Abstract
RePORT International is a collaborative research network of investigators from multiple countries and institutions with the goal of establishing a bio-repository of specimens and clinical data for the study of active TB and latent TB infection (LTBI). During the first meeting of RePORT International in Boston, Massachusetts, the results of research pertinent to TB control and eradication were presented, including advances in the research of Mycobacterium tuberculosis (MTB) persistence and drug resistance, TB diagnostics, drug and vaccine development.
Keywords: RePORT, tuberculosis, research, advances
The Mission of RePORT
RePORT (Regional Prospective Observational Research on Tuberculosis) International is a research network that serves as a platform for new discoveries in tuberculosis (TB). RePORT sites currently exist in India, Brazil, South Africa and Indonesia. Funding is provided jointly by the US Government (the Division of AIDS (DAIDS), National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), and the Office of AIDS Research (OAR)) and the governments of India (Division of Biotechnology, the Indian Council of Medical Research (ICMR)), Brazil (Fundação Oswaldo Cruz (FioCruz)), South Africa (Medical Research Council) and Indonesia (The Center of Applied Health Technology and Clinical Epidemiology, National Institute of Health Research and Development (NIHRD)). Central to the activities of the RePORT International Consortium is a Common Protocol that includes standardized clinical definitions and standard operating procedures (SOPs) for collecting data and specimens. The RePORT International Common Protocol is a prospective observational non-interventional study that enrolls individuals with active TB or latent TB infection (LTBI). Its activities include collection of clinical information and establishment of a bio-repository for blood, sputum, urine and saliva specimens for potential national and international collaborations. It establishes a common framework, set of standards, and definitions that can be integrated with eligible country-specific protocols to facilitate collaborations between institutions worldwide. The primary objective is to provide specimens and linked clinical data to researchers and their collaborators to discover host biomarkers that characterize risk of treatment failure and TB recurrence, as well as progression from LTBI to active TB. The Common Protocol will also support research focusing on better understanding the pathogenesis and prognosis of TB. The RePORT network is well positioned to conduct clinical research on new TB diagnostics, therapeutics and vaccines. A description of the organizational structure and aims of RePORT International has been published.[1]
In India, there are site-specific protocols as well as the Common Protocol. The site-specific protocols address the impact of co-morbidities such as diabetes, helminth infection, HIV infection, malnutrition, smoking, indoor air pollution, alcoholism, and anemia on TB pathogenesis and the modulation of host biomarkers. The approaches taken include immunogenetics, transcriptomics and assessment of modulation of the severity of disease by eicosanoid pathways.
The current activities in Brazil focus on biomarker discovery for TB infection and disease, the social determinants of TB and HIV infection, and the pharmacogenomic predictors of treatment toxicity, treatment failure and relapse in HIV-related tuberculosis. There is also interest in applying whole genome sequencing to elucidate the mechanisms of emergence of drug resistance. RePORT Brazil also has developed a secondary focus on TB-diabetes comorbidity, in close collaboration with investigators from RePORT-India.
In Indonesia, the RePORT partners are studying the proportion of multi-drug-resistant tuberculosis (MDR-TB) amongst new and previously treated cases of active TB, while evaluating the accuracy of clinical diagnostic methods by comparing clinically-defined and laboratory-confirmed cases. The group also is evaluating how treatment success is affected by the individual’s demographics, contact history, smoking history, treatment-seeking behaviors, co-morbidities, prior drug resistance, symptoms, cavity presence, nutritional status, treatment compliance, and specific Mycobacterium tuberculosis (MTB) strain.
The first meeting of RePORT International coincided with the fourth RePORT India Joint Leadership meeting and was held in Boston Massachusetts, USA, September 23rd - 25th, 2015. It was hosted by the Department of Medicine of Boston University School of Medicine, Boston Medical Center and the Lifespan/Tufts/Brown Center for AIDS Research (CFAR). The purpose of this meeting was to present the state of the art in TB research in areas pertinent to RePORT and to encourage collaboration among RePORT teams and with other investigators.
Introduction
TB is the leading infectious cause of death in the world. Despite vast efforts and major scientific advances, understanding is limited with regards to critical aspects of host and pathogen biology and interactions that result in containment or progression of latent TB infection and that affect cure of disease. Much is unknown about the dynamics of MTB transmission, what determines active versus latent infection, as well as progression from the latent stage to subclinical TB to active symptomatic disease. Notification systems and active case finding processes often fail to reach all those who are at risk of disease, and imperfect levels of sensitivity and specificity of diagnostic tests account for undetected cases, particularly in pediatric and extra-pulmonary TB cases. The current vaccine, Mycobacterium bovis-derived Bacillus Calmette-Guerin (BCG), is partially effective in preventing TB in infants and children, but has no impact on the public health problem of TB because it fails to protect against infectious TB in adults. The 6-month multi-drug therapeutic regimen is complicated to administer programmatically, leading to loss of follow-up, treatment failure and emergence of drug resistance. Further, cases of relapse and reinfection occur even amongst patients with successfully completed treatment courses. To add to these many barriers to effective control of TB, emergent strains with different levels of drug resistance challenge the current drug options and choice of treatment duration.
Drug Resistance and MTB Persistence
Standard management of TB disease consists of two months of intensive induction therapy with isoniazid (INH), rifampin (RIF), pyrazinamide (PZA), and ethambutol (EMB), followed by four months of continuation treatment with isoniazid and rifampin. Among cases of drug-susceptible TB (DS-TB) who are appropriately treated and complete a full course of active anti-TB drugs, the relapse rate within the first year after treatment completion is 1.8 to 2.1 relapse rate per 100 person-years, depending on risk factors such as presence of cavitation and HIV infection status.[2,3]
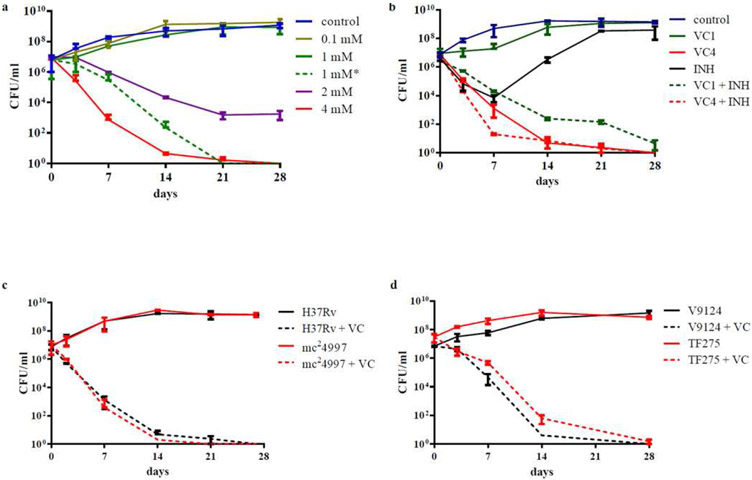
The general belief is that relapse is attributable to the incomplete eradication of bacteria. Persisting MTB evade rapid killing by chemotherapeutic agents and are the reservoir for treatment failure and relapse. Isoniazid kills up to 99.9% of MTB in vitro. Although this represents a high killing rate, the remaining 0.1% of bacteria, when re-grown, are not killed by the drug and instead represent a population of “persister” organisms. In vitro, populations of persister cells are observable as displaying distinct phenotypes. When treated with a differentiating stain (Ziehl-Neelson) followed by an acidic wash, the mycolic acid within the mycobacterial cell wall retains the dye. Unlike other bacteria, in which the acid wash removes the red stain, mycobacteria retain it and are therefore considered acid-fast positive. Acid-fast positive MTB are sterilized more quickly than acid-fast negative MTB, whereas acid-fast negative MTB are more prone to entering dormancy and showing “persister” phenotypes. These observations suggest that acid fastness may be associated with different patterns of behavior and survival. Acid fastness is regulated by a signal transduction pathway with many intervening factors – some of them potentially key in determining drug resistance and the persister phenotype. Persister cells are killed at a reduced rate in vivo, raising the question of whether a drug can completely sterilize MTB. If such a drug was to exist, which mechanisms would it have to target in order to kill actively growing cells and persister cells? Dr. William Jacobs’ presentation dealt with the epigenetic variations of MTB and the “persister” populations. He and his research team have developed an animal model using CBaJ mice and MTB strain H37Rv to identify drugs that can target actively growing and persister MTB cells. The group has reported vitamin C to have an in vitro bactericidal effect on colonies of drug-susceptible, multi drug-resistant, and extensively drug-resistant MTB (figure 1).[4] Following a dose-dependent response, vitamin C eliminated MTB with a minimum inhibitory concentration (MIC) of 1 mM. Visible bactericidal activity was seen with 2 mM, and complete sterilization of MTB colonies was achieved by three weeks when 4 mM were added. Although a one-time administration of 1 mM of vitamin C resulted only in mild and reversible bacteriostatic activity, the group found that daily addition of 1 mM for 4 days achieved similar outcomes as a single dose of 4 mM. Compared to treatment with isoniazid, which resulted in the selection of isoniazid-resistant mutants, vitamin C produced continued eradication of MTB. The combination of vitamin C and isoniazid also resulted in increased and prolonged killing rates. Interestingly, the bactericidal effect of vitamin C was higher for MTB than for other strains of mycobacteria, and non-tuberculous mycobacteria required concentrations as great as 32 times the MIC for MTB.[4]
Figure 1 -.
Vitamin C sterilizes drug-susceptible and drug-resistant M. tuberculosis strains (a) M. tuberculosis H37Rv cultures grown to an OD600nm of ≈ 0.75 were diluted 1/20 and treated with increasing amounts of vitamin C (VC, from 0.1 mM to 4 mM). 1 mM* represents an experiment where 1 mM of vitamin C was added to the culture daily for the first 4 days of treatment. (b) M. tuberculosis H37Rv was treated with INH (7 μM, 20× MIC), vitamin C (1 or 4 mM) and a combination of INH and vitamin C (1 and 4 mM). (c) mc24997, a RIF- and INH-resistant M. tuberculosis H37Rv strain was treated with vitamin C (4 mM). (d) Vitamin C (4 mM) was added to a drug-susceptible strain (V9124) and to an extensively drug-resistant strain (TF275) of M. tuberculosis from the Kwa-Zulu Natal province of South Africa. Growth was followed and CFUs were determined by plating 10- fold serial dilutions and incubating the plates at 37°C for 4 weeks. The experiments were done at leas t in triplicate and the average with standard deviation is plotted.
Vilchèze C, Hartman T, Weinrick B, Jacobs WR. Mycobacterium tuberculosis is extraordinarily sensitive to killing by a vitamin C-induced Fenton reaction. Nat Commun 2013;4:1881. doi:10.1038/ncomms2898.
Although vitamin C is both an anti-oxidant and a pro-oxidant, its ability to eradicate MTB most likely derives from its pro-oxidant properties. By promoting the reduction of ferric ions to the more reactive ferrous ion form, it increases the production of reactive oxygen species (ROS), leading to lipid alterations, redox imbalance, and DNA damage. Supporting evidence that iron excess is a mechanism through which vitamin C kills MTB is the reversion of its bactericidal effect when an iron-chelating agent is added. Similarly, vitamin C effects were inhibited when cultures were kept in anaerobic conditions that impaired the formation of ROS. Mutant strains with defective synthesis of the reducing agent mycothiol (N-acetylcysteine glucosamine inositol) were also more susceptible to cellular damage by vitamin C, suggesting that the unopposed oxidative stress is the direct mechanism of action. Mycothiol is an actinobacteria-specific thiol with multiple functions in protein structure, regulation of cysteine reserves, protection against oxidative stress, and detoxification of alkylating agents and antibiotics.[5] It is required for MTB killing by isoniazid and ethionamide. MTB strains with co-resistance to these drugs have mutations in the mshA gene, which encodes for glycosyltransferase MshA involved in the synthesis of mycothiol. Inhibition of mycothiol biosynthesis leads to accumulation of cysteine, conferring drug resistance.[6]
Multiple studies have shown a benefit to adding vitamin C to the current standard anti-tuberculous medications. Rifampin, isoniazid and vitamin C seem to act synergistically, enhancing MTB eradication.[7] Despite promising laboratorial data and studies in animal models, human clinical trials will be needed for validation and standardization of vitamin C use in tuberculosis treatment regimens.
The persistent survival of some organisms reflects an ability to adapt to different host conditions and environmental changes. Dr. Kathleen McDonough reported on the mechanisms through which MTB can integrate and respond to multiple signals, maximize survival, and modify host responses. Succinate dehydrogenase is an enzyme of the oxidative phosphorylation complex and the TCA cycle. It is involved in energy production pathways used by MTB and its secretion and accumulation is essential for adaptation to hypoxia. Modulation of the regulators of this response can greatly impact TB pathogenesis. Adenosine 3′, 5′-cyclic monophosphate (cAMP) is a signal molecule that contributes to global regulation of gene expression in MTB, in part, by affecting the activity of transcription factors such as Rv1675c, also known as Cmr (cAMP and macrophage regulator),[8] and Rv3676, also known as CRP (cAMP-responsive protein). CRP has been implicated in the activation and regulation of succinate dehydrogenase gene expression, determining the capacity for metabolic adaptation.[9] It is of interest that Cmr has multiple binding sites adjacent to members of the DosR (DevR) dormancy regulon, suggesting that it may play a role in TB dormancy and persistence.[10] In addition to cAMP, Cmr regulates MTB gene expression in response to the intra-macrophage environment.[8] Among other factors, the efficiency with which macrophages can phagocytize and clear MTB can determine progression to latent or active infection. Understanding how transcription factors like Cmr and CRP influence gene expression in response to the host environment has clinical relevance for the biology of infection, drug targets, virulence determinants, and biomarkers.
Dr. David Sherman reported on genome-scale experimental approaches and modeling tools to systematically map MTB gene regulatory mechanisms, including those implicated in anti-mycobacterial drug evasion. By cloning and over-expressing each individual transcription factor (TF) in what are called transcription factor over-expressing (TFOE) strains, his laboratory was able to identify unique DNA binding sites as well as the genes differentially regulated by each TF and determine specific regulatory signatures. Using this system[11] on 206 MTB DNA-binding proteins, the Sherman group catalogued over 10,000 unique DNA binding events[12] that collectively produce more than 9000 individual gene expression changes.[13] Eleven percent (11%) of MTB TFs are not associated with any transcriptional changes when over-expressed, suggesting that some TFs may be expressed at saturating levels under baseline conditions, or that they require specific activating factors. Most transcription factors, however, are expressed below their physiological maximum in this system. Indexing of TFOE regulatory pathways has led to the creation of a searchable map of the transcriptional regulatory pathways in MTB. The TFOE framework can help form and test hypotheses of gene and regulatory function.
Dr. Sherman’s team used TFOE to predict and validate the phenotype of a regulator that dramatically alters susceptibility to isoniazid.[13] Isoniazid is converted from its pro-drug form to the active compound by catalase – an enzyme encoded by the gene katG gene (Rv1908c). KatG is essential for isoniazid activity. Changes in its expression potentially lead to decreased levels of the active drug. The TFOE dataset identified the repressor furA (Rv1909c) as the only transcriptional regulator of katG. Over-expressed furA leads to suppressed levels of katG, in turn leading to reduced levels of catalase and decreased conversion of isoniazid. The role of furA is confirmed by the fact that MTB strains that have been manipulated to overexpress this repressor can survive and grow in cultures with isoniazid concentrations that completely inhibited growth of un-induced strains.[13]
The Sherman group also has applied genome-scale modeling tools to the TFOE network. One of these tools, the Environmental Gene Regulatory Influence Network (EGRIN), detects specific sets of genes that are coordinately up- or down-regulated under some but not all conditions.[14] It identifies conditionally co-regulated genes (bi-clusters) and predicts relevant regulators and motifs. In addition, they have integrated 104 TFs from their regulatory network with a genome-scale metabolic model. Using the MTB Probabilistic Regulation of Metabolism 2.0 (MTBPROM2.0) framework, this transcriptional regulatory network was linked to an in silico metabolic model that can predict bacterial viability under different metabolic conditions.[15] The model can be used to simulate the consequences of knocking out and/or over-expressing any one of the 104 transcription factors. Both EGRIN and PROM were successful in predicting MTB strains with altered susceptibility to the drug bedaquiline as well as drug:drug synergy. Interrogation of gene regulatory and metabolic networks with new in silico tools such as EGRIN and PROM is likely to become an important resource for modeling the action of new drug targets.
Dr. Bruno B. Andrade reported on the interplay between oxidative stress and immune responses in TB. Infection with MTB is associated with increased production of free radicals that cause oxidative stress and augment lipid peroxidation, leading to inflammation and tissue destruction. Matrix metalloproteinases (MMP) degrade lung tissue and are expressed in high levels in TB. In fact, MMP levels in sputum or plasma samples correlated with severity of disease. Heme oxygenase-1 (HO-1), in turn, is a cyto-protective antioxidant highly expressed in the lungs. Dr. Andrade reported high levels of metalloproteinase-1 (MMP-1) and HO-1 in the blood of adult and pediatric patients with active pulmonary TB in Brazil and India, when compared to those with latent infection.[16,17] The data suggest that measuring the two enzymes can serve as a biomarker of active disease. However, the group also detected an inverse correlation in the expression of these biomarkers – high levels of HO-1 were accompanied by low levels of MMP-1, and vice-versa.[18] Two patterns were identified in the TB population: one group of patients showed very high levels of HO-1 but low levels of MMP-1, whereas the other showed very high levels of MMP-1 but low levels of HO-1. Interestingly, these two patterns were also associated with different clinical presentations and inflammatory profiles. Dr. Andrade and collaborators have elucidated the mechanism underlying this inverse relationship. HO-1-mediated production of carbon monoxide (CO) suppresses MMP-1 expression at the transcriptional level by inhibiting c-Jun/AP-1 activation in MTB-infected macrophages. Suppressed levels of MMP-1, in turn, increase the activation of HO-1 transcription factors.[18] The inverse correlation between HO-1 and MMP-1 has not been observed in non-tuberculous lung conditions, demonstrating specificity to TB. CO has been ascribed antimicrobial effects before, as the resistance of MTB to CO increased its pathogenicity and survival. By inhibiting MMP-1 expression, CO also proved to have regulatory effects on collagen degradation.[18] HO-1 and MMP-1 levels may be of use in the diagnosis of active TB, and cross-talk between the two may determine distinct clinical phenotypes. Pharmacological induction of HO-1 leading to CO production could be used as a strategy for MMP-1 inhibition, potentially changing the treatment paradigm and ultimately the course of disease in TB patients.
Drug resistance can be primary (transmitted) or secondary (acquired). Whereas acquired resistance often is the result of inadequate treatment, poor adherence or malabsorption of drugs, primary resistance is produced by transmission of an intrinsically resistant strain. Dissemination of primary resistance is influenced by the transmissibility of the strain, the clinical characteristics of the host (cough frequency and intensity), and the time to detection and treatment of the index case. Dr. Megan Murray reported on the transmissibility of drug-susceptible (DS-) and drug-resistant (DR-) TB and the risk factors associated with the two different types of resistance. Conducted in Lima, Peru, the EpiStudy enrolled index cases with DS- and DR-TB and their household contacts, who were prospectively followed for 12 months for the development of active TB disease and drug resistance. The aim of the study was to determine whether characteristics of the index case, the contact, or the environment correlated with the risk and type of resistance. The study has shed light on the determinants of TB transmission within a household. Recent findings include the observation that household contacts of HIV-infected TB patients with a CD4 count ≤250 cells/μL were less likely to become infected (as defined by post-exposure TST ≥10 mm or ≥5 mm if HIV-infected, without a previous diagnosis of TB or a history of TST positivity).[19] Among DS-TB household contacts, the group also found that protection against active TB was associated with BCG vaccination in HIV-uninfected children younger than 10 years, and with IPT in HIV-uninfected contacts younger than 30. The risk of incident TB associated with LTBI was greatest for children younger than 5 years and decreased with age.[20]
By observing the properties of the environmental context of each participant, the study assessed the influence of social determinants. The results showed that in households of higher socioeconomic status the risk of transmitted resistance was higher compared to acquired resistance. Conversely, acquired resistance occurred more often than transmitted resistance in households with a lower socioeconomic status, where quality of housing was poorer.[21] Though counter-intuitive, the results mirror those of previous studies and raise interesting questions regarding the dynamics of TB transmission. Many factors cause and derive from specific socioeconomic statuses. Employment, housing conditions, ventilation, nutritional needs, smoking and drinking habits, level of education, number of co-morbidities, access to health care systems and many other factors correlate with socioeconomic status. Further studies are needed to discern exactly which contributors correlate with risk of TB.
Dr. Barry Bloom reviewed the mechanisms of innate immune defense against TB. T cells are crucial in the response to MTB infection. The secreted IFN-γ triggers macrophage activation; macrophages, in turn, secrete IL-15 and induce an antimicrobial pathway that is vitamin D-dependent.[22] The killing of intracellular MTB is attributed to the pathway’s antimicrobial end-product – cathelicidin.[22] Interestingly, this pathway is inhibited by type I interferons (IFN-α and IFN-β).[23] Gene co-expression analysis has showed that this pathway is mediated by IL-32, a pleiotropic pro-inflammatory cytokine secreted by macrophages upon MTB stimulation. IL-32 enhances monocyte differentiation into dendritic cells that cross-present antigens to CD8+ T cells[24] and trigger an anti-microbial response in macrophages. It induces cathelicidin depending on the presence of adequate 25-hydroxyvitamin D levels.[25] The finding of an association between ability to produce vitamin D and susceptibility to microbial infection is supported by the observation that levels of cathelicidin messenger RNA and 25-hydroxyvitamin D are low in the sera of African-American individuals, who are known to have increased susceptibility to tuberculosis.[22,26] Studies of gene expression by cell populations from different clinical settings (active versus latent TB versus healthy controls versus individuals undergoing TB treatment) have shown that expression of IL-32 and the IL-15-induced networks are associated with latency, thus representing a molecular marker of immune protection against progression to active disease.[25]
From an evolutionary perspective, killing the host halts transmission of the pathogen. MTB therefore benefits from disease states that promote transmission. In the pathogenesis of MTB this is accomplished through granuloma formation. Dr. Igor Kramnik used a mouse model that develops human-like TB to study the mechanisms involved. He and his group discovered that the sst1 (super-susceptibility to tuberculosis) locus is a genetic determinant of host resistance to infection with intracellular pathogens such as MTB, Chlamydia pneumoniae, and Listeria monocytogenes.[27,28] Sst1 controls the progression of pulmonary TB in immunocompetent individuals by modulating the stress response of TNF-α-activated macrophages, up-regulating type I IFN pathways, and ultimately inducing granuloma necrosis. It is involved in the regulation of an NO synthase-independent mechanism of innate immunity that also mediates the anti-tuberculosis effects of BCG vaccination.[27,29] Compared to wild type mice, mice that express the sst1 susceptible locus and are infected with Chlamydia pneumoniae show increased levels of lung inflammation, exaggerated macrophage and neutrophil influx, and development of fibrosis.[27] Similarly, tuberculosis-infected mice with the sst1 susceptible locus show an increased number of lung granulomas.[30] Within the sst1 locus, a nuclear protein of intracellular pathogen resistance (Ipr1) was found to be the mediator of this effect by regulating macrophage activation and death pathways.[31] Ipr1 is an IFN-inducible protein; its expression is regulated by type I and II interferons at the transcriptional and posttranscriptional level. The Ipr1 gene is up-regulated in sst1 resistant macrophages, but is not expressed in sst1 susceptible macrophages.[32]
Similar to TNF-α pathway manipulation, augmenting the macrophage response to IFN-γ holds the potential for maximizing the intracellular response against MTB. IFN-γ activates macrophages and stimulates the production of highly toxic NO and reactive oxygen species (ROS). It also reduces inflammation and tissue damage during infection. Many pathogens target these protective mechanisms by suppressing T cell production of IFN-γ, producing IFN-γ-neutralizing antibodies, or down-regulating the responsiveness of macrophages. Recombinant IFN-γ and compounds that can amplify macrophage sensitivity to IFN-γ would be able to restore normal immunity.[32]
Accumulation of Ipr1 was used as a surrogate of IFN-γ-inducible macrophage activation during a high-throughput screen (HTS) to detect molecules that synergize with low levels of IFN-γ. HTS was applied to the Boston University Center for Molecular Discovery (BU-CMD) chemical database of 3840 compounds that included novel and diverse chemotypes. Three compounds showed appropriate activity and specificity - C9433, C8808 and C5557 - all belonging to the flavagline family – derivatives of the natural product rocaglamide A, also known as rocaglates.[32] Rocaglamide derivatives had been previously described to have anti-leukemic and cytotoxic properties. These compounds were observed to act synergistically with low doses of IFN-γ within the macrophage to activate mRNA expression of Irf1 – a transcription factor essential for host resistance to TB. They were also observed to induce stress responses and autophagy, and to decrease expression of nitric oxide synthase and type I IFN.[32]
Pharmacological manipulation of immune pathways, such as those of TNF-α and IFN-γ, constitutes a form of host-directed therapy (HDT), which, rather than eradicating the pathogenic organism, potentiates the mechanisms of effector immunity in the host to alter the pathogenesis of disease. Rocaglate derivatives may therefore be potential agents of HDT, allowing for fine-tuning of macrophage effector functions.
Much remains to be discovered about the relationship between type I IFN and the development of host tolerance to intracellular pathogens, but the mechanisms involved may constitute new biomarkers and therapeutic targets.[27] At the same time, the successful experiments performed by the Kramnik group highlight the importance of animal models, as they suggest that mice may be suitable to reproduce a spectrum of pulmonary tuberculosis to more accurately predict the efficacy of new interventions in genetically heterogeneous human populations.[33]
TB Diagnostics
The challenges in diagnosing TB have spurred new and innovative methods. One potential approach is the systematic analysis of host RNA gene expression in response to TB. Several studies have demonstrated that specific transcriptional signatures can distinguish between TB and other inflammatory and infectious diseases, and between active TB and latent TB infection. Dr. Robert J. Wilkinson discussed the study conducted in the United Kingdom and South Africa that identified the 86-gene whole blood signature specific for TB.[34] The signature was found to correlate with disease extent and to diminish in response to adequate TB treatment, reverting towards that of healthy controls. It allowed differentiation between active TB and other diseases with a sensitivity ranging from 90-92% and a pooled specificity of 83%. Interestingly, a few (5-10%) patients with a diagnosis of LTBI shared the same transcriptomic signature as those with active disease, suggesting that the presence of this signature may predict progression from latency to the active stage. Using pathway analysis, the investigators determined that neutrophil-driven interferon (IFN) signaling is the dominant pathway in this signature, as genes downstream of both IFN-γ and type I IFN-α/β receptor signaling were significantly over-represented.[34] Knowledge of the specific genetic expression and response to TB has implications for the development of new therapeutic targets and vaccines, as well as new diagnostic tests.
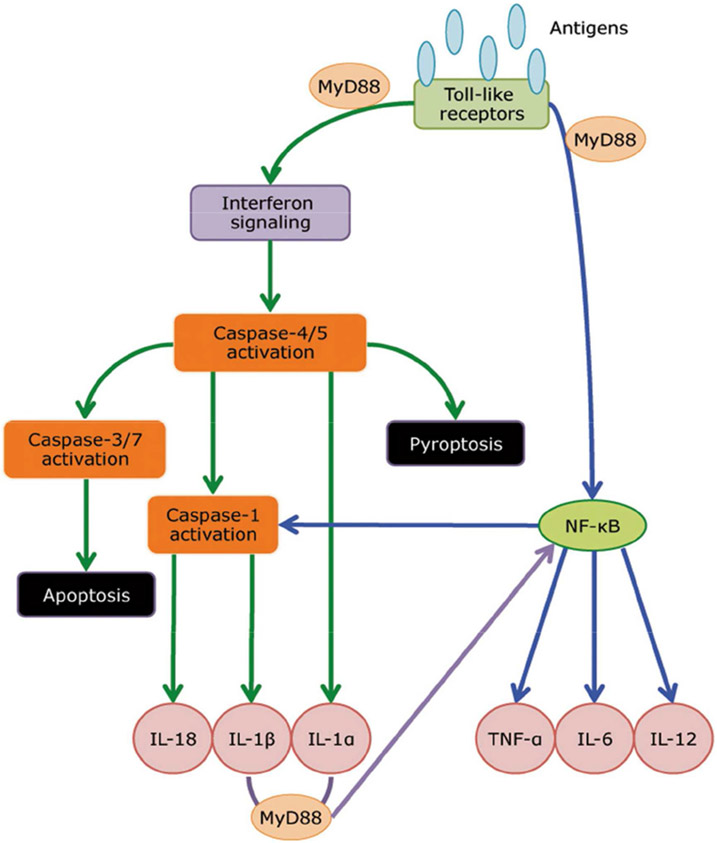
Transcriptomic profiling has also been applied in cases where a previously stable patient with HIV-associated TB initiates antiretroviral therapy (ART) and, as a consequence, develops immune reconstitution inflammatory syndrome (IRIS) with exacerbated inflammatory features of TB. To date it had been impossible to predict who will develop IRIS with ART initiation. Wilkinson’s group examined the transcriptomic signature of patients with HIV-associated TB with the goal of detecting any potential associations with the risk of developing IRIS.[35] Although initially there was no difference in the transcriptomes of patients who went on to develop TB-IRIS and those who did not, within 2 to 5 days after ART initiation there was a detectable transcript signature of 22 transcripts. By 2 weeks, a 43-transcript signature was identified as characteristic of TB-IRIS. Functional analysis of the identified genes showed downstream over-expression of predominantly innate immune signaling pathways, including TREM-1, TLR, IL-1, and type I IFN, with ultimately increased levels of pro-inflammatory cytokines and chemokines, including TNF-α, IL-1β, IL-6, IL-8, IL-12, IL-18, granulocyte–macrophage colony-stimulating factor (GM-CSF), monocyte chemoattractant protein-1, and macrophage inflammatory protein-2. Based on these findings, the cause of TB-IRIS appears to be a change in innate recognition of a pre-existing pathogen load with downstream inflammatory consequences (Figure 2). Identification and interruption of the pathways involved in this phenomenon may help prevent the condition. The biomarker also could be used as an indication for patients that would benefit from preventive measures, such as corticosteroids.
Figure 2 -.
A model of innate receptor signaling in mediating TB-IRIS pathogenesis. Microarray profiling revealed that TLR signaling and inflammasome activation are critical in mediating TB-IRIS pathogenesis. Our proposed model begins with M. tuberculosis antigen recognition by surface-expressing TLRs, which triggers the downstream signaling cascade with adaptor molecules such as MyD88 and IRAK4 to activate IRF7, thereby triggering the production of type I IFN. Paracrine signaling of Type I IFN to IFNAR recruits and phosphorylates STAT1/2 dimers, leading to further recruitment of IRF9 and the formation of ISGF3, thereby inducing pro-caspase-11 (caspase-4/5 in human) and AIM-2 inflammasome (caspase-1). Caspase-11 cleaves IL-1α into its mature form and can lead to pyroptosis. The noncanonical inflammasome (caspase-11) can also activate the canonical inflammasome (caspase-1), which cleaves IL-1β and IL-18 into their mature form. Alternatively, TLR signaling via MyD88 can also activate NF-κb via the TAK1/IKK complex. Activation of NF-κb triggers the production of an array of cytokines, including TNF-α, IL-6 and IL-12. In addition, NF-κb can also activates NLRP1/3 inflammasomes and subsequently leads to the production of IL-1β and IL-18.
Lai RPJ, Meintjes G, Wilkinson RJ. HIV-1 tuberculosis-associated immune reconstitution inflammatory syndrome. Semin Immunopathol 2016;38:185–98. doi:10.1007/s00281-015-0532-2.
Dr. Robert Husson reported on the potential application of urinary proteins as biomarkers for TB in pediatric populations. TB diagnosis can be particularly challenging in children, given the low rates of microbiological confirmation. Biomarkers measurable in the urine are of particular interest, as they represent an easy, quick, and inexpensive method with the potential for point-of-care, adequate for this population. Small volumes of urine are needed (as little as 300 uL), and samples are easily obtained in a non-invasive manner that carries little biohazard risk when compared to blood draws. Different proteins and combinations of proteins can be identified through mass spectrometry with the goal of screening, confirming a diagnosis, or evaluating a therapeutic response.
Another innovative approach to TB markers is the analysis of pathogen-specific lipid profiles, also known as lipidomics. This includes the identification and quantification of lipid molecules, their associated pathways, and relationship to other metabolites within an organism. A relatively new field, the study of metabolites largely relies on nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS) as detection methods. The new technologies allow for separation, identification and profiling of lipid repertoires into comprehensive databases, generating an organism’s lipidome.[36] Dr. Branch Moody described progress in the lipidomic dissection of MTB. A lipid profile unique to MTB would have the potential to serve as a diagnostic target. Comparison between the lipidome of BCG and MTB strain H37Rv revealed more than 1,000 molecular differences. Significantly high concentrations of the previously unknown 1-tuberculosinyladenosine (1-TbAd) were found in MTB, but not in BCG.[37] Shedding of 1-TbAd from MTB has been detected in vitro and in the lungs of infected mice in vivo.[38] Detection of pathogen-shed antigens is used in the diagnosis of other infections, such as Legionella and Cryptococcus, but has never been used in tuberculosis. It can be a highly specific method provided that the target is a well-known species-specific pathogen. Because lipids have relative evolutionary conservation across different species, they have been overlooked as potential candidates. However, the MTB lipidome is an exception, containing more than 100 subclasses of species-specific lipids. The promising MTB-specific 1-TbAD could be an important target in TB diagnostics.
Drug Development
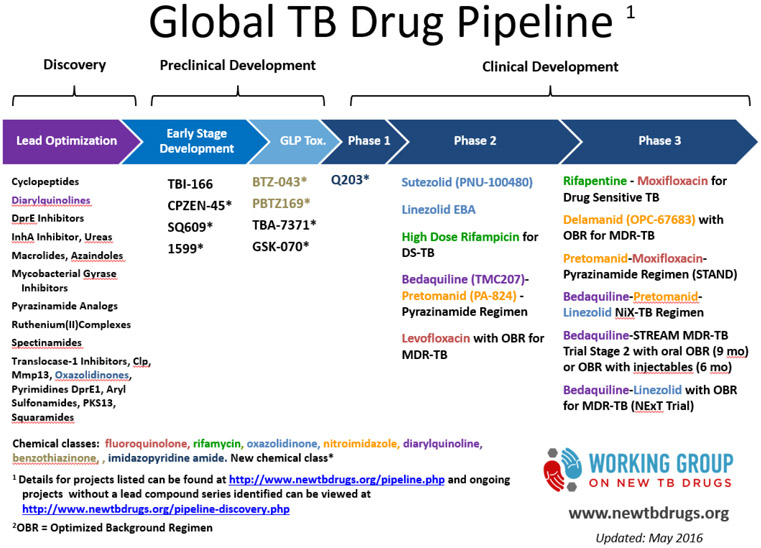
Despite pharmacological advances and the undeniable benefits of current anti-tuberculous drugs, TB treatment remains sub-optimal. The long course duration poses a challenge to patient compliance that increases the risk of failure and relapse. Even among those that complete adequate treatment there are a small number of cases that relapse, which reflects incomplete bacterial clearance by the active drugs. There is great need for drugs that can increase the treatment success rates by improving MTB killing and shortening treatment regimens. Dr. Susan Dorman provided an update on the current efforts in anti-TB drug development. There are a number of drug candidates currently in preclinical development, and approximately one dozen drugs/drug combinations undergoing clinical testing (Figure 3). There is a need for a greater number of later stage drug candidates, trials that include children and adolescents, trials for non-pulmonary TB (especially TB meningitis), and studies of surrogate markers of sterilizing activity that can help identifying “true cure”. A surrogate marker is a test that can serve as a substitute for a clinically meaningful endpoint and is expected to predict the effect of a therapy.
Figure 3 -.
Current TB drug pipeline (http://www.newtbdrugs.org/pipeline.php, accessed on July 7th, 2016)
In order to meet the challenges of effectively clearing MTB infection, new approaches will be needed. Dr. Eric Rubin discussed the role of genetics in drug discovery. To date no antibiotics have arisen from genetic approaches. Current drugs act on a limited group of well-known bacterial targets with well-described mechanisms of resistance. The possibilities of genetic analysis include identification of new targets and pathways, drug resistance mechanisms, mechanisms of action, and new compounds. Genome sequencing is used to identify specific genes required for bacterial growth. Interestingly, disruption of many MTB genome sequences rarely produces a significant impact on bacterial growth and fitness. This may be due to redundancy of genes and pathways, or to the differential activation of genes in specific stages of disease (activity versus latency).[39] Once an essential gene has been identified, inhibition must be proven to affect bacterial viability in both in vitro and in vivo conditions. It must also be deemed a “drugable” target.[39] Genetic sensitization is a target-based whole cell screening (TB-WCS) discovery tool that uses mutant strains that under-express essential genes to discover new drug targets. Laboratorial models must mimic the conditions of MTB infection in the host by creating stress conditions of hypoxia, low pH, NO stress, and short chain fatty acids as the source of carbon. Under these conditions, high throughput screening (HTS) can be used to model and manipulate the host-pathogen system as a whole. HTS is a data processing software that can rapidly identify active compounds, antibodies, or genes that modulate a particular biomolecular pathway.
The combination of these methods has yielded a number of drug-target pair candidates. Diarylquinolines (R207910) were found to inhibit the proton pump of adenosine triphosphate (ATP) synthase, killing both drug-sensitive and drug-resistant MTB. Compared to standard treatment regimens, diarylquinolines (bedaquiline) accelerated bactericidal activity and led to complete culture conversion in 2 months.[40] After proving efficacious in human volunteers, and despite remaining questions regarding its safety, the class received approval from the Food and Drug Administration (FDA) for the treatment of adults with pulmonary MDR-TB.[41]
Another promising class, 1,3-benzothiazin-4-ones (BTZs) were identified as inhibitors of the enzyme decaprenylphosphoryl-beta-d-ribose 2'-epimerase – a key precursor of arabinans, essential components of the cell wall.[42] BTZ043, a compound of the BTZ class, is able to eradicate MTB populations in vitro, ex vivo, and in the mouse model.[43] The candidate is currently in late-stage preclinical development. Further studies will help characterize its profile of action in tuberculosis treatment.[42]
Similarly, PA-824 (or pretomanid), a prodrug of the nitroimidazole class, was found to have an antitubercular effect on both actively replicating and latent bacteria when reduced to its active form by the enzyme deazaflavin-dependent nitroreductase (Ddn).[44,45]
Decreasing the expression of an essential gene will often increase the cell's sensitivity to inhibitors that act, directly or indirectly, on the same biochemical pathway.[39] As an example, PanC (pantothenate synthetase), a promising recently discovered enzymatic target, is required for the synthesis of membrane fatty acids. It is essential for MTB growth, but absent in mammals.[46,47] Inhibition of PanC directly limits mycobacterial growth, but also renders the cells more susceptible to drugs targeting cell wall biosynthesis, such as isoniazid and ethambutol.[39] Out of a molecular library, HTS has identified 3-biphenyl-4-cyanopyrrole-2-carboxylic acids as a class of compounds that effectively inhibits PanC and MTB growth.[46]
Ioerger et al used whole-cell screening and HTS to discover compounds that inhibit growth and subsequently identified in vitro mutants with intrinsic resistance to them. Whole-genome sequencing of these strains identified resistance-associated polymorphisms whose roles were then tested using recombinant strains in which the single point mutations were introduced. Using this method, the group was able to identify resistance-linked genes for eight anti-tubercular compounds, including three novel candidates: Pks13 – polyketide synthase involved in mycolic acid biosynthesis, AspS – aspartyl-tRNA synthetase, and EccB3 – a component of the ESX-3 type VII secretion system. These targets are not targeted by currently available drugs, but whole-cell screening and HTS hold the potential for discovery of new agents.[48]
HTS has the potential to discover not only new compounds, but also new mechanisms of synergy. Classically, synergistic compounds act on the same targets. Physiological synergy, on the other hand, occurs when different molecules preferentially act on distinct metabolic states of the bacterium. Most desirably, compounds can accentuate the anti-bacterial activity of the host immune system and other drugs, in a host-pathogen synergistic manner, by enhancing drug delivery or penetration, and altering bacterial state. A variety of compounds has been proposed for investigation, including modulators of immunity, ion channels, kinase cascades, inflammation, autophagy, and lipid metabolism.[39]
Dr. James Collins described the application of engineering principles at the cellular and molecular level in order to create networks of synthetic biology. The models include extensive circuits of DNA sequences, signaling pathways, transcription regulators, and gene products. The different system components are programmable into modeling the pathways discovered and mapped in real life cell systems. This network biology approach can be applied to various cell systems, including those of pathogens. The goal is to enable computational simulation of how manipulating different components or events (“turning on and off different switches”) can affect different mechanisms. For example, with MTB infection antibiotics often target more than one mechanism of bacterial structure and metabolism. Some of these mechanisms remain unknown, as do the mechanisms underlying the emergence of resistance. Ultimately, the more or less efficacious action of a drug can result in bacterial lethality, stasis, or tolerance. Dr. Collins research aims to ascertain which switches are fundamental for the optimization of antibiotic activity by targeting and boosting specific processes in cellular damage.
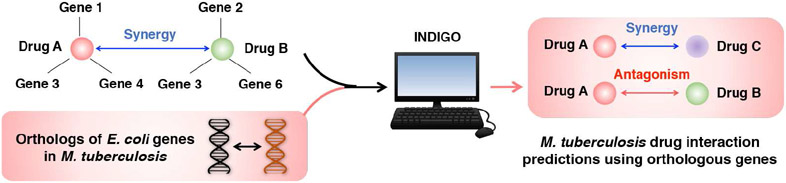
Dr. Collins’ group has developed a computational approach called INDIGO, which uses chemogenomics data to predict antibiotic combinations that interact synergistically or antagonistically in inhibiting bacterial growth (Chandrasekaran et al., 2016, in press). Combination antibiotic therapies are being increasingly used in the clinic to enhance potency and counter drug resistance. However, the large search-space of candidate drugs and dosage regimes makes the identification of effective combinations highly challenging. INDIGO addresses this challenge by making use of chemogenomic profiling, a high-throughput approach that identifies genes that when deleted, can enhance or reduce sensitivity to an individual drug. INDIGO quantifies the influence of individual chemical-genetic interactions on drug synergy and antagonism. To train INDIGO, they experimentally measured interaction outcomes for a large panel of antibiotic combinations and compiled chemogenomics data from literature for the model organism E. coli. Their analysis revealed a core set of genes and pathways (e.g., central metabolism) that are predictive of antibiotic interactions in E. coli. By identifying interactions that are associated with orthologous genes, they successfully estimated drug-interaction outcomes in the bacterial pathogens Mycobacterium tuberculosis and Staphylococcus aureus, using the E. coli INDIGO model. Novel predictions were then experimentally validated in E. coli and S. aureus. By applying the approach to over a million potential combination therapies against tuberculosis, they have identified novel combinations that are more effective, i.e. more synergistic, than currently-used combinations for treating tuberculosis (Chandrasekaran et al. unpublished data). INDIGO thus enables the discovery of effective combination therapies in less-studied pathogens by leveraging chemogenomics data in model organisms.
The metabolic state of bacteria influences their susceptibility to antibiotics. At the same time, bacteria can enhance their tolerance to antibiotics by altering their metabolic state. Data suggest that antibiotic efficacy is linked to bacterial cellular respiration, as uncoupling of ATP synthesis accelerates respiration and results in enhanced bactericidal efficacy. If through network biology one could determine how metabolic pathways of the host and bacterial cells are controlled, new therapeutic compounds could be developed that target and manipulate the right switches towards successful eradication of MTB.
Vaccine Development
Disease control remains a challenge in tuberculosis due, in part, to the lack of an approach to prevent infection, prevent progression of the latent stage to active TB, prevent recurrence after treatment or prevent reinfection. The search for a vaccine includes agents that can act at three levels: pre-exposure (preventing infection), post-exposure (preventing disease), and therapeutic (optimizing treatment). Dr. Edward Nardell discussed the importance of vaccines, prioritizing the development of agents that can prevent infection rather than disease. This approach would minimize the costs associated with the larger vaccine trials necessary to study prevention of disease. There is great demand not only for preventative vaccines, but for therapeutic vaccines as well. The addition of a therapeutic vaccine that can optimize the current treatment regimen could mean a reduction in treatment duration, increase in compliance, and increase in efficacy in clearing MTB and avoiding transmission. This would be particularly important in the context of MDR-TB.
Dr. Nardell described BCG vaccination in the guinea pig natural transmission model. In human populations, BCG is mainly effective for the prevention of extrapulmonary complications of TB in neonates. There is limited evidence and discordant results on how BCG impacts the risk of disease and modulates disease severity in adult populations. In the guinea pig animal model, BCG slows the progression and reduces the severity of TB disease, especially when combined with drug-therapy. In one study,[49] vaccinated and unvaccinated guinea pigs received drug treatment. The vaccinated group developed fewer granulomas, and therefore less residual infection and reactivation disease.[49] To better understand why, Ordway et al used flow cytometry to monitor changes in cell populations in the lung and lymph nodes of vaccinated and unvaccinated guinea pigs that were exposed to a virulent strain of MTB.[50] Comparatively, the vaccinated group showed a reduced influx of neutrophils, associated with an early and significantly increased influx of CD4 and CD8 T cells, B cells, and MHCII-expressing macrophages into the lungs.[50] The robust response of CD4 T cells induced by BCG was shown to occur in response to specific highly immunogenic mycobacterial lipopeptides, highlighting the importance of unique lipid patterns in the immune response against MTB.[51] The efficacy of BCG vaccination varied with inoculum size, strain virulence, duration of exposure, and reinfection occurrence.
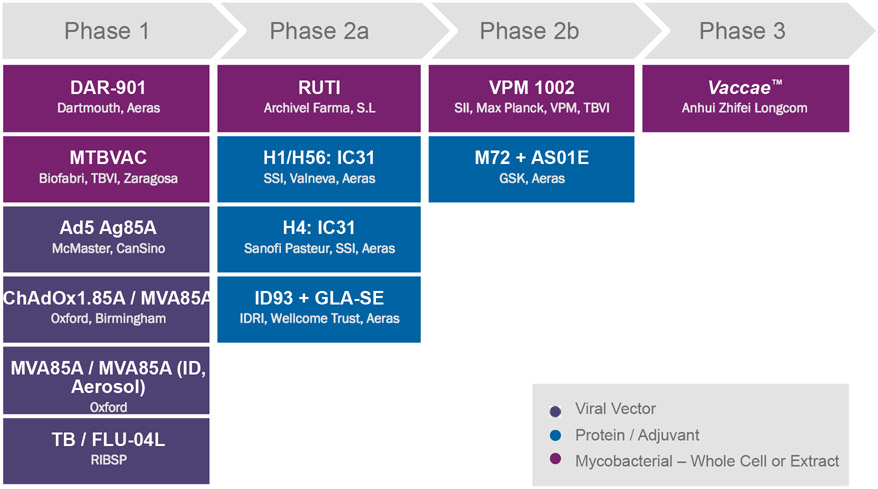
Dr. Lewis Schrager discussed the outcome of the MVA85A trial, which failed to show efficacy beyond that provided by BCG in a phase IIb study of TB disease prevention in BCG-vaccinated infants. Strategies to address the ongoing challenges to TB vaccine development include the development of more rigorous criteria for selection of vaccine candidates for large-scale trials, the creation of better animal and human models, selecting vaccine candidates that generate a greater diversity of immune responses beyond the classical CD4+ and CD8+ T-cell responses manifest by the current candidates, and the development of relevant functional assays.[52] Dr. Schrager concluded with an overview of the 13 candidates in the vaccine clinical pipeline. Currently there are six TB vaccine candidates in phase I, four in phase IIa, two in phase IIb, and one in phase III clinical trials (Figure 3).
VPM1002, a prime recombinant vaccine, was discussed by Dr. Umesh Shaligram. VPM1002 is a recombinant mutant of BCG that includes a gene of Listeria monocytogenes coding for the protein listeriolysin. Listeriolysin forms pores in the membrane of endocytic phagosomes that allow for the bacterium-derived peptides to be presented to MHC I pathways in the cytosol. In contrast with MHC II pathways occurring within the phagosome, MHC I triggers CD8 cell populations, enhancing the mechanisms of protective immunity. The vaccine’s action is optimized by acidic environments in which listeriolysin is preferentially active. For this reason, and unlike BCG, the gene for urease C has been inactivated in VPM1002, as hydrolysis of urea into carbon dioxide and ammonia causes an increase in pH. VPM1002 is intended to prevent disease after MTB infection, but as a second indication, it can also be administered post-exposure to prevent relapse after TB treatment completion. It was developed by investigators at the Max Planck Institute for Infection Biology and is currently being produced by the Serum Institute of India. Much like BCG, its target population is composed of newborns of high endemic areas. However, compared to BCG, VPM1002 has superior safety and efficacy, inducing and expanding central memory T cells (TCM) and follicular helper T cells (TFH) populations.
VPM1002 has successfully completed preclinical studies in animal models and a phase Ia trial in Germany that included a cohort of healthy males with and without past exposure to BCG. In South Africa, a phase Ib trial enrolled healthy males and females with pre-exposure to BCG living in a TB endemic area, and phase IIa followed BCG-unexposed newborns. In all trials VPM1002 was reported to be safe and highly immunogenic, inducing multifunctional T cells.
In the post-exposure setting, a clinical trial will be conducted in India, including RePORT India sites. The study population will be a cohort of 2000 TB patients who successfully completed TB treatment. They will be randomized into a vaccine or placebo group and followed for 12 months. RePORT India sites will participate in this trial. Interestingly, VPM1002 is also being tested for its potential benefits in preventing bladder cancer. This immunotherapeutic property is yet another characteristic shared with BCG
Dr. Steve Reed described one of the candidates currently undergoing development at the Infectious Disease Research Institute, Seattle, WA, USA. ID93 is a recombinant vaccine that combines four antigens associated with virulence and latency (stress-induced) factors of Mycobacterium tuberculosis: Rv2608, Rv3619, Rv3620, and Rv1813. All four antigens are found in both MTB and BCG. The genome sequences that encode pathogenic antigens can be cloned, purified, and manipulated so as to be expressed by recombinant DNA technology. Recombinant vaccines use this technology to express one or multiple antigens of a certain pathogen on a plasmid or bacterial/viral vector and induce host immunity against it. Because they present weak immunogenicity when given alone, an adjuvant is often administered in combination for an improved long-lasting immune response. ID93 was originally intended for pre-exposure prevention of infection and disease, although it could also be applied post-infection to prevent recurrence. It will soon also be tested as a therapeutic vaccine adjunct to antibiotic treatment. Generally, the goal is to boost the response to previously administered BCG, preventing disease and/or disease recurrence in BCG-immunized individuals and/or individuals with prior TB disease. The vaccine will be administered with glucopyranosyl lipid adjuvant-stable emulsion (GLA-SE), a toll-like receptor-4 agonist, as the adjuvant.
In animal models, ID93 showed promising results. MTB-infected mice and cynomolgus macaques that received ID93 in addition to an antibiotic course showed reduced bacterial loads, improved lung pathology and survival rates. The treatment elicited a more robust Th1 immune response, shortening the duration of antibiotic therapy. ID93 has been administered to healthy adults in randomized double-blind phase I trials. The subjects were aged 18 to 45 years old, HIV-uninfected, and BCG-vaccinated. ID93 showed good immunogenicity and safety profile on both phase Ia and Ib studies, with only mild and transient adverse events, and no serious adverse effects (SAEs) or fatalities reported. All subjects receiving the vaccine showed an increase in CD4 T cell response, as measured by interferon-gamma (IFN-γ) Enzyme-Linked ImmunoSpot (ELISPOT) analysis and intracellular cytokine staining of peripheral blood mononuclear cells (PBMCs). A phase IIa study of safety and prevention of disease recurrence is currently ongoing in a population of TB patients who completed treatment. Planning is in progress to test the vaccine on a cohort of TB patients at different times during the course of antibiotic treatment. Given that reducing treatment duration is one of the desired goals for a successful vaccine, this will help elucidate the optimal time of vaccination.
The H56:IC31 fusion protein, originally designed to boost BCG responses, is a promising candidate for a post-exposure and therapeutic vaccine as well. It expresses antigens Ag85B, ESAT-6, and Rv2660c – the latter being a dormancy-specific antigen useful in targeting latent forms of the bacteria. Dr. Morten Ruhwald reported that in phase I and IIa trials the vaccine was well tolerated and immunogenic in adults and adolescents, inducing T-central memory (TCM) cells with polyfunctional (IFN-γ+/IL-2+/TNF-α+) and bi-functional (IL-2+/TNF-α+) profiles irrespective of pre-existing MTB infection and HIV status. When administered to cynomolgus macaques as a booster to BCG, this vaccine reduced clinical disease after challenge with MTB and prevented reactivation of latent foci.
The scientific rationale for vaccine development to date has focused on the ability of host T cells to provide lasting immunity against MTB infection. The memory response from natural TB infection is not always sufficient to protect from subsequent challenge, as evidenced by the occurrence of re-infection. Provided that T cells have preserved function, this is presumably due to the bacteria’s ability to evade immunity. Both CD4+ and CD8+ T cells play important roles in the pathogenesis of TB. Whereas depletion of CD4+ cells causes a more pronounced risk of developing active TB following initial infection, CD8+ cells are also recruited to the site of infection and are important in controlling the latent stage of disease.[53] The immunodominant MTB antigen TB10.4 (EsxH) elicits a robust memory CD8+ T cell response,[54] but only small numbers of TB10.4-specific cells are activated upon new exposure. Dr. Samuel Behar and his team compared naïve and memory CD8+ T cell populations during stimulation with TB10.4 and found that the primary (naïve) response outnumbered the secondary (memory) response.[55] Normally, memory T cells respond more rapidly than the primary (naïve) response, which suggests that impairment of adequate memory T cell expansion may be the reason for the inefficacy of some vaccine candidates against MTB. Expansion of memory T cell populations relies on T cell receptor (TCR)-associated factors and is dependent on adequate exposure to significant levels of antigen. This research group proposes that the reduced fitness of memory CD8+ T cells may be due to low sensitivity to the antigen, a consequence of decreased antigen presentation. This can be associated with the mechanical obstacles posed by granulomas and the limited function of infected antigen-presenting cells (APC) in the lung, which can mimic a “low antigen” state and lead to suboptimal T cell activation.[54] Another possibility is that chronic inflammation leads to T cell dysfunction.[56] T cell sensitivity to antigens can be improved by increasing TCR affinity – a potential goal of future vaccine candidates. Vaccines that can elicit high affinity T cells would probably be more effective than ones that elicit large numbers of T cells.
Conclusions
Challenges remain on all fronts of TB research. A better understanding of the pathogenesis of infection is the cornerstone for the development of better diagnostic methods, drug regimens, and vaccines. At the first meeting of the RePORT International network the activity, scope and promise of recent basic and translational research on TB were highlighted. The translational advances already have yielded promising new vaccine, diagnostic and drug candidates for prevention, detection and treatment of TB. The basic research has fostered new paradigms and concepts that in the long run could lead to fundamental shifts in approaches relevant to TB eradication.
Building capacity among investigators in high-burden countries to do cutting-edge TB research requires more than just training investigators and study staff, and getting laboratories to perform at high standard. It also requires them to be engaged in hearing about and critically appraising the newly presented data and conclusions from in vitro and animal model studies, and engage in the back-and-forth of digesting new data, and the back-and-forth of scientific discourse. The meeting in Boston presented a continuum of ideas, some of which are directly applicable to programs in the near future, and others of which form the basis for the next important questions to pose in the field research setting.
The goal of the RePORT network is to enable and facilitate research that promotes understanding of TB pathogenesis, the creation of TB diagnostics, and the development of TB drugs and vaccines. Initial RePORT consortia are situated in high-burden middle-income countries, which can benefit from the research findings while simultaneously developing into mature clinical research sites. The International Common Protocol provides standardized clinical definitions and SOPs for collecting data and specimens, hopefully making use, re-use and sharing for future investigations both feasible and informative. RePORT continues to expand in both collaborations, geographic locations, and expertise. A RePORT International Coordinating Center (RICC) has been created that will facilitate communication and collaboration between RePORT networks. The RICC will develop, curate, and improve RePORT SOPs, provide seed money to advance cross-consortia scientific priorities, host scientific workshops, and coordinate data and specimen sharing. With the expansion of its coordinated efforts, the RePORT sites will have unprecedented opportunities to contribute to the research necessary to overcome remaining obstacles to TB eradication. The RICC will work to ensure that the global TB research community is aware of the RePORT network, views it as a global resource, and seeks collaborations as appropriate.
Figure 4 -.
INDIGO takes in known drug-interaction and chemogenomics data in model organisms (E. coli), and enables large-scale prediction of drug interaction outcomes. By analyzing the evolutionary conservation of genes identified by INDIGO to be predictive of drug interactions in E. coli, we successfully predicted drug synergy and antagonism in the bacterial pathogens Mycobacterium tuberculosis and Staphylococcus aureus.
Figure 5 -.
Current TB vaccine pipeline (information self-reported by vaccine sponsors to AERAS, last revised on December 17th, 2015)
Acknowledgments
Research reported in this publication was, in part, supported by the National Institute Of Allergy And Infectious Diseases of the National Institutes of Health (under Award Number U19AI111276), and by the Lifespan/Tufts/Brown Center for AIDS Research (Grant Number P30AI042853 from the National Institute Of Allergy And Infectious Diseases). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of Allergy And Infectious Diseases or the National Institute of Health.
Information in this manuscript was, in part, collected as part of the Regional Prospective Observational Research for Tuberculosis (RePORT) India Consortium Activities. This project has been funded in whole or in part with Federal funds from the Government of India’s (GOI) Department of Biotechnology (DBT), the Indian Council of Medical Research (ICMR), the United States National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID), Office of AIDS Research (OAR), and distributed in part by CRDF Global. The contents of this publication are solely the responsibility of the authors and do not represent the official views of the DBT, the ICMR, the NIH, or CRDF Global. Any mention of trade names, commercial projects, or organizations does not imply endorsement by any of the sponsoring organizations.
Appendix
Participating institutions§, Principal Investigators†, co-principal investigators°, DBT, ICMR, and NIAID staff include:
Byramjee Jeejeebhoy Medical College (BJMC)§: Vidya Mave†, Dileep B. Kadam°;
National Institute for Research in Tuberculosis (NIRT)§: Padmapriyadarsini Chandrasekaran†;
Johns Hopkins University (JHU)§: Amita Gupta†;
Blue Peter Public Health and Research Centre (BPHRC)-LEPRA§: Vijaya Valluri†;
University of Texas Health Center§: Ramakrishna Vankayalapati†;
Christian Medical College (CMC)§: Devasahayam Christopher†;
University of Cambridge§: Lalita Ramakrishnan†;
University of Washington§: John Szumowski†;
Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER)§: Gautam Roy†;
Boston Medical Center§: Jerrold Ellner†;
Boston University§: Natasha Hochberg°;
Rutgers, New Jersey Medical School§: Padmini Salgame°;
M.V. Diabetes Research Centre (MVDRC)§: Vijay Viswanathan†;
University of Massachusetts Medical School§: Hardy Kornfeld†;
The Centre for Health Research and Development - Society for Applied Studies (CHRD-SAS)§: Sunita Taneja;
Westat§: Sonia K Stoszek, Georgine Price, Fatima Jones, Amanda Fournier, Bob Harris;
FHI 360§: Carol Dukes Hamilton;
DBT§: T.S. Rao, Jyoti Logani;
ICMR§: Soumya Swaminathan and Rashmi Arora;
NIAID§: Peter Kim, Sudha Srinivasan; and
CRDF Global§: Tara Caton
Revision and approval of the scientific content of this manuscript was made possible with collaboration from the speakers of the First Meeting of RePORT International:
Bruno B. Andrade, Unidade de Medicina Investigativa, Laboratório Integrado de Microbiologia e Imunorregulação, Instituto Gonçalo Moniz, Fundação Oswaldo Cruz, Salvador, Brazil
Samuel Behar, University of Massachusetts Medical School, Worcester, Massachusetts, USA
James Collins, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA
Susan Dorman, Johns Hopkins School of Medicine, Baltimore, Maryland, USA
Robert Husson, Harvard Medical School and Boston Children’s Hospital, Boston, Massachusetts, USA
Igor Kramnik, Boston University School of Medicine and National Emergent Infectious Diseases Laboratories, Boston, Massachusetts, USA
William Jacobs, Albert Einstein College of Medicine, New York City, New York, USA
Kathleen McDonough, Wadsworth Center, Albany, New York, USA
Megan Murray, Harvard Medical School and Brigham and Women’s Hospital, Boston, Massachusetts, USA
Branch Moody, Harvard Medical School and Brigham and Women’s Hospital, Boston, Massachusetts, USA
Edward Nardell, Harvard Medical School and Brigham and Women’s Hospital, Boston, Massachusetts, USA
Steve Reed, Infectious Disease Research Institute, Seattle, Washington, USA
Eric Rubin, Harvard University School of Public Health, Boston, Massachusetts, USA
Morten Ruhwald, Statens Serum Institute in Denmark, Copenhagen, Denmark
Umesh Shaligram, Serum Institute of India, Pune, India
David Sherman, Center for Infectious Disease Research, Seattle, Washington, USA
Lewis Schrager, AERAS
Robert Wilkinson, University of Cape Town, Cape Town, South Africa
The First Meeting of RePORT International was made possible with support from the Boston University School of Medicine, Department of Medicine, and help from David Hom, Rachelle Joseph and Valine Valbrun, from Boston Medical Center, Section of Infectious Disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hamilton CD, Swaminathan S, Christopher DJ, Ellner J, Gupta A, Sterling TR, et al. RePORT International: Advancing Tuberculosis Biomarker Research Through Global Collaboration. Clin Infect Dis 2015;61Suppl 3:S155–9. doi: 10.1093/cid/civ611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lambert M-L, Hasker E, Van Deun A, Roberfroid D, Boelaert M, Van der Stuyft P. Recurrence in tuberculosis: relapse or reinfection? Lancet Infect Dis 2003;3:282–7. [DOI] [PubMed] [Google Scholar]
- [3].Crampin AC, Mwaungulu JN, Mwaungulu FD, Mwafulirwa DT, Munthali K, Floyd S, et al. Recurrent TB: relapse or reinfection? The effect of HIV in a general population cohort in Malawi. AIDS 2010;24:417–26. doi: 10.1097/QAD.0b013e32832f51cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Vilchèze C, Hartman T, Weinrick B, Jacobs WR. Mycobacterium tuberculosis is extraordinarily sensitive to killing by a vitamin C-induced Fenton reaction. Nat Commun 2013;4:1881. doi: 10.1038/ncomms2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Newton GL, Buchmeier N, Fahey RC. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol Mol Biol Rev 2008;72:471–94. doi: 10.1128/MMBR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vilchèze C, Av-Gay Y, Attarian R, Liu Z, Hazbón MH, Colangeli R, et al. Mycothiol biosynthesis is essential for ethionamide susceptibility in Mycobacterium tuberculosis. Mol Microbiol 2008;69:1316–29. doi: 10.1111/j.1365-2958.2008.06365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Khameneh B, Fazly Bazzaz BS, Amani A, Rostami J, Vahdati-Mashhadian N. Combination of anti-tuberculosis drugs with vitamin C or NAC against different Staphylococcus aureus and Mycobacterium tuberculosis strains. Microb Pathog 2016;93:83–7. doi: 10.1016/j.micpath.2015.11.006. [DOI] [PubMed] [Google Scholar]
- [8].Gazdik MA, Bai G, Wu Y, McDonough KA. Rv1675c (cmr) regulates intramacrophage and cyclic AMP-induced gene expression in Mycobacterium tuberculosis-complex mycobacteria. Mol Microbiol 2009;71:434–48. doi: 10.1111/j.1365-2958.2008.06541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Knapp GS, Lyubetskaya A, Peterson MW, Gomes ALC, Ma Z, Galagan JE, et al. Role of intragenic binding of cAMP responsive protein (CRP) in regulation of the succinate dehydrogenase genes Rv0249c-Rv0247c in TB complex mycobacteria. Nucleic Acids Res 2015;43:5377–93. doi: 10.1093/nar/gkv420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ranganathan S, Bai G, Lyubetskaya A, Knapp GS, Peterson MW, Gazdik M, et al. Characterization of a cAMP responsive transcription factor, Cmr (Rv1675c), in TB complex mycobacteria reveals overlap with the DosR (DevR) dormancy regulon. Nucleic Acids Res 2016;44:134–51. doi: 10.1093/nar/gkv889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, Sweet L, et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 2013;499:178–83. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Minch KJ, Rustad TR, Peterson EJR, Winkler J, Reiss DJ, Ma S, et al. The DNA-binding network of Mycobacterium tuberculosis. Nat Commun 2015;6:5829. doi: 10.1038/ncomms6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rustad TR, Minch KJ, Ma S, Winkler JK, Hobbs S, Hickey M, et al. Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression-derived regulatory network. Genome Biol 2014;15:502. doi: 10.1186/PREACCEPT-1701638048134699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Peterson EJR, Reiss DJ, Turkarslan S, Minch KJ, Rustad T, Plaisier CL, et al. A high-resolution network model for global gene regulation in Mycobacterium tuberculosis. Nucleic Acids Res 2014;42:11291–303. doi: 10.1093/nar/gku777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ma S, Minch KJ, Rustad TR, Hobbs S, Zhou S-L, Sherman DR, et al. Integrated Modeling of Gene Regulatory and Metabolic Networks in Mycobacterium tuberculosis. PLoS Comput Biol 2015;11:e1004543. doi: 10.1371/journal.pcbi.1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Andrade BB, Pavan Kumar N, Amaral EP, Riteau N, Mayer-Barber KD, Tosh KW, et al. Heme Oxygenase-1 Regulation of Matrix Metalloproteinase-1 Expression Underlies Distinct Disease Profiles in Tuberculosis. J Immunol 2015;195:2763–73. doi: 10.4049/jimmunol.1500942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Andrade BB, Pavan Kumar N, Mayer-Barber KD, Barber DL, Sridhar R, Rekha VVB, et al. Plasma heme oxygenase-1 levels distinguish latent or successfully treated human tuberculosis from active disease. PLoS One 2013;8:e62618. doi: 10.1371/journal.pone.0062618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Andrade BB, Pavan Kumar N, Amaral EP, Riteau N, Mayer-Barber KD, Tosh KW, et al. Heme Oxygenase-1 Regulation of Matrix Metalloproteinase-1 Expression Underlies Distinct Disease Profiles in Tuberculosis. J Immunol 2015;195:2763–73. doi: 10.4049/jimmunol.1500942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Huang C-C, Tchetgen ET, Becerra MC, Cohen T, Hughes KC, Zhang Z, et al. The effect of HIV-related immunosuppression on the risk of tuberculosis transmission to household contacts. Clin Infect Dis 2014;58:765–74. doi: 10.1093/cid/cit948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zelner JL, Murray MB, Becerra MC, Galea J, Lecca L, Calderon R, et al. Bacillus Calmette-Guérin and isoniazid preventive therapy protect contacts of patients with tuberculosis. Am J Respir Crit Care Med 2014;189:853–9. doi: 10.1164/rccm.201310-1896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Odone A, Calderon R, Becerra MC, Zhang Z, Contreras CC, Yataco R, et al. Acquired and Transmitted Multidrug Resistant Tuberculosis: The Role of Social Determinants. PLoS One 2016;11:e0146642. doi: 10.1371/journal.pone.0146642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 2006;311:1770–3. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- [23].Teles RMB, Graeber TG, Krutzik SR, Montoya D, Schenk M, Lee DJ, et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science 2013;339:1448–53. doi: 10.1126/science.1233665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schenk M, Krutzik SR, Sieling PA, Lee DJ, Teles RMB, Ochoa MT, et al. NOD2 triggers an interleukin-32-dependent human dendritic cell program in leprosy. Nat Med 2012;18:555–63. doi: 10.1038/nm.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Montoya D, Inkeles MS, Liu PT, Realegeno S, Teles RMB, Vaidya P, et al. IL-32 is a molecular marker of a host defense network in human tuberculosis. Sci Transl Med 2014;6:250ra114. doi: 10.1126/scitranslmed.3009546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fabri M, Stenger S, Shin D-M, Yuk J-M, Liu PT, Realegeno S, et al. Vitamin D is required for IFN-gamma-mediated antimicrobial activity of human macrophages. Sci Transl Med 2011;3:104ra102. doi: 10.1126/scitranslmed.3003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].He X, Berland R, Mekasha S, Christensen TG, Alroy J, Kramnik I, et al. The sst1 resistance locus regulates evasion of type I interferon signaling by Chlamydia pneumoniae as a disease tolerance mechanism. PLoS Pathog 2013;9:e1003569. doi: 10.1371/journal.ppat.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Boyartchuk V, Rojas M, Yan B-S, Jobe O, Hurt N, Dorfman DM, et al. The host resistance locus sst1 controls innate immunity to Listeria monocytogenes infection in immunodeficient mice. J Immunol 2004;173:5112–20. [DOI] [PubMed] [Google Scholar]
- [29].Yan B-S, Pichugin AV, Jobe O, Helming L, Eruslanov EB, Gutiérrez-Pabello JA, et al. Progression of pulmonary tuberculosis and efficiency of bacillus Calmette-Guérin vaccination are genetically controlled via a common sst1-mediated mechanism of innate immunity. J Immunol 2007;179:6919–32. [DOI] [PubMed] [Google Scholar]
- [30].Kramnik I, Beamer G. Mouse models of human TB pathology: roles in the analysis of necrosis and the development of host-directed therapies. Semin Immunopathol 2016;38:221–37. doi: 10.1007/s00281-015-0538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pan H, Yan B-S, Rojas M, Shebzukhov YV, Zhou H, Kobzik L, et al. Ipr1 gene mediates innate immunity to tuberculosis. Nature 2005;434:767–72. doi: 10.1038/nature03419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bhattacharya B, Chatterjee S, Devine WG, Kobzik L, Beeler AB, Porco JA, et al. Fine-tuning of macrophage activation using synthetic rocaglate derivatives. Sci Rep 2016;6:24409. doi: 10.1038/srep24409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pichugin AV, Yan B-S, Sloutsky A, Kobzik L, Kramnik I, Via L, et al. Dominant role of the sst1 locus in pathogenesis of necrotizing lung granulomas during chronic tuberculosis infection and reactivation in genetically resistant hosts. Am J Pathol 2009;174:2190–201. doi: 10.2353/ajpath.2009.081075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Berry MPR, Graham CM, McNab FW, Xu Z, Bloch SAA, Oni T, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010;466:973–7. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lai RPJ, Meintjes G, Wilkinson KA, Graham CM, Marais S, Van der Plas H, et al. HIV-tuberculosis-associated immune reconstitution inflammatory syndrome is characterized by Toll-like receptor and inflammasome signalling. Nat Commun 2015;6:8451. doi: 10.1038/ncomms9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Layre E, Al-Mubarak R, Belisle JT, Branch Moody D. Mycobacterial Lipidomics. Microbiol Spectr 2014;2. doi: 10.1128/microbiolspec.MGM2-0033-2013. [DOI] [PubMed] [Google Scholar]
- [37].Layre E, Lee HJ, Young DC, Martinot AJ, Buter J, Minnaard AJ, et al. Molecular profiling of Mycobacterium tuberculosis identifies tuberculosinyl nucleoside products of the virulence-associated enzyme Rv3378c. Proc Natl Acad Sci U S A 2014;111:2978–83. doi: 10.1073/pnas.1315883111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Young DC, Layre E, Pan S-J, Tapley A, Adamson J, Seshadri C, et al. In vivo biosynthesis of terpene nucleosides provides unique chemical markers of Mycobacterium tuberculosis infection. Chem Biol 2015;22:516–26. doi: 10.1016/j.chembiol.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Baer CE, Rubin EJ, Sassetti CM. New insights into TB physiology suggest untapped therapeutic opportunities. Immunol Rev 2015;264:327–43. doi: 10.1111/imr.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Andries K, Verhasselt P, Guillemont J, Göhlmann HWH, Neefs J-M, Winkler H, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 2005;307:223–7. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- [41].Chahine EB, Karaoui LR, Mansour H. Bedaquiline: a novel diarylquinoline for multidrug-resistant tuberculosis. Ann Pharmacother 2014;48:107–15. doi: 10.1177/1060028013504087. [DOI] [PubMed] [Google Scholar]
- [42].Neres J, Pojer F, Molteni E, Chiarelli LR, Dhar N, Boy-Röttger S, et al. Structural basis for benzothiazinone-mediated killing of Mycobacterium tuberculosis. Sci Transl Med 2012;4:150ra121. doi: 10.1126/scitranslmed.3004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Makarov V, Manina G, Mikusova K, Möllmann U, Ryabova O, Saint-Joanis B, et al. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science 2009;324:801–4. doi: 10.1126/science.1171583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Manjunatha UH, Boshoff H, Dowd CS, Zhang L, Albert TJ, Norton JE, et al. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 2006;103:431–6. doi: 10.1073/pnas.0508392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Somasundaram S, Anand RS, Venkatesan P, Paramasivan CN. Bactericidal activity of PA-824 against Mycobacterium tuberculosis under anaerobic conditions and computational analysis of its novel analogues against mutant Ddn receptor. BMC Microbiol 2013;13:218. doi: 10.1186/1471-2180-13-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kumar A, Casey A, Odingo J, Kesicki EA, Abrahams G, Vieth M, et al. A high-throughput screen against pantothenate synthetase (PanC) identifies 3-biphenyl-4-cyanopyrrole-2-carboxylic acids as a new class of inhibitor with activity against Mycobacterium tuberculosis. PLoS One 2013;8:e72786. doi: 10.1371/journal.pone.0072786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Abrahams GL, Kumar A, Savvi S, Hung AW, Wen S, Abell C, et al. Pathway-Selective Sensitization of Mycobacterium tuberculosis for Target-Based Whole-Cell Screening. Chem Biol 2012;19:844–54. doi: 10.1016/j.chembiol.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ioerger TR, O’Malley T, Liao R, Guinn KM, Hickey MJ, Mohaideen N, et al. Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PLoS One 2013;8:e75245. doi: 10.1371/journal.pone.0075245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Shang S, Shanley CA, Caraway ML, Orme EA, Henao-Tamayo M, Hascall-Dove L, et al. Drug treatment combined with BCG vaccination reduces disease reactivation in guinea pigs infected with Mycobacterium tuberculosis. Vaccine 2012;30:1572–82. doi: 10.1016/j.vaccine.2011.12.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ordway D, Henao-Tamayo M, Shanley C, Smith EE, Palanisamy G, Wang B, et al. Influence of Mycobacterium bovis BCG vaccination on cellular immune response of guinea pigs challenged with Mycobacterium tuberculosis. Clin Vaccine Immunol 2008;15:1248–58. doi: 10.1128/CVI.00019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kaufmann E, Spohr C, Battenfeld S, De Paepe D, Holzhauser T, Balks E, et al. BCG Vaccination Induces Robust CD4+ T Cell Responses to Mycobacterium tuberculosis Complex-Specific Lipopeptides in Guinea Pigs. J Immunol 2016;196:2723–32. doi: 10.4049/jimmunol.1502307. [DOI] [PubMed] [Google Scholar]
- [52].Ndiaye BP, Thienemann F, Ota M, Landry BS, Camara M, Dièye S, et al. Safety, immunogenicity, and efficacy of the candidate tuberculosis vaccine MVA85A in healthy adults infected with HIV-1: a randomised, placebo-controlled, phase 2 trial. Lancet Respir Med 2015;3:190–200. doi: 10.1016/S2213-2600(15)00037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Behar SM. Antigen-specific CD8(+) T cells and protective immunity to tuberculosis. Adv Exp Med Biol 2013;783:141–63. doi: 10.1007/978-1-4614-6111-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Nunes-Alves C, Booty MG, Carpenter SM, Rothchild AC, Martin CJ, Desjardins D, et al. Human and Murine Clonal CD8+ T Cell Expansions Arise during Tuberculosis Because of TCR Selection. PLoS Pathog 2015;11:e1004849. doi: 10.1371/journal.ppat.1004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Carpenter SM, Nunes-Alves C, Booty MG, Way SS, Behar SM. A Higher Activation Threshold of Memory CD8+ T Cells Has a Fitness Cost That Is Modified by TCR Affinity during Tuberculosis. PLoS Pathog 2016;12:e1005380. doi: 10.1371/journal.ppat.1005380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Behar SM, Carpenter SM, Booty MG, Barber DL, Jayaraman P. Orchestration of pulmonary T cell immunity during Mycobacterium tuberculosis infection: Immunity interruptus. Semin Immunol 2014;26:559–77. doi: 10.1016/j.smim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]