Abstract
MicroRNAs are short, evolutionarily conserved noncoding RNAs that are critical for the control of normal cellular physiology. In the retina, photoreceptors are highly specialized neurons that transduce light into electrical signals. Photoreceptors, however, are unable to process visual stimuli without the support of the retinal pigment epithelium (RPE). The RPE performs numerous functions to aid the retina, including the generation of visual chromophore and metabolic support. Recent work has underscored how microRNAs enable vision through their contributions to RPE functions. This review focuses on the biogenesis and control of microRNAs in rodents and humans, the roles microRNAs play in RPE function and degeneration, and how microRNAs could serve as potential therapeutics and biomarkers for visual diseases.
The retinal pigment epithelium’s critical functions
The RPE is a postmitotic epithelial monolayer that is located directly adjacent to the photoreceptor outer segments (POS) of the outer neural retina. Both the RPE and retina develop from the optic neuroepithelium, and the RPE is necessary for both proper retinal development and function throughout the human life span [1]. Furthermore, the retina is the only directly viewable portion of the central nervous system (CNS), and numerous noninvasive tools for investigating the structure, function, and physiology of the retina have been developed, allowing for direct assessment in vivo of neural function. Thus, study of the RPE and retina is likely to yield valuable insights on the structure and function of the CNS more generally.
The RPE and photoreceptors are deeply intertwined, both physically and functionally. Multiple functions of the RPE directly support photoreceptor health, including critical steps in the visual (retinoid) cycle, transport of nutrients, maintenance of the blood–retinal barrier, absorption of stray light, and daily phagocytosis of shed POS [2]. Of note, the RPE secretes growth factors such as the pigment epithelium-derived factor (PEDF) that maintain normal neural function [3]. The RPE also has a glial-like role in the regulation of ion balances necessary for photoreceptor excitability [4]. Critically, the RPE and photoreceptors are metabolically coupled. As the outer retina is highly hypoxic, photoreceptors rely almost exclusively on glycolysis of glucose supplied by the choroidal vasculature via the RPE, while the RPE largely avoids the use of glucose, relying instead primarily on oxidative metabolism of spent metabolites and macromolecules from photoreceptors such as lactate, succinate, proline, and materials from phagocytosed POS [2,5,6]. The RPE is heavily dependent on mitochondrial metabolism, and RPE metabolic derangement from mitochondrial damage is detrimental to RPE and photoreceptor alike. Thus, persistent dysfunction of the RPE can lead to the decline of photoreceptors; indeed, RPE deficits are hypothesized to contribute to the early pathogenesis of age-related macular degeneration (AMD; see Glossary). One major contributor to RPE genetic and functional regulation is its system of microRNAs (miRs). In this review, we first provide a brief overview of miR biogenesis and physiology as well as summarize recent work in the field of rodent and human miR RPE biology in various contexts, from tissue development to normal function and disease. Lastly, we discuss methods for studying miR biology, offer perspective on the potential utilization of miR in diagnostics and therapeutics, and suggest potential new avenues for advancement of the field.
miR biogenesis and physiology
First discovered in Caenorhabditis elegans, miRs were identified to be noncoding RNAs that suppress mRNA expression through complementary base pairing [7,8]. Since their discovery, miRs have been found in a diverse array of organisms, from eukaryotes to viruses. Their nucleotide sequences and functions are often broadly conserved across species, supporting their hypothesized role in fine-tuning and controlling gene expression. This review focuses specifically on recent reports of miRs in the eyes of humans and in rodent models, and of the RPE in particular, though due to the focused scope of this review, we are unable to discuss several important reports of other eye-related miR findings.
In most instances, eukaryotic miRs are transcribed by RNA polymerase II in the nucleus to form a primary miR (pri-miR), which assumes a stem-loop structure (Figure 1). Pri-miRs are then usually processed by Drosha and DCGR8 to form a pre-miR and then exported to the cytoplasm by exportin 5 and RAN-GTP [9]. The pre-miR undergoes further processing by Dicer and forms an RNA complex composed of the miR and its passenger strand. The duplex is then loaded into an RNA-induced silencing complex (RISC), of which the main effectors are the Argonaute proteins, mainly Argonaute 2 (Ago2) [10]. The passenger strand is then removed from the RISC and the mature miR, which is roughly 20–22 nucleotides long, remains bound to Ago2 [11]. Of note, either strand can be loaded into the RISC; the sequences closer to the 5′-end of the original pri-miR are called the 5p miRs, and the sequences closer to the 3′-end are called the 3p miRs. Once bound to Ago2, the miR–RISC complex is relatively stable, with reported half-lives on the order of days, though in some specialized compartments, such as the CNS, neuronal miR expression is tightly regulated by activity, and turnover happens more rapidly, on the order of hours [12].
Figure 1. Canonical microRNA biogenesis and function.
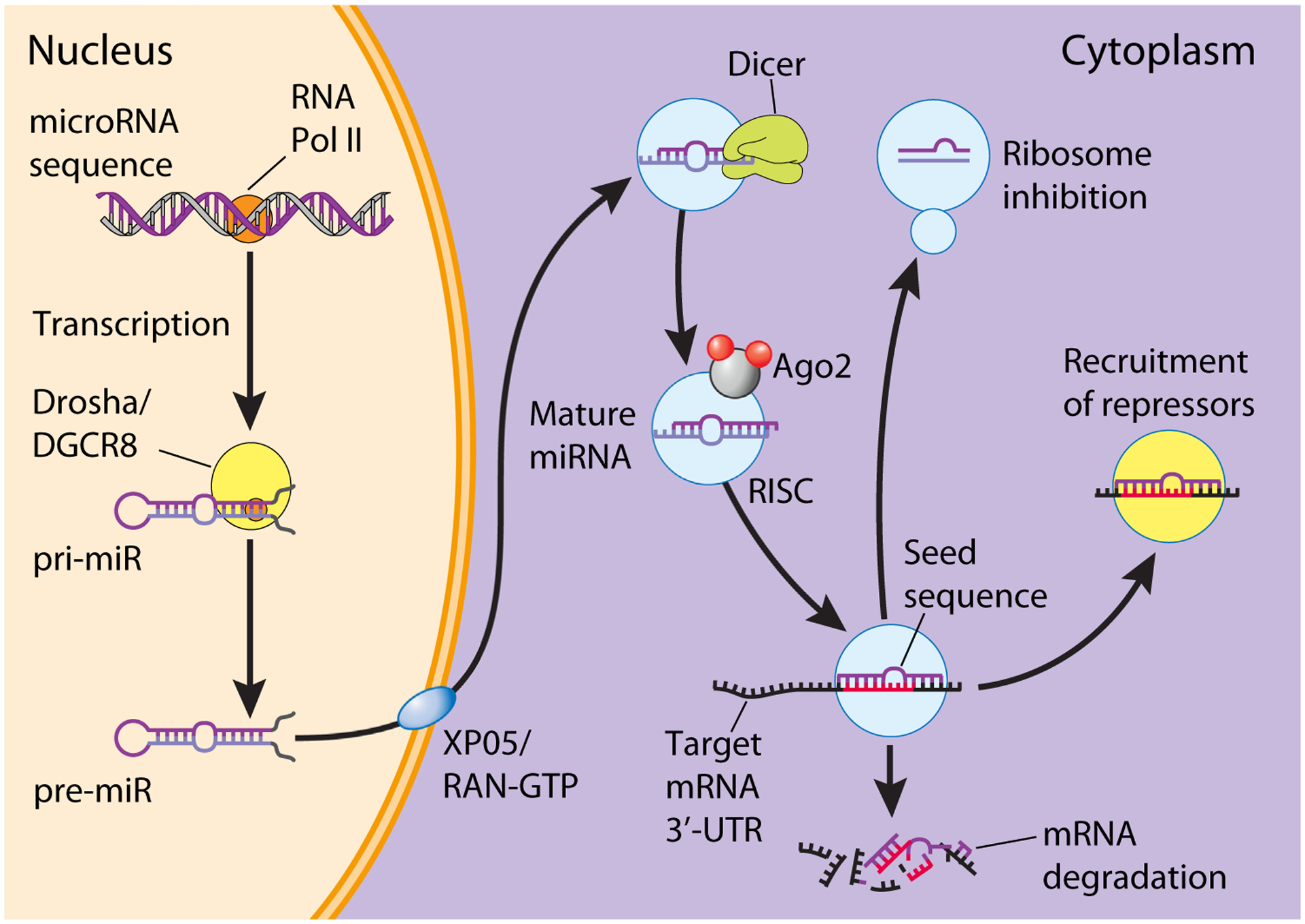
Most microRNAs (miRs) are transcribed by RNA polymerase II in the nucleus to form a primary miR (pri-miR). The pri-miR is processed by Drosha/DGCR8 to form a pre-miR, which is then exported to the cytoplasm by exportin 5 (XPO5) and RAN-GTP. The pre-miR is cleaved by Dicer and loaded into the RNA-induced silencing complex (RISC), where a mature miR pairs to the 3′-UTR of its target mRNA through complementary base pairing to the seed sequence. Expression of the target mRNA is suppressed by the RISC through either mRNA degradation or translational repression, mediated through recruitment of repressors or ribosomal inhibition. Abbreviations: 3′-UTR, 3′-untranslated region; Ago2, Argonaute 2.
miRs canonically repress mRNA expression through base pairing of the seed sequence to a complementary site in the 3′-untranslated region (3′-UTR) of the target mRNA molecule [8,10]. The seed sequence is a generally well-conserved 7–8-nucleotide region at the 5′-end of the miR, starting with the second nucleotide. Tight complementarity of the target mRNA to the seed sequence of the miR loaded into the RISC is thought to be the basis for the specificity of miR regulatory functions [13]. Accordingly, most miR target prediction algorithms heavily weight the presence of a seed sequence binding region [14,15]. Once bound to their target, vertebrate miRs generally induce downregulation of mRNA expression through two main mechanisms: translational repression or deadenylation and destabilization of the mRNA [16,17]. Although most miR interactions lead to decreased gene expression, there are limited reports of miR expression leading to increased translation and upregulation of target genes under specific conditions, such as cell cycle quiescence and altered metabolism [18,19]. Another layer of complexity is found in miR families, which share identical or similar seed sequences. These so-called iso-miRs are thought to offer redundant control over mRNAs, as they can target the same mRNAs, and some knockdown experiments only show an effect when multiple members of a miR family are disrupted [20].
Although the canonical pathway of miR biogenesis and function has been well described, knowledge of other mechanisms of miR-mediated regulation is still evolving. For example, miRs can be generated through pathways independent of Drosha/DCGR8 or Dicer [21–23]. Similarly, while seed sequence binding to the 3′-UTR is heavily weighted in predicting miR interactions, there are multiple reports of noncanonical binding events. These include interactions with an incomplete seed sequence, pairing with the 3′-end of the miR, and pairing with mRNAs in their 5′-untranslated region and open reading frames. Indeed, up to half of the miR–mRNA pairs isolated by various methods of cross-linking or copurifying with Argonaute show promiscuous and noncanonical binding [24,25]. The relevance of these interactions is unclear, suggesting that the RISC spends time transiently sampling and binding to potential targets. Hence, it seems reasonable to speculate that additional pathways controlling miR expression and function may be characterized, and these could potentially lead to the discovery of novel functions of these noncoding RNAs. Alternatively, the RISC could have additional regulatory functions in certain contexts that have not been fully explored and cannot be fully described by current models.
miRs in the RPE
As a nonrenewing tissue, the RPE must carefully coordinate and regulate its functions in support of photoreceptors. The additional layer of control offered by miRs allows the RPE to regulate its metabolism, phagocytosis, and other pathways in a coordinated manner. Recent work on RPE miRs, discussed later, has highlighted both the critical roles of miRs in the RPE and how miRs may enable intercellular communication (Figure 2). Collectively, these studies have advanced current understanding of both the regulatory mechanisms of RPE physiology during healthy conditions and how dysregulation of the RPE contributes to retinal diseases.
Figure 2. MicroRNA interactions and functions in the RPE and retina.
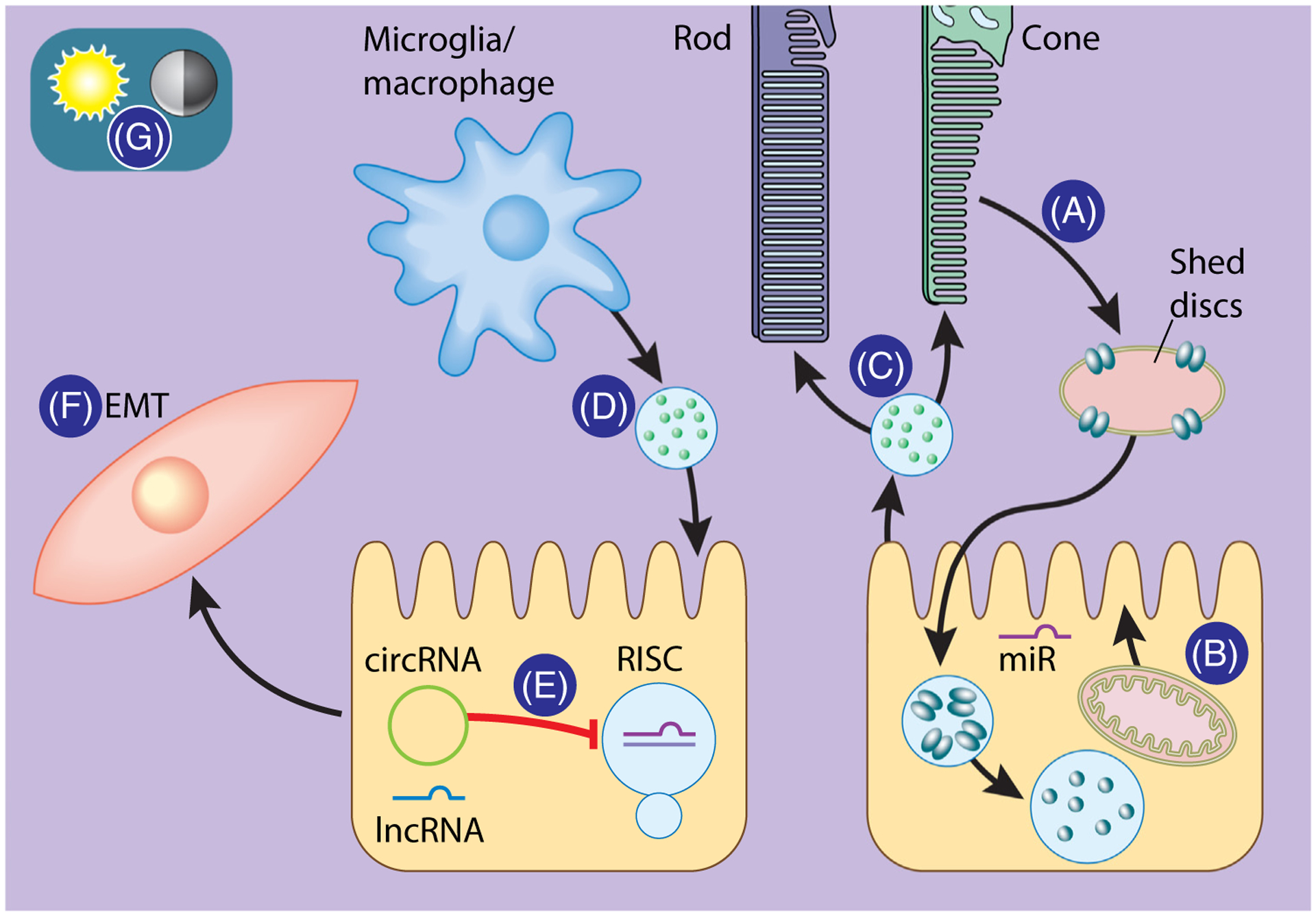
MicroRNAs (miRs) can facilitate RPE-intrinsic functions and intercellular communication within the eye. The daily release of photoreceptor outer segments (POS) as shed discs from rods and cones is regulated by RPE miRs (A). miRs contribute to the RPE response to oxidative and mitochondrial stress (B). Extracellular vesicles (EVs) from the RPE can influence the function and gene expression in rod and cone photoreceptors (C). The RPE, in turn, can be modulated by EVs from microglia and macrophages found in the retina (D). The RPE can also regulate miR levels through the expression of other noncoding RNAs, such as long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs), which contain miR binding sites (E). Lastly, miR expression can induce or suppress epithelial–mesenchymal transition (EMT), a feature of RPE pathology and degeneration (F). Expression of miRs in the retina is tightly controlled by diurnal and circadian patterns (G). Abbreviations: RISC, RNA-induced silencing complex; RPE, retinal pigment epithelium.
There are several critical features of the miRs found in the eye. For example, miRs in photoreceptors and other CNS neurons were shown to be translated rapidly and degraded in a rapid and direct manner by light [12]. This transient, cyclical behavior contrasts with the longer half-lives shown for miRs in other tissues, suggesting unique functions and circadian regulation for miRs in photoreceptors and the RPE [26,27]. miRs are also indispensable for RPE and eye development and maintenance. In mature murine RPE, loss of Dicer1 or DCGR8 resulted in severe disorganization of the RPE and retinal dysfunction, and similar defects can be found in photoreceptors lacking Dicer1 and retinas lacking Ago2 [28–30]. Similarly, decreased expression of Dicer1 is correlated with both dry and wet AMD, though the role of Dicer1 in this pathology is disputed and may also be related to the processing of Alus and other short RNA species [31,32]. While the importance of miRs and their processing machinery in the RPE is not in doubt, their exact roles need further elucidation.
RPE development and tissue identity
miRs play a pivotal role in RPE development. For example, master transcription factors such as Pax6 and Mitf regulate RPE differentiation in part through miRs [33–36]. Several miRs are differentially regulated in ophthalmic development, such as miR-204 and miR-302 [37]. During human RPE development, miR-302 was shown to be highly downregulated, and overexpression of miR-302 induced dedifferentiation of RPE through the targeting of CDKN1A [38]. Recent work has identified other miRs thought to play roles during development, including miR-125n, let-7a, and miR-382 [39,40]. Not only do miRs regulate normal RPE development, they also regulate transdifferentiation. A major phenotype of RPE stress in AMD and other diseases is epithelial–mesenchymal transition (EMT), where transdifferentiation of RPE cells results in a loss of epithelial phenotype and an increased ability to migrate and proliferate, to the detriment of the retina and its vasculature [41–43]. For example, in vitro, EMT was repressed in cultured human RPE through the expression of miR-27b-3p, which targeted HOXC6 and downregulated transforming growth factor (TGF)-β2 signaling [44]. By contrast, when EMT was induced by exogenous TGF-β2 or mechanical stretch in the transformed ARPE-19 human RPE line, miR-29b was downregulated [45]. In streptozotocin-induced diabetes, EMT of murine RPE was promoted by miR-195 through downregulation of Smurf2, resulting in increased vascular endothelial growth factor A (VEGF-A) expression [46]. Further support for this hypothesis can be found in animal model studies of EMT. A major focus of the study of EMT and RPE dysfunction is in proliferative vitreoretinopathy, where inflammatory and dedifferentiation signaling through molecules such as Yap1, galectin-1, TGF-β, and caveolin-1 drives RPE EMT and subretinal fibrosis [47–52]. Similarly, EMT in a mouse model of diabetic retinopathy (DR) was inhibited through either overexpression of miR-2094 or inhibition of its target, the long noncoding RNA (lncRNA) NEAT1 [53]. Further study of miRs in normal and aberrant development of the RPE will deepen our understanding of the basic processes that drive differentiation and will identify targetable miRs and pathways that could be manipulated in RPE dysfunction in order to maintain cellular and tissue identity.
Phagocytosis and endolysosomal function
One critical role for the RPE is the circadian phagocytosis of POS. Up to 10% of POS are shed each day, necessitating their daily removal and processing [54]. miRs regulate the phagocytosis of POS and their subsequent breakdown in the lysosomal pathway. For instance, overexpression of miR-302d suppressed RPE phagocytosis [38]. Other miRs such as miR-184, miR-29, and miR-1273g modulate autophagy, a process through which the RPE breaks down and recycles POS [55–57]. Further understanding of autophagy could lead to a new targetable pathway for AMD treatment [58]. Integration of our knowledge of the general phagocytic pathway and identification of miRs that regulate various steps in that pathway have resulted in the identification of numerous miR–mRNA pairs in the RPE, such as miR-204 targeting Rab22a, a regulator of vesicle trafficking [59]. Additionally, mice that lack miR-211 globally exhibit defects in RPE-mediated phagocytosis and endolysosomal function; miR-211 was shown to downregulate the cytoskeletal protein Ezrin, which is highly expressed in the RPE as a coordinator of phagocytosis. Confirming the role of Ezrin in RPE phagocytic function, pharmacological inhibition of Ezrin with the small molecule NSC668394 by blocking Ezrin actin binding through phosphorylation at T567 rescued the lysosomal dysfunction phenotype in miR-211-knockout mice [60–62]. Further identification of miR regulators of phagocytosis can point to novel risk factors for RPE dysfunction in the context of AMD and suggest potential targets for treatment or augmentation of RPE function.
Exosome-derived miRs
A large proportion of cell and tissue types secrete exosomes and extracellular vesicles (EVs). These EVs are theorized to enable intercellular communication and can contain DNA, RNA, proteins, and other metabolites [63]. A growing body of literature has investigated the EVs released by the RPE in both homeostasis and disease [64–66]. EVs frequently carry miRs as cargo, and EVs from RPE may help regulate retinal and RPE physiology.
In AMD, inflammation is often implicated in RPE and retinal stress, and signaling from the immune system plays a role. Retinal and RPE degeneration often leads to the migration of macrophages and microglia into the outer retina. To investigate the role of EV secretion between the RPE and macrophages, human macrophages and iPSC-derived human RPE were cocultured in transwell plates. EV communication between the two cell types was associated with the secretion of proinflammatory cytokines such as interleukin (IL)-6 and VEGF and miR-494-3p from the RPE [67]. Another study reported that RPE samples from elderly humans released miR-21 from EVs that were then taken up by microglia, leading to a change in expression of genes in the p53 signaling pathway, implicating microglia in regulation of aging and AMD [68].
EMT can also be induced by exosomes that carry miRs. In one model, EMT was induced in ARPE-19 cells via administration of TGF-β. When EVs were isolated from these induced ARPE-19 cells, the EVs were themselves able to induce EMT in naive ARPE-19 cells, and miR-543 was identified as the most significantly enriched miR in those EVs [64]. Similarly, exosomes derived from ARPE-19 cells in high-glucose conditions which contained miR-202-5p were able to suppress vascular proliferation and EMT through direct targeting of TGF-βR2 [69]. While these preliminary results indicate that EV-derived miRs could modulate EMT, further work in primary cultures and animal models are needed to confirm these findings. Nonetheless, the identification of critical miRs contained in EVs could be leveraged as a treatment platform for a variety of RPE diseases and offer a potential delivery system for miRs.
Inflammation, oxidative stress, and neovascularization
As a postmitotic monolayer, the RPE must last a lifetime to provide support to photoreceptors. Inflammation and oxidative stress can result in RPE deterioration, and miRs can help the RPE respond to such stressors. For instance, in ARPE-19 cells, inflammation caused by the Nlrp3 inflammasome is directly counteracted by targeting of NLRP3 mRNA by miR-22-3p or miR-191-5p [70,71]. In primary human RPE cultures, apoptosis triggered through the Fas receptor can be abrogated by targeting of Fas receptor by miR-374 [72].
Oxidative stress and lipid dysregulation in the RPE potentially contribute to AMD and other RPE and retinal disorders. This can be through direct modulation of mitochondria or through interaction with downstream effectors [73,74]. miR-33 was recently shown to modulate expression of ABCA1, a cholesterol pump in the RPE, and suppression of miR-33 via antisense oligonucleotides in mouse and non-human primate models alleviated the dry AMD phenotype [75]. Similarly, the H-RPE cell line treated with oxidized low-density lipoproteins exhibited altered expression of miRs and genes known to cause retinitis pigmentosa [76]. Additionally, hyperglycemia is thought to cause oxidative and metabolic stress in the RPE, leading to apoptosis. Preliminary data from the ARPE-19 and RPE-1 cell lines suggest that protection from high-glucose conditions is mediated in part through miRs such as miR-27a, which inhibits Toll-like receptor (TLR) 4, and miR-25, which inhibits PTEN and Akt signaling [39,77,78]. Two studies independently concluded that miR-125b inhibits oxidative stress through binding to hexokinase 2, a key enzyme in the glycolysis pathway, though they reached opposite conclusions about the role of hexokinase 2 in the ARPE-19 cell line. While one study argued that inhibition of hexokinase 2 resulted in decreased oxidative stress from hyperglycemia, according to the second study, disordered glucose metabolism was restored and protective against hydrogen peroxide treatment [79,80]. Further work in human and animal samples is necessary to reconcile these opposing ideas. Other identified mediators of oxidative stress and their miR regulators include Nrf2, regulated by miR-626, miR-141, and miR-144 [81–84], αV integrin (IGTAV) and PEDF through miR-25 [85], and sirtuin 1 (SIRT1) through miR-204 [86].
The RPE is thought to play an important role in the development of vascular complications such as choroidal neovascularization (CNV) and DR. Dysregulated RPE miRs and other signals can lead to the promotion of angiogenesis, in opposition to the RPE’s normal antiangiogenic interactions [87]. Similarly, though DR has classically been considered as a vascular disease, the RPE is thought to modulate DR, in part through the decreased secretion of antiangiogenic factors such as PEDF, increased secretion of proinflammatory cytokines, barrier dysfunction, production of reactive oxygen species, potential modulation of the visual cycle, and various other mechanisms [88]. For in-depth coverage of the role of the RPE in DR, we refer the readers to several reviews that cover this topic [88–92]. As inflammation and oxidative stress are major mechanisms of RPE stress and damage, augmentation of the ability of the RPE to withstand such stressors can offer avenues to stop RPE and retinal degeneration. Similarly, vascular abnormalities found in the retina and choroid can be modulated by the RPE and offer new tools to target vascularization in a vessel-independent manner.
Other noncoding RNAs and their interactions with miRs
miRs are not the only type of noncoding RNA present in the RPE. Other RNA species, such as lncRNAs and circular RNAs (circRNAs), can dampen the activity of miRs by providing complementary binding sites that divert miRs from their mRNA targets [93]. For instance, the lncRNA NEAT1 expressed in the RPE in one study was shown to interact with miR-148a-3p to suppress PTEN and macrophage polarization in a mouse model of CNV, and in another study, it was shown to decrease the development of DR in a streptozotocin-induced mouse model of diabetes [53,94]. Another study of PTEN signaling in the RPE identified the circRNA NR3C1 as a miR-382-5p modulator in human AMD samples, protecting iPSC-derived human RPE from stress and dysfunction [39]. Similarly, the lncRNA LINC00167 was downregulated in human AMD samples and played an important role in modulating SOCS3 signaling by binding miR-203a-3p to maintain RPE differentiation and function [95]. Lastly, a study in a mouse CNV model identified 100 differentially regulated circRNAs and explored their regulatory networks and interactions with miRs and mRNAs [96]. In all, these results emphasize the continuing need to further characterize interactions between miRs and noncoding RNAs in the RPE. The ongoing identification of other noncoding RNA species that may modulate RPE physiology promises to be an exciting new frontier in RNA biology.
Novel methods to investigate miRs
Computational tools used for the prediction of miR–mRNA interactions rely on finding seed sequence homology in mRNA 3′-UTRs. Results from these predictive algorithms imply that a single miR could regulate hundreds, if not thousands, of genes [14,15]. Similarly, many genes could potentially be regulated by numerous miRs. However, not every predicted interaction is a bona fide interaction, and as such must be verified by orthogonal techniques such as the luciferase assay [10]. Additionally, the local composition and concentration of miRs and mRNA targets varies widely by tissue type and conditions, further adding complexity and nuance to the interpretation of predicted interactions.
However, with the advent of next-generation sequencing (NGS) platforms, the ability to interrogate the composition of miRs in specific tissues has been greatly advanced. Numerous techniques to capture miR–mRNA and Argonaute–miR interactions have been pioneered, which do not rely on prepredicted targets. These approaches often rely on the immunoprecipitation of Argonaute ribonucleoprotein complexes and cross-linking of Argonaute and miRs [often referred to as cross-linking immunoprecipitation sequencing (CLIP-seq)] [97,98]. Common pitfalls of these techniques include high background, incomplete UV cross-linking, dependence on high-quality immunoprecipitation antibodies, reliance on cell lines, and artificial manipulation of Argonaute or miR levels. Thus, the development of more physiologically relevant methods is necessary for tissue- and disease-specific miR profiling. For example, the newly described Halo-enhanced Ago2 pull-down (HEAP) protocol sidesteps many of the aforementioned limitations [99]. In the HEAP mouse model, a conditional Ago2 allele covalently tagged with the 33-kDa HaloTag is expressed from the endogenous Ago2 locus only upon expression of Cre recombinase in a particular tissue. This technique allows for the specific isolation of Ago2-containing RISCs from tissues collected from an appropriate tissue-specific Cre mouse line, although it still relies on UV cross-linking and has a downstream workflow similar to previously described CLIP-seq workflows. Combined with the generation of new RPE-specific Cre lines, such as the recently described RPE65-ERT2-cre line [100], an RPE-specific unbiased analysis of miRs and their mRNA binding is now easier and more feasible. Another recent innovation has been the use of RNA immunoprecipitation sequencing (RIP-seq) for the in vivo unbiased isolation of Ago2–miR complexes [101]. Using an Argonaute-binding antibody, RIP-seq takes advantage of the direct pull down of these complexes by digesting unprotected and unbound RNA before elution and library construction for NGS, though this approach retains some of the specific limitations described earlier. As the investigation of the networks of miRs and their target genes is critical for a fuller understanding of ophthalmic genetic regulation, development of new tools to further probe miR biology will be needed for uncovering novel miRs and their targets in a wide variety of ophthalmic contexts.
Potential applications of miRs as therapeutics and biomarkers
Because they play a critical role in the regulation of RPE physiology and disease, miRs are an attractive target for therapeutic and biomarker development [102]. A careful understanding of miR genetics and mutations can lead to a better understanding of risk factors and biomarker identification. For instance, a familial mutation in miR-204 was described as the cause of an inherited retinal dystrophy and defective iris development, highlighting the contributions of miR-204 both to ophthalmic development and eye disease [103]. This mutation is one of the first identified miR mutations that were found to lead to a syndromic inherited retinal disease, suggesting that other miR variants could lead to both inherited disease and susceptibility to other pathologies. For other ophthalmic diseases, various serum miRs can serve as accessible biomarkers (Figure 3). For example, miR-19a, miR-126, and miR-410 are differentially expressed in the serum of dry AMD patients [104]. miRs also have been described as serum markers for CNV and DR. These include miR-486a-5p and miR-92a-3p in murine laser-induced CNV [105]; miR-34-p, miR-126-3p, miR-145-5p, and miR-205-5p, which are correlated with VEGF-A expression in human wet AMD [106]; and miR-423-5p in human DR [107].
Figure 3. MicroRNAs for potential clinical applications.
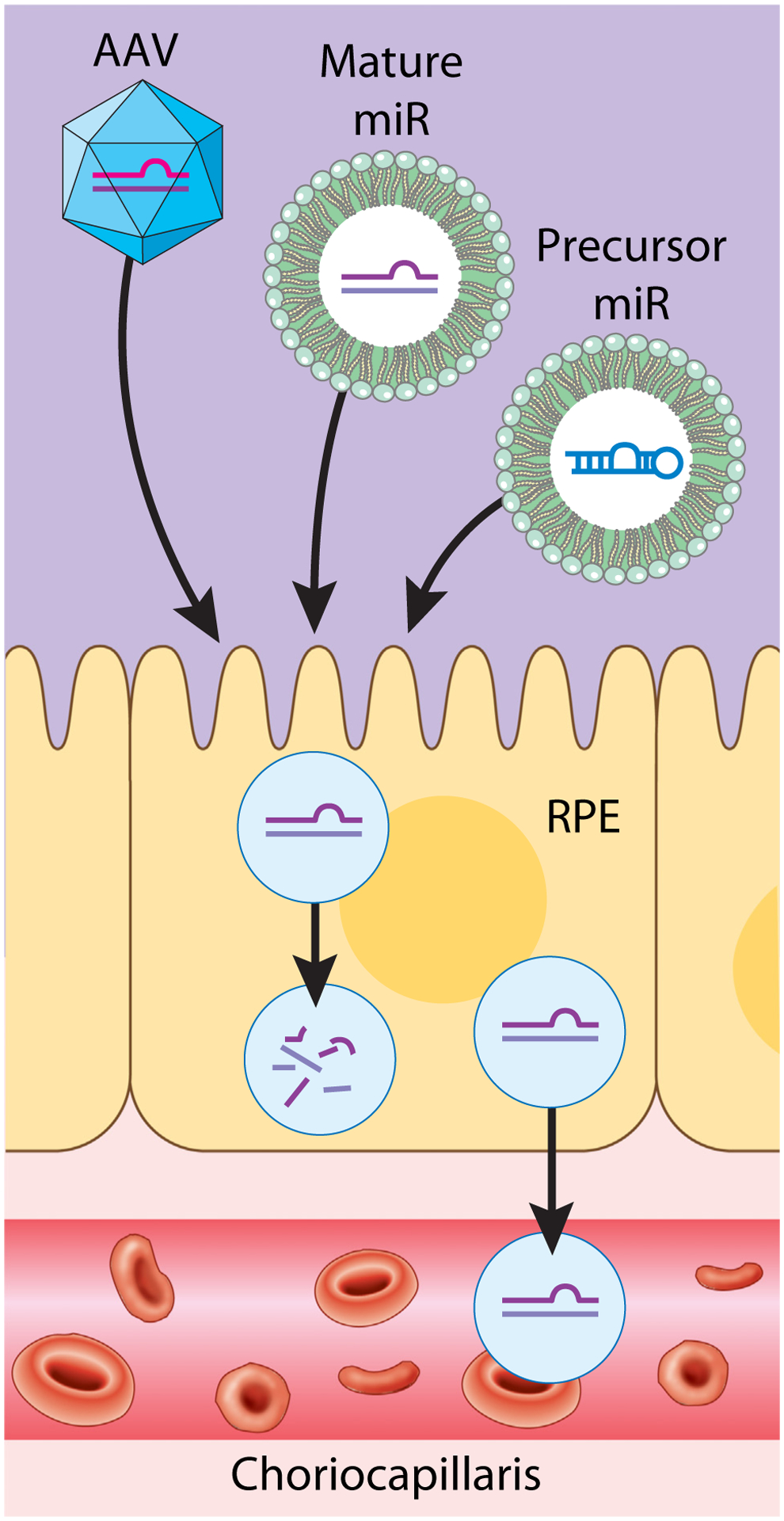
MicroRNAs (miRs) can be delivered to the RPE for therapeutic purposes through different modalities, such as through viral vectors (AAV, lentivirus, etc.) or through lipid nanoparticles. These can be delivered as mature miRs or as precursor miRs. miRs that are released into the systemic circulation in exosomes and other extracellular vesicles can be detected in blood and could potentially serve as biomarkers of diseases such as AMD or CNV. Abbreviations: AAV, adeno-associated virus; AMD, age-related macular degeneration; CNV, choroidal neovascularization; RPE, retinal pigment epithelium.
Because of their role in regulating disease, miRs could be exploited to target various pathologies. Antisense oligonucleotides and other interfering nucleic acid molecules are already being tested in clinical and preclinical studies for their efficacy in treating various disorders, suggesting that miRs could also potentially be employed to treat retinal and RPE diseases [108]. For example, subretinal injection of lentiviruses expressing antiangiogenic miRs in mice suppressed CNV [109]. Similarly, an adeno-associated virus (AAV) encoding miR-204 was shown to counteract retinal degeneration and microglial activation in the rhodopsin P347S and the AIPL1-knockout mouse models, while intraocular injection of a miR-126-expressing plasmid suppressed VEGF-A expression in a rat model of retinopathy of prematurity [110,111]. We hypothesize that the engineered and targeted expression of miRs could prove particularly beneficial as a gene therapy or augmentation modality, as miRs that target multiple genes and pathways can be administered to a broader patient pool, in contrast to other gene therapy treatments that must be tailored to specific gene mutations. Intriguingly, miRs have even been shown in vitro to induce the transdifferentiation of RPE into photoreceptors through miR-124 and the miR-183/96/182 cluster, potentially offering another route to regenerate photoreceptors and other neural cell types through the targeted reprogramming of RPE via various miRs [112,113]. Further preclinical development of miR therapy is needed through comparison of preformed precursor or mature miRs or constructs that express miRs. Additional refinement of both delivery modalities, such as viral or lipid nanoparticle systems, and stabilizing or regulatory modifications of RNA species must be accomplished before widespread application to clinical needs can be implemented (Figure 3) [114]. Alternatively, careful identification of miR targets or agents that modify miR expression in common degenerative or pathological pathways could lead to development of biologics or small molecules that target either miRs or their downstream targets [115].
Concluding remarks and future perspectives
miRs regulate and influence the development and function of the RPE. They are also intricately involved in the development of diseases of the RPE and, by extension, diseases of the neural retina, such as AMD, DR, and CNV. Accordingly, miRs could also serve as potential targets in the design of treatments for these diseases. While the broad strokes of miR–mRNA cross talk have thus far been outlined, the specific roles of individual miRs and their targets remain more elusive, as most studies focus on one miR and its most important putative target. Moreover, studies typically address a specific context, and in a different context, another target of the same miR might be the more relevant one. Many identified miR–mRNA pairs have also not been validated in other studies, pointing to the need for further work to confirm these interactions.
A number of preliminary studies on miR roles in the RPE have thus far been conducted in immortalized cell lines, such as the widely used ARPE-19 line. However, the clinical relevance of these studies must be corroborated with rigorous studies in primary cultured RPE, iPSC-derived RPE cultures, animal models, and human samples before these findings can be more generally studied for application in the clinic. In addition, while miRs have so far been studied with regard to their targeting of specific genes, in our view, they should also be studied more broadly using bioinformatics and systems biology approaches to dissect their basic biology and effects on broader signaling pathways and gene regulation (see Outstanding questions). Considering redundancy in miR systems and various homologous members of miR families that could have overlapping functions, we opine that care should be taken to validate the effects of miR deletion and overexpression. Furthermore, given that many miRs are not tissue or cell specific, the generation and interpretation of animal models for the effect of miRs on the RPE and other tissues must carefully consider the critical roles of miRs in development and tissue homeostasis; ideally, suppression or overexpression of various miRs should be done in a temporally and spatially controlled manner.
Outstanding questions.
How does the RPE sense changes in light, and what signals are required to up- and downregulate microRNA (miR) expression in response to light? Are these miR-regulatory signals intrinsic to the RPE or do they originate in the neural retina, and what other purposes do these signals serve? What drives the circadian rhythm in the RPE? Does the RPE have circadian-independent light sensitivity?
Is iso-miR expression in the eye tissue specific? Do iso-miRs have tissue-specific roles and targets? Do they define tissue specificity?
Does the RPE utilize miRs to communicate with photoreceptors to mediate phagocytosis, metabolism, and other interconnected functions? If so, how? Is it through exosomes, through the regulation of miR target proteins, or through other mechanisms?
What are the molecular regulators of miR expression in RPE development?
Do complementary miR networks within the RPE cross-compensate for changes in miR expression?
How do miRs interact with circular RNAs and other noncoding RNAs in the RPE?
How does the miR content of the RPE influence gene expression and translation in photoreceptors and the neural retina?
We believe that miRs could serve as effective biomarkers of disease, and engineered expression of miRs could be a powerful approach to therapeutic intervention in RPE diseases. A deeper understanding of miRs in the RPE and retina will also lead to insights into the regulation of neural function and health and how miRs serve to modulate important RPE roles such as phagocytosis. Indeed, deeper understanding of miR regulation of RPE phagocytosis may even lead to more efficient and selective RPE uptake of lipid nanoparticles and other gene transfer modalities that could enable transformative gene editing and gene delivery for inherited diseases. Further study of miRs will hopefully lead to insights that allow for early detection of and intervention for RPE and retinal diseases.
Highlights.
The retinal pigment epithelium (RPE) is crucial for maintaining photoreceptor and neural retinal health. Dysregulation of this postmitotic monolayer leads to retinal degeneration in various diseases such as age-related macular degeneration.
MicroRNAs have been shown to regulate the gene expression of many vital RPE pathways. These include oxidative stress and metabolism, phagocytosis and endolysosomal function, exosomes and intercellular signaling, and vascular homeostasis.
RPE development has been shown to be heavily dependent on the correct expression of microRNAs and their processing machinery, and altered microRNA levels can suppress differentiation or promote dedifferentiation of the RPE.
Perturbations in the levels or composition of RPE microRNAs lead to RPE dysfunction and the progression of disease, revealing potential targets for therapeutic and biomarker development.
Acknowledgments
We thank our colleagues at the University of California, Irvine Center for Translational Vision Research and Gavin Herbert Eye Institute for their insightful comments. We also thank Kelly O’Neil of Penumbra Design, Inc. for creating the figures. Funding for this work came in part from National Institutes of Health grants R01EY009339, R24EY027283 (K.P.), and T32GM008620 (S.W.D.), the Research to Prevent Blindness Stein Innovation Award (K.P.), and unrestricted grants from Research to Prevent Blindness to the Department of Ophthalmology at the University of California, Irvine.
Glossary
- Age-related macular degeneration (AMD)
the macula is an area of the eye with an overrepresentation of cones; it is responsible for high-acuity vision necessary for activities such as reading, facial recognition, and driving. Degeneration of the macula in AMD leads to significant visual impairment. Risk factors include advanced age, smoking, genetics, and diet.
- Choroidal neovascularization (CNV)
a pathology found in ‘wet’/neovascular age-related macular degeneration, CNV is the abnormal growth of vasculature in the choroid, which is found under the RPE. Major complications of CNV include vascular leakage and hemorrhage, which can lead to vision loss. Though the RPE plays a role in this pathology, it is thought that the choriocapillaris initiates CNV through blood vessel signaling.
- Diabetic retinopathy (DR)
a late-stage complication of diabetes mellitus is DR, a disease of the vasculature of the retina due to dysfunction of the retinal endothelial cells, and it is divided into nonproliferative and proliferative disease. It is a major cause of acquired blindness. Current treatments include anti-VEGF therapy, laser photocoagulation, steroid treatment, and surgery. Recent work has uncovered an important role of the RPE and neural retina in contributing to the progression of this vascular disease.
- Epithelial–mesenchymal transition (EMT)
epithelial cells under stress can deprogram away from healthy cells through mechanisms such as downregulation of adhesion molecule expression, detachment from their basement membranes, and loss of apical–basal polarization. These mesenchymal-like cells can proliferate and invade and play a role in numerous pathologies, such as cancer and tissue degeneration.
- Extracellular vesicle (EV)
usually 40–160 nm in diameter, is formed by invagination of intracellular cellular membranes to form multivesicular bodies, which are subsequently released extracellularly upon fusion with the plasma membrane. They can contain DNA, protein, RNA, and other cargoes and are hypothesized to enable intercellular and long-distance communication and regulation.
Footnotes
Declaration of interests
K.P. is the Chief Scientific Officer of Polgenix, Inc. Polgenix, Inc. activities are unrelated to the work described here.
References
- 1.Fuhrmann S et al. (2014) Retinal pigment epithelium development, plasticity, and tissue homeostasis. Exp. Eye Res 123, 141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lakkaraju A et al. (2020) The cell biology of the retinal pigment epithelium. Prog. Retin. Eye Res 78, 100846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tombran-Tink J and Barnstable CJ (2003) PEDF: a multifaceted neurotrophic factor. Nat. Rev. Neurosci 4, 628–636 [DOI] [PubMed] [Google Scholar]
- 4.Strauss O (2005) The retinal pigment epithelium in visual function. Physiol. Rev 85, 845–881 [DOI] [PubMed] [Google Scholar]
- 5.Bisbach CM et al. (2020) Succinate can shuttle reducing power from the hypoxic retina to the O2-rich pigment epithelium. Cell Rep 31, 107606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du J et al. (2021) Proline metabolism and transport in retinal health and disease. Amino Acids 53, 1789–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee RC et al. (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 [DOI] [PubMed] [Google Scholar]
- 8.Wightman B et al. (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 [DOI] [PubMed] [Google Scholar]
- 9.Ha M and Kim VN (2014) Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol 15, 509–524 [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP (2018) Metazoan microRNAs. Cell 173, 20–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krol J et al. (2010) Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141, 618–631 [DOI] [PubMed] [Google Scholar]
- 13.Ameres SL et al. (2007) Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130, 101–112 [DOI] [PubMed] [Google Scholar]
- 14.Agarwal V et al. (2015) Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y and Wang X (2020) miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 48, D127–D131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonas S and Izaurralde E (2015) Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet 16, 421–433 [DOI] [PubMed] [Google Scholar]
- 17.Chen C-YA and Shyu A-B (2011) Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA 2, 167–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ørom UA et al. (2008) MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30, 460–471 [DOI] [PubMed] [Google Scholar]
- 19.Truesdell SS et al. (2012) MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci. Rep 2, 842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutsche LK et al. (2018) Combined experimental and system-level analyses reveal the complex regulatory network of miR-124 during human neurogenesis. Cell Syst 7, 438–452.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cifuentes D et al. (2010) A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328, 1694–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheloufi S et al. (2010) A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465, 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J-S et al. (2010) Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. U. S. A 107, 15163–15168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loeb GB et al. (2012) Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell 48, 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helwak A et al. (2013) Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153, 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou L et al. (2021) A genome-wide microRNA screen identifies the microRNA-183/96/182 cluster as a modulator of circadian rhythms. Proc. Natl. Acad. Sci. U. S. A 118, e2020454118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiménez E et al. (2021) Regulation of the glycine transporter GLYT1 by microRNAs. Neurochem. Res Published online January 23, 2021. 10.1007/s11064-021-03228-x [DOI] [PubMed] [Google Scholar]
- 28.Sundermeier TR et al. (2017) MicroRNA-processing enzymes are essential for survival and function of mature retinal pigmented epithelial cells in mice. J. Biol. Chem 292, 3366–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aldunate EZ et al. (2019) Conditional Dicer1 depletion using Chrnb4-Cre leads to cone cell death and impaired photopic vision. Sci. Rep 9, 2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X-J et al. (2021) Retinal degeneration caused by Ago2 disruption. Invest. Ophthalmol. Vis. Sci 62, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaneko H et al. (2011) DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471, 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright CB et al. (2020) Chronic Dicer1 deficiency promotes atrophic and neovascular outer retinal pathologies in mice. Proc. Natl. Acad. Sci. U. S. A 117, 2579–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaham O et al. (2013) Pax6 regulates gene expression in the vertebrate lens through miR-204. PLoS Genet 9, e1003357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adijanto J et al. (2012) Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression. J. Biol. Chem 287, 20491–20503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conte I et al. (2010) miR-204 is required for lens and retinal development via Meis2 targeting. Proc. Natl. Acad. Sci. U. S. A 107, 15491–15496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma X et al. (2018) Regulation of cell proliferation in the retinal pigment epithelium: differential regulation of the death-associated protein like-1 DAPL1 by alternative MITF splice forms. Pigment Cell Melanoma Res 31, 411–422 [DOI] [PubMed] [Google Scholar]
- 37.Li W-B et al. (2012) Development of retinal pigment epithelium from human parthenogenetic embryonic stem cells and microRNA signature. Invest. Ophthalmol. Vis. Sci 53, 5334–5343 [DOI] [PubMed] [Google Scholar]
- 38.Jiang C et al. (2018) c-Jun-mediated microRNA-302d-3p induces RPE dedifferentiation by targeting p21Waf1/Cip1. Cell Death Dis 9, 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X et al. (2020) Circular noncoding RNA NR3C1 acts as a miR-382-5p sponge to protect RPE functions via regulating PTEN/AKT/mTOR signaling pathway. Mol. Ther 28, 929–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shahriari F et al. (2020) MicroRNA profiling reveals important functions of miR-125b and let-7a during human retinal pigment epithelial cell differentiation. Exp. Eye Res 190, 107883. [DOI] [PubMed] [Google Scholar]
- 41.Xie L et al. (2021) The HIF-1α/p53/miRNA-34a/Klotho axis in retinal pigment epithelial cells promotes subretinal fibrosis and exacerbates choroidal neovascularization. J. Cell. Mol. Med 25, 1700–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou M et al. (2020) Role of epithelial-mesenchymal transition in retinal pigment epithelium dysfunction. Front. Cell Dev. Biol 8, 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jun JH et al. (2020) Regulation of Ras homolog family member G by microRNA-124 regulates proliferation and migration of human retinal pigment epithelial cells. Sci. Rep 10, 15420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li D et al. (2021) Human umbilical cord mesenchymal stem cell-derived exosomal miR-27b attenuates subretinal fibrosis via suppressing epithelial-mesenchymal transition by targeting HOXC6. Stem Cell Res. Ther 12, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao Q et al. (2019) The role of mechanical stretch and TGF-β2 in epithelial-mesenchymal transition of retinal pigment epithelial cells. Int. J. Ophthalmol 12, 1832–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu S-H et al. (2021) MiR-195 inhibits the ubiquitination and degradation of YY1 by Smurf2, and induces EMT and cell permeability of retinal pigment epithelial cells. Cell Death Dis 12, 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X et al. (2020) Interleukin-6 promotes proliferative vitreoretinopathy by inducing epithelial-mesenchymal transition via the JAK1/STAT3 signaling pathway. Mol. Vis 26, 517–529 [PMC free article] [PubMed] [Google Scholar]
- 48.Wu D et al. (2019) Galectin-1 promotes choroidal neovascularization and subretinal fibrosis mediated via epithelial-mesenchymal transition. FASEB J 33, 2498–2513 [DOI] [PubMed] [Google Scholar]
- 49.Lu Q et al. (2020) Yap1 is required for maintenance of adult RPE differentiation. FASEB J 34, 6757–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohlmann A et al. (2016) Epithelial-mesenchymal transition of the retinal pigment epithelium causes choriocapillaris atrophy. Histochem. Cell Biol 146, 769–780 [DOI] [PubMed] [Google Scholar]
- 51.Nagasaka Y et al. (2017) Role of caveolin-1 for blocking the epithelial-mesenchymal transition in proliferative vitreoretinopathy. Invest. Ophthalmol. Vis. Sci 58, 221–229 [DOI] [PubMed] [Google Scholar]
- 52.Kobayashi Y et al. (2021) Inhibition of epithelial-mesenchymal transition in retinal pigment epithelial cells by a retinoic acid receptor-α agonist. Sci. Rep 11, 11842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y et al. (2021) LncRNA NEAT1 regulated diabetic retinal epithelial-mesenchymal transition through regulating miR-204/SOX4 axis. PeerJ 9, e11817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young RW and Bok D (1969) Participation of the retinal pigment epithelium in the rod outer segment renewal process. J. Cell Biol 42, 392–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye Z et al. (2017) miRNA-1273g-3p involvement in development of diabetic retinopathy by modulating the autophagy-lysosome pathway. Med. Sci. Monit 23, 5744–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murad N et al. (2014) miR-184 regulates ezrin, LAMP-1 expression, affects phagocytosis in human retinal pigment epithelium and is downregulated in age-related macular degeneration. FEBS J 281, 5251–5264 [DOI] [PubMed] [Google Scholar]
- 57.Cai J et al. (2019) MicroRNA-29 enhances autophagy and cleanses exogenous mutant αB-crystallin in retinal pigment epithelial cells. Exp. Cell Res 374, 231–248 [DOI] [PubMed] [Google Scholar]
- 58.Hyttinen JMT et al. (2021) MicroRNAs in the regulation of autophagy and their possible use in age-related macular degeneration therapy. Ageing Res. Rev 67, 101260. [DOI] [PubMed] [Google Scholar]
- 59.Zhang C et al. (2019) Regulation of phagolysosomal activity by miR-204 critically influences structure and function of retinal pigment epithelium/retina. Hum. Mol. Genet 28, 3355–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naso F et al. (2020) Light-responsive microRNA miR-211 targets Ezrin to modulate lysosomal biogenesis and retinal cell clearance. EMBO J 39, e102468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bulut G et al. (2012) Small molecule inhibitors of ezrin inhibit the invasive phenotype of osteosarcoma cells. Oncogene 31, 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Çelik H et al. (2016) Ezrin inhibition up-regulates stress response gene expression. J. Biol. Chem 291, 13257–13270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalluri R and LeBleu VS (2020) The biology, function, and biomedical applications of exosomes. Science 367, eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y et al. (2020) Exosomes mediate an epithelial-mesenchymal transition cascade in retinal pigment epithelial cells: Implications for proliferative vitreoretinopathy. J. Cell. Mol. Med 24, 13324–13335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shah N et al. (2018) Extracellular vesicle-mediated long-range communication in stressed retinal pigment epithelial cell monolayers. Biochim. Biophys. Acta Mol. basis Dis 1864, 2610–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Knickelbein JE et al. (2016) Modulation of immune responses by extracellular vesicles from retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci 57, 4101–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mukai A et al. (2021) Mitochondrial miRNA494-3p in extracellular vesicles participates in cellular interplay of iPS-Derived human retinal pigment epithelium with macrophages. Exp. Eye Res 208, 108621. [DOI] [PubMed] [Google Scholar]
- 68.Morris DR et al. (2020) Exosomal miRNA transfer between retinal microglia and RPE. Int. J. Mol. Sci 21, 3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gu S et al. (2020) Retinal pigment epithelial cells secrete miR-202-5p-containing exosomes to protect against proliferative diabetic retinopathy. Exp. Eye Res 201, 108271. [DOI] [PubMed] [Google Scholar]
- 70.Hu Z et al. (2019) Protective effects of microRNA-22-3p against retinal pigment epithelial inflammatory damage by targeting NLRP3 inflammasome. J. Cell. Physiol 234, 18849–18857 [DOI] [PubMed] [Google Scholar]
- 71.Chen J et al. (2021) MicroRNA-191-5p ameliorates amyloid-β 1–40–mediated retinal pigment epithelium cell injury by suppressing the NLRP3 inflammasome pathway. FASEB J 35, e21184. [DOI] [PubMed] [Google Scholar]
- 72.Tasharrofi N et al. (2017) Survival improvement in human retinal pigment epithelial cells via Fas receptor targeting by miR-374a. J. Cell. Biochem 118, 4854–4861 [DOI] [PubMed] [Google Scholar]
- 73.Indrieri A et al. (2019) miR-181a/b downregulation exerts a protective action on mitochondrial disease models. EMBO Mol. Med 11, e8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jun S et al. (2019) The impact of lipids, lipid oxidation, and inflammation on AMD, and the potential role of miRNAs on lipid metabolism in the RPE. Exp. Eye Res 181, 346–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gnanaguru G et al. (2021) Targeting of miR-33 ameliorates phenotypes linked to age-related macular degeneration. Mol. Ther 29, 2281–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Donato L et al. (2018) miRNAexpression profile of retinal pigment epithelial cells under oxidative stress conditions. FEBS Open Bio 8, 219–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zha X et al. (2020) Overexpression of METTL3 attenuates high-glucose induced RPE cell pyroptosis by regulating miR-25-3p/PTEN/Akt signaling cascade through DGCR8. Aging (Albany NY) 12, 8137–8150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang X et al. (2018) MicroRNA-27a protects retinal pigment epithelial cells under high glucose conditions by targeting TLR4. Exp. Ther. Med 16, 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang J-F et al. (2018) MicroRNA-125b protects hyperglycemia-induced, human retinal pigment epithelial cells (RPE) from death by targeting hexokinase 2. Int. J. Clin. Exp. Pathol 11, 3111–3118 [PMC free article] [PubMed] [Google Scholar]
- 80.Liu G et al. (2018) Inhibition of the oxidative stress-induced miR-125b protects glucose metabolic disorders of human retinal pigment epithelium (RPE) cells. Cell. Mol. Biol. (Noisy-le-grand) 64, 1–5 [PubMed] [Google Scholar]
- 81.Jadeja RN et al. (2020) Inhibiting microRNA-144 potentiates Nrf2-dependent antioxidant signaling in RPE and protects against oxidative stress-induced outer retinal degeneration. Redox Biol 28, 101336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheng L-B et al. (2017) miRNA-141 attenuates UV-induced oxidative stress via activating Keap1-Nrf2 signaling in human retinal pigment epithelium cells and retinal ganglion cells. Oncotarget 8, 13186–13194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tang C-Z et al. (2018) Activation of Nrf2 by ginsenoside Rh3 protects retinal pigment epithelium cells and retinal ganglion cells from UV. Free Radic. Biol. Med 117, 238–246 [DOI] [PubMed] [Google Scholar]
- 84.Xu X-Z et al. (2019) Targeting Keap1 by miR-626 protects retinal pigment epithelium cells from oxidative injury by activating Nrf2 signaling. Free Radic. Biol. Med 143, 387–396 [DOI] [PubMed] [Google Scholar]
- 85.Zhang J et al. (2017) miR-25 mediates retinal degeneration via inhibiting ITGAV and PEDF in rat. Curr. Mol. Med 17, 359–374 [DOI] [PubMed] [Google Scholar]
- 86.Peng Q-H et al. (2020) Astragalus polysaccharide attenuates metabolic memory-triggered ER stress and apoptosis via regulation of miR-204/SIRT1 axis in retinal pigment epithelial cells. Biosci. Rep 40, BSR20192121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gu C et al. (2021) Adipose mesenchymal stem cells-secreted extracellular vesicles containing microRNA-192 delays diabetic retinopathy by targeting ITGA1. J. Cell. Physiol 236, 5036–5051 [DOI] [PubMed] [Google Scholar]
- 88.Tonade D and Kern TS (2021) Photoreceptor cells and RPE contribute to the development of diabetic retinopathy. Prog. Retin. Eye Res 83, 100919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xi L (2020) Pigment epithelium-derived factor as a possible treatment agent for choroidal neovascularization. Oxidative Med. Cell. Longev 2020, 8941057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ţălu Ş and Nicoara SD (2021) Malfunction of outer retinal barrier and choroid in the occurrence and progression of diabetic macular edema. World J. Diabetes 12, 437–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xia T and Rizzolo LJ (2017) Effects of diabetic retinopathy on the barrier functions of the retinal pigment epithelium. Vis. Res 139, 72–81 [DOI] [PubMed] [Google Scholar]
- 92.Ponnalagu M et al. (2017) Retinal pigment epithelium-secretome: a diabetic retinopathy perspective. Cytokine 95, 126–135 [DOI] [PubMed] [Google Scholar]
- 93.Lu M (2020) Circular RNA: functions, applications and prospects. ExRNA 2, 1 [Google Scholar]
- 94.Zhang P et al. (2020) LncRNA NEAT1 sponges MiRNA-148a-3p to suppress choroidal neovascularization and M2 macrophage polarization. Mol. Immunol 127, 212–222 [DOI] [PubMed] [Google Scholar]
- 95.Chen X et al. (2020) LINC00167 regulates RPE differentiation by targeting the miR-203a-3p/SOCS3 axis. Mol. Ther. Nucleic Acids 19, 1015–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu X et al. (2020) Investigation of circRNA expression profiles and analysis of circRNA-miRNA-mRNA networks in an animal (mouse) model of age-related macular degeneration. Curr. Eye Res 45, 1173–1180 [DOI] [PubMed] [Google Scholar]
- 97.Li J and Zhang Y (2019) Current experimental strategies for intracellular target identification of microRNA. ExRNA 1, 6 [Google Scholar]
- 98.Thomson DW et al. (2011) Experimental strategies for microRNA target identification. Nucleic Acids Res 39, 6845–6853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li X et al. (2020) High-resolution in vivo identification of miRNA targets by Halo-enhanced Ago2 pull-down. Mol. Cell 79, 167–179.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Choi EH et al. (2021) An inducible Cre mouse for studying roles of the RPE in retinal physiology and disease. JCI Insight 6, e146604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Petri R and Jakobsson J (2018) Identifying miRNA targets using AGO-RIPseq. Methods Mol. Biol 1720, 131–140 [DOI] [PubMed] [Google Scholar]
- 102.Oda S and Yokoi T (2021) Recent progress in the use of microRNAs as biomarkers for drug-induced toxicities in contrast to traditional biomarkers: a comparative review. Drug Metab. Pharmacokinet 37, 100372. [DOI] [PubMed] [Google Scholar]
- 103.Conte I et al. (2015) MiR-204 is responsible for inherited retinal dystrophy associated with ocular coloboma. Proc. Natl. Acad. Sci. U. S. A 112, E3236–E3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.ElShelmani H et al. (2021) The role of deregulated microRNAs in age-related macular degeneration pathology. Transl. Vis. Sci. Technol 10, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kiel C et al. (2020) A circulating microRNA profile in a laser-induced mouse model of choroidal neovascularization. Int. J. Mol. Sci 21, 2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Blasiak J et al. (2019) Expression of VEGFA-regulating miRNAs and mortality in wet AMD. J. Cell. Mol. Med 23, 8464–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xiao Q et al. (2019) NFE2/miR-423-5p/TFF1 axis regulates high glucose-induced apoptosis in retinal pigment epithelial cells. BMC Mol. Cell Biol 20, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Crooke ST et al. (2021) Antisense technology: a review. J. Biol. Chem 296, 100416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Askou AL et al. (2017) Suppression of choroidal neovascularization in mice by subretinal delivery of multigenic lentiviral vectors encoding anti-angiogenic microRNAs. Hum. Gene Ther. Methods 28, 222–233 [DOI] [PubMed] [Google Scholar]
- 110.Karali M et al. (2020) AAV-miR-204 protects from retinal degeneration by attenuation of microglia activation and photoreceptor cell death. Mol. Ther. Nucleic Acids 19, 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fan Y-Y et al. (2021) MicroRNA-126 inhibits pathological retinal neovascularization via suppressing vascular endothelial growth factor expression in a rat model of retinopathy of prematurity. Eur. J. Pharmacol 900, 174035. [DOI] [PubMed] [Google Scholar]
- 112.Li B et al. (2021) Direct conversion of adult human retinal pigmented epithelium cells to neurons with photoreceptor properties. Exp. Biol. Med. (Maywood) 246, 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Davari M et al. (2017) Overexpression of miR-183/−96/−182 triggers neuronal cell fate in human retinal pigment epithelial (hRPE) cells in culture. Biochem. Biophys. Res. Commun 483, 745–751 [DOI] [PubMed] [Google Scholar]
- 114.Pilsl S et al. (2020) Optoribogenetic control of regulatory RNA molecules. Nat. Commun 11, 4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gu J et al. (2021) Geniposide alleviates choroidal neovascularization by downregulating HB-EGF release from RPE cells by downregulating the miR-145-5p/NF-κB axis. Exp. Eye Res 208, 108624. [DOI] [PubMed] [Google Scholar]


