Abstract
Mercury exists in organic, inorganic, and elemental forms; all of them are highly toxic. A sensor which could detect all forms of mercury below the permissible level in environmental and biological samples would be advantageous. A facile method to synthesize N-acetyl cysteine capped cadmium selenide quantum dots (CdSe QDs) with an emission at 554 nm was reported. CdSe QDs showed high sensitivity and selectivity toward Hg in aqueous media as well as biological fluids like simulated cerebrospinal fluid, saliva, and urine, and also in natural fluids like juices of tomato, sugarcane, and lime. The sensing mechanism is attributed to the interactions between Hg and CdSe QDs inducing fluorescence quenching. The limit of detection is 1.62, 0.75, and 1.27 ppb for organic, inorganic and elemental mercury, respectively, which is below WHO guidelines. The suitability of the sensor for estimating Hg in biological fluids was demonstrated by recovery experiments. Besides sensing, a two color cell imaging method was developed employing CdSe QDs and acridine orange. Using this method, the uptake of Hg in living cells was demonstrated.
CdSe QDs fluorescence is highly selective and sensitive to mercury.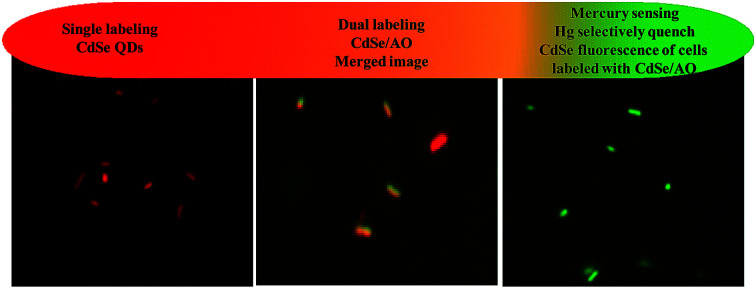
1. Introduction
Mercury is one of the hazardous metals which exist in elemental, inorganic and organic forms.1 It can be absorbed through the skin and mucous membranes and affects the brain, kidneys, and lungs.2 Over the years, the anthropogenic mercury emission has increased rapidly by the processes of coal combustion, waste combustion, electronics, gold production, non-ferrous metal production, cement production, caustic soda production, iron and steel production, and so-forth.3 Mercury discharged into the environment was converted into organic forms by microbes which will enter into the human food chain at toxic concentrations. For examples, methyl mercury formed by bacteria in an aquatic ecosystem was biomagnified within the aquatic food chain and reached higher levels in the top level aquatic predators.4 Other than unknown exposure and inhalation of discharged mercury inland, the consumption of these aquatic species also becomes a source for mercury exposure for humans. According to world health organization (WHO) guidelines, a maximum permissible level of mercury in food and drinking water is ∼2 ppb.5 Concerns over the human safety directed researchers to develop a variety of sensors based on electrochemical response, conductivity, or a change in color and fluorescence.6,7 Hitherto, many efficient sensors are developed by testing with inorganic mercury especially Hg2+, whereas only limited studies are available for direct sensing of elemental mercury and organic mercury in aqueous environments.8–12 Hence, developing an ultrasensitive sensor for monitoring all forms of mercury in aqueous environments and biological fluids is of great interest.
Nanocrystalline semiconductor or quantum dots (QDs) have been extensively studied for their excellent optical and electronic properties.13 Particularly, QDs with water-solubility, photostability, high fluorescence intensity and narrow emission spectra with low toxicity are useful for biological applications as biomarkers.14 the most popular approach to get water-soluble QDs is to cap or functionalize the QDs with designer ligands. It has been known that the fluorescence of QDs can be modulated by capping agents to detect heavy metals in aqueous solution. For example, N-acetyl-l-cysteine (NAC) functionalized copper sulfide QDs detects Hg2+, Ag+ and Au3+ by the mechanism of aggregate-induced quenching and electron transfer with cation exchange.15 Cadmium sulfide QDs capped with hyper branched graft copolymers detects Hg2+ with limit of detection (LOD) of 15 nM.16 CdS QDs capped with thioglycollic acid detects dopamine with LOD of 0.68 μM.17 Pencilliamine capped CdTe QDs detects Cu2+ ions by fluorescence quenching.18 Cysteamine capped CdTe QDs prepared by one pot synthetic method senses Hg2+ with LOD of 4 nM.19 Fluorescent carbon dots prepared through electrochemical carbonization of urea and citric acid selectively detects Hg2+ with LOD of 3.3 nM.20 CdSe QDs conjugated with silver nanoparticles were reported for sensing glucose with limit of detection of 1.86 mM.21 CdSe QDs surfaces modified with triethanolamine detects Hg2+ ions and I− with high selectivity and sensitivity.22 Other than QDs, metal nanoparticles also useful for sensing biomolecules and heavy metals.23 Even though many efficient sensors are reported for Hg2+ ions, there has been no report focusing on the determination of elemental and organic mercury using CdSe QDs.
In this paper, we report the use of NAC as the stabilizer to synthesize CdSe QDs in aqueous phase using cadmium chloride as precursor and sodium hydrogen selenide in dimethylformamide as the source of selenium. The choice of NAC is based on the fact that the good water solubility and biocompatibility, moreover, the available functional group can provide better surface passivation and ensures high fluorescence from the QDs. The synthesized CdSe QDs display emission at 554 nm and capable of selectively sense mercury in aqueous solution and biological fluids based on fluorescence quenching. The limit of detection for organic, inorganic and elemental mercury is 1.62, 0.75, and 1.27 ppb, respectively. In addition, we illustrate the unique advantage of labelling live bacterial cells by CdSe QDs and demonstrated the accumulation of mercury in bacterial cells through quenching the fluorescence of labelled cells.
2. Results and discussion
2.1. Characterization of cadmium selenide quantum dots
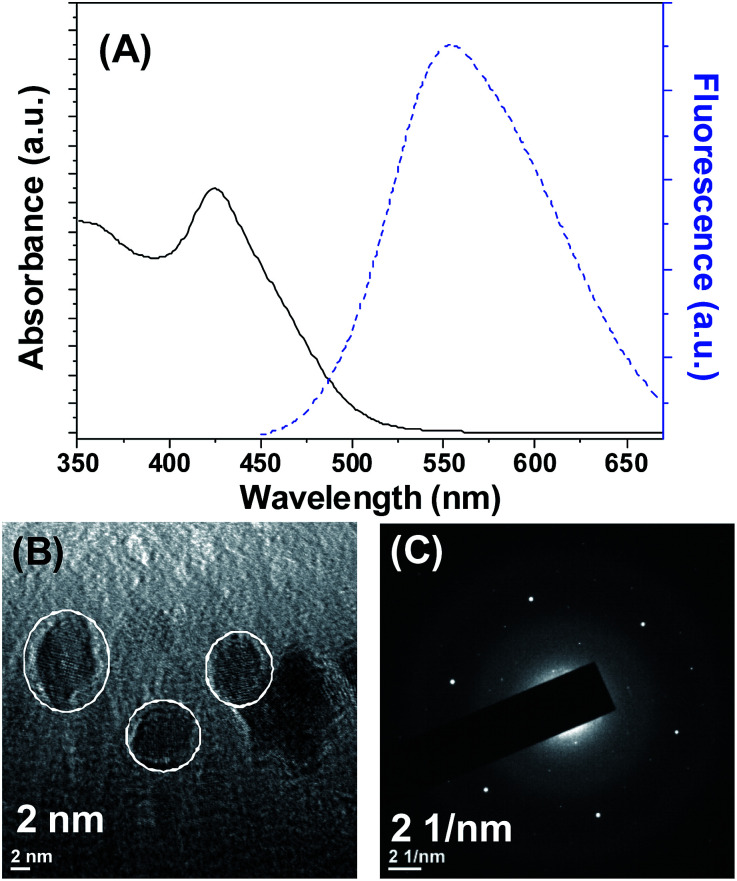
Cadmium selenide quantum dots (CdSe QDs) were synthesized by refluxing cadmium chloride and sodium hydrogen selenide in the presence of N-acetylcysteine. The UV-vis spectrum and fluorescence emission spectrum of the CdSe QDs were shown in Fig. 1. An absorption peak for CdSe QDs was observed at 425 nm (Fig. 1A). The band gap computed from the absorption peak for CdSe QDs was 2.92 eV, whereas the band gap of bulk is 1.7 eV.24 The difference in the band gap is attributed to the size effect. The fluorescence emission spectra of CdSe QDs measured at different excitation wavelength revealed that the maximum fluorescence intensity was observed while excitation fixed between 360–420 nm (Fig. S1†). Noteworthy, irrespective of excitation wavelength CdSe QDs fluorescence emission maximum was observed around 554 nm (Fig. 1A and S1†). The quantum yield (QY) of CdSe QDs was determined as 11.03% using quinine sulfate as the standard reference. The lower QY as compared to standard can be attributed to the defects caused by the surface related trap states, which is common in aqueous phase synthesis.24 The size analyzed by transmission electron microscopy (TEM) revealed that the average size of CdSe QDs is 4.8 ± 1.6 nm (Fig. 1B). The lattice fringe patterns observed in TEM micrograph and selected area electron diffraction (SAED) pattern (Fig. 1C) supports the crystalline nature of the CdSe QDs.
Fig. 1. (A) UV visible and fluorescence emission spectra of CdSe QDs. (B) Transmission electron microscopy image of CdSe QDs. (C) SAED pattern of CdSe QDs.
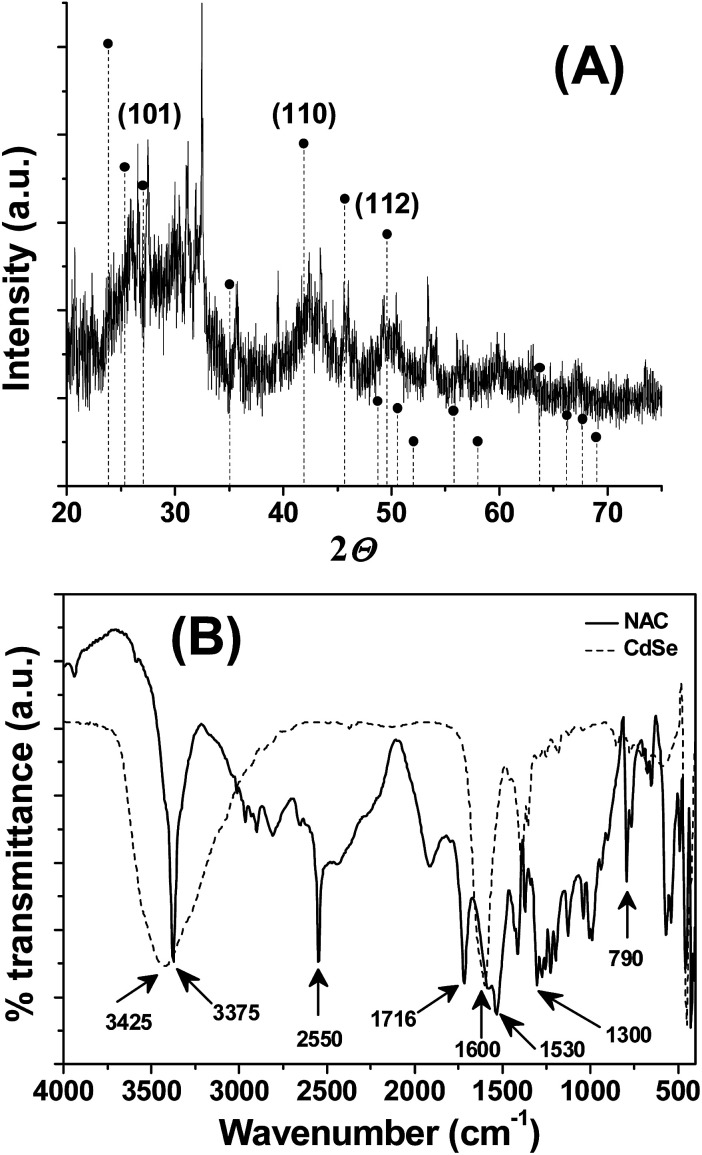
The powdered XRD pattern showed three distinct diffraction peaks, situated at 27.03, 42.35, and 49.59°, corresponding to the crystal faces of (1 0 1), (1 1 0) and (1 1 2) planes, respectively (Fig. 2A). The diffraction peaks in this pattern are in agreement with a hexagonal structure of CdSe (PDF-65-3415) and are consistent with the literature.25,26 The broadened XRD peaks accounts for the formation of crystalline nanodimensional CdSe. The noise in XRD pattern can be attributed either to surface defects or to the smaller size of QDs. Attempts to synthesis CdSe QDs either in the presence of l-cysteine or l-cysteine ethyl ester or l-methionine did not yield the expected luminophore. Hence, the contribution of the NAC functional group in the stabilization of CdSe QDs was analyzed through comparing the FTIR spectra of NAC and CdSe QDs (Fig. 2B). As noted from Fig. 2B, NAC shows IR peaks corresponds to the S–H stretching at 2550 cm−1, C O stretching of COOH at 1716 cm−1, C O stretching of CONH at 1600 cm−1, C–N, N–H bending at 1530 cm−1, N–H bending at 1300 cm−1, out of plane N–H bending at 790 cm−1. In the case of CdSe QDs, the IR peaks at 2550 cm−1, 1716 cm−1, 1530 cm−1, 1300 cm−1, 790 cm−1 was absent, suggests that they may involve in capping the QDs. Moreover, the O–H, N–H stretching observed at 3375 cm−1 of NAC was broadened in the synthesized CdSe QDs, indicating the strong interaction between the O–H group and CdSe QDs. These observations support that the –COOH, –SH and –CONH– groups of NAC are involved in the stabilization of CdSe QDs and prevents the QDs from agglomeration and fluorescence self quenching.
Fig. 2. (A) Powered XRD of CdSe QDs. The vertical dotted lines corresponds to the reference, hexagonal CdSe (PDF-65-3415), (B) FTIR spectra of NAC (solid line) and NAC stabilized CdSe QDs (dotted lines).
2.2. Mercury sensing
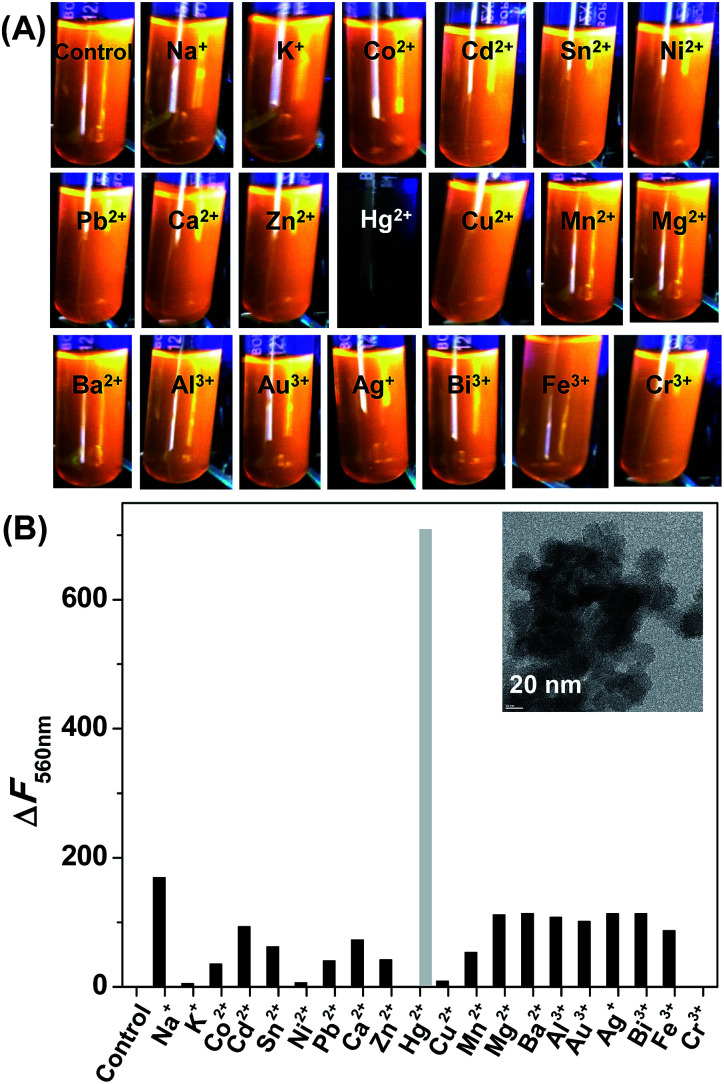
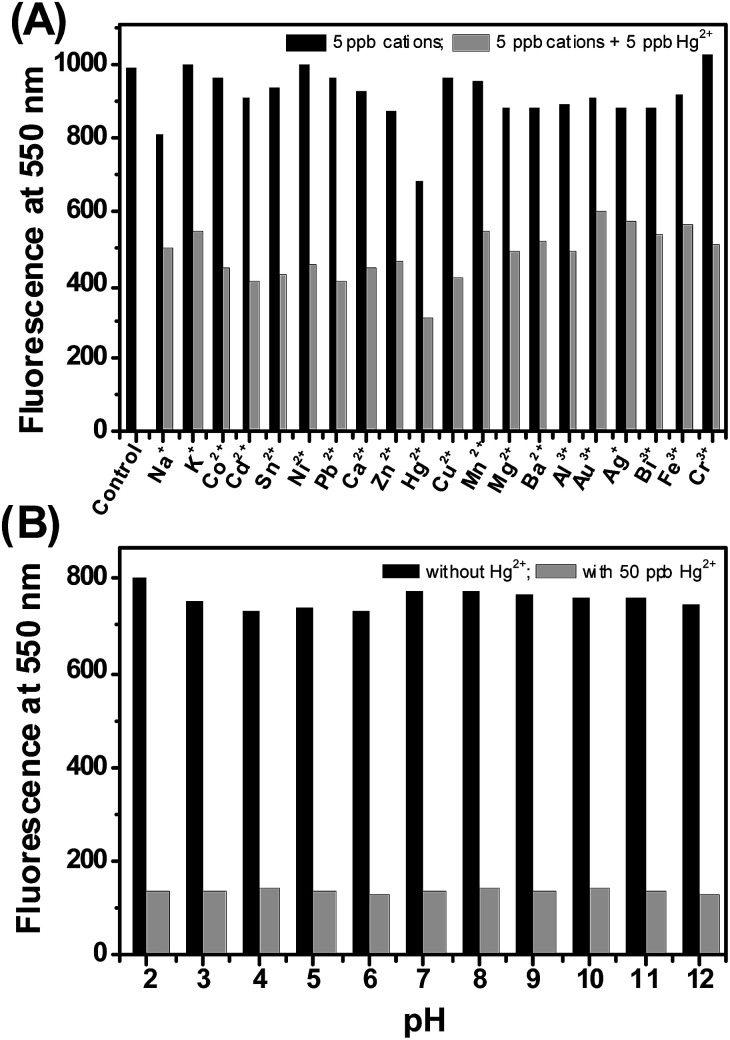
The sensing experiments were first illustrated with inorganic salts. The fluorescence responsiveness of CdSe QDs were monitored in the presence of listed metal ions – Na+, K+, Co2+, Cd2+, Sn2+, Ni2+, Pb2+, Ca2+, Zn2+, Hg2+, Cu2+, Mn2+, Mg2+, Ba2+, Al3+, Au3+, Ag+, Bi3+, Fe3+, Cr3+. The addition of 50 ppb Hg2+ to the CdSe QDs rapidly quench the fluorescence and causing aggregation of CdSe QDs, whereas the fluorescence of CdSe QDs remains unaffected in the presence of other metal ions (Fig. 3). The aggregation was confirmed by TEM, which showed that CdSe QDs aggregated irregularly and larger clusters were formed (inset Fig. 3B).
Fig. 3. (A) CdSe QDs photos taken after the addition of 50 ppb metal ions. (B) Change in fluorescence of CdSe QDs in the presence of 50 ppb metal ions. Only Hg2+ ions drastically quench the fluorescence from CdSe QDs. Inset: TEM image showed Hg2+ induced changes in the aggregation state of CdSe QDs.
The mechanism for the aggregation was investigated by adding excess EDTA to CdSe QDs–Hg2+ aggregate. Though, EDTA has a stronger affinity to Hg2+ and other metal ions, but the addition of EDTA did not cause fluorescence recovery from the aggregate (Fig. S2†). This suggests that the aggregation most likely arises because of the formation of a shell of mercury with or without amalgamation on the QDs surface, and the impact of the mercury shell quenches the fluorescence of CdSe QDs. At the same time, the higher selectivity of CdSe QDs for mercury over other metal ions is not known with certainty, but the theoretical calculation by others indicated that the dispersion forces between closed-shell metal atoms are highly specific and strong, particularly when heavy ions such as Ag+ (4d10), Au+ (5d10) and Hg2+ (5d10) are involved.27–29
The difference in the standard reduction potential of Cd2+ (E0 = −0.40 V) and Hg2+ (E0 = +0.85 V) also suggest that the Hg2+ has a greater tendency to get reduced and favours Cd–Hg metallophilic interactions, which may result in the CdSe QDs–Hg2+ aggregates. Energy dispersive X-ray diffraction and elemental mapping confirms the presence of cadmium, selenium, and mercury in the aggregate (Fig. S3 and S4†), and supports the conclusions drawn above.
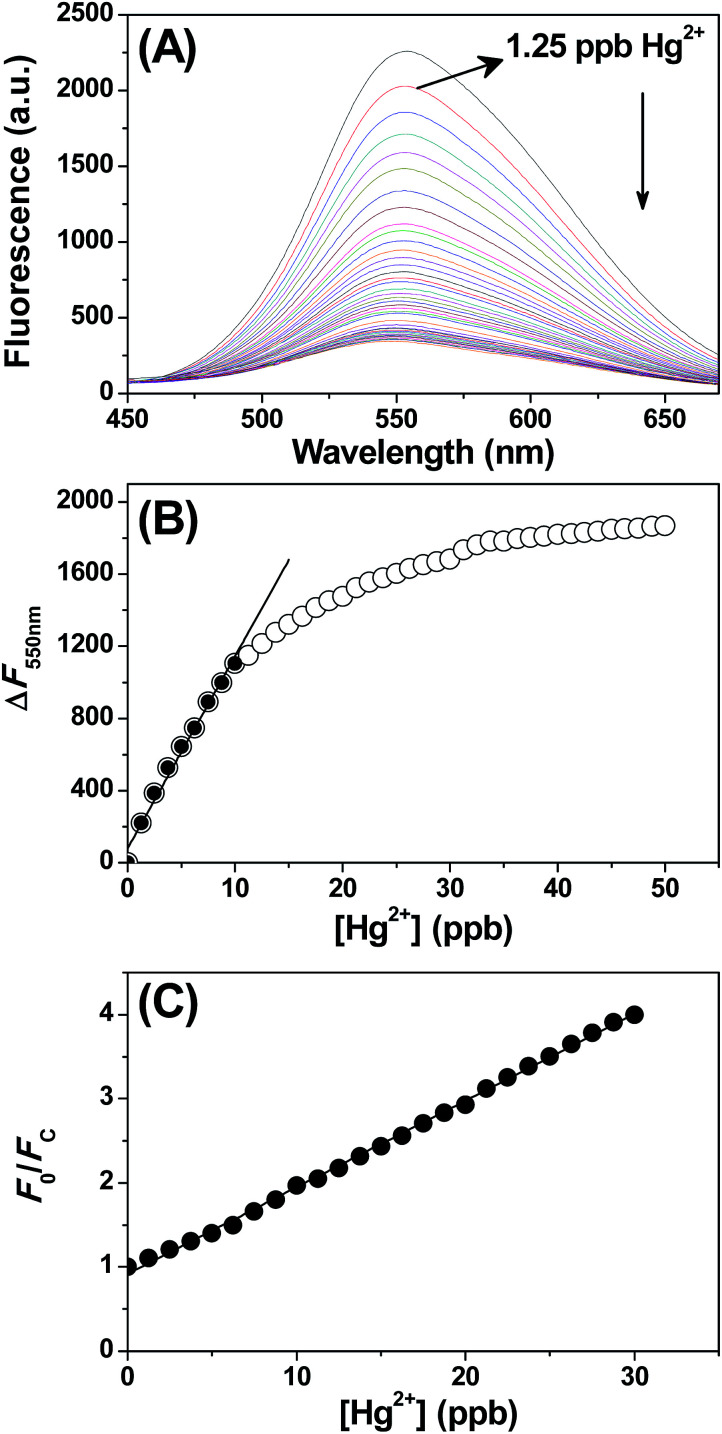
Having observed the high sensitivity of the sensor towards Hg2+ ion, we have investigated the effectiveness of the CdSe QDs in the detection of organic mercury and elemental mercury. The details of the preparation of phenyl mercury chloride and elemental mercury were described in the Experimental section. The aqueous solubility of phenyl mercury chloride and elemental mercury was facilitated by adding trace amount of polyvinylalcohol. The absolute concentration of mercury was determined by AAS analysis and the solutions were diluted appropriately to prepare lower concentrations of mercury. The addition of either phenyl mercury chloride or elemental mercury quenches the CdSe QDs fluorescence (Fig. S5A and S6A†) and showed trends similar to Fig. 4. The plot of ΔF versus the concentration of phenylmercury chloride and elemental mercury was shown in Fig. S5 and S6,† respectively. From this linear plot, LOD for phenyl mercury chloride and elemental mercury were determined as 1.62 ppb and 1.27 ppb, respectively. According to the World Health Organization (WHO), the maximum acceptable level of Hg in drinking water is less than 2 ppb, and thus the proposed sensor also met the requirement of the WHO guidelines.
Fig. 4. (A) Fluorescence response of CdSe QDs upon addition of Hg2+ ions. The arrow indicates the decrease in the fluorescence maximum with increasing concentration of Hg2+ ions. (B) Plot of change in fluorescence intensity at 550 nm versus Hg2+ ions. Linearity was observed in the concentration range of Hg2+ ions from 0–10 ppb. (C) Stern–Volmer plot display linearity between 0–30 ppb, suggesting the formation CdSe QDs–Hg2+ complex.
The mercury detection ability of the CdSe QDs could be affected by common interfering cations like Na+, K+, Co2+, Cd2+, Sn2+, Ni2+, Pb2+, Ca2+, Zn2+, Cu2+, Mn2+, Mg2+, Ba2+, Al3+, Au3+, Ag+, Bi3+, Fe3+, Cr3+. The sensitivity of the sensor investigated in the presence of interfering cations illustrated that the background cations have no effect on CdSe QDs fluorescence, but significant quenching was observed with the mixture of mercury and background cations (Fig. 5A). These results suggest that CdSe QDs has excellent ability to sense mercury, even in the presence of co-existing interfering cations. Next, the potential of the sensor to work on a wide range of pH was investigated. As noted from Fig. 5B, CdSe QDs display good fluorescence intensity irrespective of the pH and the addition of Hg2+ resulted in fluorescence quenching. The results indicate that CdSe QDs is stable at a pH range of 2–12 and has good selectivity to detect Hg2+ at a wide pH range.
Fig. 5. (A) Interference study. The fluorescence responses of CdSe QDs in the presence of background metal ions. (B) pH influences on the fluorescence of CdSe QDs, without and with 50 ppb Hg2+ ions. λex = 360 nm, λem = 550 nm.
2.3. Detecting mercury in biological fluids
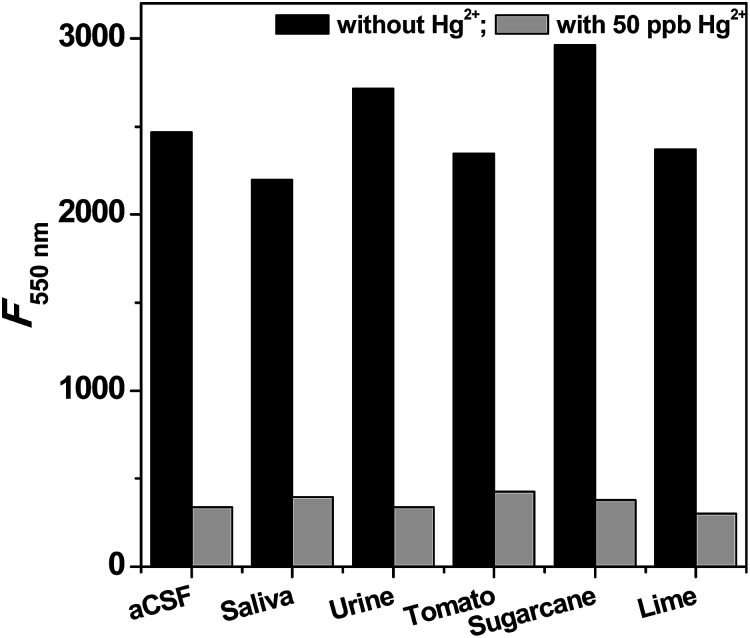
The exposure to high levels of mercury may lead to accumulation in biological fluids. Therefore, the performance of the sensor was tested in simulated biological fluids like saliva, urine and artificial cerebrospinal fluid (aCSF) and also in natural fluid obtained from sugarcane, lemon and tomato (Fig. 6). The addition of CdSe QDs to these fluids displays fluorescence as similar to water solublized CdSe QDs. This indicates that the chosen fluids are free from mercury, and thus, the fluids are spiked with standard solutions of mercury for testing the potential of the proposed sensor. Strikingly, the fluorescence was quenched in the fluids containing mercury, suggesting that CdSe QDs has high selectivity and sensitivity for mercury even with interference from higher concentrations of other ions (Fig. 6). Similar fluorescence responses are elicited by biological fluids containing phenylmercury chloride and elemental mercury. The practical use of the proposed sensor system was further investigated by recovery experiments. The chosen biological fluid has no Hg, thus, we spiked them with known concentration of Hg and analyzed with standard plot. The obtained results are summarized in Table 1, which suggests that the recovery rate of the spiked mercury were in acceptable range of 88–113%, indicating the applicability and reliability of the proposed method. The advantage of the CdSe QDs is that the naturally occurring metabolites in biological fluids do not interfere in this assay.
Fig. 6. Selective sensing of Hg2+ions in the simulated biological fluids and real biological fluids. Prior to measurement, the samples were spiked with 50 ppb of Hg2+ ions.
Recovery experiment. Estimation of mercury in the simulated biological fluids and real fluids, S – spiked, F – found, R – recovery. Experiments are performed in triplicate and the average value is presented.
| S (ppb) | Simulated biological fluids | Real fluids | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aCSF | Saliva | Urine | Tomato juice | Sugarcane juice | Lime juice | |||||||
| F (ppb) | R (%) | F (ppb) | R (%) | F (ppb) | R (%) | F (ppb) | R (%) | F (ppb) | R (%) | F (ppb) | R (%) | |
| Spiked with metallic Hg | ||||||||||||
| 2 | 2.07 | 103.5 | 2.16 | 108.0 | 2.23 | 111.5 | 2.12 | 106.0 | 2.19 | 109.5 | 1.89 | 94.5 |
| 4 | 4.39 | 109.8 | 4.19 | 104.8 | 4.09 | 102.3 | 3.78 | 94.5 | 3.92 | 98.0 | 4.13 | 103.3 |
| 6 | 6.34 | 105.7 | 6.14 | 102.3 | 6.24 | 104.0 | 6.04 | 100.7 | 5.82 | 97.0 | 5.88 | 98.0 |
| 8 | 8.66 | 108.3 | 8.44 | 105.5 | 7.95 | 99.3 | 7.5 | 93.8 | 7.46 | 93.3 | 8.13 | 101.6 |
| 10 | 10.53 | 105.3 | 10.3 | 103.0 | 9.53 | 95.3 | 9.4 | 94.0 | 10.23 | 102.3 | 9.87 | 98.7 |
| Spiked with Ph-Hg | ||||||||||||
| 2 | 2.27 | 113.5 | 2.17 | 108.5 | 2.24 | 112.0 | 2.027 | 101.4 | 2.127 | 106.4 | 2.07 | 103.5 |
| 4 | 4.33 | 108.3 | 4.03 | 100.8 | 4.13 | 103.3 | 4.23 | 105.8 | 4.313 | 107.8 | 3.933 | 98.3 |
| 6 | 6.23 | 103.8 | 6.03 | 100.5 | 5.83 | 97.2 | 5.93 | 98.8 | 6.13 | 102.2 | 6.33 | 105.5 |
| 8 | 7.75 | 96.95 | 7.95 | 99.4 | 7.84 | 98.0 | 7.65 | 95.6 | 8.15 | 101.9 | 8.22 | 102.8 |
| 10 | 8.76 | 87.6 | 8.88 | 88.8 | 8.96 | 89.6 | 9.75 | 97.5 | 9.87 | 98.7 | 10.2 | 102.0 |
| Spiked with Hg 2+ ions | ||||||||||||
| 2 | 1.85 | 92.6 | 1.92 | 96.1 | 1.98 | 98.8 | 1.89 | 94.2 | 1.86 | 92.8 | 2.15 | 107.6 |
| 4 | 3.79 | 94.8 | 3.86 | 96.6 | 3.65 | 91.2 | 3.91 | 97.9 | 3.46 | 86.6 | 3.77 | 94.2 |
| 6 | 5.54 | 92.3 | 5.86 | 97.7 | 5.32 | 88.6 | 5.46 | 91.0 | 5.85 | 97.4 | 5.62 | 93.6 |
| 8 | 7.78 | 97.2 | 7.08 | 88.6 | 7.49 | 93.6 | 7.64 | 95.5 | 7.42 | 92.7 | 7.52 | 94.0 |
| 10 | 9.37 | 93.7 | 9.34 | 93.4 | 9.79 | 97.9 | 9.72 | 97.2 | 9.22 | 92.1 | 9.41 | 94.1 |
2.4. Mercury recognition by CdSe QDs in living cells
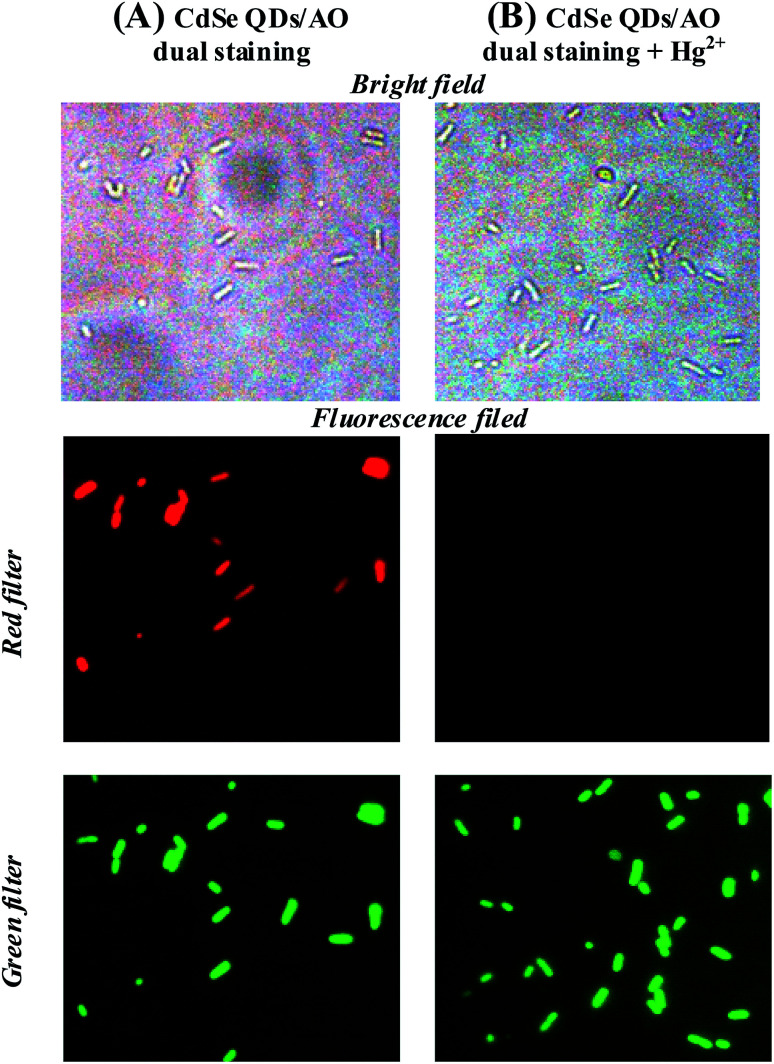
The possibility of using CdSe QDs for the recognition of the mercury in living cells was demonstrated through fluorescence imaging study. Escherichia coli were chosen as a live cell model. For imaging studies, live cells preexposed to CdSe QDs were washed with PBS buffer, and viewed through a fluorescence microscope using a red filter. A bright red fluorescent was observed in E. coli cells, which indicates that the cells are stained with CdSe QDs [Fig. S7†]. To know that CdSe QDs stained the live cells, a dual staining method was developed using acridine orange (AO) and CdSe QDs. AO, a membrane permeable nucleic acid-selective fluorescent dye, commonly used to monitor viable cells. The cells were treated with the combination of CdSe QDs and AO, and viewed through a fluorescence microscope using green and red filter as an excitation source for AO and CdSe QDs, respectively. As shown in Fig. 7A, the intact cells are visible in bright field and dark field. The cells showed both CdSe QDs and AO fluorescence at appropriate excitation and display corresponding red and green fluorescence signals from the cells. These results suggest that CdSe QDs were membrane permeable and suitable for live cell imaging without additives.
Fig. 7. (A) Two colour staining. E. coli cells stained with the mixture of CdSe QDs and AO. Red fluorescence and green fluorescence signals corresponds to CdSe QDs and AO, respectively. (B) Mercury sensing in E. coli cells. The cells were stained with CdSe QDs/AO and then, treated with Hg2+. AO green fluorescence indicates cells are live and the fluorescence quenching of CdSe QDs is attributed to the selective sensing of accumulated Hg2+ inside the cells (magnification – 100×).
The mercury accumulations in living cells were investigated using CdSe QDs. Typically, the cells stained with CdSe QDs showed red fluorescence as described above, but the addition of Hg2+ ion as low as 25 ppb diminishes the fluorescence (Fig. S7C†). The selectivity of CdSe QDs for Hg2+ ion was further demonstrated by dual staining method as described above. For two colours staining, the cells were treated with a solution containing AO and CdSe QDs and then, exposed to 25 ppb of Hg2+ ion. On viewing through fluorescence microscope, a bright green fluorescence was observed for AO, whereas no fluorescence from CdSe QDs. These results indicate that the accumulation of Hg2+ ion in the cells selectively diminishes the CdSe QDs fluorescence (Fig. 7B). In short, using the CdSe QDs probe and AO, a dual colour cell imaging for live cell was obtained and demonstrated the detection of Hg2+ ion uptake in living cells through selectively quenching the CdSe QDs fluorescence.
3. Experimental
3.1. Preparation of CdSe QDs
Sodium hydroselenide (NaHSe) was synthesized through dissolution of selenium powder (30 mg) and sodium borohydride (75 mg) in 9 mL dimethylformamide. The reaction was conducted in an ice bath under nitrogen atmosphere for 2 h. The complete disappearance of selenium powder confirms the formation of NaHSe. CdSe QDs were obtained by reacting cadmium chloride and NaHSe in the presence of NAC. Typically, CdCl2 (63 mg) and NAC (159 mg) was dissolved in 43 mL double distilled water in a 250 mL two-neck flask fitted with Liebig condenser. After 20 min, the pH of the solution was adjusted to 9 using NaOH. To this solution, the freshly prepared 7 mL NaHSe solution was injected slowly for 5 min and refluxed for 15 min. The obtained red colored solution showed bright orange fluorescence under UV lamp at 360 nm.
UV-visible absorption spectra of CdSe QDs were acquired in Thermo Scientific Evolution 201. Fluorescence spectra were recorded on a Jasco-FP8200 spectrofluorimeter. The spectral slit width was set to 5 nm for excitation and emission monochromators. All optical measurements were performed at room temperature under ambient conditions. The morphology, size and crystallinity of CdSe QDs were measured in the transmission electron microscope (TEM) (JEOL-JEM 1011, Japan). TEM sample was prepared by drop cast an aqueous CdSe QDs solution onto carbon-coated copper grid and dried under vacuum. Powder X-ray diffraction spectrum of CdSe QDs was measured with lyphophilized powder on a XRD Bruker D8 Advance X-ray diffractometer using monochromatic Cu Kα radiation. Energy dispersive X-ray spectrum (EDAX) was acquired in OXFORD instrument INCA penta FETX3.
3.2. Sensor study
Metal ion solutions for sensor studies were prepared by dissolving the requisite amount of metal salts in double distilled deionised water. Hg solutions were prepared as follows. 2.5 g of metallic mercury was added to 50 mL of water and sonicated for 5 min. 15 mL of this mixture was added to 80 mL of water containing 50 mg polyvinyl alcohol (PVA).9 The obtained solution is transparent and stable. Phenyl mercuric chloride was prepared by standard procedure.30 Briefly, 1 mol of aniline was mixed with 10 mL of cold concentrated hydrochloric acid (Solution A). Sodium nitrite (0.13 mol) was dissolved in ice water (Solution B). Solution B is added slowly to Solution A under stirring condition in an ice bath. The obtained diazoniam salt was allowed to react with 24.6 mmol of mercury chloride. The white precipitate formed from the reaction was collected by filtration and washed thrice with cold water to remove colored impurities. The air dried powder was dissolved in 50 mL of acetone and 100 mg copper powder was added and allowed to stand for overnight. The insoluble material is collected and extracted with 10 mL hot xylene. The filtrate was cooled to crystallize phenylmercury chloride. The phenylmercury chloride solution was prepared by dissolving requisite amount in 50 mL of water containing 50 mg PVA. The exact concentration of mercury solutions was determined by ASS (Outsourced to Tamilnadu Test House Private Limited, Chennai, India) (Fig. S8–S10†) and the solution were diluted appropriately with water to prepare analyte solutions.
Artificial body fluids like cerebrospinal fluid (aCSF), saliva and urine used in this study were prepared according to the reported procedure.31,32 Briefly, aCSF contains 119 mM NaCl, 26.2 mM NaHCO3, 2.5 mM KCl, 1 mM NaH2PO4, 1.3 mM MgCl2, 10 mM glucose in 100 mL. Artificial saliva contains 12.5 mg NaCl, 96.4 mg KCl, 18.9 mg KSCN, 65.5 mg KH2PO4, 20 mg urea, 22.9 mg CaCl2, Na2SO4, 17.8 NH4Cl mg, 63.1 mg NaHCO3 in 100 mL and pH was adjusted to 6.7 using HCl. Artificial urine contains 2.5 g urea, 0.9 g NaCl, 0.3 g NH4Cl, 0.2 g creatine, 0.25 g Na2HPO4, 0.25 g KH2PO4, 0.3 g Na2SO3 in 100 mL and pH were adjusted to 6.7 using HCl. Natural fluids like tomato juice, lime juices, sugarcane juices were prepared by squeezing the vegetables collected from local vegetable shops. The juices were diluted by adding 1 mL of juices into 100 mL of distilled water and spiked with analyte solutions.
The CdSe QDs working solution was prepared by diluting the stock solution five times in double distilled water. Sensing experiment was carried out by adding aliquots of metal ion solutions to CdSe QDs working solution and the change in fluorescence were recorded in Jasco-FP8200 spectrofluorimeter. The spectrofluorimetric titrations were performed as follows: 3 mL of the CdSe working solution was pipette into a 1 cm × 1 cm quartz cuvette and varying volumes of mercury stock solutions were added to the cuvette. The mixture was briefly mixed for a few seconds before making the fluorescence measurements.
3.3. Live cell sensing
CdSe QDs was subjected for imaging live cells. Typically, E. coli cells (1 × 105 cfu mL−1) were inoculated in 20 mL sterile LB media and cultured for 6 h at 37 °C. The live cells were collected by centrifugation at 2000 rpm and resuspended in 400 μL of freshly prepared CdSe QDs and made up to 1 mL with PBS. After incubation at 37 °C for 2 h, the bacterial cells were collected by centrifugation at 2000 rpm and washed twice with PBS buffer. The obtained CdSe QDs labeled cell suspension was drop cast on sterile glass slides and visualized under Nikon Eclipse NieU fluorescence microscope. Mercury sensing in live cell was performed by treating the CdSe QDs labeled cells with 25 ppb mercury solution and imaged in fluorescence microscopy.
In order to prove that the CdSe QDs labelled live cells, a dual staining method was developed. Briefly, log phase E. coli cells were collected and exposed to 400 μL of freshly prepared CdSe QDs and acridine orange (50 μg mL−1) and made up to 1 mL with PBS. The dual labelled cells were collected and image in fluorescence microscopy. AO was visualized using green filter, and CdSe QDs was visualized using a red filter. Mercury sensing was carried out as described above using the dual labelled cells.
4. Conclusions
In summary, we synthesized NAC functionalized CdSe QDs that selectively detects mercury in organic, inorganic and elemental forms. The sensing mechanism through fluorescence quenching is attributed to strong metallophilic interactions between the mercury and CdSe QDs. The proposed probe could selectively sense mercury in a wide range of pH, and also in natural fluids with good recovery. Results of the fluorescence microscopic studies revealed that CdSe QDs is cell permeable and could be used as safer reagents to live cell imaging. Moreover, the dual probe method developed in this study has unique potential in the detection of the uptake of mercury in live cells. The scope for further development of this dual probe concept is extensive, in view of labelling live cells and detection of metal accumulation in live cells.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
SBS earnestly acknowledges the teaching assistantship from SASTRA Deemed University. We thank Dr Vairaprakash for the help in synthesizing phenylmercury chloride. We gratefully acknowledge central research facility (R&M/0021/SCBT-007/2012-13), SASTRA Deemed University and DST-FIST grant (SR/FST/ETI-331/2013) to SCBT, SASTRA for establishing infrastructure facilities.
Electronic supplementary information (ESI) available: Emission spectra, Ph-Hg titration, elemental mercury titration, EDAX, bioimaging, amount of Hg-ASS report. See DOI: 10.1039/c9ra04753k
References
- Driscoll C. T. Mason R. P. Chan H. M. Jacob D. J. Pirrone N. Environ. Sci. Technol. 2013;47:4967. doi: 10.1021/es305071v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. D. Zheng W. J. Prev. Med. Public Health. 2012;45:344. doi: 10.3961/jpmph.2012.45.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger Chakraborty L. Qureshi A. Vadenbo C. Hellweg S. Environ. Sci. Technol. 2013;47:8105. doi: 10.1021/es401006k. [DOI] [PubMed] [Google Scholar]
- Lee C. S. Fisher N. S. Environ. Toxicol. Chem. 2017;36:1287. doi: 10.1002/etc.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan E. M. Lippard S. J. Chem. Rev. 2008;108:3443. doi: 10.1021/cr068000q. [DOI] [PubMed] [Google Scholar]
- Ariño C. Serrano N. Díaz-Cruz J. M. Esteban M. Anal. Chim. Acta. 2017;990:11. doi: 10.1016/j.aca.2017.07.069. [DOI] [PubMed] [Google Scholar]
- Chen G. Guo Z. Zeng G. Tang L. Analyst. 2015;140:5400. doi: 10.1039/C5AN00389J. [DOI] [PubMed] [Google Scholar]
- Ahmed K. B. A. Senthilnathan R. Megarajan S. Anbazhagan V. J. Photochem. Photobiol., B. 2015;151:39. doi: 10.1016/j.jphotobiol.2015.07.003. [DOI] [PubMed] [Google Scholar]
- Ramesh G. V. Radhakrishnan T. P. ACS Appl. Mater. Interfaces. 2011;3:988. doi: 10.1021/am200023w. [DOI] [PubMed] [Google Scholar]
- Deng L. Li Y. Yan X. Xiao J. Ma C. Zheng J. Liu S. Yang R. Anal. Chem. 2015;87:2452. doi: 10.1021/ac504538v. [DOI] [PubMed] [Google Scholar]
- Lin Y. W. Chang H. T. Analyst. 2011;136:3323. doi: 10.1039/C1AN15117G. [DOI] [PubMed] [Google Scholar]
- Kabir K. M. Sabri Y. M. Matthews G. I. Jones L. A. Ippolito S. J. Bhargava S. K. Analyst. 2015;140:5508. doi: 10.1039/C5AN00360A. [DOI] [PubMed] [Google Scholar]
- Wang Y. Hu R. Lin G. Roy I. Yong K. T. ACS Appl. Mater. Interfaces. 2013;5:2786. doi: 10.1021/am302030a. [DOI] [PubMed] [Google Scholar]
- Wegner K. D. Hildebrandt N. Chem. Soc. Rev. 2015;44:4792. doi: 10.1039/C4CS00532E. [DOI] [PubMed] [Google Scholar]
- Du W. Liao L. Yang L. Qin A. Liang A. Sci. Rep. 2017;7:11451. doi: 10.1038/s41598-017-10904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You F. Ya-Qian C. Hua-Ji L. Yu C. Sens. Actuators, B. 2017;251:171. doi: 10.1016/j.snb.2017.05.070. [DOI] [Google Scholar]
- Kulchat S. Boonta W. Todee A. Sianglam P. Ngeontae W. Spectrochim. Acta, Part A. 2018;196:7. doi: 10.1016/j.saa.2018.01.062. [DOI] [PubMed] [Google Scholar]
- Mohammad-Rezaei R. Razmi H. Abdolmohammad Zadeh H. Luminescence. 2013;28:503. doi: 10.1002/bio.2484. [DOI] [PubMed] [Google Scholar]
- Ding X. Qu L. Yang R. Zhou Y. Li J. Luminescence. 2015;30:465. doi: 10.1002/bio.2761. [DOI] [PubMed] [Google Scholar]
- Hou Y. Lu Q. Deng J. Li H. Zhang Y. Anal. Chim. Acta. 2015;866:69–74. doi: 10.1016/j.aca.2015.01.039. [DOI] [PubMed] [Google Scholar]
- Tang Y. Yang Q. Wu T. Liu L. Ding Y. Yu B. Langmuir. 2014;30:6324. doi: 10.1021/la5012154. [DOI] [PubMed] [Google Scholar]
- Shang Z. B. Wang Y. Jin W. J. Talanta. 2009;78:364–369. doi: 10.1016/j.talanta.2008.11.025. [DOI] [PubMed] [Google Scholar]
- Piriya V. S. A. Joseph P. Daniel S. C. G. K. Lakshmanan S. Kinoshita T. Muthusamy S. Mater. Sci. Eng., C. 2017;78:1231. doi: 10.1016/j.msec.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Kalasad M. N. Rabinal M. K. Mulimani B. G. Langmuir. 2009;25:12729. doi: 10.1021/la901798y. [DOI] [PubMed] [Google Scholar]
- Hodlur R. M. Rabinal M. K. Chem. Eng. J. 2014;244:8. doi: 10.1016/j.cej.2014.01.064. [DOI] [Google Scholar]
- Chunjin W. Jinyu L. Fang G. Shuxia G. Yongcui Z. Dan Z. J. Spectrosc. 2015:369145. [Google Scholar]
- Xie J. Zheng Y. Ying J. Y. Chem. Commun. 2010;46:961. doi: 10.1039/B920748A. [DOI] [PubMed] [Google Scholar]
- Kim M. Taylor T. J. Gabbaï F. P. J. Am. Chem. Soc. 2008;130:6332. doi: 10.1021/ja801626c. [DOI] [PubMed] [Google Scholar]
- Sculfort S. Braunstein P. Chem. Soc. Rev. 2011;40:2741. doi: 10.1039/C0CS00102C. [DOI] [PubMed] [Google Scholar]
- Nesmajanow A. N. Org. Synth. 1932;12:54. doi: 10.15227/orgsyn.012.0054. [DOI] [Google Scholar]
- Ranc V. Markova Z. Hajduch M. Prucek R. Kvitek L. Kaslik J. Safarova K. Zboril R. Anal. Chem. 2014;86:2939. doi: 10.1021/ac500394g. [DOI] [PubMed] [Google Scholar]
- Monika P. Adam V. Microchem. J. 2017;134:197. doi: 10.1016/j.microc.2017.06.004. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.