Abstract
Resistance to peptide deformylase inhibitors in Escherichia coli or Staphylococcus aureus is due to inactivation of transformylase activity. Knockout experiments in Streptococcus pneumoniae R6x indicate that the transformylase (fmt) and deformylase (defB) genes are essential and that a def paralog (defA) is not. Actinonin-resistant mutants of S. pneumoniae ATCC 49619 harbor mutations in defB but not in fmt. Reintroduction of the mutated defB gene into wild-type S. pneumoniae R6x recreates the resistance phenotype. The altered enzyme displays decreased sensitivity to actinonin.
The formylation-deformylation cycle of the translation-initiating methionine is a characteristic feature of bacterial protein synthesis. Deformylation of the nascent protein is catalyzed by an iron metalloenzyme, peptide deformylase (PDF) (10, 21). The deformylase gene is essential in Escherichia coli and Staphylococcus aureus (13–15). Novel antibacterials that are PDF inhibitors, such as actinonin, recently have been discovered by screening compound collections and combinatorial libraries (1, 5, 7, 9, 11).
Resistance to PDF inhibitors has been reported in S. aureus and E. coli (7, 13). The mechanism of resistance is based on the loss of transformylase activity, which renders deformylase nonessential. However, loss of transformylation comes at a cost to the bacteria: S. aureus fmt mutants are slow growers, and the virulence of resistant mutants is attenuated (13). In the present work, resistance to this new class of antibiotics is examined in Streptococcus pneumoniae.
(Part of this research was presented at the 40th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Ontario, Canada, 17 to 20 September, 2000.)
MATERIALS AND METHODS
Strains and plasmids used in this study are listed in Table 1. Spontaneous PDF inhibitor-resistant mutants were isolated by plating an exponentially growing culture of S. pneumoniae ATCC 49619 on blood agar containing 100 μg of actinonin (Sigma, St. Louis, Mo.) per ml. Growth was determined spectrophotometrically at 600 nm using Mueller-Hinton broth with lysed horse blood. MICs were determined as described elsewhere (5).
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| VECO2068 | F−araD139 Δ(ara, leu)7697 galE15 galK16 Δ(lac)X74 rpsL hsdR2 (rk− mk+) mcrA mcrB1 ΔtolC ΔPdef::PBAD = def; PBAD-regulated def gene | 13 |
| BL21(DE3)/pLysS | F−ompT hsdSB (rB− mB−) dcm gal λ(DE3) (pLysS); expression strain for PT7-regulated gene expression | Novagen, Madison, Wis. |
| S. pneumoniae | ||
| R6x | Unencapsulated strain for laboratory genetics | 24 |
| ATCC 49619 | American Type Culture Collection, Manassas, Va. | |
| VSPN6501 | ATCC 49619 defB(Q172K); actinonin-resistant | This study |
| VSPN6503 | ATCC 49619 defB(A123D); actinonin-resistant | This study |
| VSPN6504 | ATCC 49619 defB(A123D); actinonin-resistant | This study |
| VSPN7011 | R6x defA::pR326defA; defA disruption | This study |
| VSPN7035 | R6x defB+::pPV302-7; expresses wild-type defB | This study |
| VSPN7036 | R6x defB::pPV303-7; expresses defB (Q172K) | This study |
| VSPN7037 | R6x defB::pPV304-5; expresses defB (A123D) | This study |
| Plasmids | ||
| pET20b(+) | PT7; overexpression vector | Novagen, Madison, Wis. |
| pET20bSpn defBopt (C-His) | pET20b(+) carrying Spn defB with optimized codon usage; encodes C-terminally His-tagged PDF (wild type) | This study |
| pET20bSpn defBopt (C-His, Q172K) | pET20bSpn defBopt (C-His) mutagenized to introduce Q172K mutation | This study |
| pET20bSpn defBopt (C-His, A123D) | pET20bSpn defBopt (C-His) mutagenized to introduce A123D mutation | This study |
| pGEX-5X-3 | Ptac; overexpression vector, GST fusion protein | Amersham Pharmacia, Piscataway, N.J. |
| pGEXSpn defA | pGEX-5X-3::defA; Ptac-gst-defA fusion | This study |
| pGEXSpn defB | pGEX-5X-3::defB; Ptac-gst-defB fusion | This study |
| pR326 | cat; E. coli, S. pneumoniae shuttle vector | 6 |
| pR326rafE | pR326::'rafE'a | 23 |
| pR326defA | pR326::'defA' | This study |
| pR326defB | pR326::'defB' | This study |
| pR326fmt | pR326::'fmt' | This study |
| pPV302-7 | pR326::'defB+; downstream end of wild-type defB ORF | This study |
| pPV303-7 | pR326::'defB(Q172K); downstream end of mutated defB ORF | This study |
| pPV304-5 | pR326::'defB(A123D); downstream end of mutated defB ORF | This study |
Apostrophes indicate the truncation of the gene at the upstream and/or downstream end, as indicated.
E. coli def and fmt sequences were used in BLAST searches at NCBI (http://www.ncbi.nlm.nih.gov/blast/blast.cgi) to identify S. pneumoniae homologs. The open reading frames (ORFs) were amplified from S. pneumoniae R6x by PCR and used in subsequent experiments.
Genes were inactivated in S. pneumoniae by insertion-duplication mutagenesis (6). Fragments internal to defA (codons 43 to 126), defB (codons 77 to 172), or fmt (codons 93 to 195) were PCR amplified and cloned into pR326, and transformants were selected as described elsewhere (6, 23). The defB allele was replaced in S. pneumoniae R6x by using a truncated defB fragment (codon 77 through stop) PCR amplified from S. pneumoniae ATCC 49619 or the resistant mutants. All constructs were confirmed by PCR and sequencing.
The ability of def gene homologs to code for a functional deformylase was tested by complementation of the arabinose-dependent phenotype of E. coli VECO2068 with pGEX-5X-3 carrying def homologs or with vector alone (see Table 1), as described elsewhere (13). The E. coli VECO2068 strain has the chromosomal copy of the essential def gene under PBAD control and will grow in the absence of inducer only when an active deformylase is expressed in trans.
The defB gene (optimized for expression in E. coli by 16 silent mutations in the first 48 codons) was cloned into pET20b(+) so as to encode a His-tagged protein. This construct was modified via PCR-mediated site-specific mutagenesis at codon 172 (CAG to aAa) or codon 123 (GCT to GaT). The resulting plasmids were introduced into E. coli BL21 for protein expression. Transformants were grown at 37°C in 500 ml of Luria broth supplemented with 100 μg of ampicillin per ml to an optical density at 600 nm of 0.5, at which point IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a concentration of 1 mM. After 3 h of induction, the cells were harvested, resuspended in 35 ml of 10 mM NaCl–20 mM Tris-HCl buffer (pH 8), and then disrupted, in the presence of catalase, by a French press. His-tagged deformylase was purified from the cell lysates by passage over a cobalt affinity column according to the manufacturer's instructions (Clontech, Palo Alto, Calif.).
Deformylase activity was determined by using a deformylase-formate dehydrogenase (FDH) coupled assay (12). Experiments were carried out at room temperature in a buffer containing 10 mM NaCl, 0.2 mg of bovine serum albumin per ml, and 50 mM HEPES (pH 7.2). The reaction was initiated by adding a mixture of 0.5 U of FDH per ml, 1 mM NAD+, and fMAS at 4 mM (5, 12). Deformylation was followed by monitoring the reduction of NAD due to the oxidation of formate by FDH. For inhibition studies to determine the 50% inhibitory concentration (IC50) values, enzyme was preincubated at different concentrations of actinonin for 10 min prior to the addition of the substrate (5).
RESULTS AND DISCUSSION
Identification of deformylase in S. pneumoniae.
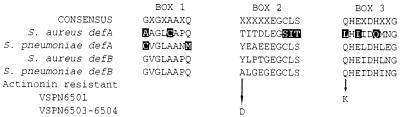
A BLAST search using the E. coli def sequence identified two S. pneumoniae def homologs, defA and defB (Fig. 1). In contrast to many other bacteria (13), neither of these homologs were adjacent to an fmt gene. Several pieces of evidence indicate that defB, and not defA, encodes the S. pneumoniae R6x PDF. The predicted DefA protein contains two substitutions (G41C and Q48M) at strictly conserved residues of a key catalytic domain, GXGXAAXQ (Fig. 1). Substitutions at either of the analogous residues of E. coli PDF dramatically impair enzyme activity (3, 4, 8, 18, 20). The S. aureus defA gene, which also contains two substitutions in this motif (Fig. 1), encodes a protein lacking PDF activity (13).
FIG. 1.
Alignment of conserved domains of deformylase proteins. Partial sequences of the predicted products of the defA and defB homologs of S. aureus and S. pneumoniae are shown aligned with consensus PDF domains (13). Residues that diverge from the consensus are highlighted. Modifications in the deformylase enzyme in the resistant S. pneumoniae mutants VSPN6501, VSPN6503, and VSPN6504 are indicated by arrows. The positions of the motifs in the S. pneumoniae DefB are as follows: box 1, G69 to Q76; box 2, A123 to S132; and box 3, Q172 to G182. The sequences of the S. pneumoniae def homologs from strain R6x have been submitted to GenBank (accession numbers: defA, AY014508; defB, AY014509). Sequences of the S. pneumoniae def and fmt homologs from S. pneumoniae ATCC 49619 were also submitted to GenBank (accession numbers: defA, AY014510; defB, AY014511; fmt, AY014512).
Despite several attempts, defB could not be inactivated by insertion-duplication mutagenesis. However, defA or rafE, a nonessential gene (23), were readily disrupted. This result implies that defB is essential, although the experiment does not exclude the possibility of a polar effect on a downstream gene. A plasmid encoding a GST-DefB fusion protein, but not one encoding GST-DefA, was able to complement the arabinose-dependent phenotype of E. coli VECO2068. Expression of the gst-defA and gst-defB fusions was confirmed by Western blotting (data not shown). Purified GST-DefB was associated with a strong PDF activity (1,300 μmol min−1 mg of protein−1). Taken together, these results argue that defB codes for a true essential deformylase, whereas defA is a paralog of unknown but nonessential function or has marginal deformylase activity unable to complement the arabinose-dependent mutant. This is similar to S. aureus RN4220, which also harbors two deformylase homologs, only one of which, defB, encodes a true PDF (13).
Isolation and characterization of actinonin-resistant mutants.
The frequency of resistance in S. pneumoniae ATCC 49619 was 10−8, 2 orders of magnitude lower than that obtained with S. aureus ATCC 25923 or S. aureus 1–63 (13). Three S. pneumoniae mutants, VSPN6501, VSPN6503, and VSPN6504, were chosen for further studies. The mutants grew at slower rates, with doubling times approximately 20% longer, when cultured in broth (Table 2). The slow-growth phenotype is less pronounced than in S. aureus actinonin-resistant strains derived from S. aureus ATCC 25923, where doubling times increase approximately 80% (13). These mutants of S. pneumoniae ATCC 49619 showed resistance to PDF inhibitors (Table 2) but were unchanged in susceptibility to penicillin, ampicillin, chloramphenicol, erythromycin, trimethoprim, or tetracycline. These results suggest that resistance occurred by a specific mechanism distinct from that for these other antibiotics and further indicates that the slower growth rate did not change the overall susceptibility of the mutant strains. No such differences in susceptibility to PDF inhibitors or doubling time were observed for strain VSPN7011, which carries a disrupted defA gene (not shown).
TABLE 2.
Genotypes and susceptibilities to PDF inhibitors, and doubling times of S. pneumoniae actinonin-resistant and -susceptible strainsa
| Strain | defB | Mean T2 in min (SD) | MIC (μg/ml)
|
|
|---|---|---|---|---|
| Act | VRC3375b | |||
| ATCC49619 | wt | 51 (1.4) | 32 | 16 |
| VSPN6501 | Q172K | 60 (2.9) | 128 | 256 |
| VSPN6503 | A123D | 64 (2.6) | 64 | 128 |
| VSPN6504 | A123D | 66 (2.3) | 128 | 128 |
wt, wild type; T2, doubling time; Act, actinonin
PDF inhibitor (D. Chen, C. Hackbarth, Z. J. Ni, W. Wang, C. Wu, D. Young, R. J. White, J. Trias, D. V. Patel, and Z. Yuan, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 2175, 2000).
Mutation of fmt leads to resistance in S. aureus or E. coli (7, 13). The fmt gene from S. pneumoniae ATCC 49619 actinonin-resistant strains was PCR amplified and sequenced. None of the resistant strains carried a mutation in fmt or flanking DNA. The inactivation of fmt was attempted to assess whether the lack of transformylase activity could provide resistance in S. pneumoniae R6x. Despite multiple attempts, the fmt gene could not be inactivated. In contrast, the homologous gene can be readily inactivated in strains of E. coli, Pseudomonas aeruginosa, or S. aureus (13, 14, 19). The inability to inactivate fmt suggests that the transformylase gene itself is essential in S. pneumoniae R6x, although it is possible that strains disrupted in fmt were not obtained because of a polar effect on a downstream gene.
The sequences and flanking DNA of both defA and defB were PCR amplified and sequenced from S. pneumoniae ATCC 49619 and mutant strains. No change was observed among the sequences of the defA homolog. However, each of the resistant strains possessed a single missense mutation in defB (Fig. 1). For S. pneumoniae VSPN6501, a Q172K substitution occurs at a residue that is strongly conserved among PDF proteins (Fig. 1). This position lies immediately upstream of the 173HEXXH177 (S. pneumoniae numbering) motif shared by all PDF proteins and characteristic of zinc hydrolases (22). A nonconservative substitution (Q131A) at the equivalent position in the E. coli enzyme has been shown to decrease enzyme activity (8). The analogous pair of His residues in the E. coli PDF has been shown by genetic and structural studies to bind the metal ion in the catalytic pocket (2–4, 8, 16–18). S. pneumoniae VSPN6503 and VSPN6504 have a A123D substitution in the predicted protein. This residue is not conserved among PDF proteins. However, the mutation introduces a charged amino acid five residues upstream of the 128EGCLS132 motif, which has been shown to be involved in binding the metal ion (Fig. 1) (2–4, 7, 8, 16–18). The mutated residue corresponds to a position two residues upstream of an Ile involved in defining a substrate-binding pocket of the E. coli enzyme (7). Thus, in all three cases, a mutation in S. pneumoniae defB is predicted to cause a substitution close to conserved domains involved in binding the metal ion or the substrate, essential for PDF activity.
Mutated defB leads to resistance.
Genes encoding the wild-type and mutated PDFs were introduced into S. pneumoniae R6x (6). Strains expressing either of the two mutated defB genes displayed reduced susceptibility to actinonin (Table 3). In addition, the mutated enzymes are indeed less sensitive to inhibition than the wild-type parent PDF (Table 3). These results confirm that, in these mutants, resistance is due to modification of the target rather than the lack of transformylation activity.
TABLE 3.
Susceptibilities to actinonin of S. pneumoniae R6x strains transformed with wild-type or mutated defB and inhibition of purified His-tagged PDF enzymes by actinonin
| S. pneumoniae strain | Expressed defB | MIC (μg/ml) | Enzyme | IC50 (nM) |
|---|---|---|---|---|
| VSPN7035 | defB | 16 | Wild type | 53 ± 6 |
| VSPN7036 | defB (Q172K) | 64 | Q172K | 70 ± 8 |
| VSPN7037 | defB (A123D) | ≥64 | A123D | 136 ± 16 |
Resistance to PDF inhibitors can occur by at least two distinct mechanisms, with different consequences predicted in each case. Resistant mutants of S. aureus or E. coli, obtained in vitro, lack transformylase activity, bypassing the essentiality of the def gene (7, 13). Mutation of fmt should lead to cross-resistance to any antibiotic for which PDF is the major target, because inhibition of deformylase would have no consequence for protein synthesis if nascent peptides were not formylated. However, mutation of fmt does have consequences for cell growth, as seen in S. aureus and E. coli fmt mutants. Notably, S. aureus fmt mutants have attenuated virulence in abscess or septicemia models, decreasing the chance that such mutants would survive during infection (7, 13). In S. pneumoniae R6x, fmt cannot be disrupted; instead, resistance to PDF inhibitors derives from modification of the target. In contrast to strains resistant via a lack of transformylation activity, it is possible that these S. pneumoniae defB mutants will not be cross-resistant to all PDF inhibitors. More potent inhibitors, or compounds that bind differently to PDF, should be active against these resistant S. pneumoniae strains. The essentiality of fmt makes PDF an attractive target for the discovery and development of novel antibiotics active against S. pneumoniae.
ACKNOWLEDGMENTS
Sequencing of S. pneumoniae was accomplished with support from TIGR, the National Institute for Allergy and Infectious Diseases, and the Merck Genome Research Institute.
We thank Dennis Young and Carsten Rosenow for advice.
REFERENCES
- 1.Apfel C, Banner D W, Bur D, Dietz M, Hirata T, Hubschwerlen C, Locher H, Page M G, Pirson W, Rosse G, Specklin J L. Hydroxamic acid derivatives as potent peptide deformylase inhibitors and antibacterial agents. J Med Chem. 2000;43:2324–2331. doi: 10.1021/jm000018k. [DOI] [PubMed] [Google Scholar]
- 2.Becker A, Schlichting I, Kabsch W, Groche D, Schultz S, Wagner A F. Iron center, substrate recognition and mechanism of peptide deformylase. Nat Struct Biol. 1998;5:1053–1058. doi: 10.1038/4162. [DOI] [PubMed] [Google Scholar]
- 3.Becker A, Schlichting I, Kabsch W, Schultz S, Wagner A F. Structure of peptide deformylase and identification of the substrate binding site. J Biol Chem. 1998;273:11413–11416. doi: 10.1074/jbc.273.19.11413. [DOI] [PubMed] [Google Scholar]
- 4.Chan M K, Gong W, Rajagopalan P T, Hao B, Tsai C M, Pei D. Crystal structure of the Escherichia coli peptide deformylase. Biochemistry. 1997;36:13904–13909. doi: 10.1021/bi9711543. [DOI] [PubMed] [Google Scholar]
- 5.Chen D Z, Patel P, Hackbarth C J, Wang W, Dreyer G, Young D C, Margolis P S, Wu C, Ni Z-J, Trias J, White R J, Yuan Z. Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry. 2000;39:1256–1262. doi: 10.1021/bi992245y. [DOI] [PubMed] [Google Scholar]
- 6.Claverys J P, Dintilhac A, Pestova E V, Martin B, Morrison D A. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene. 1995;164:123–128. doi: 10.1016/0378-1119(95)00485-o. [DOI] [PubMed] [Google Scholar]
- 7.Clements J M, Beckett R P, Brown A, Catlin G, Lobell M, Palan S, Thomas W, Whittaker M, Wood S, Salama S, Baker P J, Rodgers H F, Barynin V, Rice D W, Hunter M G. Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor. Antimicrob Agents Chemother. 2001;45:563–570. doi: 10.1128/AAC.45.2.563-570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dardel F, Ragusa S, Lazennec C, Blanquet S, Meinnel T. Solution structure of nickel-peptide deformylase. J Mol Biol. 1998;280:501–513. doi: 10.1006/jmbi.1998.1882. [DOI] [PubMed] [Google Scholar]
- 9.Giglione C, Pierre M, Meinnel T. Peptide deformylase as a target for new generation, broad spectrum antimicrobial agents. Mol Microbiol. 2000;36:1197–1205. doi: 10.1046/j.1365-2958.2000.01908.x. [DOI] [PubMed] [Google Scholar]
- 10.Groche D, Becker A, Schlichting I, Kabsch W, Schultz S, Wagner A F. Isolation and crystallization of functionally competent Escherichia coli peptide deformylase forms containing either iron or nickel in the active site. Biochem Biophys Res Commun. 1998;246:342–346. doi: 10.1006/bbrc.1998.8616. [DOI] [PubMed] [Google Scholar]
- 11.Huntington K M, Yi T, Wei Y, Pei D. Synthesis and antibacterial activity of peptide deformylase inhibitors. Biochemistry. 2000;39:4543–4551. doi: 10.1021/bi992452y. [DOI] [PubMed] [Google Scholar]
- 12.Lazennec C, Meinnel T. Formate dehydrogenase-coupled spectrophotometric assay of peptide deformylase. Anal Biochem. 1997;244:180–182. doi: 10.1006/abio.1996.9910. [DOI] [PubMed] [Google Scholar]
- 13.Margolis P S, Hackbarth C J, Young D C, Wang W, Chen D, Yuan Z, White R, Trias J. Peptide deformylase in Staphylococcus aureus: resistance to inhibition is mediated by mutations in the formyltransferase gene. Antimicrob Agents Chemother. 2000;44:1825–1831. doi: 10.1128/aac.44.7.1825-1831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazel D, Pochet S, Marliere P. Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation. EMBO J. 1994;13:914–923. doi: 10.1002/j.1460-2075.1994.tb06335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meinnel T, Blanquet S. Characterization of the Thermus thermophilus locus encoding peptide deformylase and methionyl-tRNA (fMet) formyltransferase. J Bacteriol. 1994;176:7387–7390. doi: 10.1128/jb.176.23.7387-7390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meinnel T, Blanquet S, Dardel F. A new subclass of the zinc metalloproteases superfamily revealed by the solution structure of peptide deformylase. J Mol Biol. 1996;262:375–386. doi: 10.1006/jmbi.1996.0521. [DOI] [PubMed] [Google Scholar]
- 17.Meinnel T, Lazennec C, Blanquet S. Mapping of the active site zinc ligands of peptide deformylase. J Mol Biol. 1995;254:175–183. doi: 10.1006/jmbi.1995.0609. [DOI] [PubMed] [Google Scholar]
- 18.Meinnel T, Lazennec C, Villoing S, Blanquet S. Structure-function relationships within the peptide deformylase family. Evidence for a conserved architecture of the active site involving three conserved motifs and a metal ion. J Mol Biol. 1997;267:749–761. doi: 10.1006/jmbi.1997.0904. [DOI] [PubMed] [Google Scholar]
- 19.Newton D T, Creuzenet C, Mangroo D. Formylation is not essential for initiation of protein synthesis in all eubacteria. J Biol Chem. 1999;274:22143–22146. doi: 10.1074/jbc.274.32.22143. [DOI] [PubMed] [Google Scholar]
- 20.Ragusa S, Mouchet P, Lazennec C, Dive V, Meinnel T. Substrate recognition and selectivity of peptide deformylase. Similarities and differences with metzincins and thermolysin. J Mol Biol. 1999;289:1445–1457. doi: 10.1006/jmbi.1999.2832. [DOI] [PubMed] [Google Scholar]
- 21.Rajagopalan P T, Yu X C, Pei D. Peptide deformylase: a new type of mononuclear iron protein. J Am Chem Soc. 1997;119:12418–12419. [Google Scholar]
- 22.Rawlings N D, Barrett A J. Evolutionary families of metallopeptidases. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 23.Rosenow C, Maniar M, Trias J. Regulation of the alpha-galactosidase activity in Streptococcus pneumoniae: characterization of the raffinose utilization system. Genome Res. 1999;9:1189–1197. doi: 10.1101/gr.9.12.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiraby G, Fox M S. Marker discrimination in transformation and mutation of pneumococcus. Proc Natl Acad Sci USA. 1973;70:3541–3545. doi: 10.1073/pnas.70.12.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]