Abstract
It is still a challenge to develop gene replacement therapy for retinal disorders caused by mutations in large genes, such as Stargardt disease (STGD). STGD is caused by mutations in ABCA4 gene. Previously, we have developed an effective non-viral gene therapy using self-assembled nanoparticles of a multifunctional pH-sensitive amino lipid ECO and a therapeutic ABCA4 plasmid containing rhodopsin promoter (pRHO–ABCA4). In this study, we modified the ABCA4 plasmid with simian virus 40 enhancer (SV40, pRHO–ABCA4–SV40) for enhanced gene expression. We also prepared and assessed the formulations of ECO/pDNA nanoparticles using sucrose or sorbitol as a stablilizer to develop consistent and stable formulations. Results demonstrated that ECO formed stable nanoparticles with pRHO–ABCA4–SV40 in the presence of sucrose, but not with sorbitol. The transfection efficiency in vitro increased significantly after introduction of SV40 enhancer for plasmid pCMV-ABCA4–SV40 with a CMV promoter. Sucrose didn’t affect the transfection efficiency, while sorbitol resulted in a fluctuation of the in vitro transfection efficiency. Subretinal gene therapy in Abca4−/− mice using ECO/pRHO–ABCA4 and ECO/pRHO–ABCA4–SV40 nanoparticles induced 36% and 29% reduction in A2E accumulation respectively. Therefore, the ECO/pABCA4 based nanoparticles are promising for non-viral gene therapy for Stargardt disease and can be expended for applications in a variety of visual dystrophies with mutated large genes.
Keywords: Non-viral gene delivery, ECO, Plasmid DNA, Gene therapy, SV40 enhancer, Stargardt disease
1. Introduction
Stargardt disease (STGD) is characterized as gradual bilateral decline in central vision and visual acuity that is commonly caused by mutations in ABCA4 gene, which encodes a 210-kDa ATP-dependent flippase importer [1,2]. Gene replacement therapy, that delivers a healthy copy of a mutated gene into the targeted cells and expresses the encoded functional protein to restore its normal function, has shown the promise for effective treatment for STGD and other genetic ocular diseases [3–5]. The first FDA approved gene therapy is adeno-associated virus (AAV) expressing hRPE65 for treating Leber’s congenital amaurosis type 2 (LCA2) [6]. The recent success has re-energized the enthusiasm in developing gene therapy to treat previously untreatable genetic disorders. Numerous gene therapies have been developed and some are now in various phases of clinical trials [7]. Most of the gene therapies under clinical development are based on AAVs. However, the broad application of AAV-based gene therapy is limited by its cargo capacity [8,9]. This greatly restricted the application of viral gene therapies to treat ocular genetic diseases caused by mutations in large genes, such as Stargardt disease (STGD) and Usher Syndrome [10–12]. For viral systems, strategies such as dual-AAV, multi-AAV vectors, and lentiviral vectors have been tested to overcome the limitations [13–16]. Clinical application of these therapies is hindered by various limitations, including poor expression of whole functional proteins and immunogenicity. Non-viral gene delivery systems do not have limitations in gene packaging capacity and have also been developed for treating various retinal genetic diseases covering wide range of gene sizes [17,18].
A challenge for gene therapy in retinal genetic diseases is to maintain prolonged stable protein expression to retain normal visual functions. Clinically, one subretinal administration of AAV therapy could last for years in LCA2 patients, but the clinical findings indicated declining therapeutic effect over time [4,19,20]. Repetitive administrations may be needed to sustain the rescuing effect in retinal structure and function. Unfortunately, immune response development after the initial viral gene therapy renders the following repeated injection of the viral vector ineffective [21]. Non-viral gene therapy exhibits low immunogenicity and can be repeatedly administered for prolonged therapeutic efficacy. However, non-viral gene delivery systems may suffer from low efficiency [22]. The functional segments on the therapeutic gene construct can be modified to enhance the therapeutic efficacy. Because of the unlimited gene loading capacity of the non-viral system, therapeutic plasmids can be modified with tissue specific promoters to achieve specific gene expression in different retinal tissues or cells [23,24]. Tissue specific promoters have been incorporated in plasmid DNA to enhance specific gene expression, while universal promoters used in AAV-based gene therapy tend to facilitate non-specific transient protein expression in all cell types [25,26]. Tissue specific promoters can also minimize off-target transgene expression and improve the safety of gene therapy. Therapeutic gene constructs can also be modified with expression enhancers for enhancing exogenous expression in mammalian cells with the non-viral delivery systems [27].
Development of stable and reproducible formulation of nanoparticle based non-viral gene therapy is an essential step for clinical translation based on the requirements from the regulatory agencies. The formulation or drug product of gene therapy should have consistent physicochemical properties, including acceptable stability and shelf life, consistent particle size, size distribution and surface charges [28]. It is costly to establish Good Manufacturing Practice (GMP) for viral gene therapies due to expensive biological production process, which leads to high prices for the therapies [29,30]. The GMP production of non-viral gene therapies can be achieved with conventional chemical process, which is more cost-effective than viral systems. In order to produce consistent and stable nanoparticle formulations, stabilizers have been used in the formulations to prevent particle aggregations [31,32]. Most stabilizers are able to form hydrogen bonds on the surface of nanoparticles to stabilize nanoparticle formulations in aqueous solution by either exchanging with water molecules or forming an inert and rigid glass matrix for an acceptable shelf-life [33]. The stabilizers are generally biocompatible substances with minimal effect on the nanoparticle functions. Sucrose, trehalose, sorbitol and hydroxyethyl starch have been used as stabilizers in a wide variety of nanoparticle formulations [34–36].
We have developed a multifunctional pH-sensitive amino lipid (1-aminoethyl)iminobis[N-(oleoylcysteinyl-1-amino-ethyl)propionamide) (ECO) based nanoparticle platform for efficient intracellular delivery of therapeutic nucleic acids, including siRNA and plasmid DNA. ECO has demonstrated excellent efficacy for cytosolic delivery of therapeutic nucleic acids of different sizes in a wide range of applications [25,26,33,37–44]. In a previous study, we demonstrated that ECO facilitated efficient subretinal delivery of a large rhodopsin promoter (photoreceptor specific) modified therapeutic ABCA4 plasmid (pRHO-ABCA4, 12 kb), and induced long-term ABCA4 expression in the retina of a mouse model of Stargardt disease [25,26]. In this study, we optimized the therapeutic plasmid with a simian virus 40 enhancer (SV40) (pRHO-ABCA4-SV40) for prolonged gene expression. We also explored and developed stable ECO/pRHO–ABCA4–SV40 nanoparticle formulations using sucrose and sorbitol to improve stability and shelf-life for clinical translation. The formulations of ECO/pCMV–ABCA4–SV40 nanoparticles were also developed as a non-specific control. The nanoparticles were characterized and tested for gene expression in vitro and in vivo. Gene replacement therapy of ECO/pRHO–ABCA4–SV40 nanoparticle formulations was also performed to evaluate the efficacy in Abca4−/− mice, the orthologous rodent model to STGD.
2. Results
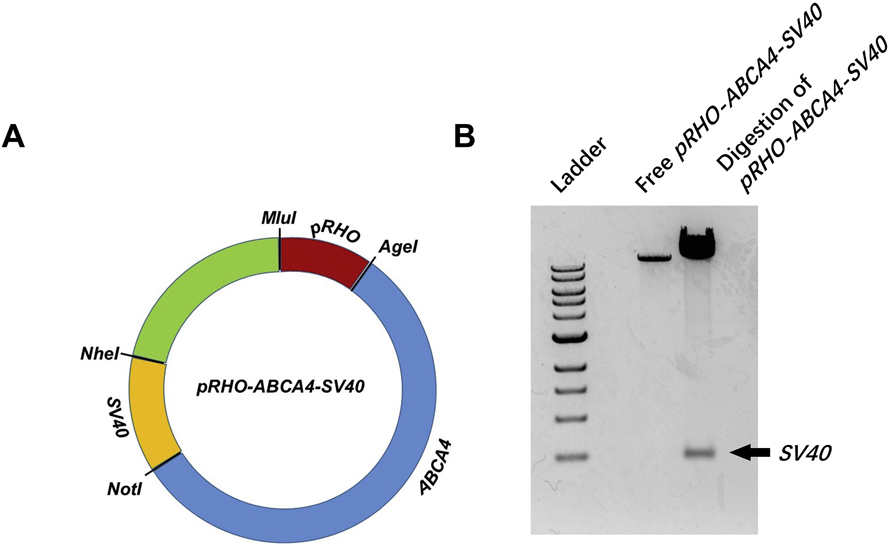
In order to enhance the expression of ABCA4 gene, a simian virus 40 enhancer (SV40) was incorporated into the pRHO–ABCA4 vector, which was used in our previous publications [25,26]. As shown in Fig. 1A, the SV40 enhancer was inserted in pRHO–ABCA4 between NheI and NotI restriction sites. The successful preparation of pRHO–ABCA4–SV40 plasmid was confirmed by agarose gel electrophoresis showing the polyA SV40 enhancer band after digestions at NheI and NotI restriction sites (Fig. 1B). Similarly, pCMV–ABCA4–SV40 was also prepared as a control plasmid using the plasmid in our previous publications [25,26].
Fig. 1.
Plasmid pRHO–ABCA4–SV40 vector map demonstrating the insertion of SV40 enhancer between NheI and NotI restriction cites (A) and agarose gel electrophoresis confirming the success of SV40 insertion (B).
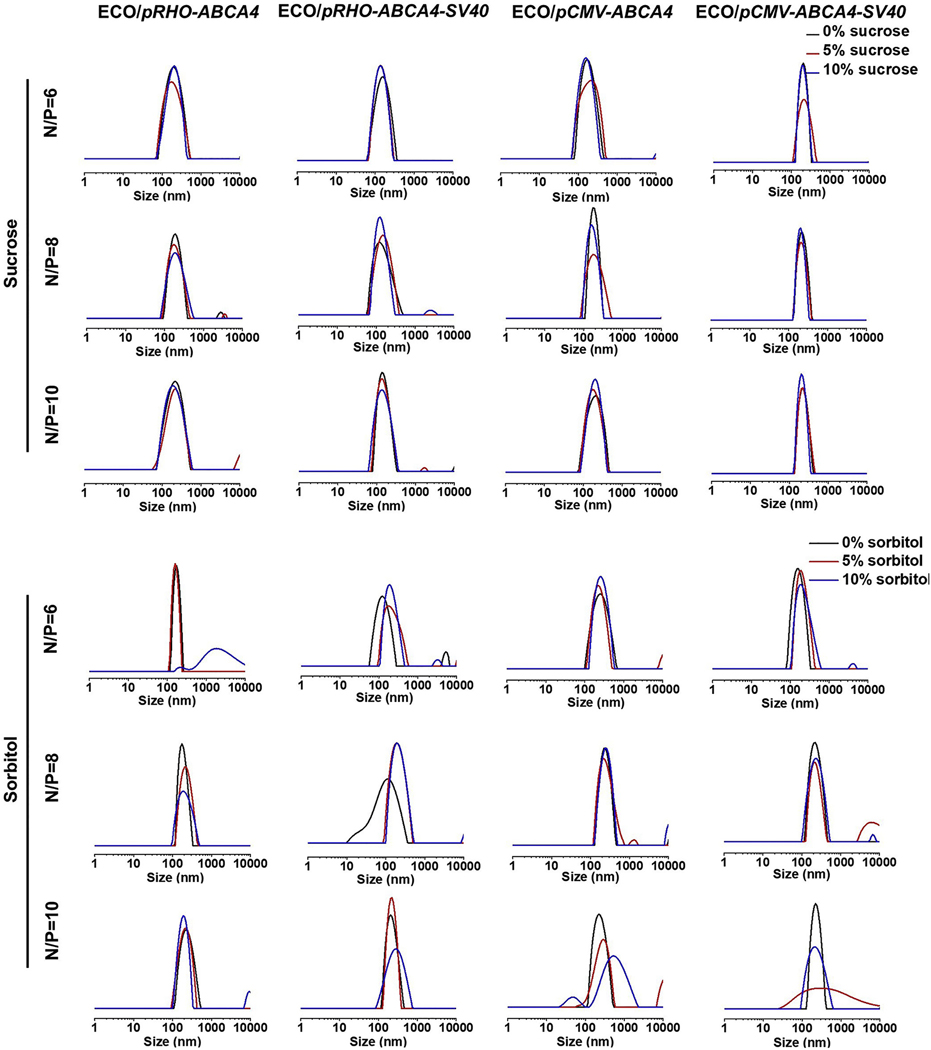
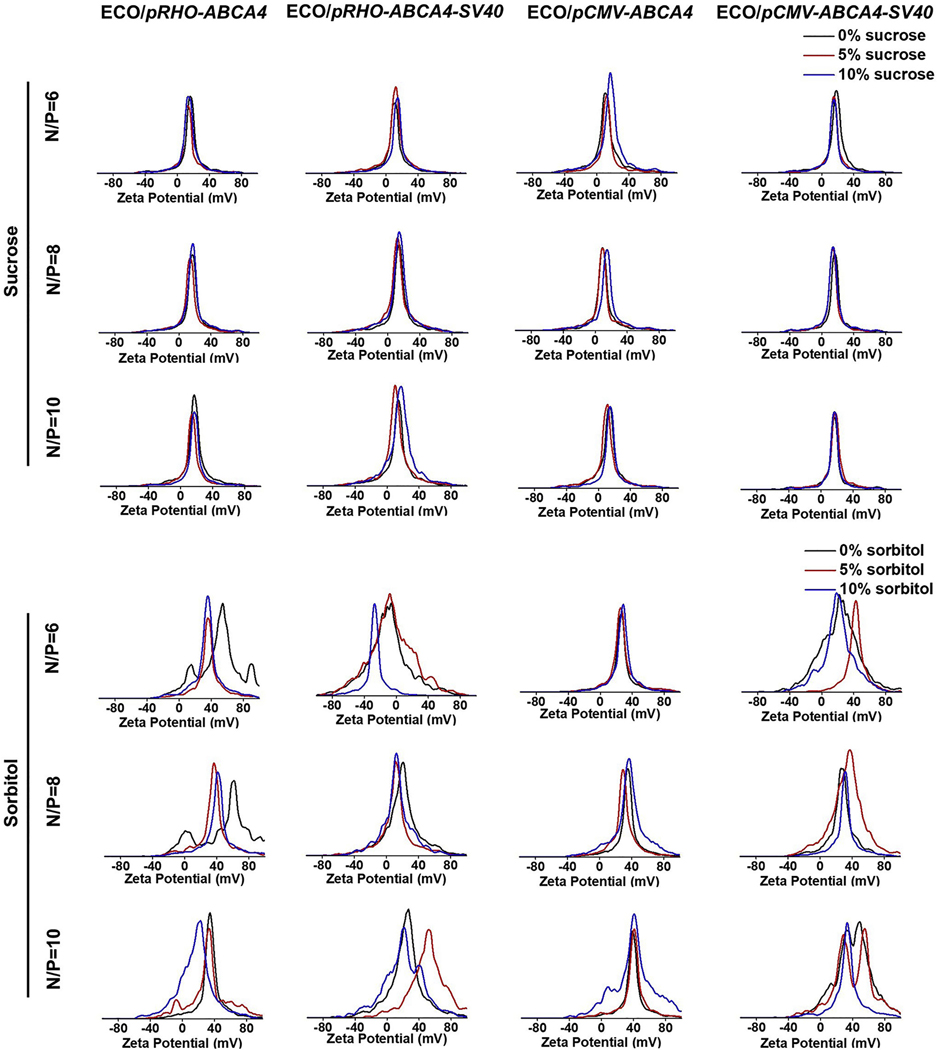
ECO/pDNA nanoparticles of pRHO–ABCA4–SV40 and pCMV–ABCA4–SV40 were first prepared by self-assembly of ECO with the plasmids at amine/phosphate (N/P) ratios of 6, 8 and 10 [25,26]. The plasmids pRHO–ABCA4 and pCMV–ABCA4 were used as controls. Sucrose or sorbitol were then added as an excipient with a concentration of 5% or 10% to stabilize the nanoparticles. The nanoparticles were characterized by dynamic light scattering (DLS) for their size distributions (Fig. 2) and zeta potential distributions (Fig. 3) under different excipient conditions. As shown in Fig. 2, ECO and pABCA4s formulated stable nanoparticles at all the N/P ratios in the presence of sucrose at both 5% and 10% contents. The sizes were between 220 nm and 250 nm and the distributions were seen as single peaks with negligible aggregation peaks and were not affected by the excipient. However, sorbitol addition affected some of the ECO/pABCA4 formulations. For example, ECO/pRHO–ABCA4 at N/P ratio of 6, ECO/pRHO–ABCA4–SV40 at N/P ratio of 8, ECO/pCMV–ABCA4 at N/P ratio of 10, and ECO/pCMV–ABCA4–SV40 at N/P ratio of 10 showed broadened size distributions after sorbitol addition. Zeta potential distributions demonstrated no noticeable change after sucrose additions for the ECO/pABCA4 nanoparticle formulations (Fig. 3). ECO/pABCA4 nanoparticles demonstrated uniformed distributions around +20 mV across all the N/P ratios under sucrose conditions. However, sorbitol addition to ECO/pABCA4 nanoparticle formulations resulted in some noticeable changes in zeta potential distributions.
Fig. 2.
Size distributions of the nanoparticles of ECO with pCMV–ABCA4, pCMV–ABCA4–SV40, pRHO–ABCA4, and pRHO–ABCA4–SV40 in the presence of sucrose and sorbitol as measured by dynamic light scattering.
Fig. 3.
Zeta potential distributions of the nanoparticle formulations of ECO with pCMV–ABCA4, pCMV–ABCA4–SV40, pRHO–ABCA4, and pRHO–ABCA4–SV40 in existence with sucrose and sorbitol as stabilizers.
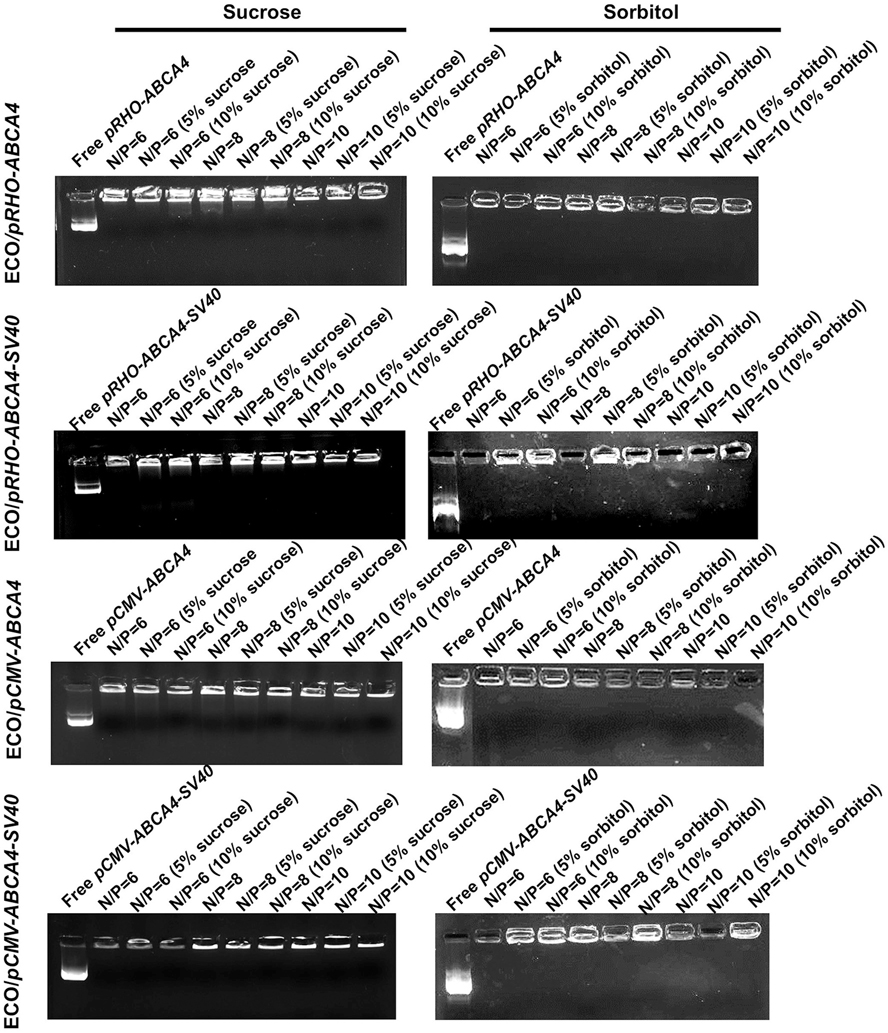
Encapsulation of pABCA4s in the ECO/pABCA4 nanoparticle formulations under different excipient conditions (sucrose and sorbitol, 5% or 10%) was verified by agarose gel electrophoresis (Fig. 4). ECO and pABCA4s formulated stable nanoparticles at all N/P ratios for all the excipients conditions, demonstrated by visible bands remaining on the top of the gels. As shown in our previous studies, ECO could efficiently encapsulate pABCA4s or other large plasmids with nanoparticle stability observed through the voltage conditions of electrophoresis [25,26,44]. Unlike the effects on the size and zeta potential distributions, sorbitol did not affect the stability and encapsulations of ECO/pABCA4 nanoparticles. Taken together, sorbitol has some unpredictable effects on the stability of the ECO/pABCA4 nanoparticles and may not be a suitable excipient for ECO/pDNA nanoparticles despite that it has commonly used on pharmaceutical formulations. Sucrose demonstrates no effect on the size, zeta potential and stability of ECO/pABCA4 nanoparticles and is a promising excipient for the formulations of ECO/pABCA4 nanoparticles.
Fig. 4.
Agarose gel electrophoresis showing the encapsulation of plasmid DNA in the nanoparticles formulated by ECO with pCMV–ABCA4, pCMV–ABCA4–SV40, pRHO–ABCA4, and pRHO–ABCA4–SV40 in the presence of sucrose and sorbitol.
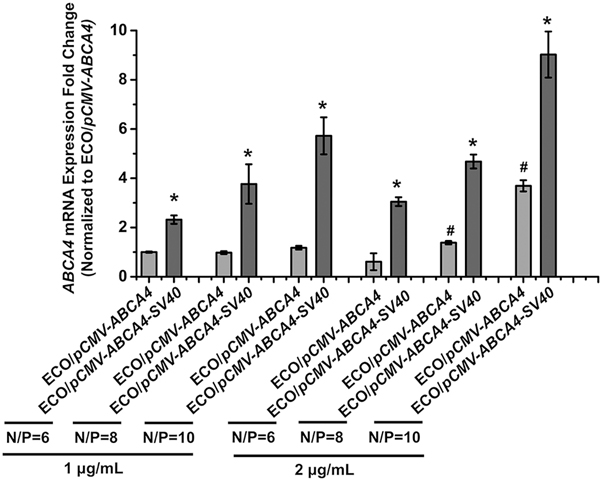
To assess the introduction of SV40 enhancer on transfection efficiency, ECO/pCMV–ABCA4 and ECO/pCMV–ABCA4–SV40 nanoparticles (N/P = 6, 8, 10; pDNA concentration 1 μg/mL and 2 μg/mL) were investigated in ARPE-19 cells. ABCA4 expressions at the mRNA level were analyzed using qRT-PCR 48 h after transfection (Fig. 5). ECO/pCMV–ABCA4–SV40 nanoparticles demonstrated significantly more ABCA4 mRNA expression than ECO/pCMV–ABCA4 without the SV40 enhancer for both doses. ABCA4 mRNA expression also increased with the N/P ratios and pDNA doses. Therefore, introduction of the SV40 enhancer is able to significantly enhance the in vitro expression of ECO/pDNA nanoparticle formulations.
Fig. 5.
In vitro transfection of ECO/pCMV–ABCA4 and ECO/pCMV–ABCA4–SV40 nanoparticles in ARPE-19 cells demonstrated by qRT-PCR of ABCA4 mRNA expression 48 h after transfections. ABCA4 mRNA expressions were normalized to the expression of ECO/pCMV–ABCA4 at N/P ratio of 6 and 1 μg/mL dose. (*p < 0.05, relative to the mRNA expression of ECO/pCMV–ABCA4 at the same condition. #p < 0.05, relative to the same nanoparticle formulation at 1 μg/mL.)
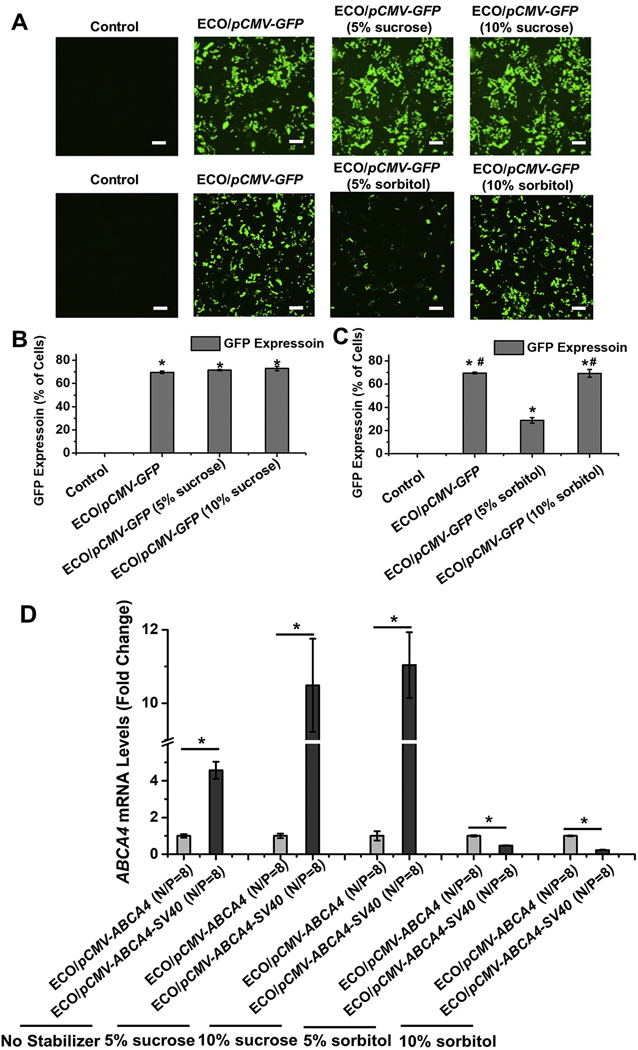
The effect of sucrose and sorbitol on gene expression was also evaluated using pCMV–ABCA4 (with and without SV40 enhancer) and a reporter plasmid pCMV–GFP in ARPE-19 cells. The GFP expression was evaluated using confocal microscope and flow cytometry 48 h after transfection (Fig. 6). Significant GFP signals were observed under confocal for all the conditions compared with untreated control using ECO/pCMV–GFP nanoparticles (Fig. 6A). The flow cytometry results also demonstrated around 70% of cells showing GFP expression for ECO/pCMV–GFP nanoparticles in 5%, 10% sucrose and 10% sorbitol (Fig. 6B and C). However, GFP expression was significantly reduced when 5% sorbitol was added (Fig. 6C). ABCA4 expression were not affected by sucrose addition as demonstrated in Fig. 6D. ECO/pCMV–ABCA4–SV40 nanoparticles demonstrated significantly higher ABCA4 mRNA expression than ECO/pCMV–ABCA4 due to the effect of the introduction of SV40 enhancer. However, sorbitol addition significantly reduced the ABCA4 mRNA expression for ECO/pCMV–ABCA4–SV40 nanoparticles (Fig. 6D). The results indicated that SV40 enhancer could enhance ABCA4 expression in vitro and sucrose had no effect on the transfection efficiency of ECO/pDNA nanoparticles, while sorbitol demonstrated unpredictable effects on ECO/pDNA transfection efficiency.
Fig. 6.
In vitro transfection of ECO/pDNA nanoparticles in ARPE-19 cells in the presence of sucrose or sorbitol. Confocal images (A) and flow cytometry (B, C) of GFP expression in ARPE-19 cells 48 h after transfection using ECO/pCMV–GFP nanoparticles (N/P = 8, DNA concentration of 1 μg/mL, under 0%, 5% and 10% sucrose or sorbitol). (D) ABCA4 mRNA expression 48 h after transfections demonstrated by qRT-PCR using ECO/pCMV–ABCA4 and ECO/pCMV–ABCA4–SV40 nanoparticles (N/P ratios of 6, 8 and 10, DNA concentration of 1 μg/mL, under 0%, 5% and 10% sucrose or sorbitol). ABCA4 mRNA expressions of ECO/pCMV–ABCA4–SV40 nanoparticles were normalized to those of ECO/pCMV–ABCA4 nanoparticles. (Scale bars represent 20 μm. *p < 0.05, relative to the non-treated control in B and C. #p < 0.05, relative to the GFP expression in 5% sorbitol. *p < 0.05, between the two groups under the line in D).
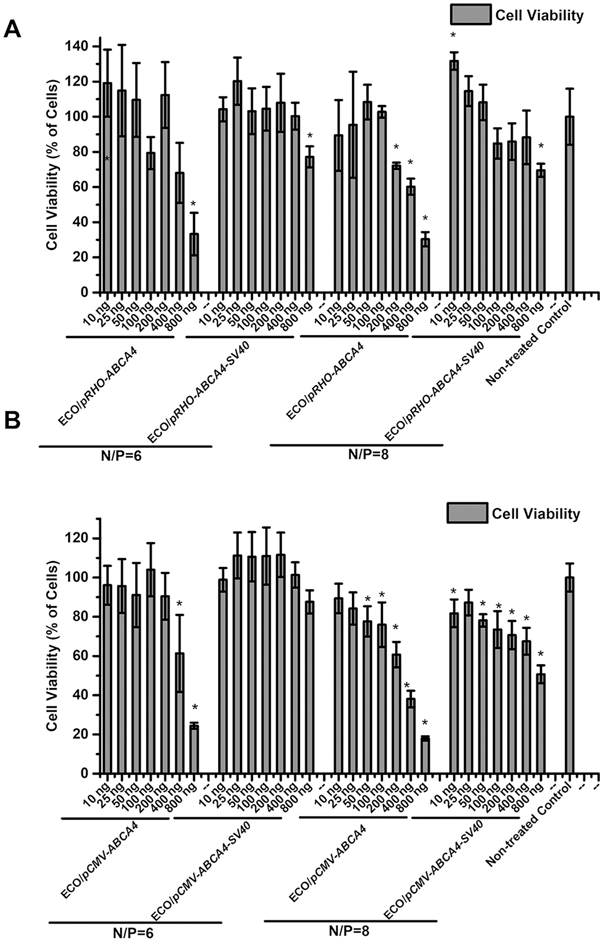
The cytotoxicity of ECO/pDNA nanoparticles were evaluated using pRHO–ABCA4 and pCMV–ABCA4 (with or without enhancer SV40) at different N/P ratios (6 and 8) and different doses in ARPE-19 cells. As shown in Fig. 7, ECO/pRHO–ABCA4 (with and without enhancer) demonstrated better cell viability than ECO/pCMV–ABCA4 (with and without enhancer). Higher pDNA doses were correlated with lower cell viabilities across all the nanoparticles. At N/P ratio of 6, almost all nanoparticles demonstrated more than 80% cell viability, except for higher doses groups. At N/P ratio of 8, reduced cell viability was observed for higher doses, especially for pCMV–ABCA4. Overall, ECO/pABCA4 nanoparticles demonstrated good cell viability and low cytotoxicity.
Fig. 7.
CCK-8 assay of cytotoxicity of (A) ECO/pRHO–ABCA4, ECO/pRHO–ABCA4–SV40, (B) ECO/pCMV–ABCA4, and ECO/pCMV–ABCA4–SV40 nanoparticles in ARPE-19 cells 48 h after transfection (*P < 0.05 relative to non-treated control).
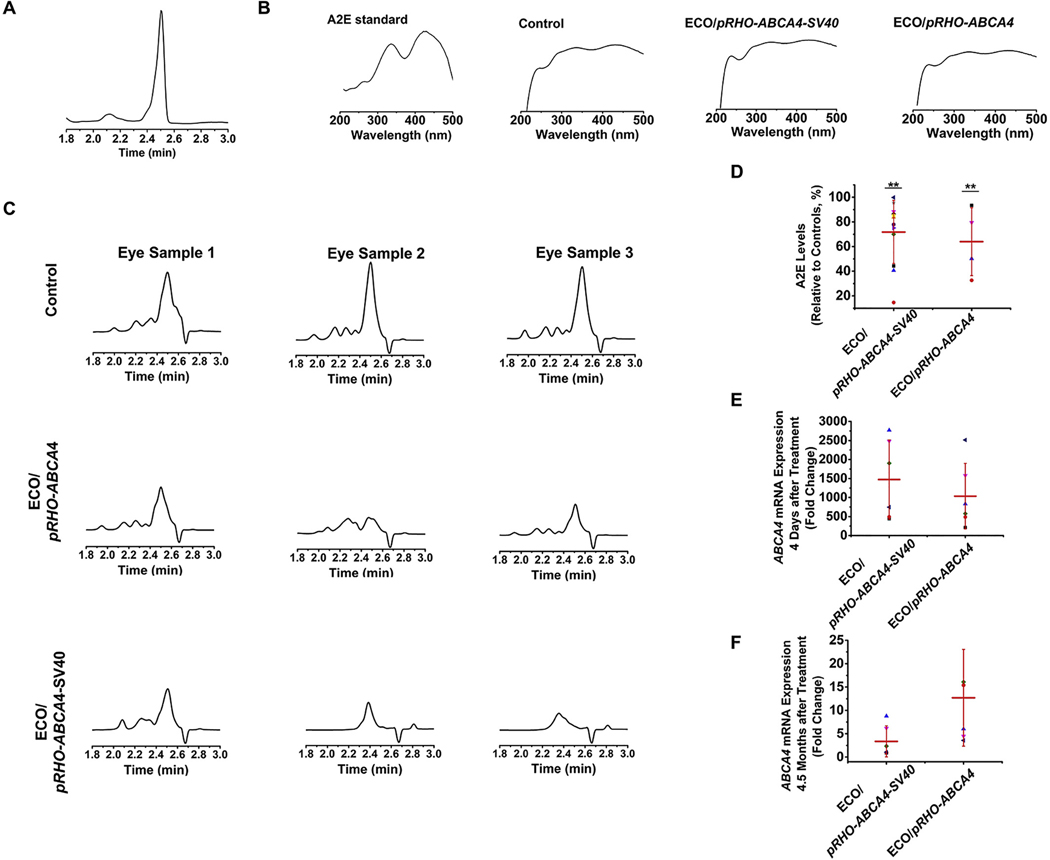
The efficacy of preventing Stargardt disease (STGD) progression using gene therapy with ECO/pRHO–ABCA4 and ECO/pRHO–ABCA4–SV40 was assessed based on the accumulation of A2E in the retinal pigmented epithelium (RPE) of Abca4−/− mice. A2E, a photo-toxic dimer of vitamin A, is a main component of lipofuscin. A2E accumulation is commonly used as an indicator of STGD progression [45,46]. One of the therapeutic strategies for treating STGD is to slow down the production of A2E to minimize chronic oxidative damage that ultimately leads to atrophy. In previous work, we demonstrated prolonged ABCA4 expression in Abca4−/− mice and reduced A2E accumulation [25]. Similarly, the Abca4−/− mice (1-month-old) received a single subretinal injection of ECO/pRHO–ABCA4 or ECO/pRHO–ABCA4–SV40 with PBS as a control. All the treated mice were euthanized 8 months after injection and the A2E levels were analyzed using high-performance liquid chromatography (HPLC). A2E was identified as the peak at 2.5 min in the chromatograms under current HPLC running conditions, which was ensured by the analysis of the synthesized A2E standard (Fig. 8A). The spectra of A2E standard, A2E from control, ECO/pRHO–ABCA4 treated, and ECO/pRHO–ABCA4–SV40 treated mice were compared and found to contain the same peaks at 336 nm and 439 nm (Fig. 8B). The area of A2E peaks reduced significantly after treatments of ECO/pRHO–ABCA4 and ECO/pRHO–ABCA4–SV40 nanoparticles compared with the control group (Fig. 8C). Quantitative analysis demonstrated an average A2E level of 71.78 ± 25.60% (ca. 29% reduction) for ECO/pRHO–ABCA4–SV40 and 63.95 ± 27.54% (ca. 36% reduction) for ECO/pRHO–ABCA4 treated mice (Fig. 8D). A large variation of in vivo efficacy was observed with ECO/pRHO–ABCA4–SV40 nanoparticles. The modification of pRHO–ABCA4 with the SV40 enhancer did not result in improvement of in vivo efficacy than the unmodified plasmid. This could be explained from the ABCA4 mRNA expression after subretinal treatments using ECO/pRHO–ABCA4 and ECO/pRHO–ABCA4–SV40 nanoparticles. ECO/pRHO–ABCA4–SV40 resulted in slightly higher ABCA4 mRNA expression than ECO/pRHO–ABCA4 at 4 days although difference was not significant (Fig. 8E). In contrast, ABCA4 mRNA expression with ECO/pRHO–ABCA4–SV40 was slightly less than ECO/pRHO–ABCA4 at 4.5 months (Fig. 8F). It implies that the plasmid pRHO–ABCA4–SV40 with the viral enhancer might be cleared faster than the one without the enhancer.
Fig. 8.
Efficacy of ECO/pRHO–ABCA4 and ECO/pRHO–ABCA40–SV40 nanoparticles for preventing A2E accumulation in Abca4−/− mice. (A) HPLC analysis of synthesized A2E standard. (B) A2E spectra of synthesized A2E standard, and samples from nanoparticle treated and control mice. (C) HPLC chromatograms of A2E from control, ECO/pRHO–ABCA4, and ECO/pRHO–ABCA40–SV40 nanoparticles treated Abca4−/− mice 8 months after subretinal injections. (D) Quantitative A2E levels of ECO/pRHO–ABCA4 and ECO/pRHO–ABCA40–SV40 nanoparticles treated Abca4−/− mice relative to PBS control mice 8 months after subretinal injections. ABCA4 mRNA expression (E) 4 days and (F) 4.5 months after subretinal treatments of ECO/pRHO–ABCA4 and ECO/pRHO–ABCA4–SV40 nanoparticles in Abca4−/− mice. (error bars = ± SD, **P < 0.05 relative to control eyes. Statistical analysis was conducted with one-way ANOVA).
3. Discussion
Gene therapy holds great promise for the treatment of monogenic retinal disorders, but still faces challenges for efficient and specific delivery of therapeutic genes, especially large genes. The clinical applications of adeno-associated viruses (AAVs) are limited to the genes that can fit in the viral cavities. The multifunctional pH-sensitive amino lipid ECO can encapsulate genetic materials of unlimited sizes from siRNAs and miRNAs to large plasmid DNAs [25,26,37–44]. ECO has demonstrated high transfection efficiency both in vitro and in vivo for various applications from cancer therapies to retinal gene therapies [25,26,37–44]. ECO based nanoparticles can facilitate pH-sensitive amphiphilic endosomal membrane destabilization and endosomal escape as well as reductive dissociation of the nanoparticles to release nucleic acids in cytoplasm, the PERC effect, which allows highly efficient cytosolic delivery of therapeutic nucleic acids [25,26,37–44]. Here, ECO readily formed stable nanoparticles with ABCA4 plasmids via self-assembly with uniformed size distributions and complete encapsulations, and mediated efficient intracellular gene delivery and expression.
Clinical translation of nanoparticle-based non-viral gene therapies for retinal genetic disorders requires the development of stable nanoparticle formulations to ensure safety and efficacy of drug products. Excipients are commonly used in nanoparticle formulations to preserve the nanoparticles with consistent physicochemical properties. Sucrose and sorbitol are the commonly used excipients for the stabilization of the nucleic acid nanoparticle formulations. Interestingly, we found in this work that they behaved differently on the stability of the ECO/pDNA nanoparticles. Although the presence of both excipients resulted in complete encapsulation of the DNA plasmids in the nanoparticles, sorbitol caused substantial aggregation and change of zeta potentials of the nanoparticles in aqueous solution, while sucrose did not have a noticeable effect on particle formulations (Figs. 2, 3 and 4). Sucrose has been tested previously as a stabilizer for ECO/siRNA nanoparticles, which also demonstrated no negative effects on particle formulations, retained transfection efficiency, and improvements in nanoparticle long-term storage [33]. The formulations with sucrose also resulted in higher expression of a reporter gene and ABCA4 than those with sorbitol. The difference between sucrose and sorbitol may be associated with their structures. Sucrose is a disaccharide of glucose and fructose, which can form an inert and rigid glass matrix that can immobilize the nanoparticles to stabilize the formulation [47]. Sorbitol is linear polyols but possibly facilitates the nanoparticles via hydrogen binding [48].
The pRHO–ABCA4 plasmid was modified by adding an enhancer SV40 with an expectation of augmenting the gene expression for efficacious treatment of Stargardt disease. We have shown previously that ECO/pRHO–ABCA4 was effective to mediate gene expression specifically in the outer segments of the photoreceptor cells. The tissue specific bovine rhodopsin specific promoter (RHO) in the plasmid facilitates specific expression of ABCA4 in the outer segment (OS) previously [25]. Enhancer sequences are often incorporated in the plasmid DNA to increase gene expression with non-viral nanoparticles. The SV40 enhancer containing a 72 base pair repeat could enhance the expression of nonviral genes in cells and in vivo [49,50]. It was incorporated in pRHO-ABCA4–SV40 to explore the potential for enhanced long-term expression of ABCA4 for treating Stargardt disease. Since it is difficult to culture photoreceptor cells, we constructed a different plasmid with a common CMV promotor and SV40 enhancer (pCMV–ABCA4–SV40) to assess the effect of SV40 enhancer in vitro with ARPE-19 cells. Significantly augmented gene expression was observed for ECO/pCMV–ABCA4–SV40 when compared to ECO/pCMV–ABCA4 (Fig. 5).
We have shown previously that ECO/pRHO–ABCA4 nanoparticles can induce up to 8-month expression of ABCA4 in mice and can delay the disease progression by reducing A2E accumulation in Abca4−/− mice [25]. The efficacy of ECO/pRHO–ABCA4–SV40 was assessed in comparison with ECO/pRHO–ABCA4 at 8 months after a single subretinal injection in Abca4−/− mice. Both nanoparticles demonstrated about 29% and 36% A2E reduction on average compared to the PBS injected controls (Fig. 8). No significant difference was observed between ECO/pRHO–ABCA4–SV40 and ECO/pRHO–ABCA4 as was shown in the in vitro experiments. The inclusion of the SV40 enhancer did not demonstrate better efficacy, possibly due to the cell type preference of SV40 and the fluctuating nature of the virus origin [51]. Another explanation could be fast clearance of the pRHO–ABCA4–SV40 plasmid with the viral enhancer from the eye based on the reduction of mRNA expression in the eye over time, (Fig. 8E,F). The other cause could be the variability for subretinal injections, which is a challenging task as well. Nevertheless, the observation is helpful to explore different strategies on the optimization of the ABCA4 plasmid for enhanced therapeutic efficacy. Currently, we are exploring modifications of ABCA4 plasmid with a human promoter and a human enhancer for clinical translation.
In conclusion, multifunctional pH-sensitive amino lipid ECO and ABCA4 plasmids form stable nanoparticle formulations through self-assembly in both 5% and 10% sucrose stabilizer. The addition of sucrose as stabilizer produced no effect on the transfection efficiency of ECO/pABCA4 nanoparticles. Modification of ABCA4 plasmids with an SV40 enhancer induced significantly higher gene expression in ARPE-19 cells, and demonstrated a similar level of A2E reduction in treated Abca4−/− mice as ECO/pRHO–ABCA4 nanoparticles. Therefore, the ECO/pABCA4 nanoparticles with a modified enhancer sequence and the stabilizer can be a promising, reliable, and safe non-viral gene therapy platform to deliver large therapeutic genes for the treatment of Stargardt disease.
4. Materials and methods
4.1. Reagents
All reagents ordered from vendors were directly used without extra purification unless they were otherwise detailed in this section. Organic solvents such as, acetonitrile (ACN), ethanol and methanol were ordered from Thermo Fisher Scientific (Hampton, NH). The synthesis of Lipid ECO followed the procedures reported previously [37,38]. For cell culture, penicillin fetal bovine serum, and streptomycin were purchased from Invitrogen (Carlsbad, CA). The ABCA4 plasmid (pCMV–ABCA4) was kindly gifted by Dr. Robert S. Molday (University of British Columbia), which included human ABCA4 cDNA sequence of full-length (NCBI Accession # NM_000350.2) on a pCEP4 backbone. pRHO–ABCA4 was prepared as previously described [25].
4.2. Plasmid construction
The pRHO–ABCA4 plasmid was constructed by molecular cloning of the cDNA for the ABCA4 gene into the linearized pRHO–DsRed plasmid with the DsRed reporter gene removed [25]. The cDNA for the ABCA4 gene was amplified by polymerase chain reaction (PCR) with the Q5 High-Fidelity DNA Polymerase enzyme. The forward primer was 5'-AATACCGGTATGGGCTTCGTGAGACAGATA-3′ and the reverse primer was 5′-TATATAGCGGCCGCTAGCTCAGTCTGCTGTTT-3′ for adding the AgeI and NotI restriction sites to the 5′- and 3′-ends of ABCA4, respectively. All enzymes were purchased from New England Biolabs (Ipswich, MA). SV40 polyA and SV40 enhancer was copied from plasmid pGL3-control vector (Promega, Madison, WI) by PCR with forward primer 5′-ATGCGGCCGCTACCACATTTGTAGAGGTTTTAC and reverse primer 5′-AAGCTAGCGCTGTGGAATGTGTGTCAG containing NotI and NheI sites on both ends. Digested and purified SV40 polyA and enhancer fragment was ligated to pRHO–ABCA4 plasmid. Final plasmid stocks were confirmed by Sanger sequencing.
4.2.1. Cell culture
ARPE-19 (ATCC, Manassas, Virginia) cells were passaged and maintained in a Dulbecco’s modified Eagle’s medium containing fetal bovine serum (10%), streptomycin (100 μg/mL), and penicillin (100 units/mL). Cells were kept in a humidified incubator at 37 °C and 5% CO2.
4.2.2. Animal
Pigmented Abca4−/− knockout mice were obtained as described previously and maintained with mixed backgrounds of 129Sv/Ev or C57BL/6 [25,26]. Animals were housed and bred in the Animal Resource Center at CWRU. All procedures followed approved protocols by the CWRU Institutional Animal Care and Use Committee (IACUC#2014–0053), which were also in compliance with recommendations from the Association for Research for Vision and Ophthalmology and the American Veterinary Medical Association Panel on Euthanasia.
4.2.3. Preparation of ECO/pDNA nanoparticles
The preparation of ECO/pDNA nanoparticles was as previously described [25,26]. Briefly, ECO (25 mM) in ethanol was added and mixed with a plasmid DNA (0.5 mg/mL) aqueous solution at predetermined volume from the N/P ratio (amine to phosphate ratio) of 6, 8 or 10. The mixture was first vortexed at 3000 rpm for 1 min and left on a shaker for 30 min. For some of the nanoparticles, sucrose (5% or 10%) or sorbitol (5% or 10%) was added after the first shaking and further shaken for another 20 min. The final DNA concentration in the nanoparticle for characterization and in vivo experiments was 200 ng/μL. For in vitro transfection, an ECO stock solution of 2.5 mM was used to formulate nanoparticles. Encapsulations of pDNA by lipid ECO in nanoparticle formulations were characterized by an agarose gel electrophoresis method. Agarose gel (0.7%) in TBE buffer (0.5%) was performed at 120 V for 25 min.
4.2.4. Dynamic light scattering
The sizes and zeta potentials were characterized for nanoparticle formulations of ECO/pDNA using a dynamic light scattering method with an Anton Paar Litesizer 500 (Anton Paar USA Inc., Ashland, VA). Each sample was analyzed at 25 °C.
4.2.5. In vitro transfection
Transfections were performed on 12-well plates, where ARPE-19 cells were seeded at a concentration of 4 × 104 cells/well. Cells were allowed to grow for 24 h before transfection. Nanoparticles of different N/P ratios at pDNA concentrations of 1 or 2 μg/mL in DMEM with 10% serum were added to ARPE-19 cells and incubated for 8 h at 37 °C. The media containing nanoparticles was then replaced with fresh DMEM (10% serum). ARPE-19 cells were further incubated for an additional 48 h. Expression of ABCA4 was evaluated by qRT-PCR at mRNA level.
Transfection of ECO/pCMV–GFP nanoparticles of N/P = 8 was also conducted similarly in ARPE-19 cells, where DMEM (10% serum) and a pDNA concentration of 1 μg/mL were used. The transfection was performed in a 12-well plate with ARPE-19 cell concentration of 4 × 104 cells per well. The nanoparticles were incubated with ARPE-19 cells as previously described. After 48 h, fluorescence images of GFP expression were acquired using an Olympus FV1000 confocal microscope.
4.2.6. Cytotoxicity
Cytotoxicity of ECO/pABCA4 nanoparticles was investigated using a CCK-8 assay (Dojindo Molecular Technologies, Inc., Washington, D.C.). Cell viability was evaluated using ARPE-19 cells on 96-well plates, where cells were seeded at a concentration of 1 × 104 cells per well. ARPE-19 cells were incubated with ECO/pABCA4 nanoparticles at different DNA doses of (10 ng, 25 ng, 50 ng, 100 ng, 200 ng, 400 ng, and 800 ng) in 100 μL DMEM (10% serum) medium for 8 h at 37 °C. Then the nanoparticle containing DMEM was replaced with fresh DMEM (10% serum). The cells were allowed to grow until 48 h and washed with PBS. The CCK-8 reagent was added to each well followed by an incubated of 1.5 h at 37 °C. The absorbance at 450 nm was recorded using a plate reader. Cell viability was characterized by normalizing to the absorption of non-treated control.
4.2.7. qRT-PCR
The analysis was performed as previously described [25,26]. A scraper was used for cell lysis and homogenization. The RNA extractions for cell samples were conducted using a QIAGEN RNeasy kit following the manufacturer’s instructions. cDNAs were synthesized from mRNA transcripts using a QIAGEN miScriptII reverse transcriptase kit (Germantown, MD). For animal experiments, the eye tissue samples were homogenized using a glass tube loaded with 0.6 mL of the lysis buffer on ice. The RNA extractions and cDNAs synthesis were performed using the same kits as described previously for cell samples. The qRT-PCR analysis was performed in a Mastercycler instrument (Eppendorf, Hauppauge, NY) using a SYBR Green Master mix (AB Biosciences, Allston, MA). Fold changes of mRNA levels were determined by normalization to 18S. Primers for ABCA4 can be found in previous work [25].
4.2.8. In vivo subretinal transfection with ECO/pDNA nanoparticles
Subretinal injection was performed as previously described [25]. The nanoparticle solution (1 μL) was injected by a pump with a steady speed of 150 nL/s into the mouse eye. Successful administration was confirmed by bleb formation in the subretinal space. A total of 200 ng plasmid or pABCA4 was delivered. Mice injected with 1 μL of PBS were used as controls.
4.3. Synthesis and HPLC analysis of A2E
Synthesis of A2E standard was the same as described previously [25]. A mixture of all-trans-retinal (100 mg, 352 μmol) and ethanolamine (9.5 mg, 155 μmol) in ethanol (3.0 mL) was stirred in the presence of acetic acid (9.3 μL, 155 μmol) at room temperature with a sealed cap in the dark for 2 days. After the mixture was concentrated in vacuo, the residue was purified by silica gel column chromatography. After elution with MeOH:CH2Cl2 (5:95), further elution with MeOH: CH2Cl2: trifluoroacetic acid (8:92:0.001) gave A2E. Pure samples were obtained by HPLC purification [ZORBAX 300 SB-C18, 9.4 × 250 mm, 84–100% water/acetonitrile for 30 min, 1.0 mL/min flow detected at UV 430 nm]. A2E was detected at retention time (tR = 35.2 min). Collection of the fraction provided pure A2E for further analysis. A2E was characterized by mass spectrometry.
A2E samples were extracted from the deep-frozen (−80 °C) eye samples. The extraction was performed in 1 mL of acetonitrile after homogenization with a Brinkmann Politron homogenizer (Kinematica, Lucerne, Switzerland). After evaporation of solvent, extracts were dissolved in 120 μL acetonitrile with 0.1% TFA. Samples (100 μL) were loaded on a C18 column (Gemini® 5 μm 110 Å HPLC Column 250 × 4.6 mm) (Phenomenex, Torrance, CA) and analyzed by an Agilent 1260 infinity II reverse-phase HPLC (Agilent Technologies, Santa Clara, CA). A2E was eluted with the following gradients of acetonitrile in water (containing 0.1% trifluoroacetic acid): 85–96% (10 min), 96% (5 min), 96–100% (2 min), and 100% (13 min) (flow rate, 1 mL/min), and they were monitored at 439 nm. For A2E quantification by reverse phase HPLC, areas of the A2E peaks from the treated mice were normalized to the average of controls. Total of 4 ECO/pRHO–ABCA4 treated eyes, 5 ECO/pRHO–ABCA4–SV40 treated eyes and 10 control eyes were analyzed.
4.3.1. Statistical analysis
Experiments were performed in triplicate and the number of animals is listed in the figure captions. Experimental data are presented as averages with standard deviations. Statistical analysis was performed with one-way ANOVA and two-tailed Student’s t-tests. A 95% confidence interval was used and P ≤ 0.05 was accepted as statistically significant.
Acknowledgements
This project was supported by the Gund-Harrington Scholars Award from the Harrington Discovery Institute and by the Foundation Fighting Blindness. Further support was obtained through grants from the NIH (NEI R24-EY-024864 and NEI R24-EY-027283), the Canadian Institute for Advanced Research (CIFAR), the Alcon Research Institute (ARI), unrestricted grants from Research to Prevent Blindness to the Departments of Ophthalmology at UCI, Grant BX003604 from the Department of Veterans Affairs. K.P. is Chief Scientific Officer of Polgenix Inc. Z.R.L. is an M. Frank Rudy and Margaret Domiter Rudy Professor of Biomedical Engineering.
Footnotes
Declaration of competing interest
The authors have declared that no conflict of interest exists.
References
- [1].Sullivan JM, Focus on molecules: ABCA4 (ABCR)–an import-directed photoreceptor retinoid flipase, Exp. Eye Res. 89 (5) (2009) 602–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wiszniewski W, Zaremba CM, Yatsenko AN, Jamrich M, Wensel TG, Lewis RA, Lupski JR, ABCA4 mutations causing mislocalization are found frequently in patients with severe retinal dystrophies, Hum. Mol. Genet. 14 (19) (2005) 2769–2778. [DOI] [PubMed] [Google Scholar]
- [3].Schimmer J, Breazzano S, Investor outlook: significance of the positive LCA2 gene therapy phase III results, Human Gene Therapy Clin. Dev 26 (4) (2015) 208–210. [DOI] [PubMed] [Google Scholar]
- [4].Simonelli F, Maguire AM, Testa F, Pierce EA, Mingozzi F, Bennicelli JL, Rossi S, Marshall K, Banfi S, Surace EM, Sun J, Redmond TM, Zhu X, Shindler KS, Ying G-S, Ziviello C, Acerra C, Wright JF, McDonnell JW, High KA, Bennett J, Auricchio A, Gene therapy for Leber’s congenital amaurosis is safe and effective through 1.5 years after vector administration, Mol. Ther. 18 (3) (2010) 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Koirala A, Conley SM, Makkia R, Liu Z, Cooper MJ, Sparrow JR, Naash MI, Persistence of non-viral vector mediated RPE65 expression: case for viability as a gene transfer therapy for RPE-based diseases, J. Control. Release 172 (3) (2013) 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Prado DA, Acosta-Acero M, Maldonado RS, Gene therapy beyond luxturna: a new horizon of the treatment for inherited retinal disease, Curr. Opin. Ophthalmol. 31 (3) (2020). [DOI] [PubMed] [Google Scholar]
- [7].Lipinski DM, Thake M, MacLaren RE, Clinical applications of retinal gene therapy, Prog. Retin. Eye Res 32 (2013) 22–47. [DOI] [PubMed] [Google Scholar]
- [8].Wu Z, Yang H, Colosi P, Effect of genome size on AAV vector packaging, Mol. Therapy 18 (1) (2010) 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Santiago-Ortiz JL, Schaffer DV, Adeno-associated virus (AAV) vectors in cancer gene therapy, J. Control. Release 240 (2016) 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schulz HL, Grassmann F, Kellner U, Spital G, Rüther K, Jägle H, Hufendiek K, Rating P, Huchzermeyer C, Baier MJ, Weber BHF, Stöhr H, Mutation Spectrum of the ABCA4 gene in 335 Stargardt disease patients from a multicenter German cohort-impact of selected deep Intronic variants and common SNPs, Invest. Ophthalmol. Vis. Sci. 58 (1) (2017) 394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Williams DS, Usher syndrome: animal models, retinal function of usher proteins, and prospects for gene therapy, Vis. Res 48 (3) (2008) 433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Delplace V, Payne S, Shoichet M, Delivery strategies for treatment of age-related ocular diseases: from a biological understanding to biomaterial solutions, J. Control. Release 219 (2015) 652–668. [DOI] [PubMed] [Google Scholar]
- [13].Palfi A, Chadderton N, McKee AG, Blanco Fernandez A, Humphries P, Kenna PF, Farrar GJ, Efficacy of Codelivery of dual AAV2/5 vectors in the murine retina and Hippocampus, Hum. Gene Ther. 23 (8) (2012) 847–858. [DOI] [PubMed] [Google Scholar]
- [14].Maddalena A, Colella P, Trapani I, Minopoli R, Iodice C, Auricchio A, Triple AAV vectors to expand AAV cargo capacity in the retina, Invest. Ophthalmol. Vis. Sci. 56 (7) (2015) 3631. [Google Scholar]
- [15].Allocca M, Garcia-Hoyos M, Doria M, Petrillo M, Kim S, Maguire A, Vicino UD, Sparrow JR, Bennett J, Auricchio A, Gene therapy of a mouse model of Stargardt’s disease using Adeno-associated viral vectors packaging large genes, Invest. Ophthalmol. Vis. Sci. 49 (13) (2008) 1129. [Google Scholar]
- [16].Miyoshi H, Takahashi M, Gage FH, Verma IM, Stable and efficient gene transfer into the retina using an HIV-based lentiviralvector, Proc. Natl. Acad. Sci. 94 (19) (1997) 10319–10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bordet T, Behar-Cohen F, Ocular gene therapies in clinical practice: viral vectors and nonviral alternatives, Drug Discov. Today 24 (8) (2019) 1685–1693. [DOI] [PubMed] [Google Scholar]
- [18].Mashal M, Attia N, Martinez-Navarrete G, Soto-Sanchez C, Fernandez E, Grijalvo S, Eritja R, Puras G, Pedraz JL, Gene delivery to the rat retina by nonviral vectors based on chloroquine-containing cationic niosomes, J. Control. Release 304 (2019) 181–190. [DOI] [PubMed] [Google Scholar]
- [19].Bennett J, Ashtari M, Wellman J, Marshall KA, Cyckowski LL, Chung DC, McCague S, Pierce EA, Chen Y, Bennicelli JL, Zhu X, Ying G.-s., Sun J, Wright JF, Auricchio A, Simonelli F, Shindler KS, Mingozzi F, High KA, Maguire AM, AAV2 gene therapy readministration in three adults with congenital blindness, Sci. Transl. Med 4 (120) (2012), 120ra15–120ra15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pierce EA, Bennett J, The status of RPE65 gene therapy trials: safety and efficacy, Cold Spring Harbor Persp. Med 5 (9) (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Federico M, Katherine AH, Immune responses to AAV in clinical trials, Curr. Gene Therapy 11 (4) (2011) 321–330. [DOI] [PubMed] [Google Scholar]
- [22].Zulliger R, Conley SM, Naash MI, Non-viral therapeutic approaches to ocular diseases: an overview and future directions, J. Control. Release 219 (2015) 471–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ye G-J, Budzynski E, Sonnentag P, Nork TM, Sheibani N, Gurel Z, Boye SL, Peterson JJ, Boye SE, Hauswirth WW, Chulay JD, Cone-specific promoters for gene therapy of Achromatopsia and other retinal diseases, Hum. Gene Ther. 27 (1) (2015) 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Beltran WA, Cideciyan AV, Boye SE, Ye G-J, Iwabe S, Dufour VL, Marinho LF, Swider M, Kosyk MS, Sha J, Boye SL, Peterson JJ, Witherspoon CD, Alexander JJ, Ying G-S, Shearman MS, Chulay JD, Hauswirth WW, Gamlin PD, Jacobson SG, Aguirre GD, Optimization of retinal gene therapy for X-linked retinitis Pigmentosa due to RPGR mutations, Mol. Ther. 25 (8) (2017) 1866–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sun D, Schur RM, Sears AE, Gao S-Q, Vaidya A, Sun W, Maeda A, Kern T, Palczewski K, Lu Z-R, Non-viral gene therapy for Stargardt disease with ECO/pRHO-ABCA4 self-assembled nanoparticles, Mol. Ther. 28 (1) (2020) 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sun D, Schur RM, Sears AE, Gao S-Q, Sun W, Naderi A, Kern T, Palczewski K, Lu Z-R, Stable retinoid analogue targeted dual pH-sensitive smart lipid ECO/pDNA nanoparticles for specific gene delivery in the retinal pigment epithelium, ACS Appl. Bio. Mater. 3 (5) (2020) 3078–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ostad-Saffari E, Moosajee M, Wong SP, Tracey-White DC, Tolmachova T, Harbottle RP, Seabra MC, Persistent expression of non-viral S/MAR vectors in the RPE for Choroideremia gene therapy, Invest. Ophthalmol. Vis. Sci. 51 (13) (2010) 3122. [Google Scholar]
- [28].Ragelle H, Danhier F, Préat V, Langer R, Anderson DG, Nanoparticle-based drug delivery systems: a commercial and regulatory outlook as the field matures, Exp. Opin. Drug Deliv 14 (7) (2017) 851–864. [DOI] [PubMed] [Google Scholar]
- [29].Rodrigues GA, Shalaev E, Karami TK, Cunningham J, Slater NKH, Rivers HM, Pharmaceutical development of AAV-based gene therapy products for the eye, Pharm. Res. 36 (2) (2018) 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Clément N, Grieger JC, Manufacturing of recombinant adeno-associated viral vectors for clinical trials, Mol. Therapy Methods Clin. Dev. 3 (2016) 16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Abdelwahed W, Degobert G, Stainmesse S, Fessi H, Freeze-drying of nanoparticles: formulation, process and storage considerations, Adv. Drug Deliv. Rev. 58 (15) (2006) 1688–1713. [DOI] [PubMed] [Google Scholar]
- [32].Fonte P, Reis S, Sarmento B, Facts and evidences on the lyophilization of polymeric nanoparticles for drug delivery, J. Control. Release 225 (2016) 75–86. [DOI] [PubMed] [Google Scholar]
- [33].Ayat NR, Sun Z, Sun D, Yin M, Hall RC, Vaidya AM, Liu X, Schilb AL, Scheidt JH, Lu Z-R, Formulation of biocompatible targeted ECO/siRNA nanoparticles with long-term stability for clinical translation of RNAi, Nucleic Acid Therap 29 (4) (2019) 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bonaccorso A, Musumeci T, Carbone C, Vicari L, Lauro MR, Puglisi G, Revisiting the role of sucrose in PLGA-PEG nanocarrier for potential intranasal delivery, Pharm. Dev. Technol 23 (3) (2018) 265–274. [DOI] [PubMed] [Google Scholar]
- [35].Doktorovova S, Shegokar R, Souto EB, Chapter 30 - Role of Excipients in formulation development and biocompatibility of lipid nanoparticles (SLNs/NLCs), in: Ficai D, Grumezescu AM (Eds.), Nanostructures for Novel Therapy, Elsevier, 2017, pp. 811–843. [Google Scholar]
- [36].Molavi F, Barzegar-Jalali M, Hamishehkar H, Polyester based polymeric nano and microparticles for pharmaceutical purposes: a review on formulation approaches, J. Control. Release 320 (2020) 265–282. [DOI] [PubMed] [Google Scholar]
- [37].Malamas AS, Gujrati M, Kummitha CM, Xu R, Lu Z-R, Design and evaluation of new pH-sensitive amphiphilic cationic lipids for siRNA delivery, J. Control. Release 171 (3) (2013) 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gujrati M, Malamas A, Shin T, Jin E, Sun Y, Lu Z-R, Multifunctional cationic lipid-based nanoparticles facilitate Endosomal escape and reduction-triggered cytosolic siRNA release, Mol. Pharm. 11 (8) (2014) 2734–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sun D, Sahu B, Gao S, Schur RM, Vaidya AM, Maeda A, Palczewski K, Lu ZR, et al. , Mol. Therapy Nucleic Acids 7 (2017) 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sun D, Maeno H, Gujrati M, Schur R, Maeda A, Maeda T, Palczewski K, Lu ZR, Self-assembly of a multifunctional lipid with Core–Shell Dendrimer DNA nanoparticles enhanced efficient gene delivery at low charge ratios into RPE cells, Macromol. Biosci 15 (12) (2015) 1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gujrati M, Vaidya A, Lu Z-R, Multifunctional pH-sensitive amino lipids for siRNA delivery, Bioconjug. Chem 27 (1) (2016) 19–35. [DOI] [PubMed] [Google Scholar]
- [42].Sun D, Schur RM, Lu Z-R, A novel nonviral gene delivery system for treating Leber’s congenital amaurosis, Ther. Deliv 8 (10) (2017) 823–826. [DOI] [PubMed] [Google Scholar]
- [43].Vaidya AM, Sun Z, Ayat N, Schilb A, Liu X, Jiang H, Sun D, Scheidt J, Qian V, He S, Gilmore H, Schiemann WP, Lu Z-R, Systemic delivery of tumor-targeting siRNA nanoparticles against an oncogenic LncRNA facilitates effective triplenegative breast Cancer therapy, Bioconjug. Chem 30 (3) (2019) 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sun D, Sun Z, Jiang H, Vaidya AM, Xin R, Ayat NR, Schilb AL, Qiao PL, Han Z, Naderi A, Lu Z-R, Synthesis and evaluation of pH-sensitive multifunctional lipids for efficient delivery of CRISPR/Cas9 in gene editing, Bioconjug. Chem 30 (3) (2019) 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Brunk UT, Terman A, Lipofuscin: mechanisms of age-related accumulation and influence on cell function12 1Guest Editor: Rajindar S. Sohal 2 This article is part of a series of reviews on “Oxidative Stress and Aging.” The full list of papers may be found on the homepage of the journal, Free Radic. Biol. Med. 33 (5) (2002) 611–619. [DOI] [PubMed] [Google Scholar]
- [46].Radu RA, Mata NL, Nusinowitz S, Liu X, Sieving PA, Travis GH, Treatment with isotretinoin inhibits lipofuscin accumulation in a mouse model of recessive Stargardt’s macular degeneration, Proc. Natl. Acad. Sci. 100 (8) (2003) 4742–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Anhorn MG, Mahler H-C, Langer K, Freeze drying of human serum albumin (HSA) nanoparticles with different excipients, Int. J. Pharm. 363 (1) (2008) 162–169. [DOI] [PubMed] [Google Scholar]
- [48].Piedmonte DM, Summers C, McAuley A, Karamujic L, Ratnaswamy G, Sorbitol crystallization can Lead to protein aggregation in frozen protein formulations, Pharm. Res. 24 (1) (2007) 136–146. [DOI] [PubMed] [Google Scholar]
- [49].Moreau P, Hen R, Wasylyk B, Everett R, Gaub MP, Chambon P, The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants, Nucleic Acids Res. 9 (22) (1981) 6047–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Xu Z-L, Mizuguchi H, Ishii-Watabe A, Uchida E, Mayumi T, Hayakawa T, Strength evaluation of transcriptional regulatory elements for transgene expression by adenovirus vector, J. Control. Release 81 (1) (2002) 155–163. [DOI] [PubMed] [Google Scholar]
- [51].de Villiers J, Olson L, Tyndall C, Schaffner W, Transcriptional ‘enhancers’ from SV40 and polyoma virus show a cell type preference, Nucleic Acids Res. 10 (24) (1982) 7965–7976. [DOI] [PMC free article] [PubMed] [Google Scholar]