Abstract
Superresolution microscopy enables probing of cellular ultrastructures. However, its widespread applications are limited by the need for expensive machinery, specific hardware, and sophisticated data processing. Expansion microscopy (ExM) improves the resolution of conventional microscopy by physically expanding biological specimens before imaging and currently provides ~70-nm resolution, which still lags behind that of modern superresolution microscopy (~30 nm). Here, we demonstrate a ninefold swelling (NIFS) hydrogel, that can reduce ExM resolution to 31 nm when using regular traditional microscopy. We also design a detachable chip that integrates all the experimental operations to facilitate the maximal reproducibility of this high-resolution imaging technology. We demonstrate this technique on the superimaging of nuclear pore complex and clathrin-coated pits, whose structures can hardly be resolved by conventional microscopy. The method presented here offers a universal platform with superresolution imaging to unveil cellular ultrastructural details using standard conventional laboratory microscopes.
We demonstrate a swollen hydrogel that acts like magnifying glass to vividly reflect the cell’s fine structures.
INTRODUCTION
Fluorescence microscopy has been widely applied to obtain detailed molecular distribution and structural information in cells and tissues. With simple sample preparation and inexpensive equipment requirements, fluorescence microscopy has become a mainstream setup in biological research such as mRNA imaging, material characterization, and pathology diagnosis (1–6). However, the elucidation of superstructure details with conventional fluorescence microscope is limited by the resolution at approximately 200 nm due to the diffraction of light (7, 8).
In recent years, novel superresolution techniques provide breakthroughs to the resolution limit. One approach is through innovations in instrumentations, such as STED (stimulated emission depletion) microscopy, SOFI (superresolution optical fluctuation imaging), and STORM (stochastic optical reconstruction microscopy) (9–15), which could reach resolutions of 30 to 50 nm. However, their widespread applications are still limited by expensive machinery, specific hardware, laborious operation, and sophisticated data processing (7, 16–18). Another approach is through innovations in sample preparation, for example, expansion microscope (ExM) (19–21), which embeds the specimen into a hydrogel that could expand four times when dialyzed in water, realizing ~70-nm resolution. The ExM technique does not require special machinery or particular data processing, therefore providing affordable approaches for superresolution imaging in most laboratories (22–28), and has been applied in neuroscience, pathology, and mRNA discovery (29–36). However, current ExM methods only manage to reach a resolution of about 70 nm, which still lags behind that in modern superresolution instruments (4, 16, 37, 38). Thus, it is imperative to develop a novel sample-embedded material with enhanced expansion factor for higher resolution of ExM.
Recently, a few efforts have been made to enhance the expansion ability of ExM (39–42). Chang et al. (40) proposed the iterative ExM (iExM) method, which consists of two rounds of sample embedment and expansion. This method naturally requires additional time consumption and specialized probe designation. Truckenbrodt et al. (43) described a protocol termed X10 microscopy, which expands the specimen ~10 times. This method needs to carefully eliminate oxygen from the monomer gelling solution for about 45 min before gelation to achieve the ideal resolution (44). To date, a convenient, time-saving, and high-resolution method for ExM still remains a major challenge.
Here, we demonstrated an expedient and efficient method to achieve superresolution imaging with a conventional microscope. To enhance swelling expansion, we developed a new hydrogel, which completely gelled within 2 hours and swelled 729 (93)-fold in volume when dialyzed in water. We termed the hydrogel ninefold swelling hydrogel (NIFS hydrogel). NIFS hydrogel used in ExM improves the resolution of conventional microscopy down to 31 nm, which is equivalent to current advanced superresolution microscopy. Moreover, we invented a detachable chip that combined almost all ExM processes, including cell culture, fixation, staining, anchoring, and gelation, to maximally reduce artificial interference errors and enhance the reproducibility of this cost-effective high-resolution imaging technology. To reduce fluorescence photobleaching during long-term post-expansion specimen imaging without losing structural resolution, we also developed a previously unidentified anti-fluorescent quenching medium that allows 90% fluorescence signal retention over 80 min of continuous exposure. This reported method offers a universal platform to unveil cellular ultrastructural details with conventional laboratory microscopes.
RESULTS
To enable 30-nm resolution in conventional microscopy, we provide a convenient approach, from material development to whole system integration, which physically expands the specimen by ninefold within a one-round expansion procedure. The expansion ability is highly dependent on the swelling capacity of materials used in ExM. We choose hydrogels as the candidate material for ExM because of their biocompatibility, light permeability, and high swell ability (45, 46). The swelling capacity of hydrogels is, in turn, highly dependent on the cross-linking agent, which is the cornerstone of the polymer network (39, 47, 48). We developed a previously unidentified cross-linking agent [namely, N,N-ethylenebisacrylamide (EBIS)] for hydrogels, which isotropically swelled 729 (93)-fold in volume when dialyzed in water.
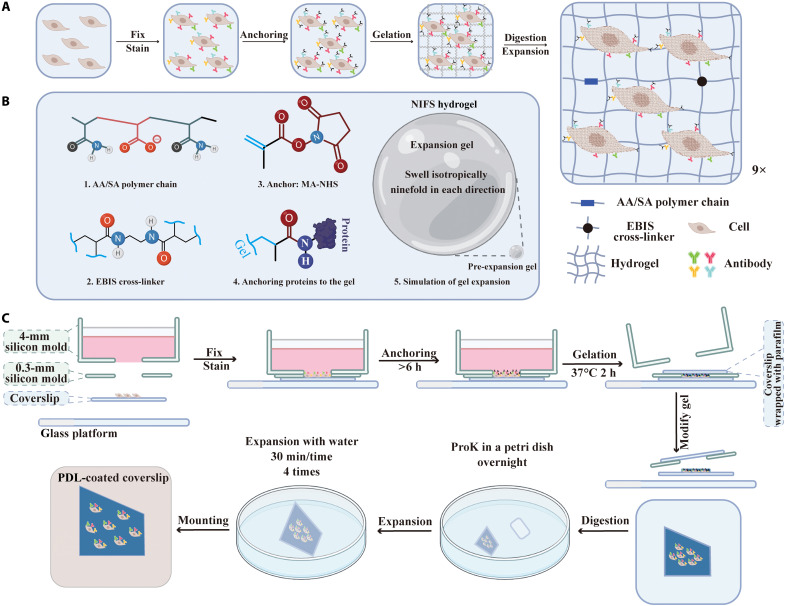
The principle of the presented ExM method is shown in Fig. 1. The expansion process of the specimen embedded with NIFS hydrogel is illustrated in Fig. 1A. The specimen was first fixed and stained with interested biomarkers via an immunostaining method. Then, the specimen was treated with an anchoring reagent [methacrylic acid N-hydroxysuccinimide ester (MA-NHS)] to assemble the methacryloyl group to all proteins of cells by reacting with the amino group (49). Next, we immersed the cells into the NIFS hydrogel monomer solution to induce complete gelation and then applied the digest buffer to loosen and mechanically homogenize the cellular structure. The digestive process facilitates the homogeneous expansion of hydrogels. Last, we expanded the sample ninefold isotropically through dialysis in water. The whole process uniformly enlarges the cell, which facilitates the observation of cellular ultrastructure in the conventional microscope without distortion.
Fig. 1. Schematic illustration of NIFS hydrogel used in ExM.
(A) Principle of ExM. (B) Chemical constitutions and expansion capacities of NIFS hydrogel. (C) Workflow of the home-developed chip used for ExM. ProK, proteinase K; PDL, poly-d-lysine.
Figure 1B displays the components of NIFS hydrogel and linkage chemistry. NIFS hydrogel has two main compositions, namely, polymer chain [consisting of SA (sodium acrylate) and AA (acrylamide)] and cross-linking agent EBIS. The cross-linking agent recruits the hydrogel structure from the polymer chain to the network. The linkage chemistry of protein retention in the gel is achieved via an anchoring reagent. The maleimide group of MA-NHS reacts with the amide group in proteins, while another reactive alkene functional group participates in the gelation process to integrate proteins into the hydrogel matrix. The simulation of NIFS hydrogel expansion (Fig. 1B, 5) implies that NIFS hydrogel could isotropically expand ninefold in each direction after expansion.
To further simplify the overall process that can be easily assembled and replicated in most laboratories, we developed an easy-to-use homemade chip in all ExM processes. The operation principle of ExM with this home-developed chip is displayed in Fig. 1C, while the real chip mold is shown in fig. S1. The chip has four components, namely, glass platform as a base to protect the fragile cover glass, cover glass for growth of adherent cells, 0.3-mm silicon mold as gelation chamber, and 4-mm silicon mold as container for culture medium, fixed buffer, and immunostaining buffer. Because of the proper adhesion and high pressure-resistance of silicon, the custom-made mold is suitable for sterile cell culture and disassemble for the subsequent ExM process. Then, we tested the expansion capacity of different lengths of cross-linkers (fig. S2); we noticed that the mitigated water affinity of cross-linkers leads to a decrease in the expansion capacity. Furthermore, the longer cross-linkers usually need additional polymerization inhibitors [such as 4-methoxyphenol (MEHQ) and 4-tert-butylcatechol (TBC)] to maintain their stability. These inhibitors will further reduce the expansion factor by participating in the gelation process (50). EBIS has appropriate affinity with water and has desired stability without additional polymerization inhibitors (48, 50). Thus, we opted to choose EBIS in our method. Then, we optimized the expansion capacity of NIFS hydrogel via the EBIS concentration, which is the cornerstone of the hydrogel. We first evaluated the expansion capacity of the sample-free hydrogel at various EBIS concentrations (fig. S3). The photographs of post-expansion hydrogels with different EBIS concentrations [0.20 to 0.06% (w/w)] in the monomer solution are shown in fig. S3A. The corresponding statistical analysis of linear expansion factor is from 4.74 to 8.92 (fig. S3B). Furthermore, the stability of expanded hydrogels has been exhaustively tested by inspecting their morphologies and expansion factor change from 1 to 10 days (fig. S4). Our results showed that the morphologies and expansion factor of expanded hydrogels were basically retained consistently in the buffer for at least 10 days (fig. S4), which means that the expanded hydrogels have the desired stability. When the EBIS concentration decreases to 0.04% (w/w), the hydrogel cannot be easily manipulated after the expansion process.
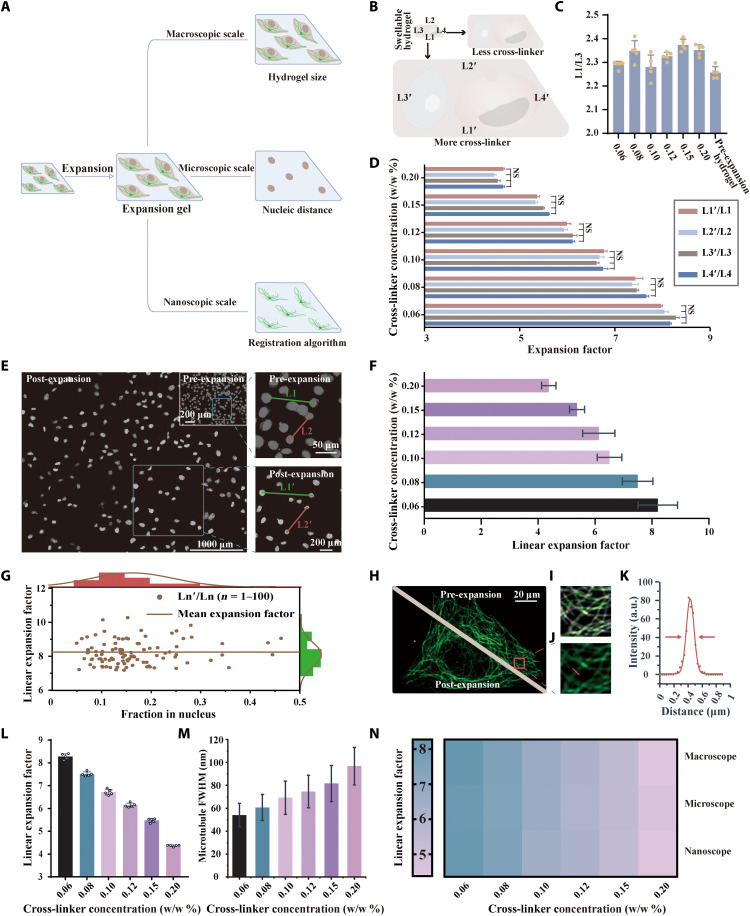
We then tested the performance of NIFS hydrogel [0.20 to 0.06% (w/w)] when biological samples were embedded in the hydrogel (Fig. 2). We evaluated the expansion ability from three scale levels: macroscopic, microscopic, and nanoscopic (Fig. 2A).
Fig. 2. Expansion ability of NIFS hydrogels with different EBIS cross-linker concentrations.
(A) Schematic diagram of three expansion factor evaluation methods. (B) The scheme highlights the swelling ability of hydrogels with different cross-linker concentrations in the macroscopic scale level. (C) The ratio between the hydrogel’s two directions (L1/L3) varies with cross-linker concentrations from 0.20% (w/w) to 0.06% (w/w) (mean ± SD; n = 5 biological replicates). (D) The macroscopic scale expansion ability of cell-embedded samples is determined by the hydrogel’s four sides’ ratio between the post- and pre-expansion hydrogels (mean ± SD; n = 5 biological replicates). (E) Cell nucleic images of pre- and post-expansion samples using NIFS hydrogels. The right images illustrate the nucleic pairwise analysis. (F) The microscopic scale expansion ability of cell-embedded samples is determined by nucleic distance. (G) Linear expansion factor [distance ratio between Ln′ and Ln (n = 1, 2, …, 100)] versus the fraction in nucleus [mean gray value of Ln′ (n = 1, 2, …, 100)]. (The brown line indicates mean expansion factor; mean ± SD = 8.21 ± 0.693.) (H) Representative microtubule images of pre- and post-expansion in the same cell. (I) Merged images of pre-expansion and algorithm-calculated post-expansion microtubule images. (Green is pre-expansion image; purple is algorithm-calculated post-expansion image.) (J) Enlarged view of the post-expansion tubulin image in the red box marked in (H). (K) Profile of red lines in (J) and the corresponding FWHM calculation by Gaussian fitting (red line). (L) Influence of the cross-linker concentrations on nanoscale expansion ability evaluated by registration algorithm of microtubule structure (mean ± SD; n = 5 biological replicates, each replicate chooses one region). (M) Microtubule’s FWHM is determined by post-expansion images with different cross-linker concentrations (mean ± SD; n = 300 microtubule profiles from three replicates; each replicate chooses one region). (N) The expansion factor comparison using three evaluation methods reaches consistent observations.
First, we analyzed the swelling ability of cell-embedded NIFS hydrogel from the macroscale level (Fig. 2B), that is, the hydrogel’s changes in size, mainly the uniformity of expansion and the coefficient of expansion. In Fig. 2C, we analyzed the uniformity of swelling from two different dimensions of a hydrogel. The ratio between the two sides of the hydrogel (L1/L3) does not vary with the ExM procedure or the cross-linker concentration variation, which indicates that the expansion of hydrogels with different concentrations of the cross-linker is isotropic. Next, the expansion factor of the macroscale level in NIFS hydrogel was determined by the distance variation of two identical points in the expansion process. Figure 2D shows the expansion tendency of NIFS hydrogels with different cross-linker concentrations, which is consistent with our anticipation. The hydrogel size expansion ability increments along with the decrease of hydrogels’ cross-linker concentration (39). Moreover, there is no obvious difference in the expansion ability between the four sides of hydrogels at different cross-linker concentrations.
Second, we analyzed the swelling ability of cell-embedded NIFS hydrogel from the microscale level, that is, the changes in the definite nucleic distance in the ExM process. The images of pre- and post-expansion cell nucleus samples are shown in Fig. 2E, where the hydrogel cross-linker concentration is 0.06% (w/w). The right side of Fig. 2E is the enlarged picture of the blue box that indicates the expansion factor calculation. The expansion factor was determined by measuring the ratio of nucleic distances between two identical cells in pre- and post-expansion samples (L1′/L1 and L2′/L2) and validated with Elastix software (Fig. 2F). For further validation of expansion uniformity, we analyzed the relationship between expansion factor and cell density (Fig. 2G). Our results indicated that the expansion factor was not influenced by the nucleus distance. Other evaluations of the swelling capacity of hydrogels at different cross-linker concentrations are shown in figs. S5 to S9. At the microscopic level, the tendency of cell-embedded hydrogels’ swelling ability at different cross-linker concentrations is consistent with the macroscale evaluation method.
Third, we analyzed the swelling ability of cell-embedded NIFS hydrogel from the nanoscale level, that is, the registration algorithm for matching the microtubule structure in ExM. Figure 2H shows images of the same cell microtubule at pre- and post-expansion stages [the hydrogel cross-linker concentration is 0.06% (w/w)]. We selected the same region in pre- and post-expansion specimens for algorithm to match these images (Fig. 2I), and the corresponding expansion factors with different cross-linker concentrations are shown in Fig. 2L. Our results showed that the nanoscopic expansion ability had the same tendency as the other two evaluation methods (macroscopic and microscopic) mentioned before. Another effective way to measure the expansion ability is to analyze the resolving capacity [full width at half maximum (FWHM)] in ExM. In Fig. 2J, an enlarged post-expansion picture of the red box in Fig. 2H is presented. We obtained the intensity profile with ImageJ along a reference line that is perpendicular to the microtubule orientation. Then, intensity profiles were fitted with Gaussian fit and calculated for FWHM (Fig. 2K). For the interpretation of FWHM, the value has a logical relationship with the microscopic resolution (43). Because of the diffraction limit of the microscope, when microscopic resolution is lower than the sample’s size, the FWHM value represents the microscopic resolution. When the resolution is higher than the samples’ size, the FWHM value represents the sample’s true size. The summary results (Fig. 2M) indicated that the mean FWHM value decreased with declining EBIS concentration (from 97.6 ± 16.4 nm to 54.0 ± 10.3 nm). That is, the resolution is improved along with declining EBIS concentration. The detailed images analyzing FWHM with different EBIS concentrations are shown in figs. S10 to S15. The heatmap (Fig. 2N) summarized the three evaluating methods (macroscopic, microscopic, and nanoscopic) and indicated that they had consistent tendency with various cross-linker concentrations. The detailed data of the cell-embedded hydrogel expansion factor evaluation methods compared to the sample-free counterpart are provided in fig. S16. These results further indicated that the tendency of expansion factors related to the EBIS concentration was consistent in macro-, micro-, and nanoscales between cell-embedded and sample-free hydrogels. The absolute value of linear expansion factor slightly decreased when cells were embedded in NIFS hydrogel. This may be caused by the additional interaction forces between cellular molecules (39).
In conclusion, this NIFS hydrogel could isotropically expand embedded cell samples and declined cross-linker concentrations that lead to enhanced expansion abilities. At the 0.06% (w/w) EBIS concentration, NIFS hydrogel improves the specimen expansion factor to 8.27-fold with minimal deformation.
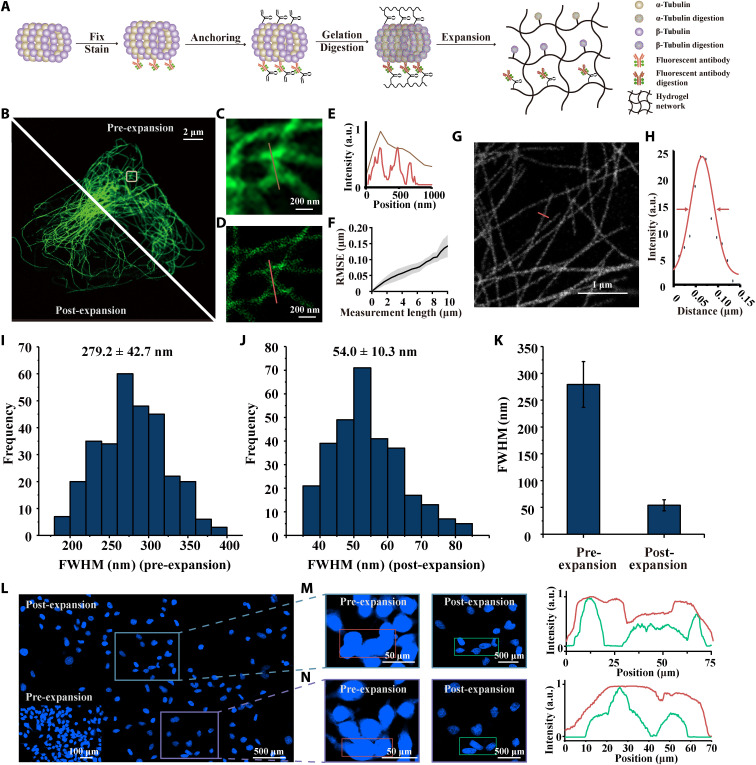
Next, we investigated the performance of the optimized NIFS hydrogel for ExM (Fig. 3). In the ExM process, the change of the cell structure can be illustrated by the microtubule protein as an example (Fig. 3A). The same cell’s microtubule images in the pre-expansion (top right) and post-expansion (bottom left) state are displayed in Fig. 3B. To observe detailed information of microtubules, we enlarged the white boxed area in Fig. 3B as shown in Fig. 3 (C and D) (pre- and post-expansion images). Figure 3E shows the corresponding line profiles marked in Fig. 3 (C and D) and unveils the precise microtubule structure, which is hardly observed in pre-expansion images. We then evaluated microtubule distortion in ExM. Figure 3F shows the root mean square error (RMSE) of ~120 nm at a distance over 10 μm. We also revalidated the distortion of this method on a large scale with the cell nucleus (fig. S17). This distortion is consistent with reported ExM methods (20, 49). Next, the microtubule image [63×/1.4 numerical aperture (NA) oil lens post-expansion sample] is shown in Fig. 3G, and we further acquired the Gaussian fitting along the red line for FWHM calculation (Fig. 3H). We measured the FWHM of microtubule in the pre- and post-expansion process (Fig. 3, I and J), and the comparison is displayed in Fig. 3K. The apparent microtubule complex FWHM with optimized NIFS hydrogel decreased from 279.2 ± 42.7 nm to 54.0 ± 10.3 nm (mean ± SD; n = 300) through the ExM examination, which was consistent with the actual size of microtubules (fig. S18) (40). Furthermore, the cell nucleus structure, which barely elaborates in conventional microscopy, was further refined in NIFS hydrogel ExM (Fig. 3, L to N). Intuitively, the profiles and images of the original structure are readily enhanced with NIFS hydrogel applied for cell analysis.
Fig. 3. The performance of optimized NIFS hydrogel for ExM.
(A) Schematic illustration of microtubule imaging pipeline using ExM. (B) Pre- and post-expansion microtubule images with the optimized NIFS hydrogel. The top right panel represents pre-expansion image, and the bottom left panel represents post-expansion image. (C) Magnified pre-expansion image of the box marked area in (B). (D) Magnified post-expansion image of the box marked area in (B). (E) Intensity profile of lines in (C) (brown line) and (D) (red line). (F) RMSE of NIFS hydrogel used in the expansion process. (Black line, mean value; gray scale, SD of three biological replicates). (G) Representative image (63×/1.40 NA) of post-expansion microtubules using optimized NIFS hydrogel. (H) Gaussian fitting of red line in (G) for FWHM calculation. (I) FWHM of microtubules (n = 300 microtubule profiles) in pre-expansion image. (J) FWHM of microtubules (n = 300 microtubule profiles) in post-expansion image. (K) Comparison of FWHM in pre- and post-expansion images. (L) Representative nucleic imaging using optimized NIFS hydrogel. The bottom left inset represents the pre-expansion image. The magnified views of the box marked areas (pre- and post-expansion images) and the corresponding profiles in (L) are shown in (M) and (N). All scales are converted to pre-expansion dimension.
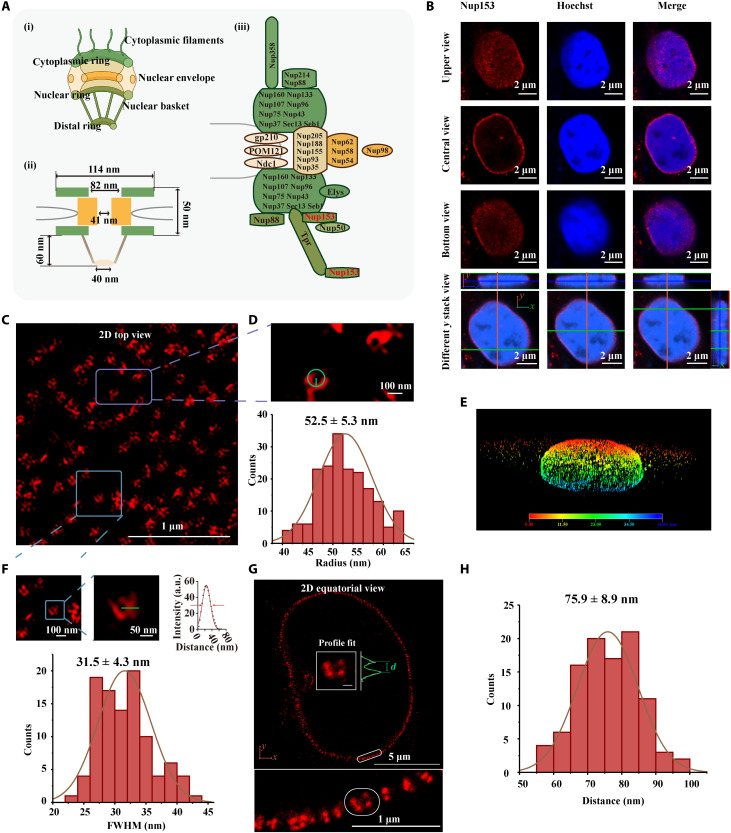
We then applied NIFS hydrogel with ExM to examine the ultrastructures that were concealed in conventional microscopy (Fig. 4). We demonstrated the superimaging of nuclear pore complex (NPC), whose structure cannot be elaborated in regular conventional light microscopy and has to be visualized in superresolution microscopes (51, 52). Figure 4A (i) shows the NPC structure, while the corresponding size distributions are provided in Fig. 4A (ii). The definite protein position in NPC is shown in Fig. 4A (iii) (53). In theory, Nup153 (a kind of NPC proteins) forms the central-null circle structure full in two-dimensional (2D) top view (parallels the cell nucleus) and a circle around the nucleus in 2D equatorial view (51, 54). The structure of Nup153 in pre-expansion images was largely concealed (Fig. 4B). However, the images of post-expansion Nup153 (Fig. 4, C to G) in a regular confocal microscope could unveil these detailed structures that previously could only be observed in superresolution microscopes.
Fig. 4. NPC images in ExM with NIFS hydrogel.
(A) (i) Overall structure of NPC, (ii) size distribution of the NPC structure, and (iii) distribution of antigens in the NPC structure. (B) Pre-expansion images of Nup153. The top and bottom views indicate that Nup153 is parallel to the nucleus. The center view shows Nup153 display around the nucleus. The different y positions cross-verify Nup153 display around the nucleus. (C) The post-expansion images illustrate the landscape spatial distribution of Nup153 (2D top view). (D) Magnified views of the purple box marked area in (C) and radius evaluation method in the individual ring of Nup153. The histogram is the radius distribution of Nup153 (n = 150 from three replicates; each replicate chooses one region). (E) 3D image of Nup153 in post-expansion confocal image. (F) Magnified views of the blue box marked area in (C) and FWHM evaluation method in the individual ring of Nup153. The histogram of FWHM distribution (n = 100 from three replicates; each replicate chooses one region). (G) 2D equatorial view of Nup153 image. The bottom is the enlarged view of the white boxed area in top images. The inside profile fit of the top image is the Nup153 ring distance analysis method. Scale bar, 100 nm. (H) Histogram of the separated Nup153 ring distance (n = 100 from three replicates; each replicate chooses one region). All scales are converted to pre-expansion dimensions.
Another critical issue about ExM is that the post-expansion imaging needs a long time for data acquisition because of larger area to scan, especially in 3D imaging where all three directions are expanded. Longer exposure poses a special challenge for fluorescence quenching. Therefore, we have developed a previously unidentified fluorescence decay-resistant medium [1% DABCO (1,4-diazabicyclo [2.2.2] octane) in water], which could retain 90% of the fluorescence signal against photobleaching without resolution decrease during the ExM imaging process for 80 min (figs. S19 and S20). For detailed analysis of NIFS hydrogels in ExM, we analyzed the two nanostructures of the Nup153 complex, namely, the circle complex in 2D top view and the parallel structures in 2D equatorial view. In Fig. 4C, the x-y plate parallels the nucleus for the analysis of the circle structure, which was completely concealed in the pre-expansion specimen in conventional microscopy. We then examined the radius of the circle structure in the post-expansion specimen (Fig. 4D). Our results showed that the Nup153 complex radius was 52.5 ± 5.3 nm, which was consistent with an early report (54). Furthermore, we analyzed the resolving ability by measuring the FWHM of Nup153 (Fig. 4F). The average of Nup153 FWHM is 31.5 ± 4.3 nm, which represents the resolution of this method. This estimate resolution is consistent with the theoretical calculation of microscopy resolution (29 nm; 63×/1.4 NA oil lens) through the expansion process (~8.24×). The detailed estimating resolution method is in fig. S21. For the Nup153 image in 2D equatorial view (Fig. 4, G and H), the distance of parallel structure was 64.5 to 82 nm in theory (combined with antibody tagging) (51), which was also consistent with our result (75.9 ± 8.9 nm). The other target, clathrin-coated pits (CCPs), was also unveiled for the ultrastructure in post-ExM, which was concealed in pre-expansion samples (fig. S22). Apparently, the central-null circle structure of CCP was clarified after expansion. Intensity profile showed improved profiling details through NIFS hydrogel ExM (fig. S22B). These results presented the practical improvement in lateral resolution after NIFS hydrogel expansion in ExM.
DISCUSSION
This study describes a strategy for enabling ~30-nm resolution imaging of biological samples with conventional laboratory microscopes by a novel superabsorbent hydrogel that enlarges the specimen on a home-developed chip that integrates all pipeline procedures. The home-developed chip simplifies the operation by integrating all the critical processes, such as cell culture, fixation, immunostaining, anchoring, and gelation for ExM. Moreover, the application of the fluorescence decay-resistant medium reduces 90% photobleaching without decreasing resolution during the imaging process for 80 min. A new proposed high-swelling material (namely, NIFS hydrogel) uniformly expands about ninefold in each direction. When high-swelling materials are combined with ExM technology, the resolution of conventional microscopy in imaging hydrogel-embedded biological sample improves down to 31 nm. This resolution enhancement enables delicate conventional microscopy investigation of the fine structure of cells (nucleus pore complex as illustration), which previously could only be observed in superresolution microscopy. Therefore, this overall pipeline is expected to serve as a user-friendly method for superresolution imaging of specimens on conventional microscopy in most laboratories.
In principle, NIFS hydrogel is compatible with the other superresolution fluorescence microscopy technologies, allowing further enhanced resolution down to several nanometers (55–59). The combination of NIFS hydrogel and superresolution microscope could potentially provide even better refined detailed structures and biomolecular distributions in optical microscopy, instead of that recently observed or has to be obtained in expensive and complicated electronic microscopy.
MATERIALS AND METHODS
Materials
Mouse anti–α-tubulin antibody (30304ES40, Yeason, Shanghai, China), anti-clathrin heavy chain antibody (ab172958, Abcam, Cambridge, MA, USA), anti-Nup153 antibody (ab24700, Abcam, Cambridge, MA, USA), and Alexa Fluor 488 donkey anti-rabbit immunoglobulin G (IgG) (406416, BioLegend, San Diego, CA, USA) were used. Alexa Fluor 594 goat anti-mouse IgG (H+L) (34112ES60) and Alexa Fluor 488 goat anti-mouse IgG (H+L) (33206ES60) were bought from Yeason (Shanghai, China). Hoechst 33342 was bought from Yuanyebio (Shanghai, China). Paraformaldehyde (4%), magnesium chloride (MgCl2), tris-HCl buffer (pH 8), Triton X-100, and phosphate-buffered saline (PBS) were bought from Sangon Biotech (Shanghai, China). Bovine serum albumin (BSA), anhydrous dimethyl sulfoxide (DMSO), sodium borohydride (NaBH4), Pipes, sodium chloride, EDTA, and AA were supplied by Innochem (Beijing, China). Proteinase K was obtained from Yeasen Biotech (Shanghai, China). MA-NHS, SA, ammonium persulfate (APS), and N,N,N′,N′-tetramethylethylenediamine (TEMED) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Glutaraldehyde (50%) was obtained from Macklin (Shanghai, China). N,N-Methylenebisacrylamide (BIS), EBIS, N,N-(propane-1,3-diyl) diacrylamide (TBIS), and N,N-(butane-1,4-diyl) diacrylamide (FBIS) were obtained from Bide Pharmatech (Shanghai, China).
Immunostaining of cultured cells
Hela cells were cultured at homemade molds in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and 1% penicillin-streptomycin. Cells were maintained at 37°C with 5% CO2. Hela cells were seeded at a density of ~10,000 cells per mold and incubated overnight. For tubulin, cells were optionally extracted for 30 s with an extraction buffer [PEM buffer (100 mM Pipes, 1 mM EDTA, and 1 mM MgCl2) (pH 7), supplemented with 0.5% Triton X-100] immediately before fixation. Specimens were fixed with 3.2% paraformaldehyde and 0.1% glutaraldehyde in PBS buffer for 10 min, followed by reduction with 0.1% NaBH4 in PBS for 7 min and 100 mM glycine in PBS for 10 min. For Nup153 and clathrin, cells were briefly washed with PBS and fixed with 3.2% paraformaldehyde in PBS for 10 min, followed by reduction with 10-min glycine in PBS for 10 min. After reduction, cells were washed three times in PBS for 5 min per each wash and then incubated with blocking/permeabilization buffer (1× PBS containing 3% BSA and 0.5% Triton X-100) for 30 min. Cells were incubated with primary antibodies in blocking/permeabilization buffer overnight (>6 hours) at 4°C and washed three times with PBS. Then, specimens were incubated for 2 hours with fluorophore-conjugated secondary antibodies in blocking/permeabilization buffer. After three more washes with PBS, the specimen was ready for pre-expansion imaging.
Anchoring to specimen
The stained cells were treated with MA-NHS (1 mg/ml) overnight at room temperature and then washed three times with PBS. MA-NHS was resuspended in anhydrous DMSO at a final concentration of 50 mg/ml. The stock solution was divided into 10-μl aliquots and stored up to 2 months in a sealed container with drying agents at −20°C.
Gelation
A layer of silicone pad was removed, and then cells were immersed in the monomer solution (Table 1) for 10 min at room temperature before gelation. The stock solution of TEMED accelerator [10% (w/w)] and APS initiator [10% (w/w)] was sequentially added to the monomer solution to a final concentration of 0.2% (w/w). The gelling solution was added to the homemade mold quickly and covered with a slide to isolate from air. After 5 min on ice, the homemade mold was placed at 37°C for 2 hours for complete gelation.
Table 1. Monomer solution.
| Component | Stock solution concentration* | Amount (ml) | Final concentration* |
| SA | 38 | 2.25 | 8.6 |
| AA | 50 | 0.5 | 2.5 |
| EBIS | 2 | 1–0.3 | 0.20–0.06 |
| Sodium chloride (NaCl) | 29.2 | 4 | 11.7 |
| PBS | 10× | 1 | 1× |
| Water | 0.65–1.35 | ||
| Total | 9.4 |
*All concentrations in g/100 ml except for PBS.
Digestion and expansion
The digest buffer was modified from E. S. Boyden’s recipe list in Table 2. To easily find the same area of expansion gel, the gel was cut in asymmetrical shape. The asymmetrical gel was immersed in the digest buffer overnight, then aspirated from the petri dish, and placed in 20 ml of deionized water to expand. Water was exchanged four times for 30 min per each exchange for the hydrogel to expand completely. Using imaging buffer [1% (v/v) DABCO in deionized water], the gel was treated before imaging.
Table 2. Digest buffer.
| Component | Stock solution concentration | Amount (ml) | Final concentration |
| Triton X-100 | 10% (w/v) | 5 | 0.5% (w/v) |
| EDTA | 0.5 M (pH 8.0) | 0.2 | 1 mM |
| Tris-HCI | 1 M (pH 8.0) | 5 | 50 mM |
| Sodium chloride (NaCl) | 5 M | 20 | 1 M |
| Proteinase K* | 800 U/ml | 1 | 8 U/ml |
| Water | 68.8 | ||
| Total | 100 |
*Proteinase K was freshly added to the digest buffer.
Mounting and imaging
Most of the imaging buffer was carefully removed from the petri dish, and then the gels were mounted on poly-d-lysine–coated cover glasses. All images were acquired on an LSM 800 confocal microscope (Zeiss, German). The pre-expansion sample was imaged with 63×/1.40 NA oil immersion objective. The expansion gel was acquired with 25×/0.8 NA water immersion objective or 63×/1.40 NA oil immersion objective.
Data analysis
Measuring expansion factor
Estimating sample-free gel expansion factors was done by measuring the radii of gels in both pre- and post-expansion images with ImageJ and then calculating the ratio. For cell-embedded expansion gels, expansion factors were evaluated from three levels: macroscopic scale, microscopic scale, and nanoscopic scale. The macroscopic scale expansion ability was determined by the hydrogel’s four sides’ ratio between the post- and pre-expansion hydrogels. The data were acquired and analyzed in ImageJ, and then the ratio was calculated. The microscopic expansion factor was determined by analyzing the distance of two identical cell nuclei in both pre- and post-expansion samples. The data were acquired and analyzed in ImageJ, and then the ratio was calculated. The nanoscopic expansion factor was determined by analyzing the size of identical microtubule zones between the pre- and post-expansion samples with registration algorithm. The data were acquired and analyzed with Elastix.
Measuring microtubule FWHM
Microtubule FWHM was measured by ImageJ. Intensity profiles perpendicular to microtubule orientation was measured. Then, the plot data were fit to a Gaussian function by ImageJ, and the FWHM was calculated from Gaussian fitting by R (Analyze→Plot profile→Data→Add fit→Gaussian fitting→Fit).
Evaluation of pre- and post-expansion image distortion and RMSE
The pre- and post-image distortion was analyzed using B-spline transformation with Elastix, and RMSE was analyzed by the Mathematica script written by Vaughan’s group (49).
Measuring NPC 153 (Nup153) radii
The radii of Nup153 were determined by circle fitting of each Nup153 complex signal in ImageJ. All sample preparation and imaging conditions are listed in fig. S24.
Acknowledgments
We thank W. Su for excellent technical assistance.
Funding: This work was supported by the National Natural Science Foundation of China (nos. 32071405, 31771088, and 31971271) and the Biomedical and Engineering Multidisciplinary Funding of Shanghai Jiao Tong University (SJTU) (no. YG2019QNB22).
Author contributions: H.L. and G.S. designed the experiments. H.L. and J.H. performed the experiments and analyzed the data. H.L., A.R.W., G.S., and X.D. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S23
Table S1
REFERENCES AND NOTES
- 1.Kyriazi M. E., Giust D., el-Sagheer A. H., Lackie P. M., Muskens O. L., Brown T., Kanaras A. G., Multiplexed mRNA sensing and combinatorial-targeted drug delivery using DNA-gold nanoparticle dimers. ACS Nano 12, 3333–3340 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Mao C., Lee M. Y., Jhan J.-R., Halpern A. R., Woodworth M. A., Glaser A. K., Chozinski T. J., Shin L., Pippin J. W., Shankland S. J., Liu J. T. C., Vaughan J. C., Feature-rich covalent stains for super-resolution and cleared tissue fluorescence microscopy. Sci. Adv. 6, eaba4542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W., Douglas D., Zhang J., Kumari S., Enuameh M. S., Dai Y., Wallace C. T., Watkins S. C., Shu W., Xing J., Live-cell imaging and analysis reveal cell phenotypic transition dynamics inherently missing in snapshot data. Sci. Adv. 6, eaba9319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddig S., Aufmkolk S., Doose S., Jobin M.-L., Werner C., Sauer M., Calebiro D., Super-resolution imaging reveals the nanoscale organization of metabotropic glutamate receptors at presynaptic active zones. Sci. Adv. 6, eaay7193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J., Zaccara S., Khuperkar D., Kim H., Tanenbaum M. E., Jaffrey S. R., Live imaging of mRNA using RNA-stabilized fluorogenic proteins. Nat. Methods 16, 862–865 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Syed A. M., MacMillan P., Ngai J., Wilhelm S., Sindhwani S., Kingston B. R., Wu J. L. Y., Llano-Suárez P., Lin Z. P., Ouyang B., Kahiel Z., Gadde S., Chan W. C. W., Liposome imaging in optically cleared tissues. Nano Lett. 20, 1362–1369 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Schermelleh L., Ferrand A., Huser T., Eggeling C., Sauer M., Biehlmaier O., Drummen G. P. C., Super-resolution microscopy demystified. Nat. Cell Biol. 21, 72–84 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Gambarotto D., Zwettler F. U., le Guennec M., Schmidt-Cernohorska M., Fortun D., Borgers S., Heine J., Schloetel J. G., Reuss M., Unser M., Boyden E. S., Sauer M., Hamel V., Guichard P., Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Methods 16, 71–74 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Betzig E., Patterson G. H., Sougrat R., Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Vicidomini G., Bianchini P., Diaspro A., STED super-resolved microscopy. Nat. Methods 15, 173–182 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Rust M. J., Bates M., Zhuang X., Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heintzmann R., Huser T., Super-resolution structured illumination microscopy. Chem. Rev. 117, 13890–13908 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Dertinger T., Colyer R., Iyer G., Weiss S., Enderlein J., Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI). Proc. Natl. Acad. Sci. U.S.A. 106, 22287–22292 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu B., Chen C., di X., Liao J., Wen S., Su Q. P., Shan X., Xu Z. Q., Ju L. A., Mi C., Wang F., Jin D., Upconversion nonlinear structured illumination microscopy. Nano Lett. 20, 4775–4781 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Szalai A. M., Siarry B., Lukin J., Williamson D. J., Unsain N., Cáceres A., Pilo-Pais M., Acuna G., Refojo D., Owen D. M., Simoncelli S., Stefani F. D., Three-dimensional total-internal reflection fluorescence nanoscopy with nanometric axial resolution by photometric localization of single molecules. Nat. Commun. 12, 517 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacquemet G., Carisey A. F., Hamidi H., Henriques R., Leterrier C., The cell biologist’s guide to super-resolution microscopy. J. Cell Sci. 133, jcs240713 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Yan R., Wang B., Xu K., Functional super-resolution microscopy of the cell. Curr. Opin. Chem. Biol. 51, 92–97 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Sroda A., Makowski A., Tenne R., Rossman U., Lubin G., Oron D., Lapkiewicz R., SOFISM: Super-resolution optical fluctuation image scanning microscopy. Optica 7, 1308–1316 (2020). [Google Scholar]
- 19.Chen F., Tillberg P. W., Boyden E. S., Expansion microscopy. Science 347, 543–548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tillberg P. W., Chen F., Piatkevich K. D., Zhao Y., Yu C. C., English B. P., Gao L., Martorell A., Suk H. J., Yoshida F., DeGennaro E. M., Roossien D. H., Gong G., Seneviratne U., Tannenbaum S. R., Desimone R., Cai D., Boyden E. S., Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 34, 987–992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ku T., Swaney J., Park J. Y., Albanese A., Murray E., Cho J. H., Park Y. G., Mangena V., Chen J., Chung K., Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 34, 973–981 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faulkner E. L., Thomas S. G., Neely R. K., An introduction to the methodology of expansion microscopy. Int. J. Biochem. Cell Biol. 124, 105764 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Engerer P., Fecher C., Misgeld T., Super-resolution microscopy writ large. Nat. Biotechnol. 34, 928–930 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Liu J., Fang X., Liu Z., Li R., Yang Y., Sun Y., Zhao Z., Wu C., Expansion microscopy with multifunctional polymer dots. Adv. Mater. 33, e2007854 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Sun D. E., Fan X., Shi Y., Zhang H., Huang Z., Cheng B., Tang Q., Li W., Zhu Y., Bai J., Liu W., Li Y., Wang X., Lei X., Chen X., Click-ExM enables expansion microscopy for all biomolecules. Nat. Methods 18, 107–113 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Wen G., Vanheusden M., Acke A., Valli D., Neely R. K., Leen V., Hofkens J., Evaluation of direct grafting strategies via trivalent anchoring for enabling lipid membrane and cytoskeleton staining in expansion microscopy. ACS Nano 14, 7860–7867 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Wen G., Vanheusden M., Leen V., Rohand T., Vandereyken K., Voet T., Hofkens J., A universal labeling strategy for nucleic acids in expansion microscopy. J. Am. Chem. Soc. 143, 13782–13789 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Cho Y., Seo J., Sim Y., Chung J., Park C. E., Park C. G., Kim D., Chang J. B., FRACTAL: Signal amplification of immunofluorescence via cyclic staining of target molecules. Nanoscale 12, 23506–23513 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Gotz R., Panzer S., Trinks N., Eilts J., Wagener J., Turrà D., Pietro A. D., Sauer M., Terpitz U., Expansion microscopy for cell biology analysis in fungi. Front. Microbiol. 11, 574 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y., Bucur O., Irshad H., Chen F., Weins A., Stancu A. L., Oh E. Y., DiStasio M., Torous V., Glass B., Stillman I. E., Schnitt S. J., Beck A. H., Boyden E. S., Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nat. Biotechnol. 35, 757–764 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen F., Wassie A. T., Cote A. J., Sinha A., Alon S., Asano S., Daugharthy E. R., Chang J. B., Marblestone A., Church G. M., Raj A., Boyden E. S., Nanoscale imaging of RNA with expansion microscopy. Nat. Methods 13, 679–684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cahoon C. K., Yu Z., Wang Y., Guo F., Unruh J. R., Slaughter B. D., Hawley R. S., Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. U.S.A. 114, E6857–E6866 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park C. E., Cho Y., Cho I., Jung H., Kim B., Shin J. H., Choi S., Kwon S. K., Hahn Y. K., Chang J. B., Super-resolution three-dimensional imaging of actin filaments in cultured cells and the brain via expansion microscopy. ACS Nano 14, 14999–15010 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Zwettler F. U., Spindler M. C., Reinhard S., Klein T., Kurz A., Benavente R., Sauer M., Tracking down the molecular architecture of the synaptonemal complex by expansion microscopy. Nat. Commun. 11, 3222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drelich L., Aboulouard S., Franck J., Salzet M., Fournier I., Wisztorski M., Toward high spatially resolved proteomics using expansion microscopy. Anal. Chem. 93, 12195–12203 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Gotz R., Kunz T. C., Fink J., Solger F., Schlegel J., Seibel J., Kozjak-Pavlovic V., Rudel T., Sauer M., Nanoscale imaging of bacterial infections by sphingolipid expansion microscopy. Nat. Commun. 11, 6173 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blom H., Widengren J., Stimulated emission depletion microscopy. Chem. Rev. 117, 7377–7427 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Bayle V., Fiche J. B., Burny C., Platre M. P., Nollmann M., Martinière A., Jaillais Y., Single-particle tracking photoactivated localization microscopy of membrane proteins in living plant tissues. Nat. Protoc. 16, 1600–1628 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Hu Y., Chu X., Chen T. T., Pan Q., Liu C., Yi J., Chu X., Improving resolving ability of expansion microscopy by varying crosslinker concentration. Chem. Commun. 56, 4176–4179 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Chang J. B., Chen F., Yoon Y. G., Jung E. E., Babcock H., Kang J. S., Asano S., Suk H. J., Pak N., Tillberg P. W., Wassie A. T., Cai D., Boyden E. S., Iterative expansion microscopy. Nat. Methods 14, 593–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park H. E., Choi D., Park J. S., Sim C., Park S., Kang S., Yim H., Lee M., Kim J., Pac J., Rhee K., Lee J., Lee Y., Lee Y., Kim S. Y., Scalable and isotropic expansion of tissues with simply tunable expansion ratio. Adv. Sci. 6, 1901673 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao R., Yu C. C., Gao L., Piatkevich K. D., Neve R. L., Munro J. B., Upadhyayula S., Boyden E. S., A highly homogeneous polymer composed of tetrahedron-like monomers for high-isotropy expansion microscopy. Nat. Nanotechnol. 16, 698–707 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Truckenbrodt S., Maidorn M., Crzan D., Wildhagen H., Kabatas S., Rizzoli S. O., X10 expansion microscopy enables 25-nm resolution on conventional microscopes. EMBO Rep. 19, e45836 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Truckenbrodt S., Sommer C., Rizzoli S. O., Danzl J. G., A practical guide to optimization in X10 expansion microscopy. Nat. Protoc. 14, 832–863 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Liu X., Liu J., Lin S., Zhao X., Hydrogel machines. Mater. Today 36, 102–124 (2020). [Google Scholar]
- 46.Gradinaru V., Treweek J., Overton K., Deisseroth K., Hydrogels in tissue engineering, scope and applications. Annu. Rev. Biophys. 47, 355–376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chavda H., Patel C., Effect of crosslinker concentration on characteristics of superporous hydrogel. Int. J. Pharm. Investig. 1, 17–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee W.-F., Lin Y.-H., Swelling behavior and drug release of NIPAAm/PEGMEA copolymeric hydrogels with different crosslinkers. J. Mater. Sci. 41, 7333–7340 (2006). [Google Scholar]
- 49.Chozinski T. J., Halpern A. R., Okawa H., Kim H. J., Tremel G. J., Wong R. O. L., Vaughan J. C., Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods 13, 485–488 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cutie S. S., Henton D. E., Powell C., Reim R. E., Smith P. B., Staples T. L., The effects of MEHQ on the polymerization of acrylic acid in the preparation of superabsorbent gels. J. Appl. Polym. Sci. 64, 577–589 (1997). [Google Scholar]
- 51.Walther T. C., Fornerod M., Pickersgill H., Goldberg M., Allen T. D., Mattaj I. W., The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 20, 5703–5714 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thevathasan J. V., Kahnwald M., Cieśliński K., Hoess P., Peneti S. K., Reitberger M., Heid D., Kasuba K. C., Hoerner S. J., Li Y., Wu Y. L., Mund M., Matti U., Pereira P. M., Henriques R., Nijmeijer B., Kueblbeck M., Sabinina V. J., Ellenberg J., Ries J., Nuclear pores as versatile reference standards for quantitative superresolution microscopy. Nat. Methods 16, 1045–1053 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fahrenkrog B., Nucleoporin gene fusions and hematopoietic malignancies. N. J. Sci. 2014, 1–18 (2014). [Google Scholar]
- 54.Pesce L., Cozzolino M., Lanzano L., Diaspro A., Bianchini P., Measuring expansion from macro- to nanoscale using NPC as intrinsic reporter. J. Biophotonics 12, e201900018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao M., Maraspini R., Beutel O., Zehtabian A., Eickholt B., Honigmann A., Ewers H., Expansion stimulated emission depletion microscopy (ExSTED). ACS Nano 12, 4178–4185 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Halpern A. R., Alas G. C. M., Chozinski T. J., Paredez A. R., Vaughan J. C., Hybrid structured illumination expansion microscopy reveals microbial cytoskeleton organization. ACS Nano 11, 12677–12686 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu H., Tong Z., Ye Q., Sun T., Hong Z., Zhang L., Bortnick A., Cho S., Beuzer P., Axelrod J., Hu Q., Wang M., Evans S. M., Murre C., Lu L. F., Sun S., Corbett K. D., Cang H., Molecular organization of mammalian meiotic chromosome axis revealed by expansion STORM microscopy. Proc. Natl. Acad. Sci. U.S.A. 116, 18423–18428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zwettler F. U., Reinhard S., Gambarotto D., Bell T. D. M., Hamel V., Guichard P., Sauer M., Molecular resolution imaging by post-labeling expansion single-molecule localization microscopy (Ex-SMLM). Nat. Commun. 11, 3388 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li R., Chen X., Lin Z., Wang Y., Sun Y., Expansion enhanced nanoscopy. Nanoscale 10, 17552–17556 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S23
Table S1