Here, Wu et al. demonstrate that the DNA repair factor HMCES strongly suppresses deletions without significantly affecting other parameters of somatic hypermutation in mouse and human B cells, thereby facilitating the production of antigen-specific antibodies. Their findings lead to a novel model for the protection of Ig gene integrity during SHM in which abasic site cross-linking by HMCES intercedes at a critical juncture during processing of vulnerable gapped DNA intermediates by BER and MMR enzymes.
Keywords: somatic hypermutation, HMCES, AID, base excision repair, mismatch repair, antibody affinity maturation
Abstract
Somatic hypermutation (SHM) produces point mutations in immunoglobulin (Ig) genes in B cells when uracils created by the activation-induced deaminase are processed in a mutagenic manner by enzymes of the base excision repair (BER) and mismatch repair (MMR) pathways. Such uracil processing creates DNA strand breaks and is susceptible to the generation of deleterious deletions. Here, we demonstrate that the DNA repair factor HMCES strongly suppresses deletions without significantly affecting other parameters of SHM in mouse and human B cells, thereby facilitating the production of antigen-specific antibodies. The deletion-prone repair pathway suppressed by HMCES operates downstream from the uracil glycosylase UNG and is mediated by the combined action of BER factor APE2 and MMR factors MSH2, MSH6, and EXO1. HMCES's ability to shield against deletions during SHM requires its capacity to form covalent cross-links with abasic sites, in sharp contrast to its DNA end-joining role in class switch recombination but analogous to its genome-stabilizing role during DNA replication. Our findings lead to a novel model for the protection of Ig gene integrity during SHM in which abasic site cross-linking by HMCES intercedes at a critical juncture during processing of vulnerable gapped DNA intermediates by BER and MMR enzymes.
Somatic hypermutation (SHM) introduces mutations, predominantly single-nucleotide point mutations, into the variable (V) region exon of assembled immunoglobulin (Ig) heavy and light chain genes in antigen-stimulated germinal center B cells (French et al. 1989; Di Noia and Neuberger 2007; Victora and Nussenzweig 2012). SHM enables the generation of populations of B cells whose surface Ig receptors vary in their antigen binding affinity. This variation is critical for the process of antibody affinity maturation in which B cells with higher-affinity Ig receptors are iteratively selected for preferential survival, proliferation, and additional rounds of SHM and affinity selection (Neuberger 2008; Victora and Nussenzweig 2012). SHM is therefore critical for effective humoral immunity and vaccine responses. SHM also generates deletions and, to a lesser extent, insertions, which are strongly selected against in the Ig V region, presumably because of their propensity to damage the integrity of the open reading frame or protein folding (Yeap et al. 2015). The mechanisms that generate deletions during SHM are not well characterized, although it is plausible that they involve double-strand DNA (dsDNA) breaks, a particularly dangerous DNA lesion (Sale and Neuberger 1998; Bross et al. 2000; Papavasiliou and Schatz 2000; Alt et al. 2013; Yeap et al. 2015; Casellas et al. 2016; Yeap and Meng 2019). Whether dedicated factors or mechanisms exist to regulate the frequency of deletions during SHM and whether they might also regulate the generation of insertions are unknown.
SHM is initiated when the activation-induced deaminase (AID), a single-strand DNA (ssDNA)-specific cytidine deaminase, converts a cytosine base (C) to uracil (U) in an Ig V region (Fig. 1A; Di Noia and Neuberger 2007). AID can target C residues on either DNA strand, acts preferentially at hotspots corresponding to the sequence RGYW or its inverse complement WRCY (R = A, G; Y = C, T; and W = A, T), and is thought to access its ssDNA substrate in the context of the transcription bubble created by paused or stalled RNA polymerase 2 (Sun et al. 2013; Methot and Di Noia 2017). Using a similar mechanism, AID deaminates C residues in Ig heavy chain gene (IGH) switch regions to initiate class switch recombination (CSR), a reaction that replaces one IGH constant region with another, thereby altering antibody effector function (Chaudhuri and Alt 2004; Xu et al. 2012).
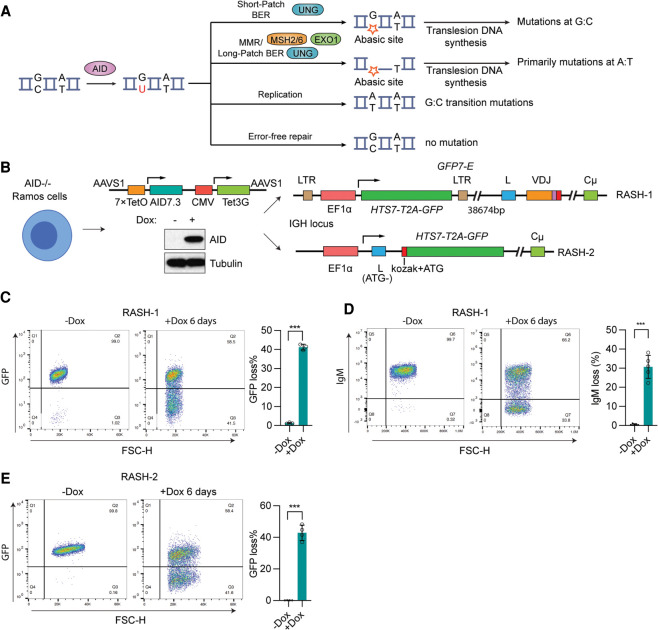
Figure 1.
Establishing a rapid assay for SHM. (A) Schematic of SHM reaction. During SHM, AID deaminates C to generate uracil, which can act as a template for replication, leading to a C:G-to-T:A transition mutation. Alternatively, noncanonical (error-prone) base excision repair (BER) initiated by UNG can create an abasic site that, if copied by a translesion DNA polymerase, yields mutations at G:C. Finally, U or U:G mismatches can trigger the generation of ssDNA gaps through the action of UNG and mismatch repair (MMR) factors MSH2, MSH6, and EXO1, which primarily yield mutations at A:T upon translesion polymerase synthesis. (B) Scheme for creating RASH-1 and RASH-2. Creation of RASH involved several steps of CRISPR/Cas9 gene targeting in Ramos. AICDA (encoding AID) was disrupted, and a construct was inserted into the AAVS1 safe harbor locus that expresses AID7.3 (AID hyperactive mutant with threefold increased catalytic activity) upon doxycycline (dox)-induced binding of Tet3G to a promoter containing seven copies of its binding site, TetO (7xTetO). Western blot showing AID protein levels in AID7.3in cells without or with 200 ng/mL dox for 24 h. The GFP7-E reporter with EF1α promoter driving transcription of HTS7 (enriched with AID hotspots)-T2A-GFP was integrated into IGH either 38,674 bp upstream of the V leader sequence (RASH-1) or by replacing IGH-VDJ coding sequences so as to retain the leader exon (which now lacks its ATG start codon; RASH-2). Kozak sequence was added to HTS7-T2A-GFP in RASH-2. (C,D) Representative flow cytometry plots of and bar graphs quantifying GFP loss (C) or IgM loss (D) in RASH-1 cells treated with dox for 6 d. (E) Representative flow cytometry plots of and bar graph quantifying GFP loss in RASH-2 treated with dox for 6 d. Throughout the figure, data are presented with the bars representing mean and the error bars as ±SD. Statistical significance was calculated using two-tailed Student's t-test. (***) P value < 0.001.
The U created by AID is the source of the mutations generated during SHM (Fig. 1A). It can be replicated over in S phase to generate C/G-to-A/T transition mutations or it (or the U:G mismatch it creates) can be recognized and processed by enzymes in the base excision repair (BER) or mismatch repair (MMR) pathways. In most cells, these pathways process U (U:G) in a high-fidelity manner to prevent mutations, but downstream from AID in germinal center B cells they act both individually and in concert—through multiple error-prone repair pathways—to generate mutations at the site at which AID acted and at surrounding G/C and A/T residues (Liu and Schatz 2009; Methot and Di Noia 2017; Pilzecker and Jacobs 2019).
The critical enzymes that initiate these error-prone repair mechanisms are the BER factor UNG (uracil DNA glycosylase) and the MMR heterodimer MSH2/MSH6 (Rada et al. 2004; Di Noia and Neuberger 2007) (Fig. 1A). UNG encodes mitochondrial (UNG1) and nuclear (UNG2) isoforms through the use of alternative promoters and splicing (Nilsen et al. 1997). UNG recognizes and removes the U base from ssDNA or dsDNA to create an abasic site, which can either serve as a noninstructive template for translesion polymerases or trigger endonucleolytic cleavage of the deoxyribose–phosphate backbone by apurinic/apyrimidinic endonucleases APE1 or APE2. APE2 has been implicated in the generation of mutations and deletions in SHM (Sabouri et al. 2009; Stavnezer et al. 2014). MSH2/6 recognizes the U:G mismatch created by the action of AID and can trigger resection of one DNA strand by the exonuclease EXO1. The DNA strand lesions created by BER and MMR factors can then be resolved by one of several pathways involving error-prone translesion polymerases to create the full spectrum of mutations associated with SHM (Methot and Di Noia 2017; Pilzecker and Jacobs 2019; Yeap and Meng 2019; Feng et al. 2020). Extensive evidence indicates that BER and MMR factors act collaboratively to create mutations during SHM and to convert ssDNA breaks and gaps into the dsDNA breaks required for CSR, with loss of either pathway causing a severe reduction in the efficiency of both SHM and CSR (Methot and Di Noia 2017; Pilzecker and Jacobs 2019; Feng et al. 2020, 2021). It is plausible that BER and MMR factors collaborate to generate deletions during SHM, although this idea has not been tested.
HMCES (5-hydroxymethylcytosine binding, embryonic stem cell-specific protein) is the sole known mammalian member of the superfamily of proteins containing a SRAP (SOS response-associated peptidase) domain, an evolutionarily ancient domain found in E. coli yedK and in many other species (Aravind et al. 2013). HMCES is recruited to stalled DNA replication forks where its SRAP domain forms a covalent cross-link with abasic sites in ssDNA, thereby shielding the abasic site from the action of endonucleases and translesion polymerases and protecting against DNA breaks and mutations (Mohni et al. 2019; Mehta et al. 2020; Srivastava et al. 2020). In this mode of action, HMCES is a suicide enzyme that is removed from DNA by ubiquitin-mediated proteolysis (Mohni et al. 2019). Structural and biochemical analyses have revealed the basis of HMCES's preference for binding to ssDNA extensions or gaps and identified a thiazolidine linkage formed between an invariant cysteine at position 2 (C2) of HMCES and the deoxyribose of the abasic site (Halabelian et al. 2019; Thompson et al. 2019; Wang et al. 2019). HMCES is highly expressed in germinal center B cells, and Hmces−/− mice exhibit a significant defect in CSR despite normal cellular parameters of the germinal center response (Shukla et al. 2020). The CSR defect is due to a selective failure of the alternative end-joining DNA repair pathway, which works in tandem with classical nonhomologous end joining to complete the CSR reaction (Boboila et al. 2010; Alt et al. 2013; Chang et al. 2017; Shukla et al. 2020). Notably, HMCES function in alternative end joining appears to involve the protection of ssDNA overhangs against excessive deletion and does not require C2, revealing a second mode of action that is independent of cross-linking to abasic sites (Shukla et al. 2020). The role of HMCES in SHM has not been explored.
In this study, we report that HMCES plays a highly selective role during SHM to limit the accumulation of deletions in SHM target regions. In both mice and a human cell line model system, loss of HMCES leads to significant increases in SHM-associated deletions with little or no significant effect on point mutation accumulation or spectrum or the frequency of insertions. Taking advantage of a new rapid SHM assay system, we demonstrate that HMCES function during SHM requires C2 and an intact ssDNA binding groove, is completely dependent on UNG, and protects against a deletion pathway that involves APE2 (but not APE1), MSH2, MSH6, and EXO1. Our results lead to a model in which cross-linking of HMCES to abasic sites generated by the combined action of AID and UNG inhibits DNA cleavage of a vulnerable ssDNA repair intermediate by APE2 and indicate that BER and MMR can act in a coordinated manner to create deletions during SHM.
Results
Establishing a rapid assay for SHM (RASH)
To facilitate the analysis of SHM factors and mechanisms, we sought to create a fast and robust cellular assay for SHM that provided high levels of AID activity, a sensitive readout of AID-dependent mutation accumulation, and temporal control. We used the human germinal center-derived lymphoma cell line Ramos because of its extensive prior validation as a SHM model system, including efficient AID-dependent mutation of its Ig heavy (IGH) and λ (IGL) chain variable regions, ability to support Ig enhancer-driven mutation of reporter vectors, and substantial mapping of sites of “off-target” SHM activity in its genome (Sale and Neuberger 1998; Papavasiliou and Schatz 2000; Ronai et al. 2007; Qian et al. 2014; Senigl et al. 2019). Ramos has been found to mirror many aspects of physiological SHM with the exception that its mutation spectrum is biased toward mutations at G/C (75%–80% as opposed to the ∼45% seen in germinal center B cells) (Sale and Neuberger 1998).
Creation of cell lines in which SHM could be assessed rapidly and sensitively (Fig. 1B) began with our previously generated AID7.3in cells, in which endogenous AICDA (encoding AID) is disrupted and AID7.3 (AID mutant K10E/E156G/T82I, with threefold increased catalytic activity) (Wang et al. 2009) is expressed from an integrated doxycycline (dox)-inducible construct (Dinesh et al. 2020). AID7.3 is tightly regulated and expressed at levels well above those of endogenous AID upon dox treatment (Fig. 1B; Supplemental Fig. S1A). We infected AID7.3in Ramos cells with a lentiviral SHM reporter vector (GFP7-E), in which the constitutive EF1α promoter drives transcription of a fusion HTS7-T2A-GFP open reading frame. HTS7 contains a dense array of SHM hotspots designed to yield stop codons upon mutation, allowing efficient detection of SHM activity by virtue of loss of GFP fluorescence (referred to here as GFP loss) (Senigl et al. 2019). As expected (Senigl et al. 2019), most cell clones with integrated GFP7-E gave rise to very little (<1%) GFP loss after 6 d of dox induction (279 of 306 clones examined), while one clone exhibited particularly strong GFP loss (Supplemental Fig. S1B, arrow). We found that this clone contained GFP7-E integrated ∼38.6 kb upstream of the rearranged IGH variable region (Fig. 1B) and exhibited robust (>40%) GFP loss with 6 d of dox treatment (Fig. 1C). AID induction also led to substantial accumulation of cells lacking surface IgM expression (referred to here as IgM loss) (Fig. 1D), which has been shown to be caused by deleterious mutations in the rearranged IGH and IGL variable regions (Neuberger et al. 1998; Papavasiliou and Schatz 2000). This clone is referred to here as RASH-1 (rapid assay for SHM).
A second cell line (RASH-2) was generated from AID7.3in Ramos cells by targeted integration of HTS7-T2A-GFP into the IGH locus, replacing the rearranged IGH variable region exon and placing the cassette under control of the EF1α promoter (Fig. 1B). The ATG start codon in the L exon was mutated and splicing between the L exon and HTS7-T2A-GFP was retained. RASH-2 cells yielded robust GFP loss upon AID induction, similar in magnitude to RASH-1 cells (Fig. 1E). Insertion of HTS7-T2A-GFP into the IGH locus in RASH-1 and RASH-2 provides a physiological genomic context for SHM, which is thought to be driven by IGH enhancer elements (Rouaud et al. 2013; Buerstedde et al. 2014; Senigl et al. 2019; Dinesh et al. 2020). Cas9-expressing derivatives of RASH-1 and RASH-2 (RASH-1C and RASH-2C) support efficient Cas9-mediated gene disruption (data not shown) and most of the cell line experiments described below were performed in RASH-1C cells or derivatives thereof.
HMCES deficiency leads to increased deletions in SHM target regions
HMCES is required for efficient CSR (Shukla et al. 2020), leading us to ask whether it might also play a role in facilitating SHM; if so, one might expect disruption of HMCES to decrease the extent of GFP loss in our RASH lines. Unexpectedly, however, when HMCES was disrupted in bulk RASH-1C cells using two independent sgRNAs delivered by electroporation (Fig. 2A,B), GFP loss increased significantly upon AID induction. Increased GFP loss was also observed in two independent RASH-2C clones depleted of HMCES and in HMCES-deficient RASH-1C single-cell clones (Fig. 2C,D; Supplemental Fig. S1C,D). Furthermore, loss of HMCES led to increased IgM loss in RASH-1C clones, with the magnitude of the increase correlating well with the increase in GFP loss (Fig. 2E; Supplemental Fig. S1E). To rule out the possibility that this phenotype was due to use of a hyperactive AID mutant, we engineered Cas9-expressing derivatives (Supplemental Fig. S1F) of Ramos line A23, which overexpresses WT human AID from a retroviral vector (Supplemental Fig. S1G; Dinesh et al. 2020). Analysis of bulk HMCES knockout (KO) populations or single-cell HMCES KO clones of A23-Cas9 revealed increased IgM loss relative to the parental A23-Cas9 cells (Supplemental Fig. S1H–J). Finally, to rule out the possibility that the phenotype was due to ectopic overexpression of AID, HMCES was knocked out in six independent WT Ramos single-cell clones, which exhibited a strong increase in IgM loss relative to WT Ramos clones (Fig. 2F,G). Disruption of HMCES did not alter levels of AID expression in these analyses (Fig. 2C,F; Supplemental Fig. S1H). Together, these results demonstrated that loss of HMCES leads to an increase in AID-dependent GFP and IgM loss and suggested the possibility that HMCES is a negative regulator of SHM.
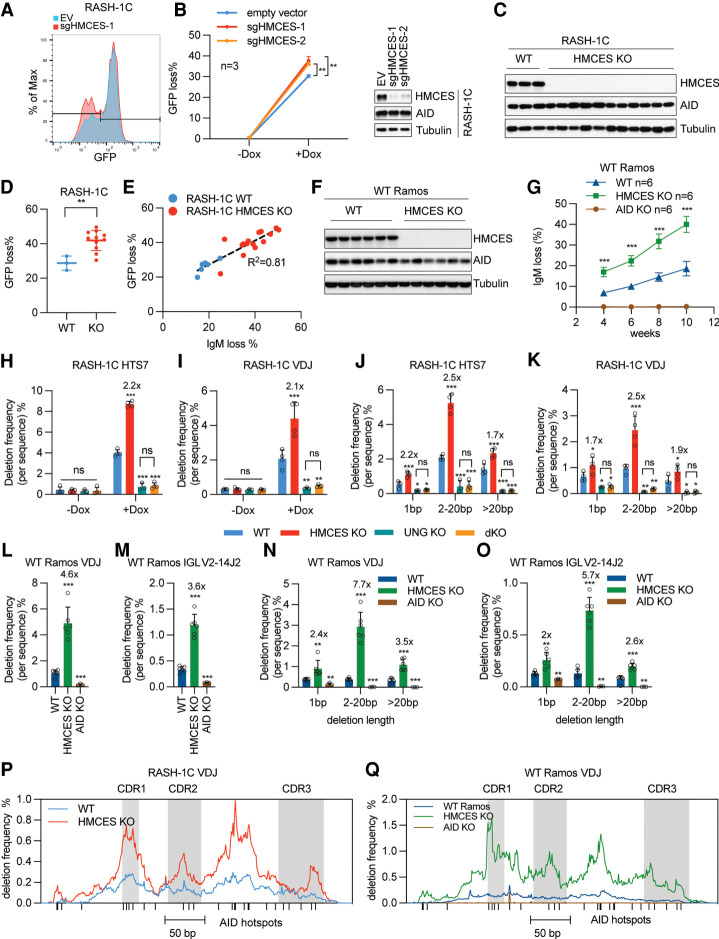
Figure 2.
HMCES deficiency leads to increased deletions in SHM target regions. (A) Representative flow cytometry histogram plot of and graph quantitating GFP loss in RASH-1C cells treated with empty vector or a HMCES sgRNA without or with 4-d dox treatment. (B) Western blot showing HMCES protein levels in RASH-1C cells treated with two different HMCES sgRNAs. (C) Western blot showing HMCES protein levels in 11 independent HMCES KO RASH-1C cell clones. (D) Dot plot of GFP loss in cells shown in C treated with dox for 4 d. (E) Scatter plot of GFP loss versus IgM loss from WT and HMCES KO RASH-1C cell clones treated with dox for 4 d. (F) Western blot showing HMCES protein levels in six independent HMCES KO Ramos cell clones. (G) IgM loss from WT, HMCES KO, or AID KO Ramos cell clones at different time points of culture. (H,I) Deletion frequency in the HTS7 (H) and VDJ (I) regions from WT, HMCES KO, UNG KO, and HMCES/UNG dKO RASH-1C cells with dox for 4 d. Deletion frequency was calculated as (number of deletions/number of sequences) × 100; fold changes compared with WT are marked above the bar. (J,K) Distribution of deletion lengths in HTS7 (J) and VDJ (K) regions from WT, HMCES KO, UNG KO, and HMCES/UNG dKO RASH-1C cells treated with dox for 4 d. Deletions were divided into three groups based on length as indicated and the percentage of sequences with indicated deletion length are shown. Fold changes compared with WT are marked above the bar. (L,M) Deletion frequency in the IGH VDJ (L) and IGL VJ (M) regions from WT, HMCES KO, and AID KO Ramos cells. Single cells were seeded and grown for 6 wk and then harvested for analysis. Fold changes compared with WT are marked above the bar. (N,O) Distribution of deletion lengths in the IGH VDJ (N) and IGL VJ (O) regions from WT, HMCES KO, and AID KO Ramos cells. Deletions were divided into three groups based on length as indicated and the percentage of sequences with the indicated deletion length are shown. Fold changes compared with WT are marked above the bar. (P) Deletion profile in the VDJ region from WT and HMCES KO RASH-1C cells treated with dox for 4 d. AID hotspots are indicated with a vertical line at the X-axis. The CDR1, CDR2, and CDR3 regions are shaded. (Q) Deletion profile in the VDJ region from WT, HMCES KO, and AID KO Ramos cells. AID hotspots are indicated with a vertical line at the X-axis. The CDR1, CDR2, and CDR3 regions are shaded. Throughout the figure, data are presented with the bars representing mean and the error bars as ±SD. Statistical significance was calculated using two-tailed Student's t-test for A and D and using one way ANOVA with Dunnett's post-test for G–O. (***) P-value < 0.001, (**) P-value < 0.01, (*) P-value < 0.05, (ns) not significant.
To ascertain the basis for increased GFP and IgM loss in HMCES-deficient cells, the HTS7 (366-bp) and IGH VDJ (389-bp) regions were PCR-amplified and subjected to high-throughput sequencing. Surprisingly, loss of HMCES in RASH-1C cells did not increase—and in fact tended to decrease—the frequency of point mutations in both HTS7 and VDJ (Supplemental Fig. S2A,B). Instead, deletions were significantly (more than twofold) increased upon knockout of HMCES (Fig. 2H,I). This increase was not observed without dox induction of AID, indicating that loss of HMCES did not detectably increase background (AID-independent) levels of deletions (Fig. 2H,I). HMCES deficiency increased 1-bp deletions, intermediate length deletions (2–20 bp), and long deletions (>20 bp) in both HTS7 and VDJ, with the largest fold increase observed for intermediate length deletions (Fig. 2J,K). Very similar results were obtained from sequencing of IGH VDJ and IGL VJ from WT and HMCES KO Ramos cells (expressing endogenous, WT AID), with the fold increase in deletions caused by loss of HMCES even greater in this context than in RASH-1C (Fig. 2L,M). Essentially all deletions were eliminated in AID KO Ramos cells, with the residual 1-bp deletions likely representing sequencing errors (Fig. 2N,O). As in RASH-1C, loss of HMCES in Ramos did not increase the point mutation frequency, and in this context, little or no significant decrease was detected (Supplemental Fig. S2C,D).
In both RASH-1C and Ramos, deletion frequencies were increased across the entire VDJ region and VJ region and peaked in several regions containing clusters of AID hotspots (RGYW or its inverse complement, WRCY; R = A, G; Y = C, T; W = A, T), consistent with the idea that the deletions originate from AID-mediated deamination events (Fig. 2P,Q; Supplemental Fig. S2E). The overall similarities in the shape of the deletion profiles in HMCES-deficient and -proficient cells (Fig. 2P,Q; Supplemental Fig. S2E) argue that loss of HMCES does not strongly perturb AID targeting. These data demonstrate that loss of HMCES results in an AID-dependent increase in deletions and that this increase is observed with WT endogenous AID and overexpressed AID7.3.
To determine how this increase in deletions influenced the distribution of events leading to GFP loss, the HTS7-GFP region was amplified from sorted GFP-negative WT and HMCES KO RASH-1C cells and subjected to Sanger sequencing. The percentage of sequences in which GFP loss could be explained by a deletion event increased from 14% in WT to 37%–44% in HMCES KO cells (Supplemental Fig. S2F), providing a ready explanation for the increase in GFP loss seen in HMCES KO cells.
A detailed examination of point mutations in the presence and absence of HMCES revealed few if any consistent changes. In RASH-1C, loss of HMCES increased the proportion of transition mutations at G/C at the expense of transversion mutations, but this change was less evident in Ramos (Supplemental Fig. S2G–N). In both RASH-1C and Ramos, loss of HMCES did not substantially alter the distribution of mutations across the IGH VDJ region, and mutations occurred preferentially near AID hotspots (Supplemental Fig. S3A–C). The frequency of insertions was significantly increased in Ramos but not in RASH-1C when HMCES was knocked out (Supplemental Fig. S3D–G). Together, these data demonstrate that disruption of HMCES in Ramos and Ramos derivatives leads to an increase in GFP loss and IgM loss, a phenotype readily explained by a consistent large increase in deletions in SHM target regions, but that changes in mutation frequency and spectrum and in insertion frequency tend to be modest or not significant and are influenced by whether the cells express hyperactive or WT AID.
The function of HMCES in SHM is dependent on abasic site cross-linking and ssDNA binding residues
HMCES (Fig. 3A) contains a well-conserved N-terminal SRAP domain (amino acids 1–270) and a C-terminal region with three proliferating cell nuclear antigen (PCNA)-interacting protein (PIP) motifs implicated in interaction with PCNA and possibly other DNA repair factors (Boehm and Washington 2016) and recruitment of HMCES to DNA replication forks (Aravind et al. 2013; Mohni et al. 2019). The invariant cysteine at position 2 (C2) is required for HMCES to cross-link to abasic sites in DNA and does so by forming a thiazolidine linkage with the deoxyribose of the abasic site (Halabelian et al. 2019; Mohni et al. 2019; Thompson et al. 2019; Wang et al. 2019). Cross-linking requires prior removal of the amino acid N-terminal to C2, which can be accomplished by HMCES autopeptidase activity using catalytic residues C2, E127, and H210 and likely by other peptidases as well (Kweon et al. 2017; Halabelian et al. 2019; Thompson et al. 2019; Wang et al. 2019). Structural analysis of the HMCES SRAP domain in complex with 3′ overhang DNA revealed that R98 and R212 form a single-strand (ss) DNA binding cleft, that E127 and H210 are closely juxtaposed to C2 and the site of cross-linking, and that W81, F92, and R106 contribute to the interaction of HMCES with DNA near the ssDNA–dsDNA junction (Halabelian et al. 2019).
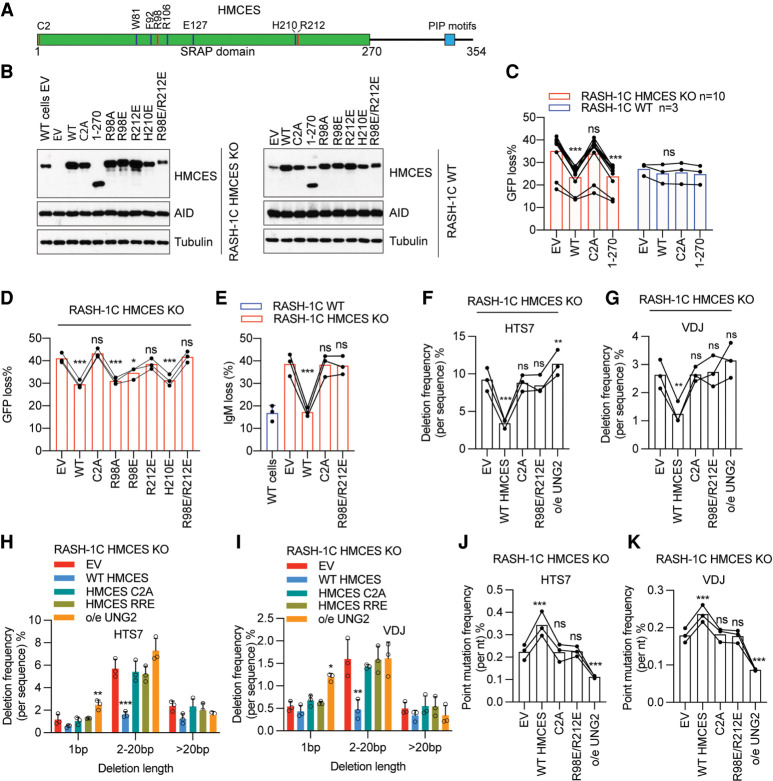
Figure 3.
The function of HMCES in SHM is dependent on abasic site cross-linking and ssDNA binding residues. (A) Schematic of human HMCES protein. (B) Western blot of HMCES and AID in WT or HMCES KO RASH-1C cells reconstituted with WT HMCES or its mutants and induced with dox for 4 d. (C,D) WT HMCES or the indicated HMCES mutants were expressed in RASH-1C cells in which HMCES was knocked out (RASH-1C HMCES KO) or intact (RASH-1C WT). Cells induced with dox for 4 d were assayed for GFP loss. (E) WT HMCES or the indicated HMCES mutants were expressed in HMCES KO RASH-1C cells. Cells induced with dox for 4 d were assayed for IgM loss. (F,G) Deletion frequencies in HTS7 (F) and VDJ (G) from HMCES KO RASH-1C cells reconstituted with WT HMCES or its mutants or overexpressing UNG2, as indicated. (H,I) Distribution of deletion lengths in the HTS7 (H) and VDJ (I) regions from HMCES KO RASH-1C cells reconstituted with WT HMCES or its mutants or overexpressing UNG2. (J,K) Point mutation frequencies in the HTS7 (J) and VDJ (K) regions from HMCES KO RASH-1C cells reconstituted with WT HMCES or its mutants or overexpressing UNG2. Throughout the figure, data are presented with the bars representing mean and the error bars as ±SD. Statistical significance was calculated using one-way ANOVA with Dunnett's post-test. (***) P-value < 0.001, (**) P-value < 0.01, (*) P-value < 0.05, (ns) not significant.
To investigate the mechanism by which HMCES functions in SHM, we expressed WT HMCES or HMCES variants in WT or HMCES KO RASH-1C cells (Fig. 3B). We found that the HMCES KO GFP loss phenotype could be fully rescued by reconstitution with WT HMCES or a C-terminal truncated protein containing only the SRAP domain (amino acids 1–270) (Fig. 3C), indicating that the C-terminal region is not required for HMCES function in SHM, as is also the case for HMCES function in CSR (Shukla et al. 2020). However, no reconstitution was observed with C2A point mutant HMCES (Fig. 3C), in striking contrast to CSR in which HMCES function is unaffected by the C2A mutation (Shukla et al. 2020). Overexpression of HMCES or the C2A or 1–270 mutants in WT cells had no significant effect on GFP loss, suggesting that protein levels of HMCES are saturating in these cells and arguing against a dominant-negative phenotype for the C2A mutant (Fig. 3C).
Analysis of DNA binding and autopeptidase catalytic residue mutants revealed a range of phenotypes. While R98A, R98E, and R212A single mutants exhibited substantial reconstitution activity, mutation of R212 to E strongly diminished activity, and double mutations of R98 and R212 to A or E eliminated HMCES activity in the GFP loss assay, indicating that the ssDNA binding cleft is important for HMCES function in SHM, as also observed for HMCES function in CSR (Fig. 3D; Supplemental Fig. S4A,B). Further analyses confirmed the lack of activity for C2A and R98E/R212E HMCES mutants, demonstrating that they cannot reconstitute the IgM loss (Fig. 3E), deletion frequency (Fig. 3F–I), point mutation frequency (Fig. 3J,K), or point mutation spectrum (Supplemental Fig. S4C–F) phenotypes in HMCES KO RASH-1C cells. Neither single mutation of W81, F92, or R106 nor triple mutation of these residues affected HMCES activity in the GFP loss assay (Supplemental Fig. S4G,H), arguing that the ssDNA–dsDNA junction binding region of HMCES is not essential for SHM function. Similarly, neither E127A nor H210E mutations compromised HMCES function in the GFP loss assay (Fig. 3D; Supplemental Fig. S4I), indicating that HMCES autopeptidase activity is not required and that a cellular peptidase activity would be predicted to be able to remove the first amino acids of HMCES to enable C2 cross-linking activity. We did not detect an effect on GFP loss of overexpression of any of the mutants in WT RASH-1C cells except for E127A, which reduced GFP loss (Supplemental Fig. S4I–K), an effect that might be attributable to the increased DNA binding observed when this residue is mutated (see the Discussion). In summary, mutational analysis of HMCES demonstrates that its function in SHM in Ramos cells requires the abasic site cross-linking residue C2, is supported by its ssDNA binding surface, and is independent of mutations that alter ssDNA–dsDNA binding residues and of autopeptidase activity.
The function of HMCES in SHM is dependent on the activity of UNG
UNG is the major uracil glycosylase in SHM and should be the primary source of abasic sites during the reaction (Fig. 1A; Di Noia et al. 2006; Methot and Di Noia 2017). If HMCES functions in SHM by cross-linking to abasic sites, as suggested by the requirement for C2, then HMCES should act downstream from UNG and be strongly dependent on UNG to exert its phenotype. To test this prediction, we compared the phenotypes of UNG KO and UNG/HMCES double-KO (dKO) cells. UNG deficiency led to increases in GFP and IgM loss in RASH-1C cells (Fig. 4A,B), as expected from prior studies showing that mutation frequencies increase when UNG activity is reduced or eliminated (Di Noia and Neuberger 2002; Rada et al. 2004; Liu et al. 2008; Feng et al. 2021; Rogier et al. 2021). The increases were comparable in magnitude with those observed in HMCES KO cells. Elimination of UNG decreased deletion frequencies dramatically, almost to the level seen in cells without AID induction (Fig. 2H,I). Strikingly, relative to UNG single-KO cells, HMCES/UNG dKO cells showed no increase in GFP or IgM loss (Fig. 4A,B), no increase in deletion frequency (Fig. 2H–K), no alteration in mutation frequency or spectrum (Supplemental Fig. S2A,B,G–J), and no difference in insertion frequency (Supplemental Fig. S3D,E). Indeed, UNG/HMCES dKO cells closely phenocopied UNG KO cells in all parameters measured.
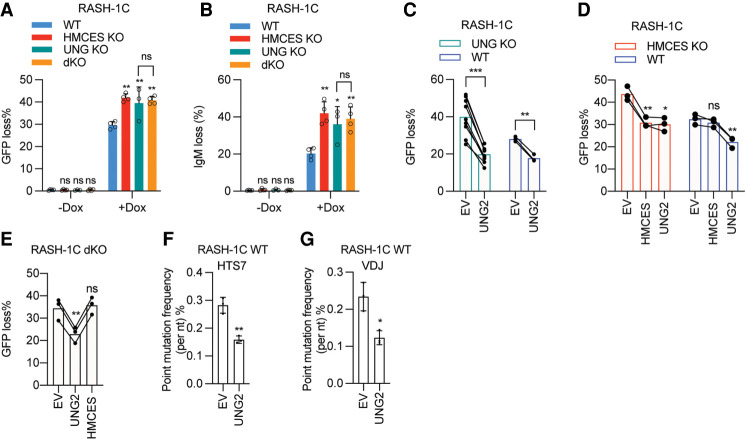
Figure 4.
The function of HMCES in SHM is dependent on the activity of UNG. (A,B) WT, HMCES KO, UNG KO, and HMCES/UNG dKO (double-KO) RASH-1C cells with dox for 4 d were assayed for GFP loss (A) and IgM loss (B). (C) UNG KO or WT RASH-1C cells overexpressing UNG2 induced with dox for 4 d were assayed for GFP loss. (D) HMCES or UNG2 was overexpressed in WT or HMCES KO RASH-1C cells. Cells induced with dox for 4 d were assayed for GFP loss. (E) HMCES and UNG dKO RASH-1C cells overexpressing UNG2 or HMCES induced with dox for 4 d were assayed for GFP loss. (F,G) Point mutation frequencies in the HTS7 (F) and VDJ (G) regions from WT RASH-1C cells overexpressing UNG2. Throughout the figure, data are presented with the bars representing mean and the error bars as ±SD. Statistical significance was calculated using one-way ANOVA with Dunnett's post-test for A, B, D, and E and using two-tailed Student's t-test for C, F, and G. (***) P-value < 0.001, (**) P-value < 0.01, (*) P-value < 0.05, (ns) not significant.
Reconstitution and overexpression experiments in RASH-1C cells further emphasized the lack of a HMCES phenotype in the absence of UNG and demonstrated that UNG exerts its effects in the absence of HMCES. Ectopic expression of UNG2 (the nuclear form of UNG) in WT, UNG KO, HMCES KO, and UNG/HMCES dKO cells reduced GFP loss (Fig. 4C–E; Supplemental Fig. S5A–C). In contrast, HMCES re-expression in UNG/HMCES dKO cells had no effect on the frequency of GFP loss (Fig. 4E) while it reduced GFP loss in HMCES single-KO cells, as expected (Fig. 4D). Ectopic UNG2 expression reduced mutation frequencies (particularly for G/C transitions) in both WT and HMCES KO cells (Figs. 3J,K, 4F,G; Supplemental Figs. S4C,D, S5D,E), demonstrating that the high-fidelity repair pathways initiated by UNG are not HMCES-dependent. UNG hyperactivity or overexpression has been shown to reduce mutation frequencies in mice and cell lines (Feng et al. 2021; Rogier et al. 2021). Ectopic UNG2 expression showed a tendency to increase deletion frequencies in both WT and HMCES KO cells, but 1-bp deletions were selectively and significantly increased only in HMCES KO cells, suggesting that HMCES protects against 1-bp deletions initiated by UNG (Fig. 3H,I; Supplemental Fig. S5F–I). We conclude that HMCES function in SHM is entirely dependent on UNG, a conclusion consistent with a model in which HMCES protects against deletions by acting on the abasic sites created by UNG. In contrast, UNG is able to alter mutation frequency and spectrum in the absence of HMCES.
APE2 contributes to deletions that arise in the absence of HMCES
By cross-linking to abasic sites, HMCES is thought to protect abasic sites from the action of endonucleases and translesion polymerases during DNA replication and, in biochemical assays, blocks cleavage by AP endonuclease APE1 (Halabelian et al. 2019; Mohni et al. 2019; Thompson et al. 2019; Wang et al. 2019; Srivastava et al. 2020). Our data raise the possibility that HMCES performs a similar function during SHM. If this is the case, then the excessive deletions that occur in the absence of HMCES would be predicted to be dependent on the action of APEs. APE2 is up-regulated in germinal center B cells and contributes to efficient point mutation accumulation and the generation of indels during SHM, while APE1, the more efficient endonuclease, is down-regulated in germinal center B cells and functions in high-fidelity short patch BER (Hegde et al. 2008; Sabouri et al. 2009; Stavnezer et al. 2014; Roco et al. 2019).
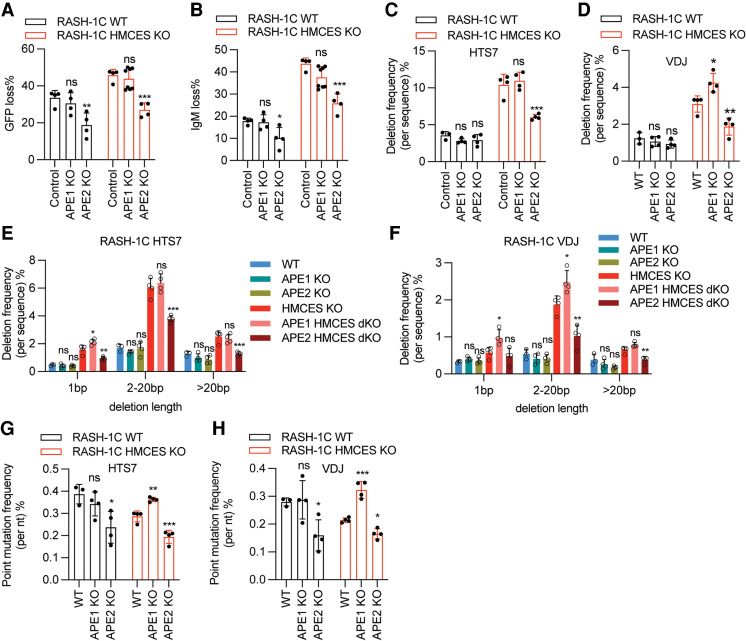
To test the prediction that APEs contribute to the excessive deletions that occur in the absence of HMCES, we knocked out APE1 and APE2 in WT and HMCES KO RASH-1C cells. Knockout of APE2 but not APE1 significantly reduced GFP loss and IgM loss in both the WT and HMCES KO context (Fig. 5A,B). Strikingly, deficiency for APE2 but not APE1 led to a significant reduction in deletion frequencies in HTS7 and VDJ in HMCES KO but not WT cells (Fig. 5C,D). This decrease was consistently observed and significant for intermediate length and long deletions but not for 1-bp deletions (Fig. 5E,F). In some cases, APE1 KO actually resulted in a significant increase in deletions, again observed only in the HMCES KO context (Fig. 5C–F). APE2 deficiency significantly reduced point mutation frequencies in WT and HMCES KO RASH-1C cells (Fig. 5G,H), with the decreases most significantly affecting G/C transversions and A/T mutations (Supplemental Fig. S6A,B). In contrast, APE1 KO led to an increase in mutation frequency specifically in the HMCES KO context (Fig. 5G,H; Supplemental Fig. S6A,B). Insertion frequencies decreased in some contexts but not others upon knockout of APE1 or APE2 (Supplemental Fig. S6C,D). These findings demonstrate that APE2 but not APE1 contributes to the deletions that accumulate in the absence of HMCES. They further suggest that APE1 protects against even higher levels of deletions in HMCES KO cells, which we speculate might occur by virtue of competition between the high-fidelity action of APE1 and the more deletion-prone action of APE2. Competition between APE1 and APE2 in the HMCES-deficient context is also consistent with our observation that while APE2 KO leads to reduced point mutations, APE1 KO has the opposite effect. Finally, to our knowledge, these findings provide the first analysis of SHM in the complete absence of APE1, revealing that it does not contribute significantly to deletions or point mutation frequency or spectrum in HMCES-sufficient cells.
Figure 5.
APE2 contributes to deletions that arise in the absence of HMCES. (A,B) APE1 or APE2 were knocked out in WT or HMCES KO RASH-1C cells, and cells were assayed for GFP loss (A) or IgM loss (B) after being induced with dox for 4 d. (C,D) Deletion frequencies in the HTS7 (C) and VDJ (D) regions from WT and HMCES KO RASH-1C cells with either APE1 or APE2 knocked out. (E,F) Distribution of deletion lengths in the HTS7 (E) and VDJ (F) regions from WT and HMCES KO RASH-1C cells with either APE1 or APE2 knocked out. (G,H) Point mutation frequencies in the HTS7 (G) and VDJ (H) regions from WT and HMCES KO RASH-1C cells with either APE1 or APE2 knocked out. Throughout the figure, data are presented with the bars representing mean and the error bars as ±SD. Statistical significance was calculated using one-way ANOVA with Dunnett's post-test. (***) P-value < 0.001, (**) P-value < 0.01, (*) P-value < 0.05, (ns) not significant.
Mismatch repair factors contribute to deletions caused by HMCES deficiency
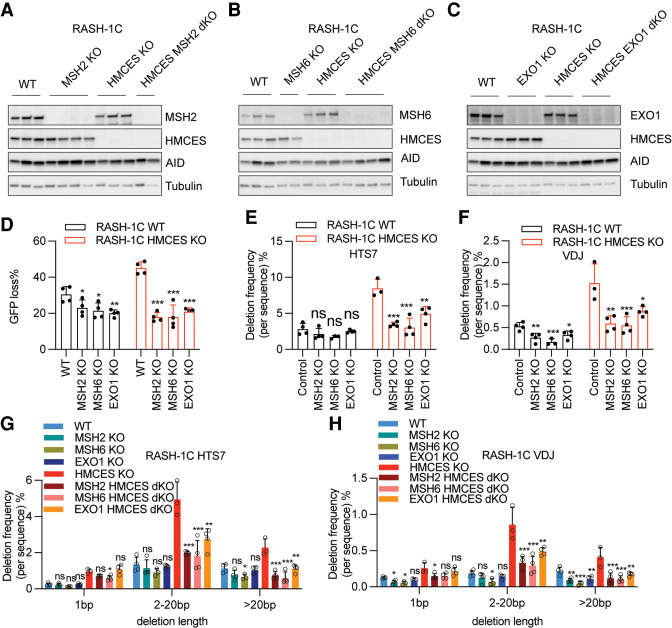
The data presented above indicate that a pathway involving UNG and APE2 contributes to the excess deletions that arise in the absence of HMCES. Because UNG and MMR factors act in concert in some SHM repair pathways to generate G/C transversion and A/T mutations and in CSR to generate DSBs (Frieder et al. 2009; Schrader et al. 2009; Methot and Di Noia 2017; Pilzecker and Jacobs 2019), we considered the possibility that they might also collaborate in the creation of deletions. We therefore assessed the consequences of knocking out MSH2 and MSH6 (which together mediate U:G mismatch recognition to initiate MMR) as well as EXO1 (which acts downstream from MSH2/6 to resect one DNA strand and create ssDNA gaps) (Fig. 6A–C). In both WT and HMCES KO RASH-1C cells, MSH2, MSH6, and EXO1 deficiency each reduced GFP loss, with larger effects observed in the HMCES KO cells (Fig. 6D). In line with these results, knockout of MSH2, MSH6, and EXO1 significantly and consistently reduced deletion frequencies in the HMCES-deficient context, with the effect strongest for MSH2 and MSH6 loss (Fig. 6E,F). Loss of these factors also appears to reduce deletions in HMCES-sufficient cells (Fig. 6E,F). As was the case for APE2, MSH2, MSH6, and EXO1 contributed strongly to intermediate and long deletions but had modest or insignificant effects on 1-bp deletions (Fig. 6G,H). Loss of the MMR factors reduced total point mutation frequencies as expected, primarily by reducing the frequency of G/C transversion mutations with a less consistent effect on A/T mutations (Supplemental Fig. S7A–H). Hence, the MMR pathway contributes less strongly to A/T mutagenesis in Ramos than in germinal center B cells (Bardwell et al. 2004; Rada et al. 2004; Shen et al. 2006), providing a plausible explanation for the known underrepresentation of A/T mutations in the Ramos SHM mutation spectrum (Sale and Neuberger 1998). Interestingly, we observed consistent decreases in insertion frequencies upon knockout of MSH2, MSH6, or EXO1 in HMCES-deficient RASH-1 cells, suggesting that the MMR pathway contributes to insertions as well as deletions when HMCES is absent (Supplemental Fig. S7I,J). In summary, our results demonstrate that MSH2, MSH6, and EXO1 contribute strongly to the generation of excessive deletions >1 bp in length that occurs in the absence of HMCES.
Figure 6.
Mismatch repair factors contribute to deletions caused by HMCES deficiency. (A–C) MSH2, MSH6, or EXO1 was knocked out in WT or HMCES KO RASH-1C cells. Western blots of MSH2 (A), MSH6 (B), and EXO1(C) in WT or HMCES KO RASH-1C cells. (D) MSH2, MSH6, or EXO1 was knocked out in WT or HMCES KO RASH-1C cells, and cells induced with dox for 4 d were assayed for GFP loss. (E,F) Deletion frequencies in the HTS7 (E) and VDJ (F) regions from WT and HMCES KO RASH-1C cells with MSH2, MSH6, or EXO1 knocked out. (G,H) Distribution of deletion lengths in the HTS7 (G) and VDJ (H) regions from WT or HMCES KO RASH-1C cells with MSH2, MSH6, or EXO1 knocked out. Throughout the figure, data are presented with the bars representing mean and the error bars as ±SD. Statistical significance was calculated using one-way ANOVA with Dunnett's post-test. (***) P-value < 0.001, (**) P-value < 0.01, (*) P-value < 0.05, (ns) not significant.
HMCES deficiency leads to increased deletions in mouse germinal center B cells
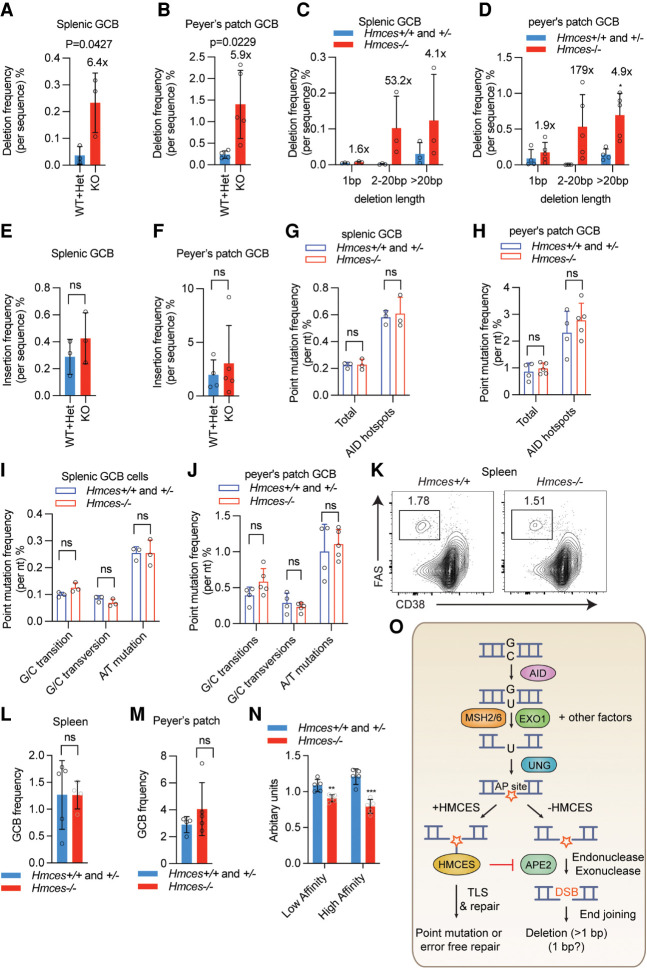
Hmces−/− mice were previously shown to exhibit normal parameters of hematopoiesis and normal numbers of splenic B cells, T cells, and germinal center B cells after immunization, but have a B cell-intrinsic defect in CSR (Shukla et al. 2020). We characterized SHM in Hmces−/− and control Hmces+/− and Hmces+/+ mice by analyzing mutations that accumulated in the region downstream from the IGH J4 gene segment in germinal center B cells from spleens of sheep red blood cell-immunized mice and Peyer's patches (PPs) of unimmunized mice. This analysis revealed an approximately sixfold increase in deletions in both the splenic and PP-derived B cells that was due almost entirely to increases in deletions >1 bp in length (Fig. 7A–D). This striking increase in deletions occurred in the absence of significant changes in the frequency of insertions or point mutations or significant alterations in the point mutation spectrum (Fig. 7E–J).
Figure 7.
HMCES deficiency leads to increased deletions in mouse germinal center B cells. (A,B) Deletion frequencies in the region downstream from the Igh J4 gene segment (JH4 intron) in germinal center B (GCB) cells from either the spleen (A) or Peyer's patch (B) of Hmces−/− and control Hmces+/− and Hmces+/+ mice. (C,D) Distribution of deletion lengths in the JH4 intron in splenic (C) and Peyer's patch GCB (D) cells from Hmces−/− and control Hmces+/+ and Hmces+/− mice. (E,F) Insertion frequencies in the JH4 intron in splenic (E) and Peyer's patch GCB (F) cells from Hmces−/− and control Hmces+/+ and Hmces+/− mice. (G,H) Point mutation frequencies in the JH4 intron in splenic (G) and Peyer's patch GCB (H) cells from Hmces−/− and control Hmces+/+ and Hmces+/− mice. (I,J) Mutation frequencies for G/C transitions, G/C transversions, and A/T mutations in the JH4 intron in splenic (I) or Peyer's patch GCB (J) cells from Hmces−/− and control Hmces+/+ and Hmces+/− mice. (K) Flow cytometry plots of GCB cells gated as the FAS+ CD38− population among total B cells in spleens of Hmces+/+ and Hmces−/− mice 14 d after immunization with NP-CGG. (L,M) Bar graphs showing the frequencies of GCB cells in Hmces+/+ or Hmces+/− and Hmces−/− mice in spleens (L) or Peyer's patches (M). (N) Bar graphs quantifying the levels of low- and high-affinity IgM antibodies measured by ELISA in the sera of Hmces−/− and control Hmces+/+ and Hmces+/− mice. NP-BSA with a conjugation ratio of 27 and 2 was used to detect low-and high-affinity antibodies, respectively. (O) Working model for the mechanism by which HMCES suppresses deletions during SHM. After AID acts, MSH2/6, EXO1, and likely other factors can resect one strand to expose a region of ssDNA containing a U residue, on which UNG can act to create an abasic site (AP site). Cleavage of this vulnerable intermediate by APE2 would result in a DNA double-strand break (DSB), which would be prone to end resection by exonucleases (including APE2 itself) and to generating a deletion. HMCES protects against this outcome by forming a covalent cross-link with the abasic site, thereby blocking cleavage by APE2. The events that occur subsequent to HMCES cross-linking are not known but could involve translesion synthesis (TLS) by an error-prone polymerase. The pathway depicted here is just one of the pathways depicted in Figure 1A (the second from the top). Throughout the figure, data are presented with the bars representing mean and the error bars as ±SD. Statistical significance was calculated using two-tailed Student's t-test. (***) |P-value < 0.001, (**) P-value < 0.01, (ns) not significant.
We reasoned that the generation of excessive deletions in Hmces-deficient mice might compromise the humoral immune response and therefore tested the antigen-specific immune response. While the frequency of germinal center B cells was normal in 4-hydroxyl nitrophenyl (NP)-conjugated ovalbumin (NP-OVA) immunized Hmces−/− mice (Fig. 7K–M), the NP hapten-specific serum antibody response was blunted, particularly for high-affinity antibody (Fig. 7N). We conclude that HMCES protects mouse germinal center B cells and human Ramos B cells from the accumulation of deletions in SHM target regions during SHM, thereby facilitating the antigen-specific antibody response. Furthermore, we conclude that HMCES exerts this protective effect while having modest or insignificant effects on other aspects of the SHM reaction.
Discussion
CSR targets long, repetitive, AID hotspot-rich switch regions and is designed to generate dsDNA breaks and deletions. These deletions can occur productively between two switch regions or nonproductively within a single switch region, with the latter readily tolerated because switch regions lie within introns (Chen et al. 2001; Reina-San-Martin et al. 2003; Chaudhuri and Alt 2004). While SHM also generates deletions, they accumulate to much lower levels in an in-frame, productive IGH V region than in a matched, out-of-frame V region, indicating that they are strongly selected against, presumably because of the damage they inflict on the open reading frame (Yeap et al. 2015). Insertions occur at lower frequencies than deletions during SHM and therefore appear to be less of a threat to reading frame integrity (Yeap et al. 2015). Evolution of a mechanism to specifically limit deletions during SHM without interfering with mutation accumulation would therefore potentially be advantageous for effective humoral immunity. Our findings demonstrate that such a mechanism does indeed exist in both human and mouse B cells, that it is mediated by HMCES acting downstream from UNG, and that in the absence of this mechanism, antibody affinity maturation is compromised. A particularly intriguing aspect of our findings is that the primary mechanisms of action of HMCES in SHM and CSR are distinct—the former but not the latter requiring the abasic site cross-linking residue C2. Hence, as discussed below, the function of HMCES during SHM is likely to resemble more closely its abasic site-protective function during DNA replication than its end-joining-promoting function during CSR.
The RASH cell lines described here provide multiple features that should facilitate future studies of SHM. Tightly regulated expression of a highly active form of AID combined with insertion of a sensitized SHM reporter cassette into the IGH locus supports robust, facile, and rapid analysis of SHM in a physiological genomic context. These features, combined with efficient CRISPR/Cas9 targeting of the Ramos genome, allow for rapid generation and analysis of mutant phenotypes and should enable genome-wide screens for factors involved in SHM. Use of a constitutive, heterologous promoter to drive HTS7-GFP expression protects reporter transcription against manipulations that alter the transcriptional activity of endogenous IGH locus control elements and distinguishes RASH from the Ramos IGH locus reporter system of Wang et al. (2014). Inefficient mutation of A/T residues and the reduced contribution of MMR factors to A/T mutagenesis in Ramos cells relative to GCB cells represents one potential limitation of the RASH assay system.
Our analysis of HMCES mutants revealed several differences between the protein features necessary for its function in SHM as compared with CSR and at DNA replication forks. The HMCES C-terminal region and the PIP motifs harbored therein are dispensable for SHM and CSR but required for HMCES function in DNA replication, while C2 is required for SHM and function in DNA replication but not CSR (Mohni et al. 2019; Shukla et al. 2020). The ssDNA binding surface is required for all identified HMCES activities but is more resilient to mutation in SHM than in CSR or DNA replication. R212E and double mutants of R98 and R212 to A or E reduce or eliminate HMCES function in SHM, demonstrating the essential contribution of these ssDNA binding residues. However, R98A, R98E, and R212A single mutants retain substantial function in SHM but show defects in ssDNA binding in vitro (Halabelian et al. 2019; Mohni et al. 2019), and R98E loses function in DNA repair after ionizing radiation (Mohni et al. 2019), while R212A fails to reconstitute CSR (Shukla et al. 2020). Furthermore, several residues involved in binding the ssDNA–dsDNA junction in the HMCES–DNA crystal structure (W81, F92, and R106) (Halabelian et al. 2019) are not required for SHM despite the fact that mutation of the corresponding residues in yedK compromises DNA binding (Wang et al. 2019). These comparisons argue that the HMCES–DNA interaction is strongly context-dependent and suggest the possibility that during SHM, other interactions or factors render HMCES less dependent on canonical modes of DNA binding. We cannot rule out that protein overexpression observed in our reconstitution system augments the function of some HMCES mutants, although, with one exception discussed below, overexpression of WT or mutant HMCES proteins has no discernable effect on GFP loss in cells expressing endogenous HMCES (Fig. 3C; Supplemental Fig. S4J,K).
Mutation of autopeptidase catalytic residues C2, E127, and H210 yielded strikingly divergent results, with C2 required and E127 and H210 dispensable for HMCES function in SHM. Importantly, of these three residues, only C2 is required for abasic site cross-linking, since E. coli yedK mutants at residue E105 or H160 (which correspond to HMCES E127 and H210) retain substantial cross-linking activity (Thompson et al. 2019; Wang et al. 2019). Hence, our data argue strongly that HMCES cross-linking activity but not autopeptidase activity is required for function during SHM and that a cellular peptidase activity can remove the N-terminal amino acid of HMCES to expose the amino group on C2 required for cross-linking. Interestingly, HMCES E127A and yedK E105A mutants exhibit strongly increased ssDNA binding activity in vitro, apparently through neutralization of charge repulsion between the glutamate side chain and DNA phosphate backbone (Halabelian et al. 2019; Wang et al. 2019). The significant reduction in GFP loss that we observed due to overexpression of HMCES E127A in WT cells (Supplemental Fig. S4I) might therefore be explained by increased HMCES ssDNA binding and protection of abasic sites. Avid ssDNA binding might help HMCES displace UNG from abasic sites, which are bound by UNG even more tightly than its U substrate (Parikh et al. 1998). Release of glycosylases from abasic sites is the rate-limiting step in BER (Jacobs and Schär 2012).
HMCES has previously been demonstrated to work through two distinct mechanisms. In the first, HMCES cross-links to abasic sites at stalled DNA replication forks, thereby preventing the genome-destabilizing action of endonucleases and translesion polymerases (Mohni et al. 2019; Mehta et al. 2020; Srivastava et al. 2020). In the second, HMCES facilitates alternative end joining by a mechanism that appears to involve ssDNA end binding and protection (Shukla et al. 2020). Multiple aspects of our findings indicate that the primary function of HMCES during SHM more closely parallels the first mechanism than the second. First, HMCES function in SHM is completely dependent on abasic site cross-linking residue C2. Second, HMCES deficiency has no detectable phenotype in the absence of UNG, consistent with HMCES acting on abasic sites. Third, during SHM, HMCES protects against a deletion pathway that is strongly dependent on APE2, which cleaves DNA at abasic sites. The HMCES–abasic site cross-link blocks endonucleolytic cleavage (e.g., by APE1) (Mohni et al. 2019), and APE2 is the dominant AP endonuclease involved in point mutation and deletion generation during SHM (Sabouri et al. 2009; Stavnezer et al. 2014). Hence, by cross-linking to abasic sites generated by UNG, HMCES could regulate the action of APE2 to protect against deletions. Fourth, our finding that HMCES selectively regulates deletion formation during SHM is fully consistent with the established role for HMCES–abasic site cross-links in suppressing dsDNA breaks during DNA replication (Mohni et al. 2019), including dsDNA breaks triggered by the replication-coupled action of the cytidine deaminase APOBEC3A (Mehta et al. 2020).
The function of HMCES in SHM and CSR is united in one respect: its independence from the HMCES C-terminal region and presumably from recruitment by PCNA to replication forks. AID initiates both SHM and CSR in the G1 phase of the cell cycle, and UNG is also thought to act predominantly in G1 during SHM (Sharbeen et al. 2012; Casellas et al. 2016; Wang et al. 2017). It is plausible that HMCES also acts in G1 during SHM, although this remains to be determined.
The excess deletions caused by loss of HMCES are almost completely eliminated by deficiency in APE2, MSH2, or MSH6, with the exception of 1-bp deletions (Figs. 5, 6), indicating that the mechanisms that generate most deletions in the absence of HMCES involve all three of these factors. Cooperation between the BER and MMR pathways is well established for both CSR and SHM (Methot and Di Noia 2017). In CSR, MMR-dependent exonuclease activity is thought to facilitate the conversion of ssDNA nicks created by BER enzymes into dsDNA breaks (Stavnezer and Schrader 2006; Methot and Di Noia 2017). In SHM, a noncanonical hybrid BER/MMR pathway appears to be responsible for about half of transversion mutations at G/C and is proposed to involve MMR-mediated strand resection to create ssDNA gaps in which the retained DNA strand contains U (Frieder et al. 2009; Thientosapol et al. 2017, 2018; Pilzecker and Jacobs 2019; Feng et al. 2020). The importance of recruitment of MMR factors for efficient SHM was highlighted in recent studies of Fam72-deficient mice and cells in which hyperactive UNG results in dramatic reductions in SHM and CSR (Feng et al. 2021; Rogier et al. 2021), apparently by quickly eliminating U:G mismatches and thereby subverting MMR engagement (Feng et al. 2021).
Based on our findings and these considerations, we propose a model in which HMCES acts to protect a particularly vulnerable ssDNA intermediate created during SHM (Fig. 7O). In this model, uracils generated by AID trigger MMR factor-dependent processing to generate an exposed ssDNA region that either contains U or is deaminated by AID to generate U. After UNG converts the U to an abasic site, HMCES binds the ssDNA region and forms a covalent cross-link with the abasic site, thereby preventing cleavage by APE2 and the generation of a dsDNA break, which could readily yield a deletion upon repair. Notably, APE2 possesses robust 3′-to-5′ exonuclease activity (Burkovics et al. 2006, 2009) that could contribute to the generation of deletions. How the HMCES–abasic site cross-link is subsequently processed during SHM and with what outcome is not known; ubiquitin-mediated proteolysis of HMCES followed by translesion polymerase gap filling is one plausible possibility. This model, while speculative, accommodates the strong ssDNA binding preference of HMCES, the requirement for HMCES C2, HMCES's dependency on UNG to exert its effects, and the major contribution made by both APE2 and MMR factors to the deletions that accumulate in the absence of HMCES. The model is not meant to exclude the participation of HMCES in other pathways and in particular might not explain the origin of many of the 1-bp deletions that arise in the absence of HMCES. One previously proposed source of deletions during SHM is dsDNA breaks created by closely opposed nicks on opposite DNA strands (Yeap and Meng 2019).
Our data and that of Shukla et al. (2020) indicate that the principal function of HMCES during SHM is a C2-dependent suppression of deletions and during CSR is a C2-independent facilitation of alternative end joining. These functions reflect the different mechanistic imperatives of SHM and CSR, with the former proceeding primarily through ssDNA lesions and seeking to avoid dsDNA breaks and the latter requiring a dsDNA break intermediate. Existing data are consistent, however, with the possibility that HMCES can cross-link to abasic sites in switch regions and suppress deletions during CSR. Similarly, we cannot rule out the possibility that HMCES sometimes facilitates end joining of dsDNA breaks during SHM.
The SRAP domain is evolutionarily ancient and traces its association with DNA repair back to prokaryotes (Aravind et al. 2013). Our findings demonstrate that HMCES, which contains the only known SRAP domain in mammals, has been co-opted for a specific purpose in SHM: that of reducing the frequency of deletions. Elimination of HMCES has no consistent effect on any other parameter of SHM that we have examined. Deletions are selected against and, as suggested by our findings, might be detrimental to the humoral immune response if too frequent. However, they can make important contributions to novel antibody specificities such as broadly neutralizing antibodies against HIV-1 (Kepler et al. 2014), suggesting that their complete suppression might not be advantageous. It will now be important to determine whether HMCES plays a role in protecting non-Ig portions of the genome from AID-initiated deletions and in suppressing other types of instability such as chromosomal translocations that are associated with the action of AID (Casellas et al. 2016).
Materials and methods
Mice
Hmces-deficient mice were generated as previously described (Kweon et al. 2017; Shukla et al. 2020) and maintained on a C57BL/6J genetic background for at least eight generations. All mice used in the studies here were 8–21 wk of age and were housed in the animal facility at the La Jolla Institute for Immunology. Male mice were used in the studies. All procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee of the La Jolla Institute for Immunology.
Cell culture
Ramos cells and cell lines derived from Ramos were grown in RPMI-1640 (Gibco 11875) supplemented with 10% FBS (Gemini) and 0.5 mg/mL penicillin–streptomycin–glutamine (Gibco 10378016) at 37°C and 5% CO2. 293T cells were grown at 37°C and 5% CO2 in DMEM (Gibco 10566) supplemented in the same fashion as Ramos cell media.
Plasmids and antibodies
px458 and lenti-SpCas9-hygro were obtained from AddGene (48138 and 104995). px458-mcherry was generated by replacing GFP in px458 with mCherry. pLW44 was generated by removing Cas9 from px458-mCherry, resulting in a 3.9-kb plasmid expressing a single guide RNA (sgRNA) and mCherry. pLW44 could be delivered into ∼20% of RASH-1C cells, compared with ∼1% when a larger plasmid (px458-mCherry, 9.3 kb) expressing Cas9, sgRNA, and mCherry was used (data not shown), allowing gene targeting with high efficiency in RASH-1C cells. Full-length HMCES was amplified from pLV-EF1a-IRES GFP-human HMCES FL (Shukla et al. 2020) and cloned into retroviral expression vector pMSCV (Papavasiliou and Schatz 2002). Point mutations and truncations of HMCES were introduced using In-Fusion cloning according to the manufacturer's protocol. The GFP7-E vector was generated by replacing the CMV promoter of GFP7 (Senigl et al. 2019) with EF1a promoter. cDNA encoding human UNG2 was amplified by PCR from Ramos cDNA and cloned into pMSCV.
The following primary antibodies were used for Western blotting: anti-AID monoclonal antibody (Thermo Fisher 39-2500), anti-HMCES (Sigma HPA044968), antitubulin (Sigma T5168), anti-MSH2 (D24B5; Cell Signaling Technology 2017), anti-MSH6 (P150; Cell Signaling Technology 3995), anti-EXO1 (Proteintech 16253-1-AP), and anti-UNG (Novus Biologicals NBP1-49985).
Lentiviral and retroviral transduction
293T cells in six-well plates were grown to 50%–80% confluence and transfected with 1 μg of lentiviral plasmid, 0.6 μg of psPAX2 (AddGene 12260), and 0.4 μg of pMD2.G (AddGene 12259) or 1 μg of retroviral plasmid and 1 μg of packaging plasmid pkat2 with JetPrime reagent (Polyplus 114-07) according to the manufacturer's protocol. At 48 h after transfection, lentivirus or retroviral-containing media were collected and filtered through a 0.45-μm filter before being used to infect cells. Ramos cells were transduced as reported previously (Dinesh et al. 2020).
Generation of cell lines
Generation of RASH-1 was performed according to the method described by Senigl et al. (2019) with slight modifications. One million AID7.3in cells (Dinesh et al. 2020) were infected with virus expressing GFP7-E vector at MOI < 0.01. Two days after infection, blasticidin (final concentration 5 μg/mL) was added. Two weeks later, GFP-positive cells were sorted in a single-cell sort mode into 96-well plates to obtain single-cell clones. Clones were cultured for 18 d in 96-well plates and then 40 µL of cells was transferred to a new plate and treated with 200 ng/mL dox for 6 d, followed by GFP loss analysis. Clones that yielded a high percentage of GFP loss were analyzed for their GFP7-E integration site using splinkerette-PCR as described by Senigl et al. (2019).
Generation of RASH-2 was performed according to the protocol described by Koch et al. (2018) with modifications. px458-mcherry expressing an sgRNA (ATAACACCAACCATACACCC) targeting upstream of the IGH variable region and donor plasmid containing EF1a promoter-HTS7-T2A-GFP (see Fig. 1A) flanked by ∼1000-bp homolog arms of the rearranged IGH variable region were delivered into AID7.3in cells by electroporation using Amaxa Nucleofector II (Lonza) with program O-006 according to the manufacturer's instructions. Homemade 1M buffer was used for electroporation according to Parreno et al. (2016). One day later, mCherry-positive cells were sorted and then expanded for 12 d, and GFP-positive cells were sorted into a 96-well plate to obtain single-cell clones. Clones were cultured for 15 d in a 96-well plate and then 50 µL of cells was transferred to a new plate and treated with 200 ng/mL dox for 6 d, followed by GFP loss analysis. Clones that yielded a high percentage of GFP loss were selected and validated by Sanger sequencing of the PCR product generated using primers (forward: ATGGTCCTGGTGGAGTTCGTGAC, and reverse: GCCGCATCGGTGATTCGGAACAGAAT) surrounding the target region.
RASH-1, RASH-2, and A23 (Dinesh et al. 2020) were infected with lentivirus expressing spCas9-hygro. Two days after infection, 0.5 mg/mL hygromycin B was added. Two weeks later, single cells were sorted into a 96-well plate, and clones expressing Cas9 were identified by Western blot. RASH-1 clones, RASH-2 clones, and A23 clones constitutively expressing Cas9 were named RASH-1C, RASH-2C, and A23-Cas9, respectively.
CRISPR/Cas9 editing in Ramos and Ramos derivative lines
Guide RNAs targeting human AICDA, HMCES, UNG, APEX1, APEX2, MSH2, MSH6, and EXO1 were designed by the Broad Institute's sgRNA designer (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design). Guide RNA sequences were as follows: sgAICDA (ACAGCCTCTTGATGAACCGG), sgHMCES-1 (GCAGTCGAGACAGAAGCACT), sgHMCES-2 (TGCGCCTACCAGGATCGGCG), sgUNG (GCGGCCCGCAACGTGCCCGT), sgAPEX1 (TAACGGGAATGCCGAAGCGT), sgAPEX2 (TAACCCTCAACGATAGCCAG), sgMSH2 (ACAGTGCGCCTTTTCGACCG), sgMSH6 (CCTGGCTAACTATAGTCGAG), and sgEXO1 (TCAGGGGGTAGATTGCCTCG).
Guide RNAs were cloned into pLW44 or px458 and delivered into cells by electroporation. One to three days later, for RASH-Cas9 cells, GFP and mCherry double-positive cells were sorted either into 15-mL tubes as bulk KO cells or sorted into 96-well plates for single-KO clone selection. For Ramos and A23-Cas9 cells, GFP-positive cells were sorted either into 15-mL tubes as bulk KO cells or sorted into 96-well plates for single-knockout clone selection.
IgM and GFP loss assay
For the IgM loss assay in RASH cells, ∼1.0 × 106 cells treated with or without 200 ng/mL dox were stained with APC mouse antihuman IgM (BD 551062) diluted 1:100 in FACS buffer (1× PBS with 2% FBS) and incubated for 20 min at room temperature. The starting point for the IgM loss assay in A23 cells and Ramos was a sorted IgM+ population. Cells were cultured and then harvested at different time points and were again stained, and percentages of IgM+ and IgM− cells were measured in accordance with published guidelines on flow cytometry (Cossarizza et al. 2021). For the GFP loss assay, RASH cells and RASH-Cas9 cells treated with or without 200 ng/mL dox for 2–6 d as indicated were assayed for the percentage of GFP+ and GFP− cells in accordance with published guidelines on flow cytometry (Cossarizza et al. 2021).
High-throughput sequencing
Analyses of mutations, deletions, and insertions were performed using a protocol modified from Illumina protocol “16S metagenomic sequencing library preparation.” Genomic DNA was extracted using DNeasy blood and tissue kit (Qiagen) according to the manufacturer's instructions. HTS7 was amplified using the oligos (forward) TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGTTCCAAAGTAGACCCAGCCTTCTAA and (reverse) GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGATTCTCCTCCACATCACCACAG. IGH VDJ was amplified using the oligos (forward) TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGGAATGCGGATATGAAGATATGAG and (reverse) GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGAGTAGCAGAGAACAGAGGCCCTAGA. IGLV2-14J2 was amplified using the oligos (forward) TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCACTGACTCACTGGCATGTATTTCT and (reverse) GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGCTGACCACAAGTTGAGACAAGATA.
PCR reactions were performed using NEBNext high-fidelity 2× PCR master mix (New England Biolabs) under the following conditions: 3 min at 98°C, followed by 25 cycles of 10 sec at 98°C, 30 sec at 60°C, 30 sec at 72°C, and finally 2 min at 72°C in a 25-µL reaction.
The JH4 intron was amplified using nested PCR. The first amplification reaction was performed under the following conditions: 3 min at 98°C, followed by 13 cycles of 10 sec at 98°C,30 sec at 66°C, 30 sec at 72°C, and finally 2 min at 72°C using the oligos (forward) GGAATTCGCCTGACATCTGAGGACTCTGC and (reverse) CTGGACTTTCGGTTTGGTG in a 50-µL reaction. Five microliters was used as a template for the second PCR, which was performed under the following conditions: 3 min at 98°C, followed by 18 cycles of 10 sec at 98°C, 30 sec at 56°C, 30 sec at 72°C, and finally 2 min at 72°C, using the oligos (forward) TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGGTCAAGGAACCTCAGTCA and (reverse) GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTCTCTAGACAGCAACTAC.
PCR products were multiplexed using Nextera XT index kit (Illumina). Sequencing on the Illumina Miseq platform (two 300-bp paired end sequencing) using Miseq reagent kit V3 was performed at the Yale Center for Genome Analysis.
Bioinformatic analysis of DNA sequence data
Sequence reads were analyzed using a custom pipeline. Paired-end reads were joined using the fastq-join tool requiring that the reads from both sides have at least a 10-bp overlap with mismatch rate ≤8%. Joined reads were aligned to their respective reference sequence using Burrows-Wheeler aligner (BWA) using default parameters. Python utility Pysamstats was then used to calculate statistics against genome positions based on sequence alignments from the aligned BAM file. Position-wise statistics were generated for reads with mapping quality of at least 60 and minimum base quality of 30.
For each sample, the aligned file (binary encoded BAM format) was converted to TSV using Java-based utility sam2tsv. A custom AWK script was used to generate the per-read-based summary files for each observed variation (deletions, insertions, and point mutations). For insertions and point mutations, only nucleotide bases with a phred quality score ≥30 were considered. For deletion events, only reads with average phred score ≥30 were considered, since the bases in question were deleted.
A custom R script was used to identify AID hotspots from the position-based stats files generated by Pysamstats.
ELISA
For NP-specific enzyme-linked immunosorbent assay (ELISA), polystyrene plates were coated overnight with NP2-BSA (conjugation ratio of 2 for high affinity) or NP27-BSA (conjugation ratio of 27 for low affinity) diluted to 10 µg/mL with PBS (Biosearch). The following day, 200 µL of blocking solution (1% BSA, 0.05% sodium azide in PBS) was added, and the plates were incubated for 2 h at 37°C in an incubator, or overnight at 4°C. After blocking, 50 µL of threefold serially diluted serum (starting dilution 1:20) in dilution buffer (0.1% BSA, 0.05% sodium azide in PBS) was added to each well, and plates were incubated for 2 h at 37°C. Plates were then washed three times with deionized water, 50 µL of the alkaline phosphatase-conjugated antimouse IgM secondary antibody (Bethyl antibody A90-101AP) diluted at 1:2500 was added, and plates were incubated for 2 h at 37°C. Plates were then washed three times with deionized water and 100 µL of p-nitrophenyl phosphate, disodium salt (PNPP from Sigma) substrate was added, and the reaction was allowed to develop for ∼20 min at room temperature before the absorbance was read at 405 nm (M2 spectramax spectrophotometer, Spectral Labs).
Statistical analysis
Data were subjected to statistical analysis and plotted using GraphPad Prism. Single comparisons were performed using two-tailed Student's t-test, whereas multiple comparisons were assessed by one-way ANOVA with Dunnett's multiple comparison test. For all analyses, P-value < 0.05 (*), P-value < 0.01 (**), and P-value < 0.001 (***). Results are reported as mean ± SD as indicated in the figure legends.
Data availability
The mutation sequencing data sets generated during this study are available under the NCBI Bioproject PRJNA820976.
Supplementary Material
Acknowledgments
This work was funded by research funds from a Gruber Science Fellowship and a National Science Foundation Graduate Research Fellowship (to R.K.D.), Leukemia and Lymphoma Society grant 5463-18 (to V.S.), and National Institutes of Health grants R01 AI127642 (to D.G.S.), K99/R00 CA248835 (to V.S.), and R35 CA210043 (to A.R.).
Author contributions: L.W., V.S., D.G.S., and A.R. designed experiments, which were performed by L.W., V.S., R.K.D., and D.X. Computational analyses were performed by A.D.Y. L.W., V.S., and D.G.S. assembled the figures and wrote the manuscript with input from the other authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349438.122.
Competing interest statement
The authors declare no competing interests.
References
- Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. 2013. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 152: 417–429. 10.1016/j.cell.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Anand S, Iyer LM. 2013. Novel autoproteolytic and DNA-damage sensing components in the bacterial SOS response and oxidized methylcytosine-induced eukaryotic DNA demethylation systems. Biol Direct 8: 20. 10.1186/1745-6150-8-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell PD, Woo CJ, Wei K, Li Z, Martin A, Sack SZ, Parris T, Edelmann W, Scharff MD. 2004. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat Immunol 5: 224–229. 10.1038/ni1031 [DOI] [PubMed] [Google Scholar]
- Boboila C, Yan C, Wesemann DR, Jankovic M, Wang JH, Manis J, Nussenzweig A, Nussenzweig M, Alt FW. 2010. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J Exp Med 207: 417–427. 10.1084/jem.20092449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm EM, Washington MT. 2016. R.I.P. to the PIP:PCNA-binding motif no longer considered specific: PIP motifs and other related sequences are not distinct entities and can bind multiple proteins involved in genome maintenance. Bioessays 38: 1117–1122. 10.1002/bies.201600116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bross L, Fukita Y, McBlane F, Démolliére C, Rajewsky K, Jacobs H. 2000. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity 13: 589–597. 10.1016/S1074-7613(00)00059-5 [DOI] [PubMed] [Google Scholar]
- Buerstedde JM, Alinikula J, Arakawa H, McDonald JJ, Schatz DG. 2014. Targeting of somatic hypermutation by immunoglobulin enhancer and enhancer-like sequences. PLoS Biol 12: e1001831. 10.1371/journal.pbio.1001831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkovics P, Szukacsov V, Unk I, Haracska L. 2006. Human Ape2 protein has a 3′–5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucl Acids Res 34: 2508–2515. 10.1093/nar/gkl259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkovics P, Hajdu I, Szukacsov V, Unk I, Haracska L. 2009. Role of PCNA-dependent stimulation of 3′-phosphodiesterase and 3′-5′ exonuclease activities of human Ape2 in repair of oxidative DNA damage. Nucl Acids Res 37: 4247–4255. 10.1093/nar/gkp357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casellas R, Basu U, Yewdell WT, Chaudhuri J, Robbiani DF, Di Noia JM. 2016. Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nat Rev Immunol 16: 164–176. 10.1038/nri.2016.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HHY, Pannunzio NR, Adachi N, Lieber MR. 2017. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 18: 495–506. 10.1038/nrm.2017.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Alt FW. 2004. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol 4: 541–552. 10.1038/nri1395 [DOI] [PubMed] [Google Scholar]
- Chen X, Kinoshita K, Honjo T. 2001. Variable deletion and duplication at recombination junction ends: implication for staggered double-strand cleavage in class-switch recombination. Proc Natl Acad Sci 98: 13860–13865. 10.1073/pnas.241524898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossarizza A, Chang HD, Radbruch A, Abrignani S, Addo R, Akdis M, Andrä I, Andreata F, Annunziato F, Arranz E, et al. 2021. Guidelines for the use of flow cytometry and cell sorting in immunological studies (third edition). Eur J Immunol 51: 2708–3145. 10.1002/eji.202170126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh RK, Barnhill B, Ilanges A, Wu L, Michelson DA, Senigl F, Alinikula J, Shabanowitz J, Hunt DF, Schatz DG. 2020. Transcription factor binding at Ig enhancers is linked to somatic hypermutation targeting. Eur J Immunol 50: 380–395. 10.1002/eji.201948357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J, Neuberger MS. 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature 419: 43–48. 10.1038/nature00981 [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem 76: 1–22. 10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Rada C, Neuberger MS. 2006. SMUG1 is able to excise uracil from immunoglobulin genes: insight into mutation versus repair. EMBO J 25: 585–595. 10.1038/sj.emboj.7600939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Seija N, Di Noia JM, Martin A. 2020. AID in antibody diversification: there and back again. Trends Immunol 41: 586–600. 10.1016/j.it.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Li C, Stewart JA, Barbulescu P, Desivo NS, Alvarez-Quilon A, Pezo RC, Perera MLW, Chan K, Tong AHY, et al. 2021. FAM72A antagonizes UNG2 to promote mutagenic repair during antibody maturation. Nature 600: 324–328. 10.1038/s41586-021-04144-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French DL, Laskov R, Scharff MD. 1989. The role of somatic hypermutation in the generation of antibody diversity. Science 244: 1152–1157. 10.1126/science.2658060 [DOI] [PubMed] [Google Scholar]
- Frieder D, Larijani M, Collins C, Shulman M, Martin A. 2009. The concerted action of Msh2 and UNG stimulates somatic hypermutation at A · T base pairs. Mol Cell Biol 29: 5148–5157. 10.1128/MCB.00647-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halabelian L, Ravichandran M, Li Y, Zeng H, Rao A, Aravind L, Arrowsmith CH. 2019. Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition. Nat Struct Mol Biol 26: 607–612. 10.1038/s41594-019-0246-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ML, Hazra TK, Mitra S. 2008. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res 18: 27–47. 10.1038/cr.2008.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs AL, Schär P. 2012. DNA glycosylases: in DNA repair and beyond. Chromosoma 121: 1–20. 10.1007/s00412-011-0347-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler TB, Liao HX, Alam SM, Bhaskarabhatla R, Zhang R, Yandava C, Stewart S, Anasti K, Kelsoe G, Parks R, et al. 2014. Immunoglobulin gene insertions and deletions in the affinity maturation of HIV-1 broadly reactive neutralizing antibodies. Cell Host Microbe 16: 304–313. 10.1016/j.chom.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch B, Nijmeijer B, Kueblbeck M, Cai Y, Walther N, Ellenberg J. 2018. Generation and validation of homozygous fluorescent knock-in cells using CRISPR–Cas9 genome editing. Nat Protoc 13: 1465–1487. 10.1038/nprot.2018.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon SM, Zhu B, Chen Y, Aravind L, Xu SY, Feldman DE. 2017. Erasure of Tet-oxidized 5-methylcytosine by a SRAP nuclease. Cell Rep 21: 482–494. 10.1016/j.celrep.2017.09.055 [DOI] [PubMed] [Google Scholar]
- Liu M, Schatz DG. 2009. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol 30: 173–181. 10.1016/j.it.2009.01.007 [DOI] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature 451: 841–845. 10.1038/nature06547 [DOI] [PubMed] [Google Scholar]
- Mehta KPM, Lovejoy CA, Zhao R, Heintzman DR, Cortez D. 2020. HMCES maintains replication fork progression and prevents double-strand breaks in response to APOBEC deamination and abasic site formation. Cell Rep 31: 107705. 10.1016/j.celrep.2020.107705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot SP, Di Noia JM. 2017. Molecular mechanisms of somatic hypermutation and class switch recombination. Adv Immunol 133: 37–87. 10.1016/bs.ai.2016.11.002 [DOI] [PubMed] [Google Scholar]
- Mohni KN, Wessel SR, Zhao R, Wojciechowski AC, Luzwick JW, Layden H, Eichman BF, Thompson PS, Mehta KPM, Cortez D. 2019. HMCES maintains genome integrity by shielding abasic sites in single-strand DNA. Cell 176: 144–153.e13. 10.1016/j.cell.2018.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger MS. 2008. Antibody diversification by somatic mutation: from Burnet onwards. Immunol Cell Biol 86: 124–132. 10.1038/sj.icb.7100160 [DOI] [PubMed] [Google Scholar]
- Neuberger MS, Ehrenstein MR, Klix N, Jolly CJ, Yelamos J, Rada C, Milstein C. 1998. Monitoring and interpreting the intrinsic features of somatic hypermutation. Immunol Rev 162: 107–116. 10.1111/j.1600-065X.1998.tb01434.x [DOI] [PubMed] [Google Scholar]
- Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, Krokan HE. 1997. Nuclear and mitochondrial uracil–DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucl Acids Res 25: 750–755. 10.1093/nar/25.4.750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. 2000. Cell-cycle-regulated DNA double-strand breaks in somatic hypermutation of immunoglobulin genes. Nature 408: 216–221. 10.1038/35041599 [DOI] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. 2002. The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J Exp Med 195: 1193–1198. 10.1084/jem.20011858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. 1998. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil–DNA glycosylase with DNA. EMBO J 17: 5214–5226. 10.1093/emboj/17.17.5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parreno J, Delve E, Andrejevic K, Paez-Parent S, Wu PH, Kandel R. 2016. Efficient, low-cost nucleofection of passaged chondrocytes. Cartilage 7: 82–91. 10.1177/1947603515609399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilzecker B, Jacobs H. 2019. Mutating for good: DNA damage responses during somatic hypermutation. Front Immunol 10: 438. 10.3389/fimmu.2019.00438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Wang Q, Dose M, Pruett N, Kieffer-Kwon KR, Resch W, Liang G, Tang Z, Mathe E, Benner C, et al. 2014. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell 159: 1524–1537. 10.1016/j.cell.2014.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C, Di Noia JM, Neuberger MS. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell 16: 163–171. 10.1016/j.molcel.2004.10.011 [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B, Difilippantonio S, Hanitsch L, Masilamani RF, Nussenzweig A, Nussenzweig MC. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J Exp Med 197: 1767–1778. 10.1084/jem.20030569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roco JA, Mesin L, Binder SC, Nefzger C, Gonzalez-Figueroa P, Canete PF, Ellyard J, Shen Q, Robert PA, Cappello J, et al. 2019. Class-switch recombination occurs infrequently in germinal centers. Immunity 51: 337–350.e7. 10.1016/j.immuni.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogier M, Moritz J, Robert I, Lescale C, Heyer V, Abello A, Martin O, Capitani K, Thomas M, Thomas-Claudepierre AS, et al. 2021. Fam72a enforces error-prone DNA repair during antibody diversification. Nature 600: 329–333. 10.1038/s41586-021-04093-y [DOI] [PubMed] [Google Scholar]
- Ronai D, Iglesias-Ussel MD, Fan M, Li Z, Martin A, Scharff MD. 2007. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J Exp Med 204: 181–190. 10.1084/jem.20062032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaud P, Vincent-Fabert C, Saintamand A, Fiancette R, Marquet M, Robert I, Reina-San-Martin B, Pinaud E, Cogné M, Denizot Y. 2013. The IgH 3′ regulatory region controls somatic hypermutation in germinal center B cells. J Exp Med 210: 1501–1507. 10.1084/jem.20130072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouri Z, Okazaki IM, Shinkura R, Begum N, Nagaoka H, Tsuchimoto D, Nakabeppu Y, Honjo T. 2009. Apex2 is required for efficient somatic hypermutation but not for class switch recombination of immunoglobulin genes. Int Immunol 21: 947–955. 10.1093/intimm/dxp061 [DOI] [PubMed] [Google Scholar]
- Sale JE, Neuberger MS. 1998. TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity 9: 859–869. 10.1016/S1074-7613(00)80651-2 [DOI] [PubMed] [Google Scholar]
- Schrader CE, Guikema JE, Wu X, Stavnezer J. 2009. The roles of APE1, APE2, DNA polymerase β and mismatch repair in creating S region DNA breaks during antibody class switch. Philos Trans R Soc Lond 364: 645–652. 10.1098/rstb.2008.0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senigl F, Maman Y, Dinesh RK, Alinikula J, Seth RB, Pecnova L, Omer AD, Rao SSP, Weisz D, Buerstedde JM, et al. 2019. Topologically associated domains delineate susceptibility to somatic hypermutation. Cell Rep 29: 3902–3915.e8. 10.1016/j.celrep.2019.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharbeen G, Yee CW, Smith AL, Jolly CJ. 2012. Ectopic restriction of DNA repair reveals that UNG2 excises AID-induced uracils predominantly or exclusively during G1 phase. J Exp Med 209: 965–974. 10.1084/jem.20112379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Tanaka A, Bozek G, Nicolae D, Storb U. 2006. Somatic hypermutation and class switch recombination in Msh6−/−Ung−/− double-knockout mice. J Immunol 177: 5386–5392. 10.4049/jimmunol.177.8.5386 [DOI] [PubMed] [Google Scholar]
- Shukla V, Halabelian L, Balagere S, Samaniego-Castruita D, Feldman DE, Arrowsmith CH, Rao A, Aravind L. 2020. HMCES functions in the alternative end-joining pathway of the DNA DSB repair during class switch recombination in B cells. Mol Cell 77: 384–394.e4. 10.1016/j.molcel.2019.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Su D, Zhang H, Chen Z, Tang M, Nie L, Chen J. 2020. HMCES safeguards replication from oxidative stress and ensures error-free repair. EMBO Rep 21: e49123. 10.15252/embr.201949123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Schrader CE. 2006. Mismatch repair converts AID-instigated nicks to double-strand breaks for antibody class-switch recombination. Trends Genet 22: 23–28. 10.1016/j.tig.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Linehan EK, Thompson MR, Habboub G, Ucher AJ, Kadungure T, Tsuchimoto D, Nakabeppu Y, Schrader CE. 2014. Differential expression of APE1 and APE2 in germinal centers promotes error-prone repair and A:T mutations during somatic hypermutation. Proc Natl Acad Sci 111: 9217–9222. 10.1073/pnas.1405590111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Rothschild G, Pefanis E, Basu U. 2013. Transcriptional stalling in B-lymphocytes: a mechanism for antibody diversification and maintenance of genomic integrity. Transcription 4: 127–135. 10.4161/trns.24556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thientosapol ES, Sharbeen G, Lau KKE, Bosnjak D, Durack T, Stevanovski I, Weninger W, Jolly CJ. 2017. Proximity to AGCT sequences dictates MMR-independent versus MMR-dependent mechanisms for AID-induced mutation via UNG2. Nucl Acids Res 45: 3146–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thientosapol ES, Bosnjak D, Durack T, Stevanovski I, van Geldermalsen M, Holst J, Jahan Z, Shepard C, Weninger W, Kim B, et al. 2018. SAMHD1 enhances immunoglobulin hypermutation by promoting transversion mutation. Proc Natl Acad Sci 115: 4921–4926. 10.1073/pnas.1719771115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PS, Amidon KM, Mohni KN, Cortez D, Eichman BF. 2019. Protection of abasic sites during DNA replication by a stable thiazolidine protein-DNA cross-link. Nat Struct Mol Biol 26: 613–618. 10.1038/s41594-019-0255-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. 2012. Germinal centers. Annu Rev Immunol 30: 429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Wang M, Yang Z, Rada C, Neuberger MS. 2009. AID upmutants isolated using a high-throughput screen highlight the immunity/cancer balance limiting DNA deaminase activity. Nat Struct Mol Biol 16: 769–776. 10.1038/nsmb.1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Fan M, Kalis S, Wei L, Scharff MD. 2014. A source of the single-stranded DNA substrate for activation-induced deaminase during somatic hypermutation. Nat Commun 5: 4137. 10.1038/ncomms5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Kieffer-Kwon KR, Oliveira TY, Mayer CT, Yao K, Pai J, Cao Z, Dose M, Casellas R, Jankovic M, et al. 2017. The cell cycle restricts activation-induced cytidine deaminase activity to early G1. J Exp Med 214: 49–58. 10.1084/jem.20161649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Bao H, Chen L, Liu Y, Li Y, Wu B, Huang H. 2019. Molecular basis of abasic site sensing in single-stranded DNA by the SRAP domain of E. coli yedK. Nucl Acids Res 47: 10388–10399. 10.1093/nar/gkz744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Zan H, Pone EJ, Mai T, Casali P. 2012. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat Rev Immunol 12: 517–531. 10.1038/nri3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap LS, Meng FL. 2019. Cis- and trans-factors affecting AID targeting and mutagenic outcomes in antibody diversification. Adv Immunol 141: 51–103. 10.1016/bs.ai.2019.01.002 [DOI] [PubMed] [Google Scholar]
- Yeap LS, Hwang JK, Du Z, Meyers RM, Meng FL, Jakubauskaite A, Liu M, Mani V, Neuberg D, Kepler TB, et al. 2015. Sequence-intrinsic mechanisms that target AID mutational outcomes on antibody genes. Cell 163: 1124–1137. 10.1016/j.cell.2015.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mutation sequencing data sets generated during this study are available under the NCBI Bioproject PRJNA820976.