Abstract
Essential hypertension (EH) is a prevalent chronic medical condition and a major risk factor for cardiovascular morbidity and mortality. Neurotransmitters are involved in the physiological process of blood pressure regulation in the body. Studies have shown that oleanolic acid (OA) can effectively regulate neurotransmitter-related metabolic disorders caused by EH, but the mechanism is still unclear. Here, we studied the neurotransmitter metabolic profiles in five brain regions by targeted metabolomics approaches in spontaneously hypertensive rats (SHRs) treated with OA and vehicle. Samples from five brain regions (hippocampus, striatum, hypothalamus, temporal lobe, and frontal lobe) were collected from the control group, the spontaneously hypertensive rat (SHR) group, and the OA group. Targeted metabolomics based on UPLC-Q-Exactive-MS was employed to characterize the dramatically changed neurotransmitters in the brain regions of SHRs treated with OA and vehicle. The expressions of the key enzymes involved in the neurotransmitter metabolism were detected by the reverse transcription-polymerase chain reaction (RT-PCR). The metabolomic profiles of SHRs pre-protected by OA were significantly different from those of unprotected SHRs. A total of 18 neurotransmitters could be confirmed as significantly altered metabolites, which were involved in tyrosine and glutamate metabolism as well as other pathways. The results showing seven key enzymes in neurotransmitter metabolism further validated the changes in the metabolic pathways. OA could effectively restore tyrosine metabolism in the striatum and hypothalamus, glutamate metabolism in the hippocampus, striatum and temporal lobe, cholinergic metabolism in the striatum, and histidine metabolism in the hypothalamus due to its inhibition of inflammatory reactions, structural damage of the neuronal cells, and increase in sedative activity. This study indicated that brain region-targeted metabolomics can provide a powerful tool to further investigate the possible mechanism of OA in EH.
Essential hypertension (EH) is a prevalent chronic medical condition and a major risk factor for cardiovascular morbidity and mortality.
1. Introduction
Essential hypertension (EH) is one of the most common cardiovascular diseases in middle-aged and elderly people. It is also a crucial risk factor for cardiovascular and cerebrovascular diseases. According to the WHO study reported in 2004, the number of deaths caused by EH worldwide is 7.5 million per year and the annual loss of disability-adjusted life is ranked 5th.1,2 The molecular mechanism and treatment of EH have received substantial research attention. A large number of studies have confirmed that blood pressure can be regulated by the central nervous system.3–6 The pathogenesis of hypertension often shows dysfunction of the complex network constituted by the nervous system, endocrine system, cardiovascular system, and immune system.7 Recent research has indicated that there are multiple receptors for neurotransmitters in each brain region.8 A variety of neurotransmitters in the brain are the material basis of neuromodulation.9 Neurotransmitters regulate the performance of the cardiovascular system through the activities of the cardiovascular central neurons. In addition, neurotransmitters can directly or indirectly affect the function of the vascular endothelium and proliferation of smooth muscles.10,11 The proliferation of smooth muscle cells plays an important role in cardiovascular proliferative diseases.12 The ventrolateral medulla, nucleus, amygdala, and other limbic structures closely related to this study, such as the striatum, frontal lobe, temporal lobe, hippocampus, and hypothalamus, all have targets for blood pressure regulation.13,14 Studies have shown that antihypertensive effects are related to the levels of neurotransmitters and cytokines in the brain.15,16 However, the dynamic variations in the levels of the neurotransmitters in different brain regions of spontaneously hypertensive rats (SHRs) pre- and post-intervention are still unclear.
Oleanolic acid (OA) is one of the most common biologically active ingredients in plants. It has a wide range of biological activities such as anti-microbial, anti-inflammatory, anti-allergic, anti-diabetes, blood-lipid lowering, and anti-microbial effects. It has been shown to improve learning ability and memory and has other neuroprotective effects.17 Recent studies have shown that OA has important physiological functions for improving the vascular endothelial cell function, such as regulating the vascular tone, maintaining the balance between coagulation and the fibrinolysis system, regulating the inflammatory cell aggregation, and inhibiting the platelet aggregation and activity.18,19 Endothelial cell damage and dysfunction cause many cardiovascular diseases such as atherosclerosis and hypertension. Although OA has been confirmed to cause hypotension and protect the endothelial function, there is no report about its antihypertensive mechanisms in the disorders of neurotransmitter metabolism.
Metabolomics is a technique of collecting and analyzing endogenous metabolites caused by pathophysiological stimulations or genetic alterations in biological systems. It is a novel method to detect metabolite changes comprehensively and holistically in the biological matrix of organisms under disease states or treatments. This makes it especially suitable for the evaluation of drugs.20 At present, the targeted metabolomic technology is much more accurate and sensitive than non-targeted metabolomics. There is no need for any cumbersome process of the identification of metabolites.21 Currently, targeted metabolomics methods are mostly based on triple quadrupole mass spectrometers and Q-Exactive high resolution mass spectrometry. This is becoming more popular for the qualitative and quantitative study of low-abundance compounds such as neurotransmitters.22,23
In this study, targeted metabolomics methods in combination with molecular biology analysis methods were developed. These methods were utilized to investigate the translation of the neurotransmitter metabolic pathway in five brain regions (hippocampus, striatum, hypothalamus, temporal lobe, and frontal lobe) of SHRs treated with OA and vehicle.24,25 This study was designed to provide complete identification of the metabolites associated with the OA treatment, resulting in a more comprehensive understanding of the intervention mechanism of OA on EH.
2. Materials and methods
2.1. Chemicals and reagents
Chromatographic grade isopropanol, acetonitrile, methanol, and formic acid (batch numbers: 172208, 156607, 166817, and 155798) were purchased from Fisher Company, USA. Ultrapure water was purchased from China Watsons Co., Ltd. We obtained 10% chloral hydrate (batch number: 20160925) from Tianjin Damao Chemical Reagent Factory Ltd. Standard OA (batch number: ST004401) was purchased from Shidande Company (Shanghai, China). Standard proline, 5-hydroxyindoleacetic acid, l-citrulline, l-arginine, l-tyrosine, l-ornithine hydrochloride, l-hydroxyproline, l-threonine, l-histidine, homovanillic acid, and l-dopa (batch numbers: A06A6L2156, K10A88B33603, 040M7629V, MKBD3032V, SM0503GE13, NJ0731TC14, S30J6G1, Z26M7H11979, S01F4G1, 060M8201V, and S07J6G1, respectively) were purchased from Yuanye Biotechnology Co., Ltd. (Shanghai, China). Standard l-leucine, l-glutamate, l-glutamine, l-serine, γ-aminobutyric acid, diazepam-d5 solution as the internal standard (IS), choline chloride, and acetylcholine chloride (batch numbers: 13022EH, SLBL8200V, SLBS8600, SLBS8540, BCBT3442, BCBW1702, LRAB0912, and O08J6K1, respectively) were purchased from Sigma Aldrich (Milan, Italy). Standard l-phenylalanine, epinephrine hydrochloride, and histamine (batch numbers: No. 412B021, No. 1225A021, and No. 925A021, respectively) were purchased from Soleil Bao Company (Beijing, China). Standard dopamine hydrochloride (batch number: 40214) was purchased from Dr Ehrenstorfer GmbH (Germany). Analysis grade chloroform and ethanol (batch numbers: 6055847 and 5644885, respectively) were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). The DEPC Water (batch number: 20160925) was purchased from Suo Laibao Biotechnology Co., Ltd. (Shanghai, China). The SYBR Premix Ex Taq™ II (Tli RNaseH Plus), PrimeScript™ RT reagent Kit With gDNA Eraser (Perfect Real Time), and Trizol Total RNA Extraction Kit (Trizol RNA Kit) were purchased from Bao Biological Engineering Co., Ltd. (Dalian, China).
2.2. Experimental animal handling procedure
We purchased 12 male Specific Pathogen-Free SHRs weighing 183.75 ± 11.17 g and 6 male Kyoto Wistar rats weighing 198.47 ± 5.81 g (provided by Beijing Weitong Lihua Experimental Animal Technology Co., Ltd). The Wistar rats were set as the control group, while the 12 SHRs were assigned randomly to two groups of 6 animals each: an SHR group and an OA group (pre-treated with OA at 1.08 mg kg−1, i.g.). The certificate of conformity for these animals was SCXK (Beijing) 2012-0001. The experimental animals were kept in the animal laboratory of Shandong University of Traditional Chinese Medicine, and the laboratory met Good Laboratory Practice requirements. The following conditions were maintained: four rats per cage, laboratory temperature 22 ± 2 °C, humidity 55 ± 5%, and a quiet room. All the rats were allowed to acclimate for 1 week before experiments. This study was performed in strict accordance with the Guide for the Care and Use of Laboratory Animals (published by the US National Institutes of Health) and was approved by the institutional animal care and research advisory committee of the Shandong University of Traditional Chinese Medicine (Jinan, China).
The OA rats were administered their assigned treatments for 28 consecutive days. The control group and the SHR group were administered the same dose of physiological saline. During the experiment, the body weights were measured weekly, and the amount administered was adjusted according to the body weight.
The non-invasive tail–tail method was used to detect the blood pressure of the tail artery in the resting state before OA intervention and 1–4 weeks after intervention. Each rat was continuously measured 3 times, and the average value was taken as the actual measurement value.
The whole rat brains (n = 6 per group) were directly removed following perfusion with physiological saline solutions and decapitation. The hypothalamus, temporal lobe, frontal lobe, hippocampus, and striatum were stored at −80 °C for metabolomic analysis and Reverse Transcription-Polymerase Chain Reaction (RT-PCR) analysis.
2.3. Metabolomics based on LC-MS
2.3.1. Sample preparation
The preparation of brain samples was performed as follows: briefly, for each 10 mg frozen brain sample, a 500 μL pre-chilled (4 °C) acetonitrile–isopropanol–water (3 : 3 : 2, v/v/v) mixture was added, and the mixtures were homogenized for 2 min on ice using a high-throughput tissue grinding machine (Xinzhi Biotechnology Co., Ltd., Ningbo, China). After centrifugation (13 000g, 4 °C) for 15 min, the supernatant was dried at an ambient temperature. The residue was re-dissolved with a 100 μL methanol–water (1 : 1, v/v) mixture. After centrifugation for 15 min, the supernatant was collected into vials for analysis.26 All the parts of the brain tissues were treated in the same way.
2.3.2. Standards and QC samples
The primary reference standard stock solutions of 23 reference substances were used at 1 mg mL−1 concentration. These solutions were stored at −20 °C until use. The stock solutions of diazepam-d5 internal standard and vitamin C (VC) were prepared in methanol : water (1 : 1) at a concentration of 1 mg mL−1 and were stored at −20 °C.
To prepare the standard curve, 10 μL of the IS solution and 10 μL of the specific working solutions were used to generate the calibration samples of 2.5, 5, 10, 50, 200, and 1000 ng mL−1 for l-threonine, histamine, l-serine, l-arginine, 5-hydroxyindoleacetic acid, acetylcholine chloride, l-ornithine hydrochloride, l-proline, and l-citrulline containing 0.1% VC. Six calibration standards of 100, 200, 500, 1000, 2000, and 5000 ng mL−1 were prepared for l-tyrosine, dopamine hydrochloride, l-leucine, l-glutamine, choline chloride, l-phenylalanine, epinephrine hydrochloride, l-histidine, 3,4-dihydroxyphenylacetic acid, l-threonine, homovanillic acid, and levodopa containing 0.1% VC. Six calibration standards of 50, 100, 500, 1000, 5000, and 20 000 ng mL−1 for l-glutamate and γ-aminobutyric acid containing 0.1% VC were prepared. Quality Control (QC) samples were prepared following the same procedure to give low, medium, and high analyte concentrations.
2.4. UPLC-QE-MS analysis
Chromatographic analysis was accomplished on an UltiMate 3000 Ultra High Performance Liquid Chromatograph (Thermo Fisher Scientific, USA) Series HPLC system coupled to a Quadrupole-Electrostatic Field Orbitrap Ultra High Resolution Mass Spectrometer (Thermo Fisher Scientific, USA). Data acquisition was done by the Xcalibur 4.1 software operated in the positive mode.
Chromatographic separation was studied on a Halo C18 column (100 × 2.1 mm, 2.7 μm) from Advanced Materials Technologies (California, USA). The column was kept at 30 ± 0.8 °C. A binary solvent system consisting of 0.05% formic acid in acetonitrile (A) and 0.05% formic acid in water (B) was used for the elution gradient. The following gradient program was used: 0–5 min 4% eluent B, 5–7 min linear gradient increase from 4% to 15% eluent B, 7–9 min linear gradient increase from 15% to 30% eluent B, 9–11 min linear gradient increase from 30 to 50% eluent B, 11–13 min linear gradient increase from 50% to 80% eluent B, 13–14 min linear gradient increase from 80% to 90% eluent B; finally, 90% eluent B was held for 1 min. The flow rate was 200 μL min−1, and the injected volume was 15 μL.
The MS parameters were as follows: capillary voltage 3500 V; capillary temperature 350 °C; sheath gas 45 arb; auxiliary gas 10 arb; source temperature 350 °C; mass spectrometry acquisition range 50–750 m/z; resolution 70 000; S-Lens RF level 50.0.
3. Validation procedures
3.1. Standard curve establishment, detection limit, and limit of quantitation
A series of standard curve samples were taken for continuous injection from low to high concentrations, and LC-MS/MS analyses was performed with 2 injections per concentration. The calibration curves were drawn by plotting the pick area ratio of the analyte to IS versus the nominal concentration of each analyte. The limit of detection (LOD) and the limit of quantitation (LOQ) were determined based on the signal-to-noise ratio, i.e., S/N = 3 and minimum concentration of the standard curve. We adhered to the rule that the limit of quantitation accuracy should be within 80–120% and the day-to-day variation should be less than or equal to 20%.
3.2. Accuracy and precision
Precision and accuracy were evaluated using three concentration points of the analyte: low, medium, and high. Six replicates of each point were analyzed to determine the intra- and inter-day accuracy and precision. This process was repeated over 2 days in order to determine the inter-day accuracy and precision. Accuracy was determined by the recovery of each QC, and precision was expressed as the coefficient of variation of the determination of QCs.
3.3. Stability
In this study, the stability of QC samples was evaluated under different storage conditions. The stability of QC samples was tested after they were stored at 4 °C for 0–24 h. The inter-day precision was determined by analyzing the three concentrations in two replicates on two separate days.
4. Molecular biology verification
The expression of the rate-limiting enzymes dopa decarboxylase, catecholoxymethyltransferase, monoamine oxidase, glutamate decarboxylase, dopamine beta hydroxylase, tyrosine hydroxylase, and acetylcholinesterase (DDC, COMT, MAO, GAD67, DBH, TH, and ACHE) related to neurotransmitter anabolism were detected by RT-PCR.
Total RNA was extracted from the remaining brain tissues of the four rats (n = 4 per group) with Trizol reagents (Bao Bioengineering, Dalian, China) according to the manufacturer's protocols. We homogenized the samples in liquid nitrogen before adding Trizol (1 mL Trizol per 20 mg of the tissue sample). The concentration and purity were measured by a Quawell Q-5000 Ultra Micro UV Visible Spectrophotometer (Quawell Technology Inc., USA). The RNA samples were dissolved in 25 μL of RNase-free H2O and stored at −80 °C until later use. First-strand cDNAs were synthesized by a Promega reverse transcription system (Promega, Madison, WI, USA). The RT-PCR primers (Shenggong, Shanghai, China) used for the above rate-limiting enzymes are listed in Table S1.† Initially, we used a 30 s pre-denaturation step at 95 °C. Each cycle consisted of a 5 s denaturation step at 95 °C, a 34 s annealing step, and an extension step at 60 °C.
5. Data and statistical analysis
The sample delivery to the mass spectrometer was managed through the LC-MS/MS software of Xcalibur, version 4.1 (Thermo Fisher, Massachusetts, USA), which was also used for tuning and data acquisition. The peak integration and data analysis were performed using Xcalibur, version 4.1 and MassFrontier 4.0 (Thermo Fisher, Massachusetts, USA). The area under the peak was used as the analytical signal for the quantitative measurement of the assay performance regarding linearity, sensitivity, and reproducibility.
To study the differences between the three groups, analysis of variance (ANOVA) was used for the data sets that were normally distributed, whereas the Kruskal–Wallis test was applied to the data sets that were not normally distributed. The significance levels for the metabolomics data were considered at p-value < 0.05, and the values were presented as mean ± standard deviation. The chemometric analysis was carried out using principal component analysis (PCA) and orthogonal partial least-squares projection to latent structure-discriminant analysis (OPLS-DA), which was achieved using the SIMCA-P 11.5 software package (Umetrics AB, Umea, Sweden).27,28 OPLS-DA is a supervised multiple regression analysis for discrimination between different datasets. The loading plots from the OPLS-DA models were obtained to identify the variables that strongly contribute to the separation of classes. Potential biomarkers were comprehensively analyzed and filtered by the OPLS-DA loading plots and ANOVA. These were represented with variable importance in the projection values (VIP) > 1 and p-value < 0.05.29 The validation and robustness of each model were evaluated by R2X (cum), R2Y (cum), and Q2 (cum) parameters, cross validation-analysis of variance (CV-ANOVA),30 and the permutation test. The bar graphs were generated using Graph Pad Prism V6. The p-values are represented in the figures as 0.01 (**) and 0.05 (*).
Statistical analyses were performed using SPSS 17.0. ANOVA was used for the data sets that are normally distributed and multiple comparisons were performed using the LSD-t test. The Kruskal–Wallis test was applied to the data sets that are not normally distributed and multiple comparisons were performed using the Dunnett T3 test. To avoid false positive events, the P values were corrected using the false discovery rate method and the fold change (FC) values were combined. These tests were used for the comparison of the content of the metabolites and the rate-limiting enzyme expression among the 3 groups. The differences between the groups were considered statistically significant at P < 0.05 and FC > 2. The results are presented as mean ± standard deviation. The error bars correspond to the standard deviation.
6. Results
6.1. Validation of quality control and the analytical method
The system stability during the instrumental analysis process was evaluated using the QC samples. The variances of the first principal component of all the QC samples were no more than two-fold of the standard deviations, indicating that the analysis system was stable and all the data were under control.
The correlation coefficient was >0.99, which revealed that the method was highly linear (Table S2†). The limits of detection ranged from 0.1 ng mL−1 to 1.5 ng mL−1 for different neurotransmitters. The intra- and inter-day precision and stability were assessed and the data are listed in Tables S3 and S4.† The results showed that the reproducibility levels of the method (coefficient of variations < 26.38%) for all the neurotransmitters were satisfactory. The recovery was assessed and the data are listed in Tables S3 and S4.† The mean recovery ranged from 70.1% to 126.31%. The results showed very quantitative recoveries for all the neurotransmitters.
6.2. Blood pressure levels
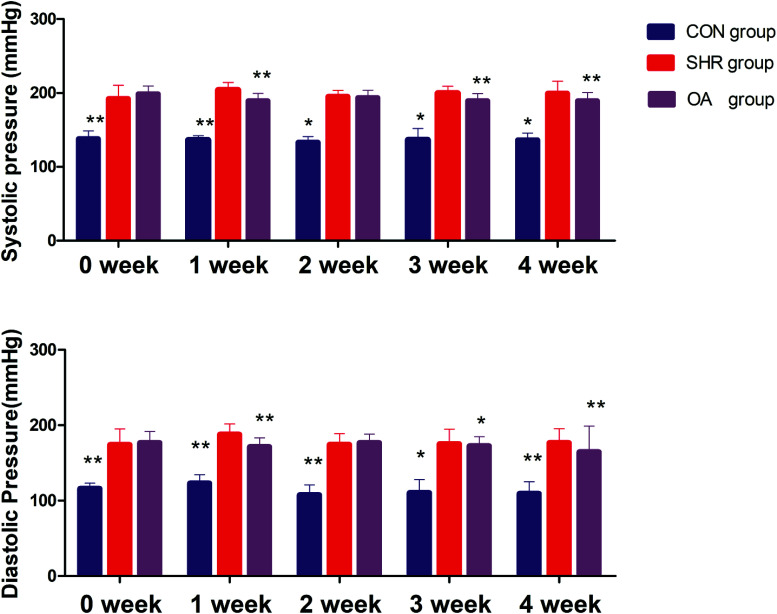
Before intervention, the systolic blood pressure and diastolic blood pressure of the model group and OA group were significantly higher than those of the control group (P < 0.01). After OA intervention, compared with the results for the model group, the systolic blood pressure and the diastolic blood pressure showed a downward trend except for that at the second week (P < 0.05, P < 0.01) (Fig. 1).
Fig. 1. Systolic blood pressure and diastolic blood pressure before and after 1–4 weeks of intervention (*P < 0.05, **P < 0.01 compared to the SHR group).
6.3. Analysis of brain metabolite profiling
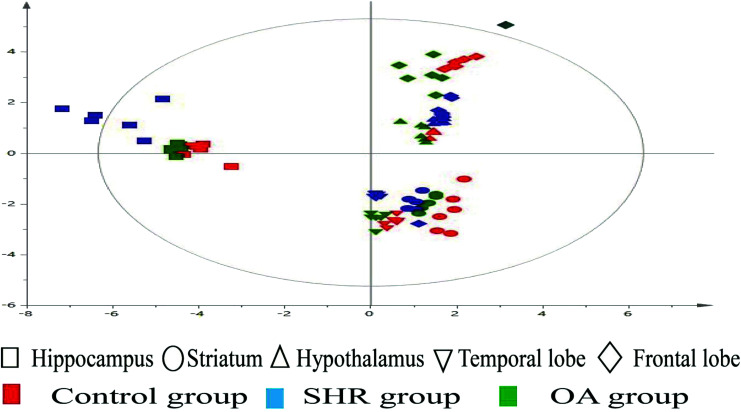
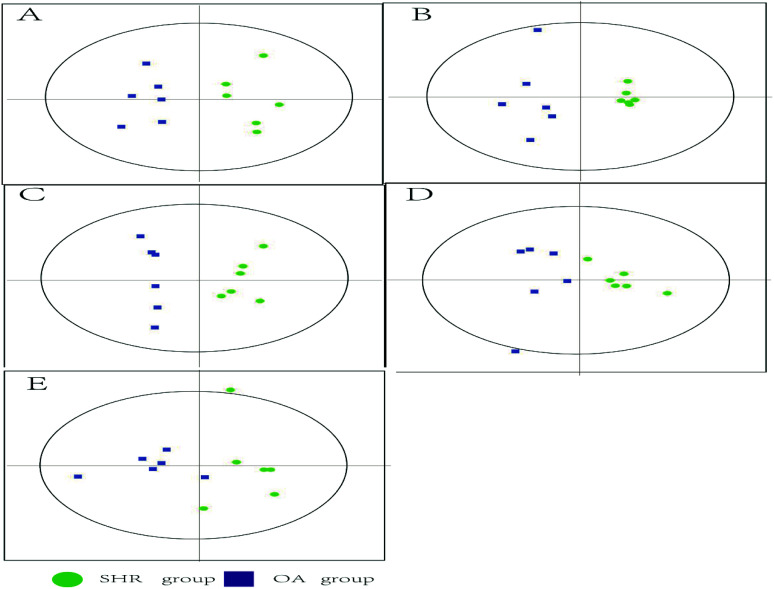
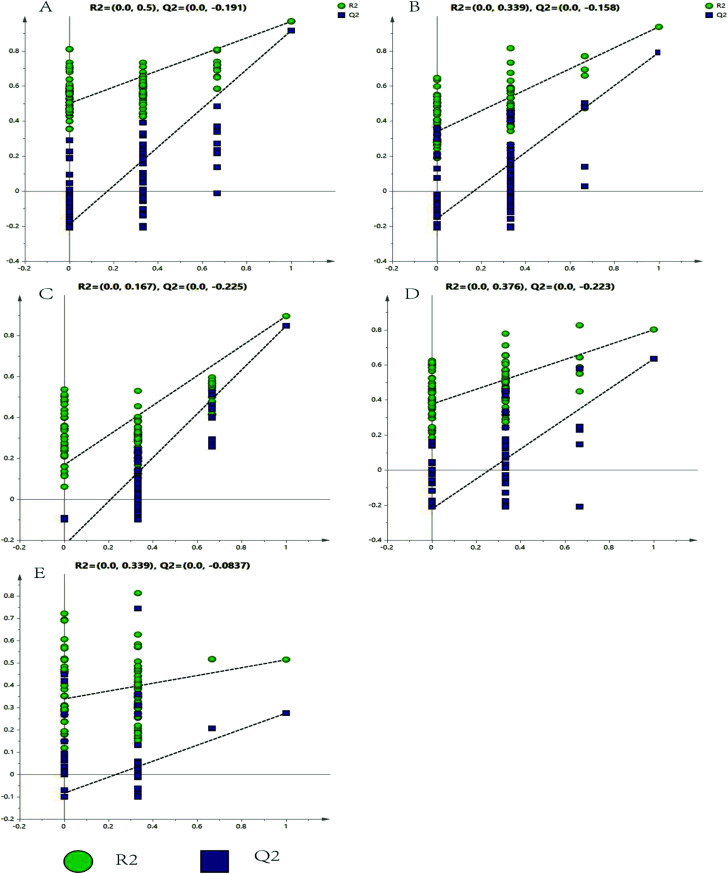
The representative LC/MS total ion current chromatograms of each brain region from the SHR group, OA group, and control group are displayed in Fig. S1A–E.† Univariate analysis revealed that differential neurotransmitters existed among the three groups in the five brain regions. Subsequently, the data were analyzed in order to identify the potential biomarkers using multivariate statistics including PCA and OPLS-DA. Initially, a PCA model was obtained from the LC/MS data of all the brain tissue samples (Fig. 2). A 2D-PCA score plot (PC1 vs. PC2) was used to reflect the hypertension-associated metabolic differences among the brain regions. The subjects naturally clustered into five broad classes, indicating that the metabolic alterations were strongly correlated with the brain tissue region. To better understand the hypertension-associated metabolic alterations by the brain region, the resulting data were divided into five subsets by the brain regions. A clear separation of the five brain regions' metabolic profiles between the SHR group and the OA group was observed, indicating that significant metabolic perturbations were treated by OA in these five brain regions. Using an OPLS-DA model, a supervised projection method, we verified the class separation for each brain region (Fig. 3A–E). In particular, we confirmed the robustness of the model through cross-validation, and the model parameters R2X, R2Y, and Q2 were all within the requirements. Finally, the results of CV-ANOVA variant suggested that the models were significant with the corresponding p values (Fig. 4A–E, Table 1).
Fig. 2. 2D-PCA scores map (PC1 versus PC2) of the LC/MS data derived from all the brain regions of the control, SHR, and OA groups (n = 6 per group).
Fig. 3. 2D cross-validated OPLS-DA score plot of the LC/MS data derived from (A) hippocampus, (B) striatum, (C) hypothalamus, (D) temporal lobe, and (E) frontal lobe obtained from the SHR and OA group brain tissues (n = 6 per group). The OPLS-DA score plots show a clear discrimination between the SHR and OA groups in all five brain regions. Each dot represents an individual sample.
Fig. 4. Validated model plots of the LC/MS data derived from the SHR group and the OA group: (A) hippocampus, (B) striatum, (C) hypothalamus, (D) temporal lobe, and (E) frontal lobe.
The model parameters R2X, R2Y, and Q2 and the CV-ANOVA results of the five brain regions.
| Hippocampus | Striatum | Hypothalamus | Temporal lobe | Frontal lobe | |
|---|---|---|---|---|---|
| R2X | 0.728 | 0.701 | 0.579 | 0.602 | 0.622 |
| R2Y | 0.723 | 0.948 | 0.905 | 0.961 | 0.848 |
| Q2 | 0.555 | 0.862 | 0.837 | 0.846 | 0.51 |
| P | 0.0472972 | 0.00392291 | 0.00032891 | 0.0479591 | 0.0228696 |
6.4. Comparison of different brain metabolites
After the validation of the analytical method, the analysis of real samples was performed. From the targeted experiments, 18 of the 23 neurotransmitters were detected in the brain samples with the exceptions of methionine, hydroxyproline, histamine, serine, and threonine. The corresponding MS parameters and retention times of 23 measured neurotransmitters are listed in Table S5.† To monitor the metabolic changes in the circulating neurotransmitters in the 3 groups of rats more comprehensively and accurately, the absolute concentrations of the other 18 neurotransmitters in the brain samples were quantified through targeted metabolomics analysis using a special pretreatment protocol.
As shown in Table S6,† compared to the control group, the SHR group demonstrates significant decrease in tyrosine, ornithine, proline, and arginine in the hippocampus and hypothalamus and a reverse trend is seen in the OA intervention. The contents of epinephrine, dopamine, and homovanillic acid increased significantly in the SHR group in the striatum and hypothalamus, whereas levodopa was found only in the striatum. The above metabolites exhibited tendencies to respond to normal groups after OA intervention. Besides, the histamine and histidine levels showed opposite trends in the hypothalamus in SHRs treated with OA and vehicle.
The contents of acetylcholine and choline were elevated in the striatum and temporal lobe in the SHR group compared with the results for the control group. After OA intervention, the levels of the neurotransmitters mentioned above tended to return to normal levels. In addition, only in the frontal lobe, 5-hydroxyindoleacetic acid showed an obvious upward trend after OA intervention.
6.5. Biomarkers in brain analysis and identification of the effects of OA on SHRs
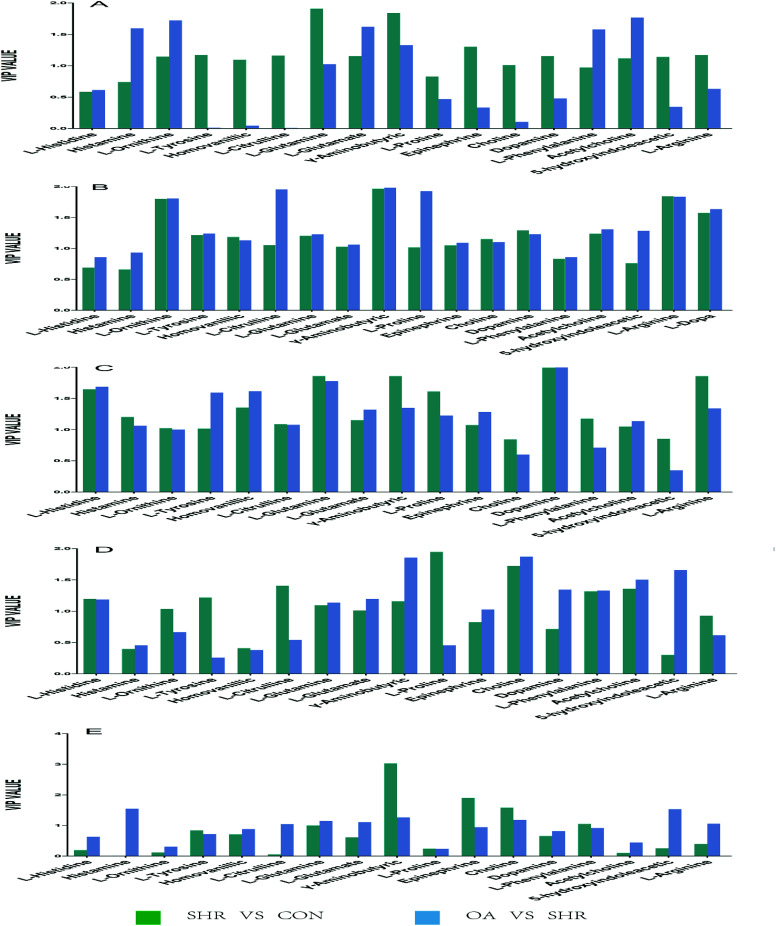
Based on the variable importance in the projection value threshold from the OPLS-DA models of the five brain regions, 18 significant variables were ultimately obtained. ANOVA, with an adjusted p value of 0.05 and FC value of 2, was performed to determine the significant differences on a metabolite-by-metabolite basis. Metabolomics profiling revealed significant perturbations. All the significant metabolites responding to EH and OA intervention were quantified and summarized (Tables 2, 3, S6 and Fig. 5).
Potential biomarkers discovered by the adjusted p values in the five brain regions.
| Hippocampus | Striatum | Hypothalamus | Temporal lobe | Frontal lobe | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Adjusted p value | Adjusted p value | Adjusted p value | Adjusted p value | p value | ||||||
| CON/SHR | OA/SHR | CON/SHR | OA/SHR | CON/SHR | OA/SHR | CON/SHR | OA/SHR | CON/SHR | OA/SHR | |
| l-Histidine | 0.582 | 0.815 | 0.035 | 0.028 | 0.05 | 0.046 | 0.009 | 0.589 | 0.158 | 1.82 |
| Histamine | 0.194 | 0.127 | 0.532 | 0.045 | 0.015 | 0.043 | 0.916 | 0.922 | 0.855 | 0.203 |
| Ornithine | 0.009 | 0.005 | 0.038 | 0.037 | 0.042 | 0.025 | 0.049 | 0.829 | 0.025 | 1.49 |
| l-Tyrosine | 0.046 | 0.038 | 0.000 | 0.009 | 0.002 | 0.013 | 0.525 | 0.029 | 0.95 | 1.568 |
| Homovanillic acid | 0.000 | 0.712 | 0.000 | 0.000 | 0.013 | 0.024 | 0.82 | 0.095 | 0.13 | 0.924 |
| Citrulline | 0.013 | 0.036 | 0.000 | 0.005 | 0.007 | 0.038 | 0.008 | 0.888 | 0.881 | 0.729 |
| l-Glutamine | 0.000 | 0.000 | 0.000 | 0.000 | 0.041 | 0.453 | 0.001 | 0.042 | 0.05 | 0.555 |
| l-Glutamate | 0.000 | 0.018 | 0.002 | 0.000 | 0.000 | 0.004 | 0.013 | 0.033 | 1.325 | 1.291 |
| γ-Aminobutyric acid | 0.034 | 0.009 | 0.000 | 0.007 | 0.049 | 0.967 | 0.029 | 0.049 | 0.012 | 1.049 |
| l-Proline | 0.025 | 0.000 | 0.000 | 0.015 | 0.003 | 0.000 | 0.05 | 0.892 | 0.55 | 1.324 |
| Epinephrine | 0.992 | 0.589 | 0.000 | 0.008 | 0.005 | 0.000 | 0.009 | 0.014 | 1.29 | 1.493 |
| Choline | 0.000 | 0.922 | 0.001 | 0.002 | 0.047 | 0.882 | 0.042 | 0.046 | 0.005 | 1.998 |
| Dopamine | 0.000 | 0.833 | 0.000 | 0.005 | 0.005 | 0.014 | 0.589 | 0.539 | 0.023 | 0.851 |
| l-Phenylalanine | 0.000 | 0.009 | 0.042 | 0.82 | 0.036 | 0.168 | 0.045 | 0.962 | 0.012 | 0.852 |
| Acetylcholine | 0.000 | 0.000 | 0.006 | 0.013 | 0.018 | 0.032 | 0.009 | 0.001 | 0.653 | 0.924 |
| 5-Hydroxyindoleacetic acid | 0.000 | 0.107 | 0.58 | 1.98 | 0.016 | 0.991 | 1.277 | 0.002 | 0.045 | 0.007 |
| l-Arginine | 0.000 | 0.000 | 0.009 | 0.032 | 0.023 | 0.004 | 0.049 | 0.853 | 0.86 | 1.222 |
| l-Dopa | — | — | 0.049 | 0.045 | — | — | — | — | — | — |
Potential biomarkers discovered by the FC values in the five brain regions.
| Hippocampus | Striatum | Hypothalamus | Temporal lobe | Frontal lobe | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FC value | FC value | FC value | FC value | FC value | ||||||
| CON/SHR | OA/SHR | CON/SHR | OA/SHR | CON/SHR | OA/SHR | CON/SHR | OA/SHR | CON/SHR | OA/SHR | |
| l-Histidine | 3.558 | 2.202 | 3.846 | 2.466 | 3.841 | 3.953 | 3.526 | 2.026 | 0.927 | 2.126 |
| Histamine | 2.922 | 2.628 | 2.194 | 2.225 | 3.728 | 3.25 | 2.299 | 3.051 | 2.738 | 1.178 |
| Ornithine | 2.462 | 2.796 | 3.813 | 3.648 | 2.24 | 3.269 | 2.488 | 2.119 | 2.863 | 2.781 |
| l-Tyrosine | 2.785 | 2.884 | 2.075 | 2.518 | 2.282 | 3.853 | 3.341 | 2.645 | 2.789 | 3.201 |
| Homovanillic acid | 2.06 | 2.748 | 2.236 | 2.501 | 3.433 | 2.357 | 2.771 | 3.231 | 2.458 | 3.974 |
| Citrulline | 3.376 | 2.527 | 3.812 | 1.65 | 3.491 | 2.878 | 2.102 | 3.553 | 3.144 | 1.764 |
| l-Glutamine | 1.625 | 3.675 | 2.478 | 2.806 | 2.719 | 2.807 | 3.131 | 2.2 | 3.182 | 2.53 |
| l-Glutamate | 2.092 | 3.72 | 2.099 | 3.886 | 2.054 | 3.331 | 3.525 | 2.639 | 3.191 | 2.567 |
| γ-Aminobutyric acid | 2.418 | 4.731 | 2.085 | 2.988 | 2.231 | 2.982 | 2.377 | 3.192 | 3.183 | 2.615 |
| l-Proline | 2.434 | 2.102 | 2.493 | 2.901 | 4.395 | 2.41 | 2.435 | 2.339 | 2.471 | 1.977 |
| Epinephrine | 2.991 | 3.928 | 3.881 | 2.222 | 3.683 | 3.932 | 3.976 | 3.319 | 1.151 | 2.895 |
| Choline | 3.503 | 3.214 | 2.9 | 3.796 | 2.789 | 1.612 | 2.377 | 2.205 | 2.05 | 2.6 |
| Dopamine | 2.065 | 3.836 | 3.203 | 2.985 | 2.283 | 2.855 | 3.055 | 2.294 | 3.243 | 3.563 |
| l-Phenylalanine | 3.375 | 1.307 | 1.308 | 2.765 | 3.073 | 2.53 | 0.779 | 2.518 | 2.224 | 2.317 |
| Acetylcholine | 3.934 | 2.721 | 4.831 | 1.534 | 2.179 | 3.708 | 2.433 | 3.145 | 2.843 | 2.264 |
| 5-Hydroxyindoleacetic acid | 2.536 | 1.049 | 2.102 | 1.28 | 2.075 | 2.015 | 2.7 | 3.984 | 3.793 | 2.432 |
| l-Arginine | 3.376 | 2.303 | 3.099 | 2.69 | 3.958 | 3.302 | 3.103 | 3.012 | 1.661 | 3.095 |
| l-Dopa | — | — | 2.926 | 3.541 | — | — | — | — | — | — |
Fig. 5. Potential biomarkers discovered by VIP values in the five brain regions: (A) hippocampus, (B) striatum, (C) hypothalamus, (D) temporal lobe, and (E) frontal lobe.
6.6. RT-PCR results
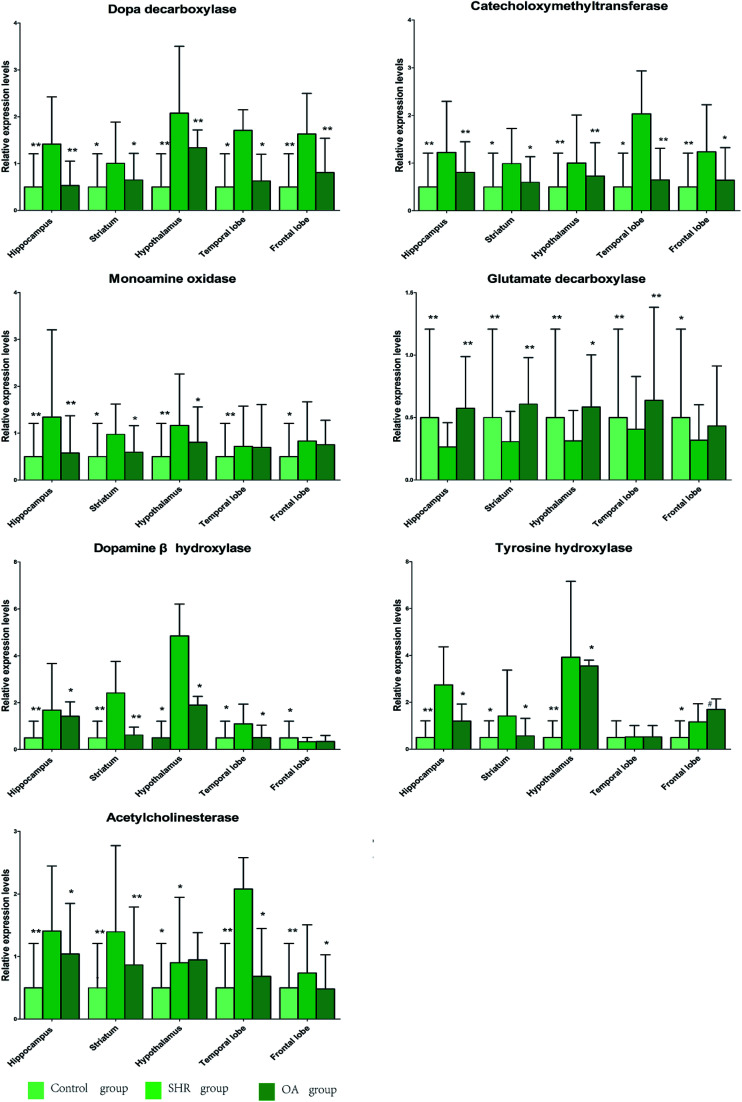
The results of 7 RT-PCR from five rat brains are listed in Fig. 6. As shown in the figure, the levels of COMT, DβH, MAO, TH, ACHE, and DDC mRNA expressions significantly increase in the SHR group in the five brain regions. These mRNA levels were significantly suppressed by OA, while GAD67 exhibited an increasing trend. The metabolism-related enzyme results are consistent with those of the neurotransmitter metabolites.
Fig. 6. The relative expression levels of DDC, COMT, MAO, GAD67, DβH, TH, and ACHE in the five brain regions of rats treated with OA (P < 0.05, **P < 0.01 compared to the model group).
7. Discussion
EH is one of the major causes of death worldwide. It is associated with a variety of pathological disorders including neurotransmitter changes, disordered angiotensin levels, impaired endothelial functions, and overexcited sympathetic nerves as well as other processes such as inflammatory factors and oxidative stress.31–33 OA has a significant antihypertensive effect on hypertensive animals, and its antihypertensive mechanism mainly involves the inhibition of vascular smooth muscle cell proliferation and inflammation of the blood vessel wall, thus reducing blood vessel stenosis caused by balloon catheter injury and other causes.34–36 Despite extensive studies, its mechanism associated with neuropathology remains completely unclear.
In order to monitor the systemic metabolic changes in SHR brains treated with OA, an LC/MS-based metabolomics approach was employed to characterize the metabolites present in five selected brain regions: the hippocampus, striatum, hypothalamus, frontal lobe, and temporal lobe. In this study, 18 significantly changed neurotransmitters related to EH were defined. These biomarkers have been reported to participate in the pathogenesis of hypertension and are closely related to the multiple etiologies and pathogenesis of the related diseases. Interestingly, not all metabolites demonstrated significant differences consistently across all brain regions, which suggested that the metabolic variations in response to EH were correlated with different brain regions. As discussed below, several metabolic pathways were perturbed in the EH brain tissues: most notably, the tyrosine metabolism, cholinergic metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, histidine metabolism, arginine and proline metabolism, and glutamate metabolism. After treatment with OA, the disturbed metabolic profiles were restored back to control-like levels, which suggested that the protective effects of OA in hypertension progress are exerted through these pathways.
7.1. Tyrosine metabolism
Tyrosine and its metabolites play critical roles in the development of EH.
The hypothalamic–pituitary–adrenal (HPA) axis and the sympathetic center are important pathways for blood pressure regulation.37 In addition, the hypothalamus is the sympathetic center and the central axis of the HPA axis.38 The striatum can regulate blood pressure through the frontal cortex–striatum–cerebellar nerve regulation pathway.39,40 Under stress conditions, EH can break down into tyrosine, impairing the cell function and causing damage to the vascular endothelium.41 After EH, the levels of tyrosine in the striatum and hypothalamus are markedly lower, while OA can increase them, which has been recognized as the regulatory signal implicated in the control of metabolism, oxidative stress response, and other cellular functions. The metabolite content is affected by synthetase and decomposing enzymes. MAO is a key enzyme in the synthesis of tyrosine. It is mainly found in adrenergic neurons.42,43 Also, major increased MAO and minor increased TH expose tyrosine sharply reduced and increase the content of levodopa.44,45 Levodopa is a precursor of synthetic catecholamines.46 A high content of levodopa exacerbates the synthesis of catecholamines through significantly increased DDC, DβH, MAO, and COMT levels.47 Catecholamines mainly promote the structural damage of the neuronal cells through sympathetic activity to raise the blood pressure.48,49 All the catecholamines were down-regulated after OA, indicating that OA could appropriately regulate the source of catecholamine synthesis.
7.2. Glutamate metabolism
Glutamate, glutamine, and γ-GABA are of great significance in the maintenance of normal brain function for blood pressure regulation. The hippocampus regulates the blood pressure mainly through the negative feedback mechanism of the HPA axis.50 As a vital component of the basal ganglia, the blood pressure regulation pathway of the striatum is a cortical–basal–thalamic–cortical loop.51 Glutamate and γ-GABA, as excitatory and inhibitory neurotransmitters, have been identified as the top two neurotransmitters that cause EH.52 A high content of glutamate can activate Ca2+ in the endothelial cells to protect the ischemic myocardium, inhibiting increasing secondary damage to the nerve function.53 In the present study, in the SHR group, glutamate and glutamine in the temporal lobe, striatum, and hippocampus were significantly higher than those in the control group, while γ-GABA showed the opposite trend; OA could regulate them effectively. Glutamate could ultimately transform into γ-aminobutyric acid through a series of reactions accompanied by GAD67.54 γ-GABA plays an important role in cardiovascular diseases since its decreased content enhances the angiotensin-converting enzyme activity.55 In addition, γ-GABA is an inhibitory neurotransmitter found in the nervous system; it is used as a sedative.56 In this study, we found that GABA decreased in SHRs, while OA could elevate it, which indicated that it may inhibit the conversion of angiotensin and increase the sedative activity to exert the intervention effect.
7.3. Cholinergic metabolism
Both acetylcholine and choline are synthesized and released in the dendrites of the nigrostriatal cells.57 EH is characterized by severe inflammatory responses to infection. The levels of acetylcholine and choline in the striatum increased in the SHR group compared with that in the control group, which was consistent with the activities of ACHE, a synthetic enzyme used to synthesize acetylcholine. The acetylcholine content increased due to the pro-inflammatory responses from increasing the expression of factors such as the cell adhesion factor and angiotensin II in EH.58 Meanwhile, as important components of the cholinergic anti-inflammatory pathway and inflammatory mediators in the acute phase, the specifically changed levels of acetylcholine might reflect the well-known anti-inflammatory effects of OA.59,60 The metabolites and synthetases returned to normal levels after OA intervention. The unique function of OA in affecting the levels of acetylcholine and choline demonstrated its exceptional anti-inflammatory effects possibly through its regulation of the cholinergic anti-inflammatory pathway, which would favorably protect the body against the EH-induced uncontrolled systemic inflammatory response.
7.4. Histidine metabolism
The hypothalamic nodule nucleus is the main site for the synthesis of histidine and histamine.61 Histidine is a common amino acid neurotransmitter and it is involved in clearing intracellular oxides and improving oxidative stress; moreover, it has anti-inflammatory activity.62,63 The mechanism for this is that it can affect the arachidonic acid metabolism and reduce the prostaglandin synthesis as prostaglandin is an important inflammatory factor causing vasoconstriction.64,65 We saw that the level of histidine in SHRs in the hypothalamus increased remarkably, which could be significantly lowered by OA, thus indicating positive feedback regulation induced by the inflammatory and oxidative stress responses during EH. Histamine is one of the earliest discovered inflammatory mediators and it is produced by the decarboxylation of histidine.66 As compared with the control group, the SHR group showed a significantly increased level of histamine, which could be only effectively decreased by the OA treatment, thus showing the exceptional anti-oxidative and anti-inflammatory abilities of OA.
8. Concluding remarks
This is the first study of the antihypertensive effect of OA from the perspective of metabolic regulation. The use of SHRs pre-treated with OA uncovered the antihypertensive effect of OA on neurotransmitters. In conclusion, OA could affect SHRs mainly via affecting the tyrosine metabolism in the striatum and hypothalamus. Glutamate metabolism activities in the hippocampus, striatum, and temporal lobe were also affected. Significant neurotransmitters and neurotransmitter rate-limiting enzymes were confirmed in the SHRs treated with OA and vehicle by metabolomics based on LC/MS. These regulation effects were closely related to its inhibition of the inflammatory reaction, structural damage of the neuronal cells, and increase in the sedative activity. Further investigations of the affected metabolic pathways that respond to OA treatment are necessary.
Conflicts of interest
On behalf of all the authors, I declare that we have no conflicts of interest.
Abbreviations
- EH
Essential hypertension
- OA
Oleanolic acid
- SHRs
Spontaneously hypertensive rats
- RT-PCR
Reverse transcription-polymerase chain reaction
- ANOVA
Analysis of variance
- PCA
Principal component analysis
- OPLS-DA
Orthogonal partial least-squares projection to latent structure-discriminant analysis
- QC
Quality control
- LOD
Limit of detection
- LOQ
Limit of quantitation
- DDC
Dopa decarboxylase
- COMT
Catecholoxymethyltransferase
- MAO
Monoamine oxidase
- GAD67
Glutamate decarboxylase
- DBH
Dopamine beta hydroxylase
- TH
Tyrosine hydroxylase
- ACHE
Acetylcholinesterase
- FC
Fold change
- HPA
Hypothalamic–pituitary–adrenal
- VC
Vitamin C
Supplementary Material
Acknowledgments
This work was founded by foundation (No. 81473653, 81774173) from the National Natural Science Foundation of China, (No. 2017CXGC1307) project funded by Shandong Province Major Scientific and Technological Innovation and (No. 2018GSF119007) Shandong Province Key Research and Development Plan.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra02377a
Notes and references
- Mills K. T. Bundy J. D. Kelly T. N. Reed J. E. Kearney P. M. Reynolds K. Chen J. He J. Circulation. 2016;134:441. doi: 10.1161/CIRCULATIONAHA.115.018912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing W. Saleh M. A. Kirabo A. Itani H. A. Montaniel K. R. C. Liang X. Wei C. Mernaugh R. L. Hua C. Bernstein K. E. J. Clin. Invest. 2016;126:50–67. doi: 10.1172/JCI80761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan Talukder W. M. J. M. A. Varadharaj S. Lian J. Kearns P. N. El-Mahdy M. A. Liu X. Zweier J. L. Am. J. Physiol.: Heart Circ. Physiol. 2016;300:H388–H396. doi: 10.1152/ajpheart.00868.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi G. Ram V. S. J. Am. Soc. Hypertens. 2016;10:457–466. doi: 10.1016/j.jash.2016.02.015. [DOI] [PubMed] [Google Scholar]
- Rubin L. J. Eur. Respir. J. 2016;48:617. doi: 10.1183/13993003.01333-2016. [DOI] [PubMed] [Google Scholar]
- Xu Q. Jensen D. D. Hua P. Feng Y. Pharmacol. Ther. 2016;164:126–134. doi: 10.1016/j.pharmthera.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimov L. O. Ershov N. I. Efimov V. M. Markel A. L. Redina O. E. BMC Genet. 2016;17:S13. doi: 10.1186/s12863-015-0307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique S. A. Tamilselvan T. Vishnupriya M. Balamurugan E. Neurochem. Res. 2018;43:1–11. doi: 10.1007/s11064-018-2533-8. [DOI] [PubMed] [Google Scholar]
- Sparks D. W. Tian M. K. Sargin D. Venkatesan S. Intson K. Lambe E. K. Front. Neural Circuits. 2017;11 doi: 10.3389/fncir.2017.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B. Zhao Y. N. Wang X. Yu X. J. Li Y. Yang H. Y. Su Q. Kang Y. M. Yang Z. M. RSC Adv. 2018;8 doi: 10.1039/C7RA09136B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S. Nagy J. A. Pal S. Vasile E. Eckelhoefer I. A. Bliss V. S. Manseau E. J. Dasgupta P. S. Dvorak H. F. Mukhopadhyay D. Nat. Med. 2001;7:569–574. doi: 10.1038/87895. [DOI] [PubMed] [Google Scholar]
- Perassa L. A. Graton M. E. Potje S. R. Troiano J. A. Lima M. S. Vale G. T. Pereira A. A. Nakamune A. C. Sumida D. H. Tirapelli C. R. Vasc. Pharmacol. 2016;87 doi: 10.1016/j.vph.2016.06.005. doi: 10.1016/j.vph.2016.06.005. [DOI] [PubMed] [Google Scholar]
- Wells A. M. Janes A. C. Liu X. Deschepper C. F. Kaufman M. J. Kantak K. M. Hippocampus. 2010;20:787–797. doi: 10.1002/hipo.20681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y. Gao H. M. Int. J. Clin. Exp. Med. 2015;8:19471. [PMC free article] [PubMed] [Google Scholar]
- Yi L. T. Li J. Liu Q. Geng D. Zhou Y. F. Ke X. Q. Chen H. Weng L. J. J. Psychopharmacol. 2013;27:459–468. doi: 10.1177/0269881112467090. [DOI] [PubMed] [Google Scholar]
- Vukelic M. Qing X. Redecha P. Koo G. Salmon J. E. J. Immunol. 2013;191:1800–1807. doi: 10.4049/jimmunol.1203467. [DOI] [PubMed] [Google Scholar]
- Kashyap D. Sharma A. Tuli H. S. Punia S. Sharma A. K. Recent Pat. Inflammation Allergy Drug Discovery. 2016;10 doi: 10.2174/1872213X10666160711143904. [DOI] [PubMed] [Google Scholar]
- Long C. Yang J. Yang H. Li X. Wang G. Mol. Med. Rep. 2016;13 doi: 10.3892/mmr.2016.5128. [DOI] [PubMed] [Google Scholar]
- Bachhav S. S. Patil S. D. Bhutada M. S. Surana S. J. Phytother. Res. 2011;25:1435–1439. doi: 10.1002/ptr.3431. [DOI] [PubMed] [Google Scholar]
- Wishart D. S. Nat. Rev. Drug Discovery. 2016;15:473–484. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- Varma V. R. Oommen A. M. Varma S. Casanova R. An Y. Andrews R. M. O'Brien R. Pletnikova O. Troncoso J. C. Toledo J. PLoS Med. 2018;15:e1002482. doi: 10.1371/journal.pmed.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha H. Cai Y. Yin Y. Wang Z. Li K. Zhu Z. J. Anal. Chem. 2018;90 doi: 10.1021/acs.analchem.7b05318. doi: 10.1021/acs.analchem.7b05318. [DOI] [PubMed] [Google Scholar]
- Bingol K. High-Throughput. 2018;7:9. doi: 10.3390/ht7020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J. X. Xia J. J. Deng F. L. Liang W. W. Wu J. Yin B. M. Dong M. X. Chen J. J. Ye F. Wang H. Y. Transl. Psychiatry. 2018;8:130. doi: 10.1038/s41398-018-0183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob M. Malkawi A. Albast N. Al S. B. Lopata A. Dasouki M. Abdel A. R. Anal. Chim. Acta. 2018 doi: 10.1016/j.aca.2018.03.058. [DOI] [PubMed] [Google Scholar]
- Feng L. Yue X. F. Chen Y. X. Liu X. M. Wang L. S. Cao F. R. Wang Q. Liao Y. H. Pan R. L. Chang Q. J. Pharm. Biomed. Anal. 2016;125:329–338. doi: 10.1016/j.jpba.2016.04.002. [DOI] [PubMed] [Google Scholar]
- Katajamaa M. Orešič M. J. Chromatogr. A. 2007;1158:318–328. doi: 10.1016/j.chroma.2007.04.021. [DOI] [PubMed] [Google Scholar]
- Want E. Masson P. Methods Mol. Biol. 2011;708:277–298. doi: 10.1007/978-1-61737-985-7_17. [DOI] [PubMed] [Google Scholar]
- Benito S. Sánchez A. Unceta N. Andrade F. Aldámiz-Echevarria L. Goicolea M. A. Barrio R. J. Anal. Bioanal. Chem. 2016;408:747–760. doi: 10.1007/s00216-015-9153-9. [DOI] [PubMed] [Google Scholar]
- Eriksson L. Trygg J. Wold S. J. Chemom. 2008;22:594–600. doi: 10.1002/cem.1187. [DOI] [Google Scholar]
- Edwards E. Dipette D. J. Trends Cardiovasc. Med. 2016;26:707–708. doi: 10.1016/j.tcm.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Gonzalez A. A. Lara L. S. Prieto M. C. Curr. Hypertens. Rep. 2017;19:62. doi: 10.1007/s11906-017-0763-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R. K. Oliveira A. C. Kim S. Rigatto K. Zubcevic J. Rathinasabapathy A. Kumar A. Lebowitz J. J. Khoshbouei H. Lobaton G. Hypertension. 2018;71 doi: 10.1161/HYPERTENSIONAHA.118.10934. doi: 10.1161/HYPERTENSIONAHA.118.10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D. Zhang X. Zhang J. Guo X. Zheng Y. Sui S. Zheng J. Oncol. Lett. 2016;12:3519. doi: 10.3892/ol.2016.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlala H. P. Metzinger T. Heerden F. R. V. Musabayane C. T. Mubagwa K. Dessy C. PLoS One. 2016;11:e0147395. doi: 10.1371/journal.pone.0147395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. Jiang Q. Wang Y. Li W. Geng M. Han Z. Chen X. Cell Biol. Int. 2017 doi: 10.1002/cbin.10838. [DOI] [PubMed] [Google Scholar]
- Daimon M. Kamba A. Murakami H. Takahashi K. Otaka H. Makita K. Yanagimachi M. Terui K. Kageyama K. Nigawara T. J. Clin. Endocrinol. Metab. 2016;101:889–897. doi: 10.1210/jc.2015-3568. [DOI] [PubMed] [Google Scholar]
- Schommer N. C. Hellhammer D. H. Kirschbaum C. Psychosom. Med. 2003;65:450–460. doi: 10.1097/01.PSY.0000035721.12441.17. [DOI] [PubMed] [Google Scholar]
- Gao C. L. Xu H. E. Xia Z. K. Ren X. G. Chun-Lei W. U. Liu G. L. J. Med. Postgrad. 2010;23(1):35–38. [Google Scholar]
- Yousfi-Alaoui M. A. Hospital S. Garcia-Sanz A. Badia A. Clos M. V. Neurochem. Res. 2001;26:1271–1275. doi: 10.1023/A:1014237215200. [DOI] [PubMed] [Google Scholar]
- Kumai T. Asoh K. Tateishi T. Tanaka M. Watanabe M. Shimizu H. Kobayashi S. Life Sci. 2000;67:1993–1999. doi: 10.1016/S0024-3205(00)00787-6. [DOI] [PubMed] [Google Scholar]
- Cheah T. B. Geffen L. B. J. Neurobiol. 2010;4:443–452. doi: 10.1002/neu.480040505. [DOI] [PubMed] [Google Scholar]
- Schoepp D. D. Azzaro A. J. J. Neurochem. 2010;37:527–530. doi: 10.1111/j.1471-4159.1981.tb00490.x. [DOI] [PubMed] [Google Scholar]
- Lockley O. E. Fregly M. J. Fater D. C. Pharmacology. 2008;31:132–149. doi: 10.1159/000138108. [DOI] [PubMed] [Google Scholar]
- Giulivi C. Davies K. J. J. Biol. Chem. 2001;276:24129–24136. doi: 10.1074/jbc.M010697200. [DOI] [PubMed] [Google Scholar]
- Cools R. Neurosci. Biobehav. Rev. 2006;30:1–23. doi: 10.1016/j.neubiorev.2005.03.024. [DOI] [PubMed] [Google Scholar]
- Andreou D. Söderman E. Axelsson T. Sedvall G. C. Terenius L. Agartz I. Jönsson E. G. Behav. Brain Funct. 2014;10:26. doi: 10.1186/1744-9081-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase M. Dringenberg T. Allelein S. Willenberg H. S. Schott M. Horm. Metab. Res. 2017;49:748–754. doi: 10.1055/s-0043-117179. [DOI] [PubMed] [Google Scholar]
- Snoep J. D. Hovens M. M. Pasha S. M. Frölich M. Pijl H. Tamsma J. T. Huisman M. V. Hypertension. 2009;54:1136–1142. doi: 10.1161/HYPERTENSIONAHA.109.134825. [DOI] [PubMed] [Google Scholar]
- Zhang Q. Ma X. Yang Y. Zhao H. Wang Y. Yao X. Wang L. Chang J. Zou H. Afr. J. Tradit., Complementary Altern. Med. 2017;14:33–46. [Google Scholar]
- Brodnik Z. D. Double M. España R. A. Jaskiw G. E. Neuropharmacology. 2017;123 doi: 10.1016/j.neuropharm.2017.05.030. doi: 10.1016/j.neuropharm.2017.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krnjević K. Nature. 1970;228:119–124. doi: 10.1038/228119a0. [DOI] [PubMed] [Google Scholar]
- Takashi M. Saori O. Yuki H. Takako O. S. Atsu A. Dianqing W. Keizo W. Takayuki S. Masanobu K. J. Neurosci. 2005;25:6826–6835. doi: 10.1523/JNEUROSCI.0945-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimvall K. Sheikh S. N. Martin D. L. J. Neurochem. 1993;60:714. doi: 10.1111/j.1471-4159.1993.tb03206.x. [DOI] [PubMed] [Google Scholar]
- Jang E. K. Kim N. Y. Ahn H. J. Ji G. E. J. Microbiol. Biotechnol. 2015;25:1315. doi: 10.4014/jmb.1412.12038. [DOI] [PubMed] [Google Scholar]
- Kish S. J. Rajput A. Gilbert J. Rozdilsky B. Chang L. J. Shannak K. Hornykiewicz O. Ann. Neurol. 2010;20:26–31. doi: 10.1002/ana.410200106. [DOI] [PubMed] [Google Scholar]
- Grady S. R. Salminen O. Laverty D. C. Whiteaker P. Mcintosh J. M. Collins A. C. Marks M. J. Biochem. Pharmacol. 2007;74:1235–1246. doi: 10.1016/j.bcp.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadakol A. Malek V. Goru S. K. Pandey A. Bagal S. Gaikwad A. B. Biochem. Biophys. Res. Commun. 2015;461:342–347. doi: 10.1016/j.bbrc.2015.04.036. [DOI] [PubMed] [Google Scholar]
- Downs A. M. Bond C. E. Hoover D. B. Neuroscience. 2014;266:178–185. doi: 10.1016/j.neuroscience.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Jonge W. D. Zanden E. V. D. The F. O. Boeckxstaens G. E. Eur. J. Gastroenterol. Hepatol. 2006;18:A7. doi: 10.1097/00042737-200601000-00045. [DOI] [Google Scholar]
- Diewald L. Heimrich B. Busselberg D. Watanabe T. Haas H. L. Eur. J. Neurosci. 2010;9:2406–2413. doi: 10.1111/j.1460-9568.1997.tb01657.x. [DOI] [PubMed] [Google Scholar]
- Lopachev A. V. Lopacheva O. M. Abaimov D. A. Koroleva O. V. Vladychenskaya E. A. Erukhimovich A. A. Fedorova T. N. Biochemistry. 2016;81:511–520. doi: 10.1134/S0006297916050084. [DOI] [PubMed] [Google Scholar]
- Akdis C. A. Blaser K. J. Allergy Clin. Immunol. 2003;112:15–22. doi: 10.1067/mai.2003.1585. [DOI] [PubMed] [Google Scholar]
- Yun H. L. Lee S. J. Seo M. H. Chang J. K. Sang S. S. Arch. Pharmacal Res. 2001;24:552–556. doi: 10.1007/BF02975164. [DOI] [PubMed] [Google Scholar]
- Steinke J. W. Huyett P. Borish L. J. Allergy Clin. Immunol. 2010;125:AB179. doi: 10.1016/j.jaci.2009.12.699. [DOI] [Google Scholar]
- Kashiwakura J. I. Ando T. Kawakami T. J. Pharm. Soc. Jpn. 2017;137:517–521. doi: 10.1248/yakushi.16-00239-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.