Here, Gualdrini et al. found that loss of H3K9me3 caused by SETDB1 depletion was associated with increased recruitment of CTCF to >1600 DNA binding motifs contained within SINE B2 repeats, a previously unidentified target of SETDB1-mediated repression. Their findings suggest a role for H3K9me3 in restraining genomic distribution and activity of CTCF, influencing chromatin organization and gene regulation.
Keywords: histone methylation, H3K9me3, CTCF, transposable elements, topological domains, macrophages
Abstract
Six methyltransferases divide labor in establishing genomic profiles of histone H3 lysine 9 methylation (H3K9me), an epigenomic modification controlling constitutive heterochromatin, gene repression, and silencing of retroelements. Among them, SETDB1 is recruited to active chromatin domains to silence the expression of endogenous retroviruses. In the context of experiments aimed at determining the impact of SETDB1 on stimulus-inducible gene expression in macrophages, we found that loss of H3K9me3 caused by SETDB1 depletion was associated with increased recruitment of CTCF to >1600 DNA binding motifs contained within SINE B2 repeats, a previously unidentified target of SETDB1-mediated repression. CTCF is an essential regulator of chromatin folding that restrains DNA looping by cohesin, thus creating boundaries among adjacent topological domains. Increased CTCF binding to SINE B2 repeats enhanced insulation at hundreds of sites and increased loop formation within topological domains containing lipopolysaccharide-inducible genes, which correlated with their impaired regulation in response to stimulation. These data indicate a role of H3K9me3 in restraining genomic distribution and activity of CTCF, with an impact on chromatin organization and gene regulation.
By altering the physical properties of chromatin and providing platforms for the recruitment of effector proteins, histone modifications control accessibility and usage of the underlying genome (Strahl and Allis 2000; Bannister and Kouzarides 2011; Allis and Jenuwein 2016). Among them, histone H3 lysine 9 methylation (H3K9me) (Rea et al. 2000) is a repressive modification with well-defined roles in the control of heterochromatin formation and partitioning (Peters et al. 2003), as well as in gene repression and silencing of retrotransposons (Tachibana et al. 2002, 2005; Matsui et al. 2010; Rowe et al. 2010; for review, see Jambhekar et al. 2019).
In mammals, H3K9me is deposited by three distinct families of enzymes. SUV39H1 and SUV39H2 (Rea et al. 2000) work in a redundant manner to establish H3K9me3 at constitutive heterochromatin, thus accounting for the bulk of H3K9me3 (Montavon et al. 2021). G9a and GLP instead deposit H3K9me1 and H3K9me2 in euchromatin to control gene repression (Tachibana et al. 2002, 2005). Finally, SETDB1 (ESET1 or KMT1E) and SETDB2 (Schultz et al. 2002; Yang et al. 2002; Falandry et al. 2010) deposit H3K9me3 mainly outside of constitutive heterochromatin, both to repress gene expression and to silence long terminal repeat (LTR)-containing retroelements, and specifically endogenous retroviruses (ERVs) in embryonic stem cells (Matsui et al. 2010; Rowe et al. 2010; Karimi et al. 2011), germ cells (Liu et al. 2014), and somatic cells (Collins et al. 2015; Koide et al. 2016; Kato et al. 2018; Adoue et al. 2019).
Recruitment of SETDB1 to genomic target sites requires a platform composed of DNA sequence-specific KRAB domain-containing zinc finger proteins (Wolf and Goff 2009; Jacobs et al. 2014; Imbeault et al. 2017) together with their associated corepressor KAP1 (TRIM28) (Matsui et al. 2010; Rowe et al. 2010). In the absence of any of these components, expression of ERVs is reactivated, with the identity of the affected ERV family being different in different tissues, likely due to the specific repertoire of transcription factors expressed in individual cell types (Kato et al. 2018). H3K9 methyltransferases other than SETDB1 also contribute to the repression of transposable elements, as indicated by the massive derepression of multiple repeat families in compound knockout fibroblasts lacking all H3K9 methyltransferases (Montavon et al. 2021).
The role of these enzymes and H3K9me3 deposition in the control of rapidly inducible and stimulus-regulated gene expression is less studied, one exception being represented by SETDB2, which is negligibly expressed in basal conditions, strongly induced by interferon (IFN) stimulation of macrophages during acute viral infection, and able to repress many antimicrobial genes, thus contributing to virus-induced susceptibility to bacterial infection (Kroetz et al. 2015; Schliehe et al. 2015).
Here, our initial objective was to investigate the role of SETDB1 in a classical model of rapid, stimulus-inducible gene expression: macrophage stimulation by lipopolysaccharide (LPS) (Glass and Natoli 2016). The results we obtained unveiled an unexpected and hitherto overlooked role of H3K9me3 in the restriction of the accessibility of a subset of CTCF sites specifically contained in compartment A, which is enriched for open chromatin (Lieberman-Aiden et al. 2009; Rao et al. 2014). Previous work showed that SINE B2 retroelements, a rodent-specific subfamily of SINEs (short interspersed nuclear elements) repeats, contain thousands of DNA binding motifs bound by CTCF (Bourque et al. 2008). CTCF (the CCCTC-binding factor) is a sequence-specific DNA binding protein that recognizes extended DNA motifs with its 11 zinc fingers and acts as a critical regulator of interphase genome folding (Splinter et al. 2006), which underlies the formation of physical contacts among distant cis-regulatory elements, thus contributing to selectivity of enhancer–promoter communication. CTCF binding to DNA extensively overlaps with cohesin localization in interphase chromatin (Parelho et al. 2008; Wendt et al. 2008). This is a consequence of CTCF blocking the loop extrusion activity of the cohesin complex (Sanborn et al. 2015; Fudenberg et al. 2016; Wutz et al. 2017), which eventually determines the boundaries of topologically associating domains (TADs) (Dixon et al. 2012; Nora et al. 2012; for reviews, see Merkenschlager and Nora 2016; Davidson and Peters 2021).
Overall, 40% of CTCF binding sites in the mouse genome derive from transposable elements (Sundaram et al. 2014). On the one hand, embedding CTCF motifs into retroelements such as SINE B2 repeats allowed the rapid expansion of the CTCF regulatory repertoire in mammals through multiple waves of retrotransposon expansion, thus critically contributing to shaping mammalian genome folding (Schmidt et al. 2012); on the other hand, it imposed the requirement for mechanisms enforcing the usage of such a large amount of DNA binding sites contained in repeats.
In this study, we found that H3K9me3 deposited by SETDB1 specifically restrained the accessibility of >1600 CTCF sites contained in SINE B2 repeats in active TADs. The increased CTCF recruitment to such sites upon SETDB1 depletion was associated both with a gain in insulation at hundreds of sites and with the formation of novel loops within TADs, which correlated with only modest changes in basal gene expression but a pervasive impairment of rapidly inducible gene expression in LPS-stimulated macrophages.
Results
SETDB1 depletion and selective H3K9me3 loss in compartment A
After terminal differentiation, mouse bone marrow-derived macrophages were infected with purified lentiviruses expressing two nontargeting (NT) shRNAs or two distinct shRNAs specific for SETDB1, causing a depletion of RNA and protein >90% (Supplemental Fig. S1A). H3K9me3 genomic distribution in control and SETDB1-depleted macrophages was analyzed by ChIP-seq (Supplemental Table S1; Supplemental Fig. S1B). Multimapping reads associated with this chromatin mark (17%–19% of total reads) were assigned to different genomic locations based on the relative abundance of uniquely mapping reads in the regions that contained the multimaps (Johnson et al. 2016). The same strategy was applied to all the ChIP-seq and ATAC-seq samples used in this study. In cells transduced with nontargeting shRNAs, we identified 86,119 H3K9me3 blocks (Fig. 1A; Supplemental Fig. S1B) with an obvious trend of comparatively higher H3K9me3 read counts in compartment B than in A (Fig. 1B). Based on length-normalized H3K9me3 read counts, SETDB1 depletion selectively affected H3K9me3 in compartment A (Fig. 1C), which is enriched for open chromatin (Lieberman-Aiden et al. 2009; Rao et al. 2014). Overall, 7543 H3K9me3 blocks (8.8% of the total) were significantly down-regulated in SETDB1-depleted cells (FDR ≤ 0.01, log2FC ≥ ±1) (Fig. 1D; Supplemental Fig. S1C,D; Supplemental Table S2). The average magnitude of H3K9me3 down-regulation in these blocks was severe (2.7-fold), indicating limited, if any, redundancy provided by other H3K9 methyltransferases. Depending on the statistical threshold used, between ∼30 Mb (FDR ≤ 0.01, log2FC ≥ ±1) and ∼80 Mb (FDR ≤ 0.05) of genome space showed reduced H3K9me3 upon SETDB1 depletion, corresponding to ∼2.6% and ∼7% of compartment A, respectively (Fig. 1E).
Figure 1.
Effect of SETDB1 depletion on H3K9me3 profiles in untreated and LPS-stimulated macrophages. (A) Snapshot showing H3K9me3 profiles in control and SETDB1-depleted untreated mouse bone marrow-derived macrophages in a 7-Mb genomic region. Compartments A (red) and B (blue) are indicated. Normalized H3K9me3 counts are reported. Data obtained with one of two distinct nontargeting (NT) or SETDB1 targeting shRNAs are shown. (B) H3K9me3 signal as a function of Hi-C compartment eigenvalue score. Shown are the H3K9me3 read counts per kilobase per compartment on the Y-axes compared with the compartment score on the X-axes in either compartment A (red) or compartment B (blue). Compartments were identified using Hi-C contact data at 40-kb resolution. (C) Mean read counts per kilobase in inactive (left) and active (right) compartments in control (NT sh; gray) and SETDB1-depleted (SETDB1 sh; orange) macrophages. Two-way ANOVA with Tukey multiple comparison test. (**) Adjusted P-value < 0.01. (D) H3K9me3 metaprofiles at genomic regions (n = 7543) showing reduced H3K9me3 in SETDB1-depleted untreated macrophages. Shown is the metaplot of the replicate average of H3K9me3 signal per base pair in control (gray) and SETDB1-depleted (orange) conditions. (E) Cumulative megabases of genomic regions showing loss of H3K9me3 in SETDB1-depleted and untreated macrophages using different thresholds (adjusted P-value and fold change computed via DESeq2) for differential peak calling. (F) Effects of 2 h of LPS stimulation on the genomic profiles of H3K9me3 in untreated macrophages transduced with a nontargeting (NT) shRNA. (G) Effects of 2 h of LPS stimulation on the 7543 H3K9me3 blocks methylated by SETDB1. (H) H3K9me3 signal intensities at the 7543 H3K9me3 blocks methylated by SETDB1 in LPS-stimulated macrophages transduced with either a nontargeting (NT, X-axis) or a SETDB1 targeting (Y-axis) shRNA.
LPS stimulation caused only minimal changes, if any, to the H3K9me3 genomic profiles, when considering both the entire complement of H3K9me3 blocks (Fig. 1F) and the 7543 blocks affected by SETDB1 depletion (Fig. 1G). Moreover, the observed reduction in H3K9me3 caused by SETDB1 depletion in untreated macrophages occurred in a substantially similar manner in LPS-stimulated conditions (Fig. 1H).
Epigenomic consequences of SETDB1 depletion in macrophages
Having determined the selective effects of SETDB1 depletion on H3K9me3 in compartment A, we set out to measure the effects of H3K9me3 loss on the macrophage epigenome. We first analyzed the impact of SETDB1 depletion on the genomic distribution of the ETS family transcription factor PU.1, which pervasively associates with the repertoire of cis-regulatory elements active or poised for activation in macrophages (Ghisletti et al. 2010; Heinz et al. 2010), where it acts to displace nucleosomes and to maintain the accessibility of the underlying DNA (Barozzi et al. 2014; Comoglio et al. 2019). We generated PU.1 ChIP-seq data in cells depleted of SETDB1 or transduced with nontargeting shRNAs. We identified >38,000 high-confidence PU.1 peaks that were largely unaffected by the depletion of SETDB1, with only 78 peaks up-regulated and one down-regulated (FDR ≤ 0.05) in SETDB1-depleted and untreated macrophages (Fig. 2A). Therefore, the overall distribution of PU.1 was unaffected by SETDB1 deficiency.
Figure 2.
Impact of SETDB1 depletion on the macrophage epigenome. (A) Genomic occupancy of PU.1 in control (NT sh; X-axis) and SETDB1-depleted (SETDB1 sh; Y-axis) untreated macrophages. PU.1 peaks were called using MACS2 with an FDR ≤10−15 and retaining only peaks with overlap >50% in the two replicates (n = 38,580). (B) Overlap between sites showing H3K9me3 signal loss in SETDB1-depleted macrophages and enhancers active in other cell types or tissues. (C) H3K27ac genomic distribution in control and SETDB1-depleted macrophages. ChIP-seq data sets were generated using cells transduced with two nontargeting (NT) or two SETDB1 targeting shRNAs. (D) Overlap of H3K27ac gain with genomic regions showing H3K9me3 reduction in SETDB1-depleted macrophages. (E) Overlap of genomic regions characterized by reduced H3K9me3 and gained H3K27ac in SETDB1-depleted macrophages with enhancers active in other lineages (left) or PU.1 peaks (right). (F) Genomic regions showing a gain in H3K27ac were divided based on the overlap with reduced H3K9me3 and analyzed for chromatin accessibility by ATAC-seq and PU.1 genomic occupancy. Shown is the metaplot of the replicate average of H3K27ac signal per base pair for the conditions reported. (G) Effects of LPS stimulation on H3K27ac at regions showing H3K27ac gain upon SETDB1 depletion. Shown are the box plots for the normalized reads per kilobase per individual replicates. Two-way ANOVA with Tukey multiple comparison test. (****) P < 0.0001. (H) Overrepresentation of transcription factor motifs at genomic sites showing increased H3K27ac in SETDB1-depleted macrophages, divided based on the overlap with H3K9me3 loss. Enrichment was calculated using all accessible sites as background sequences.
We next analyzed whether the loss of H3K9me3 caused by SETDB1 depletion could determine widespread increase in accessibility and histone acetylation of cis-regulatory elements, including lineage-inappropriate enhancers; namely, enhancers selectively active in other cell types. We first analyzed the overlap between reduced H3K9me3 and a comprehensive catalog of mouse enhancers (Yue et al. 2014; Cusanovich et al. 2018) and found that 55% of the hypomethylated blocks matched enhancers active in other cell types (Fig. 2B). Next, we used ChIP-seq to analyze the genomic distribution of histone H3 lysine 27 acetylation (H3K27ac), a marker of active enhancers and promoters, and ATAC-seq to determine changes in chromatin accessibility. The effects of SETDB1 loss on genomic H3K27ac profiles were of remarkably limited amplitude, with only 579 out of 61,755 H3K27ac peaks showing a significant gain in signal intensity (FDR ≤ 0.01, log2FC ≥ ± 1) (Fig. 2C; Supplemental Table S3). Only 29 H3K27ac peaks underwent significant, albeit minimal, reduction in intensity (Fig. 2C). Of the 579 peaks of increased H3K27ac, 308 were contained within 257 blocks of reduced H3K9me3 (Fig. 2D). Therefore, only 3.4% (257 out of 7453) of the chromatin blocks showing reduced H3K9me3 upon SETDB1 depletion gained acetylation. Remarkably, 77% of these 257 sites overlapped enhancers active in other lineages (Fig. 2E), and 62% of them were bound by PU.1 (Fig. 2E) like the remaining repertoire of active regulatory regions in these cells.
At genomic regions showing concomitant H3K9me3 reduction and H3K27ac gain, both ATAC-seq and PU.1 signals were also enhanced, indicating increased accessibility and occupancy (Fig. 2F; Supplemental Table S2).
Interestingly, although H3K9me3 genomic profiles were unaffected by LPS stimulation (Fig. 1F,G), H3K27ac signal gains observed in cells depleted of SETDB1 were reduced by LPS treatment, albeit only moderately (Fig. 2G).
Finally, a motif enrichment analysis at accessible sites showed that hyperacetylated peaks occurring within regions with reduced H3K9me3 were characterized by the overrepresentation of ETS sites that differed from canonical PU.1 sites (Fig. 2H; Supplemental Table S4; Curina et al. 2017). Therefore, increased PU.1 recruitment to these regions may be mediated by relatively low-affinity DNA binding sites rather than by the canonical motif that accounts for its pervasive recruitment to myeloid-specific cis-regulatory elements. Hyperacetylated peaks occurring outside of regions of H3K9me3 loss were characterized by the overrepresentation of AP-1 sites and E-boxes, likely indicating a distinct mechanism of activation (Fig. 2H; Supplemental Table S4).
Overall, these data indicate that the extensive loss of H3K9me3 in compartment A was not associated with a widespread increase in acetylation or accessibility of silent euchromatic regions, possibly due to the lack of expression of transcription factors acting on them. Moreover, it indicates a limited impact of H3K9me3 on the recruitment of PU.1, the critical TF required for maintenance of the macrophage epigenome.
SETDB1 controls the accessibility of CTCF sites in SINE B2 repeats
It has been reported that in cells lacking SETDB1, repeats of multiple ERV families are activated, as indicated by the increased abundance of the corresponding RNAs, while the expression of non-LTR retrotransposons such as SINEs and LINEs is not affected (Matsui et al. 2010; Rowe et al. 2010; Karimi et al. 2011, Liu et al. 2014, Collins et al. 2015; Koide et al. 2016; Kato et al. 2018; Adoue et al. 2019; Montavon et al. 2021). We analyzed the enrichment of different repeat families at genomic sites characterized by loss of H3K9me3 in cells depleted of SETDB1. Enrichment of repeat families was evaluated by performing hypergeometrical testing considering the RepeatMasker-annotated repeats contained within H3K9me3 blocks in compartment A. This analysis revealed the expected enrichment of LTR-containing retrotransposons at regions characterized by H3K9me3 loss, which was mainly accounted for by class III ERVs of the ERVL/MaLR group (Fig. 3A; Supplemental Table S5; Stocking and Kozak 2008). When restricting the enrichment analysis to repeat elements contained within sites hyperacetylated in SETDB1 KD cells (Fig. 2C), only class III ERVs were found to be overrepresented (Supplemental Table S5). Unexpectedly, the retroelements with the strongest overall enrichment in regions that showed reduced H3K9me3 upon SETDB1 depletion were SINE repeats, and in particular those belonging to the B2 subfamily (Fig. 3A; Supplemental Table S5). SINE B2 are rodent-specific non-LTR retrotransposons that account for ∼0.7% of the mouse genome and contain thousands of CTCF binding sites that are functional and bound in vivo (Bourque et al. 2008).
Figure 3.
SETDB1 deposited H3K9me3 and control of CTCF recruitment to SINE B2 repeats. (A) Enrichment of different repeat families at genomic regions showing H3K9me3 loss in SETDB1-depleted macrophages. Reported is the Bonferroni adjusted P-value and the log2 scaled ratio between expected and observed frequencies. (B) Motif overrepresentation analysis at repeats contained in genomic regions showing H3K9me3 reduction (left) or H3K9me3 loss and concurrent H3K27ac gain (right) upon SETDB1 depletion. Data are separately shown for LTR and SINE retrotransposons. Enrichment was computed using HOMER (Heinz et al. 2010), providing distinct custom sets of background sequences: sequences of all LTRs and SINEs in the mm10 genome for the 7543 H3K9me3 set and sequences of LTRs and SINEs within the 7543 H3K9me3 sites for the 257 H3K9me3 sites with concurrent hyperacetylation. (C) CTCF genomic distribution in control and SETDB1-depleted macrophages. CTCF ChIP-seq was carried out using cells transduced with either two nontargeting (NT) shRNAs or two SETDB1 targeting shRNAs. (D) CTCF binding at genomic regions showing reduced H3K9me3 in SETDB1-depleted macrophages. (E) Quantitation of the effects shown in D reporting the normalized read counts per kilobase of CTCF in the various conditions. Two-way ANOVA with Tukey multiple comparison test. (****) P < 0.0001. (F) CTCF peaks called as up-regulated (red), unaffected (gray), or down-regulated (blue) upon SETDB1 depletion. (G) Association of constitutive CTCF peaks and peaks induced upon SETDB1 depletion with different families of repeat elements. Shown is the repeat density per base pair. (H) Distance between CTCF peaks and H3K27ac peaks induced upon SETDB1 depletion. (Left) Distance between gained H3K27ac peaks (n = 579) and constitutive (gray) or induced (red) CTCF peaks. (****) P < 0.0001, Mann–Whitney test. (Right) Bootstrap resampling test assessing the mean distance between randomly selected CTCF peaks and gained H3K27ac (P < 0.0001, Wilcoxon signed rank test). Mean distance between induced CTCF peaks and up-regulated H3K27ac sites is shown with a dashed green line. (I) Cumulative distribution of the H3K9me3 signal centered on the summits of the CTCF peaks induced upon SETDB1 depletion. (J) Heat maps displaying CTCF and H3K9me3 ChIP-seq signals for constitutive and induced CTCF peaks in both control (NT sh) and SETDB1-depleted (SETDB1 sh) cells. Enlarged view for the 1644 induced CTCF peaks is shown at the right of each heat map. (K) Inverse relationship between CTCF binding intensity (±400 bp surrounding CTCF peak summit) and H3K9me3. CTCF peaks were divided into quintiles based on increasing H3K9me3 signal. Two-way ANOVA with Tukey multiple comparison test. (****) P < 0.0001. (L) CpG DNA methylation at both CTCF-induced and not induced peaks. In case multiple CTCF motifs were associated with a single peak, only methylation data at the best motif were reported.
We next analyzed TF motifs overrepresented in SINE or LTR repeats either at all regions showing H3K9me3 reduction in SETDB1-depleted cells (n = 7543) or at those regions where H3K9me3 reduction was associated with H3K27ac gain (n = 257) (Fig. 3B; Supplemental Table S4). Cognate motifs for PU.1 and C/EBPβ, another TF with a pervasive association with the myeloid enhancer repertoire (Glass and Natoli 2016), were selectively overrepresented in the LTR retrotransposons that gained acetylation. Conversely, the most overrepresented motif when considering all repeats that showed reduced H3K9me3 was the CTCF DNA binding site, which was specifically associated with SINE repeats (Fig. 3B; Supplemental Table S4).
These data suggested the possibility that loss of H3K9me3 may bring about the selective exposure of CTCF motifs contained within SINE B2 repeats. To determine whether such sites were functional, we analyzed the genomic distribution of CTCF by ChIP-seq. We detected a total of 40,893 CTCF peaks in wild-type and SETDB1-depleted macrophages, with 78% of them in compartment A, and a clear trend toward increased occupancy in SETDB1-depleted macrophages (Fig. 3C; Supplemental Table S6). When considering the CTCF peaks that overlapped regions characterized by H3K9me3 loss, they were nearly all strongly up-regulated in SETDB1-depleted cells (Fig. 3D). Overall, SETDB1 depletion induced a broad enhancement of CTCF occupancy that was of particularly high magnitude at regions showing H3K9me3 loss (Fig. 3E). CTCF signal gains observed at other regions may be associated with levels of H3K9me3 reductions in SETDB1-depleted cells that were just below the thresholds used for differential peak calling.
Differential peak calling analysis showed that 1644 CTCF peaks (4% of total) were significantly up-regulated in cells depleted of SETDB1 while only a handful of them (0.1%) were down-regulated (Fig. 3F). Approximately 94% of the gained CTCF peaks occurred within compartment A (P < 0.0001, Wilcoxon signed rank test). To determine whether gained CTCF peaks represented de novo binding events or enhancement of pre-existing events, we first analyzed the frequency distribution of the CTCF ChIP-seq signal per base pair in wild-type macrophages and found that it was comparable at gained and not-gained peaks (Supplemental Fig. S2A). SETBD1 depletion resulted in the selective increase in read density at inducible peaks (Supplemental Fig. S2A). Importantly, the majority of the inducible CTCF peaks were also identified in wild-type macrophages (Supplemental Fig. S2B). Moreover, the CTCF motifs at gained and not-gained peaks were of comparable quality (Supplemental Fig. S2C). Altogether, these observations suggest that gained CTCF peaks in SETDB1-depleted cells arise from increased binding to optimal CTCF DNA binding motifs. Compared with all CTCF peaks, the gained ones showed almost a threefold higher density of SINE B2 elements and instead the underrepresentation of other repeat elements, thus confirming the close relationship between gained CTCF binding and this family of repeats (Fig. 3G). Moreover, as compared with constitutive CTCF peaks, those gained upon SETDB1 depletion were significantly closer to sites of gained acetylation, both by directly comparing the overall distance distribution and by performing a CTCF peak resampling test (Fig. 3H).
While only ∼60% of such gained CTCF peaks overlapped with the identified blocks of reduced H3K9me3, the H3K9me3 signal under all of the 1644 peaks showed clearly reduced intensity in SETDB1-depleted cells as compared with control cells (Supplemental Fig. S2D). Therefore, the overlap of gained CTCF peaks with regions of H3K9me3 loss is in fact underestimated because of the stringent thresholds applied to call differential H3K9me3 blocks. Importantly, increased CTCF peaks occurred at regions of pointed H3K9me3 signal (Fig. 3I), a characteristic of compartment A. Moreover, consistent with the limited impact of LPS on H3K9me3 profiles (Fig. 1G,H), CTCF binding was left substantially unaffected by LPS stimulation in both the presence and absence of SETDB1 (Supplemental Fig. S2E,F).
We further explored the relationship between H3K9me3 and CTCF binding. When comparing the signal distribution of CTCF versus H3K9me3, we noticed a clear inverse relationship between their intensities (Fig. 3J). Moreover, the gained CTCF peaks showed the highest H3K9me3 signal in wild-type macrophages (Fig. 3J). When dividing CTCF peaks into quintiles based on increasing H3K9me3 signal, CTCF binding intensity was lower in the highest H3K9me3 quintile, and SETDB1 depletion specifically led to a significant increase in CTCF signal at sites in this group (Fig. 3K), indirectly suggesting a direct role of H3K9me3 in control of CTCF binding.
Control of CTCF binding to its cognate motifs was previously reported to be mediated by two main mechanisms: CpG methylation of the CTCF DNA binding motif (Wang et al. 2012) and competition with the transcription factor ADNP for the consensus CTCF motif (Ostapcuk et al. 2018; Kaaij et al. 2019). To determine the possible involvement of these two mechanisms in the CTCF binding gain caused by SETDB1 depletion, we first analyzed DNA cytosine methylation profiles using published whole-genome bisulfite sequencing data sets in mouse macrophages (Link et al. 2018). CpG methylation of motifs associated with both constitutive and gained CTCF peaks was uniformly very low (Fig. 3L; Supplemental Fig. S2G), indicating that at least in the vast majority of cases changes in DNA methylation cannot account for gained CTCF binding in SETDB1-depleted cells.
Next, to measure ADNP occupancy, we generated ChIP-seq data sets in control and SETDB1-depleted macrophages. The ∼90,000 ADNP peaks identified were overall unaffected by SETDB1 depletion (Supplemental Fig. S2H). In particular, no change in ADNP occupancy was detected at sites where SETDB1 depletion led to H3K9me3 loss as well as at sites corresponding to the 1644 gained CTCF peaks (Supplemental Fig. S2H–K). Therefore, relief from ADNP-mediated competition for CTCF sites was not involved in gained CTCF binding.
Overall, these data hint at a direct role of H3K9me3 in the control of CTCF recruitment to cognate motifs in SINE B2 repeats. This interpretation is in keeping with experiments showing loss of CTCF binding upon focused delivery of H3K9me3 to constitutively bound CTCF motifs using the dCas9–KRAB epigenome editor (Tarjan et al. 2019).
Altered chromatin organization in SETDB1-depleted macrophages
We next analyzed the impact of SETDB1 depletion and concurrent increased CTCF binding on chromatin organization. TADs are generated by cohesin-mediated loop extrusion, which is limited by boundary elements generated by CTCF recruitment to cognate sites or clusters of sites (Sanborn et al. 2015; Fudenberg et al. 2016; Nora et al. 2017). Binding of CTCF to motifs that are usually not accessible may in principle generate novel insulation boundaries, thus disrupting pre-existing TADs and altering overall chromatin organization. To address this possibility, we generated Hi-C data in wild-type and SETDB1-depleted macrophages (quality controls are reported in Supplemental Fig. S3A,B). The overall chromosome interaction landscapes were very similar in wild-type and SETDB1-depleted macrophages when comparing the relative contact probability (RCP), the genome-wide compartment score, and the genome-wide insulation score (Supplemental Fig. S3C,D). High similarity between wild-type and SETDB1-depleted macrophages was also observed when comparing TAD number and structure (Supplemental Fig. S3E,F). Only a mild increase in intra-TAD contacts was detected, consistent with a higher contact probability at shorter distances, as also shown by assessing the RCP (Supplemental Fig. S3C).
To assess the impact of SETDB1 depletion and the associated increase in CTCF binding on local chromatin architecture, we assessed whether insulation sites, defined as point of local minima in the genome-wide insulation score, displayed significant changes upon SETDB1 depletion. A general trend toward a gain in insulation (namely, a reduction in the insulation score) in SETDB1-depleted macrophages was observed across all identified insulation sites, with 368 of them being statistically significant (Fig. 4A). Sites that gained insulation were significantly closer to inducible CTCF peaks than the unaffected ones (Fig. 4B). To further evaluate how inducible CTCF peaks affected nearby insulation sites, we assessed the insulation score profiles upstream of and downstream from inducible CTCF peaks and found a consistent increase in insulation at pre-existing insulation sites in the vicinity of CTCF peaks induced upon SETDB1 depletion (Fig. 4C). These effects are likely to arise mainly indirectly given that only ∼20% of induced CTCF peaks were found at insulation sites (Fig. 4D).
Figure 4.
Effect of SETDB1 depletion on chromatin organization in macrophages. (A) Insulation scores were measured at insulation sites in control (NT sh) and SETDB1-depleted (SETDB1 sh) macrophages, and differences were calculated. Insulation sites were identified genome-wide as point of local minima in the insulation score at 10-kb resolution. (B) CTCF peaks enhanced in SETDB1-depleted macrophages are associated with increased insulation capacity at pre-existing insulation sites. (**) P < 0.01, Mann–Whitney test. (C) Heat maps showing insulation scores in control macrophages (white/blue color palette) and the log2 fold change (white/red color palette) between control and SETDB1-depleted macrophages (log2 of the ratios between minimum–maximum normalized SETDB1 sh and NT sh bins) for genomic areas upstream of and downstream from the strand-oriented CTCF motifs. (D) Proportion of constitutive (top) and induced (bottom) CTCF peaks found at insulation sites. (E) Aggregate region analysis (ARA) showing Hi-C contact probabilities (i.e., average observed over expected contacts) in control (NT sh) and SETDB1-depleted (SETDB1 sh) macrophages. Plots are centered on either constitutive (top) or inducible (bottom) CTCF peaks matching insulation sites. Observed versus expected interaction ratios are shown together with the difference between control and SETDB1-depleted conditions. (F) Aggregate plots as in E centered on CTCF peaks elsewhere within TADs. (G) Frequency distribution (top panel) of the differences in loop intensities normalized to local background (bottom panel) in SETDB1-depleted versus control macrophages. (H) Aggregate plots (left) centered on down-regulated (top), invariant (middle), and up-regulated (bottom) loops in SETDB1-depleted versus control macrophages. (I) Box plots of the distribution of the fold changes of contact intensities per loop over the surrounding local background in control and SETDB1-depleted macrophages. Two-way ANOVA with Tukey multiple comparison test. (****) P < 0.0001, (**) P < 0.01, (*) P < 0.05. (J) Distance between induced CTCF peaks and induced loops. (Left) Distance between induced CTCF and invariant (gray), down-regulated (cyan), or induced (red) loop anchors. One-way ANOVA with Tukey multiple comparison test. (****) P < 0.0001. (Right) Bootstrap randomization test assessing the mean distance between randomly placed anchors and induced CTCF peaks (P < 0.0001, Wilcoxon signed rank test). Mean distance between induced CTCF peaks and up-regulated loops is shown with a dashed green line.
To obtain an additional description of the global trends of contact probabilities in wild-type and SETDB1-depleted macrophages, we generated aggregate plots displaying Hi-C contact probabilities (Flyamer et al. 2017; van der Weide et al. 2021). Plots were centered on the summit of induced or not-induced CTCF peaks that were either at insulation sites (Fig. 4E) or elsewhere inside TADs (Fig. 4F). While only small changes could be observed at insulation sites, a clear directional increase in contact probability was associated with inducible CTCF peaks outside of insulation sites (Fig. 4E,F).
These data prompted us to analyze whether gained CTCF binding events caused by SETDB1 depletion were associated with the formation of new loops and/or the enhancement of previously existing ones. We defined a high-confidence set of loops identified in both replicates of each experimental condition (Supplemental Table S7). More than 85% of the identified loops overlapped with those previously identified by others in macrophages (Mumbach et al. 2019), confirming the high quality and reproducibility of these data sets. To assess changes in loop intensities, we analyzed the frequency distribution of the differences in mean intensity between control and SETDB1 knockdown conditions (Fig. 4G; Supplemental Table S7). We considered as differential loops those with a difference in mean intensity between control and SETDB1 knockdown macrophages greater than one standard deviation above or below the observed mean difference (Materials and Methods; Supplemental Table S7). Overall, depletion of SETDB1 increased signal intensity at 1291 loops, while causing no changes or low-magnitude reductions at all others (Fig. 4H,I; Supplemental Table S7). Importantly, induced CTCF peaks were significantly closer to the up-regulated loops than to the invariant and the down-regulated ones (Fig. 4J, left). Moreover, the distance between up-regulated loops and induced CTCF peaks was significantly smaller than that with randomly resampled CTCF peaks (Fig. 4J, right).
Taken together, these data show that SETDB1 depletion augmented the accessibility of CTCF motifs embedded in SINE B2 repeats, thus leading to increased binding of CTCF and eventually to enhanced loop formation.
Basal and LPS-inducible gene expression in SETDB1-depleted macrophages
The results shown above highlight a potential link between induced CTCF binding, induced acetylation, and increased loop formation following SETDB1 depletion. Analysis of the H3K27ac associated with loop anchors showed that loops induced upon SETDB1 depletion brought loci with low H3K27ac in proximity to loci with high H3K27ac signal (Fig. 5A). Examination of the bulk H3K27ac signal associated with these loop anchors showed a significant increase in acetylation associated with the depletion of SETDB1 (Fig. 5B). To determine the functional impact of the observed increases in loop formation and in acetylation associated with induced CTCF peaks, we generated RNA-seq data sets in control and SETDB1-depleted macrophages. SETDB1 depletion caused overall limited effects that were compatible with its repressive role; namely, increased expression of 118 genes (log2FC ≥ ± 1 and FDR ≤ 0.05) and reduced expression of only 33 genes, with the magnitude of gene expression changes being higher for up-regulated than for down-regulated genes (Fig. 5C; Supplemental Table S8). Genes that were up-regulated in SETDB1-depleted macrophages were significantly closer than the down-regulated ones to induced CTCF peaks (Fig. 5D). Furthermore, ∼76% of the up-regulated genes were found within active contact domains containing enhanced CTCF peaks, an association significantly higher than expected by chance (P < 0.0001, Wilcoxon signed rank test, expected mean proportion 20%). In addition, induced loops and up-regulated genes were closer to each other than randomly expected (Fig. 5E), suggesting a possible cause–effect relationship between increased loop formation, increased acetylation, and gene induction. Indeed, only those loops associated with up-regulated genes (78 loops linked to 48 up-regulated genes) showed a significant increase in SETDB1-depleted cells, while loops associated with down-regulated genes showed no significant change (Fig. 5F).
Figure 5.
Basal gene expression and its correlation with altered chromatin organization in SETDB1-depleted macrophages. (A) Heat map showing H3K27ac ChIP-seq signal in control macrophages (NT sh) at anchors of induced loops centered on the strongest H3K27ac peak. Each anchor pair was oriented in order to display the highest H3K27ac signal in the heat map at the right. The two heat maps were ranked based on the mean signal in anchors 1 and 2. (B) H3K27ac at induced loops is enhanced upon deletion of SETDB1. Wilcoxon matched-pairs signed rank test. (****) P < 0.0001. (C) Differentially expressed genes in untreated control and SETDB1-depleted macrophages. Data are from n = 2 independent biological replicates, each one done in duplicate with both shRNAs (total n = 4 samples per group). (D) Distance between up-regulated and down-regulated genes in SETDB1-depleted macrophages and induced CTCF peaks. (****) P < 0.0001, Mann–Whitney test. (E) Bootstrap randomization test assessing the mean distance between randomly placed anchors and up-regulated genes. The mean distance between induced loops and up-regulated genes is shown with a dashed green line. (F) Aggregate plots showing intensities of loops associated with genes up-regulated or down-regulated in SETDB1-depleted macrophages. (Right) Box plots showing the distribution of the fold changes of contact intensities per loop over the surrounding local background in control and SETDB1-depleted macrophages. Wilcoxon matched-pairs signed rank test. (****) P < 0.0001. (G) Genomic snapshot showing a genomic interval containing the Tmem154 gene. Data shown from top to bottom include H3K27ac, RNA-seq, contact density plot generated using Tmem154 as viewpoint (yellow highlight and red dashed square), loops identified via cLoop divided into up-regulated (red) and down-regulated (cyan), identified inducible CTCF peak upstream of Tmem154 (green), and normalized contact maps in control and SETDB1-depleted macrophages. (H) Zoomed-in view of the region surrounding Tmem154 showing H3K9me3 and CTCF peaks regulated by SETDB1 depletion. The M1-type and M2-type CTCF motifs contained in the SINE B2 elements are shown.
While the mechanisms accounting for the transcriptional effects of SETDB1 depletion are likely complex, the possible direct links between altered chromatin organization and changes in gene expression were exemplified by Tmem154, one of the two most up-regulated genes in unstimulated SETDB1-depleted macrophages. Increased CTCF binding upstream of the Tmem154 gene promoter was associated with increased contacts in its close proximity as well as in the surrounding regions, which may represent indirect consequences of the previous ones (Fig. 5G). In particular, increased contacts were generated between Tmem154 and a downstream region overlapping the Fbxw7 gene, which contained an extended set of highly acetylated putative regulatory regions (Fig. 5G). The two newly generated CTCF peaks observed in SETDB1-depleted cells were associated with two distinct SINE B2 elements, both containing canonical CTCF motifs (Fig. 5H). To assess the cause–effect relationship between CTCF induction and Tmem154 up-regulation in SETDB1-depleted cells, we used CRISPR/Cas9 to delete the CTCF binding sites associated with the inducible CTCF peak upstream of Tmem154 (Supplemental Fig. S4A). A total of 15 sgRNA pairs was screened, leading on average to ∼20% deletion efficiency in primary wild-type or SETDB1-depleted macrophages (Supplemental Fig. S4B,C). Consistent with a direct involvement of the identified inducible CTCF peak upstream of the Tmem154 promoter, the increase in Tmem154 expression induced by SETDB1 depletion was reduced upon deletion of the CTCF binding sites (Supplemental Fig. S4D).
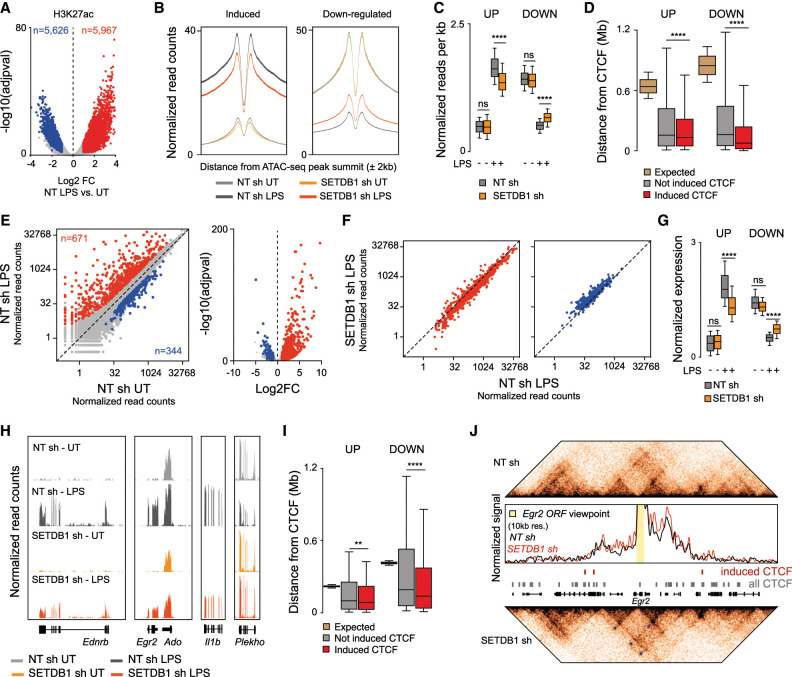
To assess whether the changes in chromatin architecture detected in resting conditions affected stimulus-induced macrophage activation, we evaluated both H3K27ac and transcriptional changes following LPS stimulation in control and SETDB1-depleted cells. LPS stimulation increased H3K27ac at 5967 genomic sites and reduced it at 5626 (Fig. 6A; Supplemental Table S3). SETDB1 depletion pervasively attenuated both LPS-induced and down-regulated H3K27ac peaks, irrespective of the magnitude of the changes induced by stimulation (Fig. 6B,C; Supplemental Fig. S6A,B). Both up-regulated and down-regulated H3K27ac peaks were significantly closer to the induced CTCF peaks than to the not-induced ones (Fig. 6D). To evaluate the consequences on LPS-driven gene expression changes (Fig. 6E), we generated RNA-seq data sets in LPS-treated control and SETDB1-depleted macrophages. All genes regulated by LPS, including the up-regulated and the down-regulated ones, displayed attenuated expression changes in SETDB1-depleted macrophages (Fig. 6F,G). The observed changes were of overall low magnitude (with a median difference of ∼1.6-fold for LPS-induced genes and 1.4-fold for LPS down-regulated genes) but high amplitude. Representative snapshots are shown in Figure 6H. We next sought evidence indirectly supporting a possible role of gained CTCF binding in the observed transcriptional changes. To this aim, we measured the distance of LPS-induced and down-regulated genes from the different groups of CTCF peaks and found that both of them were significantly closer to the CTCF peaks induced upon SETDB1 depletion than to the invariant ones (Fig. 6I). In addition, between 72% and 75% of LPS-regulated genes were found within active contact domains containing enhanced CTCF peaks. Such association was significantly higher than expected by chance (P < 0.0001, Wilcoxon signed rank test, expected mean proportion ∼21.5%). Importantly, global perturbation in LPS-regulated gene expression was not associated with detectable changes in the activation of the main signaling pathways triggered by stimulation (Supplemental Fig. S5).
Figure 6.
Effects of SETDB1 depletion on LPS-regulated gene expression and its correlation with chromatin folding. (A) H3K27ac peaks significantly changed in LPS-stimulated macrophages (2 h). Up-regulated peaks are shown in red, and down-regulated peaks are shown in blue. Data are from n = 2 biological replicates. (B,C) Effects of SETDB1 depletion on LPS-regulated H3K27ac are shown as metaplots (B) or box plots of normalized and centered read counts per kilobase across all conditions (C). Two-way ANOVA with Tukey multiple comparison test. (****) P < 0.0001. (D) Distance between LPS-induced H3K27ac peaks and CTCF peaks induced upon SETDB1 depletion (expected distances were computed through 10,000 bootstrap resamplings of all CTCF peaks). One-way ANOVA with Tukey multiple comparison test. (****) P < 0.0001. (E) Gene expression changes induced by LPS stimulation (2 h) of mouse macrophages. (F) Effect of SETDB1 depletion on both LPS-induced (red) and down-regulated (blue) genes. (G) Quantification of the effects shown in F reporting the normalized and centered read counts per kilobase across all conditions. Two-way ANOVA with Tukey multiple comparison test. (****) P < 0.0001. (H) Representative snapshots of LPS-inducible and SETDB1-regulated genes. (I) Distance between LPS-regulated genes and CTCF peaks induced in SETDB1-depleted macrophages (expected distances were computed through bootstrap resampling of all CTCF peaks). One-way ANOVA with Tukey multiple comparison test. (****) P < 0.0001. (J) Genomic snapshot showing the LPS-inducible Egr2 gene. The heat maps display normalized contact densities in control (top) and SETDB1-depleted (bottom) macrophages. The contact density plot generated using Egr2 as the viewpoint is shown together with the position of induced (red) and constitutive (gray) CTCF peaks.
As a representative example, Figure 6J shows the genomic region containing the LPS-inducible gene Egr2, which encodes an early inducible transcription factor whose expression is attenuated in SETDB1-depleted cells (Supplemental Table S8). This region contains three CTCF peaks induced upon SETDB1 depletion that were associated with an overall increase in contact density and the enhancement of several loops (Fig. 6J, arrowheads).
Discussion
In this study, we report a critical role of the H3K9 methyltransferase SETDB1 in restricting the accessibility of a large repertoire of CTCF DNA binding motifs embedded in SINE B2 retrotransposons. Current models posit that the presence of CTCF motifs in repeats allowed species-specific waves of expansion of the repertoire of genomic CTCF DNA binding sites (Bourque et al. 2008; Schmidt et al. 2012; Sundaram et al. 2014). Our results integrate these findings with evidence demonstrating that the usage of a fraction of CTCF DNA binding sites contained in SINE B2 repeats is restrained by the deposition of H3K9me3 by SETDB1.
Among the six H3K9 methyltransferases present in mammals, SETDB1 has a specific and nonredundant role in the deposition of H3K9me3 in compartment A, where it is tethered by hundreds of sequence-specific KRAB domain-containing zinc finger transcription factors (Wolf and Goff 2009; Jacobs et al. 2014; Imbeault et al. 2017) mainly to repress endogenous retroviruses (Matsui et al. 2010; Rowe et al. 2010, Karimi et al. 2011; Collins et al. 2015; Koide et al. 2016). We hypothesize that the main reason explaining the insofar overlooked role of SETDB1 in the control of the accessibility of SINE repeats is that previous analyses were mainly based on the use of RNA-seq data to identify overexpressed repeats, while our starting point in this study was the characterization of repeats overrepresented at euchromatic sites of H3K9me3 hypomethylation in SETDB1-depleted macrophages. Consistent with this interpretation, data reported in a recent study (Montavon et al. 2021) showed that several repeat types, including SINEs, displayed increased accessibility in cells lacking various H3K9 methyltransferases without necessarily being derepressed at the transcriptional level. Therefore, the scope of SETDB1-mediated repression of repeat elements is likely broader than reported before. An important issue to address in the future will be the identification of the mechanism targeting SETDB1 to SINE B2 repeats. It is reasonable to hypothesize the involvement of specific KRAB domain zinc finger proteins recognizing shared DNA sequence features of SINE B2 elements.
The presence of CTCF DNA binding sites in SINE B2 repeats may in principle generate a large excess of motifs that, if pervasively bound, may greatly impact overall interphase genome folding and usage, as indicated by the analyses of 3D genome organization and gene expression reported here. For most transcription factors, usage of the repertoire of cognate DNA binding motifs in the genome, which include bona fide functional sites on the one hand and randomly occurring nonfunctional motifs on the other, is controlled by combinatorial binding with other transcription factors recognizing adjacent motifs in the same cis-regulatory element (Barozzi et al. 2014). However, the DNA binding motif bound by CTCF, a protein with 11 zinc fingers, is long and complex, with an information content that is unusually high for eukaryotic transcription factors (Wunderlich and Mirny 2009), which makes the contribution of other transcription factors likely dispensable for efficient CTCF binding. Instead, control of the access of CTCF to its cognate DNA binding sites appears to be regulated by two main types of chromatin modifications. First, CpG methylation prevents CTCF binding to DNA (Wang et al. 2012), as exemplified by methylation-controlled CTCF recruitment to the imprinted H19/Igf2 locus (Hark et al. 2000). Second, data shown here indicate that H3K9me3 deposition by SETDB1 prevents the usage of a vast repertoire of CTCF motifs embedded in SINE B2 elements. In line with our data, deletion of SETDB1 in neurons was shown to increase CTCF binding to a 1.2-Mb TAD including >70 protocadherin genes, eventually determining its disruption (Jiang et al. 2017). An additional mechanism controlling CTCF recruitment to motifs contained in SINE B2 elements is the competition for the same motif with the homeobox protein ADNP (Ostapcuk et al. 2018; Kaaij et al. 2019). However, the genomic distribution of ADNP was largely unaffected by SETDB1 depletion, which in principle rules out its involvement.
A well-known conundrum is the discrepancy between the large effects of CTCF or cohesin subunit loss on interphase genome folding on the one hand and the overall low-magnitude effects on gene regulation on the other (Nora et al. 2017). However, the overall impact of CTCF or cohesin loss on transcription becomes more obvious in models of highly regulated or rapidly induced gene expression. Indeed, the results reported here are consistent with the previously reported role of cohesin in the widespread control of stimulus-inducible gene expression in macrophages (Cuartero et al. 2018). Overall, these results highlight the notion that control of genome folding by CTCF and cohesin may be particularly relevant in conditions of dynamic gene regulation that probably require optimal genome folding to enable fast and/or highly regulated changes in transcriptional activity. Conversely, our data do not support the notion that the stronger effects on gene expression measured in LPS-stimulated as compared with unstimulated macrophages were accounted for by the impact of LPS stimulation on CTCF genomic recruitment. Indeed, LPS stimulation did not impact either H3K9me3 distribution or CTCF binding to chromatin, and the consequences of SETDB1 depletion on H3K9me3 or CTCF genomic profiles were virtually identical in untreated and LPS-stimulated cells.
In conclusion, deposition of H3K9me3 by SETDB1 prevents the inappropriate recruitment of CTCF to sites contained within SINE B2 repeats, and by doing so it maintains a genome organization that optimizes rapidly inducible gene expression changes. It is clear, however, that additional effects of SETDB1 depletion that are not related to 3D chromatin organization may directly or indirectly contribute to the observed alterations of LPS-regulated gene transcription.
Materials and methods
Cell culture and reagents
Bone marrow isolation from male mice (age 6–8 wk, C57/BL6 strain from Charles River or Rosa26Cas9 knock-in mice) was performed in accordance with Italian law (D.lgs. 26/2014), which enforces Directive 2010/63/EU (Directive 2010/63/EU of the European Parliament and of the Council of September 22, 2010, on the protection of animals used for scientific purposes). All animal procedures were approved by the Italian Ministry of Health (346/2017-PR). Bone marrow-derived macrophage (BMDM) cultures were generated as described (Curina et al. 2017). Cells were treated with lipopolysaccharide from E. coli, serotype EH100(Ra) from Alexis (ALX-581-010-L002) at 10 ng/mL. HEK 293 cells (from ATCC) were cultured in DMEM with 10% South American serum, 1% pen/strep (Sigma P4333), and 1% L-Glutamax (Gibco 35050061). Cell lines were authenticated by the Tissue Culture Facility of the European Institute of Oncology using the GenePrint10 system (Promega) and routinely screened for mycoplasma contamination.
Lentiviral shRNA delivery in BMDMs
Infection conditions were optimized either in 48-well, six-well, or 10-cm plates for growth conditions, plate types, and viral dose. PLKO.1 lentiviral shRNA vectors targeting Setdb1 protein were purchased from Merck (TRCN0000092975 and TRCN0000305600). PLKO.1 nontargeting shRNA1 (NT1: MISSION pLKO.1-puro luciferase shRNA control) and shRNA2 (NT2: MISSION pLKO.1-puro nonmammalian shRNA control) were also purchased from Merck. Lentiviral infection was performed using the Addgene plasmid 10878 protocol version 1.0, December 2006. Briefly, 293T cells were plated at 2.5 × 106 cells per 10-cm plate the day before transfection. Transfer vector PLKO.1-shSetdb1 or PLKO.1-shNT, packaging plasmid psPAX2, and envelope plasmid pMD2.G were cotransfected by using either Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) according to the manufacturer's protocol or calcium phosphate. Transfection medium was replaced by fresh growth medium 6–8 h after transfection. The virus-containing media were collected at 36 and 48 h after transfection. Viruses were purified and concentrated by Lenti-X concentrator (Takara 631231). BMDM cells were cultured in BMDM medium for 5 d and subjected to two rounds of infection following puromycin (4 μg/mL) selection for 72 h. Setdb1 depletion was tested either by RT-qPCR or Western blot.
RT-qPCR
For RT-qPCR experiments, 250 or 500 ng of total RNA was reverse-transcribed with ImProm-II reverse transcription system (Promega A3800) following the manufacturer's instructions. RT-qPCR was assembled with Fast SYBR Green master mix (Applied Biosystems 4385614) and run on a QuantStudio 5 or a ViiA 7 real-time PCR machine (Applied Biosystems). Analysis was done on the Thermo Fisher Cloud platform, and primers were either obtained from the UPL (Universal Primer List, Roche) or designed with Primer3.
Western blots
Total cellular lysates were obtained with lysis buffer (50 mM Tris-HCl at pH 7.5, 300 mM NaCl, 1% NP-40, 1 mM EDTA) with fresh addition of a cocktail of protease inhibitors (c-Complete EDTA-free 3x20 tablets, Roche) and 1 mM PMSF, sonicated, and clarified by centrifugation. Protein extracts were resolved on SDS–polyacrylamide gel, blotted onto nitrocellulose membranes, and probed with the antibodies listed in Supplemental Table S9.
ChIP-seq
ChIP-seq was carried out as previously described (Balestrieri et al. 2018). Briefly, 106 lentiviral-infected BMDM cells (H3K27ac, H3K9me3, and PU.1) or 20 × 106 lentiviral-infected BMDM cells (CTCF) were fixed with 1% formaldehyde (Merck 252549) for 10 min at room temperature and lysed to prepare nuclear extracts. For the ADNP ChIP-seq, 40 × 106 BMDM cells (2% HCOH fixation) were used. After chromatin shearing by sonication with a Branson Ultrasonics Sonifier, lysates were incubated overnight at 4°C with protein G Dynabeads (Invitrogen 10009D) previously coupled with 1 μg of antihistone antibody or 10 μg of anti-TF antibody (all the antibodies are listed in Supplemental Table S9). After immunoprecipitation, beads were recovered using a magnetic stand and washed with RIPA buffer (50 mM HEPES-KOH at pH 7.6, 500 mM LiCl, 1 mM EDTA, 1% NP-40, 0.7% Na-deoxycholate). Immunoprecipitated chromatin was eluted and cross-link-reverted overnight at 65°C. DNA was purified with QIAquick PCR purification kit (Qiagen) and then quantified with QuantiFluor (Promega). DNA libraries were prepared for NextSeq500 sequencing as described (Austenaa et al. 2021). The purified DNA libraries were quantified with the Quantifluor reagent (Promega E2670) and the quality of the size was controlled with the Tapestation instrument (Agilent) using the high-sensitivity assay HD5000 (Agilent 5067-5592). Library DNA were diluted to a working concentration of 4 nM and 76-bp single reads sequenced on an Illumina NextSeq500 platform. Antibodies are listed in Supplemental Table S9.
ATAC-seq
Lentiviral-infected BMDM cells (5 × 104) were pelleted by centrifugation and resuspended in 50 µL of lysis buffer (10 mM Tris-HCl at pH 7.4, 10 mM MgCl2, 0.1% Igepal CA-630), gently pipetting up and down 10 times. Nuclei were pelleted by centrifugation at 500g for 10 min at 4°C and resuspended in 25 µL of reaction buffer containing 1 µL of Tn5 transposase made in-house using a described protocol (Picelli et al. 2014a), 5 µL of 5× transposase buffer (50 mM Tris-HCl at pH 8.4, 25 mM MgCl2), and 19 µL of milliQ water. The tagmentation mix was incubated for 60 min at 37°C gently shaking in order to avoid precipitation of nuclei. The reaction was stopped by the addition of 5 µL of cleanup buffer (900 mM NaCl, 30 mM EDTA), 2 µL of 5% SDS, and 2 µL of 20 µg/µL Proteinase K (Merck P2308) and incubation for 30 min at 40°C. Tagmented DNA was isolated using 2× Agencourt Ampure Xp beads (Beckman A63881) and amplified by PCR with the HotStart Kapa HiFi enzyme (Roche 07958889001). Fragments <600 bp were isolated by negative size selection (using 0.65× Agencourt Ampure Xp beads) and then purified with 1.8× Agencourt Ampure Xp beads. Libraries were single-end-sequenced (76 bp) on an Illumina NextSeq500 platform.
RNA-seq
Total RNA was extracted from 105 cells using the Zymo Quick-RNA kit (Zymo Research R1055). RNA-seq was carried out using the SMART-seq2 protocol (Picelli et al. 2014b) with minor modifications. Briefly, 5 ng of total RNA was reverse-transcribed with template switching using oligo(dT) primers and an LNA-containing template-switching oligo (TSO). The resulting cDNA was preamplified, purified, and tagmented with Tn5 transposase produced in-house using a described protocol (Picelli et al. 2014a). cDNA fragments generated after tagmentation were gap-repaired, enriched by PCR, and purified to create the final cDNA library for Illumina NextSeq500 sequencing.
Hi-C
BMDM cells (5 × 106) were detached from a 10-cm plate using Versene (0.02% EDTA; Lonza BE17-711E), washed twice with ice-cold PBS, and fixed with 1% of methanol-free formaldehyde (Thermo Fisher 28906) for 15 min at room temperature. For efficient quenching of the formaldehyde, 0.125 M glycine was added to the single-cell suspension for 5 min at room temperature. After two washes with PBS, cells were lysed in 500 μL of freshly prepared ice-cold lysis buffer supplemented with protease inhibitors (10 mM Tris-HCl at pH 8.0, 10 mM NaCl, 0.2% NP40, 1× Protease inhibitors [Roche 11873580001]) and incubated for 20 min on ice. The integrity of the nuclei was checked under the microscope.
After centrifugation, nuclei were washed once with 1× NEBuffer2 (NEB B7002S) and the pellet was resuspended with 200 μL of 1× NEBuffer2 containing 10 μL of 10% SDS (final concentration 0.5%) and incubated for 60 min at 37°C, shaking at 900 rpm. The nucleus membrane permeabilization was then quenched by adding 400 μL of 1× NEBuffer2 and then 120 μL of 10% Triton X-100. Nuclei were washed once and then gently resuspended in 300 μL of 1× NEBuffer2. To digest the chromatin, 400 U of MboI (NEB BA0147M) was added and the incubation was left at 700 rpm overnight at 37°C. A second round of digestion was performed, adding 200 U of MboI for 120 min. MboI inactivation was performed for 15 min at 65°C.
To biotinylate the free chromatin ends, a mix (1.5 μL of 10 mM dCTP, 1.5 μL of 10 mM dGTP, 1.5 μL of 10 mM dTTP, 37.5 μL of 0.4 mM Biotin-11-dATP, 1 μL of 50 U/μL DNA polymerase I large [Klenow] fragment [NEB M0210M]) was added and incubated at 750 rpm for 45 min at 37°C. After enzyme inactivation at 65°C and centrifugation at 3000 rpm, free chromatin ends were ligated in a total volume of 1200 μL by adding T4 DNA ligase buffer (1× final; NEB B0202S), 10% Triton (0.8% final), 120 μg of BSA, and 2000 U of ligase (NEB M0202S) overnight at 16°C. Following the incubation, nuclei were pelleted at 3000 rpm for 5 min and resuspend in 200 μL of 1× NEBuffer2. RNA and proteins were digested upon addition of first 5 μL of RNase A for 15 min at 37°C and then 400 μg of Proteinase K for 30 min at 55°C. The chromatin was then reverse-cross-linked overnight at 65°C. The next day, DNA was purified with phenol/chloroform/isoamyl alcohol and precipitated by adding 3 M NaAc (pH 5.2) and 100% EtOH. DNA was sheared on the Diagenode Bioruptor Pico and size was controlled on a Tapestation (Agilent). DNA fragments were in the range of 200–300 bp. Ligated and biotinylated fragments were enriched through biotin pull-down using magnetic streptavidin T1 beads (Thermo Fisher 65601). End repair, A-tailing, and ligation of the adapters were performed on beads, and streptavidin beads containing the biotinylated DNA fragments were used as a template for the PCR amplification step, following the NEB Next Ultra protocol (NEB E7370S). The Hi-C library was then purified with 1.8× Agencourt Ampure Xp beads and quality was controlled on a Tapestation. The Hi-C library was sequenced on an Illumina NovaSeq platform for 100 cycles with two 51-bp paired-end reads.
CRISPR/Cas9-mediated genomic deletion
BMDM cells obtained from Rosa26-Cas9 knock-in mice (Platt et al. 2014) were coinfected with two lentiGuide-Puro (Addgene plasmid 52963) plasmids, each one expressing one sgRNA of 15 pairs targeting the CTCF motif upstream of the Tmem154 locus (chromosome 3: 84,657,704–84,659,779) or two irrelevant target guides and the PLKO.1 lentiviral shRNA vectors targeting Setdb1 or PLKO.1 nontargeting. sgRNAs were designed using the CRISPR design tool CHOPCHOP (https://chopchop.cbu.uib.no) and cloned into the lentiGuide-Puro plasmid according to the manufacturer's instruction. After infection and 72 h of puromycin selection (4 μg/mL), RNA and genomic DNA were collected. Genomic DNA was extracted using the NucleoSpin tissue kit (Macherey-Nagel) following the manufacturer's instructions. To quantitatively assess the efficiency of cutting, a qPCR reaction was performed as described above. Primers amplifying an unaffected region of DNA were used for normalization. Tmem154 expression was assessed by RT-qPCR. Guides and primers are listed in Supplemental Table S9.
ChIP-seq data analysis
Single-end reads were mapped to the mouse mm10 genome (Illumina's iGenomes reference annotation downloaded from UCSC, http://support.illumina.com/sequencing/sequencing_software/igenome.html) by exploiting the mapping steps of ShortStack (Axtell 2013; Johnson et al. 2016). Briefly, ShortStack exploits Bowtie to identify all best-matched alignments for each read. It computes a probability for each alignment to different loci in the genome. To do so, the frequency of uniquely aligned reads mapping within the vicinity of multimapping reads is used to redistribute multimapping reads accordingly. While this methodology has little impact for some types of ChIP-seq samples such as H2K27ac and PU.1 ChIP-seq, ∼15% more reads were retrieved for H3K9me3 ChIP-seq and ∼8% more were retrieved for CTCF ChIP-seq samples. Mapped reads were filtered in order to remove randomly placed multimappers and unmapped reads using SAMtools standard procedure (version 1.9) (Li et al. 2009). In order to identify high-confidence genome-wide sites of enrichment per antibody, we implemented three strategies accordingly to the type of signal distribution evaluated after visual inspection of Bigwig tracks on IGV (Robinson et al. 2011):
First, for narrow TF-like peaks such as PU.1, CTCF, and ADNP, we implemented MACS2 version 2.2.6 (Zhang et al. 2008) with parameters “‐‐keep-dup ‘all’ ‐‐call-summits ‐‐nomodel ‐‐nolambda ‐‐extsize 300.” Each ChIP was compared with the input DNA from bone marrow-derived macrophages. A reference set of peaks per antibody was generated by selecting significant peaks in each samples/replicate (q-value ≤ 10−5 for CTCF and ADNP; q-value ≤ 10−15 for PU.1) and being consistent between replicates (overlapping area of at least 50% between replicates).
Second, for H2K27ac, we used peaks identified in ATAC-seq samples (see below), expanding the ATAC peaks by 1.5 kb on both sides.
Third, for H3K9me3, we exploited SICER v1.1 (Zang et al. 2009) with parameters “mm10 1 250 300 0.80 1000 .01” against the input DNA from macrophages. To increase the signal to noise, given the lower and more dispersed signal of H3K9me3 in euchromatic active compartments, we combined the samples by conditions and selected significant blocks with an FDR ≤0.01.
Read counts per sample at the identified loci were retrieved using the R/Bioconductor packages GenomicRanges and GenomicAlignment (Lawrence et al. 2013). Sample normalization was achieved by selecting invariant peaks across samples (Gualdrini et al. 2016). Briefly, we modeled the frequency distribution of the differences in read counts across samples with a log-normal distribution. Peaks laying within 1σ of the best-fitted mean difference were considered as invariant and used to normalize the samples. Differentially regulated peaks were selected using DESeq2 (R/Bioconductor package version 1.26.0; R version 3.6.2) (Love et al. 2014) after turning off the default normalization that DESeq2 applies.
Repeat enrichment analysis was carried out by assessing the overlap between groups of peaks and the mm10 RepeatMasker track (RepeatMasker v. 4.0.7) downloaded from the UCSC table browser. The hypergeometrical test was used to compute the probability of systematically retrieved sets of repeats under H3K9me3 down-regulated peaks considering the observed frequency in all H3K9me3 found within active compartments. Bonferroni correction for multiple testing was applied.
Motif enrichment analysis at either peaks or associated repeats were conducted using HOMER (Heinz et al. 2010) with default parameters. Dedicated backgrounds were used per comparison. In particular, motif enrichments at SINE and LTRs contained in H3K9me3 regions were calculated using as background sequences either all SINE repeats or all LTRs, respectively.
The RegionR Bioconductor library was used to perform iterative resampling and randomization of genomic features in order to estimate the expected mean distance between the elements under analysis (see individual figures) (Gel et al. 2016).
ATAC-seq data analysis
Following adapter trimming, single-end reads were trimmed with Trimmomatic (version 0.39 flags: EADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:38) (Bolger et al. 2014). Resulting reads were mapped with ShortStack (Johnson et al. 2016). Bam files were then filtered using SAMtools (version 1.9) in order to remove unmapped reads, failing quality, or mapping to mitochondrial chromosomes. Reads were then shifted 4 nt on the positive strand and −5 nt on the negative strand using basic awk operations (Buenrostro et al. 2013). Peak calling was performed using MACS2 (version 2.2.6; options ‐‐nomodel ‐‐extsize 146 ‐‐nolambda ‐‐keep-dup “all” ‐‐call-summits). A reference set of peaks was created by selecting peaks called in each sample/replicate with q-value ≤ 0.01 and being consistent between replicates (overlapping area of at least 50% between replicates). The resulting set of peaks was used to count reads in each sample using the R/Bioconductor packages GenomicRanges and GenomicAlignment (Lawrence et al. 2013). Sample normalization was achieved by selecting invariant ATAC peaks across samples as described in “ChIP-seq Data Analysis” (see above). Differentially regulated peaks were selected using DESeq2 (R/Bioconductor package version 1.26.0; R version 3.6.2) after turning off the default normalization that DESeq2 applies.
RNA-seq data analysis
Single-end reads were mapped after adapter trimming to the mouse genome assembly mm10 (Illumina's iGenomes reference annotation downloaded from UCSC, http://support.illumina.com/sequencing/sequencing_software/igenome.html) and the RefSeq transcript annotation (ncbiRefSeqCurated November 16, 2017) using topHat2 (TopHat v2.1.1) (Trapnell et al. 2012) with parameters ‐‐max-multihits 1 ‐‐b2-very-sensitive. Reads mapping to the ENCODE blacklist regions (https://github.com/Boyle-Lab/Blacklist) (Amemiya et al. 2019) were removed using standard Bedtools operations (Bedtools v2.29.2). Per-gene read counts were retrieved using standard R/Bioconductor packages (e.g., GenomicRanges and GenomicAlignment together with the proper GFF RefSeq annotation ncbiRefSeqCurated, November 16, 2017). Sample normalization was achieved by selecting invariant genes across samples/conditions (Gualdrini et al. 2016) similarly to the strategy applied to ChIP-seq and ATAC-seq. Differentially regulated genes were selected using DESeq2 (R/Bioconductor package version 1.26.0; R version 3.6.2) after turning off the default normalization that DESeq2 applies.
Hi-C data analysis
In situ Hi-C paired-end reads were analyzed using Hi-C-Pro (Servant et al. 2015) version 3.0.0 with parameters “‐‐very-sensitive -L 25 ‐‐score-min L,-0.6,-0.2 ‐‐end-to-end ‐‐reorder” for both local and global alignment. Unique and valid pairs were kept and contact maps were generated at 10- and 40-kb resolutions. Individual contact matrices were normalized using the iterative correction normalization (ICE) method provided by Hi-C-Pro and previously presented by Imakaev et al. (2012).
Relative contact probability (RCP), aggregate region analysis (ARA), aggregate peak analysis (APA), and aggregate TAD analysis (ATA) were performed using the R library GENOVA (van der Weide et al. 2021) after importing individual iced normalized contact matrices at different resolutions. As specified in each figure legend, either individual samples or the average contact matrices between replicates were used and displayed.
Compartment score was computed exploiting the “compartment_score” function part of the GENOVA library at 40-kb resolution. The score was determined by computing observed/expected matrices per chromosomes followed by eigen decomposition. Finally, the first eigenvector of the matrix was multiplied by the square root of the corresponding eigenvalue (Lieberman-Aiden et al. 2009). Compartment scores were correlated to ATAC-seq signal to assign positive compartment score to accessible active chromatin regions. Contiguous bins belonging to the same compartment are referred to as “contact domains.” These elements were considered when assessing the co-occurrence of inducible CTCF peaks and differentially expressed genes.
Insulation scores were computed exploiting the “insulation_score” function part of the GENOVA library. Briefly, average signal intensity per Hi-C bin was computed for a sliding window along the diagonal. The calculated score was scaled to the genome-wide average signal leading to the insulation score per Hi-C bin. Insulation profiles around viewpoints were produced using insulation scores computed at 10-kb resolution with a sliding square window of 25 bins. Local minima inflections in the insulation scores were identified using a custom R function that identifies local minima with a sliding window of 10 bins. The identified local minima were further filtered considering bins with an insulation score greater than −2. Changes in insulation score at the identified potential insulation sites was evaluated by linear modeling fitting using the R Bioconductor library limma. Insulation log2 fold change heat maps comparing SETDB1-depleted and control cells was carried out by first jointly scaling the two data sets (joint minimum–maximum normalization leading to insulation scores ranging between 0 and 1) and then computing the ratios per bin.
Hi-C loops were identified per sample and replica using cLoops (Cao et al. 2020) using default preset for Hi-C loop calling (option -m 3). We then determined a high-confidence set of loops by selecting per sample/replica significant loops with mild FDR cutoff (≤0.1). Anchors defined by the selected loops were reframed by combining anchors within 5 kb. We then selected those loops that appeared to be reproducible across replicates for the control and SETDB1 KD condition. The union of the selected loops per condition was considered high confidence. Indeed ∼85% of the identified loops overlapped with loops previously identified in macrophage in situ Hi-C samples (GSE115524) (Mumbach et al. 2019). To assess changes in loop intensities, we assessed the frequency distribution of the differences in mean intensity (expressed as the log2FC of the loop signal compared with the local background surrounding the loop spots) between control and SETDB1 knockdown conditions. Differentially controlled loops were then defined as those with a difference in mean intensity between control and SETDB1 knockdown conditions greater than one standard deviation above or below the observed mean difference.
TADs were identified using CaTCH (Lee et al. 2013). Briefly reciprocal insulation cutoff was selected in order to maximize CTCF peak enrichment at TAD boundaries. TAD calling was performed on each individual replicate. TADs showing at least 90% reciprocal overlap between replicates were considered for further analysis.
Bisulfate sequencing analysis
Bone marrow-derived macrophage (C57/BL6) bisulfate sequencing data were retrieved from GEO (GSM2974655) and analyzed using Bismark (Krueger and Andrews 2011). Bismark (Bowtie2)-aligned BAM files were deduplicated using the function “deduplicate_bismark.” A full cytosine methylation report was extracted using the “bismark_methylation_extractor” function with options “‐‐cytosine_report ‐‐comprehensive ‐‐merge_non_CpG” and used for downstream analysis.
Data and software availability
Raw and processed sequencing data were deposited in the Gene Expression Omnibus (GEO) repository under accession number GSE189975.
Supplementary Material
Acknowledgments
We thank Luca Giorgetti (Friedrich Miescher Institute, Basel) and Iros Barozzi (University of Wien) for critical comments on the manuscript. This work was supported by the European Research Council (Advanced ERC grant no. 692789 to G.N.) and Marie Sklodowska-Curie Actions (MSCA-IF; “MetChromTx” ID 789792 to F.G.). F.P. is a PhD student in the European School of Molecular Medicine (SEMM).
Author Contributions: F.G., S.P., and G.N. conceived the study. S.P., M.S., E.P., and F.P. generated the data. F.G. analyzed the data. G.N., F.G., and S.P. wrote the manuscript. G.N. acquired the funding.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349282.121.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Adoue V, Binet B, Malbec A, Fourquet J, Romagnoli P, van Meerwijk JPM, Amigorena S, Joffre OP. 2019. The histone methyltransferase SETDB1 controls T helper cell lineage integrity by repressing endogenous retroviruses. Immunity 50: 629–644.e8. 10.1016/j.immuni.2019.01.003 [DOI] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T. 2016. The molecular hallmarks of epigenetic control. Nat Rev Genet 17: 487–500. 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- Amemiya HM, Kundaje A, Boyle AP. 2019. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep 9: 9354. 10.1038/s41598-019-45839-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austenaa LMI, Piccolo V, Russo M, Prosperini E, Polletti S, Polizzese D, Ghisletti S, Barozzi I, Diaferia GR, Natoli G. 2021. A first exon termination checkpoint preferentially suppresses extragenic transcription. Nat Struct Mol Biol 28: 337–346. 10.1038/s41594-021-00572-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. 2013. Shortstack: comprehensive annotation and quantification of small RNA genes. RNA 19: 740–751. 10.1261/rna.035279.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrieri C, Alfarano G, Milan M, Tosi V, Prosperini E, Nicoli P, Palamidessi A, Scita G, Diaferia GR, Natoli G. 2018. Co-optation of tandem DNA repeats for the maintenance of mesenchymal identity. Cell 173: 1150–1164.e14. 10.1016/j.cell.2018.03.081 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21: 381–395. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, Natoli G. 2014. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell 54: 844–857. 10.1016/j.molcel.2014.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, et al. 2008. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 18: 1752–1762. 10.1101/gr.080663.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Chen Z, Chen X, Ai D, Chen G, McDermott J, Huang Y, Guo X, Han JJ. 2020. Accurate loop calling for 3D genomic data with cLoops. Bioinformatics 36: 666–675. [DOI] [PubMed] [Google Scholar]
- Collins PL, Kyle KE, Egawa T, Shinkai Y, Oltz EM. 2015. The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proc Natl Acad Sci 112: 8367–8372. 10.1073/pnas.1422187112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoglio F, Simonatto M, Polletti S, Liu X, Smale ST, Barozzi I, Natoli G. 2019. Dissection of acute stimulus-inducible nucleosome remodeling in mammalian cells. Genes Dev 33: 1159–1174. 10.1101/gad.326348.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuartero S, Weiss FD, Dharmalingam G, Guo Y, Ing-Simmons E, Masella S, Robles-Rebollo I, Xiao X, Wang YF, Barozzi I, et al. 2018. Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat Immunol 19: 932–941. 10.1038/s41590-018-0184-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curina A, Termanini A, Barozzi I, Prosperini E, Simonatto M, Polletti S, Silvola A, Soldi M, Austenaa L, Bonaldi T, et al. 2017. High constitutive activity of a broad panel of housekeeping and tissue-specific cis-regulatory elements depends on a subset of ETS proteins. Genes Dev 31: 399–412. 10.1101/gad.293134.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, DeWitt WS, et al. 2018. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174: 1309–1324.e18. 10.1016/j.cell.2018.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson IF, Peters JM. 2021. Genome folding through loop extrusion by SMC complexes. Nat Rev Mol Cell Biol 22: 445–464. 10.1038/s41580-021-00349-7 [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falandry C, Fourel G, Galy V, Ristriani T, Horard B, Bensimon E, Salles G, Gilson E, Magdinier F. 2010. CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J Biol Chem 285: 20234–20241. 10.1074/jbc.M109.052399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, Tachibana-Konwalski K. 2017. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544: 110–114. 10.1038/nature21711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2016. Formation of chromosomal domains by loop extrusion. Cell Rep 15: 2038–2049. 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gel B, Diez-Villanueva A, Serra E, Buschbeck M, Peinado MA, Malinverni R. 2016. Regioner: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32: 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32: 317–328. 10.1016/j.immuni.2010.02.008 [DOI] [PubMed] [Google Scholar]
- Glass CK, Natoli G. 2016. Molecular control of activation and priming in macrophages. Nat Immunol 17: 26–33. 10.1038/ni.3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdrini F, Esnault C, Horswell S, Stewart A, Matthews N, Treisman R. 2016. SRF Co-factors control the balance between cell proliferation and contractility. Mol Cell 64: 1048–1061. 10.1016/j.molcel.2016.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489. 10.1038/35013106 [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA. 2012. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods 9: 999–1003. 10.1038/nmeth.2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeault M, Helleboid PY, Trono D. 2017. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543: 550–554. 10.1038/nature21683 [DOI] [PubMed] [Google Scholar]
- Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. 2014. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516: 242–245. 10.1038/nature13760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A, Dhall A, Shi Y. 2019. Roles and regulation of histone methylation in animal development. Nat Rev Mol Cell Biol 20: 625–641. 10.1038/s41580-019-0151-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Loh YE, Rajarajan P, Hirayama T, Liao W, Kassim BS, Javidfar B, Hartley BJ, Kleofas L, Park RB, et al. 2017. The methyltransferase SETDB1 regulates a large neuron-specific topological chromatin domain. Nat Genet 49: 1239–1250. 10.1038/ng.3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NR, Yeoh JM, Coruh C, Axtell MJ. 2016. Improved placement of multi-mapping small RNAs. G3 6: 2103–2111. 10.1534/g3.116.030452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaaij LJT, Mohn F, van der Weide RH, de Wit E, Bühler M. 2019. The ChAHP complex counteracts chromatin looping at CTCF sites that emerged from SINE expansions in mouse. Cell 178: 1437–1451.e14. 10.1016/j.cell.2019.08.007 [DOI] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. 2011. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8: 676–687. 10.1016/j.stem.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Takemoto K, Shinkai Y. 2018. A somatic role for the histone methyltransferase Setdb1 in endogenous retrovirus silencing. Nat Commun 9: 1683. 10.1038/s41467-018-04132-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide S, Oshima M, Takubo K, Yamazaki S, Nitta E, Saraya A, Aoyama K, Kato Y, Miyagi S, Nakajima-Takagi Y, et al. 2016. Setdb1 maintains hematopoietic stem and progenitor cells by restricting the ectopic activation of nonhematopoietic genes. Blood 128: 638–649. 10.1182/blood-2016-01-694810 [DOI] [PubMed] [Google Scholar]
- Kroetz DN, Allen RM, Schaller MA, Cavallaro C, Ito T, Kunkel SL. 2015. Type I interferon induced epigenetic regulation of macrophages suppresses innate and adaptive immunity in acute respiratory viral infection. PLoS Pathog 11: e1005338. 10.1371/journal.ppat.1005338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27: 1571–1572. 10.1093/bioinformatics/btr167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9: e1003118. 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AY, Bauersachs R, Janas MS, Jarner MF, Kamphuisen PW, Meyer G, Khorana AA, CATCH Investigators. 2013. CATCH: a randomised clinical trial comparing long-term tinzaparin versus warfarin for treatment of acute venous thromboembolism in cancer patients. BMC Cancer 13: 284. 10.1186/1471-2407-13-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link VM, Duttke SH, Chun HB, Holtman IR, Westin E, Hoeksema MA, Abe Y, Skola D, Romanoski CE, Tao J, et al. 2018. Analysis of genetically diverse macrophages reveals local and domain-wide mechanisms that control transcription factor binding and function. Cell 173: 1796–1809.e17. 10.1016/j.cell.2018.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Brind'Amour J, Karimi MM, Shirane K, Bogutz A, Lefebvre L, Sasaki H, Shinkai Y, Lorincz MC. 2014. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev 28: 2041–2055. 10.1101/gad.244848.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931. 10.1038/nature08858 [DOI] [PubMed] [Google Scholar]
- Merkenschlager M, Nora EP. 2016. CTCF and cohesin in genome folding and transcriptional gene regulation. Annu Rev Genomics Hum Genet 17: 17–43. 10.1146/annurev-genom-083115-022339 [DOI] [PubMed] [Google Scholar]
- Montavon T, Shukeir N, Erikson G, Engist B, Onishi-Seebacher M, Ryan D, Musa Y, Mittler G, Meyer AG, Genoud C, et al. 2021. Complete loss of H3K9 methylation dissolves mouse heterochromatin organization. Nat Commun 12: 4359. 10.1038/s41467-021-24532-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach MR, Granja JM, Flynn RA, Roake CM, Satpathy AT, Rubin AJ, Qi Y, Jiang Z, Shams S, Louie BH, et al. 2019. HiChIRP reveals RNA-associated chromosome conformation. Nat Methods 16: 489–492. 10.1038/s41592-019-0407-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169: 930–944.e22. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapcuk V, Mohn F, Carl SH, Basters A, Hess D, Iesmantavicius V, Lampersberger L, Flemr M, Pandey A, Thomä NH, et al. 2018. Activity-dependent neuroprotective protein recruits HP1 and CHD4 to control lineage-specifying genes. Nature 557: 739–743. 10.1038/s41586-018-0153-8 [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433. 10.1016/j.cell.2008.01.011 [DOI] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589. 10.1016/S1097-2765(03)00477-5 [DOI] [PubMed] [Google Scholar]
- Picelli S, Björklund AK, Reinius B, Sagasser S, Winberg G, Sandberg R. 2014a. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res 24: 2033–2040. 10.1101/gr.177881.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. 2014b. Full-length RNA-seq from single cells using Smart-Seq2. Nat Protoc 9: 171–181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. 2014. CRISPR–Cas9 knockin mice for genome editing and cancer modeling. Cell 159: 440–455. 10.1016/j.cell.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599. 10.1038/35020506 [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240. 10.1038/nature08674 [DOI] [PubMed] [Google Scholar]
- Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci 112: E6456–E6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliehe C, Flynn EK, Vilagos B, Richson U, Swaminanthan S, Bosnjak B, Bauer L, Kandasamy RK, Griesshammer IM, Kosack L, et al. 2015. The methyltransferase Setdb2 mediates virus-induced susceptibility to bacterial superinfection. Nat Immunol 16: 67–74. 10.1038/ni.3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Gonçalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT. 2012. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148: 335–348. 10.1016/j.cell.2011.11.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ III. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16: 919–932. 10.1101/gad.973302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, Barillot E. 2015. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16: 259. 10.1186/s13059-015-0831-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. 2006. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev 20: 2349–2354. 10.1101/gad.399506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocking C, Kozak CA. 2008. Murine endogenous retroviruses. Cell Mol Life Sci 65: 3383–3398. 10.1007/s00018-008-8497-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403: 41–45. 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. 2014. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res 24: 1963–1976. 10.1101/gr.168872.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779–1791. 10.1101/gad.989402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. 2005. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 19: 815–826. 10.1101/gad.1284005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarjan DR, Flavahan WA, Bernstein BE. 2019. Epigenome editing strategies for the functional annotation of CTCF insulators. Nat Commun 10: 4258. 10.1038/s41467-019-12166-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weide RH, van den Brand T, Haarhuis JHI, Teunissen H, Rowland BD, de Wit E. 2021. Hi-C analyses with GENOVA: a case study with cohesin variants. NAR Genom Bioinform 3: lqab040. 10.1093/nargab/lqab040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, et al. 2012. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res 22: 1680–1688. 10.1101/gr.136101.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801. 10.1038/nature06634 [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204. 10.1038/nature07844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich Z, Mirny LA. 2009. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet 25: 434–440. 10.1016/j.tig.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz G, Várnai C, Nagasaka K, Cisneros DA, Stocsits RR, Tang W, Schoenfelder S, Jessberger G, Muhar M, Hossain MJ, et al. 2017. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36: 3573–3599. 10.15252/embj.201798004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xia L, Wu DY, Wang H, Chansky HA, Schubach WH, Hickstein DD, Zhang Y. 2002. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene 21: 148–152. 10.1038/sj.onc.1204998 [DOI] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364. 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. 2009. A clustering approach for identification of enriched domains from histone modification ChIP-seq data. Bioinformatics 25: 1952–1958. 10.1093/bioinformatics/btp340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed sequencing data were deposited in the Gene Expression Omnibus (GEO) repository under accession number GSE189975.