Abstract
Background
Identification of residual disease in patients with localized non-small cell lung cancer (NSCLC) following treatment with curative intent holds promise to identify patients at risk of relapse. New methods can detect circulating tumour DNA (ctDNA) in plasma to fractional concentrations as low as a few parts per million, and clinical evidence is required to inform their use.
Patients and methods
We analyzed 363 serial plasma samples from 88 patients with early-stage NSCLC (48.9%/28.4%/22.7% at stage I/II/III), predominantly adenocarcinomas (62.5%), treated with curative intent by surgery (n = 61), surgery and adjuvant chemotherapy/radiotherapy (n = 8), or chemoradiotherapy (n = 19). Tumour exome sequencing identified somatic mutations and plasma was analyzed using patient-specific RaDaR™ assays with up to 48 amplicons targeting tumour-specific variants unique to each patient.
Results
ctDNA was detected before treatment in 24%, 77% and 87% of patients with stage I, II and III disease, respectively, and in 26% of all longitudinal samples. The median tumour fraction detected was 0.042%, with 63% of samples <0.1% and 36% of samples <0.01%. ctDNA detection had clinical specificity >98.5% and preceded clinical detection of recurrence of the primary tumour by a median of 212.5 days. ctDNA was detected after treatment in 18/28 (64.3%) of patients who had clinical recurrence of their primary tumour. Detection within the landmark timepoint 2 weeks to 4 months after treatment end occurred in 17% of patients, and was associated with shorter recurrence-free survival [hazard ratio (HR): 14.8, P <0.00001] and overall survival (HR: 5.48, P <0.0003). ctDNA was detected 1-3 days after surgery in 25% of patients yet was not associated with disease recurrence. Detection before treatment was associated with shorter overall survival and recurrence-free survival (HR: 2.97 and 3.14, P values 0.01 and 0.003, respectively).
Conclusions
ctDNA detection after initial treatment of patients with early-stage NSCLC using sensitive patient-specific assays has potential to identify patients who may benefit from further therapeutic intervention.
Key words: non-small cell lung cancer (NSCLC), liquid biopsy, minimal residual disease (MRD), cell-free DNA (cfDNA), circulating tumour DNA (ctDNA), early detection
Highlights
-
•
Sensitive personalized assays were used to detect ctDNA in 363 plasma samples from 88 patients with early-stage NSCLC.
-
•
Exome sequencing of the primary tumour was used to design personalized assays targeting 48 variants unique to each patient.
-
•
ctDNA was detected pretreatment in 51% of patients and after treatment in 64.3% who had recurrence of their primary tumour.
-
•
Detection at a landmark timepoint after treatment was associated with shorter recurrence-free and overall survival times.
-
•
Detection of minimal residual disease after treatment of early-stage NSCLC can identify patients for further intervention.
Introduction
Lung cancer is the leading cause of cancer mortality worldwide, with nearly 1.8 million deaths in 2020.1 Approximately 85% of cases are non-small cell lung cancer (NSCLC), with lung adenocarcinoma and lung squamous cell carcinoma being the most prevalent subtypes. The majority of patients present with stage III-IV disease, with limited treatment options and survival rates.2 Patients with early-stage (I-II) disease are generally offered treatment with curative intent, surgical resection or radical radiotherapy, while those with locally advanced disease (IIIA/B) are treated with surgery and adjuvant chemotherapy or chemoradiotherapy (CRT) ± immune checkpoint inhibitors (ICIs).3 In these patients, routine surveillance after treatment is carried out using serial radiological imaging to detect macroscopic disease recurrence, with limited sensitivity. Biomarkers with high specificity and sensitivity for detection of minimal residual disease (MRD) are needed to better identify patients at risk of relapse, and those who could benefit most from additional adjuvant and/or maintenance therapy while avoiding overtreatment of patients who have been successfully cured.
Recent research has shown that circulating tumour DNA (ctDNA) in blood can be used as a liquid biopsy, providing a minimally invasive diagnostic tool to assess tumour burden in cancer patients.4, 5, 6 Fragments of DNA are released into the circulation as cell-free DNA through apoptosis, necrosis or active release. In cancer patients, DNA is also released from tumour cells, and this ctDNA may represent a small fraction of the total cell-free DNA. Initial studies analyzed individual somatic mutations that were common to specific cancer types, using methods such as digital PCR and BEAMing.7,8 Subsequent studies utilized next-generation sequencing (NGS) to monitor multiple mutations in parallel.5,9, 10, 11 Sensitivity for ctDNA detection can be increased by error suppression techniques, analysis of a larger number of somatic variants or larger volume of blood.12, 13, 14, 15, 16, 17 Using prior tumour sequencing information to identify mutations, highly sensitive patient-specific assays have recently been developed to detect ctDNA down to a variant allele fraction (VAF) of 0.003% or lower.14,15,17,18
Such improvements have enabled rapid advancement of ctDNA as a liquid biopsy for clinical applications, where it is now being investigated or implemented for cancer diagnosis, to guide treatment selection, to monitor therapeutic response and to identify newly emerging resistance mutations.4 With the development of highly-sensitive patient-specific assays, ctDNA has been used to detect MRD in patients with breast, colorectal, lung cancer and other solid tumours.16,19, 20, 21, 22, 23, 24, 25, 26 In NSCLC, Chaudhuri et al. 21 demonstrated that residual ctDNA could be detected in 94% of stage I-III patients treated predominantly (35/40 patients) by radiotherapy or CRT who subsequently recurred. Post-treatment detection of ctDNA preceded progression on imaging in 72% of patients by 5.2 months (median). Others recently demonstrated that following CRT, patients with residual disease had superior outcomes when treated with consolidation ICIs27 while patients with undetectable ctDNA following CRT had good outcomes irrespective of further immunotherapy. This highlights the potential application of ctDNA to guide clinicians to escalate or de-escalate therapy.28,29
Here, we assess ctDNA levels in plasma from 88 patients with stage IA to IIIB NSCLC who underwent treatment with curative intent, by surgery (69 patients, 78.4%) and/or radical (chemo)radiotherapy (19 patients, 21.6%). We used patient-specific assays tracking up to 48 somatic variants for each patient, with the aim to determine whether ctDNA detection after treatment could predict patient outcomes.
Methods
Additional details for the Methods are provided in Supplementary Methods, available at https://doi.org/10.1016/j.annonc.2022.02.007.
Patient cohort and samples
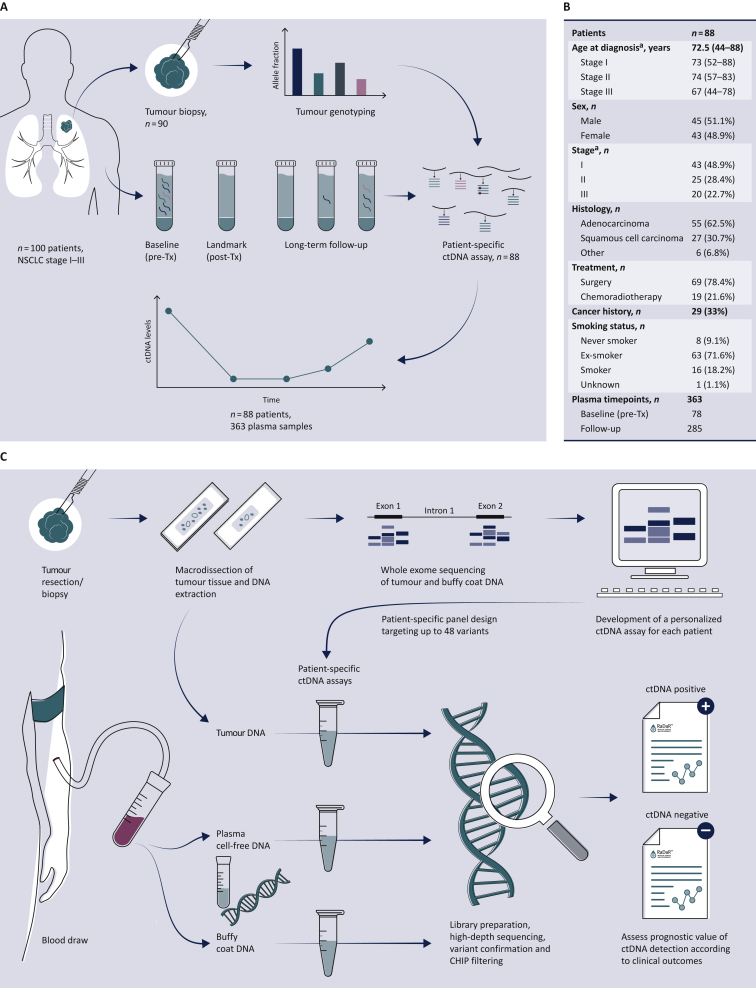
A total of 100 treatment-naive patients with stage IA-IIIB NSCLC, scheduled to undergo treatment with curative intent, were recruited to the LUng cancer CIrculating tumour DNA (LUCID) Study (Figure 1A, Supplementary Table S1, available at https://doi.org/10.1016/j.annonc.2022.02.007). The study was approved by the local research ethics committee (REC: 14/WM/1072, ClinicalTrials.gov Identifier: NCT04153526). Patients were recruited at Royal Papworth Hospital or Addenbrooke’s Hospital (Cambridge, UK). All patients provided written informed consent for participation in the study and collection of tissue and blood specimens. Cancer stages were classified according to the 7th TNM classification system. For 90 of these patients, formalin-fixed, paraffin-embedded (FFPE) tumour tissue specimens were available from surgical or diagnostic primary tumour biopsies (Supplementary Table S1, available at https://doi.org/10.1016/j.annonc.2022.02.007), including 70/70 patients who underwent surgery (100%) and 20/30 patients who were treated by chemotherapy and/or radiotherapy (66.7%). For 88 of those (Figure 1B), patient-specific assays were successfully designed and validated (see below).
Figure 1.
Study overview.
(A) Study design: 100 patients with NSCLC stage I-III were recruited to the LUCID study. For 90 patients, tumour specimens were available and were analyzed by whole exome sequencing (WES), to identify somatic single nucleotide variants for design of patient-specific assays. Assays were successfully developed (panel B) for 88 patients. A total of 363 plasma samples collected from these patients at multiple timepoints before and after treatment were analyzed using those personalized assays to detect ctDNA and estimate its fractional concentration. The prognostic value of ctDNA detection was assessed by comparing ctDNA data with clinical outcomes to evaluate the relapse-free survival and overall survival for patient subgroups. A CONSORT flow diagram is provided in Supplementary Figure S1, available at https://doi.org/10.1016/j.annonc.2022.02.007. (B) Patient demographics of the 88 patients enrolled in the LUCID study who had available tumour tissue for sequencing and a patient-specific ctDNA assay successfully designed.
ca., carcinoma; ctDNA, circulating tumour DNA; NSCLC, non-small cell lung cancer; Tx, treatment.
aCancer stage was defined at the time of diagnosis according to diagnostic pathways,30 and was updated after surgery based on pathological analysis of the tumour specimen for patients who underwent surgery. (C) Overview of the workflow involved in the development of the RaDaR™ sequencing assays used in this study. Personalized sequencing assays were designed based on WES data from tumour and buffy coat samples, targeting 48 somatic variants for each assay. These were used to amplify regions in plasma cell-free DNA, tumour DNA and buffy coat DNA using multiplex PCR and high-depth next generation sequencing. Data from tumour DNA and buffy coat samples were used to confirm the detection of somatic variants and to exclude variants which may derive from clonal hematopoiesis of indeterminate potential (CHIP). Plasma samples were classified as ctDNA positive (detected) or ctDNA negative (not detected) based on the RaDaR™ assay sequencing data and analytical pipeline, and the estimated variant allele fraction (eVAF) was calculated.
A total of 363 plasma samples were collected from these 88 patients (see CONSORT flow diagram in Supplementary Figure S1, available at https://doi.org/10.1016/j.annonc.2022.02.007). Plasma samples were collected before the start of treatment, during treatment (depending on treatment schedule) and after the end of treatment, and at up to three regular clinical follow-up visits (scheduled at ∼3-monthly intervals). For a subset of patients, an additional plasma sample was collected after disease recurrence or diagnosis of a second primary tumour. Plasma specimens were collected into 9 ml S-Monovette® K3EDTA tubes (Sarstedt, Nümbrecht, Germany), centrifuged at 1600 g for 10 min within 1 h of venipuncture with further centrifugation of the supernatant at 20 000 g for 10 min. Plasma aliquots were stored at −80°C. Whole blood specimens for germline analysis were collected into 2.6 ml K3EDTA tubes and frozen at −80°C.
Patients were followed for a median of 3 years (range: 42 days to 5 years), and clinical outcomes recorded (Supplementary Table S1, available at https://doi.org/10.1016/j.annonc.2022.02.007).
Tissue analysis by whole exome sequencing
DNA was extracted from FFPE tissue sections using the QIAamp® DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) with modifications and DNA repair. Buffy coat samples were extracted using the QIAamp DNA Blood Mini Kit or QIAsymphony DSP Circulating DNA kit (Qiagen). Whole exome sequencing (WES) of sheared DNA was carried out as previously described.30 Mutation calling was carried out using Mutect2 (MuTect2 v3.831,32), or Mutect2, VarDict v1.4.1033 and Freebayes v1.1.34
For eight of the patients who had clinical progression or a second primary tumour diagnosed, additional tumour biopsies were collected at the time of progression (Supplementary Table S1, available at https://doi.org/10.1016/j.annonc.2022.02.007) and were similarly analyzed by WES. Patient 277 had a clinical diagnosis of a second primary cancer; however, sequencing of biopsies from the first primary tumour and the suspected second primary indicated that the suspected second primary was in fact a recurrence of the first primary tumour. For the other seven cases, data from WES of tumour biopsies agreed with the clinical assessment of either recurrence of the first primary cancer or a confirmed second primary cancer.
Plasma analysis by personalized ctDNA sequencing assays
Personalized ctDNA ‘RaDaR™’ assays were designed and validated for 88 patients (Figure 1C). RaDaR™ NGS liquid biopsy assays are based on personalized multiplex PCR amplification of cell-free DNA. Tumour-specific variants identified by exome sequencing of the primary tumour and buffy coat DNA were ranked and prioritized for inclusion into custom panels, incorporating 48 amplicons per patient targeting patient-specific variants (except for patient 283, for whom 47 variants were used; Supplementary Table S2, available at https://doi.org/10.1016/j.annonc.2022.02.007). For each patient, a single individualized panel of primers was created covering patient-specific variants combined with a fixed primer panel covering common population-specific single nucleotide polymorphisms, for internal sample quality control (QC) during NGS testing. Multiplex PCR reactions were carried out and libraries sequenced as previously described18,35,36 to validate primer pools using between 500 and 2000 amplifiable copies of tumour DNA as well as 500 amplifiable copies of DNA extracted from buffy coat samples from the respective patient as a QC. Results were used to confirm the amplification of the target regions and to confirm the presence of selected single nucleotide variants (SNVs) in tumour DNA. Target SNVs were excluded if signal was also observed in buffy coat DNA, to reduce the potential impact of germline mutations, mosaicism or variants arising from clonal haematopoiesis of indeterminate potential37 (sequenced at depth >1000 for 88% of variants and >100 for 94% of variants; depth was <100 for most variants for patient 221 and data were missing for patient 236; Supplementary Table S2, available at https://doi.org/10.1016/j.annonc.2022.02.007).
Cell-free DNA was extracted from up to 4 ml of plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen) and quantified by droplet digital PCR (ddPCR) (BioRad QX200) as previously described.35
Multiplex PCR was carried out in replicate reactions, generating sequencing libraries of target regions from a total of 1356-20 000 amplifiable copies per plasma sample as measured by ddPCR (median 8360, mean 9937, with >2000 amplifiable copies for 99.7% of samples; Supplementary Table S3, available at https://doi.org/10.1016/j.annonc.2022.02.007). High-depth NGS of amplified libraries was carried out on the NovaSeq® 6000 system (Illumina, San Diego, CA) using PE150 sequencing, generating 2 355 500 304 total paired reads (excluding non-mappable reads and reads with Phred score <20) and a median of 6 030 997 reads per sample [interquartile range (IQR) 5 519 416 to 6 660 932]. The median coverage for each variant was 174 669 reads per sample. A median of 45 amplicons passed QC for each assay, with a minimal target read depth of 40 000 per sample per variant.
Sequencing data fastq files were demultiplexed using bcl2fastq; reads were then aligned38 to the GRCh38/hg38 assembly of the human genome and processed to identify primer pairs and count mutant and reference bases. A statistical model was used to assess the significance of the observed counts for each variant and the information was integrated over the entire set of personalized variants from an individual sample to obtain evidence of ctDNA presence (ctDNA positive) or absence (ctDNA negative) at the sample level. The model also generated an estimated VAF (eVAF) for each sample.
Statistical analysis
Survival analysis was based on three different stratifications: detection at baseline before treatment, at the ‘landmark’ timepoint, or at any time ≥2 weeks after treatment (including at disease relapse). The ‘landmark’ timepoint is the first sample collected in the time window between 2 weeks and 4 months following the end of treatment. Patients with a cancer event unrelated to the original tumour (i.e. a ‘second primary’ tumour) were considered censored for purposes of freedom from recurrence (FFR) and recurrence-free survival (RFS), but not for overall survival (OS). Death was included as an event for OS and RFS (if not preceded by a second primary tumour), but not for FFR. Hazard ratios (HRs) and significance were obtained using the R package ‘survival’ based on the time from the end of treatment. Kaplan–Meier and log rank tests were used to evaluate the predictive value of ctDNA detection.
Results
A median of 328 SNVs were identified by WES per patient [interquartile range (IQR): 205–491]. Of these, 99.8% were private mutations to a specific patient.30 The SNVs were used to design for each patient a personalized RaDaR™ assay (Figure 1), built on the InVision® platform which utilizes multiplex PCR and targeted NGS.18,35 RaDaR™ assays were successfully designed and validated for 88 of the 90 patients for whom tumour specimens were available (97.8%) and in total for 88% of the 100 patients in the LUCID study, including 69/70 (98.6%) of patients who underwent surgery and 19/30 (63.3%) of patients who did not undergo surgery. For 12 of the 100 patients recruited to LUCID, RaDaR™ assays were not available: for 10 patients, tumour samples were not available; for patient 253, tumour WES was of insufficient quality for assay design; and for patient 287, the RaDaR™ assay failed QC (Supplementary Table S1, available at https://doi.org/10.1016/j.annonc.2022.02.007).
Analysis of ctDNA levels before treatment
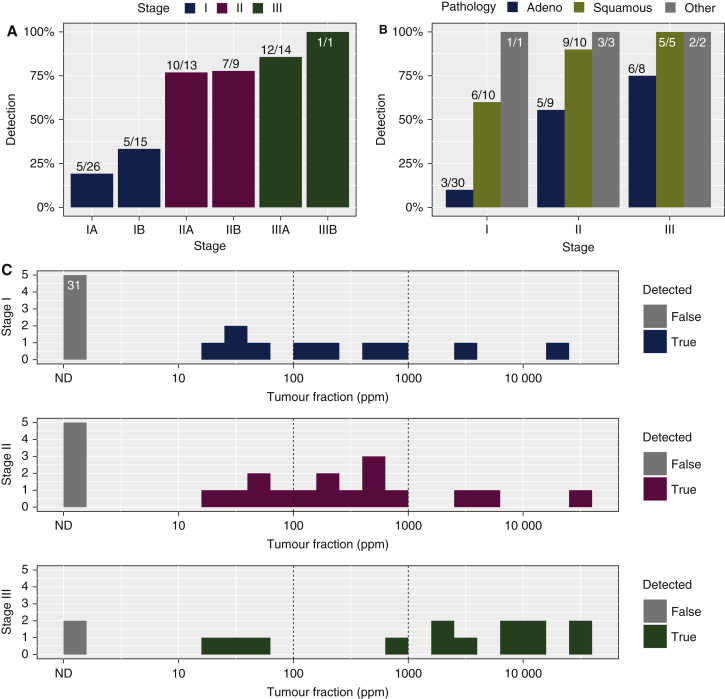
Plasma samples collected before start of treatment (‘baseline’ timepoint) were analyzed for 78 patients. ctDNA was detected in 51% (40/78) of pretreatment samples overall, including 24% (10/41) of patients with stage I, 77% (17/22) with stage II and 87% (13/15) with stage III disease (Figure 2A). ctDNA was detected before treatment in a higher proportion of patients with lung squamous cell carcinomas (80%, 20/25) compared with adenocarcinomas (29.8%, 14/47) (Figure 2B). Median levels of ctDNA detected pretreatment were eVAF = 0.049%, 25 of 40 (62.5%) of samples with ctDNA detected had eVAF<0.1% and 12 of 40 (30%) of samples with ctDNA detected had eVAF <0.01% (Figure 2C).
Figure 2.
ctDNA detection before treatment.
Histograms showing detection rates of ctDNA before treatment of 78 patients where plasma samples were available pretreatment, (A) according to disease stage and (B) according to disease stage and histological subtype. Detection rates are shown in percentages, and the numbers of samples in each group are indicated above the bars (detected/total). (C) ctDNA levels, shown as estimated variant allele fraction (eVAF) in parts per million (ppm) of tumour DNA to total DNA, in plasma samples collected before treatment from patients with non-small cell lung cancer at stage I (top), II (middle) and III (bottom). Samples with ctDNA not detected are shown in grey bars, including 31 samples from patients with stage I in which ctDNA was not detected before treatment. Vertical dotted lines indicate eVAF of 0.01% (100 ppm) and 0.1% (1000 ppm).
ctDNA, circulating tumour DNA; ND, not detected.
Longitudinal monitoring of ctDNA to detect residual disease and recurrence
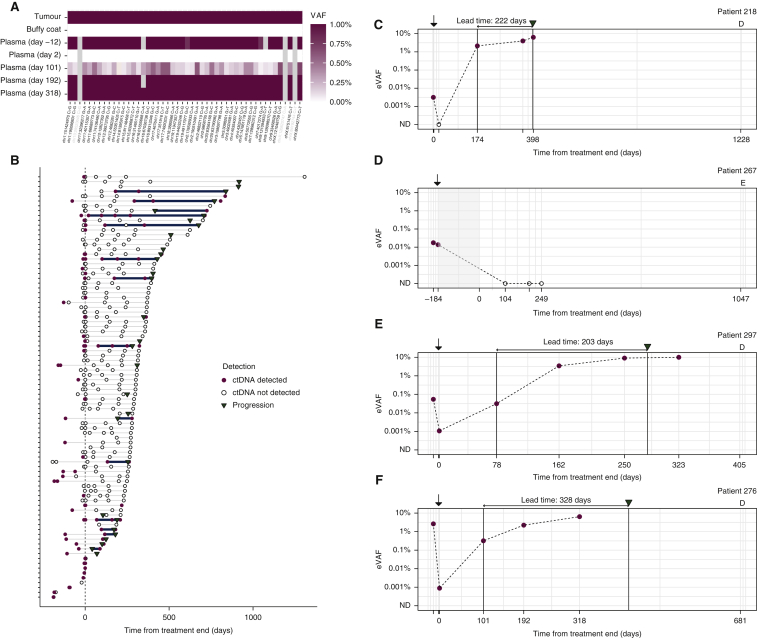
Personalized RaDaR™ assays were used to analyze serial samples from each patient (Figure 3A), analyzing a total of 363 plasma samples (Figure 3B, Supplementary Figure S2, available at https://doi.org/10.1016/j.annonc.2022.02.007). Overall, ctDNA was detected by these assays in 56% of patients (49/88), and in 26% of all samples (94/363), at a median eVAF of 0.042% for positive samples. We evaluated time courses of ctDNA detection (Supplementary Figure S3, available at https://doi.org/10.1016/j.annonc.2022.02.007) and highlight several illustrative examples (Figure 3C-F).
Figure 3.
Longitudinal monitoring of ctDNA levels in plasma.
(A) Example heat map showing the results of analysis of 48 variants in tumour DNA, buffy coat DNA and cell-free DNA from multiple plasma samples from patient 276 using a patient-specific RaDaR™ assay, using a color-coded scale of variant allele fractions (VAFs) ranging from 0% to ≥1%. Each row represents a different sample type or plasma timepoint (indicating days from end of treatment), and each column represents a single variant. Variants shaded in grey were excluded from analysis due to low read depth. (B) Summary of all 363 serial plasma samples from 88 patients indicating when ctDNA was detected (red dots) or not detected (black circles). Clinical recurrence is indicated with an inverted triangle. The lead time between ctDNA detection ≥2 weeks from end of treatment and clinical recurrence is indicated with a solid line. Time is measured from end of treatment. Supplementary Figure S2, available at https://doi.org/10.1016/j.annonc.2022.02.007 shows the data split into cases that had ctDNA detected or not detected in the baseline (pretreatment) samples. Examples of longitudinal monitoring of ctDNA in plasma from: (C) patient 218 with adenocarcinoma and stage IA disease treated by surgery; (D) patient 267 with adenocarcinoma and stage IIB disease, treated with surgery followed by adjuvant chemotherapy and radiotherapy (indicated by the shaded region)—the patient had no clinical progression until they were lost to follow-up 1047 days after the end of treatment; (E) patient 297 with adenocarcinoma and stage IIA disease treated by surgery; (F) patient 276 with squamous cell carcinoma and stage IIA disease, treated by surgery. Red dots indicate samples with ctDNA detected, and black circles indicate samples in which ctDNA was not detected, when analyzed using patient-specific RaDaR™ assays. Time is measured (horizontal axis) from the end of treatment. Black arrows above the plot indicate the time of surgery, and clinical recurrence if this occurred during the study is indicated by an inverted triangle. ‘D’ or ‘E’ above the axes indicate death, or end of follow-up. Time of these events is indicated on the horizontal axis. Bold vertical lines are a guide to the eye and indicate the lead time (shown in text above the axes) between the earliest detection of ctDNA, ≥2 weeks after the end of treatment, and when clinical progression was first recorded for each patient. Supplementary Figure S3, available at https://doi.org/10.1016/j.annonc.2022.02.007 shows time courses, with long-term outcomes, for all 88 patients.
ctDNA, circulating tumour DNA; ND, not detected.
In 48 patients with samples collected 1-3 days after surgery, ctDNA was detected in 12 patients (25%), with a median eVAF of 0.0026% (26 parts per million, ppm). Only 6 of those 12 patients had later recurrence of their primary tumour, supporting an interpretation that ctDNA may be transiently present in the blood at low concentrations in cases where the disease has been entirely resected.
We focused analysis on 230 plasma samples collected from 77 patients during observation, defined here as all timepoints ≥2 weeks after the end of treatment. ctDNA was detected during observation in 40 samples from 20 patients (26% of the 77 patients). A total of 38 of the 40 samples were from 18 patients who had recurrence of their first primary tumour. A total of 2 of the 40 samples were from the set of 152 samples collected during observation from 49 patients who did not have recurrence of their first primary tumour during observation (Supplementary Figure S1 and Supplementary Table S1, available at https://doi.org/10.1016/j.annonc.2022.02.007). Considering these two samples, where there was no confirmed recurrence of the primary tumour, as possible false positives, the positive predictive value (PPV) of ctDNA detection during observation for recurrence of the first primary tumour was 95% (38/40 positive samples), and the specificity was 98.7% (150/152 samples).
Across the cohort, 28 patients who had samples collected during observation experienced clinical recurrence of their first primary tumour. ctDNA was detected in 18 of these 28 patients (64.3%) (Supplementary Figure S1, available at https://doi.org/10.1016/j.annonc.2022.02.007) at eVAFs ranging from 0.0011% (11 ppm) to >10% (Supplementary Figure S3, available at https://doi.org/10.1016/j.annonc.2022.02.007).
For 12 patients, ctDNA was detected in samples collected during observation before recurrence, and in those cases the median lead time between ctDNA detection and clinical recurrence was 212.5 days (Supplementary Table S1, available at https://doi.org/10.1016/j.annonc.2022.02.007). Of the 20 patients who had clinical recurrence >200 days after end of treatment, ctDNA was detected before recurrence in 8 cases (40%), and in those cases the median lead time for ctDNA detection was 402.5 days. In the remaining 12 cases, the median time from the last (ctDNA negative) sample to recurrence was 192 days.
Additional observations are described in Supplementary Information, available at https://doi.org/10.1016/j.annonc.2022.02.007.
ctDNA detection ≥2 weeks after treatment is predictive of clinical disease relapse
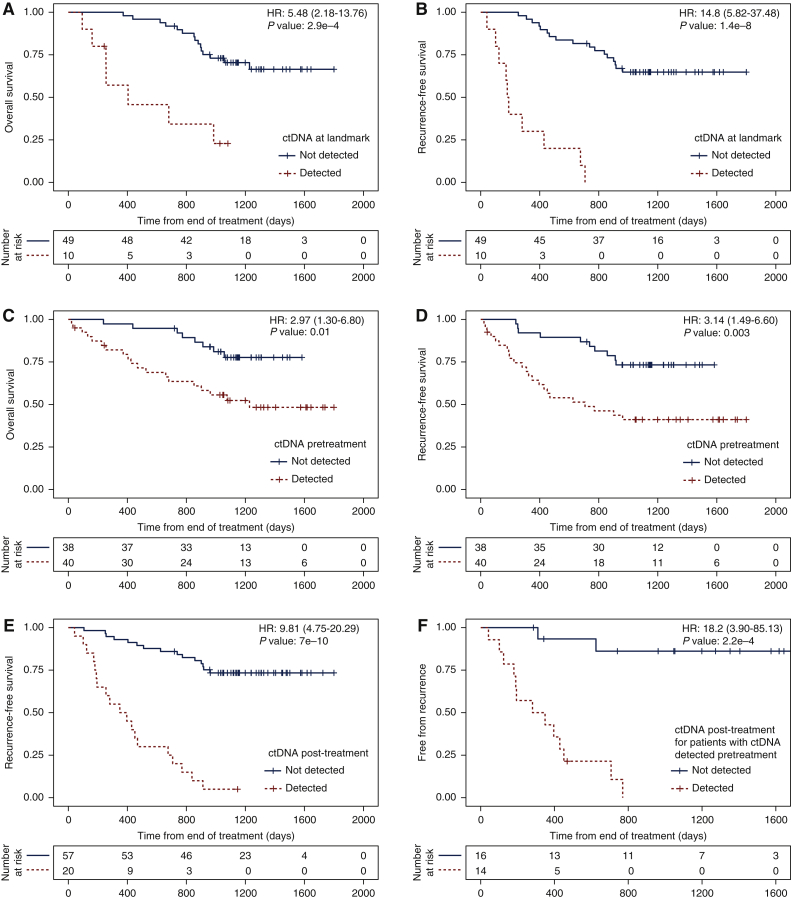
We compared the detection of ctDNA to clinical outcomes including OS (counting as events death from any cause), RFS (counting as events either recurrence of the first primary tumour, or death if not preceded by a second primary tumour) and FFR (counting as events only recurrence of the first primary tumour) (Figure 4).
Figure 4.
Survival analysis based on ctDNA detection.
Kaplan–Meier analysis showing the fraction of patients without events as a function of time. Patients right-censored due to loss of information are shown as vertical tick-marks. Patient subgroups are defined based on ctDNA detection using the RaDaR™ assay at different timepoints (see individual panels). Patients with ctDNA detected are shown in red dotted lines, and those with ctDNA not detected at the respective timepoints are shown in black solid lines. The numbers of patients remaining at risk are shown below each graph. (A) Overall survival (OS) and (B) recurrence-free survival (RFS, counting as events either recurrence of the first primary tumour or death if not preceded by a second primary tumour), for patients split by ctDNA detection at the landmark timepoint, which is the first plasma sample available in the window ≥2 weeks and ≤4 months after the end of treatment (data available for 59 patients in total). Hazard ratios: 5.48 [95% confidence interval (CI): 2.18-13.76] and 14.8 (95% CI: 5.82-37.48) for OS and RFS, respectively (P values: 2.9e-4 and 1.4e-8). (C) OS and (D) RFS for patients split by ctDNA detection before treatment (data available for 78 patients in total). Hazard ratios: 2.97 (95% CI: 1.30-6.80) and 3.14 (95% CI: 1.49-6.60) for OS and RFS, respectively (P values: 0.01 and 0.003). (E) RFS for patients split by ctDNA detection at any timepoint ≥2 weeks after the end of treatment (data available for 77 patients in total). Hazard ratio: 9.81 (95% CI: 4.75-20.29, P value: 7e-10). (F) Fraction of patients who remain free from recurrence split by ctDNA detection at any timepoint ≥2 weeks after the end of treatment, for patients for whom ctDNA was detected before treatment (data available for 30 patients in total). Hazard ratio: 18.2 (95% CI: 3.9-85.13, P value: 2.2e-4).
Of 59 patients who had a plasma sample collected at a landmark timepoint, defined as the first sample collected in a time-frame between 2 weeks and 4 months from the end of treatment, 10 (17%) had ctDNA detected, and all 10 had recurrence of their first primary tumour (Supplementary Figure S1, available at https://doi.org/10.1016/j.annonc.2022.02.007), resulting in 100% specificity and PPV for recurrence. These patients had a shorter OS and RFS compared with patients for whom ctDNA was not detected at landmark, with an HR of 5.48 and 14.8, respectively (P value 0.00029 and 1.4e-8; Figure 4A and B), a negative predictive value (NPV) of 75.5% (37 of 49) for recurrence and 65.3% (32 of 49) for recurrence or death (Supplementary Figure S4A and B, available at https://doi.org/10.1016/j.annonc.2022.02.007).
Patients for whom ctDNA was detected pretreatment also had shorter OS and RFS compared with patients for whom ctDNA was not detected at baseline; however, the HRs and P values were much lower (HR: 2.97 and 3.14, P values 0.01 and 0.003, respectively; Figure 4C and D, Supplementary Figure S4C, available at https://doi.org/10.1016/j.annonc.2022.02.007). A stronger difference in RFS was observed when comparing patients in whom ctDNA was detected versus not detected at any timepoint during observation after treatment (HR: 9.81, P value 7e-10; Figure 4E) while maintaining high specificity (18/20, 90%; Supplementary Figure S4D, available at https://doi.org/10.1016/j.annonc.2022.02.007).
In the subset of patients with ctDNA detected pretreatment, both sensitivity and specificity were high for detection of ctDNA after treatment (Figure 4F, Supplementary Figure S5, available at https://doi.org/10.1016/j.annonc.2022.02.007). Only 2 of the 16 patients for whom ctDNA was detected pretreatment, but not detected during observation, had a clinical recurrence of their first primary tumour (Figure 4F), corresponding to NPV = 87.5% (14/16). Of 15 patients who had clinical recurrence of their first primary tumour, ctDNA was detected during observation in 13 (sensitivity = 86.7%), and 13/14 patients with ctDNA detected during observation experienced clinical recurrence (PPV = 92.9%). This resulted in a significant HR for recurrence and RFS for ctDNA detection during observation (HR: 18.2, P value 0.00022; Figure 4F) and at landmark (Supplementary Figure S5, available at https://doi.org/10.1016/j.annonc.2022.02.007). For patients with ctDNA not detected at baseline, ctDNA was detected at landmark in 1/29 patients and during observation in 4/37 patients (Supplementary Table S1, available at https://doi.org/10.1016/j.annonc.2022.02.007). For additional detail see Supplementary Information, available at https://doi.org/10.1016/j.annonc.2022.02.007.
Discussion
Analysis of ctDNA before and after treatment with curative intent of patients with early-stage NSCLC (Figure 1) demonstrated that detection of ctDNA using sensitive personalized assays can inform patient prognosis and help guide patient treatment (Figure 4). Several illustrative cases demonstrate possible scenarios (Figure 3). ctDNA was detected before treatment in half of the patients (Figure 2), and those had a high risk of earlier recurrence or death (HR: 3.1 and 3.0). More informative was analysis of ctDNA after treatment. ctDNA detection in a landmark window, the first sample collected ≥2 weeks and <4 months after treatment, was associated with a 5.5-fold higher risk of death and 14.8-fold higher risk of recurrence of the primary tumour. Patients for whom ctDNA was detected before treatment, but not after treatment, had excellent outcomes with recurrence in only 2/16 patients (Figure 4), suggesting the potential for de-escalation of adjuvant treatment. This adds to accumulating evidence for the potential utility of ctDNA testing to risk-stratify patients to identify which patients may benefit most from adjuvant therapy or be at high risk of disease recurrence.
Recent results support this approach in multiple cancer types.23,26 In early-stage NSCLC treated by surgery and/or radiotherapy, several recent studies used high-sensitivity methods to test associations between ctDNA detection and patient outcomes after treatment with curative intent21,39,40 (see Supplementary Information, available at https://doi.org/10.1016/j.annonc.2022.02.007). Here we applied high-sensitivity personalized assays to study a population with earlier stage disease, of whom 70% underwent surgical treatment, and observed lead times of ∼200 days from ctDNA detection to disease recurrence.
The main limitation of our study was that the design included routine sampling only for 9 months after the end of treatment, and samples were collected beyond this in only a subset of patients at the time of recurrence. For patients who recurred later than ∼18 months after end of treatment, samples were generally not collected for >200 days before their clinical progression. Nonetheless, the RaDaR™ assays employed in this study showed the ability to detect ctDNA in plasma of patients with no overt or radiological evidence of disease, to fractional concentrations as low as 0.0011% (11 ppm) in patients who went on to experience recurrence of their tumour. The detection of ctDNA at such low concentrations resulted in a long lead time, preceding clinical identification of recurrence by 200 days or more, and this lead time was even longer in the subset of patients whose recurrence occurred later than 200 days.
The clinical specificity of the assay was high: of 152 samples collected after treatment from 49 patients who did not have a recurrence of their primary tumour, ctDNA was detected in only 2 samples from 2 patients. For those two patients, the last follow-up was at 251 days and 313 days after ctDNA detection, and later recurrence cannot be ruled out. Counting these as false positives, the clinical specificity achieved was 98.7% at the sample level, and the PPV for recurrence was 95% (38/40 samples positive for ctDNA were associated with recurrence). This high clinical specificity was observed despite a high analytical sensitivity: more than one-third of the samples with ctDNA detected had eVAFs <100 ppm (<0.01%), a higher proportion than observed in previous studies, highlighting the high sensitivity needed to detect ctDNA in patients with low disease burden to provide a longer lead time before clinical identification of recurrence. Using an assay with high specificity supports the ability to carry out repeat surveillance testing for detection of MRD, which increases the chance of detecting ctDNA positive cases who may otherwise be missed by testing at a single post-treatment MRD timepoint. Of 22 patients who recurred and had samples available at the landmark timepoint, 10 were positive for ctDNA (sensitivity = 45.4%), compared with 18 of 28 patients who recurred (64.3%) for whom ctDNA was detected at landmark or later.
Analysis of samples collected before initiation of treatment was predictive and can help increase the NPV of ctDNA detection during long-term observation. In patients for whom ctDNA was detected before treatment, ctDNA was also detected during long-term observation for 13 of 15 cases who had clinical recurrence of their primary tumour. Of 16 patients for whom ctDNA was detected pretreatment but not detected during observation, only 2 had a clinical recurrence of their first primary tumour, resulting in high sensitivity, PPV and NPV. Only few patients with ctDNA not detected pretreatment experienced clinical recurrence. Analysis of pretreatment samples alone, however, was not as predictive as analysis during observation or analysis of a single sample collected at a landmark timepoint after end of treatment (lower HR and P value; Figure 4).
A key consideration for deploying such assays is the requirement to analyze tumour tissue samples to obtain a panel of mutations for the design of personalized assays. Tumour samples for sequencing were available in our study for all patients who underwent surgery, but for only 20 of 30 patients who did not undergo surgery, and we were able to design assays for 88 of 100 patients. Reliance on a tumour specimen increases the complexity of the process and it may not be available for all patients; however, if this results in improved analytical sensitivity and therefore improved clinical performance, this needs to be weighed and balanced.
We provide evidence to show that when ctDNA is detected, relapse is likely, and patients may benefit from additional treatment or participation in a clinical trial. When ctDNA is not detected, there is still a considerable chance of relapse and the current data do not provide sufficient evidence to change the standard of care when adjuvant treatment is otherwise indicated. Larger studies and prospective clinical trials are needed to determine the balance of benefit/harm in further treatment of patients following initial treatment with curative intent, when ctDNA is not detected even using highly sensitive assays. Our data suggest that a high negative predictive power could be achieved by analysis of multiple samples both before and after treatment using highly sensitive ctDNA assays. Our results also support that analysis for residual disease may be best delayed beyond the first few days, as we detected ctDNA 1-3 days after surgery in 25% of patients, but half of those patients did not have clinical relapse (with median follow-up of 543.5 days).
In summary, our study adds to the accumulating evidence that supports the utility of ctDNA testing for detection of residual disease and recurrence. This may be used as a sensitive tool for identifying patients at high risk of relapse who may benefit from additional adjuvant therapy or may be eligible for enrolment into clinical trials. In the future, ctDNA testing may allow identification of patients at lower risk of relapse, for whom it may be possible to consider less intensive or shorter treatment courses.
Acknowledgements
We thank the patients and their families. We also thank the Cambridge Cancer Trials Unit – Cancer Theme at Addenbrooke’s Hospital (Gail Doughton, Wendy Qian, Tim Eisen) as well as the research staff at Addenbrooke’s Hospital and Royal Papworth (Ellen Moseley, Amanda Stone, Amy Gladwell, Theresa Green, Vicky Senior, Julia Knight). ctDNA isolation and generation of tumour exome libraries from second primary cancers and relapse tumours were carried out by the Cancer Molecular Diagnostics Laboratory. We thank the Stratified Medicine Core Laboratory Next Generation Sequencing Hub, Cambridge Biomedical Research Centre for isolation of buffy coat DNA. We also thank the Cancer Research UK Cambridge Institute Compliance and Biobanking, Bioinformatics and Genomics Core facilities, and Inivata‘s Product Development, Computational Biology and Clinical Operations teams.
Funding
This work was supported by the University of Cambridge, Cancer Research UK [grant numbers A20240, C9545/A29580] and the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC Grant Agreement [grant number n.337905]. The LUCID study was supported by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre [grant number BRC-1215-20014] and the Cambridge Clinical Trials Unit (CCTU) (no grant number). The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care. Inivata provided analysis of samples using the RaDaR™ assay. The Cancer Molecular Diagnostics Laboratory is supported by NIHR Cambridge Biomedical Research Centre [BRC-1215-20014], The Mark Foundation for Cancer Research [RG95043] and the Cancer Research UK Cambridge Centre [C9685/A25177]. RCR and DMR are supported by Cambridge NIHR Biomedical Research Centre [grant number BRC-1215-20014] and Royal Papworth Hospital.
Disclosure
NR and DGa are co-founders, and NR, DGa, MP, KHo, SH, GM, CGS and GS are current or former employees/officers/consultants of Inivata Ltd. Inivata provided analysis of samples using the RaDaR™ assays. DGa and KHe are current employees of AstraZeneca Inc. All other authors have declared no conflicts of interest.
Contributor Information
R.C. Rintoul, Email: rcr39@cam.ac.uk.
N. Rosenfeld, Email: nitzan.rosenfeld@cruk.cam.ac.uk.
Supplementary data
References
- 1.Globocan 2020. World Health Organisation, International Agency for Research on Cancer. https://gco.iarc.fr/today/data/factsheets/cancers/15-Lung-fact-sheet.pdf Available at. Accessed March 18, 2020.
- 2.Cancer Research UK. 2020. https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/lung-cancer Available at. Accessed March 18, 2020.
- 3.NICE. Lung cancer in adults: treatments with curative intent. Webpage. 2021. https://www.nice.org.uk/guidance/qs17/chapter/Quality-statement-5-Treatment-with-curative-intent Available at. Accessed March 18, 2020.
- 4.Wan J.C.M., Massie C., Garcia-Corbacho J., et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 5.Dawson S.-J., Tsui D.W.Y., Murtaza M., et al. Analysis of circulating tumour DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 6.Parkinson C.A., Gale D., Piskorz A.M., et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Med. 2016;13(12) doi: 10.1371/journal.pmed.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogelstein B., Kinzler K.W. Digital PCR. Proc Natl Acad Sci U S A. 1999;96(16):9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diehl F., Schmidt K., Choti M.A., et al. Circulating mutant DNA to assess tumour dynamics. Nat Med. 2008;14(9):985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forshew T., Murtaza M., Parkinson C., et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 10.Murtaza M., Dawson S.-J., Tsui D.W.Y., et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 11.Murtaza M., Dawson S.-J., Pogrebniak K., et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun. 2015;6:8760. doi: 10.1038/ncomms9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinde I., Wu J., Papadopoulos N., Kinzler K.W., Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108(23) doi: 10.1073/pnas.1105422108. :9530-9335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newman A.M., Lovejoy A.F., Klass D.M., et al. Integrated digital error suppression for improved detection of circulating tumour DNA. Nat Biotechnol. 2016;34(5):547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan J.C.M., Heider K., Gale D., et al. ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci Transl Med. 2020;12(548) doi: 10.1126/scitranslmed.aaz8084. [DOI] [PubMed] [Google Scholar]
- 15.Mcdonald B.R., Contente-cuomo T., Sammut S.-J., et al. Personalized circulating tumour DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Transl Med. 2019;11(504) doi: 10.1126/scitranslmed.aax7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons H.A., Rhoades J., Reed S.C., et al. Sensitive detection of minimal residual disease in patients treated for early-stage breast cancer. Clin Cancer Res. 2020;26(11):2556–2564. doi: 10.1158/1078-0432.CCR-19-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurtz D.M., Soo J., Co Ting Keh L., et al. Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumour DNA. Nat Biotechnol. 2021;39(12):1537–1547. doi: 10.1038/s41587-021-00981-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsico G., Sharma G., Perry M., et al. Abstract 3097: Analytical development of the RaDaR™ assay, a highly sensitive and specific assay for the monitoring of minimal residual disease. Cancer Res. 2020;80(suppl 16):3097. [Google Scholar]
- 19.Garcia-Murillas I., Schiavon G., Weigelt B., et al. Mutation tracking in circulating tumour DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7(302):302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 20.Tie J., Wang Y., Tomasetti C., et al. Circulating tumour DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346) doi: 10.1126/scitranslmed.aaf6219. 346ra92-346ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhuri A.A., Chabon J.J., Lovejoy A.F., et al. Early detection of molecular residual disease in localized lung cancer by circulating tumour DNA profiling. Cancer Discov. 2017;7(12):1394–1403. doi: 10.1158/2159-8290.CD-17-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azad T.D., Chaudhuri A.A., Fang P., et al. Circulating tumour DNA analysis for detection of minimal residual disease after chemoradiotherapy for localized esophageal cancer. Gastroenterology. 2020;158(3):494–505.e6. doi: 10.1053/j.gastro.2019.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moding E.J., Nabet B.Y., Alizadeh A.A., Diehn M. Detecting liquid remnants of solid tumours: circulating tumour DNA minimal residual disease. Cancer Discov. 2021;11(12):2968–2986. doi: 10.1158/2159-8290.CD-21-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen K., Zhao H., Shi Y., et al. Perioperative dynamic changes in circulating tumour DNA in patients with lung cancer (DYNAMIC) Clin Cancer Res. 2019;25(23):7058–7067. doi: 10.1158/1078-0432.CCR-19-1213. [DOI] [PubMed] [Google Scholar]
- 25.Zviran A., Schulman R.C., Shah M., et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat Med. 2020;26(7):1114–1124. doi: 10.1038/s41591-020-0915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan J.C.M., Mughal T.I., Razavi P., et al. Liquid biopsies for residual disease and recurrence. Med. 2021;2(12):1292–1313. doi: 10.1016/j.medj.2021.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Moding E.J., Liu Y., Nabet B.Y., et al. Circulating tumour DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small-cell lung cancer. Nat Cancer. 2020;1(2):176–183. doi: 10.1038/s43018-019-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powles T., Assaf Z.J., Davarpanah N., et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature. 2021;595:432–437. doi: 10.1038/s41586-021-03642-9. [DOI] [PubMed] [Google Scholar]
- 29.Rostami A., Bratman S.V. Utilizing circulating tumour DNA in radiation oncology. Radiother Oncol. 2017;124(3):357–364. doi: 10.1016/j.radonc.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Heider K, Wan JCM, Gale D, et al. ctDNA detection by personalised assays in early-stage NSCLC. MedRxiv. 2021. 10.1101/2021.06.01.21258171. Available at http://medrxiv.org/content/early/2021/06/03/2021.06.01.21258171.abstract. Accessed March 18, 2022. [DOI]
- 31.Cibulskis K., Lawrence M.S., Carter S.L., et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broad Institute. Mutect2 [Internet]. GATK. https://gatk.broadinstitute.org/hc/en-us/articles/360037593851-Mutect2 Available at. Accessed March 18, 2022.
- 33.Lai Z., Markovets A., Ahdesmaki M., et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44(11) doi: 10.1093/nar/gkw227. :e108-e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing [Internet]. 2012 http://arxiv.org/abs/1207.3907 . Available at. Accessed March 18, 2022.
- 35.Gale D., Lawson A.R.J., Howarth K., et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS One. 2018;13(3) doi: 10.1371/journal.pone.0194630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plagnol V., Woodhouse S., Howarth K., et al. Analytical validation of a next generation sequencing liquid biopsy assay for high sensitivity broad molecular profiling. PLoS One. 2018;13(3) doi: 10.1371/journal.pone.0193802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbosh C., Swanton C., Birkbak N.J. Clonal haematopoiesis: a source of biological noise in cell-free DNA analyses. Ann Oncol. 2019;30(3):358–359. doi: 10.1093/annonc/mdy552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM; p. 1303.https://arxiv.org/abs/1303.3997 :arXiv. Available at. Accessed March 18, 2022. [Google Scholar]
- 39.Abbosh C., Birkbak N.J., Wilson G.A., et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545(7655):446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abbosh C., Frankell A., Garnett A., et al. Phylogenetic tracking and minimal residual disease detection using ctDNA in early-stage NSCLC: a lung TRACERx study. AACR. Cancer Res. 2020;80(suppl 16) Abstract CT023. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.