The crystal structure of the PX (phox homology) domain of the Saccharomyces cerevisiae Vps17p protein was determined. The absence of basic residues around the canonical binding site suggests an inability to bind phosphatidylinositol phosphate molecules.
Keywords: AlphaFold2, International Space Station, microgravity, phox homology, PX domain, Vps17p, Saccharomyces cerevisiae
Abstract
The structure determination of the PX (phox homology) domain of the Saccharomyces cerevisiae Vps17p protein presented a challenging case for molecular replacement because it has noncrystallographic symmetry close to a crystallographic axis. The combination of diffraction-quality crystals grown under microgravity on the International Space Station and a highly accurate template structure predicted by AlphaFold2 provided the key to successful crystal structure determination. Although the structure of the Vps17p PX domain is seen in many PX domains, no basic residues are found around the canonical phosphatidylinositol phosphate (PtdIns-P) binding site, suggesting an inability to bind PtdIns-P molecules.
1. Introduction
Recycling of receptors and transporters from the endosomes to the Golgi compartment is essential for the assembly and function of the lysosome (Cullen, 2008 ▸). Saccharomyces cerevisiae (baker’s yeast) has three major endosomal recycling pathways: retromer, Snx4 and Mvp1/Snx8 (Suzuki et al., 2021 ▸). The principal components of the three pathways are SNX–BAR proteins (van Weering et al., 2010 ▸; Yong et al., 2020 ▸). SNX (sorting nexin) is a diverse protein family that contains the PX (phox homology) domain and performs cellular trafficking functions (Hanley & Cooper, 2020 ▸). The PX domain can bind phosphatidylinositol phosphates (PtdIns-Ps; Teasdale & Collins, 2012 ▸). SNX–BAR is an SNX subfamily that contains a BAR (Bin–amphiphysin–Rvs167) domain in addition to the PX domain. The BAR domain functions to induce and stabilize membrane curvature. S. cerevisiae has at least 15 PX domain-containing proteins (Yu & Lemmon, 2001 ▸), including seven SNX–BAR proteins (Suzuki et al., 2021 ▸). Among them, Vps17p (vacuolar protein sorting-associated protein 17) functions in membrane binding/deformation as the SNX–BAR protein subunit of the membrane-associated retromer complex (Seaman & Williams, 2002 ▸). Vps5p is the other SNX–BAR protein in the same retromer complex (Seaman & Williams, 2002 ▸). The human ortholog of Vps17p is SNX5/SNX6, and that of Vps5p is SNX1/SNX2 (Cullen, 2008 ▸).
The PX domain is approximately 110 residues long and is conserved from yeast to humans (Teasdale & Collins, 2012 ▸). Despite the minimal amino-acid sequence conservation (the average percentage identity and similarity are 15% and 30%, respectively), the three-dimensional structure of the PX domain is highly conserved. It folds into a three-stranded antiparallel β-sheet followed by three α-helices, with a segment containing a PxxP motif between them. In our structural studies of PX domains, we obtained crystals of the PX domain from the Vps17p protein. In a search for crystals of diffraction quality, we used the microgravity environment on the International Space Station (ISS; Sato et al., 2006 ▸). Even though a crystal diffracted to 2.0 Å resolution, we were unable to solve the phase problem at the time. After 18 years, the structure predicted by AlphaFold2 (Jumper et al., 2021 ▸) prompted us to try molecular replacement again using the original diffraction data. Here, we report the 3D structure of the Vps17p PX domain and discuss its structural features and a possible reason for the unsuccessful molecular replacement in earlier attempts.
2. Materials and methods
2.1. Expression of the PX domain of the S. cerevisiae Vps17 protein
The PX domain (residues 100–234) of Vps17p from S. cerevisiae (UniProtKB P32913, VPS17_YEAST and YOR132W) was produced in Escherichia coli BL21(DE3) Gold cells (Novagen) using the pGEX-6P-1 vector (Amersham Biosciences). The transformed E. coli cells were cultured at 310 K in LB medium containing 50 µg ml−1 ampicillin. When the OD600 reached 0.5, the culture was supplemented with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). The cells were cultured at 310 K for 3 h for induction. After collection by centrifugation, the cell pellet was resuspended in 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM DTT. The cells were then disrupted by sonication and the resultant solution was separated into supernatant and pellet fractions by centrifugation. The supernatant was adsorbed to Glutathione Sepharose 4B resin (GE Healthcare) and the PX domain was eluted with 10 mM reduced l-glutathione. The fractions containing the PX domain were combined and concentrated by ultrafiltration (Amicon Ultra, Millipore). The fusion protein was cleaved with 3C protease to release the GST tag. The protein was further purified by chromatography on a POROS-HS cation-exchange column (Thermo Fisher Scientific) and then on a TSKgel SuperSW2000 gel-filtration column (TOSOH). The fractions containing the PX domain were combined and concentrated to 16–24 mg ml−1 by ultrafiltration in 20 mM MOPS–NaOH pH 6.5, 50 mM NaCl, 1 mM DTT.
2.2. Crystallization
Initial screening was performed using the sitting-drop vapor-diffusion method in 96-well Intelli-Plates (Art Robbins Instruments). Sitting drops were set up by mixing equal volumes (0.2 µl each) of the protein solution and the reservoir solution using an automated dispenser (Hydra II Plus One System, Apogent Discoveries). Initial screening was performed at 293 K using the Crystal Screen and Crystal Screen 2 kits (Hampton Research). Crystals were obtained with Crystal Screen solution Nos. 16, 32, 34 and 47 and Crystal Screen 2 solution Nos. 23, 32 and 36. Optimization was performed by the hanging-drop vapor-diffusion method in 24-well VDX greased plates (Hampton Research). Hanging drops were prepared manually by mixing 1 µl protein solution and 1 µl reservoir solution. Each hanging drop was placed over 0.4 ml reservoir solution. After optimization, crystals were grown in Crystal Screen solution No. 16 (1.5 M lithium sulfate, 0.1 M HEPES–NaOH pH 7.5) at 293 K. The PX domain crystals were cryoprotected by the addition of glycerol at 30% and were cryocooled in a nitrogen-gas stream (100 K).
Diffraction-quality Vps17p PX domain crystals were obtained using the counter-diffusion crystallization method (Tanaka et al., 2004 ▸) with a configuration of the gel-tube method (Sato et al., 2006 ▸). A capillary filled with the protein stock solution (16 mg ml−1) was inserted into agarose gel in a silicon tube. The length and inner diameter of the capillary were 60 and 0.5 mm, respectively, and the gel length was 6 mm. The end of the gel was soaked in reservoir solution (1.5 M lithium sulfate, 0.1 M HEPES–NaOH pH 7.5). Crystallization was performed at 293 K in the microgravity environment of the ISS during the fourth mission of JAXA–GCF (Japan Aerospace Exploration Agency–Granada Crystallization Facility, August–October 2004; Sato et al., 2006 ▸).
2.3. Data collection and processing
The Vps17p PX domain crystals grown in the microgravity environment were cryoprotected by soaking in a cryoprotectant solution (1.5 M lithium sulfate, 0.1 M HEPES–NaOH pH 7.5, 30% glycerol) and then cooled in liquid nitrogen. The crystals diffracted to 2.0 Å resolution on BL12B2 at SPring-8, Harima, Japan. 100 diffraction patterns were recorded with an ADSC Q4 detector and were processed on-site with HKL-2000 (Otwinowski & Minor, 1997 ▸) and off-site with iMosflm version 7.2.2 (Battye et al., 2011 ▸).
2.4. Structure solution and refinement
The crystal structure was solved by the molecular-replacement method with MOLREP version 11.7 (Vagin & Teplyakov, 2010 ▸) in the CCP4 suite (Winn et al., 2011 ▸) using the structure predicted by AlphaFold2 as the search model (Jumper et al., 2021 ▸). We used the ColabFold server at https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb for prediction. The input sequence was residues 108–227 (LLAKVTGLERFGSATGKKENPTIIFDCSTNLPTFRKQQYKNVKKSYEEFHQLFKYLNVAIQESFVPTLPSAYTTFGINSEEDRMKVTRNFQLWFNRLSQDPLIIRNEEVAFFIESDFNTY) of Vps17p. The model-refinement calculation was performed using Phenix version 1.18 (Liebschner et al., 2019 ▸) and the side chains and water molecules were manually fitted using Coot version 0.9.6 (Emsley et al., 2010 ▸).
In our molecular-replacement attempts with structures from the PDB as templates, we used MrBUMP version 2.2.6 (Keegan & Winn, 2008 ▸) in the CCP4 suite. MrBUMP performs automated search-model generation and automated molecular replacement.
2.5. Structural analyses
The stereochemistry of the final model was analyzed using MolProbity (Williams et al., 2018 ▸) and the secondary structures were assigned using DSSP (Kabsch & Sander, 1983 ▸). The root-mean-square deviation (r.m.s.d.) between two structures was calculated with the align command in PyMOL version 2.4.2 (Schrödinger; https://www.pymol.org/). To find similar structures in the PDB, we used the DALI server (Holm, 2020 ▸) at http://ekhidna2.biocenter.helsinki.fi/dali/. Protein illustrations were generated with PyMOL.
3. Results and discussion
3.1. Crystallization and structure determination of the Vps17p PX domain
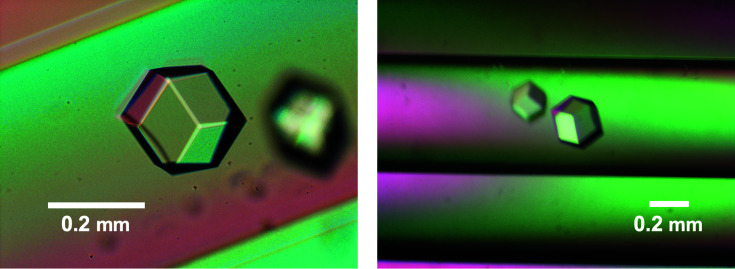
The PX domain of the S. cerevisiae Vps17 protein encompasses residues 108–227. At the beginning of our study, we prepared a 147-residue segment of the Vps17p protein (residues 88–234). Crystals with a diameter of 0.05 mm were obtained, but the diffraction was limited to 3.5 Å resolution. Because a shorter fragment was generated by limited proteolysis during prolonged storage at 4°C, we expressed a 135-residue fragment (residues 100–234) in E. coli BL21 cells as a fusion protein with the GST (glutathione S-transferase) protein. The PX domain was purified by affinity, cation-exchange and gel-filtration chromatography. The GST tag was removed by limited proteolysis after the affinity purification. The purified PX domain was subjected to crystallization screening. After optimization, we obtained crystals of 0.2 mm in diameter. Although the highest-angle spot was at 2.3 Å resolution on BL41XU at SPring-8, Harima, Japan, the poor quality of the diffraction images did not allow us to proceed to further processing. We also obtained selenomethionine-derivative crystals (2.8 Å resolution) but without successful structure solution. During these efforts, we obtained polyhedral shaped crystals under a microgravity environment (Fig. 1 ▸). A crystal with a diameter of 0.2 mm diffracted to 2.0 Å resolution in November 2004 on BL12B2 at SPring-8, Harima, Japan. The crystal belonged to space group P213, with unit-cell parameters a = b = c = 100.28 Å, α = β = γ = 90°. The data-collection statistics are summarized in Table 1 ▸. Even though the crystal was of good quality and several PX domain structures were available in the PDB, molecular replacement was unsuccessful.
Figure 1.
Crystals of the Vps17p PX domain grown in a microgravity environment. The scale bars represent 0.2 mm.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| X-ray data-collection satistics | |
| X-ray source | BL12B2, SPring-8 |
| Wavelength (Å ) | 1.000 |
| Space group | P213 |
| a, b, c (Å) | 100.28, 100.28, 100.28 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution range (Å) | 44.8–2.02 (2.07–2.02) |
| R merge | 0.178 (0.808) |
| 〈I/σ(I)〉 | 8.2 (1.5) |
| CC1/2 | 0.950 (0.548) |
| Completeness (%) | 100 (99.6) |
| Multiplicity | 7.9 (3.5) |
| Model-refinement statistics | |
| No. of reflections | 22287 |
| R work/R free † | 0.199/0.251 |
| No. of atoms | |
| Protein | 2075 |
| Sulfate | 20 |
| Water | 148 |
| Average B factors (Å2) | |
| Protein | 13.78 |
| Sulfate | 45.89 |
| Water | 18.73 |
| R.m.s.d. | |
| Bond lengths (Å) | 0.008 |
| Angles (°) | 0.91 |
| Ramachandran plot‡ | |
| Favored (%) | 98.75 |
| Allowed (%) | 1.25 |
| Outliers (%) | 0.0 |
4.8% of the reflections were excluded and used to calculate R free.
Analyzed with MolProbity.
After 18 years, we used AlphaFold2 (Jumper et al., 2021 ▸) to obtain a predicted structure of the Vps17p PX domain. In July 2021, we submitted the amino-acid sequence of the Vps17p PX domain to the ColabFold server. The predicted structure worked well as a template structure for molecular replacement. The asymmetric unit contained two PX domain molecules. The final refinement converged with R work and R free values of 0.199 and 0.251, respectively. The refinement statistics are presented in Table 1 ▸. The stereochemistry of the final model was good, and no residues were found in the outlier regions of the Ramachandran plot. The coordinates and structure factors have been deposited in the PDB under accession code 7x4o.
3.2. Overall structure of the Vps17p PX domain
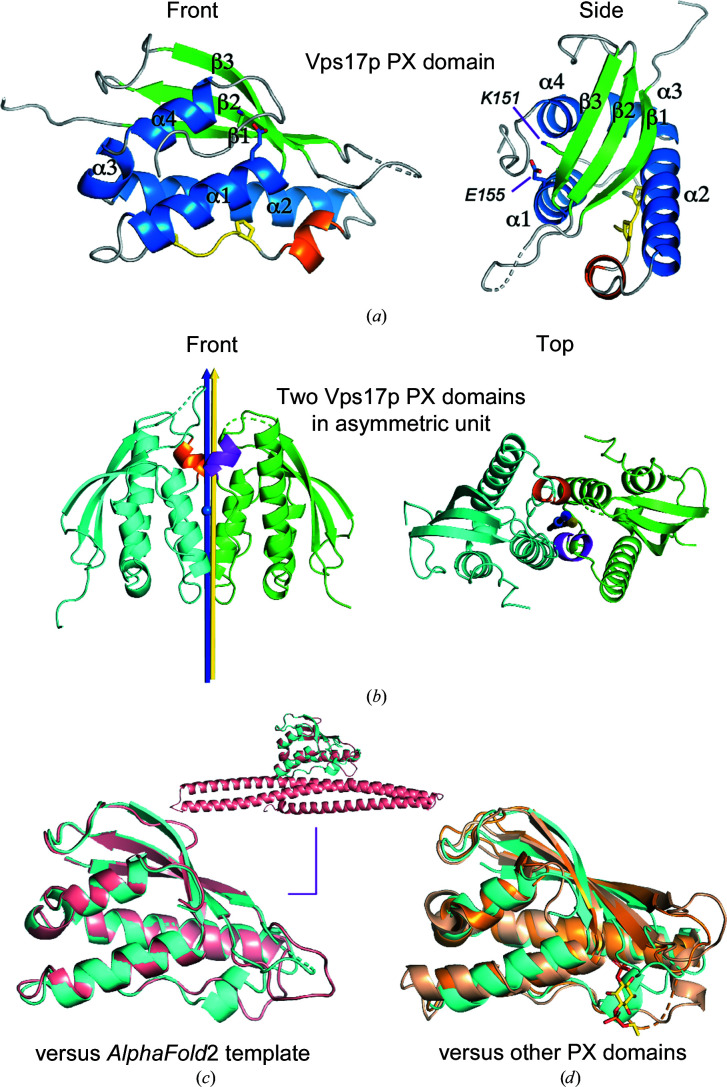
The Vps17p PX domain has a flat, compact shape and the typical main-chain topology unique to PX domains. It consists of three strands, β1 (Leu108–Glu116), β2 (Thr129–Thr136) and β3 (Gln145–Ser152), in the N-terminal third of the amino-acid sequence and four helices, α1 (Tyr153–Ala166), α2 (Glu187–Gln206), α3 (Pro208–Arg212) and α4 (Glu214–Glu221), in the C-terminal two-thirds (Fig. 2 ▸ a). The three strands form an antiparallel β-sheet structure. Pro173 and Pro176 are the two conserved proline residues in the PxxP motif, which is located between α1 and α2. The side chains of Lys151 and Glu155 form a conserved salt bridge inside the hydrophobic core (Fig. 2 ▸ a).
Figure 2.
Structure of the Vps17p PX domain and comparison with other PX domain structures. (a) Secondary structures: β-strands, green; α-helices, blue; extra short α-helix, orange; PxxP motif, yellow. The side chains of Lys151 and Glu155 are shown as stick models and form a conserved salt bridge inside the hydrophobic core. (b) The two PX domain chains (cyan and green) in the asymmetric unit are in close contact. The noncanonical α-helix, which is probably formed by crystal contacts, is shown in orange and magenta. The noncrystallographic pseudo-twofold rotation axis (blue arrow) between the two PX domain chains is almost parallel to one of the crystallographic twofold screw axes (yellow arrow). The NCS axis passes through the center of mass (blue ball) of the two chains. The angle between the two axes is 1.7°. (c) Superimposition of the Vps17p PX domain (chain B, cyan) with the template PX domain structure predicted by AlphaFold2 (salmon). The inset shows the same superimposition but includes the predicted BAR domain structure. (d) Superimposition of the Vps17p PX domain (chain B, cyan) with the S. cerevisiae Grp19p (SNX3) PX domain (PDB entry 1ocu, chain A, wheat) and the human SNX11 PX domain (PDB entry 6koi, chain P, bright orange). The Grp19p PX domain is a complex with the water-soluble analog diC4PtdIns-3-P (yellow sticks).
There is a noncanonical short helix (Ala178–Phe182) immediately after the PxxP motif. We inferred that this short helix was induced artificially by the close crystal contacts at the interface between the two PX domain molecules in the asymmetric unit (Fig. 2 ▸ b). Convincingly, the PX domain contacts the BAR domain in the AlphaFold2 structure of the Vps17p protein and this helix does not exist at the PX–BAR interface (Fig. 2 ▸ c), suggesting that the Vps17p PX domain alone is a monomer in solution.
3.3. Structural comparison with other PX domains
Molecular replacement was previously unsuccessful, and at the time we attributed the phasing failure to unusual structural features of the Vps17p PX domain. Here, we searched for the reason behind the failure of molecular replacement. Firstly, we conducted a 3D homology search using the DALI server (Holm, 2020 ▸). Against a representative subset of PDB entries (the PDB25 option), the PX domain of S. cerevisiae Grp19p (also known as SNX3; PDB entry 1ocu, chain A; Zhou et al., 2003 ▸) was at the top of the list. The Grp19p PX domain superimposed on the refined Vps17p PX domain with an r.m.s.d. of 1.7 Å over 79 Cα atoms. Against the full PDB, the top match was the PX domain of human SNX11 (PDB entry 6koi, chain P; Xu et al., 2020 ▸). The SNX11 PX domain superimposed on the refined Vps17p PX domain with an r.m.s.d. of 1.3 Å over 56 Cα atoms. Reflecting the moderately good r.m.s.d.s (1 Å < r.m.s.d. < 2 Å), the overall structures are similar to that of the Vps17p PX domain, but the details are different (Fig. 2 ▸ d). Thus, contrary to our expectation, the Vps17p PX domain has no special features that could interfere with molecular replacement. For confirmation, we attempted molecular replacement using the two PX domain structures, but no solutions were found. The best MOLREP scores were as low as corrF = 0.3209 and TF/sig = 3.08 for the Grp19p PX domain and corrF = 0.3351, TF/sig = 1.08 for the SNX11 PX domain. We also tried to remove long loops in the template structures (residues 45–57 in PDB entry 1ocu and residues 28–34 in PDB entry 6koi) and even the use of polyalanine models, but without success.
As an alternative approach, we tried MrBUMP in the CCP4 suite. MrBUMP uses phmmer to find a set of structures with similar sequences to a query sequence. At the top of the list was the PX domain of S. cerevisiae Grp19p (PDB entry 1ocs, chain A; Zhou et al., 2003 ▸). PDB entry 1ocs is the same structure as PDB entry 1ocu found in the DALI search, but PDB entry 1ocs is an apo form whereas PDB entry 1ocu is a ligand-bound form (Zhou et al., 2003 ▸). As expected, molecular replacement with PDB entry 1ocs as a template failed: the best MOLREP scores were as low as corrF = 0.3092 and TF/sig = 3.85. In sum, no PDB entries that are effective as a template structure have been deposited since our initial attempts at molecular replacement.
Finally, we compared the structure determined by molecular replacement and the AlphaFold2 structure. In accordance with the high structural similarity (0.38 Å over 97 Cα atoms; Fig. 2 ▸ c), MOLREP provided high scores of corrF = 0.5359 and TF/sig = 19.62.
3.4. Reason for the phasing failure with the PDB entries
Noncrystallographic symmetry registered in the PDB can be the true crystallographic symmetry (Wang, 2015 ▸). However, in the case of the Vps17p PX domain crystal the electron densities of the two chains in the asymmetric unit are different, particularly in the loop segment residues 116–126. The noncrystallographic pseudo-twofold rotation axis (blue in Fig. 2 ▸ b) almost overlaps with one of the crystallographic twofold screw axes (yellow in Fig. 2 ▸ b). This coincidence resulted in pseudo-centering on the intensity distribution in a native Patterson map. In addition, the two polypeptide chains in the asymmetric unit are tightly packed (Fig. 2 ▸ b). The peaks derived from the intermolecular vectors are located near the origin of the native Patterson map, and their distribution overlaps with the peaks from the intramolecular vectors. These properties of the native Patterson map of the Vps17p PX domain crystal make molecular replacement difficult without a highly accurate template structure.
3.5. Possibility of PtdIns-P binding
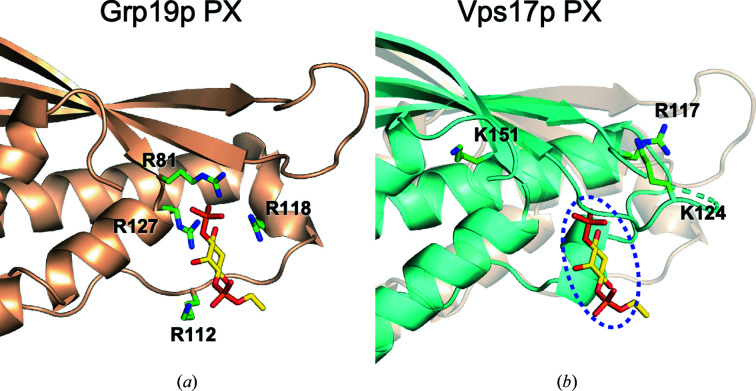
Most PX domains specifically bind to PtdIns-3-P (Yu & Lemmon, 2001 ▸). A PtdIns-3-P molecule has two negatively charged phosphate groups in the head moiety. As a natural consequence, the PX domains possess positively charged arginine and lysine residues located peripherally to the canonical PtdIns-3-P binding site. A short-chain, water-soluble analog, d-myo-phosphatidylinositol-3-phosphate (diC4PtdIns-3-P), is frequently used in crystallization. The crystal structure of the S. cerevisiae Grp19p PX domain in complex with a diC4PtdIns-3-P molecule is shown in Fig. 3 ▸(a) (Zhou et al., 2003 ▸). As expected, there are four basic residues within interaction distance of the phosphate groups of the ligand. In contrast, although the Vps17p PX domain possesses three basic residues around the PtdIns-P binding site, they appear to be too far away for direct interaction with a putative ligand in the binding site, unless a large conformational change occurs (Fig. 3 ▸ b). This notion suggests that the Vps17p PX domain is either unable to bind to or only weakly interacts with PtdIns-Ps. Indeed, a biochemical study demonstrated that the Grp19p PX domain was strongly bound to PtdIns-3-P, whereas the Vps17p PX domain had a very low affinity for PtdIns-3-P (Yu & Lemmon, 2001 ▸). The same study also showed that the PX domain of the other SNX–BAR protein, Vps5p, in the retromer bound to PtdIns-3-P very weakly. Another study confirmed the absence of binding of Vsp5p PX domain to any PtdIns-Ps (Song et al., 2001 ▸). These data argue that the binding of PtdIns-3-P by either Vps5p or Vps17p is not essential for the recruitment of the retromer complex onto endosomal membranes (Seaman & Williams, 2002 ▸).
Figure 3.
PtdIns-P binding site. (a) S. cerevisiae Grp19p (SNX3) PX domain (PDB entry 1ocu, chain A). A diC4PtdIns-3-P molecule is bound to the canonical PtdIns-3-P binding site of the PX domain. (b) The putative PtdIns-P binding site (dashed oval) of the Vps17p PX domain (cyan cartoon) is marked by the position of the diC4PtdIns-3-P molecule bound to the superimposed Grp19p PX domain (transparent wheat cartoon). In (a) and (b), positively charged residues surrounding the PtdIns-P binding sites are shown as green stick models and labeled.
4. Conclusions
The structure determination of the Vps17p PX domain represents a challenging case of molecular replacement with noncrystallographic symmetry close to a crystallographic axis. The combination of diffraction-quality crystals grown under microgravity conditions and a highly accurate template structure predicted by AlphaFold2 provided the key to successful structure determination. Although the structure of the Vps17p PX domain is seen in many PX domains, no basic residues are found around the canonical PtdIns-P binding site, suggesting that the Vps17p PX domain itself cannot bind to endosomal membranes containing PtdIns-3-P. The formation of the retromer complex on the endosomal membranes is thus mediated by other regions of Vps17p outside its PX domain.
Supplementary Material
PDB reference: PX domain of S. cerevisiae Vps17p, 7x4o
Acknowledgments
We thank Professor Takashi Ito (Kyushu University) for generation of the S. cerevisiae Vps17p PX domain expression plasmid. TO performed the purification, crystallized the protein and wrote the paper, KI supported the crystallization of the protein under microgravity conditions and collected X-ray diffraction data, and NM analyzed the data and wrote the paper with DK. All authors discussed the results and commented on the manuscript. The authors declare no competing interests.
Funding Statement
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant No. JP21H02448 and Mitsubishi Foundation (Japan) Research Grants in the Natural Sciences Grant No. 202110017 to DK.
References
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Cullen, P. J. (2008). Nat. Rev. Mol. Cell Biol. 9, 574–582. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Hanley, S. E. & Cooper, K. F. (2020). Cells, 10, 17. [DOI] [PMC free article] [PubMed]
- Holm, L. (2020). Methods Mol. Biol. 2112, 29–42. [DOI] [PubMed]
- Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., Tunyasuvunakool, K., Bates, R., Žídek, A., Potapenko, A., Bridgland, A., Meyer, C., Kohl, S. A. A., Ballard, A. J., Cowie, A., Romera-Paredes, B., Nikolov, S., Jain, R., Adler, J., Back, T., Petersen, S., Reiman, D., Clancy, E., Zielinski, M., Steinegger, M., Pacholska, M., Berghammer, T., Bodenstein, S., Silver, D., Vinyals, O., Senior, A. W., Kavukcuoglu, K., Kohli, P. & Hassabis, D. (2021). Nature, 596, 583–589.
- Kabsch, W. & Sander, C. (1983). Biopolymers, 22, 2577–2637. [DOI] [PubMed]
- Keegan, R. M. & Winn, M. D. (2008). Acta Cryst. D64, 119–124. [DOI] [PMC free article] [PubMed]
- Liebschner, D., Afonine, P. V., Baker, M. L., Bunkóczi, G., Chen, V. B., Croll, T. I., Hintze, B., Hung, L.-W., Jain, S., McCoy, A. J., Moriarty, N. W., Oeffner, R. D., Poon, B. K., Prisant, M. G., Read, R. J., Richardson, J. S., Richardson, D. C., Sammito, M. D., Sobolev, O. V., Stockwell, D. H., Terwilliger, T. C., Urzhumtsev, A. G., Videau, L. L., Williams, C. J. & Adams, P. D. (2019). Acta Cryst. D75, 861–877.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Sato, M., Tanaka, H., Inaka, K., Shinozaki, S., Yamanaka, A., Takahashi, S., Yamanaka, M., Hirota, E., Sugiyama, S., Kato, M., Saito, C., Sano, S., Motohara, M., Nakamura, T., Kobayashi, T., Yoshitomi, S. & Tanaka, T. (2006). Microgravity Sci. Technol. 18, 184–189.
- Seaman, M. N. J. & Williams, H. P. (2002). Mol. Biol. Cell, 13, 2826–2840. [DOI] [PMC free article] [PubMed]
- Song, X., Xu, W., Zhang, A., Huang, G., Liang, X., Virbasius, J. V., Czech, M. P. & Zhou, G. W. (2001). Biochemistry, 40, 8940–8944. [DOI] [PubMed]
- Suzuki, S. W., Oishi, A., Nikulin, N., Jorgensen, J. R., Baile, M. G. & Emr, S. D. (2021). eLife, 10, e69883. [DOI] [PMC free article] [PubMed]
- Tanaka, H., Inaka, K., Sugiyama, S., Takahashi, S., Sano, S., Sato, M. & Yoshitomi, S. (2004). J. Synchrotron Rad. 11, 45–48. [DOI] [PubMed]
- Teasdale, R. D. & Collins, B. M. (2012). Biochem. J. 441, 39–59. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Wang, J. (2015). Protein Sci. 24, 621–632. [DOI] [PMC free article] [PubMed]
- Weering, J. R. T. van, Verkade, P. & Cullen, P. J. (2010). Semin. Cell Dev. Biol. 21, 371–380. [DOI] [PMC free article] [PubMed]
- Williams, C. J., Headd, J. J., Moriarty, N. W., Prisant, M. G., Videau, L. L., Deis, L. N., Verma, V., Keedy, D. A., Hintze, B. J., Chen, V. B., Jain, S., Lewis, S. M., Arendall, W. B., Snoeyink, J., Adams, P. D., Lovell, S. C., Richardson, J. S. & Richardson, D. C. (2018). Protein Sci. 27, 293–315. [DOI] [PMC free article] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Xu, T., Gan, Q., Wu, B., Yin, M., Xu, J., Shu, X. & Liu, J. (2020). J. Mol. Biol. 432, 4750–4761. [DOI] [PubMed]
- Yong, X., Zhao, L., Deng, W., Sun, H., Zhou, X., Mao, L., Hu, W., Shen, X., Sun, Q., Billadeau, D. D., Xue, Y. & Jia, D. (2020). PLoS Biol. 18, e3000631. [DOI] [PMC free article] [PubMed]
- Yu, J. W. & Lemmon, M. A. (2001). J. Biol. Chem. 276, 44179–44184. [DOI] [PubMed]
- Zhou, C.-Z., Li de La Sierra-Gallay, I., Quevillon-Cheruel, S., Collinet, B., Minard, P., Blondeau, K., Henckes, G., Aufrère, R., Leulliot, N., Graille, M., Sorel, I., Savarin, P., de la Torre, F., Poupon, A., Janin, J. & van Tilbeurgh, H. (2003). J. Biol. Chem. 278, 50371–50376. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: PX domain of S. cerevisiae Vps17p, 7x4o