTTCGGCAGCACA-3′ and 5′-AACGCTTCACGAATTTGCGT-3′; SETD5, 5′-CGATCATCCCTCGTTCTGACC-3′ and 5′-CCCCTGCACTTGTCACAGTT-3′; circPTPRM, 5′-GGGCATCTTGCTGTTCGTGA-3′ and 5′-TTCAGTGGGAACAGCACCTG-3′; GAPDH, 5′-CAGGAGGCATTGCTGATGAT-3′ and 5′-GAAGGCTGGGGCTCATTT-3′.
Western Blot
We used RIPA buffer (CST 9806) to extract total proteins from cells or mouse tissues, then determined protein concentrations using the BCA protein quantification kit (Abbkine KTD3001). We separated equal numbers of protein samples via Sodium Dodecyl Sulfate PolyAcrylamide Gel Electrophoresis (SDS-PAGE) (12% polyacrylamide gel), then transferred the results to a Poly vinylidene fluoride (PVDF) membrane (Millipore IPFL07810). The membrane was blocked with 5% milk and incubated overnight at 4°C with antibodies SETD5 (1:100) and GAPDH (1:100), then incubated for 1 h at room temperature with the corresponding secondary antibody (Thermo Fisher 31431) (1:1000), and subsequently visualized using the Odyssey CLx infrared imaging system.
Statistical Analysis
Using the IBM SPSS 18.0 software package, we calculated statistical results via the Student's t-test or one-way ANOVA. Experimental data were expressed as means ± standard deviations (x ± SD) for all 3 independent experiments. Statistical significance was determined at P <.05.
Results
The Expression of circPTPRM was Upregulated in NSCLC Tissue Samples and Cell Lines While the Expression of miR-139-5p was Downregulated
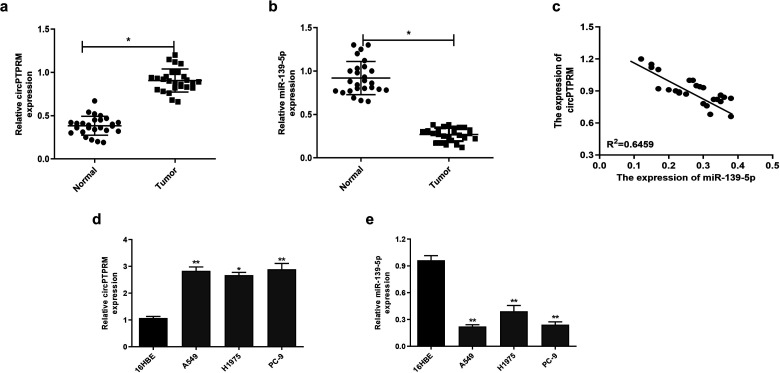
We examined the correlation between circPTPRM and mir-139-5p in NSCLC progression by evaluating thier expression of them in NSCLC patient tissues and NSCLC cell lines. Our experiments showed that circPTPRM and mir-139-5p were significantly upregulated (Figure 1a) and downregulated (Figure 1b) in NSCLC tissues, respectively, compared with adjacent normal tissues. As shown in Figure 1c, the expression of circPTPRM was significantly negatively correlated with miR-139-5p, suggesting that circPTPRM and mir-139-5p may be involved in the development of NSCLC. Compared to 16HBE cells, we also found that the respective expressions of circPTPRM and miR-139-5p were upregulated (Figure 1d) and downregulated (Figure 1e) in NSCLC cell lines, including A549, H1975, and PC-9 . This strengthens the evidence that circPTPRM and mir-139-5p are associated with NSCLC pathologic processes.
Figure 1.
circPTPRM was upregulated and miR-139-5p is downregulated in the pulmonary adenocarcinoma tissue samples and cell lines. (a and b) The expression levels of circPTPRM and miR-139-5p were measured by qRT-PCR in the NSCLC patient tissues (n = 26) and the adjacent normal tissues (n = 26). (c) The correlation relationship of circPTPRM with miR-139-5p was analyzed by qRT-PCR in the NSCLC tissues sample. (d and e) The expression levels of circPTPRM and miR-139-5p were assessed by qRT-PCR in pulmonary adenocarcinoma cell lines (eg A549, H1975, and PC-9) and normal human bronchial epithelial 16HBE cells. Data are presented as mean ± SD.
Statistical significant differences were indicated as *P < .05, **P < .01. Abbreviations: circPTPRM: circ RNA PTPRM; qRT-PCR: quantitative real-time polymerase chain reaction.
The Depletion of circPTPRM; Inhibited Proliferation, Migration, and Promoted Apoptosis in NSCLC Cell Lines
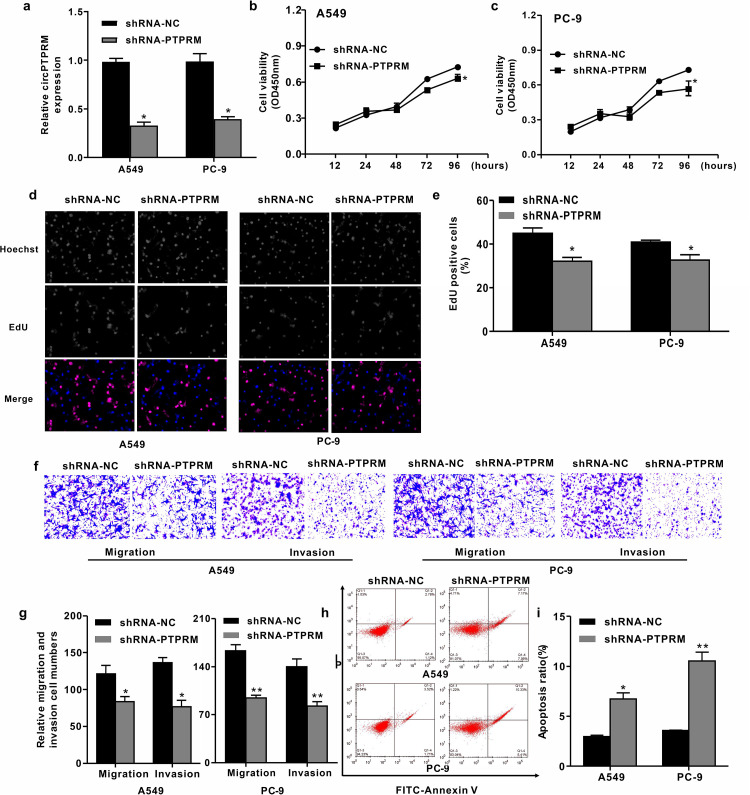
We conducted a loss-of-function experiment to examine the biological function of circPTPRM in NSCLC. Here, NSCLC cell lines A549 and PC-9 were infected with a lentiviral plasmid carrying either (dele) circPTPRM shRNA (PTPRM) or the corresponding control shRNA (shRNA-NC), and we verified the interference efficiency of circPTPRM via qRT-PCR (Figure 2a). The (dele) CCK8 results showed that knocking down circPTPRM significantly reduced the viability of A549 and PC-9 cells (Figure 2b and c). Meanwhile, circPTPRM inhibition also inhibited EdU-positive cells (Figure 2d and e), indicating that circPTPRM is necessary for the proliferation of NSCLC cells. Further, transwell testing showed that a circPTPRM knockout weakened the migration and invasion ability of A549 and PC-9 cells (Figure 2f and g), suggesting that circPTPRM is involved in the in vitro progression of NSCLC. Similarly, inhibition of circPTPRM significantly increased the apoptosis of A549 and PC-9 cells (Figure 2h and i), thus confirming the role of circPTPRM in the development and progression of NSCLC.
Figure 2.
Downregulation of circPTPRM inhibited cell proliferation, migration, invasion, and promoted apoptosis in pulmonary adenocarcinoma cells. (a) The expression levels of circPTPRM were measured by qRT-PCR assays in A549 and PC-9 cells. (b and c) The cell viability was tested by CCK8 assays in A549 and PC-9 cells. (d and e) The cell proliferation was tested by EdU assays (Apollo Fluorochromes) in A549 and PC-9 cells (magnification 10X). (f and g) The cell migration and invasion ability were examined by transwell assays in A549 and PC-9 cells (magnification 10X). (h and i) The cell apoptosis was measured by flow cytometry analysis in the cells, in which the x-axis represented the Annexin V-FITC and the y-axis represented the propidium iodide (PI). Data are presented as mean ± SD.
Statistical significant differences were indicated as *P < .05, **P < .01. Abbreviations: circPTPRM: circ RNA PTPRM; qRT-PCR: quantitative real-time polymerase chain reaction; SD: standard deviation.
circPTPRM Reversely Modulated the miR-139-5p Expression by Acting as its ceRNA
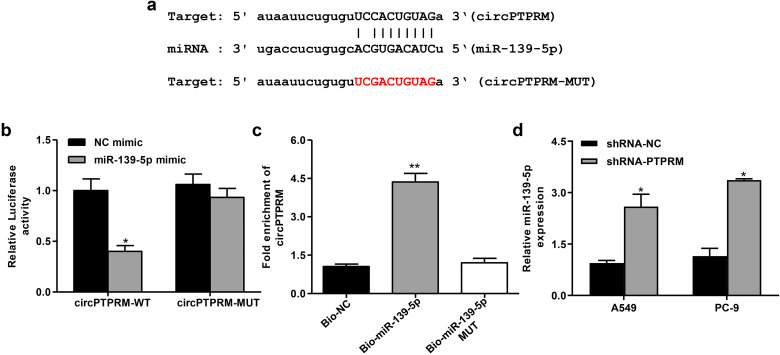
To further evaluate the relationship between circPTPRM and mir-139-5p, we performed a bioinformatics analysis on a public database. Therefore, we found that circPTPRM shares a binding site with miR-139-5p, indicating that miR-139-5p may be associated with circPTPRM interaction (Figure 3a). Dual-luciferase reporter gene assay showed that miR-139-5p mimics can significantly inhibit the expression of WT-PTPRM, but cannot affect the expression of MUT-PTPRM (Figure 3b). Subsequently, RNA pull-down experiments showed that WT-miR-139-5p binds to circPTPRM was more capable of binding to circPTPRM than MUT-mir-139-5p, showing that miR-139-5p could directly bind to circPTPRM (Figure 3c). We transduced sh-PTPRM into A549 and PC-9 cells to evaluate the regulatory effect of circPTPRM on mir-139-5p, and found that the downregulation of circPTPRM could significantly promote the expression of mir-139-5p in A549 and PC-9 cells (Figure 3d). These results indicate that circPTPRM may play the spongy effect of miR-139-5p in NSCLC cells.
Figure 3.
circPTPRM interacts with miR-139-5p in vitro. (a) Potential interaction between circPTPRM and miR-139-5p was identified by the bioinformatic analysis using Starbase v2.0 (http://starbase.sysu.edu.cn/index.php). (b) Luciferase activities of PTPRM (PTPRM-WT) and PTPRM with the miRNA-binding site mutant (PTPRM-MUT) were determined by luciferase reporter gene assays in the A549 cells treated with NC mimics, miR-139-5p mimic, respectively. (c) The interaction of PTPRM with wild-type (WT) miR-139-5p (bio-miR-139-5p), miR-139-5p with the PTPRM-binding site mutant (bio-miR-139-5p MUT) was examined by RNA pull-down followed by qRT-PCR in the A549 cells. (d) The expression levels of miR-139-5p were measured by qRT-PCR in the A549 cells infected with the lentiviral plasmids carrying PTPRM shRNA (sh-PTPRM) or corresponding control shRNA (sh-NC). Data are presented as mean ± SD.
Statistical significant differences were indicated as *P < .05, **P < .01. Abbreviations: circPTPRM: circ RNA PTPRM; PTPRM-WT, wild-type reporter vectors PTPRM; PTPRM-MU, mutant-type reporter vectors PTPRM; qRT-PCR: quantitative real-time polymerase chain reaction; SD: standard deviation.
circPTPRM Promotes NSCLC Progression by Targeting miR-139-5p in Vitro
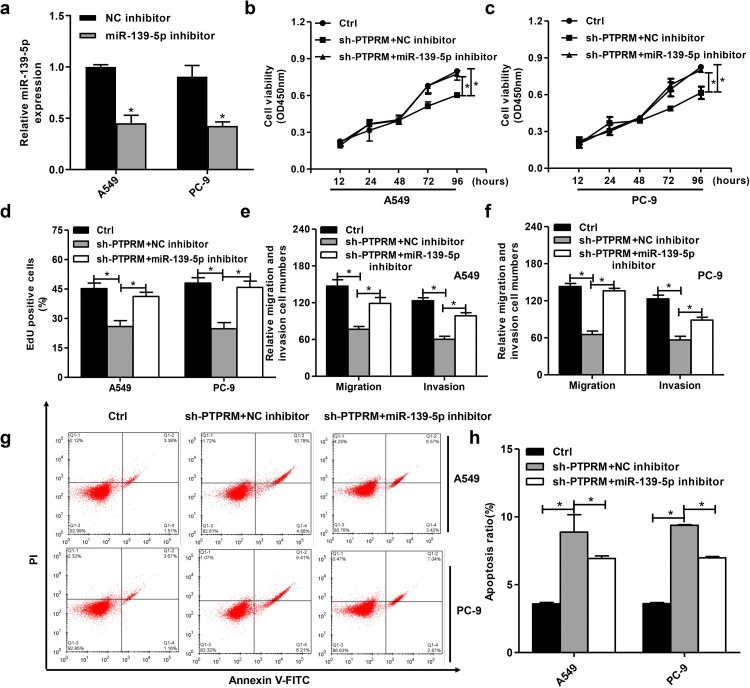
We then investigated the roles of the circPTPRM/miR-139-5p axis in NSCLC development in vitro (Figure 4a). CCK8 test showed that PTPRM consumption reduced; miR-139-5p inhibitors enhanced the viability of A549, PC-9 cells and weakened the effects of PTPRM knockdown (Figure 4b,c). Second, EdU experiments showed that PTPRM consumption was reduced, while miR-139-5p inhibitors, respectively, enhanced the proliferation of A549, PC-9 cells and reduced the effects of inhibiting PTPRM (Figure 4d). Moreover, the consumption of PTPRM was inhibited; miR-139-5p inhibitors promoted the migration and invasion of A549, PC-9 cells and weakened the effects of PTPRM consumption (Figure 4e and f). Finally, a flow cytometry analysis showed that apoptosis increased after PTPRM removal. Meanwhile, miR-139-5p inhibitors reduced the effects of this removal (Figure 4g and h). These data indicate that circPTPRM may induce the progression of NSCLC by targeting miR-139-5p in vitro.
Figure 4.
circPTPRM promotes NSCLC cell proliferation, migration, invasion, and promoted apoptosis by targeting miR-139-5p in vitro. (a) The expression levels of miR-139-5p were measured by qRT-PCR assays in A549 and PC-9 cells. (b and c) The cell viability was tested by CCK8 assays in A549 and PC-9 cells. (d) The cell proliferation was tested by EdU assays in A549 and PC-9 cells. (e and f) The cell migration and invasion were examined by transwell assays in A549 and PC-9 cells. (g and h) The cell apoptosis was measured by flow cytometry analysis in the cells, in which the x-axis represented the Annexin V-FITC and the y-axis represented the propidium iodide (PI). Data are presented as mean ± SD.
Statistical significant differences were indicated as *P < .05. Abbreviations: circPTPRM: circ RNA PTPRM; qRT-PCR: quantitative real-time polymerase chain reaction; SD: standard deviation.
MiR-139-5p Inhibits NSCLC Progression by Targeting SETD5 in Vitro
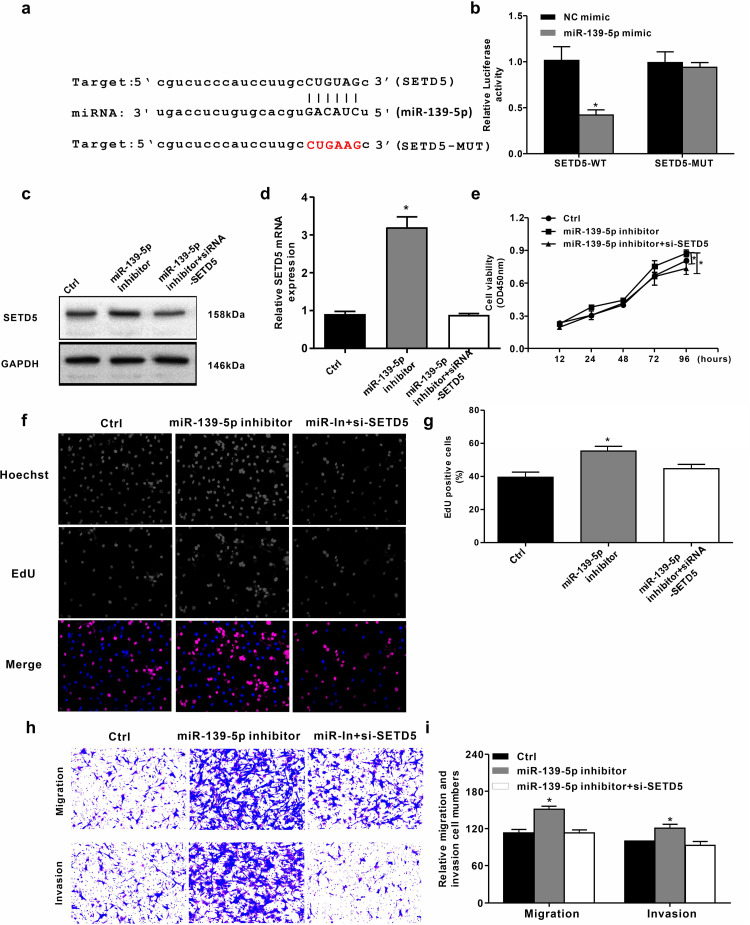
We attempted to locate the target gene of miR-139-5p in NSCLC development in vitro. We used Starbase v2.0 (http://starbase.sysu.edu.cn/) to identify the target site of miR-139-5p in SETD5 3′- UTR (see online Supplemental material for the other targets of miR-139-5p) (Figure 5a). Notably, miR-139-5p mimics inhibited luciferase activity for WT SETD5, but does not affect SETD5-MUT in A549 cells (Figure 5b). We then transfected the SETD5 siRNA and control siRNA into A549 cells and verified them, expression of the SETD5 protein was upregulated by the miR-139-5p inhibitor, suggesting that miR-139-5p targets SETD5 (Figure 5c). Similarly, the miR-139-5p inhibitor enhanced the expression of SETD5 mRNA (Figure 5d). Further, CCK8 and EdU analyses showed that SETD5 consumption impaired cell proliferation, while the miR-139-5p inhibitor enhanced it (Figure 5f and g). Meanwhile, the miR-139-5p inhibitor promoted the migration and invasion of A549 cells, but the SETD5 knockout could block these effects (Figure 5h and i). These results may suggest that miR-139-5p inhibits the progression of NSCLC by targeting SETD5 in vitro.
Figure 5.
MiR-139-5p inhibits NSCLC progression by targeting SETD5 in vitro. (a) The interaction of miR-139-5p and SETD5 3′’ UTR was identified by bioinformatic analysis using Starbase v2.0 (http://starbase.sysu.edu.cn/index.php). (b) The luciferase activities of wild-type (WT) SETD5 (SETD5-WT) and SETD5 with the miRNA-binding site mutant (SETD5-MUT) were determined by luciferase reporter gene assays in the A549 cells treated with control mimic (Ctrl), miR-139-5p mimic, respectively. (c) The protein expression of SETD5 and GAPDH was tested by Western blot analysis in the A549 cells treated with control inhibitor (NC inhibitor), miR-139-5p inhibitor, or SETD5 siRNA (SETD5-siRNA), respectively. (d) The mRNA expression of SETD5 was measured by qRT-PCR in the A549 cells transfected with control siRNA (NC-siRNA) or SETD5 siRNA (SETD5-siRNA). (e to i) The A549 cells were treated with control inhibitor (Ctrl), miR-139 inhibitor, or co-treated with SETD5 siRNA (SETD5-siRNA) and inhibitors of miR-139-5p, respectively. (e) The cell viability was analyzed by CCK8 assays in the cells. (f and g) The cell proliferation was tested by EdU assays in the cells (magnification 10X). (h and i) The cell migration and invasion were examined by transwell assays in the cells (magnification 10X). Data are presented as mean ± SD.
Statistical significant differences were indicated as *P < .05. Abbreviations: SD: standard deviation; SETD5: SET domain containing 5; SETD5-WT, wild-type reporter vectors SETD5; SETD5-MU, mutant-type reporter vectors SETD5; qRT-PCR: quantitative real-time polymerase chain reaction.
CircPTPRM Contributes to Tumor Growth of NSCLC via miR-139-5p/SETD5 Axis in Vivo
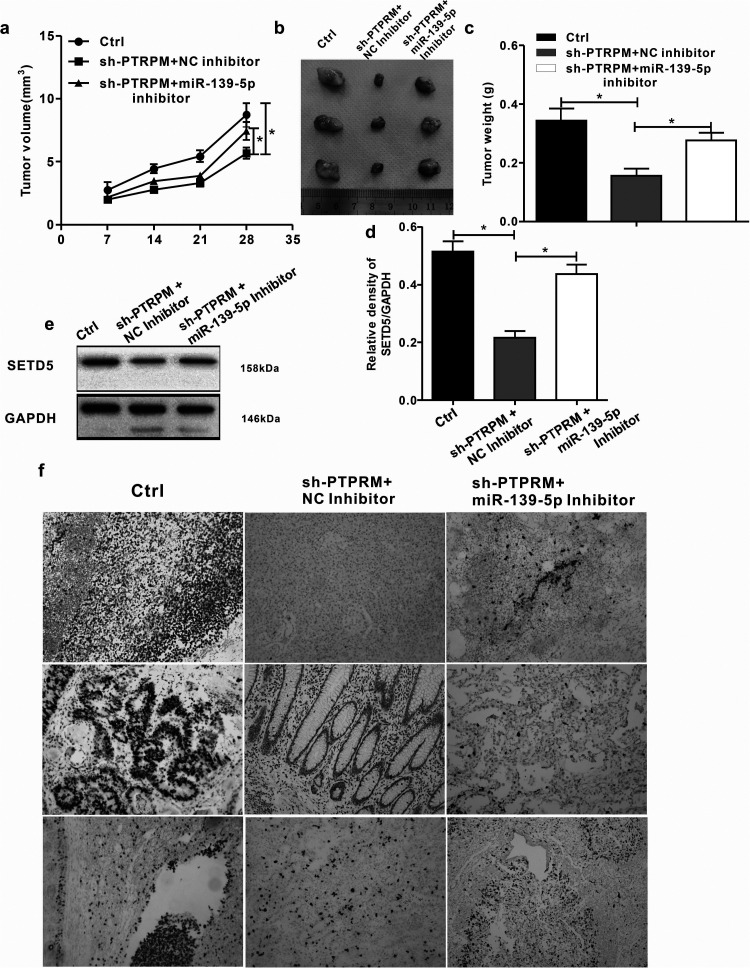
We further evaluated how the circPTPRM/miR-139-5p axis affected NSCLC development in vivo through a tumorigenicity analysis on nude mice injected with A549 cells, which were either treated with the control shRNA and PTPRM shRNA or PTPRM shRNA and miR-139-5p inhibitor. The depletion of PTPRM significantly reduced tumor growth in A549 cells. As an inhibitor of tumor growth, miR-139-5p was able to rescue the phenotype. We then measured tumor growth volume (Figure 6a), tumor size (Figure 6b), tumor weight (Figure 6c), and STED5 expression in the lung tissue of the mice (Figure 6f). We also demonstrated that the miR-139-5p inhibitor enhanced SETD5 expression in the lung tissue of the mice by inhibiting PTPRM (Figure 6e), while the miR-139-5p inhibitor inhibited the expression of miR-139-5p by inhibiting PTPRM, which was attenuated by the miR-139-5p inhibitor (Figure 6d). In conclusion, these data suggest that circPTPRM promotes NSCLC tumor growth in vivo via the miR-139-5p/SETD5 axis.
Figure 6.
circPTPRM contributes to tumor growth of NSCLC via miR-139-5p/SETD5 axis in vivo. (a to f) The effect of circPTPRM–miR-139-5p axis on tumor growth of NSCLC cells in vivo was analyzed by nude mice tumorigenicity assay. The A549 cells were treated with control shRNA (Ctrl), PTPRM shRNA (sh-PTPRM ), or co-treated with sh-PTPRM , and the inhibitors of miR-139-5p and injected into the nude mice. (a) Tumor volumes were calculated and shown. (b) Representative images of dissected tumors from nude mice were presented. (c) Tumor weight was calculated and shown. (d) The expression levels of miR-139-5p were tested by qRT-PCR in the tumor tissues. (e) The protein expression levels of SETD5 and GAPDH were examined by Western blot analysis in the tumor tissues. (f) The expression levels of SETD5 of the tumor tissues were measured by immunohistochemical staining (magnification 10X). Data are presented as mean ± SD.
Statistical significant differences were indicated as *P < .05.Abbreviations: circPTPRM: circ RNA PTPRM; SD: standard deviation; SETD5: SET domain containing 5; qRT-PCR: quantitative real-time polymerase chain reaction.
Discussion
NSCLC is the main type of lung cancer, with high incidence and high mortality. CircRNA, as an important regulatory factor in cancer development, is also involved in the regulation of NSCLC.
Previous research has shown that circRNA_101237 has a carcinogenic effect in NSCLC by regulating the miR-490-3p/MAPK1 axis. 38 Further, circRNA_0000429 regulates the development of NSCLC by acting as a sponge of miR-1197 to control MADD, 39 circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p, 40 and circRNA_103993 promotes the progression of NSCLC by regulating the miR-1271/ERG signal. 41 There are few studies on circPTPRM, but some studies have shown that circPTPRM is related to the occurrence and development of hepatocellular carcinoma. 12
This study found increased expression levels of circPTPRM in NSCLC patients and NSCLC cell lines. The deletion of circPTPRM reduced the proliferation, migration, and invasion of NSCLC cells by targeting miR-139-5p in vitro, while circPTPRM promoted the growth of NSCLC tumors through the miR-139-5p/SETD5 axis in vivo. Our experiments revealed previously unknown functions of circPTPRM in the development of NSCLC, thus producing valuable evidence for the role of circRNA in this field. These findings on the mechanism of the relationship between circPTPRM, mir-139-5p, and SETD5 enrich our understanding of circPTPRM, and provide a basis for future studies on the role of circPTPRM.
As the main component of ncRNA, miRNA critically interacts with other ncRNAs in the physiological and pathological processes of NSCLC. MiR-99b inhibits NSCLC cell invasion and migration by targeting NIPBL. 42 Hydrophobically modified let7b miRNA enhances biodistribution to NSCLC and downregulates HMGA2 in vivo. 43 MiR-597 inhibits NSCLC progression by negatively regulating CDK2 expression. 44 MiR-126 targeting PIK3R2 inhibits NSCLC A549 cell proliferation, migration, and invasion by regulating the PTEN/PI3K/AKT pathway. 45 As a key regulator, miR-139-5p is also involved in the regulation of cancer development through interactions with circRNA and targeted genes. Circular RNA circBACH2 plays a role in papillary thyroid carcinoma by sponging miR-139-5p and regulating LMO4 expression. 26 LncRNA RP3-439F8.1 promotes GBM cell proliferation and progression by sponging miR-139-5p to upregulate NR5A2. 46 MiR-139-5p reverses CD44 + /CD133 + -associated multidrug resistance by downregulating Notch1 in colorectal carcinoma cells. 47 MiR-139-5p-ZEB1 is a molecular regulator of the growth, invasion, and epithelial-to-mesenchymal transition of cervical cancer. 48 Moreover, reduced expression levels of miR-139-5p have been found in NSCLC patients, which is related to the diagnosis and prognosis of NSCLC. 49
This study found decreased expression levels of miR-139-5p in NSCLC patients and NSCLC cell lines. Our mechanical studies further prove that miR-139-5p inhibits the progression of NSCLC by targeting SETD5; here, miRNA can be absorbed by the circPTPRM sponge. These data highlight a previously unreported role of miR-139-5p in NSCLC development while identifying the new upstream circPTPRM and downstream target SETD5 in NSCLC regulation. Many studies have confirmed that SETD5 is active in the development of several cancers. For example, high expressions of SETD5 are related to poor clinical outcomes in human gastric cancer. 50 It is also a potential prognostic biomarker that promotes esophageal squamous cell carcinoma stemness. 51 Whole transcriptome sequencing has shown that increased SETD5 expression levels are present in metastatic castration-resistant prostate cancer. 33 SETD5-coordinated chromatin reprogramming regulates adaptive resistance to targeted pancreatic cancer therapy. 52 SETD5 facilitates tumor growth and pulmonary metastasis through the upregulation of AKT1 signaling in breast cancer. 53 In addition, it may play a key role in the development and progression of bladder cancer. 54 As for research on NSCLC, SETD5 facilitates tumor cell invasion and is associated with poor prognosis; 34 it also enhances cell stemness via the PI3K/Akt/mTOR pathway. 55
However, there are still some limitations in our research. Based on online databases, we found a series of miRNAs that had potential binding sites with circPTPRM. We finally chose miR-139-5p because it has a targeting relationship with circPTPRM and SETD5, as well as a significant negative correlation in tumor tissues. And in the end, we did confirm their relationship. In the subsequent studies, we will search for more evidence to support the relationship of them by performing sequencing analysis or from TCGA or other databases. In terms of animal trials, due to the limitations of experiments and funds, we chose a small sample. Thus, a further study based on large sample size is needed to confirm all speculations in this study.
Our data further show that SETD5 is involved in the progression of NSCLC. Specifically, we found that miR-139-5p targets SETD5, which may be sponged by circPTPRM in the system. This study, therefore, provides new evidence showing that SETD5 is a crucial factor in the development of NSCLC.
Conclusions
This study found that circPTPRM affects NSCLC. Here, miR-139-5p can inhibit the promoting effect of circPTPRM on NSCLC progression, while circPTPRM enhances the expression of SETD5 by inhibiting miR-139-5p. Our experiments produced evidence for the regulation of the circPTPRM/miR-139-5p/SETD5 axis in NSCLC development. Therefore, circPTPRM may become a promising target for NSCLC treatments.
Key Points
Significant findings of this study:
circPTPRM plays a key role in the development of NSCLC.
What this study adds:
Although it is known that circular RNAs (circRNAs) are important regulators in the progression of a variety of cancers, there was heretofore a lack of information on this in NSCLC development.
Supplemental Material
Supplemental material, sj-xlsx-1-tct-10.1177_15330338221090090 for CircRNA PTPRM Promotes Non-Small Cell Lung Cancer Progression by Modulating the miR-139-5p/SETD5 Axis by Zeyong Jiang, Jian Zhao, Hanlin Zou and Kaican Cai in Technology in Cancer Research & Treatment
Acknowledgments
First and foremost, I would like to show my deepest gratitude to my supervisor, Dr Cai Kaican, a respectable, responsible, and resourceful scholar, who has provided me with valuable guidance in every stage of the writing of this thesis. Without his enlightening instruction, impressive kindness, and patience, I could not have completed my thesis. His keen and vigorous academic observation enlightens me not only in this thesis but also in my future study. Last but not least, I would like to thank all my friends, especially my three lovely schoolmates, for their encouragement and support.
Abbreviations
- ATCC
American Type Tissue Culture Collection
- ceRNAs
competitive endogenous RNAs
- circRNAs
circular RNAs
- circPTPRM
cicrc RNA PTPRM
- IARC
International Agency for Research on Cancer
- MREs
microRNA recognition elements
- ncRNA
non-coding RNA
- PI
propidium iodide
- PTP
Protein Tyrosine Phosphatase
- PTPRM
Protein Tyrosine Phosphatase receptor type M
- PTPRM-WT
wild-type reporter vectors PTPRM
- PTPRM-MU
mutant-type reporter vectors PTPRM
- qRT-PCR
quantitative real-time polymerase chain reaction
- SD
standard deviation
- SEDT5
SET domain containing 5
- sh-PTPRM
PTPRM shRNA
- SETD5-WT
wild-type reporter vectors SETD5
- SETD5-MU
mutant-type reporter vectors SETD5, SETD5
- WT
wild type
- 3′UTR
3′ untranslated region.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Guangzhou Science and Technology Program, Guangdong Basic and Applied Basic Research Foundation (grant numbers 202002030006 and 2019A1515010026).
Ethics Approval: All animals were kept in a pathogen-free environment and fed ad-lib. The procedures for care and use of animals were approved by the Ethics Committee of the Institutional Animal Care and Use Committee of Guangzhou Medical University (approval number: GD2019-046) in June 10, 2019 and all applicable institutional and governmental regulations concerning the ethical use of animals were followed.
We collected 28 clinical specimens of NSCLC from the Affiliated Cancer Hospital and Institute of Guangzhou Medical University between September 2019 and June 2020.
Our study was approved by The Affiliated Cancer Hospital and Institute of Guangzhou Medical University Ethics Committee (approval number: 201466). All patients provided written informed consent prior to enrollment in the study.
Supplemental Material: Supplemental material for this article is available online.
ORCID iD: Zeyong Jiang, MD https://orcid.org/0000-0002-8976-6701
Hanlin Zou https://orcid.org/0000-0003-4280-5782
References
- 1.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Lung Cancer and Personalized Medicine. 2016;893:1-19. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Bratová M, Karlínová B, Skrickova J, et al. Non-small cell lung cancer as a chronic disease–A prospective study from the Czech TULUNG registry. in Vivo. 2020;34(1):369-379. doi: 10.21873/invivo.11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr Kumarakulasinghe N, Zanwijk Nv, Soo RA. Molecular targeted therapy in the treatment of advanced stage non-small cell lung cancer (NSCLC). Respirology 2015;20(3):370-378. doi: 10.1111/resp.12490 [DOI] [PubMed] [Google Scholar]
- 5.Moro-Sibilot D, Smit E, de Castro Carpeño J, et al. Outcomes and resource use of non-small cell lung cancer (NSCLC) patients treated with first-line platinum-based chemotherapy across Europe: FRAME prospective observational study. Lung Cancer. 2015;88(2):215-222. doi: 10.1016/j.lungcan.2015.02.011 [DOI] [PubMed] [Google Scholar]
- 6.Ettinger DS, Aisner DL, Wood DE, et al. NCCN Guidelines insights: non-small cell lung cancer, version 5.2018. J Natl Compr Cancer Network. 2018;16(7):807-821. doi: 10.6004/jnccn.2018.0062 [DOI] [PubMed] [Google Scholar]
- 7.Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7(7):1566-1576. [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Xiao Y, Wu L, Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. 2018;52(3):743-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tautz L, Critton DA, Grotegut S. Protein tyrosine phosphatases: structure, function, and implication in human disease. Methods Mol Biol. 2013;1053:179-221. doi: 10.1007/978-1-62703-562-0_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bollu LR, Mazumdar A, Savage MI, Brown PH. Molecular pathways: targeting protein tyrosine phosphatases in cancer. Clinical Cancer Research : an Official Journal of the American Association for Cancer Research. 2017;23(9):2136-2142. doi: 10.1158/1078-0432.CCR-16-0934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun PH, Lin Y, Mason M. Protein tyrosine phosphatase kappa (PTPRK) is a negative regulator of adhesion and invasion of breast cancer cells, and associates with poor prognosis of breast cancer. Journal of Cancer Research & Clinical Oncology. 2013;139(7):1129-1139. doi: 10.1007/s00432-013-1421-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao Z, Cui XW. Circular RNA expression and circPTPRM promotes proliferation and migration in hepatocellular carcinoma. 2019;36(10):86. [Google Scholar]
- 13.Wang Q, Liu X, Zhu R. Long noncoding RNAs as diagnostic and therapeutic targets for ischemic stroke. Curr Pharm Des. 2019;25(10):1115-1121. doi: 10.2174/1381612825666190328112844 [DOI] [PubMed] [Google Scholar]
- 14.Tiedt S, Dichgans M. Role of non-coding RNAs in stroke. Stroke. 2018;49(12):3098-3106. doi: 10.1161/STROKEAHA.118.021010 [DOI] [PubMed] [Google Scholar]
- 15.Karapetyan AR, Buiting C, Kuiper RA, Coolen MW. Regulatory roles for long ncRNA and mRNA. Cancers (Basel). 2013;5(2):462-490. doi: 10.3390/cancers5020462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Niu L, Jiang S, et al. Comprehensive analysis of aberrantly expressed profiles of lncRNAs and miRNAs with associated ceRNA network in muscle-invasive bladder cancer. Oncotarget. 2016;7(52):86174-86185. doi: 10.18632/oncotarget.13363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang K, Li Q, Kang X, Wang Y, Wang S. Identification and functional characterization of lncRNAs acting as ceRNA involved in the malignant progression of glioblastoma multiforme. Oncol Rep. 2016;36(5):2911-2925. doi: 10.3892/or.2016.5070 [DOI] [PubMed] [Google Scholar]
- 18.Wei C, Luo T, Zou S, Zhou X, Wu A. Differentially expressed lncRNAs and miRNAs with associated ceRNA networks in aged mice with postoperative cognitive dysfunction. Oncotarget. 2017;8(34):55901-55914. doi: 10.18632/oncotarget.18362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet. 2014;5:8-8. doi: 10.3389/fgene.2014.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79-79. doi: 10.1186/s12943-018-0827-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202-1207. doi: 10.1016/j.jaci.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin Y, Zhou X, Cheng H, Li L, Liu H. Serum miR-342-3p is a novel diagnostic and prognostic biomarker for non-small cell lung cancer. Int J Clin Exp Pathol. 2018;11(5):2742-2748. [PMC free article] [PubMed] [Google Scholar]
- 23.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discovery. 2017;16(3):203-222. doi: 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 24.G, Zhang Y. MicroRNA-340-5p suppresses non-small cell lung cancer cell growth and metastasis by targeting ZNF503. 2019;24:34. [Google Scholar]
- 25.Jin RH, Yu DJ, Zhong M. MiR-1269a acts as an onco-miRNA in non-small cell lung cancer via down-regulating SOX6. Eur Rev Med Pharmacol Sci. 2018;22(15):4888-4897. [DOI] [PubMed] [Google Scholar]
- 26.Cai X, Zhao Z, Dong J, et al. Circular RNA circBACH2 plays a role in papillary thyroid carcinoma by sponging miR-139-5p and regulating LMO4 expression. Cell Death Dis. 2019;10(3):184. doi: 10.1038/s41419-019-1439-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grouse L. Translational genetic research of complex diseases. Journal of Translational Internal Medicine. 2015;3(4):137-143. doi: 10.1515/jtim-2015-0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osipovich AB, Gangula R, Vianna PG, Magnuson MA. Setd5 is essential for mammalian development and the co-transcriptional regulation of histone acetylation. Development. 2016;143(24):4595-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuechler A, Zink AM, Wieland T, et al. Loss-of-function variants of SETD5 cause intellectual disability and the core phenotype of microdeletion 3p25.3 syndrome. Eur J Hum Genet. 2015;23(6):753-760. doi: 10.1038/ejhg.2014.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lossignol D. A little help from steroids in oncology. Journal of Translational Internal Medicine. 2016;4(1):52-54. doi: 10.1515/jtim-2016-0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loïc P, Gaëlle V, Fabrice S, Virginie M. miR126-5p repression of ALCAM and SetD5 in endothelial cells regulates leucocyte adhesion and transmigration. Cardiovasc Res. 2014;102(3):436-447. [DOI] [PubMed] [Google Scholar]
- 32.Dmitriev AA, Rosenberg EE, Krasnov GS, et al. Identification of novel epigenetic markers of prostate cancer by NotI-microarray analysis. Dis Markers. 2015;2015:241301. doi: 10.1155/2015/241301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sowalsky AG, Xia Z, Wang L, et al. Whole transcriptome sequencing reveals extensive unspliced mRNA in metastatic castration-resistant prostate cancer. Mol Cancer Res. 2015;13(1):98-106. doi: 10.1158/1541-7786.MCR-14-0273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu H, Sun J, Zhao C, et al. SET Domain containing protein 5 (SETD5) enhances tumor cell invasion and is associated with a poor prognosis in non-small cell lung cancer patients. BMC Cancer. 2019;19(1):736. doi: 10.1186/s12885-019-5944-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Percie du Sert N, Hurst V, Ahluwalia A, et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. Br J Pharmacol. 2020;177(16):3617-3624. doi: 10.1111/bph.15193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong KL, Lu MY, Liu F, et al. Isosteviol sodium protects neural cells against hypoxia-induced apoptosis through inhibiting MAPK and NF-kappaB pathways. Journal of Stroke and Cerebrovascular Diseases : the Official Journal of National Stroke Association. 2019;28(1):175-184. doi: 10.1016/j.jstrokecerebrovasdis.2018.09.020 [DOI] [PubMed] [Google Scholar]
- 37.National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals , et al. Guide for the Care and Use of Laboratory Animals. 8th dition. National Academies Press (US); 2011; DOI: 10.17226/12910. [DOI] [PubMed] [Google Scholar]
- 38.Zhang ZY, Gao XH, Ma MY, Zhao CL, Zhang YL, Guo SS. CircRNA_101237 promotes NSCLC progression via the miRNA-490-3p/MAPK1 axis. Sci Rep. 2020;10(1):9024. doi: 10.1038/s41598-020-65920-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang JY, Zhang F, Hong L, Wei SJ. CircRNA_0000429 regulates development of NSCLC by acting as a sponge of miR-1197 to control MADD. Cancer Manag Res. 2021;13:861-870. doi: 10.2147/CMAR.S270790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang T, Wang X, Du Q, et al. The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p. Biochem Biophys Res Commun. 2019;513(4):904-911. doi: 10.1016/j.bbrc.2019.04.108 [DOI] [PubMed] [Google Scholar]
- 41.Lv YS, Wang C, Li LX, Han S, Li Y. Effects of circRNA_103993 on the proliferation and apoptosis of NSCLC cells through miR-1271/ERG signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(16):8384-8393. [DOI] [PubMed] [Google Scholar]
- 42.Xu JX, Liu CM, Ma CP. MicroRNA-99b inhibits NSCLC cell invasion and migration by directly targeting NIPBL. Eur Rev Med Pharmacol Sci. 2021;25(4):1890-1898. [DOI] [PubMed] [Google Scholar]
- 43.Segal M, Biscans A, Gilles ME, et al. Hydrophobically modified let-7b miRNA enhances biodistribution to NSCLC and downregulates HMGA2 in vivo. Molecular Therapy - Nucleic Acids. 2020;19:267-277. doi: 10.1016/j.omtn.2019.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu DJ, Li YH, Zhong M. MicroRNA-597 inhibits NSCLC progression through negatively regulating CDK2 expression. Eur Rev Med Pharmacol Sci. 2020;24(8):4288-4297. [DOI] [PubMed] [Google Scholar]
- 45.Song L, Li D, Gu Y, et al. MicroRNA-126 targeting PIK3R2 inhibits NSCLC A549 cell proliferation, migration, and invasion by regulation of PTEN/PI3K/AKT pathway. Clin Lung Cancer. 2016;17(5):e65-e75. doi: 10.1016/j.cllc.2016.03.012 [DOI] [PubMed] [Google Scholar]
- 46.Qi J, Pan L, Yu Z, Ni W. The lncRNA RP3-439F8.1 promotes GBM cell proliferation and progression by sponging miR-139-5p to upregulate NR5A2. Pathology - Research and Practice. 2021;223:153319. doi: 10.1016/j.prp.2020.153319 [DOI] [PubMed] [Google Scholar]
- 47.Ke X, Ke S, Xin L, Li Y, Yin P. MiR-139-5p reverses CD44 + /CD133 + -associated multidrug resistance by downregulating NOTCH1 in colorectal carcinoma cells. Oncotarget. 2016;7(46):75118-75129. doi: 10.18632/oncotarget.12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun J, Wang S, Liu P, Liu Y. MiR-139-5p-ZEB1 is a molecular regulator of growth, invasion, and epithelial-to-mesenchymal transition of cervical cancer. Cancer Manag Res. 2020;12:12723-12733. doi: 10.2147/CMAR.S267634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun C, Sang M, Li S, et al. Hsa-miR-139-5p inhibits proliferation and causes apoptosis associated with down-regulation of c-met. Oncotarget. 2015;6(37):39756. doi: 10.18632/oncotarget.5476 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Saha SK, Biswas PK, Gil M, Cho SG. High expression of TTYH3 is related to poor clinical outcomes in human gastric cancer. J Clin Med. 2019;8(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piao L, Li H, Feng Y, Yang Z, Kim S, Xuan Y. SET domain-containing 5 is a potential prognostic biomarker that promotes esophageal squamous cell carcinoma stemness. Exp Cell Res. 2020;389(1):111861. doi: 10.1016/j.yexcr.2020.111861 [DOI] [PubMed] [Google Scholar]
- 52.Wang Z, Hausmann S, Lyu R, et al. SETD5-Coordinated Chromatin reprogramming regulates adaptive resistance to targeted pancreatic cancer therapy. Cancer Cell. 2020;37(6):834-849.):):6): e813. doi: 10.1016/j.ccell.2020.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Z, Zhang C, Che N, Feng Y, Xuan Y. Su(var)3–9, enhancer of zeste, and trithorax domain-containing 5 facilitates tumor growth and pulmonary metastasis through up-regulation of AKT1 signaling in breast cancer. Am J Pathol. 2021;191(1):180-193. doi: 10.1016/j.ajpath.2020.10.005 [DOI] [PubMed] [Google Scholar]
- 54.Ding B, Yan L, Zhang Y, et al. Analysis of the role of mutations in the KMT2D histone lysine methyltransferase in bladder cancer. FEBS Open Bio. 2019;9(4):693-706. doi: 10.1002/2211-5463.12600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Q, Sun Z, Li J, Zhang D, Guo B, Zhang T. SET Domain-Containing protein 5 enhances the cell stemness of non-small cell lung cancer via the PI3K/akt/mTOR pathway. J Environ Pathol Toxicol Oncol. 2021;40(2):55-63. doi: 10.1615/JEnvironPatholToxicolOncol.2021036991 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xlsx-1-tct-10.1177_15330338221090090 for CircRNA PTPRM Promotes Non-Small Cell Lung Cancer Progression by Modulating the miR-139-5p/SETD5 Axis by Zeyong Jiang, Jian Zhao, Hanlin Zou and Kaican Cai in Technology in Cancer Research & Treatment