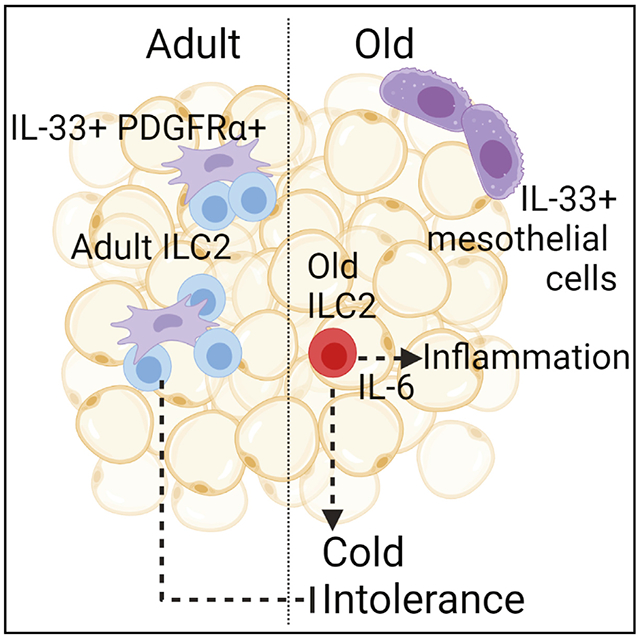
SUMMARY
Aging impairs the integrated immunometabolic responses, which have evolved to maintain core body temperature in homeotherms to survive cold stress, infections, and dietary restriction. Adipose tissue inflammation regulates the thermogenic stress response, but how adipose tissue-resident cells instigate thermogenic failure in the aged are unknown. Here, we define alterations in the adipose-resident immune system and identify that type 2 innate lymphoid cells (ILC2s) are lost in aging. Restoration of ILC2 numbers in aged mice to levels seen in adults through IL-33 supplementation failed to rescue old mice from metabolic impairment and increased cold-induced lethality. Transcriptomic analyses revealed intrinsic defects in aged ILC2, and adoptive transfer of adult ILC2s are sufficient to protect old mice against cold. Thus, the functional defects in adipose ILC2s during aging drive thermogenic failure.
In brief
Goldberg et al. find that adipose-resident ILC2s become impaired during aging, in part through dysregulated expression of their primary survival cytokine, IL-33. Aged ILC2s obtain proinflammatory and senescence-like transcriptional signatures, and this might contribute to increased cold lethality in old mice with expanded aged ILC2s.
Graphical Abstract
INTRODUCTION
Increasing evidence implicates the immune system as a key node in maintaining adipose tissue homeostasis by controlling the immunometabolic set point of inflammation. The adipose-resident immune compartment controls inflammation in visceral adipose tissue (VAT) and regulates fasting-induced lipolysis (Camell et al., 2017; Kosteli et al., 2010), cold-induced thermogenesis (Brestoff et al., 2015; Kohlgruber et al., 2018; Lee et al., 2015), and insulin resistance (Feuerer et al., 2009; Wolf et al., 2017). Although decades of research have pinpointed numerous defects in innate and adaptive arms of the anti-microbial immune response (Goldberg and Dixit, 2015; Goronzy and Weyand, 2019; Shaw et al., 2013), only a limited number of studies have examined how tissue-resident immune cells that are responsible for maintaining tissue homeostasis regulate the aging process and resilience to stress (Granot et al., 2017; Snyder et al., 2019; Thome et al., 2014).
Adipose tissue aging is characterized by heightened inflammation, mild insulin resistance, and fasting-induced lipolysis resistance. Although these phenotypes are also shared in obese adipose tissue, there are important differences that distinguish aged versus obese adipose. Among these differences are the composition and activities of different immune populations: regulatory T cells (Tregs) are protective in obesity (Feuerer et al., 2009), but their accumulation is detrimental during aging (Bapat et al., 2015); adipose B cells acquire a unique aged phenotype that is associated with cold intolerance (Camell et al., 2019); proinflammatory macrophages dominate obese adipose tissue (Lumeng et al., 2007a, 2007b; Weisberg et al., 2003), whereas aged adipose tissue displays loss of M2-like macrophages (Lumeng et al., 2011) and pathways acquisition of senescence signatures and catecholamine degradation enzymes that lead to suppression of lipolysis (Camell et al., 2017). However, the detailed description of tissue-resident immune cell transcriptome at single-cell resolution in aging adipose tissue has not yet been defined. Here, we report age-related changes in adipose tissue-resident leukocyte populations, including dramatic loss of group 2 innate lymphocyte cells (ILC2s). Through orthogonal validation approaches, we identify that during aging, IL-33 multiplies pathogenic ILC2s which have intrinsic defects that promote proinflammatory responses and lower adipose tissue resilience.
RESULTS AND DISCUSSION
Aging leads to alterations in the visceral adipose-resident immune compartment
Many of the physiological protective mechanisms described above decline with age, leading us to hypothesize that the adipose-resident immune compartment becomes dysfunctional during aging. To broadly characterize the VAT-resident immune compartments in adult and old mice, we performed intravascular (IV) labeling to isolate exclusively tissue-resident cells (Anderson et al., 2014) followed by single-cell RNA sequencing (scRNA-seq) (Figures 1A–1D, equal labeling efficiency demonstrated in Figures S1A and S1B, adult data were published previously (GSE137076) (Goldberg et al., 2020). In agreement with prior studies, we observed losses in macrophages (clusters 0 and 4) (Lumeng et al., 2011), and increases in CD4 and CD8 αβ T cells (cluster 1), B cells (cluster 10, 11) (Camell et al., 2019), and regulatory T cells (Tregs, cluster 8) (Kolodin et al., 2015). We also identified profound losses of NK (cluster 5) and NKT cells (cluster 3) in aged adipose tissue. Among the most striking changes was an almost complete loss of type-2 innate lymphoid cells (ILC2s, cluster 2) (Figures 1B and 1C), which we verified by flow cytometry (Figures 1E–1G and S1C). We were especially intrigued by the loss of ILC2s in aged VAT because of their important role in promoting metabolic health and cold tolerance (Brestoff et al., 2015; Lee et al., 2015; Molofsky et al., 2013), both of which are impaired during aging. Indeed, prior studies found that ILC2s are reduced in adipose tissue of 15-month-old mice (equivalent to approximately 45 years in humans) (Molofsky et al., 2015) and 28-week-old mice (approximately 35-year-old human) (Kohlgruber et al.,2018) suggesting that by middle age, adipose ILC2 maintenance is perturbed. Moreover, ILC2s exhibit strong tissue specificity (Ricardo-Gonzalez et al., 2018) and are uniquely regulated in different tissues during the aging process (D’Souza et al., 2019; Fung et al., 2020; Molofsky et al., 2015), indicating that specific regulatory mechanisms might be responsible for their depletion in aged adipose tissue.
Figure 1. The adipose-resident immune compartment changes during aging.
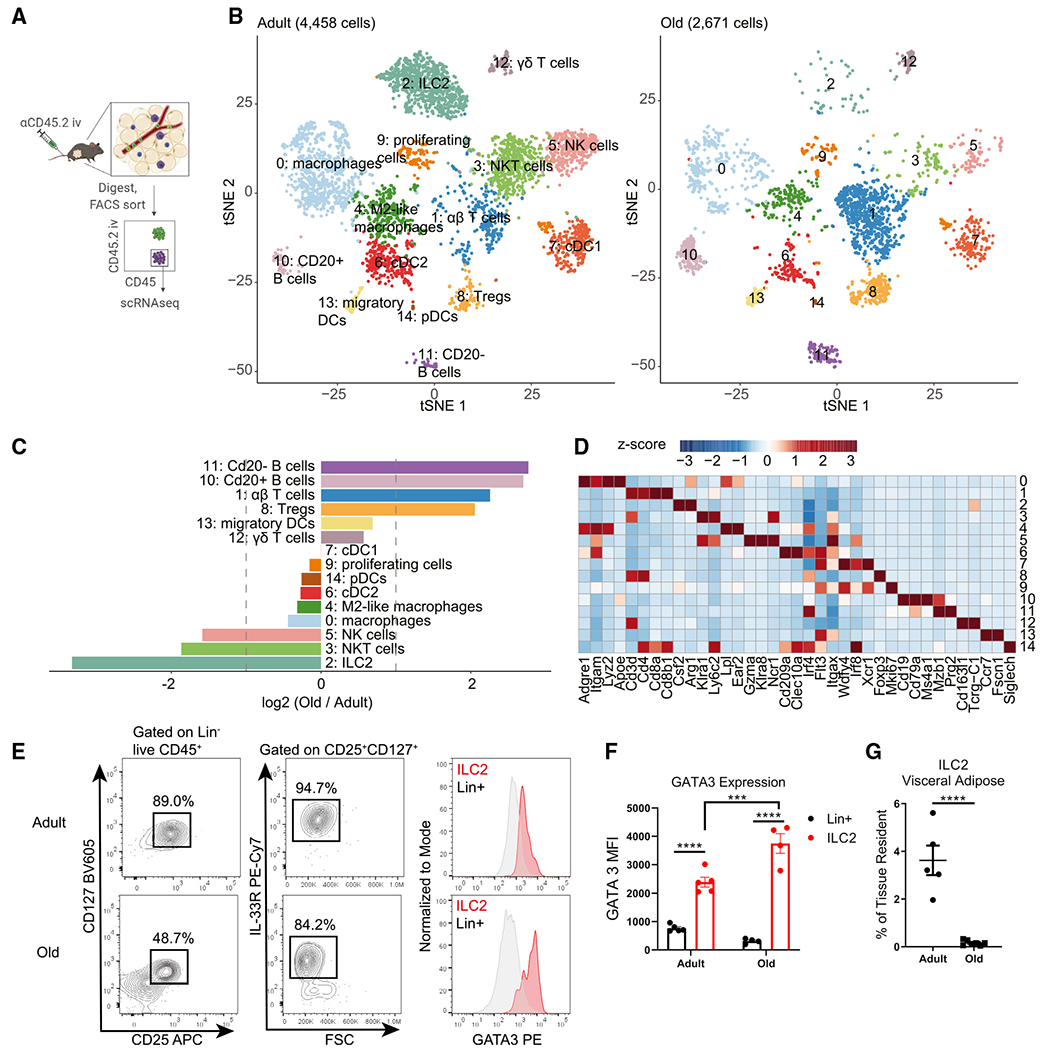
(A) Schematic depicting experimental strategy to define the adipose-resident immune compartment.
(B) tSNE plots of tissue-resident CD45+ cells from adult (left) and old (right) male visceral adipose tissue.
(C) Bar chart showing population fold changes in relative abundance of each cluster induced by aging.
(D) Heatmap showing main lineage-defining genes to identify each cluster.
For (B–D), data were generated from pooling n = 4 adult and n = 3 old male mice, so that a total of 1 g of adipose tissue was used to isolate cells for each sample.
(E) Representative flow cytometry analysis of visceral adipose tissue ILC2 gating.
(F) Quantitation of GATA3 expression in adult and old ILC2s (red) versus Lin+ cells (gray). Statistical differences were calculated by two-way ANOVA. E.rror bars are SEM.
(G) Quantification of ILC2 abundance in visceral adipose tissue in adult and old mice. Statistical difference was calculated by unpaired t test. Error bars are SEM.
(E–G) Data are from a single experiment but representative of at least three independently repeated experiments.
ILC2s are seeded early in life and then retained within their respective tissues (Schneider et al., 2019). Consistent with these data, we found that VAT ILC2s remain tissue-resident throughout lifespan (Figures 2A and 2B), and that their loss occurred in both male and female mice (Figure 2C). ILC2s are an important source of IL-5 and therefore a primary regulator of eosinophils in VAT. In agreement with a recent study (Brigger et al., 2020), we also found that eosinophils decline in both male and female mice during aging (Figure 2D). Because obesity and age-related inflammation are associated with loss of ILC2s in adipose tissue (Brestoff et al., 2015; Molofsky et al., 2015), we wondered whether the accumulation of fat mass might drive the ILC2 decline. We assessed the abundance of ILC2s in VAT in middle-aged (15-month-old male) life-long calorie-restricted (CR) mice, which do not gain weight or fat mass during aging, and compared this with ad libitum-fed age-matched controls and young mice. Indeed, we found that the proportion of ILC2s (Figure 2E, left), but not eosinophils (Figure 2F. left), was retained in the VAT of aged CR mice. The actual numbers of ILC2s and eosinophils, normalized to tissue weight, were not preserved (Figures 2E and 2F, right) presumably because CR induces peripheral leukocytopenia (Collins et al., 2019; Jordan et al., 2019; Meydani et al., 2016; Nagai et al., 2019). We have previously demonstrated that obesity and age-related adipose tissue inflammation leading to impaired metabolic health is driven by chronic low-level activation of the NLRP3 inflammasome (Camell et al., 2019; Vandanmagsar et al., 2011; Youm et al., 2013). However, aged ad libitum-fed Nlrp3−/− mice do not retain adipose ILC2s (Figure 2G), indicating that their loss may be mediated by weight gain but is independent of NLRP3-inflammasome dependent cytokines IL1β and IL-18.
Figure 2. Loss of ILC2s during aging is sex independent.

(A) Representative gating scheme defining tissue residence within indicated visceral adipose immune populations.
(B) The percentage of ILC2s that are tissue resident (iv-label negative) in males and females across lifespan. Error bars are SEM.
(C and D) The percentage of tissue-resident CD45+ cells that are (C) ILC2s and (D) eosinophils in aging male and female mice. Trend line is best-fit lines for males and females, separately.
For (A–D), data are representative of 5 independent experiments; n = 5/group and each symbol represents an individual mouse.
(E and F)The percentage of adipose immune cells that are (E) ILC2s or (F) eosinophils in 15-month-old calorie-restricted (CR) male mice (n = 7) compared with adult (n = 5) and ad libitum age-matched control male mice (n = 5). Left panels report frequencies, and right panels report absolute numbers normalized to tissue weight.
(G) Abundance of ILC2s in visceral adipose tissue from adult, old, and age-matched old Nlrp3−/− male mice. All groups were fed ad libitum.
For(E–G), each symbol represents an individual mouse and are representative of two independent experiments. Statistical differences were calculated by one-way ANOVA with Tukey’s correction for multiple comparisons. Error bars are SEM.
Dysregulation of IL-33 underlies ILC2 loss in aged adipose tissue
IL-33 is the primary survival cytokine that regulates ILC2s in VAT. IL-33 is produced by PDGFRα+ mesenchymal progenitors and perivascular and endothelial cells in young mice (Dahlgren et al., 2019; Kohlgruber et al., 2018; Mahlakõiv et al., 2019; Spallanzani et al., 2019; Zeyda et al., 2013). However, when we examined which cell types express Il33 in old mice using scRNA-seq of the entire VAT stromal-vascular fraction, rather than detecting Il33 expression in the Pdgfra+ cluster, we found that in aged mice nearly all Il33 expression was restricted to a cluster of cells expressing the mesothelial cell marker Msln (Figures 3A, S2A, and S2B). These data indicate that the cellular source of IL-33 in adipose tissue is switched during aging. Gene expression analysis by qPCR revealed an overall increase in Il33 expression in VAT, but not subcutaneous adipose tissue, from old mice compared with young adult mice (Figures 3B and 3C). These data suggest that alterations in cellular source of IL-33 may not allow ILC2s to access appropriate tonic signals required for maintenance in VAT during aging. Interestingly, concomitant with the increase in IL-33, aging led to specific increased expression of an Il1rl1 transcript variant that encodes for soluble IL-33R in aged VAT with no change in serum levels (Figures 3D–3F). Interestingly, despite important differences in the obese versus aged adipose-resident immune system (Lee and Dixit, 2020), increased soluble IL-33R was also recently reported in high-fat-diet-induced obesity (Zhao et al., 2020). Given the increase in IL-33 expression, together with higher soluble IL-33R, we considered the possibility of IL-33 signaling resistance in aged VAT that impairs ILC2 maintenance. However, intraperitoneal (i.p.) injections of exogenous IL-33 in young and old mice caused equivalent STAT3 activation, suggesting that overall IL-33 sensitivity in aged adipose tissue is intact upon IL33 supplementation (Figure 3G). Notably, the cellular source of soluble IL-33R is unknown in these studies. Additionally, whether increased soluble IL-33R is causal versus a consequence of elevated IL-33 in aged adipose remains to be determined. Regardless, these data provide a plausible mechanism by which IL-33 bioavailability in adipose niches may become reduced during aging, leading to loss of ILC2s.
Figure 3. Aged ILC2s are IL-33-responsive but not metabolically protective.
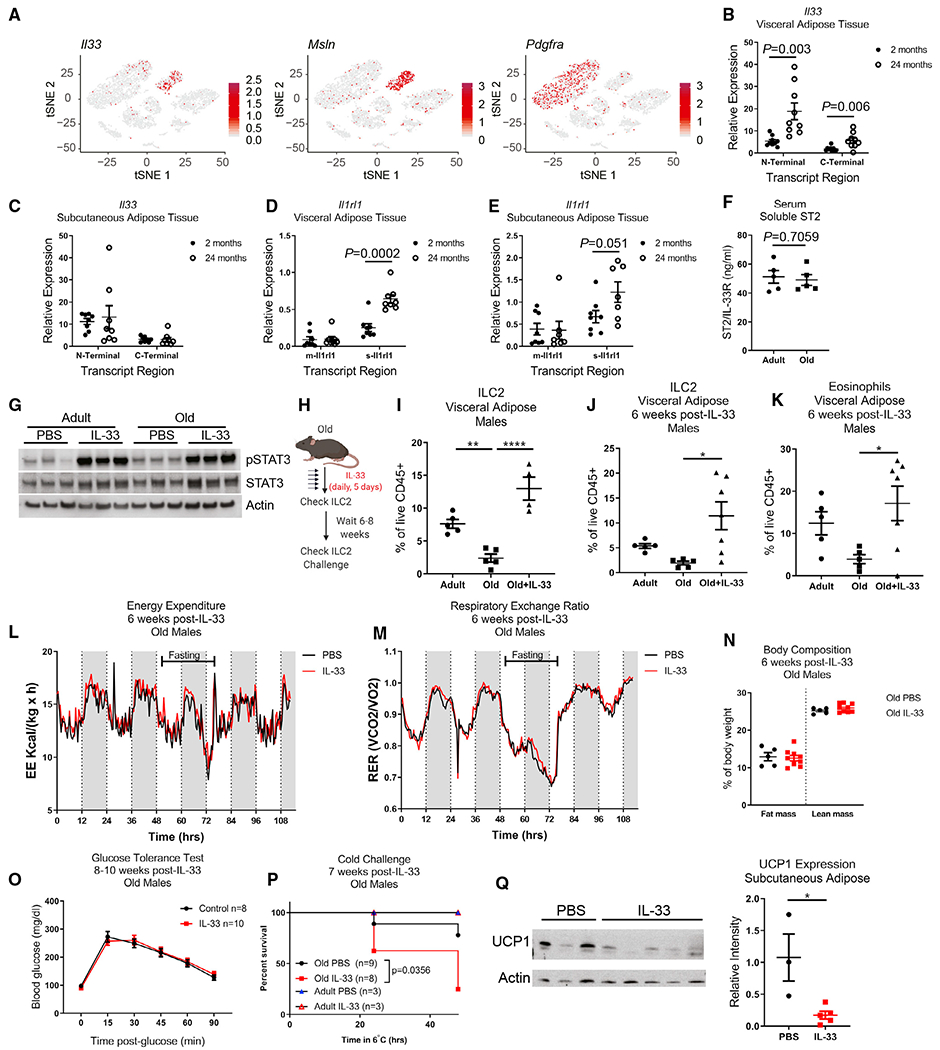
(A) tSNE plots of scRNA-seq analysis of total SVF of visceral adipose tissue obtained from a pooled sample of n = 4 old female mice. Expression of Il33, Msln, and Pdgfra, are overlaid individually in red.
(B and C) Il33 gene expression was measured by qPCR in (B) visceral adipose tissue and (C) subcutaneous inguinal adipose tissue of adult and old mice. Primers were designed to measure relative expression of both N-terminal and C-terminal regions of Il33. Data are pooled from two independent experiments in female mice, for a total of n = 9 mice/group for visceral adipose and n = 8 mice/group for subcutaneous adipose samples. Each dot represents an individual mouse. Statistical differences between adult versus old mice were calculated by unpaired t test for each transcript region analyzed.
(D and E) Il1rl1 gene expression was measured in (D) visceral adipose tissue and (E) subcutaneous inguinal adipose tissue of adult and old mice. Primers were designed to discriminate between the membrane-bound (m-Il1rl1) and soluble (s-Il1rl1) transcript variants. Data are pooled from two independent experiments in female mice, for a total of n = 9 mice/group for visceral adipose and n = 8 mice/group for subcutaneous adipose samples. Each dot represents an individual mouse. Statistical differences between adult versus old mice were calculated by unpaired t test for each transcript region analyzed.
(F) Soluble IL33R (ST2) was measured in serum by ELISA. Statistical p value was calculated by unpaired t test.
(G) Western blots of whole visceral adipose tissue lysate from adult and old female mice treated with IL-33 or PBS. Each lane represents an individual mouse, and data are representative of two independent experiments.
(H) Schematic depicting experimental design. IL-33 was given by i.p. injection daily for 5 consecutive days.
(I) ILC2 abundance in visceral adipose tissue 2 days after completing IL-33 treatments.
(J) ILC2s in visceral adipose tissue 6 weeks after completion of IL-33 treatment. Data are representative of 5 independent experiments, assessed through a range of 6–10 weeks after completing IL-33 treatments. Statistical differences were calculated by one-way ANOVA with Tukey’s correction for multiple comparisons.
(K) Eosinophils in visceral adipose tissue 6 weeks after completion of IL-33 treatment. Data are representative of 5 independent experiments, assessed through a range of 6–10 weeks after completing IL-33 treatments. Statistical differences were calculated by one-way ANOVA with Tukey’s correction for multiple comparisons.
(L and M) (L) Energy expenditure and (M) RER in the fed and fasting state, measured 6 weeks after IL-33 treatment in old male mice. Data are representative of two independent cohorts.
(N) Body composition in old male mice 6 weeks after IL-33 treatment. Data are representative of 2 independent experiments.
(O) Glucose tolerance test in old male mice 8–10 weeks after IL-33 treatment. Data are pooled from two independent experiments.
(P) Survival of male mice subjected to cold challenge at 7 weeks post-IL-33 treatment. Survival of old mice are pooled from 2 independent experiments and were analyzed by log-rank test. Adult mice were only included in one of these independent experiments.
(Q) Protein expression of UCP1 in subcutaneous (inguinal) adipose tissue was measured by western blot in adult mice that survived 48 h cold challenge. Data are representative of two independent experiments. Western blot was quantified by normalizing UCP1 band intensities to Actin bands in their respective lanes. Statistical difference was calculated by non-parametric t test.
(B–F, I–K, N, O, and Q) Error bars are SEM.
Next, we tested if providing excess IL-33 could override its apparent dysregulation to recover ILC2s in old mice (Figure 3H). Consistent with the above data (Figure 3G), treating old mice with IL-33 restored the numbers of ILC2s in aged adipose tissue (Figure 3I). Co-administration of the sphingosine-1-phosphate receptor (S1PR) antagonist, FTY720, did not block ILC2 expansion, indicating that this was due to in situ proliferation (Figures S2C and S2D). However, given that IL-33 can be an alarmin in the IL-1 cytokine family and stimulates NFκB (Schmitz et al., 2005) and that immediately following IL-33 treatment there is also substantial eosinophilia in adipose tissue (Figure S2D), we wondered whether the expanded ILC2 generates a sustained biological impact on aged VAT. Indeed, our protocol caused a sustained increase in numbers of ILC2s (Figure 3J) and eosinophils (Figure 3K) in VAT for at least 6–8 weeks following the cessation of IL-33 treatment in aged mice.
Given prior results that IL-33 induces UCP1 in adipose tissue and increases thermogenesis in young and obese mice (Brestoff et al., 2015; Lee et al., 2015), we tested potential therapeutic impact of long-term restoration of ILC2s by IL-33 treatment on correcting adipose tissue metabolic deficits seen with aging. Surprisingly, as compared with PBS-treated control old mice, IL-33 treatment did not enhance energy expenditure (EE) (Figure 3L) or affect respiratory exchange ratio (RER) (Figure 3M) despite being reported to do so in young adult animals (Brestoff et al., 2015). Moreover, unlike what has been reported in young mice (Brestoff et al., 2015), IL-33 treatment also did not change fat mass/body composition (Figure 3N) or glucose tolerance (Figure 3O) in old mice. The inconsistencies between old mice and previous reports in young and obese mice prompted us to also test the metabolic impact of IL-33 treatment in adult mice. Using comprehensive lab animal monitoring system (CLAMS), we detected no change in EE in adult mice treated with IL-33 as compared with PBS treatment (Figures S3A–S3F). In agreement with previous reports, we did measure a modest increase in RER (Figures S3G and S3H). However, we did not detect any change in UCP1 expression in subcutaneous or brown adipose tissue depots after IL-33 treatment (Figures S3I and S3J). These data indicate that assumptions about the role of the IL-33/ILC2 axis in metabolic health may be incomplete and that further studies are needed to establish the function of IL-33 in adipose biology.
Another reported function of adipose ILC2 is to protect core body temperature in response to cold challenge by inducing adaptive thermogenesis through beiging of white adipose tissue (Brestoff et al., 2015; Lee et al., 2015). To our surprise, old mice with IL-33-mediated ILC2 restoration exhibited increased mortality in response to cold challenge (Figure 3P). Lower expression of UCP1 in their subcutaneous white adipose tissue indicated that this was due to an inability to induce adaptive thermogenesis and beiging (Figure 3Q).
Consistent with above data that exogenous IL-33 does not affect fatty acid substrate availability in aged VAT, the energetic parameters and induction of fasting-induced lipolysis were also not improved following IL-33 treatment (Figures S3K–S3P). However, we cannot rule out the possibility that the IL-33-induced expansion of Tregs in these mice might antagonize any metabolic benefits of the ILC2s during aging (Kolodin et al., 2015).
We next considered the possibility that Tregs, which increase with age in male VAT and also expand in response to IL-33 (Figures S4A–S4D), might outcompete ILC2s for cytokine survival factors because both subsets have uniquely high expression of Il1rl1, Il7r, and Il2ra (Figures S4E–S4G) compared with all other adipose-resident immune cells. This hypothesis is also supported by previous studies showing that adipose Tregs disrupt metabolic health during aging (Bapat et al., 2015), in contrast to their protective role in obesity (Feuerer et al., 2009). However, we found that visceral adipose Treg expansion is largely restricted to males and not female mice (Figure S4A) while ILC2 loss and restoration by IL-33 is evident in both sexes (Figures 1C, 3I, and S2C). Moreover, the in vivo dose response experiments actually indicate that ILC2s have higher IL-33 sensitivity compared with Tregs (Figure S4D). These data collectively suggest that Treg homeostasis does not compete with ILC2 maintenance.
Cell-intrinsic defects in aged ILC2s cause impaired thermogenesis during cold challenge
Unexpected impairment of thermogenic responses and increased mortality in IL-33-treated old mice after cold challenge raised the possibility that aged ILC2s, and possibly the expanded eosinophils, were pathogenic. To test this possibility, we treated adult and old mice with IL-33 and then sorted VAT ILC2s for bulk RNA-seq (Figure 4A). Expansion of old ILC2s by IL-33 was necessary, as so few ILC2s remain in old adipose tissue that we were unable to obtain sufficient cell quality for RNA-seq from isolated cells. Gene set enrichment analysis revealed significant alterations in transcriptional programs of ILC2s in adult and old mice (Figures 4B–4D). Adult IL-33-expanded ILC2s showed enrichment of proliferation, transcription, and translation pathways, consistent with their mechanisms of self-renewal. In contrast, old IL-33-expanded ILC2s were enriched in cytokine signaling and activation pathways. We further examined the genes in the most enriched pathway, the JAK/STAT KEGG pathway (Figures 4B and 4C) in old ILC2s, and found enhanced expression of Il6, Oslm, and Lif, which all encode cytokines in the IL-6 family that are capable of inducing thermogenesis (Beretta et al., 2002; Chen et al., 2019) but are also associated with inflammatory disease and “inflammaging” (Franceschi and Campisi, 2014). These gene signatures also suggested that old ILC2s might acquire age-related senescence-associated secretory phenotype (SASP)-like features, and further analyses showed that old ILC2s also have enhanced expression of the classical senescence marker Cdkn1a, encoding p21 (log2 fold change = 0.718; padj = 3.64 × 10−11). The potential importance of acquisition of a senescent/SASP phenotype in aging immune cells was recently demonstrated to drive detrimental aging processes in multiple tissues (Yousefzadeh et al., 2021). Similarly, old ILC2 had increased expression of Il13, which is a common ILC2 cytokine associated with asthma, but can sustain alternatively activated macrophages associated with improved adipose tissue inflammation (Molofsky et al., 2013). However, macrophages within the adipose-resident compartment actually decline with age, and the proportion of proinflammatory macrophages increases (Camell et al., 2017; Lumeng et al., 2011), suggesting that despite cytokine mRNA expression, old ILC2 function is impaired and this could further compound their numerical loss. To validate the RNA-seq data, we assessed cytokine secretion from IL-33-expanded adipose ILC2 by sort-purifying cells and stimulating with PMA + ionomycin to measure their maximum cytokine secretion capacity. In agreement with the transcriptomics, old ILC2s produced more IL-10 and IL-6, but we observed no changes in IL-13 secretion (Figures 4E–4G). However, we speculate that the physiological effects of these cytokines are not borne out in aged adipose, as described above, because their ability to produce certain cytokines is negated by their depletion during aging. Notably, this premise of functionality versus numbers was recently demonstrated in adipose tissue eosinophils (Knights et al., 2020), which also become dysregulated during aging (Brigger et al., 2020) and are controlled by ILC2-derived IL-5 (Molofsky et al., 2013; Nussbaum et al., 2013). We also noted enhanced expression of Ifngr1, Ifnar1, and Ifnar2 in aged ILC2s (Figure 4C). Increased inflammation in adipose tissue, and specifically IFN-γ, has been linked to loss of ILC2s in adipose tissue of obese and middle-aged mice (Molofsky et al., 2015). As T cells were previously shown to infiltrate the aging brain and secrete IFN-γ (Dulken et al., 2019), and aging adipose tissue has increased accumulation of T cells (Figures 1B and 1C), we wondered whether T cells could be a source of IFN-γ to influence old ILC2s. Reevaluation of our scRNA-seq data showed that adipose-resident αβ T cells express high levels of Ifng, and their accumulation could contribute to the proinflammatory phenotype of aged ILC2s (Figures 4H and 4I). Similarly, increased type 1 interferon signatures during aging have been reported in several tissues (Benayoun et al., 2019; Dulken et al., 2019; Youm et al., 2013). These data demonstrate that old ILC2 may be highly susceptible to cytokine changes in aging adipose tissue and that old ILC2s might skew toward a proinflammatory SASP-like features with intrinsically altered survival mechanisms. Future studies designed to identify the interactions between tissue-resident immune cell type(s) responsible for cytokine balance in aging adipose tissue will be key to understanding mechanisms that drive age-related inflammation and metabolic dysregulation.
Figure 4. Intrinsic defects in aged ILC2s predispose to cold intolerance.
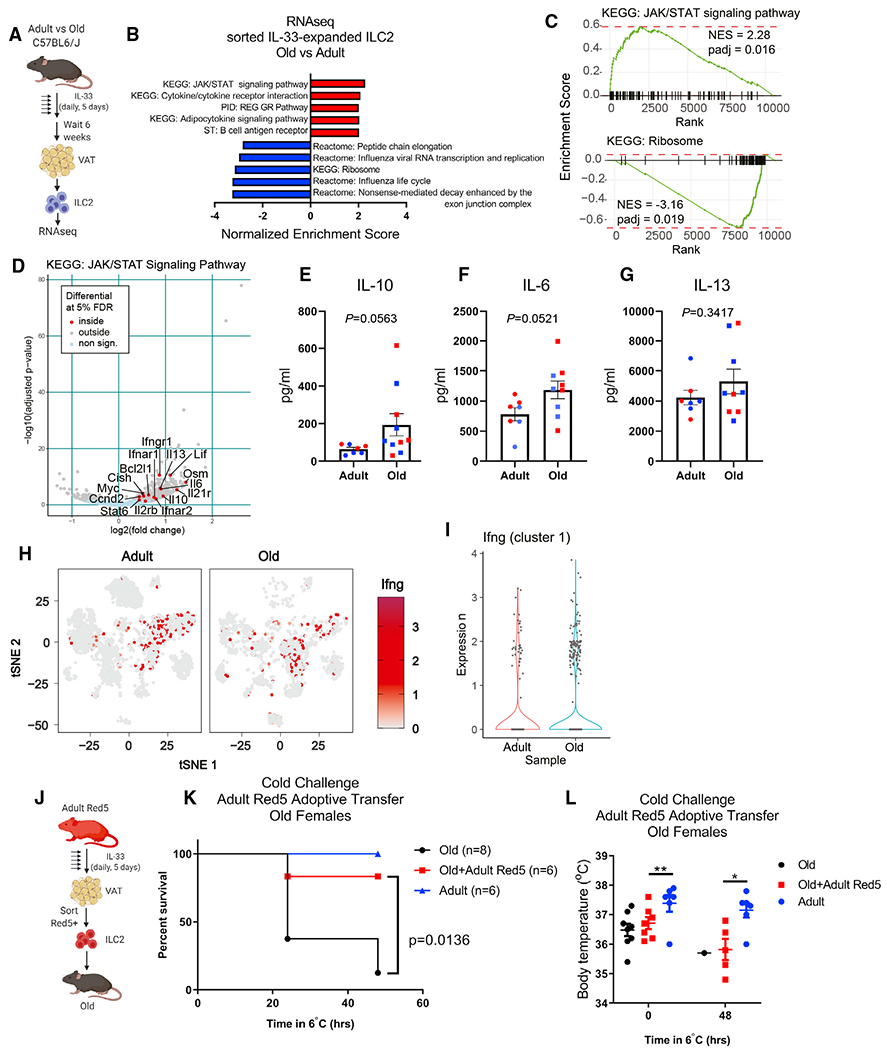
(A) ILC2 bulk RNA-seq experimental design. IL-33 was given by i.p. injection daily for 5 consecutive days and mice were rested for 6 additional weeks prior to tissue collection and ILC2 isolation.
(B) Gene set enrichment analysis of IL-33-expanded ILC2s from adult and old mice. n = 3 adult and n = 3 old samples, each containing sorted ILC2s pooled from n = 2 biological replicate male mice were used for bulk RNA-seq analyses.
(C) GSEA curves of significantly regulated pathways identified in (B).
(D) Volcano plot highlighting significantly regulated genes from the KEGG JAK/STAT signaling pathway gene list.
(E–G) (E) IL-10, (F) IL-6, and (G) IL-13 cytokine secretion was measured from sorted adult and old adipose tissue ILC2s after IL-33-mediated expansion in vivo.
Each symbol represents an individual mouse, and both age groups are pooled from male (blue) and female (red) mice. All mice were treated with IL-33 simultaneously, ILC2s were sorted from n = 1–2 mice/group and stimulated across 4 separate days, and all supernatants were analyzed together. Statistical differences were calculated by t test. Error bars are SEM.
(H) Ifng expression is depicted in red and overlaid across all tissue-resident CD45+ cell subsets in gray.
(I) Violin plots of Ifng expression within cluster 1 αβ T cells.
(J) Adoptive transfer experimental design. Red5 mice were treated with IL-33 by i.p. injection daily for 5 consecutive days.
(K) Survival of old female mice that survived 48 h cold challenge ± receiving adult female Red5+ cells. Date are pooled from two independent experiments. Statistical differences were analyzed by log-rank test.
(L) Body temperatures of old female mice that survived 48 h cold challenge ± receiving adult female Red5+ cells. Data are pooled from 2 independent experiments. Statistical differences in body temperature were calculated by two-way ANOVA. Error bars are SEM.
Next, we considered the possibility that aged ILC2s might perturb norepinephrine bioavailability, similar to adipose tissue macrophages (Camell et al., 2017; Pirzgalska et al., 2017; Wolf et al., 2017). However, our RNA-seq data indicate adipose ILC2s express negligible levels of Adrb3 (baseMean = 0.38), which encodes the β3-adrenergic receptor involved in epinephrine- and norepinephrine-mediated lipolysis and thermogenesis. This, combined with the null effects in our in vivo fasting (Figure 3L) and in vitro lipolysis assays (Figures S3O and S3P), rules out a role for ILC2s in this aspect of neuro-immune control of energy homeostasis during aging.
Finally, to directly test if old ILC2s are dysfunctional, we performed adoptive transfer experiments. We sorted adult ILC2s from VAT of adult IL-5-RFP (Red5) reporter mice after IL-33- mediated expansion in vivo (Figure 4J). To isolate the role of ILC2s independent of their activation of eosinophils (Figure 3K), we sorted ILC2s from Il5RFP/RFP genotyped mice, which do not produce IL-5 protein (Nussbaum et al., 2013). Old mice that received adult Red5 ILC2s were protected against cold challenge (Figure 4K) although even the surviving mice still exhibited some age-related impairment in maintaining their core body temperatures (Figure 4L). Importantly, transferring adult Red5 ILC2s to old mice is sufficient to rescue susceptibility to cold challenge, indicating that aged ILC2s are intrinsically defective.
In summary, our study identifies both cell-intrinsic and -extrinsic mechanisms leading to numerical and functional loss of ILC2s in aging adipose tissue. Given the important roles reported for ILC2s in mediating glucose homeostasis and inducing thermogenesis, we were surprised that restoring old ILC2s not only did not improve either of these outcomes in old mice but instead impaired tissue homeostasis and increased susceptibility to cold challenge. In addition, the transfer of IL-5-deficient Red5 ILC2s protected old mice from cold challenge despite the recipients still having relatively lower than normal core body temperature. These data indicate that IL-5 and presumable eosinophil activation is dispensable for surviving cold challenge but may be necessary for inducing thermogenesis. Interestingly, IL-5Rα is also expressed on adipose tissue B cells, which we have previously shown to expand in aged female mice, and whose depletion also improves old mouse survival during cold challenge (Camell et al., 2019). However, the B cell expansion in aged adipose tissue is predominantly noted in female mice, and its potential relationship with ILC2s and eosinophils requires further investigation.
It is unclear why a different source of IL-33 in aged adipose might detrimentally alter ILC2s. Mesothelial cells have been identified as a rare source of IL-33 that acts as an alarmin to promote peritoneal inflammation after infection (Mahlakõiv et al., 2019) But how mesothelial- versus mesenchymal-derived IL-33 could lead to ILC2 dysfunction is unclear. (Hung et al., 2020). It is likely that the different locations of these cell types within the adipose tissue might limit IL-33 availability to ILC2s, whose localization is highly enriched in perivascular adventitia (Dahlgren et al., 2019). The importance of IL-33 source is under-scored by a recent study demonstrating that epithelial-derived IL-33 stimulates protective ILC2 responses in the intestine, but CD11c+ dendritic-cell-derived IL-33 impairs protective immunity against helminth infection (Hung et al., 2020). The significance of spatial organization of IL-33 is further supported by findings that cell-cell interactions between ILC2s and IL-33-producing PDGFRα+ cells are important for ILC2 maintenance/proliferation (Rana et al., 2019). Similarly, increased gene expression of soluble IL33R could reduce bioavailability of IL-33 to adipose-resident ILC2s. Collectively, these data support a model in which loss of IL-33 availability drives the age-related loss in ILC2s. Moreover, ILC2s might adapt to this limitation, and become permanently reprogrammed in the process. In this case, when expanded with IL-33, a pool of pathogenic ILC2s multiply in aged adipose that are no longer capable of normal ILC2 functions. Alternatively, due to their acquisition of proinflammatory cytokine production, it is possible that the loss of ILC2s during aging reflects a programed adaptive protective mechanism. In conclusion, these data establish that IL-33-mediated restoration of adipose ILC2s in aged mice is pathogenic and impairs thermogenesis. These findings further highlight the critical role of adipose-tissue-resident immune compartment in mechanisms contributing to loss of tissue resilience and homeostasis during aging.
Limitations of study
Although we made our best efforts to confirm all phenotypes in both male and female mice, limitations in aged mouse availability prevented all analyses from being performed in both sexes. In particular, adoptive transfer experiments and RNA-seq experiments could only be tested in females. Similarly, we were unable to obtain healthy aged human VAT to test if mice and humans share similar patterns of ILC2 loss. While our hypothesis that reduced IL-33 bioavailability in aged adipose tissue is supported by our data in Figure 3, our RNA-seq data in Figure 4 also indicate an inflammatory environment independently of NLRP3 inflammasome drives age-related loss of ILC2s. Future studies that are dedicated to uncovering the responsible cell types and that test how long-term blockade of responsible cytokines are needed to identify the full mechanism of ILC2 decline during aging. Finally, we were unable to pinpoint the precise mechanism by which old ILC2s cause lethality during cold challenge. Functional assays beyond transcriptomics will likely be required to uncover this deleterious action of old ILC2s.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Vishwa Deep Dixit (Vishwa.dixit@yale.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
RNA sequencing data reported in this paper was deposited to the Gene Expression Omnibus and accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC anti-mouse CD3 Antibody | Biolegend | Cat#100203; RRID:AB_312660 |
| FITC anti-mouse TCR β chain Antibody | Biolegend | Cat#109205; RRID:AB_313428 |
| FITC anti-mouse CD5 Antibody | Biolegend | Cat#100605; RRID:AB_312734 |
| FITC anti-mouse/human CD11b Antibody | Biolegend | Cat#101205; RRID:AB_312788 |
| FITC anti-mouse CD19 Antibody | Biolegend | Cat#115505; RRID:AB_313640 |
| FITC anti-mouse CD11c Antibody | Biolegend | Cat#117305; RRID:AB_313774 |
| FITC anti-mouse FcεRIα Antibody | Biolegend | Cat#134305; RRID:AB_1626102 |
| FITC anti-mouse Ly-6G/Ly-6C (Gr-1) Antibody | Biolegend | Cat#108405; RRID:AB_313370 |
| FITC anti-mouse TER-119/Erythroid Cells Antibody | Biolegend | Cat#116205; RRID:AB_313706 |
| FITC anti-mouse F4/80 Antibody | Biolegend | Cat#123107; RRID:AB_893500 |
| FITC anti-mouse TCR γ/δ Antibody | Biolegend | Cat#118105; RRID:AB_313829 |
| FITC anti-mouse NK-1.1 Antibody | Biolegend | Cat#108705; RRID:AB_313392 |
| A700 anti-mouse CD3 Antibody | Biolegend | Cat#100216; RRID:AB_493697 |
| A700 anti-mouse TCR β chain Antibody | Biolegend | Cat#109224; RRID:AB_1027648 |
| A700 anti-mouse CD5 Antibody | Biolegend | Cat#100636; RRID:AB_2687002 |
| A700 anti-mouse/human CD11b Antibody | Biolegend | Cat#101222; RRID:AB_493705 |
| A700 anti-mouse CD19 Antibody | Biolegend | Cat#115528; RRID:AB_493735 |
| A700 anti-mouse CD11c Antibody | Biolegend | Cat#117320; RRID:AB_528736 |
| A700 anti-mouse FcεRIα Antibody | Biolegend | Cat#134324; RRID:AB_2566734 |
| A700 anti-mouse Ly-6G/Ly-6C (Gr-1) Antibody | Biolegend | Cat#108422; RRID:AB_2137487 |
| A700 anti-mouse TER-119/Erythroid Cells Antibody | Biolegend | Cat#116220; RRID:AB_528963 |
| A700 anti-mouse F4/80 Antibody | Biolegend | Cat#123130; RRID:AB_2293450 |
| A700 anti-mouse NK-1.1 Antibody | Biolegend | Cat#108730; RRID:AB_2291262 |
| IL-33R (ST2) Rat anti-Mouse, PE-Cyanine7, Clone: RMST2-2 | eBioscience | Cat#501123451 |
| Brilliant Violet 711 anti-mouse CD45 Antibody, Clone 30-F11 | Biolegend | Cat#103147; RRID:AB_2564383 |
| FITC anti-mouse CD45.2 Antibody, Clone 104 | Biolegend | Cat#109806; RRID:AB_313443 |
| GATA3-PE antibody | Fisher Scientific | Cat#12-9966-41; RRID:AB_1963601 |
| APC anti-mouse CD25 Antibody | Biolegend | Cat#101909; RRID:AB_961208 |
| Brilliant Violet 605 anti-mouse CD127 (IL-7Rα) Antibody | Biolegend | Cat#135025; RRID:AB_2562114 |
| phosphorylated STAT3 (Tyr705) | Cell Signaling | Cat#9145; RRID:AB_2491009 |
| UCP1 | Abcam | Cat#ab10983; RRID:AB_2241462 |
| Total STAT3 | Cell Signaling | Cat#79D7, RRID:AB_331269 |
| b-Actin | Cell Signaling | Cat#4967L; RRID:AB_330288 |
| HRP-conjugated goat-anti-rabbit | ThermoScientific | Cat#PI31460; RRID:AB_228341 |
| Fc Block | Biolegend | Cat#101302; RRID:AB_312801 |
| Chemicals, peptides, and recombinant proteins | ||
| IL-33, mouse, carrier-free | Biolegend | Cat#580508 |
| Collagenase, Type 2 | Fisher Scientific | Cat#NC9870009 |
| FTY-720 | Sigma | Cat#SML0700-25MG |
| Critical commercial assays | ||
| Glycerol measurement kit | Sigma | Cat#MAK117 |
| IL-33R ELISA | R&D | Cat#MST200 |
| Luminex Assay | R&D | custom pre-mixed multi-analyte kit |
| Deposited data | ||
| Adult adipose-resident CD45+ scRNAseq | Goldberg et al., 2020 | GEO: GSE137076 |
| Old adipose-resident CD45+ scRNAseq | This paper | GEO: GSE180910 |
| Old whole adipose tissue SVF scRNAseq | This paper | GEO: GSE180910 |
| Adult and Old IL-33-expanded ILC2 RNAseq | This paper | GEO: GSE180858 |
| Experimental models: Organisms/strains | ||
| Adult C57BL6/J mice | Jackson Labs | stock #000664 |
| Old C57BL6/J mice | NIA Aged Rodent Colony | N/A |
| Red5 mice | Jackson Labs, Nussbaum et al., 2013 | stock #030926 |
| Nlrp3−/− mice | Mariathasan et al., 2006 | Bred in Dixit colony |
| Calorie-restricted mice | NIA Aged Rodent Colony | N/A |
| Rag−/−IL-2Rγ−/− mice | Taconic | Stock# 4111 |
| Oligonucleotides | ||
| Soluble IL33R (Variant 3) Forward 5′-TGGCTAGGACCTCTGGCTAA-3′ | This Paper | N/A |
| Soluble IL33R (Variant 3) Reverse 5′-ATGGTGTGTTCACTAGGCGG-3′ | This Paper | N/A |
| Membrane-bound IL33R (Variant 1) Forward 5′-CTCTGCCCGACGTTCTTGA-3′ | This Paper | N/A |
| Membrane-bound IL33R (Variant 1) Reverse 5′-AACCCCTGATGTGTCTCAGT-3′ | This Paper | N/A |
| IL-33 N-terminal Forward 5′-AACTCCAAGATTTCCCCGGC-3′ | This Paper | N/A |
| IL-33 N-terminal Reverse 5′-TTATGGTGAGGCCAGAACGG-3′ | This Paper | N/A |
| IL-33 C-terminal Forward 5′-AGACCAGGTGCTACTACGCT-3′ | This Paper | N/A |
| IL-33 C-terminal Reverse 5′-ACGTCACCCCTTTGAAGCTC-3′ | This Paper | N/A |
| Gapdh Forward 5′-TCAACAGCAACTCCCACTCTTCCA-3′ | Youm et al., 2015 | N/A |
| Gapdh Reverse 5′-ACCCTGTTGCTGTAGCCGTATTCA-3′ | Youm et al., 2015 | N/A |
| Software and algorithms | ||
| Prism (v8.4.3) | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Biorender | Biorender | https://biorender.com/ |
| FACSDiva (v7.0) | BD | https://www.bdbiosciences.com/en-eu/products/software/instrument-software/bd-facsdiva-software#Overview |
| FlowJo (v10.6) | Tree Star | https://www.flowjo.com/solutions/flowjo/downloads |
| Cell Ranger Single-Cell Software Suite | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome |
| Seurat (v2.3) | Satija Lab, New York Genome Center | https://satijalab.org/seurat/ |
| Other | ||
| Zombie Aqua Fixable Viability dye | Biolegend | Cat#423102 |
| Foxp3 Fix/Perm Intracellular Staining Kit | eBioscience | Cat#00-5521-00 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mice were on the C57BL/6J genetic background. Adult (8-12 weeks old) and old (18-24 months old) mice were obtained from Jackson Labs and the National Institute on Aging (NIA) Aged Rodent Colony. Red5 (Il5RFP/RFP) mice were purchased from Jackson Labs (stock #030926) and were originally generated and described by Nussbaum et al (Nussbaum et al., 2013). Nlrp3−/− mice have been described previously (Mariathasan et al., 2006) and were bred and aged in the Dixit lab, as were the control wildtype mice. Calorie-restricted mice and their age-matched ad libitum controls were obtained from the NIA Aged Rodent Colony. Rag−/−IL-2Rγ−/− mice were purchased from Taconic (model #4111). Mice were housed under standard 12hr light/dark cycles under specific pathogen-free conditions. Mice were group-housed, except in cold challenge experiments, in which mice were singly-housed. Male and female mice were used throughout the study, and these details are clarified in each figure and corresponding legend. All animal procedures were approved by the Yale Institutional Animal Care and Use Committee.
METHOD DETAILS
IL-33 treatments
Mice were treated by i.p. injection of IL-33 (Biolegend) at a dose of 12.5ug/kg body weight, unless a different dose is specifically indicated in the figure, for 5 consecutive days. Mice were analyzed early (2 days) or late (at least 6-8 weeks) after completing IL-33 treatments. Where indicated, mice were co-treated with FTY-720 (1mg/kg) every other day.
Glucose tolerance tests
Mice were fasted 16hr prior to glucose tolerance test. Glucose was given by i.p. injection based on body weight (0.4g/kg) and blood glucose levels were measured by handheld glucometer.
Body composition and energy expenditure
Body composition was measured in vivo by magnetic resonance imaging (EchoMRI; Echo Medical Systems). The TSE PhenoMaster System (V3.0.3) Indirect Calorimetry System was used to monitor energy expenditure, activity, and food and water intake in individual mice at 30min intervals. Oxygen consumption and carbon dioxide production measurements were used to determine energy expenditure.
IL33R (ST2) measurements
IL33R protein levels were measured in serum by ELISA (R&D) according to the manufacturer’s instructions. Relative expression of soluble and membrane-bound IL33R expression was measured by qPCR. Primer sequences are provided in the key resources table.
Western blots
Mice were treated with IL-33 for 5 consecutive days, as described above. 30 minutes after their final IL-33 injection, visceral adipose tissue was collected and snap frozen in liquid nitrogen. Tissue was homogenized in RIPA buffer containing protease inhibitors (Pierce) and protein expression was assessed by western blot. Antibodies against phosphorylated STAT3 (pSTAT3, 1:1000, Tyr705 rabbit, Cell signaling), total STAT3 (1:1000; 79D7 rabbit, Cell Signaling), and β-actin (1:1,000 4967L; Cell Signaling) and HRP-conjugated goat-anti-rabbit secondary antibody (PI31460; Thermo Scientific) were used to visualize proteins using chemiluminescence (PI32106; Thermo Scientific).
Flow cytometry
Intravascular labeling was performed by iv injection of 2.5ug CD45.2-FITC diluted in 100ul PBS. Mice were euthanized exactly 3 minutes after injection for tissue collection. Adipose tissue was digested in HBSS + 1mg/mL Collagenase I in shaking 37°C water bath. Cells were stained with live/dead viability dye (Invitrogen), incubated with Fc block, and then stained for surface markers including CD45, CD127, CD25, IL-33R (ST2), and a dump channel that included CD3, CD19, γδ TCR, CD4, CD8, NK1.1, CD11b, F4/80, Ly6G, Ly6C, and Ter-119. All antibodies were purchased from eBioscience or Biolegend. When needed, intracellular staining for Foxp3 was performed using the eBioscience Fix/Perm nuclear staining kit, otherwise cells were fixed in 2% PFA. Samples were acquired on a custom LSR II and data was analyzed in FlowJo.
In vitro lipolysis assay
Glycerol release from adipose tissue explants was used to asses induction of lipolysis. Approximately 10mg of adipose was used for each assay. Where indicated, 103 sorted adult Red5 cells were added for the entire duration of the assay. Isoproterenol was added at a concentration of 1 μM where indicated. Glycerol release was measured according to the manufacturer’s instructions (Sigma, MAK117).
Single-cell RNA sequencing sample preparation
After iv-labeling epidydimal white adipose tissue was collected, digested, and stained for viability (live/dead Aqua, ThermoFisher) and pan-CD45 to FACS sort live tissue-resident hematopoietic cells. To control for increased lipid content of adipose tissue mass during aging, adipose tissue from n=4 adult and n=3 old mice were pooled so that 1 gram of tissue for each age group was used for isolating tissue-resident CD45+ cells for sequencing. For total SVF analyses, visceral adipose tissue from n=4 old female mice were pooled. Cells were prepared for single-cell sequencing according to the 10X Genomics protocols. Sequencing was performed on a HiSeq4000.
scRNAseq alignment, barcode assignment and unique molecular identifier (UMI) counting
The Cell Ranger Single-Cell Software Suite (v2.1.1) (available at https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger) was used to perform sample demultiplexing, barcode processing, and single-cell 3’ counting. Cellranger mkfastq was used to demultiplex raw base call files from the HiSeq4000 sequencer into sample-specific fastq files. Subsequently, fastq files for each sample were processed with cellranger counts to align reads to the mouse reference (version mm10-2.1.0). The default estimated cell count value of 10,000 was used for this experiment. Samples were subsampled to have equal numbers of confidently mapped reads per cell.
scRNAseq preprocessing analysis with Seurat package
For the analysis, the R (v3.4.2) package Seurat (v2.3) (Butler et al., 2018; Stuart et al., 2019) was used. Cell Ranger filtered genes by barcode expression matrices were used as analysis inputs. Samples were pooled together using the AddSample function. The fraction of mitochondrial genes was calculated for every cell, and cells with high (>5%) mitochondrial fraction were filtered out. Expression measurements for each cell were normalized by total expression and then scaled to 10,000, after that log normalization was performed (NormalizeData function). Two sources of unwanted variation: UMI counts and fraction of mitochondrial reads – were removed with ScaleData function.
scRNAseq dimensionality reduction and clustering
The most variable genes were detected using the FindVariableGenes function. PCA was run only using these genes. Cells are represented with t-SNE (t-distributed Stochastic Neighbor Embedding) plots. We applied RunTSNE function to normalized data, using first 10 PCA components. For clustering, we used function FindClusters that implements SNN (shared nearest neighbor) modularity optimization-based clustering algorithm on top 10 PCA components using resolution of 0.5.
scRNAseq identification of cluster-specific genes and marker-based classification
To identify marker genes, FindAllMarkers function was used with likelihood-ratio test for single cell gene expression. For each cluster, only genes that were expressed in more than 10% of cells with at least 0.1-fold difference (log-scale) were considered. For heatmap representation, mean expression of markers inside each cluster was used.
scRNAseq differential expression
To obtain differential expression between clusters, MAST test was performed, and p-value adjustment was done using the Bonferroni correction (Finak et al., 2015). Only genes that were expressed in more than 10% of cells in cluster were considered.
Bulk RNA isolation and transcriptome analysis
ILC2 were FACS sorted from epidydimal fat and RNA was isolated using the RLT method. RNA was sequenced on a HiSeq2500. Fastq files for each sample were aligned to the mm10 genome (Gencode, release M23) using STAR (v2.7.3a) with the following parameters: STAR–genomeDir $GENOME_DIR –readFilesIn $WORK_DIR/$FILE_1 $WORK_DIR/$FILE_2 –runThreadN 12 –readFiles-Command zcat –outFilterMultimapNmax 15 –outFilterMismatchNmax6 –outReadsUnmapped Fastx –outSAMstrandField intronMotif –outSAMtype BAM SortedByCoordinate –outFileNamePrefix./$ (Dobin et al., 2013). Quality control was performed by FastQC (v0.11.3), MultiQC (v1.1) (Ewels et al., 2016), and Picard tools (v2.18.4). Quantification was done using htseq-count function from HTSeq framework (v0.9.1): htseq-count -f bam -r pos -s no -t exon $BAM $ANNOTATION > $OUTPUT (Anders et al., 2015). Differential expression analysis was done using DESeq function from DeSeq2 package (Love et al., 2014) (v1.24.0) with default settings. Significance threshold was set to adjusted p-value < 0.05. Gene set enrichment analysis via fgsea R package (Sergushichev, 2016) (v1.10.0) was used to identify enriched pathways and plot enrichment curves.
Adoptive transfer
Adult Red5 mice were treated with IL-33 for 5 consecutive days as described above, rested for 2 days, and then RFP+ cells were sorted from digested visceral adipose tissue using a BD FACSAria. A total of 3x105 sorted ILC2 were transferred into old sex-matched recipients in bilateral i.p. injections (1.5x105 per injection), and recipient mice were simultaneously given a single dose of IL-33 at 12.5ug/kg body weight to facilitate engraftment. Recipient mice were rested for 1-2 weeks and then subjected to cold challenge.
Cold challenge
Mice were singly-housed in cages that contained bedding, but without nestlet, for cold challenge at 6°C. Mice were monitored twice daily for survival during cold challenge, and adipose tissue was collected from surviving mice at 48hr time point for western blot of UCP1 expression.
Cytokine secretion
Mice were treated with IL-33 as described above. Approximate 7x104 ILC2 per mouse were sorted from adipose tissue and stimulated in a total volume of 0.1mL RPMI+5% FCS with PMA+ionomycin for 4 hours at 37C. Culture supernatants were stored at −80C until analysis with Luminex assay. Samples were run on a Luminex 200.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Statistical differences were calculated and graphed in GraphPad Prism software. All statistical tests and group sizes for each experiment are indicated in the corresponding figure legend. For all experiments, p < 0.05 was considered significant. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Material
Highlights.
ILC2s are lost during aging
Expanding aged ILC2s with IL-33 increases cold lethality in old mice
Adult ILC2s transferred into old mice are sufficient to rescue cold lethality
ACKNOWLEDGMENTS
We thank all members of the Dixit lab for critical discussion and feedback related to this project. All schematics were created with BioRender.com. The Dixit lab is supported in part by NIH grants P01AG051459, AR070811, and AG068863 and Cure Alzheimer’s Fund. The Goldberg Lab is supported in part by the National Institute on Aging (R00AG058801). T.T. is supported by a postdoctoral fellowship from the Japan Society for the Promotion of Science.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cmet.2021.08.004.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Anders S, Pyl PT, and Huber W (2015). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, and Masopust D (2014). Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc 9, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapat SP, Myoung Suh J, Fang S, Liu S, Zhang Y, Cheng A, Zhou C, Liang Y, LeBlanc M, Liddle C, et al. (2015). Depletion of fat-resident Treg cells prevents age-associated insulin resistance. Nature 528, 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Singh PP, Mahmoudi S, Harel I, Casey KM, Dulken BW, Kundaje A, and Brunet A (2019). Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res. 29, 697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beretta E, Dhillon H, Kalra PS, and Kalra SP (2002). Central LIF gene therapy suppresses food intake, body weight, serum leptin and insulin for extended periods. Peptides 23, 975–984. [DOI] [PubMed] [Google Scholar]
- Brestoff JR, Kim BS, Saenz SA, Stine RR, Monticelli LA, Sonnenberg GF, Thome JJ, Farber DL, Lutfy K, Seale P, and Artis D (2015). Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature 519, 242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigger D, Riether C, van Brummelen R, Mosher KI, Shiu A, Ding Z, Zbären N, Gasser P, Guntern P, Yousef H, et al. (2020). Eosinophils regulate adipose tissue inflammation and sustain physical and immunological fitness in old age. Nat. Metab 2, 688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camell CD, Günther P, Lee A, Goldberg EL, Spadaro O, Youm YH, Bartke A, Hubbard GB, Ikeno Y, Ruddle NH, et al. (2019). Aging induces an Nlrp3 inflammasome-dependent expansion of adipose B cells that impairs metabolic homeostasis. Cell Metab. 30, 1024–1039.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camell CD, Sander J, Spadaro O, Lee A, Nguyen KY, Wing A, Goldberg EL, Youm YH, Brown CW, Elsworth J, et al. (2017). Inflammasome-driven catecholamine catabolism in macrophages blunts lipolysis during ageing. Nature 550, 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Shi P, Wang D, Liu Q, Li X, Wang Y, Zou D, Huang Z, Gao X, and Lin Z (2019). Epidermis-activated gasdermin-A3 enhances thermogenesis of brown adipose tissue through IL-6/Stat3 signaling. Am. J. Pathol 189, 1041–1052. [DOI] [PubMed] [Google Scholar]
- Collins N, Han SJ, Enamorado M, Link VM, Huang B, Moseman EA, Kishton RJ, Shannon JP, Dixit D, Schwab SR, et al. (2019). The bone marrow protects and optimizes immunological memory during dietary restriction. Cell 178, 1088–1101.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren MW, Jones SW, Cautivo KM, Dubinin A, Ortiz-Carpena JF, Farhat S, Yu KS, Lee K, Wang C, Molofsky AV, et al. (2019). Adventitial stromal cells define group 2 innate lymphoid cell tissue niches. Immunity 50, 707–722.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 20, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza SS, Shen X, Fung ITH, Ye L, Kuentzel M, Chittur SV, Furuya Y, Siebel CW, Maillard IP, Metzger DW, and Yang Q (2019). Compartmentalized effects of aging on group 2 innate lymphoid cell development and function. Aging Cell 18, e13019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulken BW, Buckley MT, Navarro Negredo P, Saligrama N, Cayrol R, Leeman DS, George BM, Boutet SC, Hebestreit K, Pluvinage JV, et al. (2019). Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature 571, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, and Käller M (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, Lee J, Goldfine AB, Benoist C, Shoelson S, and Mathis D (2009). Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med 15, 930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter K, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, and Campisi J (2014). Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A Biol. Sci. Med. Sci 69 (suppl 1), S4–S9. [DOI] [PubMed] [Google Scholar]
- Fung ITH, Sankar P, Zhang Y, Robison LS, Zhao X, D’Souza SS, Salinero AE, Wang Y, Qian J, Kuentzel ML, et al. (2020). Activation of group 2 innate lymphoid cells alleviates aging-associated cognitive decline. J. Exp. Med 217, e20190915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg EL, and Dixit VD (2015). Drivers of age-related inflammation and strategies for healthspan extension. Immunol. Rev 265, 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg EL, Shchukina I, Asher JL, Sidorov S, Artyomov MN, and Dixit VD (2020). Ketogenesis activates metabolically protective γδ T cells in visceral adipose tissue. Nat. Metab 2, 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goronzy JJ, and Weyand CM (2019). Mechanisms underlying T cell ageing. Nat. Rev. Immunol 19, 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granot T, Senda T, Carpenter DJ, Matsuoka N, Weiner J, Gordon CL, Miron M, Kumar BV, Griesemer A, Ho S-H, et al. (2017). Dendritic cells display subset and tissue-specific maturation dynamics over human life. Immunity 46, 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LY, Tanaka Y, Herbine K, Pastore C, Singh B, Ferguson A, Vora N, Douglas B, Zullo K, Behrens EM, et al. (2020). Cellular context of IL-33 expression dictates impact on anti-helminth immunity. Sci. Immunol 5, eabc6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S, Tung N, Casanova-Acebes M, Chang C, Cantoni C, Zhang D, Wirtz TH, Naik S, Rose SA, Brocker CN, et al. (2019). Dietary intake regulates the circulating inflammatory monocyte pool. Cell 178, 1102–1114.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights AJ, Vohralik EJ, Houweling PJ, Stout ES, Norton LJ, Alexopoulos SJ, Yik JJ, Mat Jusoh H, Olzomer EM, Bell-Anderson KS, et al. (2020). Eosinophil function in adipose tissue is regulated by Kruppel-like factor 3 (KLF3). Nat. Commun 11, 2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlgruber AC, Gal-Oz ST, LaMarche NM, Shimazaki M, Duquette D, Koay HF, Nguyen HN, Mina AI, Paras T, Tavakkoli A, et al. (2018). γδ T cells producing interleukin-17A regulate adipose regulatory T cell homeostasis and thermogenesis. Nat. Immunol 19, 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodin D, van Panhuys N, Li C, Magnuson AM, Cipolletta D, Miller CM, Wagers A, Germain RN, Benoist C, and Mathis D (2015). Antigen- and cytokine-driven accumulation of regulatory T cells in visceral adipose tissue of lean mice. Cell Metab. 21, 543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosteli A, Sugaru E, Haemmerle G, Martin JF, Lei J, Zechner R, and Ferrante AW Jr. (2010). Weight loss and lipolysis promote a dynamic immune response in murine adipose tissue. J. Clin. Invest 120, 3466–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, and Dixit VD (2020). Dietary regulation of immunity. Immunity 53, 510–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MW, Odegaard JI, Mukundan L, Qiu Y, Molofsky AB, Nussbaum JC, Yun K, Locksley RM, and Chawla A (2015). Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell 160, 74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng CN, Bodzin JL, and Saltiel AR (2007a). Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest 117, 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng CN, Deyoung SM, Bodzin JL, and Saltiel AR (2007b). Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes 56, 16–23. [DOI] [PubMed] [Google Scholar]
- Lumeng CN, Liu J, Geletka L, Delaney C, Delproposto J, Desai A, Oatmen K, Martinez-Santibanez G, Julius A, Garg S, and Yung RL (2011). Aging is associated with an increase in T cells and inflammatory macrophages in visceral adipose tissue. J. Immunol 187, 6208–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlakõiv T, Flamar AL, Johnston LK, Moriyama S, Putzel GG, Bryce PJ, and Artis D (2019). Stromal cells maintain immune cell homeostasis in adipose tissue via production of interleukin-33. Sci. Immunol 4, eaax0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, and Dixit VM (2006). Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440, 228–232. [DOI] [PubMed] [Google Scholar]
- Meydani SN, Das SK, Pieper CF, Lewis MR, Klein S, Dixit VD, Gupta AK, Villareal DT, Bhapkar M, Huang M, et al. (2016). Long-term moderate calorie restriction inhibits inflammation without impairing cell-mediated immunity: a randomized controlled trial in non-obese humans. Aging (Albany, NY) 8, 1416–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Nussbaum JC, Liang HE, Van Dyken SJ, Cheng LE, Mohapatra A, Chawla A, and Locksley RM (2013). Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J. Exp. Med 210, 535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Van Gool F, Liang HE, Van Dyken SJ, Nussbaum JC, Lee J, Bluestone JA, and Locksley RM (2015). Interleukin-33 and Interferon-Gamma Counter-Regulate Group 2 innate lymphoid cell activation during immune perturbation. Immunity 43, 161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M, Noguchi R, Takahashi D, Morikawa T, Koshida K, Komiyama S, Ishihara N, Yamada T, Kawamura YI, Muroi K, et al. (2019). Fastingrefeeding impacts immune cell dynamics and mucosal immune responses. Cell 178, 1072–1087.e14. [DOI] [PubMed] [Google Scholar]
- Nussbaum JC, Van Dyken SJ, von Moltke J, Cheng LE, Mohapatra A, Molofsky AB, Thornton EE, Krummel MF, Chawla A, Liang H-E, and Locksley RM (2013). Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 502, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirzgalska RM, Seixas E, Seidman JS, Link VM, Sánchez NM, Mahù I, Mendes R, Gres V, Kubasova N, Morris I, et al. (2017). Sympathetic neuron-associated macrophages contribute to obesity by importing and metabolizing norepinephrine. Nat. Med 23, 1309–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana BMJ, Jou E, Barlow JL, Rodriguez-Rodriguez N, Walker JA, Knox C, Jolin HE, Hardman CS, Sivasubramaniam M, Szeto A, et al. (2019). A stromal cell niche sustains ILC2-mediated type-2 conditioning in adipose tissue. J. Exp. Med 216, 1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo-Gonzalez RR, Van Dyken SJ, Schneider C, Lee J, Nussbaum JC, Liang HE, Vaka D, Eckalbar WL, Molofsky AB, Erle DJ, and Locksley RM (2018). Tissue signals imprint ILC2 identity with anticipatory function. Nat. Immunol 19, 1093–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, et al. (2005). IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity 23, 479–490. [DOI] [PubMed] [Google Scholar]
- Schneider C, Lee J, Koga S, Ricardo-Gonzalez RR, Nussbaum JC, Smith LK, Villeda SA, Liang HE, and Locksley RM (2019). Tissue-resident group 2 innate lymphoid cells differentiate by layered ontogeny and in situ perinatal priming. Immunity 50, 1425–1438.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergushichev A (2016). An algorithm forfast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. 10.1101/060012. [DOI] [Google Scholar]
- Shaw AC, Goldstein DR, and Montgomery RR (2013). Age-dependent dysregulation of innate immunity. Nat. Rev. Immunol 13, 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder ME, Finlayson MO, Connors TJ, Dogra P, Senda T, Bush E, Carpenter D, Marboe C, Benvenuto L, Shah L, et al. (2019). Generation and persistence of human tissue-resident memory T cells in lung transplantation. Sci. Immunol 4, eaav5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallanzani RG, Zemmour D, Xiao T, Jayewickreme T, Li C, Bryce PJ, Benoist C, and Mathis D (2019). Distinct immunocyte-promoting and adipocyte-generating stromal components coordinate adipose tissue immune and metabolic tenors. Sci. Immunol 4, eaaw3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome JJ, Yudanin N, Ohmura Y, Kubota M, Grinshpun B, Sathaliyawala T, Kato T, Lerner H, Shen Y, and Farber DL (2014). Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell 159, 814–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandanmagsar B, Youm YH, Ravussin A, Galgani JE, Stadler K, Mynatt RL, Ravussin E, Stephens JM, and Dixit VD (2011). The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat. Med 17, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, and Ferrante AW Jr. (2003). Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest 112, 1796–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf Y, Boura-Halfon S, Cortese N, Haimon Z, Sar Shalom H, Kuperman Y, Kalchenko V, Brandis A, David E, Segal-Hayoun Y, et al. (2017). Brown-adipose-tissue macrophages control tissue innervation and homeostatic energy expenditure. Nat. Immunol 18, 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youm YH, Grant RW, McCabe LR, Albarado DC, Nguyen KY, Ravussin A, Pistell P, Newman S, Carter R, Laque A, et al. (2013). Canonical Nlrp3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab. 18, 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youm YH, Nguyen KY, Grant RW, Goldberg EL, Bodogai M, Kim D, D’Agostino D, Planavsky N, Lupfer C, Kanneganti TD, et al. (2015). The ketone metabolite β-hydroxybutyrate blocks NLRP3 inflammasome-mediated inflammatory disease. Nat. Med 21, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefzadeh MJ, Flores RR, Zhu Y, Schmiechen ZC, Brooks RW, Trussoni CE, Cui Y, Angelini L, Lee K-A, McGowan SJ, et al. (2021). An aged immune system drives senescence and ageing of solid organs. Nature 594, 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyda M, Wernly B, Demyanets S, Kaun C, Hämmerle M, Hantusch B, Schranz M, Neuhofer A, Itariu BK, Keck M, et al. (2013). Severe obesity increases adipose tissue expression of interleukin-33 and its receptor ST2, both predominantly detectable in endothelial cells of human adipose tissue. Int. J. Obes. (Lond) 37, 658–665. [DOI] [PubMed] [Google Scholar]
- Zhao XY, Zhou L, Chen Z, Ji Y, Peng X, Qi L, Li S, and Lin JD (2020). The obesity-induced adipokine sST2 exacerbates adipose Treg and ILC2 depletion and promotes insulin resistance. Sci. Adv 6, eaay6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data reported in this paper was deposited to the Gene Expression Omnibus and accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC anti-mouse CD3 Antibody | Biolegend | Cat#100203; RRID:AB_312660 |
| FITC anti-mouse TCR β chain Antibody | Biolegend | Cat#109205; RRID:AB_313428 |
| FITC anti-mouse CD5 Antibody | Biolegend | Cat#100605; RRID:AB_312734 |
| FITC anti-mouse/human CD11b Antibody | Biolegend | Cat#101205; RRID:AB_312788 |
| FITC anti-mouse CD19 Antibody | Biolegend | Cat#115505; RRID:AB_313640 |
| FITC anti-mouse CD11c Antibody | Biolegend | Cat#117305; RRID:AB_313774 |
| FITC anti-mouse FcεRIα Antibody | Biolegend | Cat#134305; RRID:AB_1626102 |
| FITC anti-mouse Ly-6G/Ly-6C (Gr-1) Antibody | Biolegend | Cat#108405; RRID:AB_313370 |
| FITC anti-mouse TER-119/Erythroid Cells Antibody | Biolegend | Cat#116205; RRID:AB_313706 |
| FITC anti-mouse F4/80 Antibody | Biolegend | Cat#123107; RRID:AB_893500 |
| FITC anti-mouse TCR γ/δ Antibody | Biolegend | Cat#118105; RRID:AB_313829 |
| FITC anti-mouse NK-1.1 Antibody | Biolegend | Cat#108705; RRID:AB_313392 |
| A700 anti-mouse CD3 Antibody | Biolegend | Cat#100216; RRID:AB_493697 |
| A700 anti-mouse TCR β chain Antibody | Biolegend | Cat#109224; RRID:AB_1027648 |
| A700 anti-mouse CD5 Antibody | Biolegend | Cat#100636; RRID:AB_2687002 |
| A700 anti-mouse/human CD11b Antibody | Biolegend | Cat#101222; RRID:AB_493705 |
| A700 anti-mouse CD19 Antibody | Biolegend | Cat#115528; RRID:AB_493735 |
| A700 anti-mouse CD11c Antibody | Biolegend | Cat#117320; RRID:AB_528736 |
| A700 anti-mouse FcεRIα Antibody | Biolegend | Cat#134324; RRID:AB_2566734 |
| A700 anti-mouse Ly-6G/Ly-6C (Gr-1) Antibody | Biolegend | Cat#108422; RRID:AB_2137487 |
| A700 anti-mouse TER-119/Erythroid Cells Antibody | Biolegend | Cat#116220; RRID:AB_528963 |
| A700 anti-mouse F4/80 Antibody | Biolegend | Cat#123130; RRID:AB_2293450 |
| A700 anti-mouse NK-1.1 Antibody | Biolegend | Cat#108730; RRID:AB_2291262 |
| IL-33R (ST2) Rat anti-Mouse, PE-Cyanine7, Clone: RMST2-2 | eBioscience | Cat#501123451 |
| Brilliant Violet 711 anti-mouse CD45 Antibody, Clone 30-F11 | Biolegend | Cat#103147; RRID:AB_2564383 |
| FITC anti-mouse CD45.2 Antibody, Clone 104 | Biolegend | Cat#109806; RRID:AB_313443 |
| GATA3-PE antibody | Fisher Scientific | Cat#12-9966-41; RRID:AB_1963601 |
| APC anti-mouse CD25 Antibody | Biolegend | Cat#101909; RRID:AB_961208 |
| Brilliant Violet 605 anti-mouse CD127 (IL-7Rα) Antibody | Biolegend | Cat#135025; RRID:AB_2562114 |
| phosphorylated STAT3 (Tyr705) | Cell Signaling | Cat#9145; RRID:AB_2491009 |
| UCP1 | Abcam | Cat#ab10983; RRID:AB_2241462 |
| Total STAT3 | Cell Signaling | Cat#79D7, RRID:AB_331269 |
| b-Actin | Cell Signaling | Cat#4967L; RRID:AB_330288 |
| HRP-conjugated goat-anti-rabbit | ThermoScientific | Cat#PI31460; RRID:AB_228341 |
| Fc Block | Biolegend | Cat#101302; RRID:AB_312801 |
| Chemicals, peptides, and recombinant proteins | ||
| IL-33, mouse, carrier-free | Biolegend | Cat#580508 |
| Collagenase, Type 2 | Fisher Scientific | Cat#NC9870009 |
| FTY-720 | Sigma | Cat#SML0700-25MG |
| Critical commercial assays | ||
| Glycerol measurement kit | Sigma | Cat#MAK117 |
| IL-33R ELISA | R&D | Cat#MST200 |
| Luminex Assay | R&D | custom pre-mixed multi-analyte kit |
| Deposited data | ||
| Adult adipose-resident CD45+ scRNAseq | Goldberg et al., 2020 | GEO: GSE137076 |
| Old adipose-resident CD45+ scRNAseq | This paper | GEO: GSE180910 |
| Old whole adipose tissue SVF scRNAseq | This paper | GEO: GSE180910 |
| Adult and Old IL-33-expanded ILC2 RNAseq | This paper | GEO: GSE180858 |
| Experimental models: Organisms/strains | ||
| Adult C57BL6/J mice | Jackson Labs | stock #000664 |
| Old C57BL6/J mice | NIA Aged Rodent Colony | N/A |
| Red5 mice | Jackson Labs, Nussbaum et al., 2013 | stock #030926 |
| Nlrp3−/− mice | Mariathasan et al., 2006 | Bred in Dixit colony |
| Calorie-restricted mice | NIA Aged Rodent Colony | N/A |
| Rag−/−IL-2Rγ−/− mice | Taconic | Stock# 4111 |
| Oligonucleotides | ||
| Soluble IL33R (Variant 3) Forward 5′-TGGCTAGGACCTCTGGCTAA-3′ | This Paper | N/A |
| Soluble IL33R (Variant 3) Reverse 5′-ATGGTGTGTTCACTAGGCGG-3′ | This Paper | N/A |
| Membrane-bound IL33R (Variant 1) Forward 5′-CTCTGCCCGACGTTCTTGA-3′ | This Paper | N/A |
| Membrane-bound IL33R (Variant 1) Reverse 5′-AACCCCTGATGTGTCTCAGT-3′ | This Paper | N/A |
| IL-33 N-terminal Forward 5′-AACTCCAAGATTTCCCCGGC-3′ | This Paper | N/A |
| IL-33 N-terminal Reverse 5′-TTATGGTGAGGCCAGAACGG-3′ | This Paper | N/A |
| IL-33 C-terminal Forward 5′-AGACCAGGTGCTACTACGCT-3′ | This Paper | N/A |
| IL-33 C-terminal Reverse 5′-ACGTCACCCCTTTGAAGCTC-3′ | This Paper | N/A |
| Gapdh Forward 5′-TCAACAGCAACTCCCACTCTTCCA-3′ | Youm et al., 2015 | N/A |
| Gapdh Reverse 5′-ACCCTGTTGCTGTAGCCGTATTCA-3′ | Youm et al., 2015 | N/A |
| Software and algorithms | ||
| Prism (v8.4.3) | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Biorender | Biorender | https://biorender.com/ |
| FACSDiva (v7.0) | BD | https://www.bdbiosciences.com/en-eu/products/software/instrument-software/bd-facsdiva-software#Overview |
| FlowJo (v10.6) | Tree Star | https://www.flowjo.com/solutions/flowjo/downloads |
| Cell Ranger Single-Cell Software Suite | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome |
| Seurat (v2.3) | Satija Lab, New York Genome Center | https://satijalab.org/seurat/ |
| Other | ||
| Zombie Aqua Fixable Viability dye | Biolegend | Cat#423102 |
| Foxp3 Fix/Perm Intracellular Staining Kit | eBioscience | Cat#00-5521-00 |


