Abstract
Stem cells are distinctive cells that have self-renewal potential and unique ability to differentiate into multiple functional cells. Stem cell is a frontier field of life science research and has always been a hot spot in biomedical research. Recent studies have shown that long non-coding RNAs (lncRNAs) have irreplaceable roles in stem cell self-renewal and differentiation. LncRNAs play crucial roles in stem cells through a variety of regulatory mechanisms, including the recruitment of RNA-binding proteins (RBPs) to affect the stability of their mRNAs or the expression of downstream genes. RBPs interact with different RNAs to regulate gene expression at transcriptional and post-transcriptional levels and play important roles in determining the fate of stem cells. In this review, the functions of lncRNAs and their RBPs in self-renewal and differentiation of stem cell are summarized. We focus on the four regulatory mechanisms by which lncRNAs and their RBPs are involved in epigenetic regulation, signaling pathway regulation, splicing, mRNA stability and subcellular localization and further discuss other noncoding RNAs (ncRNAs) and their RBPs in the fate of stem cells. This work provides a more comprehensive understanding of the roles of lncRNAs in determining the fate of stem cells, and a further understanding of their regulatory mechanisms will provide a theoretical basis for the development of clinical regenerative medicine.
Keywords: LncRNAs, RBPs, Stem cells, Self-renewal, Differentiation
Introduction
Stem cells have the unique abilities to perpetuate themselves through self-renewal and to generate at least one type of highly mature cells through differentiation. According to their sources, stem cells can be divided into embryonic stem cells (ESCs), adult stem cells (ASCs) and induced pluripotent stem cells (iPSCs) [1]. Adult stem cells mainly include neural stem cells (NSCs), mesenchymal stem cells (MSCs), hematopoietic stem cells (HSCs), reproductive stem cells (RSCs), etc. [2]. iPSCs, which reduce ethical disputes in clinical practice, are the most commonly used cell lines in stem cell research [3]. According to different differentiation potentials, stem cells can be divided into totipotent stem cells (TSCs), pluripotent stem cells (PSCs), multipotent stem cells (MSCs) and unipotent stem cells (USCs) [4]. In recent years, the research of stem cell has made remarkable progresses. Because stem cells have the potential for multidirectional differentiation and self-renewal ability, they play important roles in repairing and replacing damaged cells, tissues and even organs, making them important seed cells for regenerative medicine at present and in the future. The application of stem cell depends on a thorough uncovering of the regulatory mechanisms involved in their cellular activities. For example, the transcriptional networks regulated by key transcription factors, such as Oct4, Sox2, and Nanog, have essential roles in maintaining pluripotency of stem cells [5–8]. Besides stem cell related proteins, many studies have identified that multiple species of RNAs are involved in the regulation of stem cell fate. Chepelev and Chen summarized a distinct splicing pattern through which stem cells can be maintained by specific mRNA isoforms, while they lead to proper differentiation after switching to other different isoforms [9]. There are also different RNAs that play important roles in precisely regulating the self-renewal of muscle stem cells [10, 11]. Pereira et al. observed that precise RNA degradation machinery is closely related to stem cell development [12]. Belair et al. demonstrated that exosomes can regulate human ESC (hESC) differentiation and RNA decay plays an importance in maintaining pluripotency [13]. LncRNA is a kind of transcript more than 200 nucleotides, which lacks protein coding potential and plays a regulatory role in maintaining the functions of stem cells. LncRNAs can form a complex with one or more RBPs and play important roles in the development of life. This review summarizes the functions of lncRNAs and their RBPs in stem cell self-renewal and differentiation.
Functions of long noncoding RNA in stem cells
RNA is a carrier of genetic information that exists in organisms and certain viruses. It performs the expression of genetic information encoded by DNA on proteins and controls various biological processes of organisms [14]. According to their function, RNAs can be divided into coding RNAs and ncRNAs. Coding RNAs generally refer to mRNA that encodes protein. ncRNAs are transcribed from DNA, which do not encode proteins. ncRNAs can be divided into the following two types according to their length: small ncRNA and long ncRNA [15]. Small ncRNAs refer to ncRNAs shorter than 200 nucleotides, while long ncRNAs (lncRNAs) are composed of 200 or more nucleotides. According to their function and subcellular location, small ncRNAs can be divided into microRNAs (miRNAs), transfer RNAs (tRNAs), piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), short hairpin RNAs (shRNAs), circular RNAs (circRNAs), ribosomal RNAs (rRNAs), etc. [16, 17] (Fig. 1).
Fig. 1.
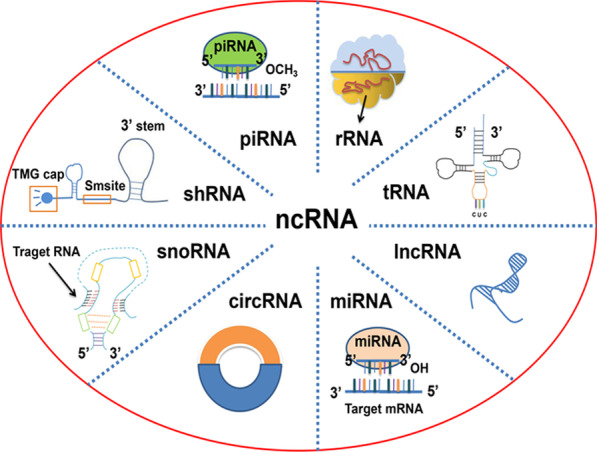
Schematic diagram of different ncRNA structures. The figure shows the classification of noncoding RNA and their structure, including piwi-interacting RNA (piRNA), short hairpin RNA (shRNA), small nucleolar RNA (snoRNA), circular RNA (circRNA), microRNA (miRNA), long noncoding RNA (lncRNA), transfer RNA (tRNA), ribosomal RNA (rRNA)
As a complex regulator in many biological processes, noncoding RNA forms an important part of the stem cell transcriptome, which is essential for the self-renewal and pluripotency maintenance of stem cells. Many studies have found that different types of ncRNAs, including lncRNAs [18, 19], microRNAs [20, 21], circRNAs [22], etc., have important influences on the fate of stem cells and may participate in different biological processes of stem cells, such as proliferation, apoptosis and differentiation. LncRNA growth arrest specific 5 (Gas5) is a host gene of small nucleolar RNA, involved in many biological functions of mESCs. Down-regulation of Gas5 expression can reduce the efficiency of somatic cell reprogramming [23]. LncRNAs are vital to the self-renewal and pluripotency maintenance of stem cells through different regulatory mechanisms. They take part in stem cell activities through regulatory mechanisms such as chromatin or histone modification around transcription factor genes, typical and atypical RNA-binding proteins, and as a sponge for microRNAs [24]. In general, lncRNAs can serve as miRNA sponge or competitive endogenous RNAs (ceRNAs) in stem cells [25]. Cyrano is a lncRNA located in the cytoplasm and nucleus of ESC, and it is a part of ncRNA regulatory network [26]. Cyrano silencing can inhibit the self-renewal and survival of ESCs, and its direct interaction with miR-7 can enhance the expression of the core pluripotency regulator Nanog to promote self-renewal of ESCs [27]. According to previous reports, lncRNAs are new regulators in the osteogenesis of MSCs [28]. The enhancement of special AT-rich sequence-binding protein 2 (SATB2) can promote the osteogenic differentiation of bone marrow mesenchymal stem cells (BMSCs) when patients have ethanol-induced osteonecrosis [29, 30]. The exosomes secreted by BMSCs contain lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), which can act as a sponge for miR-34c, increase the expression of SATB2 and help alleviate osteoporosis [31]. Wang et al. confirmed that lncRNA-ROR stabilizes the expression of the main pluripotent genes Oct4, Nanog and Sox2 by inhibiting miR-145, maintaining the pluripotency of hESCs [32].
Functions of RNA-binding protein in stem cells
RNA-binding proteins are a group of proteins involved in RNA-related metabolic regulatory processes. They can bind double or single-stranded RNAs to participate in the formation of ribonucleoprotein (RNP) complexes and influence the fate of RNA. RBPs, as trans-acting factors, are mainly mediated by specific RNA-binding domains (RBDs) [33]. They affect all aspects of post-transcriptional regulation and act in the regulation of RNA synthesis, splicing, modification, nuclear output, localization, protein stability and translation rate [34]. These RBPs functions are highly dependent on their structural characteristics, namely the presence and alignment of RNA-binding domains. Most RBPs contain multiple RNA-binding domains and RNA-binding motifs, which can recruit multiple RNAs to bind to them. Therefore, studying the interaction between RNA and protein is critical to exploring the function of RNA [35]. There is abundant research evidence that RBP plays an important role in stem cells. Most lncRNAs interact with the corresponding RBPs [36] and play irreplaceable roles in various cells and physiological processes. The interaction of different lncRNAs and their RBPs is responsible for a large number of biological events in stem cells, such as changing or stabilizing their nuclear outlet, regulating transcription or translation, especially playing an important role in stem cell self-renewal and maintenance of pluripotency [37, 38].
The functions of lncRNAs and their RBPs in stem cells
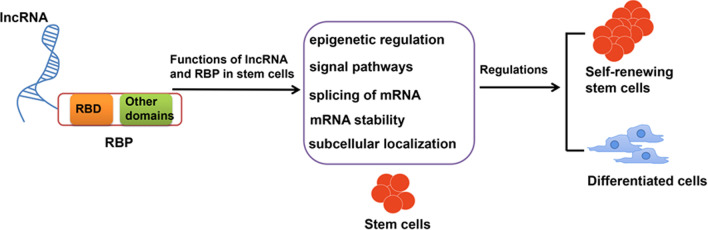
LncRNAs and RBPs are key regulators of gene expression and the core of many cells functions, such as protein synthesis, mRNA assembly, virus replication, and cells development regulation. Together, they have a profound effect on life activities of stem cells [39]. LncRNAs usually function in stem cell by binding RBPs. LncRNA Cyrano binds to signal transducer and activator of transcription 3 (STAT3), which is essential for the proliferation and self-renewal of ESCs [40]. Zhao et al. identified 39 lncRNAs binding to Klf4 by RNA immunoprecipitation and sequencing (RIP-SEQ) assay in the process of studying the role of lncRNA sites in tissue 3D chromatin structure. Considering the known role of lncRNAs in maintenance and inducing pluripotency, lncRNA 5430416N02Rik was screened. The interaction between 5430416N02Rik and Mid1 loci activates Mid1 expression and promotes Mid1 transcription, which in turn promotes the rapid proliferation of ESCs [41]. LncRNAs have been proved to play an important role in the osteogenic differentiation of MSC. Periodontal ligament stem cells (PDLSCs) are MSCs derived from oral tissues and possess multi-dimensional differentiation potential and good self-renewal ability. Osteogenic differentiation is one of the most important characteristics of oral stem cell pluripotency, which plays a significant role in periodontal tissue regeneration [42]. Lin28 is a key factor regulating development time. Lin28 has two conserved paralogs, lin28 homolog A (lin28A) and lin28 homolog B (lin28B) [43]. Previous studies have found that lin28A is involved in cells development, metabolism and stem cell maintenance [44, 45]. LncRNA taurine up-regulated gene 1 (TUG1) is mainly distributed in the nucleus of PDLSCs, and the dynamic expression of TUG1 is increased in osteogenic PDLSCs. As a regulator of PDLSCs osteogenesis, TUG1 interacts with lin28A to activate lin28A and promote the expression of multiple osteogenic genes, thereby promoting the osteogenic differentiation of PDLSCs [46]. In this review, we elaborate the specific regulatory mechanism of lncRNAs and their RBPs in regulating the fate of stem cells, which is of great significance to explore the therapeutic potential of stem cells in medicine. The functions of lncRNAs and their RBPs mainly include the following five aspects. Next, we will focus on these five regulatory mechanisms and illustrate the functions of lncRNAs and their RBPs in stem cells (Fig. 2).
Fig. 2.
Functional crosstalk between lncRNAs and their RBPs. LncRNA binds to RBP to control the self-renewal and differentiation of stem cells through five regulatory mechanisms, including epigenetic regulation, signal pathways, splicing of mRNA, mRNA stability and subcellular localization
LncRNAs and their RBPs affect epigenetic regulation in stem cells
Epigenetic regulatory factors are one of the key regulatory factors for cells differentiation. As an emerging epigenetic factor, ncRNAs have an important impact on determining the fate of stem cells. LncRNA-binding proteins can directly regulate or enhance the transcription of gene expression to maintain the state of stem cells. The function of lncRNAs will change with its cell location. Nuclear lncRNAs can recruit chromatin modifying factors and other proteins or transcription factors. The nuclear localization of lncRNAs provides evidence for epigenetic and transcriptional regulation.
Polycomb inhibitory complex 2 (PRC2) is a protein complex that has epigenetic regulation function and can maintain histone modifications [47]. The mammalian PRC2 core complex includes enhancer of zeste homolog 1/2 (EZH1 nd/or EZH2), embryonic ectoderm development (EED) and suppressor of zeste 12 (SUZ12). EZH1/2 is the catalytic subunit of PRC2 and each subunit contains a SET domain. Through this domain, the methyl portion of the cofactor s-adenosine-l-methionine (SAM) is transferred to lys27 of histone H3 (H3K27), but EZH2 itself lacks histone methyltransferase (HMTase) activity and requires the assistance of the other two core subunits of PRC2 [48]. H3K27me3 is one of the dominant parts of epigenetic regulation [49]. In human mesenchymal stem cells (hMSCs), inhibition of EZH2 can reduce adipogenesis and induce bone formation [50]. LncRNA maternally expressed 3 (MEG3) can interact with EZH2, and the down-regulation of MEG3 or EZH2 promotes the osteogenic differentiation of human dental follicle stem cells (hDFSCs) by reducing the occupation of H3K27me3 on the Wnt gene promoter [51]. Targeted degradation of EZH2 may have the potential to induce bone formation of bone marrow mesenchymal stem cells, which provides important evidence for the treatment of MSC-related diseases such as bone aging and osteoporosis. Oct4 is expressed in germ cells, the inner cell mass of preimplantation embryos, and embryonic stem cells. Among mouse ESCs (mESCs), Oct4 is essential for establishing a core transcription network that maintains pluripotency and self-renewal. The researchers found that lncRNA Oct4 pseudogene 4 (Oct4P4), which is derived from the mouse Oct4 pseudogene, is an epigenetic regulator of gene expression and has a bearing on the self-renewal of mESC. During mESC differentiation, Oct4P4 is up-regulated and combined with SUV39H1 HMTase to form a complex, so that the complex recruits H3K9me3 and HP1α to the Oct4 promoter region, resulting in Oct4 silencing and prevention of mESC self-renewal [52]. LncRNA ES1, ES2 and ES3, as nuclear lncRNAs, can interact with SUZ12 and Sox2 that are components of the PRC2 complex, to maintain the pluripotency of hESCs [53]. Large-intergenic noncoding RNAs (lincRNAs) 1614 can bind to multiple pluripotency factors, which is essential for maintaining the pluripotency of ESCs. Linc1614 interacts with Sox2 and recruits the PRC2 complex to T, Eomes, and Pitx2 and other developmental gene regions and inhibits the expression of these developmental genes through catalytic inhibition of H3K27me3 modification to maintain the pluripotency of mESCs [54]. p53 is a tumor suppressor that actively participates in the differentiation of hESCs and mESCs. Jain et al. discovered that a p53-regulated, pluripotency-specific lncRNA, p53-regulated and ESC-associated 1 (PRESS1) can interact with sirtuin6 (SIRT6) and inhibit SIRT6 from attaching to chromatin, thus maintaining the acetylation level of Histone H3K56 and H3K9 on promoters of pluripotent genes such as Oct4 and Nanog to protect hESC pluripotency [55]. A new research identified a new type of lncRNA that is expressed at a low level in bladder cancer stem cells (BCSCs). LncRNA LBCS binds to heterogeneous ribonucleoprotein K (hnRNPK) and EZH2 and acts as a scaffold to induce the formation of the hnRNPK-EZH2 complex. Next, LBCS guides the complex to the Sox2 promoter and H3K27me3 to inhibit Sox2 expression and the self-renewal of BCSCs [56]. The discovery of this regulatory mechanism not only indicates that lncRNAs have a significant epigenetic regulation role in the process of tumorigenesis, but also provides potential prognostic indicators and theoretical basis for the treatment of bladder cancer.
LncRNAs and their RBPs are involved in the regulation of signaling pathways in stem cells
Signaling pathways, including fibroblast growth factor (FGF)/extracellular signal-regulated kinase (ERK) signaling pathway, notch, bone morphogenetic protein (BMP), Wnt and other signaling pathways, are key regulatory mechanisms controlling the functions of various stem cells. These signal pathways can interact with each other to form a synergistic or antagonistic signal network, but most of them are independent of each other. Stem cells generally maintain their pluripotency and self-renewal by activating molecular pathways required for self-renewal and inhibiting genes required for differentiation.
LincU, which is localized in the cytoplasm, is highly expressed in a Nanog-dependent manner in naive stem cells. LincU can stabilize Dusp9 and inhibit MAPK/ERK signal through degradation mediated by ubiquitinated proteasome, enhancing the pluripotency of mESCs [57]. Thoc5 is an essential factor for trim71 interacting long noncoding RNA 1 (Trincrl) nuclear export and an essential component of ESCs to inhibit ERK signaling. Thoc5 promotes the export of Trincr1 to the cytoplasm. In the cytoplasm, lncRNA Trincr1 binds to tripartite motif 71 (Trim71), inhibits the stability of SHC binding and spindle associated 1 (SHCBP1), leads to down-regulation of SHCBP1 protein, phosphorylates ERK and regulates the expression of ERK pathway target genes, and then makes mESCs that depend on Trim71 can proliferate rapidly [58]. The normal differentiation of mESCs is coordinated by transcription factors, epigenetic regulatory factors, chromatin regulatory factors and signaling pathways. According to reports, the leukemia inhibitory factor (LIF)/STAT3 signaling pathway is essential for maintaining the self-renewal and pluripotency of mESCs [59]. Linc1557 is mainly located in the cytoplasm of mESCs and directly interacts with STAT3 through specific binding sites to regulate the stability of its mRNA, regulating the LIF/STAT3 signaling pathway and promoting the self-renewal of mESCs [60]. Peptidylprolyl Isomerase E (PPIE) is an RNA-binding protein located in the nucleus of hESCs [61]. PPIE can regulate lncRNA hFAST processing and export and help it get into cytoplasm. In the cytoplasm of hESCs, the hFAST binds to the WD40 domain of beta-transducin repeat containing E3 ubiquitin protein (β-TrCP) and blocks β-TrCP interaction with phosphorylated β-catenin to prevent its degradation. Thus, Wnt signaling is activated and maintains the pluripotency of hESCs [62]. LncR492 is a lineage-specific inhibitor of neuroectodermal differentiation. Human embryonic lethal abnormal vision-like protein HuR belongs to the Hu family of RNA-binding proteins. Under normal cellular and physiological conditions, HuR is mainly located in the nucleus. When pressure is applied to it, HuR can migrate to the cytoplasm and stabilize and increase the translation of target mRNA [63]. Researchers have discovered that lncR492 interacts with the mRNA-binding protein HuR and inhibits the neuroectodermal differentiation of mESCs by activating Wnt signaling [64]. Not only stem cells have the characteristics of self-renewal and rare differentiation, but cancer stem cells also have the characteristics of stemness. The Switch/Sucrose Nonfermentable (SWI/SNF) complex is a chromatin remodeling complex composed of SNF5, BRG1 and BAF170. LncRNA T cell factor 7 (TCF7) binds to BAF170 and recruits the SWI/SNF complex to the TCF7 promoter to regulate its expression, leading to the activation of Wnt signaling and promoting the self-renewal of liver cancer stem cells (liver CSCs) [65]. The Hippo-Yes-Associated Protein (YAP) signaling pathway mediates the differentiation and self-renewal capabilities of a variety of adult stem cells and affects the adipose osteogenic differentiation of hMSCs [66]. Human dental pulp stem cells (hDPSCs) are considered as MSCs due to their strong ability to proliferate and differentiate into different lineages [67]. LncRNA H19 recruits EZH2 to the large tumor suppressor 1 (LATS1) promoter region to induce H3K27me3, inhibiting the expression of LATS1, blocking the activation of the Hippo-YAP signaling pathway and enhancing the dentin differentiation and proliferation of hDPSCs [68]. BMP signaling pathway is involved in the self-renewal and differentiation process of ESCs [69, 70]. LncRNA heart and neural crest derivatives expressed 2-antisense RNA 1 (HAND2-AS1) combines with INO80 complex to promote the expression of bone morphogenetic protein receptor 1A (BMPR1A) and activate the BMP signal, thus promoting the self-renewal of liver CSCs. HAND2-AS1 promotes the self-renewal of liver CSCs and the occurrence of liver tumors, providing a potential new target for the treatment of hepatocellular carcinoma (HCC) [71].
LncRNAs and their RBPs regulate the splicing of mRNA in stem cells
Splicing is an mRNA processing event. The splicing of mRNA can be used as a gateway to control the self-renewal and differentiation of stem cells. The coordinated control of mRNA and rRNA processing controls the pluripotency and differentiation of embryonic stem cells [72]. LncRNAs and their RBPs participate in the regulation of stem cell self-renewal and pluripotency maintenance through the splicing of mRNA. Protein kinase C δ (PKCδ) is a factor that can activate the survival and proliferation of neurons. Splicing of PKCδII can increase the survival rate of neurons. The exosomes secreted by human adipose stem cells (hASCs) contain lncRNA MALAT1, and lncRNA MALAT regulates the splicing of genes mainly by regulating the activities of splicing factors. MALAT1 promotes splicing or controls the assembly of complexes that mediate gene expression to regulate the expression of downstream genes. It can be absorbed by hippocampal HT22 cells and promotes the splicing of PKCδII by binding the splicing factor serine-rich arginine splicing factor 2 (SRSF2), which leads to the proliferation of neurons in the brain injury site of hASCs. This holds great promise for the treatment of traumatic brain injury and other neurodegenerative diseases [73]. MEG3 is a kind of lncRNA that can regulate the osteogenic differentiation of MSCs through a variety of ways. Up-regulated MEG3 interacts with hnRNPI to inhibit the expression of BMP2. HnRNPI plays an active role in mRNA splicing and significantly inhibits osteogenic differentiation of human periodontal ligament cells (hPDLCs) through this regulatory mechanism [74]. This discovery also provides a new method for the treatment of periodontitis.
Splicing can occur either cis or trans, where cis splicing connects exons of a single pre-mRNA, while trans-splicing connects exons of two or more independent pre-mRNAs from the same gene (intra-trans-splicing) or two or more different genes (intergene trans-splicing). It usually occurs in single-celled organisms, nematodes, and trypanosomes [75]. Trans-splicing connects different pre-mRNA exons for splicing to generate different transcripts from a limited number of genes. This is a post-transcriptional event. Studies have found that trans-splicing lncRNAs influence on the pluripotency maintenance of stem cells [76, 77]. Trans-spliced noncoding RNA RMST (tsRMST) is a typical example of trans-splicing that occurs in normal human cells and is a trans-splicing subtype of RMST. RMST is a lncRNA gene located on the qarm of chromosome 12, with about 2.6 kb in length, which plays an important role in the process of neural differentiation [54]. The expression of RMST is critical for the binding of Sox2 to neurogenic genes, and tsRMST is an emerging regulatory lncRNA associated with human stem cell pluripotency [54, 78]. Yu et al. demonstrated that tsRMST is highly expressed in hESCs and down-regulated during in vitro differentiation. It interacts with Nanog and SUZ12 to form a complex that inhibits the expression of Wnt5A along with differentiation-related transcription factors and inhibits non-standard Wnt pathways, preventing differentiation of hESCs [79]. tsRMST inhibits gene expression through association with pleiotropic-related transcription factors, PRC2 components and subsequent trimethylation of H3K27 in the target gene promoter region [80]. Furthermore, microarray and Ingenious Pathway Analysis (IPA) have demonstrated the inhibitory effect of tsRMST on noncanonical Wnt signaling pathway. Previously, the regulatory role of lncRNA combined with RBP in some signaling pathways, especially typical Wnt signaling pathways, has been studied a lot, but the regulatory role of lncRNAs in noncanonical Wnt signaling pathway is poorly understood. Wnt proteins are classified into typical proteins and non-standard proteins. Wnt5A and Wnt5B belong to non-standard proteins [81]. A recent study found that the non-standard Wnt5A/Fzd2 pathway plays an important role in prostate cancer [82]. The discovery of the role of tsRMST in non-standard Wnt signaling pathway provides further insights into the role of lncRNA in regulating signaling pathways and provides important insights into the role of trans-splicing lncRNA in the pluripotency maintenance and lineage differentiation of hESCs.
LncRNAs and their RBPs are involved in the regulation of the stability of mRNA in stem cells
Many studies have emphasized that gene expression is precisely regulated by mRNA stability. mRNA stability largely affects the secondary and tertiary structures of the mRNAs, and the accessibility of various RBPs to the mRNAs. The stability of mRNA is regulated by diverse RNA modifications. To date, hundreds of different RNA modifications have been characterized [83]. However, the mRNA stability is also affected by lncRNAs and their RBPs in stem cells.
Heterogeneous nuclear ribonucleoproteins (hnRNPs) belong to a well-known splicing protein family, which has been widely studied in the field of stem cells, and great achievements have been made in recent years. As typical RNA-binding proteins, hnRNPs effect on mRNA stability and gene transcription [84]. HnRNPs bind to lncRNA and form a complex or directly bind to mRNA in stem cells, affecting the stability of mRNA. As a member of the hnRNP family, polypyrimidine tract binding protein 1 (PTBP1 or hnRNPI) is a multifunctional protein associated with neurogenesis and participates in all steps of RNA biogenesis. Its regulation of mRNA stability and pre-mRNA splicing has been demonstrated in previous studies [85]. Studies on the relationship between energy metabolism and the fate of stem cells have identified Lncenc1 as the first lncRNA that connects self-renewal and energy metabolism in PSCs. HnRNPK binds to the promoter of glycolysis genes and directly regulates the transcription of these genes. Lncenc1, hnRNPK and PTBP1 form a complex that regulates the transcription of glycolysis-related genes to maintain glycolytic activity and promote the self-renewal of naive embryonic stem cells (nESCs) [86, 87]. In addition, PTBP1 can perform different functions by shuttling between the nucleus and cytoplasm [88]. In the nucleus, PTBP1 forms a ribonucleoprotein complex and regulates splicing, polyadenylation and mRNA export. In the cytoplasm, PTBP1 is involved in translation initiation and mRNA stability [89]. Knockdown of PTBP1 can cause premature differentiation and impaired motor behavior of NSC in mouse brain [90]. Pnky is a conserved, neuro-specific nuclear lncRNA that plays a unique role in controlling neurogenesis. Knocking down Pnky can lead to the production of neurons. Pnky and PTBP1 specifically bind to each other, which maintains the stability of their mRNA, inhibits the differentiation of NSCs and affects neuronal development [91, 92]. A new study showed that the lncRNA anti-differentiation noncoding RNA (ANCR) located in cytoplasmic binds to PTBP1, promotes the interaction between PTBP1 and inhibitor of DNA binding 2 (ID2) mRNA, and enhances the stability of ID2 mRNA, resulting in inhibition the differentiation of human adipose-derived mesenchymal stem cells (hAMSCs) into definitive endoderm cells (DECs) [93]. When breast cancer cells are under hypoxic conditions, lncRNA KB-1980E6.3 is necessary to maintain stemness. LncRNA KB-1980E6.3 binds to insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) to form a complex under the induction of HIF-1α to recognize m6A modified c-myc coding region instability determinant (CRD) mRNA and enhance the stability of c-myc mRNA, which significantly promotes the stemness and proliferation of breast cancer stem cells (BCSCs) in hypoxic tumor microenvironment [94].
LncRNAs and their RBPs are involved in the regulation of the subcellular localization in stem cells
LncRNAs, as a kind of RNA that does not encode protein, have been shown to be distributed in both the nucleus and the cytoplasm. In the process of summarizing the functions of lncRNAs and their RBPs, we found that lncRNAs often combine with their RBPs to achieve their functions, affecting the modification level or localization of RBPs and thus affecting downstream genes. LncRNA Panct1 is a polyadenylation transcription unit located in the nucleus and involved in the maintenance of pluripotency of mESCs. Panct1 can interact with transient octamer binding factor 1 (TOBF1) and regulate its subcellular location. Overexpression of Panct1 leads to higher expression of pluripotency genes such as Oct4, Nanog, Zscan4c, Sox2, and Klf4 and leads to the pluripotency maintenance of mESCs [95]. However, the subcellular localization of lncRNAs can also regulate the fate of stem cells. Subcellular localization of lncRNA FAST is different in hESCs and mESCs. hFAST locates in the cytoplasm, while mFAST locates in nucleus. Guo, et al. certified that cytoplasmic hFAST but not nuclear mFAST promotes Wnt signaling in pluripotency maintenance of hESCs. In the cytoplasm of hESCs, the hFAST binds to WD40 domain of β- TrCP, impedes the interaction between β- TrCP and phosphorylated β-catenin and promotes Wnt activity to maintaining pluripotency of hESCs [63].
To date, the regulatory mechanisms underlying the regulation of protein localization and transformation have been rarely studied in embryonic stem cells and adult stem cells, but widely studied in tumor cells. OLA1P2 is an up-regulated lncRNA induced by aspirin. Aspirin can induce the up-regulation of forkhead box D3 (FOXD3) gene expression in tumor cells. FOXD3 protein transcriptionally up-regulates the expression of OLA1P2. Highly expressed OLA1P2 directly binds to phosphorylated STAT3 (Tyr705) protein, which inhibits its nuclear import to a large extent, blocks the formation of phosphorylated STAT3 homodimers, and significantly affects STAT3 signaling, which inhibits the proliferation and migration of cancer cells. The discovery of this regulatory mechanism provides a potential therapeutic target and new insights for cancer chemotherapy [96]. Furthermore, previous studies have shown that FOXD3 is essential for the self-renewal, survival and pluripotency maintenance of human embryonic stem cells. Studying the regulatory mechanism of interaction between OLA1P2 and phosphorylated STAT3 protein may have great value in stem cells [97]. Linc00460 is located in the cytoplasm, hnRNPK is distributed in both cytoplasm and nucleus. Linc00460 binds with and translocates hnRNPK that is located in nucleus to the cytoplasm to participate in special mRNA stability and translation regulation, and promotes cell migration and invasion through inducing EMT in lung cancer [98]. The regulation of lncRNA on the tumor suppressor p53 pathway has been a topic of great interest. Ras-GTase-activating protein SH3 domain binding protein 1 (G3BP1) is a specific binding protein of the SH3 domain of Ras GTPase activating protein (Ras-GAP), a negative feedback regulator of Ras activity. It is distributed in the cytoplasm and has an RRM domain, which can directly interact with RNA, so G3BP1 belongs to the RBP family [99]. Inactivation of p53 is a key event in tumor Linc00460 that is located in the cytoplasm, hnRNPK is distributed in both cytoplasm and nucleus. Linc00460 binds with and translocates hnRNPK that is located in nucleus to the cytoplasm to participate in special mRNA stability and translation regulation, and promotes cell migration and invasion through inducing EMT in lung cancer formation. Mutations, transcription inactivation, abnormal degradation and changes in subcellular localization of p53 may lead to the failure of p53 to perform normal functions. It has been reported that G3BP1 can bind to the RRM domain of p53 and transfer p53 from the nucleus to the cytoplasm. Thus, p53 cannot play its function in the nucleus and promotes the occurrence and development of malignant tumors [100]. However, Mao et al. demonstrated that p53 can return from the cytoplasm to the nucleus and act as a transcription factor to inhibit the proliferation of lung adenocarcinoma cells. The main reason is that lncRNA P53RRA competitively inhibits the binding of G3BP1 and p53 in the cytoplasm through specific binding with G3BP1 [101]. The discovery of this regulatory mechanism will provide a great reference value for subsequent research on self-renewal and pluripotency maintenance of stem cells.
Conclusions and perspectives
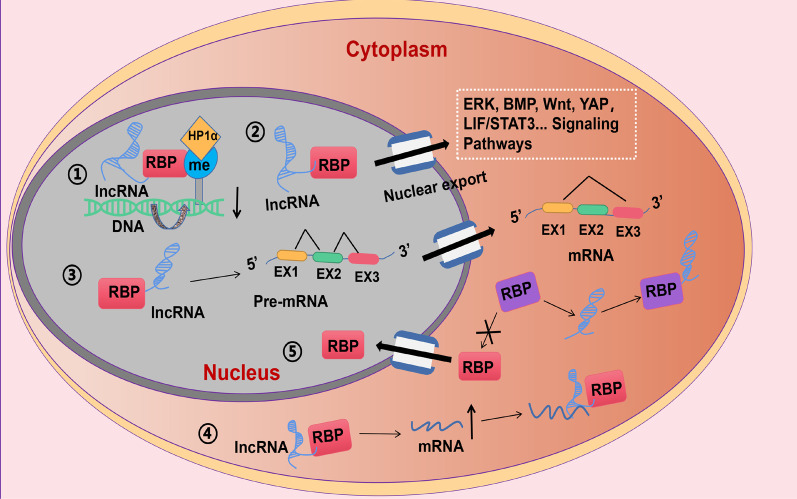
There are a large number of lncRNAs in the mammalian genome, which constitute an important part of the genome. RBPs and lncRNAs can affect the localization, stability and translation of target mRNAs. The combination of versatility and structural flexibility between the two enables RBPs and lncRNAs to form a strong regulatory network that is closely related to the function of stem cells [102]. In this review, we focus on the functions by which lncRNAs and their RBPs maintain the self-renewal and pluripotency of stem cells. The main regulatory mechanisms include (1) LncRNAs and their RBPs affect epigenetic regulation in stem cells. (2) LncRNAs and their RBPs are involved in the regulation of signaling pathways in stem cells. (3) LncRNAs and their RBPs regulate the splicing of mRNA in stem cells. (4) LncRNAs and their RBPs are involved in the regulation of the stability of mRNA in stem cells. (5) LncRNAs and their RBPs are involved in the regulation of the subcellular localization in stem cells (Fig. 3, Table 1).
Fig. 3.
Functions of lncRNAs and their RBPs in self-renewal and differentiation of stem cells. (1) LncRNAs and their RBPs affect epigenetic regulation in stem cells. For example, up-regulated Oct4P4 combines with SUV39H1 HMTase to form a complex, which recruits H3K9me3 and HP1α to the Oct4 promoter region, resulting in Oct4 silencing and preventing mESC self-renewal. (2) LncRNAs and their RBPs are involved in the regulation of signaling pathways in stem cells. For example, lncR492 interacts with the mRNA-binding protein HuR and inhibits the neuroectodermal differentiation of mESCs by activating Wnt signaling. (3) LncRNAs and their RBPs regulate the splicing of mRNA in stem cells. For example, MALAT1 can be absorbed by hippocampal HT22 cells and promotes the splicing of PKCδII by binding the splicing factor SRSF2, which leads to the proliferation of neurons in the brain injury site of hASCs. (4) LncRNAs and their RBPs are involved in the regulation of the stability of mRNA in stem cells. For example, lncRNA ANCR binds to PTBP1, promotes the interaction between PTBP1 and inhibitor of ID2 mRNA and enhances the stability of ID2 mRNA, resulting in inhibition of the differentiation of hAMSCs into DE cells. (5) LncRNAs and their RBPs are involved in the regulation of the subcellular localization in stem cells. For example, lncRNA P53RRA inhibits the binding of G3BP1 and p53 in cytoplasm through specifically binding with G3BP1, so p53 is transferred from cytoplasm to nucleus to function as a transcription factor
Table 1.
Functions of LncRNAs and their RBPs in self-renewal and differentiation of stem cells
| LncRNA | RBPs | Cell type | Biological function | Regulatory mechanism | References |
|---|---|---|---|---|---|
| MEG3 | EZH2 | hDFSC | Promote the osteogenic differentiation of hDFSC | MEG3 interacts with EZH2, down-regulation of MEG3 or EZH2 reduces the occupation of H3K27me3 on the Wnt gene promoter | [51] |
| Oct4P4 | SUV39H1 HMTase | mESC | Inhibit the self-renewal of mESC | OctP4 combines with SUV39, H1 and HMTase to form a complex, recruits H3K9me3 and HP1α to the Oct4 promoter region and results in Oct4 silencing | [52] |
| ES1, ES2, ES3 | SUZ12 and Sox2 | hESC | Maintain the pluripotency of hESC | ES1, ES2 and ES3 interact with SUZ12 and Sox2, which are the components of PRC2 complex | [53] |
| Linc1614 | PRC2 complex and Sox2 | mESC | Maintain the pluripotency of mESC | Linc1614 interacts with Sox2, recruits the PRC2 complex to T, Eomes, and Pitx2 and other developmental gene regions and inhibits their expression | [54] |
| LncPRESS1 | SIRT6 | mESC | Maintain the pluripotency of mESC | LncPRESS1 interacts with SIRT6 and inhibits SIRT6 from attaching to chromatin, maintaining the acetylation level of Histone H3K56 and H3K9 on the promoters of pluripotent genes such as Oct4 and Nanog | [55] |
| LBCS | hnRNPK and EZH2 | BCSC | Inhibit the self-renewal of BCSC | LBCS binds hnRNPK and EZH2 to form the hnRNPK-EZH2 complex, guides the complex to the Sox2 promoter and H3K27me3 to inhibit Sox2 expression | [56] |
| LincU | Dusp9 | mESC | Maintain the pluripotency of mESC | LincU binds and stabilizes ERK-specific phosphatase DUSP to restrict MAPK/ERK activity | [57] |
| Trincr1 | Trim71 | mESC | Promote the proliferation of mESC | Thoc5 regulates the export of Trincr1 to the cytoplasm, lncRNA Trincr1 binds to Trim71 in the cytoplasm, inhibits the activity of SHCBP1 and phosphorylates ERK and promotes the expression of ERK pathway target genes | [58] |
| Linc1557 | STAT3 | mESC | Promote the self-renewal of mESC | Linc1557 interacts with STAT3 through specific binding sites to regulate the stability of its mRNA, thus regulating the LIF/STAT3 signaling pathway | [60] |
| hFAST | β- TrCP | hESC | Maintain the pluripotency of hESC | hFAST binds to WD40 domain of β- TrCP, impedes the interaction between β- TrCP and phosphorylated β-catenin and promotes Wnt activity | [62] |
| LncR492 | HuR | mESC | Inhibit the neuroectodermal differentiation of mESC | LncR492 interacts with HuR and activates Wnt signaling | [64] |
| TCF7 | BAF170 and SWI/SNF complex | liver CSC | Promote the self-renewal of liver CSC | TCF binds to BAF170 and recruits the SWI/SNF complex to the TCF7 promoter to regulate its expression, leading to the activation of Wnt signaling | [65] |
| H19 | EZH2 | hDPSC | Promote the dentin differentiation and proliferation of hDPSC | H19 recruits EZH2 to the LATS1 promoter region to induce H3K27me3, inhibiting the expression of LATS1, blocking the activation of the Hippo-YAP signaling pathway | [68] |
| HAND2-AS1 | INO80 complex | live CSC | Promote the self-renewal of liver CSC | HAND2-AS1 combines with INO80 complex to promote the expression of BMPR1A and activate the BMP signaling | [71] |
| MALAT1 | SRSF2 | hASC | promote the Proliferation of neurons in the brain injury site of hASC | MALAT1 promotes the splicing of PKCδII by binding with SRSF2 | [73] |
| tsRMST | Nanog and SUZ12 | hESC | Inhibit the differentiation of hESC | tsRMST interacts with Nanog and SUZ12 to form a complex and inhibits the expression of Wnt5A through differentiation-related transcription factors, and inhibits non-standard Wnt pathway | [79] |
| Pnky | PTBP1 | NSC | Inhibit the differentiation of NSC | Pnky interacts with PTBP1 to maintain the stability of its mRNA | [91, 92] |
| ANCR | PTBP1 | hAMSC | Inhibit the differentiation of hAMSC into DE cell | ANCR binds to PTBP1, promotes the interaction between PTBP1 and ID2 mRNA and enhances the stability of ID2 mRNA | [93] |
| KB-1980E6.3 | IGF2BP1 | BCSC | Promote the stemness and proliferation of BCSC | KB-1980E6.3 binds to IGF2BP1 to form a complex under the induction of HIF-1α to recognize and enhance the stability of c-myc mRNA | [94] |
Although some progress has been made in the research field of lncRNAs, and RBPs and in-depth understanding has been obtained in the basic research on lncRNAs and their RBPs in the self-renewal and differentiation of stem cells, their regulatory mechanisms are complex and there are still many molecular mechanisms that have not been clearly elucidated, and the roles played by lncRNAs and their RBPs in stem cells deserve further investigation. To date, many achievements and breakthroughs have been obtained about the other ncRNAs and their RBPs involved in stem cell self-renewal and differentiation. For example, miR-342-5p inhibits the differentiation of NSCs/intermediate neural progenitor cells (INPCs) into astrocytes, likely mediated by directly targeting site 1 and site 2 in the 3’UTR of GFAP mRNA and inhibiting GFAP expression [103]. Up-regulation of miR-145 is essential for normal neuronal differentiation. During NSCs differentiation, miR-145 interacts with Sox2 and lin28 mRNAs to down-regulate their expression and increase let-7 levels, thereby facilitating the differentiation NSCs into neurons [104]. CircRNAs play crucial roles in the initiation and development of diseases and gain significant attention. Several circRNAs are enriched in undifferentiated hESCs, such as circBIRC6, circCORO1C and circFOXP1, which are functionally associated with stemness. They act as “sponges” to recruit RBPs to monitor the stem cell self-renewal and differentiation. Knockdown of circular RNA H19 induces adipogenic differentiation of human adipose-derived stem cells (hADSCs) via interacting with PTBP1 [105]. Basic research is an important cornerstone to promote the development of clinical medicine and scientific civilization. The above research results bring new thinking and direction for future research on the biological functions of stem cells. However, the research of other ncRNAs such as tRNA and snoRNA., and their RBPs in stem cells needs to be further explored. Therefore, studying the effects of different RNAs combined with their RBPs on stem cell functions and in-depth investigation on the regulatory mechanisms can deepen our understanding of the self-renewal and differentiation of stem cells, and of the occurrence and development of relevant diseases, provide new ideas and strategies for disease prevention, diagnosis and treatment, and offer references for applications of pluripotent stem cells in clinical biomedical research.
Acknowledgements
Not applicable.
Abbreviations
- LncRNAs
Long noncoding RNAs
- RBP
RNA-binding proteins
- ncRNAs
Noncoding RNAs
- ESCs
Embryonic stem cells
- ASCs
Adult stem cells
- iPSCs
Induced pluripotent stem cells
- NSCs
Neural stem cells
- MSCs
Mesenchymal stem cells
- HSCs
Hematopoietic stem cells
- RSCs
Reproductive stem cells
- TSCs
Totipotent stem cells
- PSCs
Pluripotent stem cells
- MSCs
Multipotent stem cells
- USCs
Unipotent stem cells
- ncRNAs
Noncoding RNAs
- hESCs
Human ESCs
- miRNAs
MicroRNAs
- siRNAs
Short interfering RNAs
- piRNAs
Piwi-interacting RNAs
- snoRNAs
Small nucleolar RNAs
- shRNAs
Short hairpin RNAs
- circRNAs
Circular RNAs
- Gas5
Growth arrest specific 5
- ceRNAs
Competitive endogenous RNAs
- SATB2
Special AT-rich sequence-binding protein 2
- BMSCs
Bone marrow mesenchymal stem cells
- MALAT1
Metastasis-associated lung adenocarcinoma transcript 1
- RNP
Ribonucleoprotein
- RBDs
RNA-binding domains
- STAT3
Signal transducer and activator of transcription 3
- PDLSCs
Periodontal ligament stem cells
- Lin28A
Lin28 homolog A
- Lin28B
Lin28 homolog B
- TUG1
Taurine up-regulated gene 1
- PRC2
Polycomb inhibitory complex 2
- EZH1
Enhancer of zeste homolog 1
- EZH2
Enhancer of zeste homolog 2
- EED
Embryonic ectoderm development
- SUZ12
Suppressor of zeste 12
- SAM
S-adenosine-l-methionine
- H3K27
Lys27 of histone H3
- HMTase
Histone methyltransferase
- hMSCs
Human mesenchymal stem cells
- MEG3
Maternally expressed 3
- hDFSCs
Human dental follicle stem cells
- mESCs
Mouse ESCs
- Oct4P4
Oct4 pseudogene 4
- LincRNAs
Large-intergenic noncoding RNAs
- PRESS1
P53-regulated and ESC-associated 1
- BCSCs
Bladder cancer stem cells
- hnRNPK
Heterogeneous ribonucleoprotein K
- FGF
Fibroblast growth factor
- ERK
Extracellular signal-regulated kinase
- BMP
Bone morphogenetic protein
- Trincrl
Trim71 interacting long noncoding RNA 1
- Trim71
Trincr1 binds to tripartite motif 71
- SHCBP1
SHC binding and spindle associated 1
- LIF
Leukemia inhibitory factor
- PPIE
Peptidylprolyl isomerase E
- β-TrCP
Beta-transducin repeat containing E3 ubiquitin protein
- SWI/SNF
Switch/sucrose nonfermentable
- TCF7
T cell factor 7
- Liver CSCs
Liver cancer stem cells
- YAP
Yes-associated protein
- hDPSCs
Human dental pulp stem cells
- LATS1
Large tumor suppressor 1
- HAND2-AS1
Heart and neural crest derivatives expressed 2-antisense RNA 1
- BMPR1A
Bone morphogenetic protein receptor 1A
- HCC
Hepatocellular carcinoma
- PKCδ
Protein kinase C δ
- hASCs
Human adipose stem cells
- SRSF2
Splicing factor serine-rich arginine splicing factor 2
- IGF2BP1
Insulin-like growth factor 2 mRNA-binding protein 1
- CRD
Coding region instability determinant
- BCSCs
Breast cancer stem cells
- tsRMST
Trans-spliced noncoding RNA RMST
- IPA
Ingenious Pathway Analysis
- hnRNPs
Heterogeneous nuclear ribonucleoproteins
- PTBP1
Polypyrimidine tract binding protein 1
- ANCR
Anti-differentiation noncoding RNA
- ID2
Inhibitor of DNA binding 2
- hAMSCs
Human adipose-derived mesenchymal stem cells
- DECs
Definitive endoderm cells
- nESCs
Naive embryonic stem cells
- hPDLCs
Human periodontal ligament cells
- TOBF1
Transient octamer binding factor 1
- FOXD3
Forkhead box D3
- G3BP1
Ras-GTase-activating protein SH3 domain binding protein 1
- Ras-GAP
Ras GTPase activating protein
- INPCs
Intermediate neural progenitor cells
- hADSCs
Human adipose-derived stem cells
Author contributions
CPR, QX and XJJ conceived and designed the study. CZ and WX collected the data and wrote the manuscript. HCZ, MZ, WDL, ZPW, LW, BZ, SSL and YZ revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (82071399, 81773179, 81472355, 81070993, 30871246), Provincial Natural Science Foundation of Hunan (2020JJ4771), National Key Research and Development Program of China (2016YFC1101502), Hunan Provincial Science and Technology Department (2014FJ6006).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Cong Zhao and Wen Xie contributed equally to this work
Contributor Information
Xingjun Jiang, Email: jiangxj@csu.edu.cn.
Qiang Xu, Email: mr.xuqiang@csu.edu.cn.
Caiping Ren, Email: rencaiping@csu.edu.cn.
References
- 1.Shahbazi MN, Siggia ED, Zernicka-Goetz M. Self-organization of stem cells into embryos: a window on early mammalian development. Science. 2019;364(6444):948–951. doi: 10.1126/science.aax0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuazon JP, Castelli V, Lee JY, Desideri GB, Stuppia L, Cimini AM, et al. Neural stem cells. Adv Exp Med Biol. 2019;1201:79–91. doi: 10.1007/978-3-030-31206-0_4. [DOI] [PubMed] [Google Scholar]
- 3.Sharlow ERGE. Introduction to the iPS cells for ischemic stroke, traumatic brain injury, and other brain-related diseases special issue. Assay Drug Dev Technol. 2020;18(2):77. [DOI] [PubMed]
- 4.Zakrzewski W, Dobrzyński M, Szymonowicz M, Rybak Z. Stem cells: past, present, and future. Stem Cell Res Ther. 2019;10(1):68. doi: 10.1186/s13287-019-1165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113(5):631–642. doi: 10.1016/S0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 6.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17(1):126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95(3):379–391. doi: 10.1016/S0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 9.Chepelev I, Chen X. Alternative splicing switching in stem cell lineages. Front Biol (Beijing) 2013;8(1):50–59. doi: 10.1007/s11515-012-1198-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xi H, Pyle A. Beyond the genome: RNA control of stem cells. Science. 2019;366(6466):684–685. doi: 10.1126/science.aaz4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boutet SC, Cheung TH, Quach NL, Liu L, Prescott SL, Edalati A, et al. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell. 2012;10(3):327–336. doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pereira P, Arraiano CM. A precision RNA degradation machinery shapes stem cell development. J Cell Biol. 2019;218(8):2437–2438. doi: 10.1083/jcb.201906115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belair C, Sim S, Kim KY, Tanaka Y, Park IH, Wolin SL. The RNA exosome nuclease complex regulates human embryonic stem cell differentiation. J Cell Biol. 2019;218(8):2564–2582. doi: 10.1083/jcb.201811148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai X, Zhang S, Zaleta-Rivera K. RNA: interactions drive functionalities. Mol Biol Rep. 2020;47(2):1413–1434. doi: 10.1007/s11033-019-05230-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie ZY, Wang P, Wu YF, Shen HY. Long non-coding RNA: the functional regulator of mesenchymal stem cells. World J Stem Cells. 2019;11(3):167–179. doi: 10.4252/wjsc.v11.i3.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127(3):761–771. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu S, Wang J, He Y, Meng N, Yan GR. Peptides/proteins encoded by non-coding RNA: a novel resource bank for drug targets and biomarkers. Front Pharmacol. 2018;9:1295. doi: 10.3389/fphar.2018.01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S, Seo HH, Lee CY, Lee J, Shin S, Kim SW, et al. Human long noncoding RNA regulation of stem cell potency and differentiation. Stem Cells Int. 2017;2017:6374504. doi: 10.1155/2017/6374504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Wang Y, Wang C, Hu JF, Li W. LncRNA functions as a new emerging epigenetic factor in determining the fate of stem cells. Front Genet. 2020;11:277. doi: 10.3389/fgene.2020.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao J, Duan FF, Wang Y. MicroRNAs and RNA binding protein regulators of microRNAs in the control of pluripotency and reprogramming. Curr Opin Genet Dev. 2017;46:95–103. doi: 10.1016/j.gde.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Heinrich EM, Dimmeler S. MicroRNAs and stem cells: control of pluripotency, reprogramming, and lineage commitment. Circ Res. 2012;110(7):1014–1022. doi: 10.1161/CIRCRESAHA.111.243394. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Jiang Z, Yu M, Yang G. Roles of circular RNAs in regulating the self-renewal and differentiation of adult stem cells. Differentiation. 2020;113:10–18. doi: 10.1016/j.diff.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Tu J, Tian G, Cheung HH, Wei W, Lee TL. Gas5 is an essential lncRNA regulator for self-renewal and pluripotency of mouse embryonic stem cells and induced pluripotent stem cells. Stem Cell Res Ther. 2018;9(1):71. doi: 10.1186/s13287-018-0813-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paci P, Colombo T, Farina L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst Biol. 2014;8:83. doi: 10.1186/1752-0509-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleaveland B, Shi CY, Stefano J, Bartel DP. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018;174(2):350–362.e317. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith KN, Starmer J, Miller SC, Sethupathy P, Magnuson T. Long noncoding RNA moderates MicroRNA activity to maintain self-renewal in embryonic stem cells. Stem Cell Rep. 2017;9(1):108–121. doi: 10.1016/j.stemcr.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tye CE, Boyd JR, Page NA, Falcone MM, Stein JL, Stein GS, et al. Regulation of osteogenesis by long noncoding RNAs: an epigenetic mechanism contributing to bone formation. Connect Tissue Res. 2018;59(sup1):35–41. doi: 10.1080/03008207.2017.1412432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu L, Xu Y, Qu H, Yu Y, Li W, Zhao Y, et al. Decrease of MiR-31 induced by TNF-α inhibitor activates SATB2/RUNX2 pathway and promotes osteogenic differentiation in ethanol-induced osteonecrosis. J Cell Physiol. 2019;234(4):4314–4326. doi: 10.1002/jcp.27210. [DOI] [PubMed] [Google Scholar]
- 30.Gu X, Li M, Jin Y, Liu D, Wei F. Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet. 2017;18(1):100. doi: 10.1186/s12863-017-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X, Yang J, Lei P, Wen T. LncRNA MALAT1 shuttled by bone marrow-derived mesenchymal stem cells-secreted exosomes alleviates osteoporosis through mediating microRNA-34c/SATB2 axis. Aging (Albany NY) 2019;11(20):8777–8791. doi: 10.18632/aging.102264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25(1):69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19(5):327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 34.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8(6):479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwakiri J, Hamada M, Asai K. Bioinformatics tools for lncRNA research. Biochim Biophys Acta. 2016;1859(1):23–30. doi: 10.1016/j.bbagrm.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 36.Li A, Ge M, Zhang Y, Peng C, Wang M. Predicting long noncoding RNA and protein interactions using heterogeneous network model. Biomed Res Int. 2015;2015:671950. doi: 10.1155/2015/671950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye J, Blelloch R. Regulation of pluripotency by RNA binding proteins. Cell Stem Cell. 2014;15(3):271–280. doi: 10.1016/j.stem.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guallar D, Wang J. RNA-binding proteins in pluripotency, differentiation, and reprogramming. Front Biol (Beijing) 2014;9(5):389–409. doi: 10.1007/s11515-014-1326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergmann JH, Li J, Eckersley-Maslin MA, Rigo F, Freier SM, Spector DL. Regulation of the ESC transcriptome by nuclear long noncoding RNAs. Genome Res. 2015;25(9):1336–1346. doi: 10.1101/gr.189027.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith KN, Starmer J, Magnuson T. Interactome determination of a long noncoding RNA implicated in embryonic stem cell self-renewal. Sci Rep. 2018;8(1):17568. doi: 10.1038/s41598-018-34864-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao T, Cai M, Liu M, Su G, An D, Moon B, et al. lncRNA 5430416N02Rik promotes the proliferation of mouse embryonic stem cells by activating Mid1 expression through 3D chromatin architecture. Stem Cell Rep. 2020;14(3):493–505. doi: 10.1016/j.stemcr.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang N, Hu X, He S, Ding W, Wang F, Zhao Y, et al. LncRNA MSC-AS1 promotes osteogenic differentiation and alleviates osteoporosis through sponging microRNA-140-5p to upregulate BMP2. Biochem Biophys Res Commun. 2019;519(4):790–796. doi: 10.1016/j.bbrc.2019.09.058. [DOI] [PubMed] [Google Scholar]
- 43.Kastenberg ZJ, Odorico JS. Alternative sources of pluripotency: science, ethics, and stem cells. Transplant Rev (Orlando) 2008;22(3):215–222. doi: 10.1016/j.trre.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12(4):395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J, et al. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012;151(4):765–777. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 46.He Q, Yang S, Gu X, Li M, Wang C, Wei F. Long noncoding RNA TUG1 facilitates osteogenic differentiation of periodontal ligament stem cells via interacting with Lin28A. Cell Death Dis. 2018;9(5):455. doi: 10.1038/s41419-018-0484-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ringrose L. Noncoding RNAs in polycomb and trithorax regulation: a quantitative perspective. Annu Rev Genet. 2017;51:385–411. doi: 10.1146/annurev-genet-120116-023402. [DOI] [PubMed] [Google Scholar]
- 48.Yu JR, Lee CH, Oksuz O, Stafford JM, Reinberg D. PRC2 is high maintenance. Genes Dev. 2019;33(15–16):903–935. doi: 10.1101/gad.325050.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ge W, Liu Y, Chen T, Zhang X, Lv L, Jin C, et al. The epigenetic promotion of osteogenic differentiation of human adipose-derived stem cells by the genetic and chemical blockade of histone demethylase LSD1. Biomaterials. 2014;35(23):6015–6025. doi: 10.1016/j.biomaterials.2014.04.055. [DOI] [PubMed] [Google Scholar]
- 50.Hemming S, Cakouros D, Isenmann S, Cooper L, Menicanin D, Zannettino A, et al. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells. 2014;32(3):802–815. doi: 10.1002/stem.1573. [DOI] [PubMed] [Google Scholar]
- 51.Deng L, Hong H, Zhang X, Chen D, Chen Z, Ling J, et al. Down-regulated lncRNA MEG3 promotes osteogenic differentiation of human dental follicle stem cells by epigenetically regulating Wnt pathway. Biochem Biophys Res Commun. 2018;503(3):2061–2067. doi: 10.1016/j.bbrc.2018.07.160. [DOI] [PubMed] [Google Scholar]
- 52.Scarola M, Comisso E, Pascolo R, Chiaradia R, Marion RM, Schneider C, et al. Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA. Nat Commun. 2015;6:7631. doi: 10.1038/ncomms8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. Embo J. 2012;31(3):522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo X, Wang Z, Lu C, Hong W, Wang G, Xu Y, et al. LincRNA-1614 coordinates Sox2/PRC2-mediated repression of developmental genes in pluripotency maintenance. J Mol Cell Biol. 2018;10(2):118–129. doi: 10.1093/jmcb/mjx041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jain AK, Xi Y, McCarthy R, Allton K, Akdemir KC, Patel LR, et al. LncPRESS1 Is a p53-regulated LncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol Cell. 2016;64(5):967–981. doi: 10.1016/j.molcel.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X, Xie R, Gu P, Huang M, Han J, Dong W, et al. Long noncoding RNA LBCS inhibits self-renewal and chemoresistance of bladder cancer stem cells through epigenetic silencing of SOX2. Clin Cancer Res. 2019;25(4):1389–1403. doi: 10.1158/1078-0432.CCR-18-1656. [DOI] [PubMed] [Google Scholar]
- 57.Jiapaer Z, Li G, Ye D, Bai M, Li J, Guo X, et al. LincU preserves naive pluripotency by restricting ERK activity in embryonic stem cells. Stem Cell Rep. 2018;11(2):395–409. doi: 10.1016/j.stemcr.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li YP, Duan FF, Zhao YT, Gu KL, Liao LQ, Su HB, et al. A TRIM71 binding long noncoding RNA Trincr1 represses FGF/ERK signaling in embryonic stem cells. Nat Commun. 2019;10(1):1368. doi: 10.1038/s41467-019-08911-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bourillot PY, Aksoy I, Schreiber V, Wianny F, Schulz H, Hummel O, et al. Novel STAT3 target genes exert distinct roles in the inhibition of mesoderm and endoderm differentiation in cooperation with Nanog. Stem Cells. 2009;27(8):1760–1771. doi: 10.1002/stem.110. [DOI] [PubMed] [Google Scholar]
- 60.Lan Y, Lu C, Yang Y, Liu X, Guo X, Xi J, et al. Linc1557 is critical for the initiation of embryonic stem cell differentiation by directly targeting the LIF/STAT3 signaling pathway. Stem Cells. 2020;38(3):340–351. doi: 10.1002/stem.3130. [DOI] [PubMed] [Google Scholar]
- 61.Bertram K, Agafonov DE, Liu WT, Dybkov O, Will CL, Hartmuth K, et al. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature. 2017;542(7641):318–323. doi: 10.1038/nature21079. [DOI] [PubMed] [Google Scholar]
- 62.Guo CJ, Ma XK, Xing YH, Zheng CC, Xu YF, Shan L, et al. Distinct processing of lncRNAs contributes to non-conserved functions in stem cells. Cell. 2020;181(3):621–636.e622. doi: 10.1016/j.cell.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 63.Matsye P, Zheng L, Si Y, Kim S, Luo W, Crossman DK, et al. HuR promotes the molecular signature and phenotype of activated microglia: Implications for amyotrophic lateral sclerosis and other neurodegenerative diseases. Glia. 2017;65(6):945–963. doi: 10.1002/glia.23137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winzi M, Casas Vila N, Paszkowski-Rogacz M, Ding L, Noack S, Theis M, et al. The long noncoding RNA lncR492 inhibits neural differentiation of murine embryonic stem cells. PLoS ONE. 2018;13(1):e0191682. doi: 10.1371/journal.pone.0191682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16(4):413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 66.Lorthongpanich C, Thumanu K, Tangkiettrakul K, Jiamvoraphong N, Laowtammathron C, Damkham N, et al. YAP as a key regulator of adipo-osteogenic differentiation in human MSCs. Stem Cell Res Ther. 2019;10(1):402. doi: 10.1186/s13287-019-1494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fujii Y, Kawase-Koga Y, Hojo H, Yano F, Sato M, Chung UI, et al. Bone regeneration by human dental pulp stem cells using a helioxanthin derivative and cell-sheet technology. Stem Cell Res Ther. 2018;9(1):24. doi: 10.1186/s13287-018-0783-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Du Z, Shi X, Guan A. lncRNA H19 facilitates the proliferation and differentiation of human dental pulp stem cells via EZH2-dependent LATS1 methylation. Mol Ther Nucleic Acids. 2021;25:116–126. doi: 10.1016/j.omtn.2021.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jain R, Li D, Gupta M, Manderfield LJ, Ifkovits JL, Wang Q et al. HEART DEVELOPMENT. Integration of Bmp and Wnt signaling by Hopx specifies commitment of cardiomyoblasts. Science. 2015;348(6242):aaa6071. [DOI] [PMC free article] [PubMed]
- 70.Múnera JO, Sundaram N, Rankin SA, Hill D, Watson C, Mahe M, et al. Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell Stem Cell. 2017;21(1):51–64.e56. doi: 10.1016/j.stem.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, Zhu P, Luo J, Wang J, Liu Z, Wu W, et al. LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. Embo J. 2019;38(17):e101110. doi: 10.15252/embj.2018101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corsini NS, Peer AM, Moeseneder P, Roiuk M, Burkard TR, Theussl HC, et al. Coordinated control of mRNA and rRNA processing controls embryonic stem cell pluripotency and differentiation. Cell Stem Cell. 2018;22(4):543-558.e512. doi: 10.1016/j.stem.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 73.El Bassit G, Patel RS, Carter G, Shibu V, Patel AA, Song S, et al. MALAT1 in human adipose stem cells modulates survival and alternative splicing of PKCδII in HT22 cells. Endocrinology. 2017;158(1):183–195. doi: 10.1210/en.2016-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Y, Zeng X, Miao J, Liu C, Wei F, Liu D, et al. Upregulation of long noncoding RNA MEG3 inhibits the osteogenic differentiation of periodontal ligament cells. J Cell Physiol. 2019;234(4):4617–4626. doi: 10.1002/jcp.27248. [DOI] [PubMed] [Google Scholar]
- 75.Hastings KE. SL trans-splicing: easy come or easy go? Trends Genet. 2005;21(4):240–247. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 76.Horiuchi T, Aigaki T. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol Cell. 2006;98(2):135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- 77.Gingeras TR. Implications of chimaeric non-co-linear transcripts. Nature. 2009;461(7261):206–211. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51(3):349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 79.Yu CY, Kuo HC. The trans-spliced long noncoding RNA tsRMST impedes human embryonic stem cell differentiation through WNT5A-mediated inhibition of the epithelial-to-mesenchymal transition. Stem Cells. 2016;34(8):2052–2062. doi: 10.1002/stem.2386. [DOI] [PubMed] [Google Scholar]
- 80.Wu CS, Yu CY, Chuang CY, Hsiao M, Kao CF, Kuo HC, et al. Integrative transcriptome sequencing identifies trans-splicing events with important roles in human embryonic stem cell pluripotency. Genome Res. 2014;24(1):25–36. doi: 10.1101/gr.159483.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiao Q, Chen Z, Jin X, Mao R, Chen Z. The many postures of noncanonical Wnt signaling in development and diseases. Biomed Pharmacother. 2017;93:359–369. doi: 10.1016/j.biopha.2017.06.061. [DOI] [PubMed] [Google Scholar]
- 82.Sandsmark E, Hansen AF, Selnaes KM, Bertilsson H, Bofin AM, Wright AJ, et al. A novel non-canonical Wnt signature for prostate cancer aggressiveness. Oncotarget. 2017;8(6):9572–9586. doi: 10.18632/oncotarget.14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boo SH, Kim YK. The emerging role of RNA modifications in the regulation of mRNA stability. Exp Mol Med. 2020;52(3):400–408. doi: 10.1038/s12276-020-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xie W, Zhu H, Zhao M, Wang L, Li S, Zhao C, et al. Crucial roles of different RNA-binding hnRNP proteins in Stem Cells. Int J Biol Sci. 2021;17(3):807–817. doi: 10.7150/ijbs.55120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit Rev Biochem Mol Biol. 2012;47(4):360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun Z, Zhu M, Lv P, Cheng L, Wang Q, Tian P, et al. The long noncoding RNA Lncenc1 maintains naive states of mouse ESCs by promoting the glycolysis pathway. Stem Cell Rep. 2018;11(3):741–755. doi: 10.1016/j.stemcr.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim JH, Hahm B, Kim YK, Choi M, Jang SK. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J Mol Biol. 2000;298(3):395–405. doi: 10.1006/jmbi.2000.3687. [DOI] [PubMed] [Google Scholar]
- 88.Knoch KP, Bergert H, Borgonovo B, Saeger HD, Altkrüger A, Verkade P, et al. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat Cell Biol. 2004;6(3):207–214. doi: 10.1038/ncb1099. [DOI] [PubMed] [Google Scholar]
- 89.Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem Soc Trans. 2008;36(Pt 4):641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 90.Shibasaki T, Tokunaga A, Sakamoto R, Sagara H, Noguchi S, Sasaoka T, et al. PTB deficiency causes the loss of adherens junctions in the dorsal telencephalon and leads to lethal hydrocephalus. Cereb Cortex. 2013;23(8):1824–1835. doi: 10.1093/cercor/bhs161. [DOI] [PubMed] [Google Scholar]
- 91.Grammatikakis I, Gorospe M. Identification of neural stem cell differentiation repressor complex Pnky-PTBP1. Stem Cell Investig. 2016;3:10. doi: 10.21037/sci.2016.03.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22(10):2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li J, Yang Y, Fan J, Xu H, Fan L, Li H, et al. Long noncoding RNA ANCR inhibits the differentiation of mesenchymal stem cells toward definitive endoderm by facilitating the association of PTBP1 with ID2. Cell Death Dis. 2019;10(7):492. doi: 10.1038/s41419-019-1738-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu P, He F, Hou Y, Tu G, Li Q, Jin T et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene. 2021;40(9):1609–27. [DOI] [PMC free article] [PubMed]
- 95.Chakraborty D, Paszkowski-Rogacz M, Berger N, Ding L, Mircetic J, Fu J, et al. lncRNA Panct1 maintains mouse embryonic stem cell identity by regulating TOBF1 recruitment to Oct-Sox sequences in early G1. Cell Rep. 2017;21(11):3012–3021. doi: 10.1016/j.celrep.2017.11.045. [DOI] [PubMed] [Google Scholar]
- 96.Guo H, Liu J, Ben Q, Qu Y, Li M, Wang Y, et al. The aspirin-induced long non-coding RNA OLA1P2 blocks phosphorylated STAT3 homodimer formation. Genome Biol. 2016;17:24. doi: 10.1186/s13059-016-0892-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arduini BL, Brivanlou AH. Modulation of FOXD3 activity in human embryonic stem cells directs pluripotency and paraxial mesoderm fates. Stem Cells. 2012;30(10):2188–2198. doi: 10.1002/stem.1200. [DOI] [PubMed] [Google Scholar]
- 98.Li K, Sun D, Gou Q, Ke X, Gong Y, Zuo Y, et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80–90. doi: 10.1016/j.canlet.2018.01.060. [DOI] [PubMed] [Google Scholar]
- 99.Bidet K, Dadlani D, Garcia-Blanco MA. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 2014;10(7):e1004242. doi: 10.1371/journal.ppat.1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim MM, Wiederschain D, Kennedy D, Hansen E, Yuan ZM. Modulation of p53 and MDM2 activity by novel interaction with Ras-GAP binding proteins (G3BP) Oncogene. 2007;26(29):4209–4215. doi: 10.1038/sj.onc.1210212. [DOI] [PubMed] [Google Scholar]
- 101.Mao C, Wang X, Liu Y, Wang M, Yan B, Jiang Y, et al. A G3BP1-interacting lncRNA promotes ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Res. 2018;78(13):3484–3496. doi: 10.1158/0008-5472.CAN-17-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: old players and new actors. Trends Cancer. 2017;3(7):506–528. doi: 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 103.Gao F, Zhang YF, Zhang ZP, Fu LA, Cao XL, Zhang YZ, et al. miR-342-5p regulates neural stem cell proliferation and differentiation downstream to notch signaling in mice. Stem Cell Reports. 2017;8(4):1032–1045. doi: 10.1016/j.stemcr.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morgado AL, Rodrigues CM, Solá S. MicroRNA-145 regulates neural stem cell differentiation through the Sox2-Lin28/let-7 signaling pathway. Stem Cells. 2016;34(5):1386–1395. doi: 10.1002/stem.2309. [DOI] [PubMed] [Google Scholar]
- 105.Zhu Y, Gui W, Lin X, Li H. Knock-down of circular RNA H19 induces human adipose-derived stem cells adipogenic differentiation via a mechanism involving the polypyrimidine tract-binding protein 1. Exp Cell Res. 2020;387(2):111753. doi: 10.1016/j.yexcr.2019.111753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.