Abstract
Populations of widespread species are usually geographically distributed through contrasting stresses, but underlying genetic mechanisms controlling this adaptation remain largely unknown. Here, we show that in Arabidopsis thaliana, allelic changes in the cis-regulatory elements, WT box and W box, in the promoter of a key transcription factor associated with oxygen sensing, RELATED TO AP 2.12 (RAP2.12), are responsible for differentially regulating tolerance to drought and flooding. These two cis-elements are regulated by different transcription factors that downstream of RAP2.12 results in differential accumulation of hypoxia-responsive transcripts. The evolution from one cis-element haplotype to the other is associated with the colonization of humid environments from arid habitats. This gene thus promotes both drought and flooding adaptation via an adaptive mechanism that diversifies its regulation through noncoding alleles.
Arabidopsis thaliana adapts to humid environments through selection acting on the oxygen-sensing system.
INTRODUCTION
Diverse environmental factors may pose different stresses on widely distributed species. Worldwide, water patterns are suffering from increasing and violent fluctuations due to accelerating global climate change (1). Over the past decades, the frequency of global drought and flood threats induced by climate change has increased markedly, and the trend is becoming serious (2, 3). For plants that have adapted to previous water patterns, this disturbance has severely affected their survival and distribution, and it is urgent to find underlying key genes and mechanisms that control the responses to such contrasted extremes related to water. Drought and flooding are typically contrasting stresses for plants. Morphological and physiological responses, signaling pathways, and regulatory mechanisms of plants to these two stresses are very different. To adapt to water scarcity, plants develop morphological architecture to maximize water uptake, physically close stomata to reduce water loss, and transmit water availability signals and photoassimilates between shoots and roots to improve plant health (4). However, to facilitate survival of submergence, including flooding and waterlogging (both of which result in reduced oxygen availability, hypoxia), plant energy metabolism switches from aerobic to anaerobic, and plants can form aerenchyma and undergo rapid shoot elongation by coordinating protein kinases, transcription factors, and plant hormones to escape from submergence (5). Oxygen sensing associated with hypoxia is controlled by the PLANT CYSTEINE OXIDASE (PCO) branch of the PROTEOLYSIS 6 (PRT6) N-degron pathway acting on Group VII ETHYLENE RESPONSE FACTOR (ERFVII) transcription factors (6, 7). Arabidopsis thaliana contains five ERFVIIs that are stabilized under low oxygen when PCO activity is inhibited (8), and stabilized ERFVIIs activate expression of a set of hypoxia-associated genes, including those encoding enzymes of anaerobic metabolism (9, 10). This pathway (that also senses nitric oxide, NO) has also been shown to influence tolerance to drought and salinity (11, 12). In A. thaliana accession Col-0, RELATED TO AP 2.12 (RAP2.12) was shown to be the predominant ERFVII in responses to hypoxia (10, 13). In Oryza sativa, several ERFVIIs, including SUBMERGENCE 1A (SUB 1A) and SNORKEL 1 (SK1) and SK2, were originally identified in different landraces, and positively enhance tolerance to flooding, although through opposite mechanisms of quiescence or escape growth strategies, and SUB1A was also shown to enhance drought tolerance (14, 15).
To explore the genetic and molecular mechanisms relevant to ecological shifts associated with water availability in plants, we analyzed two accessions of A. thaliana with contrasting humidity conditions, one from the Sichuan Basin, which is well known as the West China Rain Belt (annual precipitation, 1092.936; aridity index, 1.029), and the other from Tibet with a contrasting arid climate (annual precipitation, 395.084; aridity index, 0.339; fig. S1). We constructed a recombinant inbred line (RIL) population using the two accessions that are adapted to these contrasting extreme environments. Using this approach, we identified a key ERFVII gene, RAP2.12, which plays a role in adaptation and transformation from drought to flood tolerance. We have shown that the level of constitutive expression of RAP2.12 is highly correlated with adaptation to native environmental humidity in accessions collected globally. This adaptive change was shown to be caused by an evolutionary shift of two core cis-elements, “WT” and “W” boxes, in the promoter of RAP2.12, which may have transformed its widespread geographical colonization from environments typified by drought with narrow relict distribution, to humid habitats. Our results highlight plant genetic adaptation to environmental patterns of water availability through noncoding DNA mutations of cis-elements of the same core gene with important roles in the contrasting habitats.
RESULTS
Tibet and Sichuan accessions are highly diverged in drought and flood tolerance
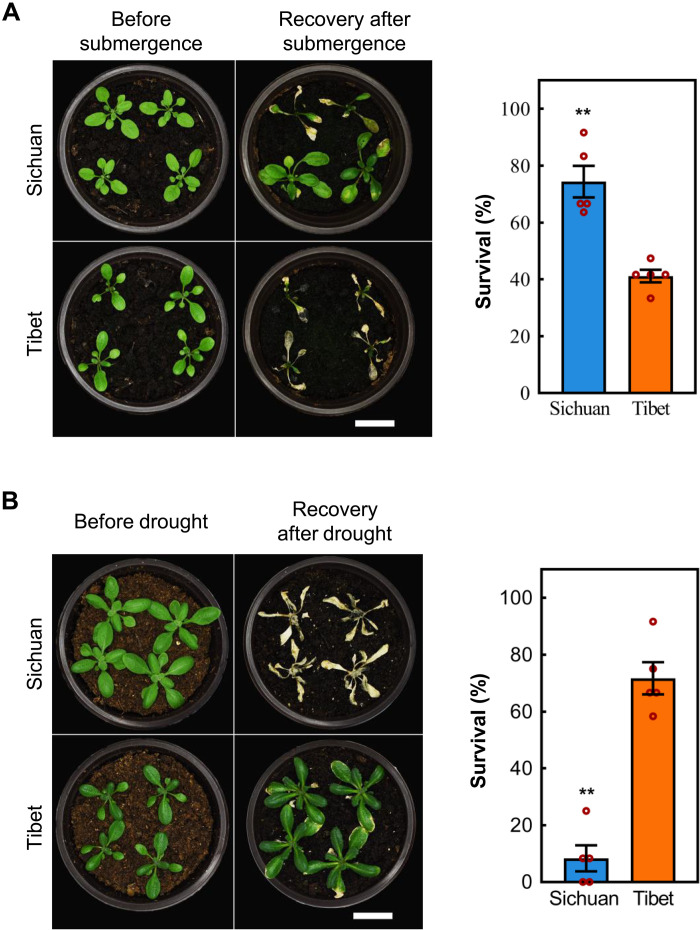
We compared the tolerances of the two accessions Sichuan and Tibet to drought and flooding extremes under controlled environmental conditions. Consistent with their native climatic adaption, Sichuan accession had higher survival, higher relative rosette dry weight (RRDW; survival rosette dry weight/control), and healthier physiological properties than the accession Tibet both at the submergence and recovery (reoxygenation) stages (Fig. 1A and fig. S2, A to C). In contrast, following cessation of watering, Tibet accession showed greater survival, higher relative water content, more sensitive stomatal regulation, and slower response to raised reactive oxygen species (ROS) (Fig. 1B and fig. S2, D to F). Therefore, Tibet and Sichuan accessions are highly diverged in tolerance to drought and flood both in overall phenotype and underlying physiology.
Fig. 1. Adaptive divergence to flood and drought in A. thaliana accessions Sichuan and Tibet.
(A and B) Representative seedlings and survival of Sichuan and Tibet accessions after 64 hours of dark/submergence and 5-day recovery (A), or without watering for 21 days, followed by 5-day rewatering (B), showing contrasting opposite tolerance to flooding and drought between Sichuan and Tibet accession. Data are means ± SE (n = 5 repeats). **P < 0.01 (Student’s t test). Scale bars, 2 cm.
Fine mapping of a submergence response QTL (quantitative trait locus) in the Sichuan accession
To identify genetic factors controlling the contrasting stress responses, we carried out a genome-wide association study (GWAS) with a population of 623 RILs constructed by crossing Sichuan and Tibet accessions. We selected as the representative traits average survival and RRDW measured 1-week recovery after submergence for 48, 60, and 72 hours at the 14-day stage (fig. S3). The GWAS for both survival and RRDW revealed the same highly associated peak on chromosome 1 (fig. S4, A and B). Single-nucleotide polymorphisms (SNPs) in a ~20-kb window around the top peak were used for haplotype analysis, and the RILs in this region were divided into three groups, Sichuan, Tibet, and heterozygous (fig. S4C). Sichuan haplotype showed a significantly higher survival and RRDW (fig. S4, D and E) than Tibet haplotype in this region. To narrow the candidate gene region, we generated the mapping population using the Tibet accession as a recurrent parent, and each generation was screened by flooding stress (fig. S5, A to D). With the overlap in results between the genome sequence of this population and haplotype analysis, the candidate region was finally narrowed in a ~7–kilobase pair (kbp) interval containing two genes, RAP2.12 (AT1G53910) and GLIP5 (AT1G53920) (fig. S4F). RAP2.12 was expressed at a significantly higher constitutive level in untreated plants of Sichuan compared to Tibet and also displayed a significantly increased expression pattern in Sichuan during flooding, while expression did not respond to flooding in the Tibet accession (fig. S4, G and H). Thus, RAP2.12 represented the most likely causal gene that underlies the variation in tolerance of the two accessions to flooding stress.
RAP2.12 directly activates expression of multiple hypoxia-responsive genes (e.g., LBD41, PCO1, HUP09, HUP43, PDC1, PGB1, SUS1, SUS4, and ADH1) that contain the consensus cis-element sequence 5′-AAACCA(G/C)(G/C)(G/C)GC-3′ [hypoxia-responsive promoter element (HRPE)] (10, 13, 16, 17). A total of 136 SNPs within the first 2 kbp upstream of the RAP2.12 ATG, 8 SNPs within two exons, and 9 SNPs within the intron were identified among Col-0, Sichuan, and Tibet accessions, which result in 62 substitutions and 19 indels in the promoter and 5 amino acid substitutions in the coding sequences (CDSs) (none of them in the DNA binding AP2 domain) (figs. S6, A and B, and S7).
Constitutive expression of RAP2.12 corresponds significantly to native climatic humidity
We constructed a reporter plasmid containing 471 bp of the hypoxia-responsive PGB1 promoter fused to luciferase (HRPE located at −172 to −183 bp from ATG) to test whether the RAP2.12 amino acid variation of Tibet affected the activation ability of hypoxic response genes. We found that three forms (Col-0, Sichuan, and Tibet) of RAP2.12 activated PGB1 promoter equally well (fig. S8A), and Tibet and Sichuan RAP2.12 complemented a pentuple mutant that removes the activity of all five A. thaliana ERFVIIs [rap2.12, rap2.2, rap2.3, hre1, and hre2, hereafter erfVII (18)] (fig. S8, B and C), indicating that the amino acid substitutions among these three accessions do not cause differentiation in flooding-induced hypoxia tolerance.
However, mRNA abundance of RAP2.12 in Col-0 and Sichuan was significantly higher under normal growth conditions and significantly more up-regulated after submergence treatments in 18-day-old rosettes than in Tibet, where expression was significantly lower and did not respond to flooding (figs. S4G and S9A). Because the other four ERFVII family members are also involved in hypoxia responsiveness (10, 13), we also tested their expression in response to submergence and found that none of them showed difference in constitutive expression, whereas RAP2.2, RAP2.3, and HRE2 in all three accessions were significantly up-regulated during submergence-induced hypoxia stress (fig. S9, B to D). Among them, we also found that HRE1 in Tibet, like RAP2.12, could not be up-regulated in response to submergence (fig. S9E).
We hypothesized that the constitutive expression level of ERFVIIs may be associated with native climatic humidity in different accessions collected from different geographical locations. We therefore analyzed correlations between ERFVII constitutive RNA expression levels in 32 accessions using data available on the eFP Browser (fig. S10A and table S1) (19), in relation to local precipitation and aridity index. For RAP2.12, a significant positive correlation with native humidity was observed (R = 0.393 and P = 0.026; fig. S10C), which was not shown for other ERFVIIs (P > 0.113; fig. S10, D to K, and table S1). These results highlight the significant association between the expression of RAP2.12 and climatic humidity adaption.
Two allelic cis-elements, the WT and W boxes, are involved in RAP2.12 differential expression
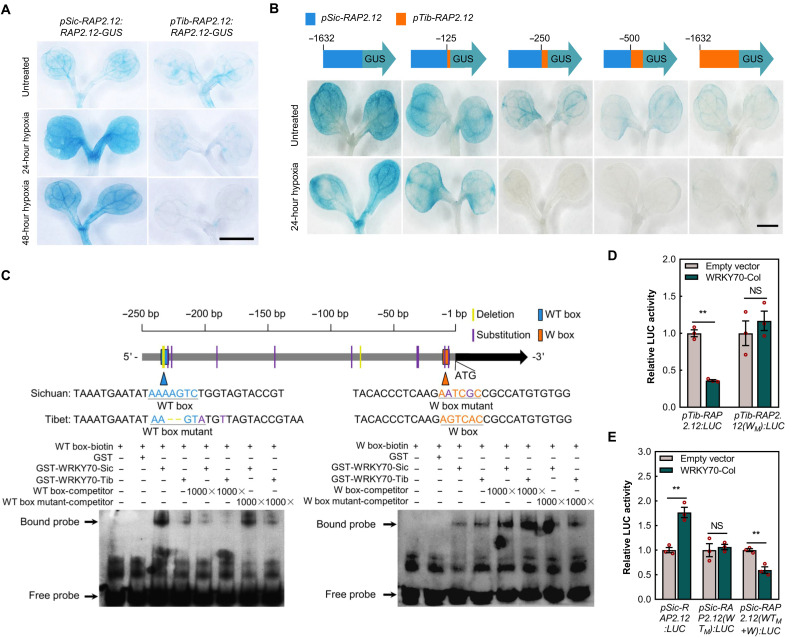
With protein-b-glucuronidase (GUS) fusion lines containing ~1.6 kb of DNA upstream of the initiating ATG [promoter (p)] and complete coding region (pERFVII:ERFVII-GUS), in the Col-0 background we could show that in response to hypoxia, transgenes with promoter-Sichuan (pSic)–RAP2.12:RAP2.12-GUS accumulated RAP2.12-GUS protein (as would be expected because of stabilization of this PCO N-degron pathway substrate in the absence of oxygen, as previously also shown for pCol-0-RAP2.12:RAP2.12-GUS) (20, 21). However, promoter-Tibet (pTib)–RAP2.12:RAP2.12-GUS could not promote RAP2.12-GUS accumulation in response to hypoxia, and RAP2.12-GUS protein almost completely disappeared after 48 hours of treatment with 5% oxygen (Fig. 2A). To confirm which SNP sites result in the divergence of RAP2.12 accumulation, we segmentally substituted the pSic promoter of pSicRAP2.12:RAP2.12-GUS with pTib-RAP2.12. Compared with normoxia, the accumulation of Sichuan RAP2.12-GUS protein under hypoxia was highly reduced when the region between −250 and −125 bp from ATG was substituted with pTib-DNA (Fig. 2B). A total of nine substitutions and three deletions were distributed in seven areas in this region (Fig. 2C and fig. S6A), among which we found two previously described core cis-element sequences, the WT box (GACTTTT) and W box (TGAC). These were dislocated in the two forms of the promoter, Sichuan containing the WT box and Tibet the W box (Fig. 2C and fig. S6A). The W box (present in some ERFVII promoters) has been shown to be bound by many plant WRKY transcription factors (22–26), whereas only WRKY70 binds to the WT box (27, 28), which is only found in the RAP2.12 promoter.
Fig. 2. Dislocation of WT and W box cis-elements in 250-bp promoter of RAP2.12 and interaction with WRKY70.
(A and B) Representative shoot images of RAP2.12-GUS reporter protein fusion lines in pSic-RAP2.12:RAP2.12-GUS and pTib-RAP2.12:RAP2.12-GUS Col-0 transgenics (A) and RAP2.12-GUS reporter lines in pSic-RAP2.12:GUS (blue), pTib-RAP2.12:GUS (orange), and piecewise substitutions of RAP2.12 promoter from Sichuan to Tibet accession (B), in 4-day-old seedlings after treatment with air (untreated) or 5% (v/v) oxygen for the indicated time. Differences of the promoters are shown in fig. S7. Scale bars, 1 mm. (C) EMSA to test for interaction of WT box and W box, with WRKY70-Sic and WRKY70-Tib. Gene structure and 30-bp-length sequences used for biotin labeling and unlabeled competitive binding are shown on the top. Core cis-element sequences of WT box and W box are in blue and orange, respectively. Residue difference between GST-WRKY70-Sic and GST-WRKY70-Tib protein is shown in fig. S11A. (D and E) Repression by WRKY70 on the promoter of pTib-RAP2.12 depends on the W box cis-element. pTib-RAP2.12(WM), W box cis-element mutant to Sichuan accession form (D). Transactivation of pSic-RAP2.12 by WRKY70 depends on the WT box cis-element. pSic-RAP2.12(WTM), WT box cis-element mutant to Tibet accession form; pSic-RAP2.12(WTM + W), adding pSic-RAP2.12(WTM) promoter to the W box cis-element corresponding to the Tibet accession (E). Data are means ± SE (n = 3 repeats). **P < 0.01; NS, not significant (Student’s t test).
WRKY70 acts to bind the RAP2.12 promoter hap-WT box
To investigate whether WRKY70 is involved in the divergence of transcript levels of RAP2.12 in Tibet and Sichuan accessions, we conducted electrophoretic mobility shift assays (EMSAs) using core cis-element sequences with their flanking sequence. Although there is one–amino acid substitution in WRKY70 between Tibet and Sichuan (fig. S11A), both WRKY70-Sic and WRKY70-Tib bound to the WT box (Fig. 2C). WRKY70 bound specifically to the WT box but much less strongly and nonspecifically to the W box cis-element (Fig. 2C). In addition, luciferase transactivation assays suggested that the three forms of WRKY70 (Col-0, Sichuan, and Tibet) enhanced expression from the Sic-RAP2.12 promoter but they all reduced expression from the Tib-RAP2.12 promoter (Fig. 2, D and E, and fig. S11B). When the W box core cis-element of Tib-RAP2.12 promoter was mutated to the Sichuan form, WRKY70 repression was removed (Fig. 2D). Similarly, when we mutated the WT box core cis-element of Sic-RAP2.12 promoter to the Tibet form, transactivation was lost (Fig. 2E). Substituting the mutated Sic-RAP2.12 promoter W box to the Tibet form resulted in reduced luciferase (LUC) activity (Fig. 2E). Because WRKY70 could not competitively bind to the W box cis-element (Fig. 2C), we believe that WRKY70 indirectly inhibited Tib-RAP2.12 expression. The proximity of the W box to the transcription start site may also interfere with transcription initiation under submergence.
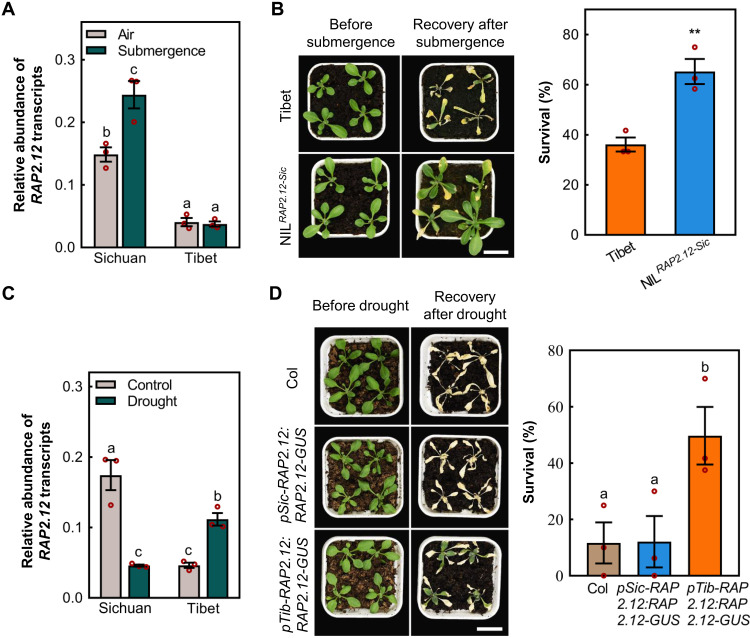
The mRNA levels of WRKY70 increased with submergence time in all three accessions (Sichuan, Tibet, and Col-0) (fig. S11C). In the Col-0 background (RAP2.12 haplotype containing the WT box but not the W box; fig. S6A), wrky70 transferred DNA (T-DNA) insertion mutant was more sensitive, and WRKY70-overexpressing line (35S:WRKY70) was more tolerant, to submergence (fig. S12, A to C) and hypoxic conditions (fig. S12, D and E). Physiological measurements of malondialdehyde (MDA) and ionic leakage during submergence and reoxygenation also revealed that the wrky70-mutant plant rosettes were more susceptible to submergence, while 35S:WRKY70 was less damaged during submergence and reoxygenation (fig. S13, A and B). Furthermore, uncoupling expression of RAP2.12 from WRKY70 (35S:RAP2.12) in the wrky70-mutant or analyzing near-isogenic lines (NILs) (replacing Tibet hap-W box RAP2.12 promoter with Sichuan hap-WT) both increased hypoxia tolerance (Fig. 3B and fig. S12, F and G), suggesting that WRKY70 activates transcription of RAP2.12 through the WT box. During dark submergence, hypoxia response marker genes were expressed lower in the wrky70 mutant than in Col-0, further indicating that WRKY70 acts through RAP2.12 (fig. S14, A to D). Of all the ERFVIIs, only constitutive expression of RAP2.12 was significantly reduced in wrky70 under normal conditions compared to Col-0, and submergence treatment could not induce its expression, similar to the behavior of RAP2.12 in the Tibet accession (Fig. 3A and fig. S15A). The normoxic expression of the other ERFVIIs was not affected in wrky70, and they were all similarly up-regulated after submergence treatment (fig. S15, B to E), indicating that WRKY70 in the Col-0 accession specifically functions to enhance expression of RAP2.12.
Fig. 3. Differential expression of RAP2.12 driven by pSic-RAP2.12 or pTib-RAP2.12 results in contrasting opposite tolerance to flooding and drought.
(A) The relative expression level of RAP2.12 in shoots after air or submergence treatments (20 hours) in Sichuan and Tibet accessions. (B) Phenotypic and survival comparison between Tibet and a Tibet NIL containing introgressed RAP2.12 (Sichuan), NILRAP2.12-Sic. Phenotypes after 48 hours of dark/submergence and 7 days recovery. Scale bar, 2 cm. (C) The differential relative expression level of RAP2.12 in shoots between Sichuan and Tibet accessions in response to drought. (D) Comparison of drought tolerance in Col-0, pSic-RAP2.12:RAP2.12-GUS, and pTib-RAP2.12:RAP2.12-GUS (all in Col-0 background). Phenotypes after 14 days without watering, followed by 2-day rewatering. Scale bar, 2 cm. Data are means ± SE (n = 3 repeats). Different letters denote significant differences according to Tukey’s post hoc test (P < 0.05). **P < 0.01 (Student’s t test).
The role of the RAP2.12 promoter in conferring tolerance to drought
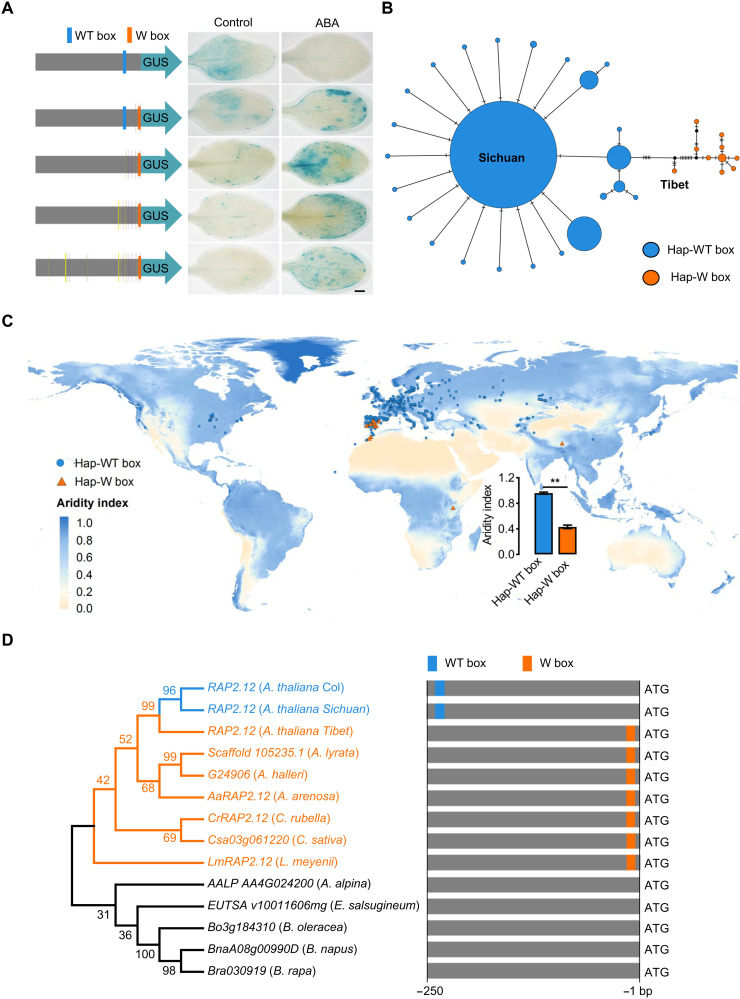
The fact that RAP2.12 is expressed in the Tibet accession even in the absence of a functional WT box suggests that other promoter elements must control expression. The expression of RAP2.12 transcripts in both in Col-0 and Sichuan accessions, which both contain the WT box (fig. S6A), showed a significant decrease under drought stress in roots and shoots (Fig. 3C and fig. S16, A to C), while on the contrary, in the Tibet accession, RAP2.12 transcripts showed a significant up-regulation (Fig. 3C). In addition, our drought treatments and previously reported results (12) showed that there was no significant difference in drought resistance among Col, rap2.12, erfVII, and pSic-RAP2.12:RAP2.12-GUS in Col-0 (Fig. 3D and fig. S17). However, pTib-RAP2.12:RAP2.12-GUS and 35S:RAP2.12 (derived from Col-0) in Col-0 background exhibited increased drought resistance (Fig. 3D and fig. S17), which is consistent with results of heterologous expression of ERFVIIs from sweet potato (IbRAP2.12) and wheat (TaERF1) in Col-0 (29, 30). By spraying abscisic acid (ABA) onto the Col-0 transgenic lines of pSic-RAP2.12:GUS, pTib-RAP2.12:GUS, and piecewise substitution of promoter transgenic lines, we found that the expression of pSic-RAP2.12 was reduced relative to control but when the Tibet-derived W box was present in the same promoter, expression was induced by ABA (Fig. 4A).
Fig. 4. Occurrence of W box and WT box in the proximal promoter of RAP2.12 in the Brassicaceae and differential association with drought and aridity index.
(A) Representative leaf images of RAP2.12-GUS reporter lines in pSic-RAP2.12:GUS, pTib-RAP2.12:GUS, and piecewise substitutions of RAP2.12 promoter from Sichuan to Tibet accession (see also Fig. 2B) in 14-day-old seedlings after spraying with 30 μM ABA for 6 hours. Scale bars, 1 mm. (B) Haplotype network of the RAP2.12 promoter in A. thaliana over the world. Each haplotype is represented by a circle. The accessions with WT box and W box are divided into Hap-WT box and Hap-W box clades. The black dots indicate missing haplotype, and circle sizes represent relative frequency. (C) Geographic distribution of Hap-WT box and Hap-W box RAP2.12 on aridity index in haplotype network testing (data S1). (D) Phylogenetic analysis of RAP2.12 promoter in Brassicaceae species by the neighbor-joining method. The gene names or numbers and species names are given in the phylogenetic tree. The distributions of WT box and W box on the RAP2.12 orthologous gene promoter are displayed on the right, and the details of the sequence are shown in fig. S19.
To analyze the distribution of haplotypes in A. thaliana accessions, we downloaded the 1- to 250-bp promoter sequences of 672 global distributed Arabidopsis accessions (table S2 and data S1), and they were perfectly divided into the hap-WT box and hap-W box in this region by haplotype network analysis (Fig. 4B). Only 12 accessions with dispersed distributions in Southwest Europe, Africa, and Asia share the hap-W box, while the other 660 accessions have the hap-WT box (Fig. 4, B and C, and table S2). Whereas most accessions with the hap-WT box are distributed in humid regions, the 12 hap-W box accessions were all sparsely and narrowly distributed in drier regions (Fig. 4, B and C). Four of these accessions, Tanz-1, IP-Cat-0, IP-Cem-0, and IP-Her-12, were found to be highly sensitive to flooding in the past comparisons (31, 32).
WT and W box cis-elements have undergone a shift in evolutionary history
The dislocated distribution of two allelic variations, WT and W box, cis-elements of RAP2.12 in arid and humid climates indicates that drought and hypoxic stresses caused by precipitation or environmental water have been a strong selection force. To better understand the effect of this selection, we analyzed whether different accessions of A. thaliana around the world are involved in this evolutionary shift by using the global nonrepetitive sites of 1475 natural inbred lines of A. thaliana (table S3), with the native precipitation, aridity index, and oxygen availability to roots data. We found that native sites with less precipitation were more prone to experience drought stress (fig. S18, A to C), and sites with heavy precipitation or high humidity were more limited in oxygen availability (fig. S18, C to F). The ecological selection attests to the significance of divergent climate adaptation of these two cis-element alleles.
To examine the origins of the two cis-element alleles, about 250 bp from ATG in RAP2.12 orthologous genes of 11 Brassicaceae species was used to construct a phylogenetic tree by the neighbor-joining method (fig. S19, A and B). Combined with the topology of the tree and the existence of two cis-elements in the sequences used, we found that the hap-WT box of RAP2.12 originated after A. thaliana had been established through diverging from closely related species (Fig. 4D and fig. S19B), and this allelic haplotype extensively expanded into humid climate areas around the world (Fig. 4C). In contrast, the hap-W box of RAP2.12 was found to be ancestral because it was present in all closely related species and genera. It first appeared even before divergences among four closely related genera, Arabidopsis, Lepidium, Capsella, and Camelina (Fig. 4D and fig. S19B). Haplotype analyses (Fig. 4B) also indicated that Tibet and the other 11 accessions of A. thaliana with the hap-W box comprise a clear clade, distinct from others with worldwide distributions. These populations may have survived as sparse relicts in Southwest Europe, Asia, and Africa before the worldwide colonization of this species after the Quaternary climatic oscillations (33). In addition, these results suggest that A. thaliana originated in dry and arid regions similar to most Brassicaceae groups and the later adaptation to flooding stress promoted its multiple colonization of high precipitation or humidity regions of Eurasia and Africa.
DISCUSSION
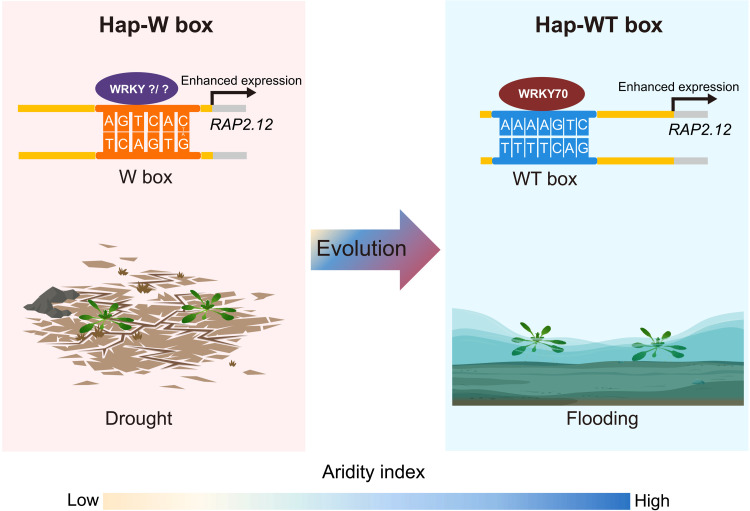
Because high precipitation and humidity lead to hypoxic stress (fig. S18, C to F), increased constitutive RAP2.12 expression may enhance the survival of local populations. WRKY70 enhances constitutive expression of RAP2.12, which is stabilized under hypoxic conditions that inhibit PCO activity, thus promoting submergence tolerance (Fig. 5). However, during drought stress, in the other extreme of climatic humidity, WRKY70 and its paralogs (WRKY46/54) are phosphorylated by BIN2 kinase, which leads to their destabilization and promotes expression of drought-inducible genes (4, 34). Despite the fact that RAP2.12 protein has been shown to promote drought tolerance by controlling the stomatal ABA response and ROS in several species (e.g., A. thaliana, rice, wheat, barley, and sweet potato) (12, 14, 30, 35), expression of RAP2.12 in the WT box haplotype does not respond to drought. However, for the alternative hap-W box populations (including Tibet), RAP2.12 can be up-regulated by other WRKY or unknown genes(s) through the W box cis-element (Figs. 4A and 5), as it was recently shown that the promoter of RAP2.2 (that also contains the W box) binds WRKY 33 and 12 (26). Therefore, a shift from W to WT box cis-element in RAP2.12 promoter is critical for humid adaptation of A. thaliana from ancestral more arid habitats. Therefore, this evolutionary trajectory reveals an unreported genetic shift for ecological adaptation between contrasting abiotic stresses (Fig. 5). Similar shifts may also exist for other key genes involved in adaptations to contrasted stresses in plants. This mechanism diversifies the upstream regulatory network of the gene in different populations, allowing adaptation to different environments with contrasting water availability. Such an evolutionary shift also suggests that crop improvement could be conducted by manipulation of cis-regulatory element combinations, to create new crop types that stabilize yield in the face of multiple abiotic stresses.
Fig. 5. A genetic model for allelic evolution of expression of RAP2.12 from drought to flooding.
In Hap-W box populations of dry areas, the expression of RAP2.12 is enhanced by other WRKY 12/33 or other unknown genes(s) through the W box cis-element to promote survival under drought stress. In Hap-WT box populations, the shift of the RAP2.12 regulatory cis-element from W to WT box allows WRKY70 to enhance its expression through WT box and promote the worldwide humid colonization in A. thaliana.
MATERIALS AND METHODS
Plant materials
The seeds of Sichuan accession [Arabidopsis Biological Resource Center (ABRC) stock no.: CS79063] were collected from Yilong County, Nanchong City, Sichuan Province, and those of Tibet accession (ABRC stock no.: CS79062) were collected from Chengguan District, Lhasa City, Tibet Autonomous Region. RILs were constructed by crossing the Sichuan with Tibet accessions and self-pollination for seven generations. All A. thaliana mutants used are in the Columbia (Col-0) accession. The pentuple mutant erfVII (rap2.12 rap2.2 rap2.3 hre1 hre2) was described previously (18). T-DNA insertion lines, wrky70 (SALK_025198) (34) and rap2.12 (SAIL_1215_H10) (21), were obtained from the Arabidopsis Biological Resource Center.
Growth conditions
Surface-sterilized (10% NaClO for 10 min) and sterile water–washed seeds were sown on half Murashige & Skoog (MS) media and chilled at 4°C for 72 hours and then germinated and vertically cultivated under a 16-hour light (137 × 100 lux, Philips F17T8/TL841 17 W)/8-hour dark (23°C) photoperiod. The seedlings cultivated on the plate and in the soil were both grown in a 23°C constant temperature growth chamber.
Stress treatments
Submergence treatment
All pots with different genotypes were staggered in the same flat, and their placement positions are exchanged once a day. Eighteen-day-old plants were completely dark submerged in a tank (W:L:H, 37 × 50 × 25) with leaves 10 cm below the water surface. After the treatment times indicated, the water in the tank was removed, and plants were returned to normal photoperiodic conditions (16/8 hours, light/dark). Flooding tolerance was assayed at least three times by using 10 to 16 plants per genotype each time. After 7 days of recovery, the rosette leaf dry weights of the surviving plants and controls were weighed to calculate RRDWs in all submergence treatments. Plant cultivation and submergence procedures were described previously (36).
Hypoxia treatment
Hypoxic equipment used in this study was described previously (37), and the hypoxic conditions were conducted by flushing the humidified 99.99% N2 gas under dark conditions. The hypoxia treatment process was as follows: Eighteen-day-old seedlings were pretreated with 5% oxygen for 24 hours and then treated with oxygen levels below 0.1% for 16 hours. After hypoxic treatment, the seedlings were returned to normoxic conditions with normal photoperiodic conditions (16/8 hours, light/dark).
Drought stress treatment
Seven-day-old plants were transferred into 200 g of soil with 65% water content in each pot. The genotypes were staggered in the same flat, and their placement positions are exchanged once a day. During drought stress, the flats were rotated frequently to minimize the effect of their growth environment (34). Survival analysis in Fig. 1B was calculated after 3 weeks of no watering and 5 days of rewatering from five biological replicates carried out at different times. Each genotype had 12 plants in three pots for each biological repeat. In addition, survival analyses in Fig. 3D and fig. S17 were carried out after 2 weeks of no watering and 2 days of rewatering for three replicates containing 10 to 16 individuals in each genotype.
GWAS and mapping
A total of 623 2-week-old RILs were submerged for 48, 60, and 72 hours, respectively, five individuals per RIL in each treatment. After 7 days of recovery under normal light conditions, the survival of each line was calculated, and the dry weights of the surviving individuals were weighed to measure the RRDW. Three-week-old rosette dry weights were used as control, and the RRDW was calculated by the following formula: average survival weight/average control weight. Subsequently, the survival and the RRDW were used for GWAS.
For SNP identification, paired-end resequencing reads of the 623 RILs in this study were mapped to the parent Sichuan accession HiFi reference genome with the Burrows-Wheeler Alignment tool (BWA version 0.7.17) (37). Next, the markdup function of Sambamba (version 0.7.1) (38) was used to mark reads that were duplicated in library preparation or sequencing. Last, SNP and indel calling were performed with the Genome Analysis Toolkit (GATK version 4.1.1.7) (39). Extra filtration steps were applied to the raw SNPs and indels using Vcftools (version 0.1.15) (40).
For GWAS, we used a total of 1,192,378 high-quality SNPs with minor allele frequencies greater than 3% and missingness rates no greater than 20%, and we included phenotypes (average survival and RRDW) from each line. The analysis was performed using GEMMA software by the univariate mixed linear model method, and the population structure was controlled using the relatedness matrix to control spurious associations (41). The GWAS significance threshold was determined by Bonferroni correction with adjusted P values = 0.05/n (n = 279,812 SNPs from the GEMMA output). The genome-wide significance level was set as 1.8 × 10−7 in both phenotypes. SNPs in a region of 20 kb around the top peak were used for haplotype analysis by hierarchical clustering method in pheatmap in R, and average survival and RRDW in each line were used to distinguish the resistance of haplotypes to flooding stress.
For mapping, as shown in fig. S5, the hybrid F1 generation of Sichuan and Tibet accessions was used for recurrent backcrossing with Tibet accession for five rounds, and then BC5F1 lines were self-bred for three generations; each generation is screened with 48 hours of submergence stress to select the most resistant lines. The submergence-resistant lines in this population were sequenced and then overlapped with the results of RILs haplotype analysis (fig. S4). The candidate region was lastly narrowed in a ~7-kb interval with two genes, RAP2.12 (AT1G53910) and GLIP5 (AT1G53920).
Plasmid construction
In the dual-luciferase assay, a 471-bp fragment upstream of the PGB1 ATG from the Col-0 or 1.6-kb fragment upstream of the RAP2.12 ATG from Sichuan or Tibet was cloned into the pGreen-0800-LUC vector as a reporter. Promoters with the replacement in the WT box and W box motif were generated by overlapping polymerase chain reaction (PCR). The constructs of p35S:RAP2.12-Col, p35S:RAP2.12-Sic, p35S:RAP2.12-Tib, p35S:WRKY70-Col, p35S:WRKY70-sic, and p35S:WRKY70-Tib were generated by inserting the coding regions of RAP2.12 or WRKY70 from three accessions into pCAMBIA1300 vector.
To construct pSic-RAP2.12:RAP2.12-GUS and pTib-RAP2.12:RAP2.12-GUS C-terminal GUS reporter protein fusion lines, 1.6-kb sequence upstream from the ATG and RAP2.12 CDSs with the stop codon removed by primers (table S4) were amplified and cloned into the pCXGUS-P vector (36) through the Xcm I enzyme sites. The pSic-RAP2.12:GUS, pTib-RAP2.12:GUS, and piecewise substitutions of promoter constructs were generated by overlapping PCR. The primers for plasmid construction were listed in table S4.
Production of transgenic Arabidopsis
To generate 35S:WRKY70 or 35S:RAP2.12 overexpressing transgenic plants, the Col-type WRKY70 or RAP2.12 CDSs were cloned into the pCAMBIA1300 by Xba I and Kpn I sites, and the wild-type plants of Col-0 were transformed with Agrobacterium tumefaciens (GV3101) carrying the constructed pCAMBIA1300. The 35S:RAP2.12-Sic/erfVII and 35S:RAP2.12-Tib/erfVII lines were generated in the erfVII background via the floral dip method (42) and identified by hygromycin screening, and transgenic lines with similar expressing levels were used in our experiments. The 35S:RAP2.12/wrky70 combination was generated in wrky70 T-DNA insertion line background via the floral dip method.
MDA measurement
MDA levels were measured according to Yuan et al. (43). Rosettes were weighed and pulverized in 1 ml of trichloroacetic acid buffer [0.1% (w/v)]. After centrifuging at 10,000g for 10 min at 4°C, 0.2 ml of supernatant was taken out to mix with 0.8 ml of concentrated trichloroacetic acid [20% (w/v)] containing 0.5% thiobarbituric acid. The mixture was boiled for 15 min and cooled quickly with ice. After centrifuging at 10,000g for 5 min at 4°C, the absorbance of the mixture was detected at 532 and 600 nm using a spectrophotometer plate reader, and the MDA contents were calculated accordingly (44).
Ion leakage assay
Ion leakage was measured as previously described (36). Rosettes of 18-day-old plants after submergence or 24-hour submergence and reoxygenation treatments were placed in 10 ml of deionized water. After vacuuming for 10 min and shaking for 1 hour, initial conductivity (S1) was measured with a conductance meter and then the rosettes were boiled for 10 min. After cooling them to room temperature with water, samples were shaken for another 10 min to measure the final conductivity (S2). Ion leakage was calculated as S1/S2 × 100%.
Relative water content measurement
Relative water content was measured and calculated with the following formula: (FW − DW)/(RW − DW) × 100%, according to Linster et al. (45). The fresh weights (FW) of 7 to 12 individual rosettes were measured and followed by 12-hour rehydration in 4°C cold water to determine the rehydrated weight (RW) of each sample. Then, the dry weights (DW) of rosettes were weighed after drying at 80°C for 24 hours.
Stomatal aperture measurement
Abaxial leaf epidermises were imaged by a confocal fluorescence microscope (Leica TCS SP5 II system, Leica, Wetzlar, Germany), and the ImageJ software was used to measure the stomatal aperture. Ten stomata were measured on the largest leaf of each individual.
Visualization of ROS
ROS accumulation was visualized by 3,3′-diaminobenzydine (DAB) staining (46). The rosettes were soaked in DAB solution (1 mg ml−1 in water, pH 3.8) for 8 hours and cleared in 95% boiling alcohol for 10 min and then photographed.
GUS staining
Seedlings or leaves were incubated in staining buffer [50 mM tris-Cl (pH 7.5), 0.2% Triton X-100, and 1 mM 5-bromo-4chloro-3-indolyl b-d-glucuronic acid (X-gluc)] at 37°C for 2 hours and then transferred into 100% alcohol to remove chlorophyll.
Luciferase activity assay
Luciferase activity assays were performed in young Nicotiana benthamiana leaves (47). The reporter constructs together with the different effector constructs were coinjected into N. benthamiana, and the Renilla luciferase (REN) gene was used as internal transfection control. The injected tobacco was incubated in dark conditions for 2 days and cultivated under normal conditions for 1 day, and then Dual-luciferase Reporter Assay System [Dual-Luciferase(R) Reporter Assay System, Promega, catalog no. E1960] was used to detect the luciferase and Renilla luciferase activities.
RNA isolation and quantitative real-time PCR
Real-time PCR was performed as previously described (36). Total RNA was isolated from rosettes using TRIzol reagent (Invitrogen Life Technologies, Shanghai, China) following the manufacturer’s instructions. Two micrograms of RNA was reverse-transcribed using PrimeScript RT reagent Kit with gDNA Eraser (Takara, catalog no. RR047A) to obtain first-strand complementary DNA synthesis. Quantitative PCR was performed with SYBR Green PCR Master Mix (TaKaRa) on a Bio-Rad CFX96 Real-Time System. ACTIN2 (At3g18780) was used as the internal control. Primers for quantitative PCR are listed in table S4.
Electrophoretic mobility shift assay
Light Shift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, catalog no. 20148) was used in all the EMSA experiments. The GST-WRKY70-Sic and GST-WRKY70-Tib proteins were expressed through 1 mM isopropyl β-d-1-thiogalactopyranoside induction in Escherichia coli strain BL21 (DE3) and then purified using Glutathione-Sepharose 4B beads (GE Healthcare). The biotin-labeled DNA was labeled at 5′ end of cis-element and synthesized by Sangon Biotech (Shanghai, China). The probe sequences are listed in table S4.
Haplotype network construction
A 253-bp promoter from ATG of RAP2.12 containing 24 SNPs in 127 accessions was used for generating the statistical parsimony network. The haplotype network was implemented by Network 10 software (www.fluxus-engineering.com) (48). Detailed sequences are available in data S1.
Phylogenetic analyses
The genomic locations and the source of each RAP2.12 orthologous sequence from 11 Brassicaceae species used in this study are listed in fig. S19. The phylogenetic tree was built using the neighbor-joining method with 500 bootstrap replicates by MEGA7 (49). Multiple sequence alignments were performed using the Muscle algorithm (50).
Statistical analysis
All statistical tests were performed using SPSS 17.0, and significance is defined by P value and marked as the following code: * < 0.05 and ** < 0.01. Statistical significance was assessed using either Student’s t test between groups or one-way analysis of variance (ANOVA) among multigroups. Quantitative data were shown as means ± SE.
Data availability
The raw sequencing dataset of RAP2.12 is available on National Center for Biotechnology Information BioProject (https://ncbi.nlm.nih.gov/bioproject) by the accession numbers PRJNA374784, PRJNA273563, and PRJEB19780 and on the 1001 Genomes (https://1001genomes.org/). The source of each sequence is listed in table S2. Annual precipitation data are downloaded from WorldClim (www.worldclim.org/) (51); the aridity index is available from the CGIAR-CSI GeoPortal (https://cgiarcsi.community/data/global-aridity-and-pet-database/) (52); oxygen availability to roots data are available from the Food and Agriculture Organization of the United Nations Soils portal (www.fao.org/soils-portal/en/) (53); and the resolution of all data is 30 arc sec (~1 km2). Absolute expression levels of ERFVII were from eFP browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) (19, 54, 55), and we did not use the expression levels of Fr-2 and Old-2 because their information is from botanic gardens without the wild collection sites. The global nonrepetitive sites of natural inbred lines of A. thaliana used in this research are listed in table S3.
Acknowledgments
We thank J. Hou, S. Wei, W. Liu W. Yang, M. Kang, L. Huang, Q. Yang, and W. Yang for assistance in RILs construction, GWAS, and the acquisition of Brassicaceae species sequences.
Funding: The work was supported by the Natural Science Foundation of China (32030006), Strategic Priority Research Program of Chinese Academy of Sciences (XDB31000000), and Fundamental Research Funds for the Central Universities (SCU2019D013 and 2020SCUNL207) to J.L. This work was also supported by the Biotechnology and Biological Sciences Research Council (grant no. BB/R002428/1) to M.J.H.
Author contributions: J.L. led the research. The experiments were conceived and designed by S.L., H.L., M.J.H., and J.L. and mainly performed by S.L. The seeds of Sichuan and Tibet accessions were sampled by Lei Zhang and Y.J. RILs were constructed by S.L. and Lushui Zhang. HiFi reference genome in Sichuan accession was assembled by P.S. S.L., Y.S., B.L., S.T., N.C., M.L., H.Z., R.L., X.F., and Y.Z. performed plasmid construction. S.L., X.G., L.L., and J.L. analyzed the data. Writing—review and editing: S.L., H.L., M.J.H., and J.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. No restrictions are placed on materials, such as material transfer agreements.
Supplementary Materials
This PDF file includes:
Figs. S1 to S19
Tables S1 and S4
Other Supplementary Material for this manuscript includes the following:
Tables S2 and S3
Data S1
REFERENCES AND NOTES
- 1.Food and Agriculture Organization of the United Nations (FAO), The impact of disasters and crises on agriculture and food security (FAO, 2021). http://www.fao.org/3/cb3673en/cb3673en.pdf.
- 2.Maurel C., Nacry P., Root architecture and hydraulics converge for acclimation to changing water availability. Nat. Plants 6, 744–749 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Zandalinas S. I., Fritschi F. B., Mittler R., Global warming, climate change, and environmental pollution: Recipe for a multifactorial stress combination disaster. Trends Plant Sci. 26, 588–599 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Gupta A., Rico-Medina A., Caño-Delgado A. I., The physiology of plant responses to drought. Science 368, 266–269 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Xie L., Zhou Y., Chen Q., Xiao S., New insights into the role of lipids in plant hypoxia responses. Prog. Lipid Res. 81, 101072 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Holdsworth M. J., Gibbs D. J., Comparative biology of oxygen sensing in plants and animals. Curr. Biol. 30, R362–R369 (2020). [DOI] [PubMed] [Google Scholar]
- 7.van Dongen J. T., Licausi F., Oxygen sensing and signaling. Annu. Rev. Plant Biol. 66, 345–367 (2015). [DOI] [PubMed] [Google Scholar]
- 8.White M. D., Kamps J. J. A. G., East S., Taylor Kearney L. J., Flashman E., The plant cysteine oxidases from Arabidopsis thaliana are kinetically tailored to act as oxygen sensors. J. Biol. Chem. 293, 11786–11795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mustroph A., Lee S. C., Oosumi T., Zanetti M. E., Yang H., Ma K., Yaghoubi-Masihi A., Fukao T., Bailey-Serres J., Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 152, 1484–1500 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasch P., Fundinger M., Muller J. T., Lee T., Bailey-Serres J., Mustroph A., Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28, 160–180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbs D. J., Md Isa N., Movahedi M., Lozano-Juste J., Mendiondo G. M., Berckhan S., Marin-de la Rosa N., Vicente Conde J., Sousa Correia C., Pearce S. P., Bassel G. W., Hamali B., Talloji P., Tome D. F., Coego A., Beynon J., Alabadi D., Bachmair A., Leon J., Gray J. E., Theodoulou F. L., Holdsworth M. J., Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 53, 369–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vicente J., Mendiondo G. M., Movahedi M., Peirats-Llobet M., Juan Y. T., Shen Y. Y., Dambire C., Smart K., Rodriguez P. L., Charng Y. Y., Gray J. E., Holdsworth M. J., The Cys-Arg/N-End rule pathway is a general sensor of abiotic stress in flowering plants. Curr. Biol. 27, 3183–3190.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bui L. T., Giuntoli B., Kosmacz M., Parlanti S., Licausi F., Constitutively expressed ERF-VII transcription factors redundantly activate the core anaerobic response in Arabidopsis thaliana. Plant Sci. 236, 37–43 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Fukao T., Yeung E., Bailey-Serres J., The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23, 412–427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroha T., Nagai K., Gamuyao R., Wang D. R., Furuta T., Nakamori M., Kitaoka T., Adachi K., Minami A., Mori Y., Mashiguchi K., Seto Y., Yamaguchi S., Kojima M., Sakakibara H., Wu J., Ebana K., Mitsuda N., Ohme-Takagi M., Yanagisawa S., Yamasaki M., Yokoyama R., Nishitani K., Mochizuki T., Tamiya G., McCouch S. R., Ashikari M., Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science 361, 181–186 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Gibbs D. J., Lee S. C., Isa N. M., Gramuglia S., Fukao T., Bassel G. W., Correia C. S., Corbineau F., Theodoulou F. L., Bailey-Serres J., Holdsworth M. J., Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Licausi F., Kosmacz M., Weits D. A., Giuntoli B., Giorgi F. M., Voesenek L. A., Perata P., van Dongen J. T., Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Abbas M., Berckhan S., Rooney D. J., Gibbs D. J., Vicente Conde J., Sousa Correia C., Bassel G. W., Marin-de la Rosa N., Leon J., Alabadi D., Blazquez M. A., Holdsworth M. J., Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr. Biol. 25, 1483–1488 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., Provart N. J., An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLOS ONE 2, e718 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weits D. A., Giuntoli B., Kosmacz M., Parlanti S., Hubberten H. M., Riegler H., Hoefgen R., Perata P., van Dongen J. T., Licausi F., Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 5, 3425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartman S., Liu Z., van Veen H., Vicente J., Reinen E., Martopawiro S., Zhang H., van Dongen N., Bosman F., Bassel G. W., Visser E. J. W., Bailey-Serres J., Theodoulou F. L., Hebelstrup K. H., Gibbs D. J., Holdsworth M. J., Sasidharan R., Voesenek L. A. C. J., Ethylene-mediated nitric oxide depletion pre-adapts plants to hypoxia stress. Nat. Commun. 10, 4020 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choura M., Rebaï A., Masmoudi K., Unraveling the WRKY transcription factors network in Arabidopsis thaliana by integrative approach. Netw. Biol. 5, 55 (2015). [Google Scholar]
- 23.Ciolkowski I., Wanke D., Birkenbihl R. P., Somssich I. E., Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68, 81–92 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Journot-Catalino N., Somssich I. E., Roby D., Kroj T., The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18, 3289–3302 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ülker B., Somssich I. E., WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 7, 491–498 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Tang H., Bi H., Liu B., Lou S., Song Y., Tong S., Chen N., Jiang Y., Liu J., Liu H., WRKY33 interacts with WRKY12 protein to up-regulate RAP2.2 during submergence induced hypoxia response in Arabidopsis thaliana. New Phytol. 229, 106–125 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Machens F., Becker M., Umrath F., Hehl R., Identification of a novel type of WRKY transcription factor binding site in elicitor-responsive cis-sequences from Arabidopsis thaliana. Plant Mol. Biol. 84, 371–385 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Zhou M., Lu Y., Bethke G., Harrison B. T., Hatsugai N., Katagiri F., Glazebrook J., WRKY70 prevents axenic activation of plant immunity by direct repression of SARD1. New Phytol. 217, 700–712 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Li Y., Zhang H., Zhang Q., Liu Q., Zhai H., Zhao N., He S., An AP2/ERF gene, IbRAP2-12, from sweetpotato is involved in salt and drought tolerance in transgenic Arabidopsis. Plant Sci. 281, 19–30 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Xu Z.-S., Xia L.-Q., Chen M., Cheng X.-G., Zhang R.-Y., Li L.-C., Zhao Y.-X., Lu Y., Ni Z.-Y., Liu L., Qiu Z.-G., Ma Y.-Z., Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 65, 719–732 (2007). [DOI] [PubMed] [Google Scholar]
- 31.X. Meng, The role of mitochondrial function and retrograde signalling for stress responses in Arabidopsis thaliana (La Trobe University, 2019); http://hdl.handle.net/1959.9/570804.
- 32.Vashisht D., Hesselink A., Pierik R., Ammerlaan J. M., Bailey-Serres J., Visser E. J., Pedersen O., van Zanten M., Vreugdenhil D., Jamar D. C., Voesenek L. A., Sasidharan R., Natural variation of submergence tolerance among Arabidopsis thaliana accessions. New Phytol. 190, 299–310 (2011). [DOI] [PubMed] [Google Scholar]
- 33.1001 Genomes Consortium , 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166, 481–491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J., Nolan T. M., Ye H., Zhang M., Tong H., Xin P., Chu J., Chu C., Li Z., Yin Y., Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29, 1425–1439 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendiondo G. M., Gibbs D. J., Szurman-Zubrzycka M., Korn A., Marquez J., Szarejko I., Maluszynski M., King J., Axcell B., Smart K., Corbineau F., Holdsworth M. J., Enhanced waterlogging tolerance in barley by manipulation of expression of the N-end rule pathway E3 ligase PROTEOLYSIS6. Plant Biotechnol. J. 14, 40–50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu B., Jiang Y., Tang H., Tong S., Lou S., Shao C., Zhang J., Song Y., Chen N., Bi H., Zhang H., Li J., Liu J., Liu H., The ubiquitin E3 ligase SR1 modulates the submergence response by degrading phosphorylated WRKY33 in Arabidopsis. Plant Cell 33, 1771–1789 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarasov A., Vilella A. J., Cuppen E., Nijman I. J., Prins P., Sambamba: Fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M. A., The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., Handsaker R. E., Lunter G., Marth G. T., Sherry S. T., McVean G., Durbin R.; 1000 Genomes Project Analysis Group , The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X., Stephens M., Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods 11, 407–409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X., Henriques R., Lin S. S., Niu Q. W., Chua N. H., Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Yuan L. B., Dai Y. S., Xie L. J., Yu L. J., Zhou Y., Lai Y. X., Yang Y. C., Xu L., Chen Q. F., Xiao S., Jasmonate regulates plant responses to postsubmergence reoxygenation through transcriptional activation of antioxidant synthesis. Plant Physiol. 173, 1864–1880 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z., Wang J., Zhang R., Huang R., The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 71, 273–287 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Linster E., Stephan I., Bienvenut W. V., Maple-Grodem J., Myklebust L. M., Huber M., Reichelt M., Sticht C., Moller S. G., Meinnel T., Arnesen T., Giglione C., Hell R., Wirtz M., Downregulation of N-terminal acetylation triggers ABA-mediated drought responses in Arabidopsis. Nat. Commun. 6, 7640 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cha J.-Y., Kim W.-Y., Kang S. B., Kim J. I., Baek D., Jung I. J., Kim M. R., Li N., Kim H.-J., Nakajima M., Asami T., Sabir J. S. M., Park H. C., Lee S. Y., Bohnert H. J., Bressan R. A., Pardo J. M., Yun D.-J., A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin biosynthesis. Nat. Commun. 6, 8041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hellens R. P., Allan A. C., Friel E. N., Bolitho K., Grafton K., Templeton M. D., Karunairetnam S., Gleave A. P., Laing W. A., Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bandelt H. J., Forster P., Röhl A., Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fick S. E., Hijmans R. J., WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315 (2017). [Google Scholar]
- 52.Trabucco A., Zomer R. J., Global Aridity Index (Global-Aridity) and Global Potential Evapo-Transpiration (Global-PET) Geospatial Database. (CGIAR Consortium for Spatial Information, 2009); http://www.csi.cgiar.org.
- 53.G. Fischer, F. Nachtergaele, S. Prieler, H. Van Velthuizen, L. Verelst, D. Wiberg, Global Agro-Ecological Zones Assessment for Agriculture (GAEZ 2008). IIASA, Laxenburg, Austria and FAO, Rome, Italy (2008). [Google Scholar]
- 54.Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K., The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Lempe J., Balasubramanian S., Sureshkumar S., Singh A., Schmid M., Weigel D., Diversity of flowering responses in wild Arabidopsis thaliana strains. PLOS Genet. 1, 109–118 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S19
Tables S1 and S4
Tables S2 and S3
Data S1
Data Availability Statement
The raw sequencing dataset of RAP2.12 is available on National Center for Biotechnology Information BioProject (https://ncbi.nlm.nih.gov/bioproject) by the accession numbers PRJNA374784, PRJNA273563, and PRJEB19780 and on the 1001 Genomes (https://1001genomes.org/). The source of each sequence is listed in table S2. Annual precipitation data are downloaded from WorldClim (www.worldclim.org/) (51); the aridity index is available from the CGIAR-CSI GeoPortal (https://cgiarcsi.community/data/global-aridity-and-pet-database/) (52); oxygen availability to roots data are available from the Food and Agriculture Organization of the United Nations Soils portal (www.fao.org/soils-portal/en/) (53); and the resolution of all data is 30 arc sec (~1 km2). Absolute expression levels of ERFVII were from eFP browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) (19, 54, 55), and we did not use the expression levels of Fr-2 and Old-2 because their information is from botanic gardens without the wild collection sites. The global nonrepetitive sites of natural inbred lines of A. thaliana used in this research are listed in table S3.