Abstract
Adolescent binge drinking is a major risk factor for psychiatric disorders later in life including alcohol use disorder. Adolescent alcohol exposure induces epigenetic reprogramming at the enhancer region of the activity-regulated cytoskeleton-associated protein (Arc) immediate-early gene, known as synaptic activity response element (SARE), and decreases Arc expression in the amygdala of both rodents and humans. The causal role of amygdalar epigenomic regulation at Arc SARE in adult anxiety and drinking after adolescent alcohol exposure is unknown. Here, we show that dCas9-P300 increases histone acetylation at the Arc SARE and normalizes deficits in Arc expression, leading to attenuation of adult anxiety and excessive alcohol drinking in a rat model of adolescent alcohol exposure. Conversely, dCas9-KRAB increases repressive histone methylation at the Arc SARE, decreases Arc expression, and produces anxiety and alcohol drinking in control rats. These results demonstrate that epigenomic editing in the amygdala can ameliorate adult psychopathology after adolescent alcohol exposure.
Causal role of epigenetic regulation in anxiety and alcohol drinking behaviors is identified in a preclinical model of AUD.
INTRODUCTION
Alcohol use disorder (AUD) is a leading preventable cause of death and has a considerable societal and economic burden (1–3). AUD has disproportionately few available therapeutic options (1, 2), and increased understanding of the risk factors that contribute to AUD could lead to therapeutic advances. A leading risk factor for developing an AUD later in life is adolescent binge alcohol consumption (4–6). Adolescence is a critical period for brain maturation, and binge drinking during this period also increases vulnerability to a higher risk of comorbid psychiatric disorders including anxiety in both rodents and humans (7–10).
The central nucleus of the amygdala (CeA) is part of the extended amygdala and is a crucial brain region in regulating both AUD and anxiety disorders (2, 9, 11–13). The brain undergoes orchestrated changes in transcription, epigenetic modifications, and intra- and interlimbic connectivity during adolescent development, which are disrupted by adolescent alcohol consumption (9–12). The immediate-early gene activity–regulated cytoskeleton-associated protein (Arc) regulates synaptic plasticity and organization and is under complex epigenetic and posttranslational regulation (14–18). Arc responds immediately and robustly to synaptic activity and is considered a key regulator of brain plasticity (11, 12, 14–18), where it orchestrates synaptic structure and tuning to regulate higher-order cognitive and affective processes as well as addiction-relevant behaviors (11, 12, 14, 18). Arc has a well-characterized enhancer region ~7 kb upstream of the Arc transcription start site (TSS) known as the synaptic activity response element (SARE), which is conserved from rodents to humans (15–18) and undergoes repressive epigenetic remodeling after adolescent alcohol exposure (11, 12).
Epigenetic dysregulation, specifically changes in histone posttranslational chemical modifications, associated with genomic regulatory regions is well documented in AUD and other psychiatric disorders (9, 19–22). The histone 3 lysine 27 acetylation (H3K27ac) epigenetic mark has largely been associated with active enhancers as opposed to poised enhancers (23, 24). Adolescent alcohol exposure decreases Arc expression in the adult amygdala of rodents and humans, likely through deposition of H3K27 trimethylation (H3K27me3), removal of H3K27ac, increases in enhancer of zeste homolog 2 (EZH2), and decreases in lysine demethylase 6B (KDM6B) and cyclic adenosine 3′,5′-monophosphate response element–binding protein (CREB)–binding protein (CBP) levels at the Arc SARE site (11, 12). To date, investigation of epigenetic consequences of developmental alcohol exposure has been largely correlative, and the causal role of specific epigenetic modifications at discrete genomic regions in the brain underlying increased risk for developing AUD and comorbid anxiety after adolescent drinking remains unexplored. Here, we leveraged epigenomic editing (25–27) using CRISPR (clustered regularly interspaced short palindromic repeats)/dCas9 construct to specifically manipulate histone acetylation and methylation at the Arc SARE site and show that bidirectional epigenetic regulation of this site in the CeA can ameliorate or mimic the detrimental effects of adolescent alcohol exposure in adulthood.
RESULTS
Activation of the Arc enhancer rescues anxiety and drinking
To test whether histone acetylation at the Arc SARE site is critical to regulating transcriptional and behavioral changes after adolescent intermittent ethanol (AIE) exposure, we used a CRISPR/dCas9 strategy that allows for targeted epigenetic modifications at discrete genomic locations (25–27). We specifically sought to determine whether restoration of histone acetylation at the Arc SARE with dCas9-P300 would ameliorate excessive drinking and anxiety phenotypes in adulthood after AIE.
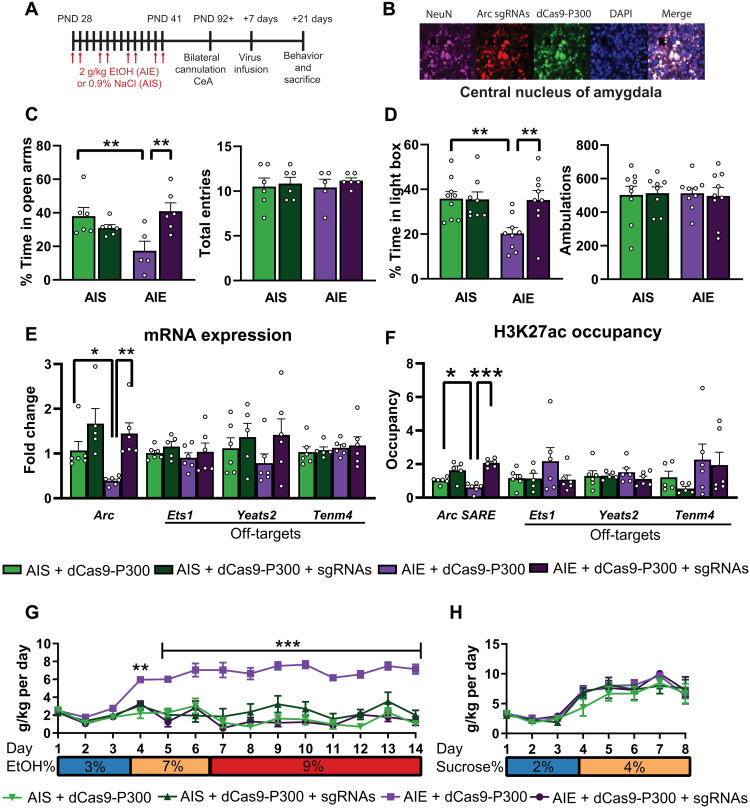
We exposed rats to binge levels of alcohol [reaching blood ethanol levels of 183 ± 4.9 mg% (28)] during adolescence [postnatal days (PNDs) 28 to 41, which is equivalent to 10 to 18 years in humans (29)] that lead to heightened anxiety and increased alcohol consumption in adulthood (11, 28–30). We then used a construct that consists of dead Cas9 with no endonuclease ability (dCas9, D10A, and H840A mutations) fused with the catalytic component of human P300 (dCas9-P300), a histone acetyltransferase (fig. S1A) that catalyzes histone acetylation, including H3K27ac (25–27, 31). We constructed four single-guide RNAs (sgRNAs) targeted to the Arc SARE in lentiviral vectors and confirmed their transduction efficacy in rat PC12 cell culture (fig. S1, A to C). Cotransduction of lentiviruses expressing with CRISPR/dCas9-P300 plus Arc SARE sgRNAs in PC12 cells increased Arc mRNA and increased H3K27ac occupancy at the Arc SARE site (fig. S1, D and E) with no effects on the predicted off-target genes (fig. S1, F and G). We then generated adolescent intermittent saline (AIS)–exposed control and AIE animals and bilaterally infused dCas9-P300 with or without sgRNAs into the CeA in adulthood (Fig. 1, A and B). We found that dCas9-P300 plus sgRNA infusion attenuated anxiety-like behavior back to control levels as measured by the elevated plus maze (EPM) and light/dark box (LDB) exploration test (Fig. 1, C and D) and decreased the excessive drinking seen in AIE-exposed rats to control levels (Fig. 1G and fig. S2A) with no effect on sucrose consumption (Fig. 1H and fig. S2C).
Fig. 1. dCas9-P300 plus sgRNAs reverses epigenetic and behavioral consequences of adolescent alcohol exposure in adulthood.
(A) Experimental timeline for AIE, AIS, CeA cannulation, lentivirus infusion, and behavioral measurement. (B) Representative immunofluorescence images showing colocalization of dCas9-P300 (anti–CRISPR-Cas9; green) and Arc sgRNAs (DsRed; red) in NeuN-positive cells (violet color) in the CeA. (C) dCas9-P300 plus sgRNA infusion ameliorates anxiety-like behavior in the EPM (two-way ANOVA, adolescent exposure and lentivirus interaction, F1,19 = 10.94, P = 0.004, n = 5 to 6 rats in each group) and (D) in the LDB (two-way ANOVA, adolescent exposure × lentivirus interaction, F1,31 = 5.19, P = 0.030, n = 8 to 9 rats in each group) exploration tests. (E) dCas9-P300 plus sgRNA infusion reverses decreased Arc mRNA expression (Arc mRNA two-way ANOVA, lentivirus infusion, F1,19 = 14.53, P = 0.001, n = 5 to 6 rats in each group) with no off-target (Ets1, Yeats2, and Tenm4) effects. (F) dCas9-P300 plus sgRNAs reverses decreased H3K27ac at the Arc SARE site (adolescent exposure × lentivirus interaction, F1,19 = 9.17, P = 0.007, n = 5 to 6 rats in each group) with no off-target effects. (G) dCas9-P300 plus sgRNA infusion into CeA attenuates excessive alcohol consumption (group × day interaction, F39,247 = 5.54, P < 0.001, n = 5 to 6 rats in each group; individual values are provided in table S3). (H) Epigenetic editing at Arc SARE site does not affect sucrose consumption (n = 5 to 6 rats in each group; individual values are provided in table S4). Two-way ANOVAs were followed by Tukey’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001. Bar and line graphs show means ± SEM. Individual values are shown with circle dots on bar graphs.
dCas9-P300 plus sgRNA infusion into CeA ameliorated AIE-induced decreases in Arc expression and associated decreased H3K27ac and increased H3K27me3 occupancy at the SARE site in adulthood without producing any effects on predicted off-target sites (Fig. 1, E and F; and fig. S3, A, B, and D). We performed additional control experiments to examine whether targeting the Arc SARE site with dCas9-P300 altered Cbp, p300, and Creb1, because they are known to be involved in increasing H3K27ac at the SARE, and their mRNA levels are decreased in the amygdala after AIE in adulthood (32). We found no effect of dCas9-P300 plus sgRNAs on any of these genes (fig. S3C), suggesting that targeted editing of the Arc SARE site by dCas9-P300 was responsible for increased H3K27ac occupancy and rescue of Arc expression. Note that because of the promiscuous histone acetyltransferase nature of P300 (31, 33), we cannot empirically rule out the involvement of other histone acetylation marks (or other epigenetic or transcriptional mechanisms). Nonetheless, these results suggest that activation of Arc SARE with dCas9-P300 in the CeA ameliorates adult anxiety and excessive alcohol drinking observed after AIE.
dCas9-P300 promotes enhancer RNA and negative elongation factor interactions
Previous reports suggest that the Arc SARE controls Arc expression through the expression of enhancer RNAs (eRNAs), which are transcribed bidirectionally from both positive and negative DNA strands flanking the Arc SARE enhancer site (15, 34). In general, both strands of eRNA have been shown to regulate gene expression and serve as indicators of active enhancers (35). Furthermore, negative elongation factor (NELF) is a protein that pauses RNA polymerase II at the Arc promoter and inhibits transcription, and studies have shown that negative-strand Arc eRNA (−) can bind to NELF, leading to the removal of NELF and increased Arc transcription (11, 15, 34). Here, we measured both eRNA species as indicators of active or repressed Arc SARE after dCas9-P300 or dCas9–KRAB (Krüppel-associated box) manipulations, respectively.
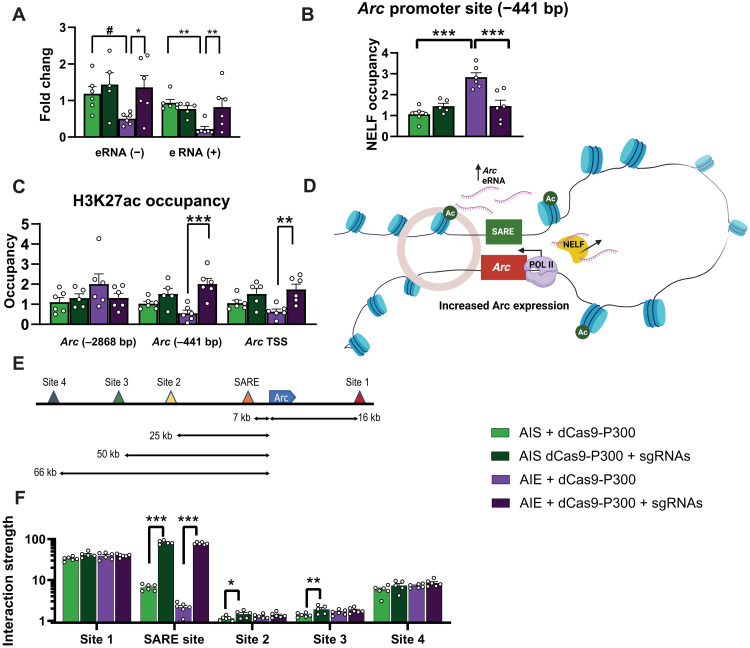
We observed that dCas9-P300 plus sgRNAs increases Arc eRNA expression in PC12 cells (fig. S1D). In vivo data also indicate that AIE decreases Arc eRNA (−) and (+) expression as reported earlier (11), and infusion of dCas9-P300 plus sgRNAs in the CeA restores normal expression of both Arc eRNAs (Fig. 2A) and blocks AIE-induced increased NELF binding to the Arc promoter (Fig. 2B). These results suggest that a P300-mediated increase in histone acetylation at the Arc SARE initiates an epigenetic circuit to rescue Arc expression in the amygdala. When we analyzed whether changes in H3K27ac were localized specifically to the enhancer, we found that dCas9-P300 infusion targeted to Arc SARE increased H3K27ac at the proximal promoter [−441 base pair (bp) upstream of the TSS] and at the TSS but not at a distal promoter region (−2868 bp upstream of the TSS) (Fig. 2C). The dCas9-P300–mediated increase in H3K27ac is also associated with a decrease in H3K27me3 occupancy at Arc SARE, Arc TSS, and Arc promoter (−441 bp) sites (fig.S3D). These results suggested that there was increased interaction between the Arc SARE and the promoter region, possibly due to enhancer-promoter chromatin looping (Fig. 2D). To test this notion, we used a chromatin conformation capture (3C) assay (34, 36) and analyzed the interaction of the Arc promoter with several sites of the Arc gene including the SARE (Fig. 2E). We found that the interactive strength of the Arc SARE and promoter site is lower in the amygdala of AIE-exposed adult rats and is then significantly increased by dCas9-P300 plus sgRNA infusion (Fig. 2F), suggesting dynamic chromatin looping (Fig. 2D) may initiate permissive histone modifications at the Arc promoter (Fig. 2C and fig. S3D). These results along with normalization of NELF binding (Fig. 2B) show that activating the enhancer using dCas9-P300 induces changes in chromatin conformation to facilitate enhancer-promoter interaction and regulate gene expression (Fig. 2D).
Fig. 2. dCas9-P300 plus sgRNAs reverses deficits in eRNA expression and increases enhancer and promoter interactions.
(A) dCas9-P300 plus sgRNAs reverses AIE-induced decreases in Arc eRNA (+) expression (adolescent exposure × lentivirus interaction F1,19 = 6.96, P = 0.016) and Arc eRNA (−) expression (lentivirus, F1,19 = 5.25, P = 0.034). (B) dCas9-P300 plus sgRNAs releases NELF from the Arc promoter (−441 bp) in the amygdala of AIE adult rats (adolescent exposure: F1,19 = 17.58, P < 0.001; adolescent exposure × lentivirus interaction: F1,19 = 17.55, P < 0.001). (C) dCas9-P300 plus sgRNAs reverses AIE-induced decrease in H3K27ac occupancy at Arc promoter (−441 bp) (adolescent exposure × lentivirus interaction: F1,19 = 5.45, P = 0.031) and Arc TSS (lentivirus: F1,19 = 14.21, P = 0.001) without any effects at distal to the TSS (−2868 bp). (D) Hypothetical model of chromatin looping most likely produced by dCas9-P300 in adult amygdala after AIE leading to increase in H3K27ac levels at Arc promoter (−441 bp and TSS) and release of NELF to increase Arc expression. (E) Arc gene showing the locations of sites including SARE that were selected to evaluate the interaction with promoter (blue color). (F) 3C assay shows that dCas9-P300 plus sgRNAs reverses lower interactive strength between Arc promoter and SARE in the amygdala of AIE adult rats (lentivirus: F1,19 = 1540.74, P < 0.001) and minimal effect of virus in AIS group at sites 2 and 3 but no change at sites 1 and 4 of the gene. Two-way ANOVAs were followed by Tukey’s post hoc test (*P < 0.05; **P < 0.01; ***P < 0.001; #P = 0.056). Bar graphs show means ± SEM (n = 5 to 6 rats in each group), and individual values are shown with circle dots on bar graphs.
Repression of the Arc enhancer promotes anxiety and drinking
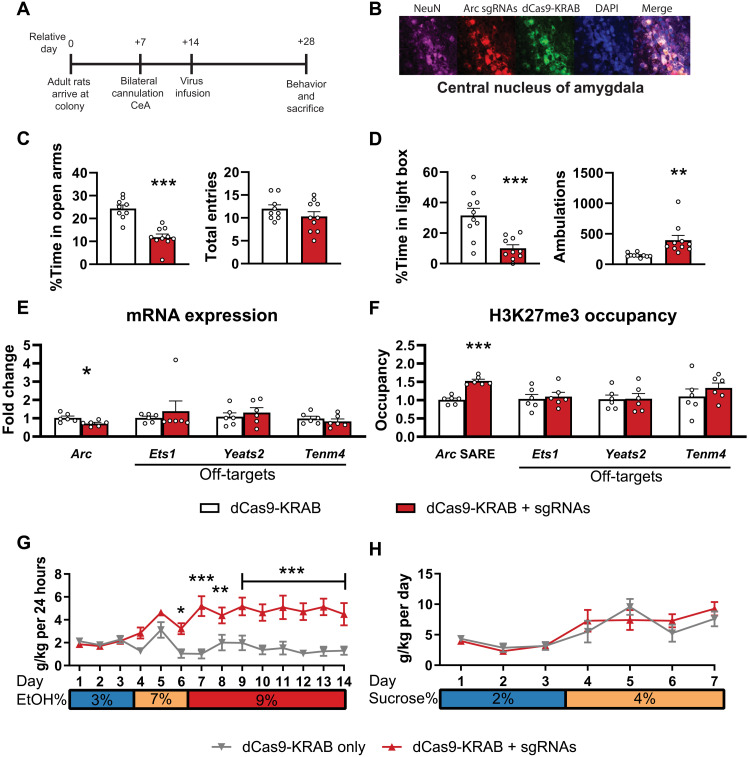
Next, we also evaluated whether infusion of CRISPR/dCas9-KRAB and Arc SARE sgRNAs into the CeA would provoke anxiety and promote drinking in alcohol-naïve control adult rats because of chromatin remodeling at the Arc SARE site (Fig. 3, A and B). dCas9-KRAB domain is a transcriptional repressor (fig. S4A) that increases repressive histone methylation marks at target genes (25, 37, 38), allowing us to causally determine whether repressing the Arc SARE increases anxiety and alcohol drinking.
Fig. 3. dCas9-KRAB plus sgRNAs induces anxiety-like behavior and higher alcohol consumption, decreases Arc expression, and causes epigenetic remodeling at the Arc SARE.
(A) Experimental timeline demonstrating the use of dCas9-KRAB in the CeA of alcohol-naïve control adult rats. (B) Representative immunofluorescence images showing colocalization of dCas9-KRAB (anti–CRISPR-Cas9; green) and Arc sgRNAs (DsRed; red) in NeuN-positive cells (violet color) in the CeA. (C) dCas9-KRAB plus sgRNAs induces anxiety like behavior in EPM (% time in open arms: t17 = 6.096, P < 0.001, n = 9 to 10 rats in each group) and (D) LDB (% time in light box: t18 = 4.101, P < 0.001; ambulation: t18 = 3.148, P = 0.006, n = 10 rats in each group) exploration tests. (E) dCas9-KRAB plus sgRNAs causes decreased Arc mRNA expression (Arc mRNA: t10 = 2.69, P = 0.022) but no change in the predicted off-target RNAs (Ets1, Yeats2, and Tenm4, n = 6 in each group). (F) dCas9-KRAB plus sgRNAs increases H3K27me3 associated with the Arc SARE site (Arc SARE: t10 = 7.78, P < 0.001) but not predicted off-target (Ets1, Yeats2, and Tenm4, n = 6 in each group) binding sites. (G) dCas9-KRAB plus sgRNAs increases ethanol consumption (lentivirus and day interaction: F13,156 = 5.19, P < 0.001, n = 6 to 8 rats in each group; individual values are provided in table S5) without any effect (H) on sucrose consumption (n = 5 to 7 in rats in each group; individual values are provided in table S6). Two-way ANOVAs were followed by Tukey’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001. Bar and line graphs show means ± SEM, and individual values are shown with circle dots on bar graphs.
First, we tested the transduction efficacy of dCas9-KRAB in PC12 cells (fig. S4, A to C) and found that cotransduction of dCas9-KRAB and Arc SARE sgRNAs decreased Arc mRNA expression and increased H3K27me3 at the Arc SARE site (fig. S4, D and F). We did not observe changes in either mRNA expression or H3K27me3 occupancy for the predicted off-target genes (fig. S4, E and G). dCas9-KRAB plus sgRNA infusion into CeA of alcohol-naïve control animals (Fig. 3, A and B) provoked anxiety-like behavior in two separate tests (EPM and LDB; Fig. 3, C and D, respectively) and increased alcohol consumption (Fig. 3G and fig. S5A) without producing any effect on sucrose intake (Fig. 3H and fig. S5C). In addition, we found that dCas9-KRAB plus sgRNA infusion into the CeA decreased Arc expression (Fig. 3E), increased H3K27me3 (Fig. 3F), and decreased H3K27ac (fig. S6C) at the Arc SARE with no observed effects on mRNA expression and H3K27me3 occupancy of off-target genes in the amygdala (Fig. 3, E and F, and fig. S6, A and B). These data further support the AIE findings that repression of the Arc SARE site in the CeA causally regulates anxiety and excessive drinking.
dCas9-KRAB attenuates eRNA and NELF interactions
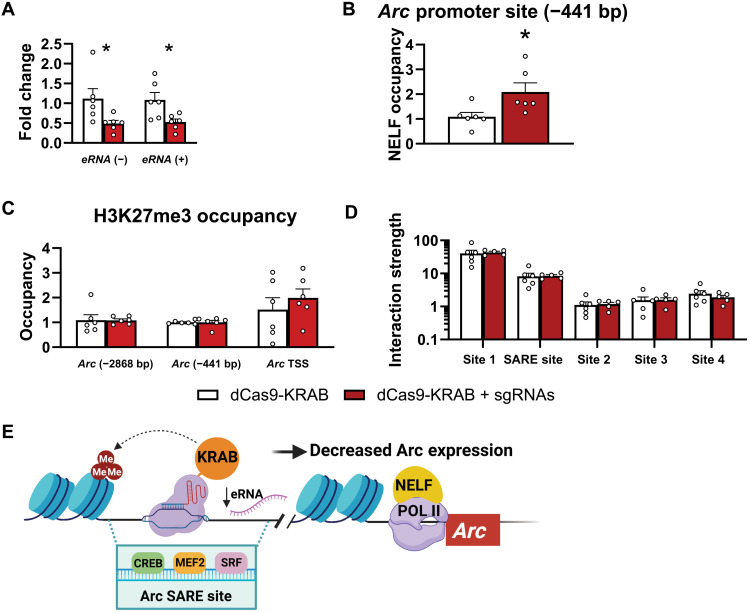
Using dCas9-KRAB, we found that repressing the Arc SARE site decreases Arc (+) and (−) eRNA expression in PC12 cells (fig. S4D) and in the amygdala (Fig. 4A). Furthermore, in rats, this corresponds to increased NELF binding at the Arc promoter (Fig. 4B). Infusion of dCas9-KRAB plus sgRNAs into the CeA increased H3K27me3 and decreased H3K27ac occupancy only at the Arc SARE site (Fig. 3F and fig. S6C) but not at the other sites (TSS; −441 and −2868 bp) of the Arc gene (Fig. 4C and fig. S6C). We also measured occupancy of another repressive histone methylation mark (H3K9me3) and found that dCas9-KRAB plus sgRNAs has no effects on the occupancy of H3K9me3 at the Arc SARE site and other sites (TSS; −441 and 2868 bp) of the Arc gene (fig. S6D). Furthermore, dCas9-KRAB plus sgRNA infusion does not alter the interactive strength of Arc SARE and promoter sites (Fig. 4D) and possibly decreases Arc expression via decreased eRNA levels and increased NELF binding at promoter (Fig. 4, A and B). These results suggest that dCas9-KRAB decreases eRNA expression, leading to increased NELF binding at the promoter and decreased Arc expression (Fig. 4E).
Fig. 4. dCas9-KRAB plus sgRNAs decreases eRNA expression and increases NELF binding at Arc promoter in the amygdala of control rats.
(A) dCas9-KRAB plus sgRNAs causes decreased Arc eRNA expression [Arc eRNA (−): Mann-Whitney U = 3.0, P = 0.015; eRNA (+): t10 = 2.715, P = 0.022, n = 6 in each group]. (B) dCas9-KRAB plus sgRNAs increases NELF binding at the Arc promoter (t10 = 2.487, P = 0.032, n = 6 in each group). (C) dCas9-KRAB plus sgRNAs produces no change in H3K27me3 occupancy at several sites of Arc (promoter −441 bp; Arc TSS, Arc distal region −2868 bp, n = 6 in each group) gene. (D) 3C assay shows that dCas9-KRAB plus sgRNAs produces no change in interactive strength between Arc promoter and SARE as well as other sites of the gene (n = 5 to 6 rats in each group). (E) Hypothetical model depicting condensed chromatin dynamics produced by dCas9-KRAB targeted at Arc SARE in adult amygdala causing decreased eRNA leading to increased NELF binding at Arc promoter and decreased Arc expression.
DISCUSSION
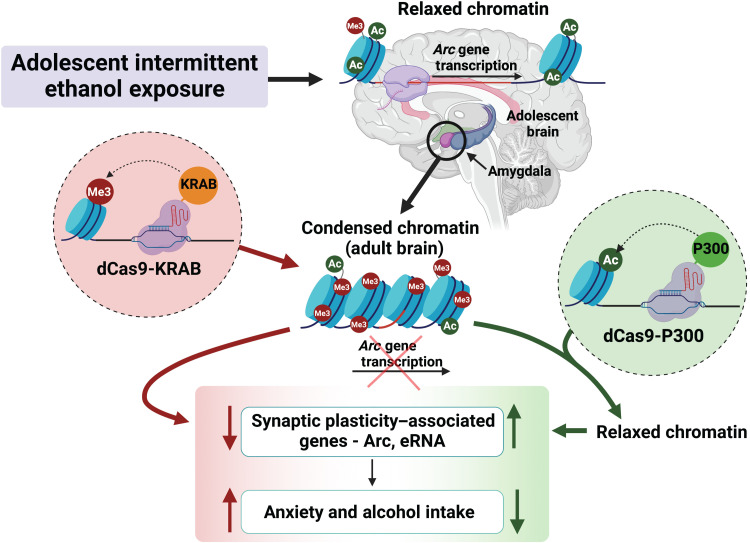
Our results indicate that targeted epigenomic editing at the Arc SARE can bidirectionally modulate behavioral changes caused by adolescent alcohol exposure (Fig. 5). Furthermore, we demonstrate that these changes are largely due to an epigenetic circuit involving transcription of eRNAs from the Arc SARE, which causes epigenetic remodeling at the Arc promoter by looping of the chromatin to allow localized release of NELF. In addition, our results indicate that activating the Arc SARE site through histone acetylation using dCas9-P300 facilitates Arc eRNA transcription and long-range promoter-enhancer interactions and modulates transcription factor binding, which is consistent with other findings (25, 39, 40). Conversely, dCas9-KRAB–mediated epigenetic suppression at Arc SARE decreases eRNA transcription and increases NELF binding at the promoter without changing enhancer and promoter interactions and promotes anxiety and excessive drinking (Fig. 5). This contrasts with other studies in which KRAB is involved in heterochromatin spreading and transcriptional repression via alternative mechanisms (41). Here, we demonstrate that dCas9-KRAB increases repressive H3K27me3, which has previously been shown to be involved in repressing eRNA transcription (35). We observed no change in H3K9me3 occupancy at the Arc SARE site after dCas9-KRAB manipulations. Other studies have shown that targeting dCas9-KRAB to specific genomic loci increases H3K9me3, but these changes have been shown to be transitory and not sufficient for transcriptional repression (38, 42). While it is likely that increased H3K27me3 at the Arc SARE contributes to repressed Arc eRNA and mRNA expression and subsequent behavioral correlates, the current study did not investigate other repressive mechanisms induced by dCas9-KRAB.
Fig. 5. Model showing that adolescent alcohol exposure produces epigenetic reprogramming in the CeA as well as anxiety and excessive alcohol intake in adulthood.
dCas9-P300 infusion in the CeA was able to increase H3K27ac at Arc SARE, increase Arc eRNA and mRNA expression, and ameliorate anxiety and excessive alcohol intake induced by AIE exposure in adulthood. On the other hand, dCas9-KRAB increased repressive epigenetic marks, H3K27me3 at Arc SARE, leading to decreased Arc eRNA and mRNA expression and development of anxiety and alcohol drinking behaviors in control adult rats. These data causally link epigenetic modifications at an enhancer region of synaptic gene Arc to adult AUD and anxiety after adolescent alcohol exposure.
Preclinical and clinical data clearly suggest that adolescent alcohol consumption can increase the susceptibility of an individual to anxiety and AUD (4–7, 9, 10). Several studies in the field have shown that various biological mechanisms, including epigenetic changes, may be involved in the persistence of the effects of adolescent alcohol exposure into adulthood (9, 10, 43). It has been shown that epigenetic drugs such as histone deacetylase (HDAC) and DNA methyltransferase (DNMT) inhibitors can attenuate adolescent alcohol exposure–induced anxiety-like and alcohol drinking behaviors in adult rats (28, 44). Similar to what was found in the current study using dCas9-P300, treatment with either a systemic HDAC inhibitor or DNMT inhibitor did not result in anxiolysis or reduce alcohol intake in AIS adult rats (28, 44). Here, there was a trend toward an increase in Arc expression and histone acetylation after dCas9-P300 treatment, but this was not statistically significant. Conversely, acute ethanol challenge in AIS adult rats increased Arc expression and produced anxiolysis that was associated with increased KDM6B/CBP, decreased H3K27me3, and increased H3K27ac occupancy at the Arc SARE site in the amygdala. These epigenetic modifications were associated with a significant increase in Arc expression in the amygdala of AIS rats (11). We therefore suggest that the dCas9-P300 manipulation did not reach the set point or biological threshold of Arc expression to induce anxiolysis, and this may be a possible explanation for the lack of observed effects of dCas9-P300 on anxiety and alcohol intake in AIS control rats.
The Arc gene is one of the most interactive genes in the altered synaptic gene network in the adult amygdala after adolescent alcohol exposure in rats (11). Arc interacts with a brain-derived neurotrophic factor (BDNF), and both are down-regulated via epigenetic mechanisms in the amygdala of rodents and humans in adulthood after adolescent alcohol consumption (11, 12, 30). Arc protein expression is lower in the CeA of alcohol-preferring rats as compared with alcohol nonpreferring rats, and this is most likely the result of increased HDAC2-mediated deficits in histone acetylation of Arc gene. Inhibition of HDAC2 expression in the CeA of preferring rats attenuated anxiety-like behaviors and excessive drinking and increased histone acetylation and Arc protein expression in the CeA (45). Furthermore, HDAC and DNMT inhibitors are also effective in preventing excessive drinking and alcohol self-administration in other animal models of AUD (46–48). These studies suggest that Arc expression is regulated via histone acetylation mechanisms and that pharmacological epigenetic agents are effective in attenuating anxiety and alcohol intake in several models of AUD. However, these studies do not establish the specific epigenetically regulated genomic regions implicated in these effects. Here, we used epigenomic editing (25–27) to modulate histone acetylation/methylation levels at a specific genomic locus regulating anxiety and excessive drinking.
AIE is associated with diminished synaptic events that are characterized by decreased mRNA and protein levels of Arc and decreased dendritic spines and synapses in the CeA in adulthood (11, 28). Our previous studies showed that up-regulation of EZH2 and down-regulation of KDM6B and CBP after AIE initiate repression of the Arc gene through increases in H3K27me3 and decreases in H3K27ac occupancy at the Arc SARE, providing an endogenous mechanism for epigenetic reprogramming in the amygdala of rodents (11) and AUD subjects with early age of onset (12). All these studies are correlative in nature, and we now causally demonstrate in detail how epigenetic activation and repression at the Arc SARE site can drive adult excessive drinking and anxiety induced by adolescent alcohol exposure. Our results further indicate that epigenetic editing using dCas9-P300 at the Arc SARE is sufficient for enhancer-mediated activation of Arc expression and can drive increased H3K27ac at the promoter site, most likely via chromatin looping (34, 39). There are few caveats to the present study. One caveat is that it was only performed in male animals, and since sex-specific effects of ethanol have been shown in the literature (49), a future study in female animals needs to be conducted. Another caveat is that we chose to mechanistically interrogate only the CeA, whereas previous studies have shown similar changes in Arc expression in the medial nucleus of amygdala (MeA) after AIE in adulthood (28). While the results of current study imply that epigenomic editing of Arc SARE in the CeA is sufficient to modulate anxiety-like and alcohol drinking behaviors, we cannot rule out the involvement of other brain regions that are implicated in these behaviors and addiction (2, 50).
Previous reports have indicated that the use of dCas9-induced epigenetic modulation at promoter regions can modulate behavior (51–53) as well as CRISPR-mediated chromosomal looping (54). Here, we demonstrate that this technology can also be used to investigate the role of noncoding RNAs, such as eRNAs, in regulating the interplay between three-dimensional chromatin structure, genetic regulatory elements, transcription factor binding, and RNA to influence adult anxiety and drinking after adolescent alcohol exposure. The current study focused on reversing epigenetic and behavioral changes in adulthood that were induced by adolescent alcohol exposure. However, we have previously observed that histone acetylation and behavioral changes also occur immediately after the AIE paradigm in adolescence and persist into adulthood (28). While the current study only focused on the long-term changes that persist until adulthood to better model what is seen in humans (4–7), the findings support the idea that activating the Arc SARE with dCas9-P300 could be applied as an intervention in adolescence. Our previous study showed that KDM6B siRNA infusion into the CeA of control adult rats produced anxiety-like behaviors, decreases in KDM6B, and increases in H3K27me3 occupancy at the Arc SARE site and suppressed Arc eRNA and mRNA expression in the amygdala (11). In addition, inhibition of Arc eRNA (−) levels in the CeA increased NELF binding, decreased Arc expression, and provoked anxiety-like behaviors in control adult rats (11). Here, we demonstrate that dCas9-KRAB infusion into CeA produces an increase in repressive H3K27me3 at the Arc SARE, decreases eRNA levels, increases NELF binding at the promoter, down-regulates Arc expression, and increases anxiety-like behavior and alcohol intake in control adult rats. Together, this suggests that epigenetic regulation at the Arc SARE site in the CeA is involved in modulating anxiety and alcohol intake. Thus, both developmental alcohol exposure–induced suppression of Arc expression and direct suppression of Arc expression in adulthood in the CeA appear to be crucial in anxiety and AUD.
Our findings regarding epigenomic editing allow for the analysis of gene- and amygdala nuclei–specific epigenetic changes that occur after adolescent alcohol exposure and persist until adulthood, driving complex behavior. The Cas9 system is easier to use and more modular than previous iterations [e.g., transcription activator–like effector nucleases (TALENs)] designed to alter precise epigenetic marks (55). In addition, the use of dCas9 to modulate gene expression, in lieu of a Cas9 enzyme with active endonuclease activity, could confer additional therapeutic and research benefit. Considerable effort has gone into reducing “off-target” double-stranded breaks that result in genome instability (56). The use of the dCas9 system avoids many of these off-target effects since DNA is not cut and not subject to error-prone nonhomologous end joining or spontaneous recombination events. The use of a dCas9 system is especially useful in the context of epigenetic regulation, and it has broad applications in the interrogation of long-lasting epigenetic changes that drive AUD and anxiety after adolescent alcohol exposure and the identification of tractable targets for the treatment of AUD and comorbid anxiety.
MATERIALS AND METHODS
Animals
Sprague-Dawley (SD) rats were acquired from Harlan Laboratories (Indianapolis, IN) and housed under a 12:12 light-dark cycle with ab libitum access to food and water. All animal experimental protocols strictly adhered to the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the University of Illinois at Chicago Institutional Animal Care and Use Committee. Number of animals was determined on the basis of our previous publications (11, 28, 30, 32). Assignment to experimental groups was random.
Adolescent intermittent alcohol exposure
SD dams with pups were acquired (arrival PND 17) from Harlan. Pups were weaned on (PND 21) and group housed (two to three pups per cage). Adolescent male rats (PND 28) were randomly assigned to receive either intermittent EtOH (2 g/kg) (20% w/v ethanol in 0.9% NaCl; AIE) or volume-matched saline (0.9% NaCl; AIS) on a 2-day on/off schedule via intraperitoneal injection until PND 41 (Fig. 1A) and then allowed to mature to adulthood (PND 92 or more) (11, 28, 32). Male adult SD rats (about 2 months old) were received from Harlan for the experiments conducted in relation to dCas9-KRAB manipulations (Fig. 3A).
In silico sgRNA design
A total of 235 bp of the rat Arc SARE region, identified previously (11, 16), were used as a template to determine sgRNA binding sites and protospacer adjacent motif (PAM) sequences at crispr.mit.edu following the algorithm described previously (55). This was further validated using the Integrated DNA Technologies (IDT; Coralville, IA, USA) sgRNA design tool. Four sgRNAs (table S1) were designed and evaluated for off-targets using IDT off-target prediction software (www.idtdna.com). Off-target evaluation was only done for locations with ≤4 bp mismatches between the genome and sgRNA sequence. Potential off-target sites were then further narrowed to genomic regions that had annotated gene encoding regions. The resulting top potential off-target sites (at least two off-targets for each sgRNA) with the lowest off-target editing score (corresponding to most likely to have off-target editing) from IDT’s prediction software were then chosen for further evaluation (one off-target gene; parp9 did not amplify in amygdala samples). Sites with greater than five mismatches are not evaluated by the software, as previous experiments that were used to design prediction algorithms have shown that the likelihood of off-target effects at these locations is extremely low (55). In addition, we have included three control genes (Cbp, p300, and Creb1) that have been shown to be decreased in the amygdala after AIE in adulthood (32).
Plasmids
Plasmids were obtained from Addgene: pLV-dCas9-KRAB-PGK-HygR (#83890; http://n2t.net/addgene:83890; RRID: Addgene_83890), pLV-dCas9-p300-P2A-PuroR (#83889; http://n2t.net/addgene:83889; RRID: Addgene_83889), and pLV-U6-gRNA-UbC-DsRed-P2A-Bsr (#83919; http://n2t.net/addgene:83919; RRID: Addgene_83919), and were gifts from C. Gersbach and have been described previously (25, 26). All plasmids were sequence verified. To construct plasmids containing sgRNAs from in silico analysis, oligonucleotides were ordered from IDT and cloned into pLV-U6-gRNA-UbC-DsRed-P2A-Bsr at BsmBI sites following methods described previously (26). Correct insertion of sgRNAs into vectors was determined using Sanger sequencing.
Lentivirus production
Lentivirus production was performed essentially as previously described (57). In brief, 293FT cells (cat. no. R70007, Invitrogen) were cultured at 37°C with 5% CO2 in Dulbecco’s modified Eagle’s medium with high glucose (cat. no. 1196511), 10% fetal bovine serum (FBS; Atlanta Biologicals), and geneticin (500 mg/ml; Invitrogen). Growth medium was removed and replaced with Opti-MEM (Invitrogen), and then packaging plasmids pxPAX2 (Addgene #12260) and pMD2.G (Addgene #12259) were cotransfected with either dCas9-P300 or sgRNA lentiviral plasmids into cells using Lipofectamine 2000 (Invitrogen). Media were removed and filtered 48 hours after transfection, the virus was concentrated using ultracentrifugation, and the viral pellet was resuspended in 20 μl of sterile phosphate-buffered saline (PBS). Lentiviral titer was determined using a p24 gag antigen enzyme-linked immunosorbent assay (ZeptoMetrix) following manufacturer’s instructions and was confirmed by transducing PC12 cells and examining red fluorescence. Titers (in picograms/milliliter gag antigen) were as follows: dCas9-P300, 3.5 × 106; dCas9-KRAB, 1.3 × 107; Arc SARE sgRNA 1, 4.6 × 107; Arc SARE sgRNA 2, 6.1 × 107; Arc SARE sgRNA 3, 4.8 × 107; and Arc SARE sgRNA 4, 4.7 × 107.
In vitro dCas9 testing
PC12 cells [cat. no. CRL-1721.1; the American Type Culture Collection (ATCC)] were grown at 37°C with 5% CO2 in F-12K medium (cat. no. 30-2004; ATCC) with 2.5% FBS and 15% horse serum. The dCas9 viral particles were mixed with four sgRNAs (1:1 ratio) and then added to the media and allowed to transduce for 72 hours. Viral transduction was confirmed using immunofluorescence microscopy on an EVOS FL microscope (Thermo Fisher Scientific). Cells were then either washed twice with PBS and processed for either mRNA analysis by the addition of TRIzol or scraped in PBS and fixed with 1% formaldehyde for 10 min at room temperature (RT) for chromatin immunoprecipitation (ChIP) analysis of target (Arc) and off-target genes using specific primers (table S1).
Stereotaxic surgery and lentiviral infusions in vivo
Adult rats were anesthetized with isoflurane (3%), bilaterally cannulated into the CeA (from bregma, posterior −2.5 mm, medial-lateral ±4.2 mm, and ventral −5.1 mm), and then allowed a 1-week recovery as we reported earlier (11, 58). Rats were singly housed after surgery. Following recovery, rats were bilaterally infused with a 1:1 mixture of viral particles (dCas9 to all four sgRNAs; total of 1.75 × 106 particles) using probes that extended 3 mm beyond the guide cannulas into the CeA. The virus was allowed to transduce for 2 weeks (Figs. 1A and 3A). Rats were then used for behavioral measurements as described below. Behavioral measurements were performed blindly. Rats were immediately anesthetized with isoflurane after behavioral testing and were decapitated to remove brains. Amygdala brain regions [predominately CeA and some surrounding MeA and basolateral amygdala (BLA) regions] were dissected out, quickly frozen, and stored at −80°C for biochemical assays (described below). The investigators performing the biochemical assays were not blinded to the group assignment.
EPM exploration test
EPM test was performed as previously described (11, 28). Before testing, rats were moved from the home colony room into a dimly lit holding room for 1 hour and then moved to the experimental room for 10 min to acclimate to the experimental environment. Rats were then placed in an EPM apparatus and allowed to explore for 5 min. Number of entries and time in the open and closed arms were recorded. Percentages of time spent on open arms were calculated from the total time spent in the open and closed arms of EPM.
LDB exploration test
LDB exploration test was performed as previously described (11, 28). The LDB (San Diego Instruments) was located in a dimly lit room with the light chamber illuminated by a 0.25 light-emitting diode. Before testing, rats were taken from the home colony room and placed in a different dimly lit room for 1 hour and then moved to the experimental room for 10 min to acclimate to the experimental environment. Rats were then placed in the LDB apparatus and allowed to freely explore for 5 min. Number of infrared beam breaks was recorded as ambulations in each compartment. Percentages of time in light box were calculated from the total time spent in light and dark boxes.
Ethanol two bottle choice drinking paradigm
Ethanol two bottle choice (2BC) drinking paradigm was performed as described previously (28, 45, 58). Rats were habituated to drink water from two bottles for 1 week. Rats then received a choice among 3% ethanol (w/v; in tap H2O) for 3 days, 7% ethanol for 3 days, and then 9% ethanol for 8 days or tap water (all 14 days). Location of ethanol and water bottles was switched daily. Consumption in volume was measured, and bottles were replaced daily. Ethanol consumption was calculated as grams/kilogram per day. Rats were weighed weekly, and an average of 3 measurements of weights was used in grams/kilogram calculations. Ethanol preference was calculated as the ratio of ethanol volume to total volume of liquid consumed.
Sucrose 2BC drinking paradigm
Two weeks after ethanol 2BC, rats received two bottles containing 2% sucrose (w/v in tap water) or tap water for 3 days and then 4% sucrose (w/v) for 4 to 5 days (45). Consumption in volume was measured, and bottles were replaced daily. Location of sucrose and water bottles was switched daily. Sucrose consumption was represented as grams/kilogram per day. Rats were weighed weekly, and an average of 2 measurements of weights was used in grams/kilogram calculations. Sucrose preference was calculated as the ratio of sucrose volume to total volume of liquid consumed.
ChIP assay
ChIPs were performed as described previously (11, 12, 59). Tissue or cells were homogenized in PBS and then cross-linked with 1% formaldehyde for 10 min at RT, quenched with 1 M glycine in 750 mM tris-HCl (pH 8.0), then centrifuged at 1600g for 10 min at 4°C, and then washed once with ice-cold PBS. Cells were lysed in lysis buffer [1% (v/v) SDS, 10 mM EDTA, and 50 mM tris-HCl (pH 8.0)]. Lysate was then sonicated using the Covaris ME220 (Covaris) to achieve sheared DNA fragments of 200 to 500 bp, which were then clarified using centrifugation (17,000g for 10 min at 4°C) to obtain a chromatin fraction. Chromatin was diluted 1:6 in ChIP washing buffer [0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM tris-HCl (pH 8.0), and 167 mM NaCl], and an aliquot was added to 2.5 volumes of 100% ethanol as an input fraction. Antibodies (table S2) were added, and chromatin and antibodies were rotated overnight at 4°C. Thirty microliters of Dynabeads A were added and rotated for 1 hour at 4°C. Samples were washed five times with ChIP washing buffer then purified using 10% w/v Chelex 100 (Bio-Rad) in sterile H2O by boiling for 95°C for 10 min followed by centrifugation. Input samples were purified by centrifugation followed by washing once with 75% ethanol and then boiled for 10 min at 95°C in 10% w/v Chelex. Purified DNA was analyzed in duplicates or triplicates using quantitative polymerase chain reaction (qPCR). The data were analyzed using the ∆∆Ct method (60), normalizing to input, and the data are expressed as fold change in protein occupancy. Primers for specific genomic locations are listed in table S1.
Quantitative real-time PCR
RNA was extracted by homogenizing tissue or cells in TRIzol and then purified using Micro Direct-zol Purification kit following the manufacturer’s instructions (Zymo Research Corporation). RNA was reverse transcribed to cDNA using MultiScribe Reverse Transcriptase (Thermo Fisher Scientific) following the manufacturer’s instructions. qPCRs were run either in duplicates or in triplicates on a CFX Connect qPCR system using PowerUp SYBR (Thermo Fisher Scientific). Gene expression was determined using the ∆∆Ct method (60) and normalized to mean of Ct values of Hprt1 housekeeping genes. Data are presented as average fold change relative to controls. Primers for specific genes are listed in table S1.
3C assay
3C qPCR was performed as previously described with some modifications (34, 36). Chromatin prepared from amygdala was cross-linked with 1% formaldehyde for 10 min at RT, and then Dpn II was used to digest DNA overnight at 37°C. Chromatin was then diluted to 2.5 ng/μl, and T4 ligase was added and incubated at 16°C for 4 hours followed by reverse transcription incubation for 30 min. Crosslinks were reversed by the addition of proteinase K and incubation at 65°C overnight. DNA was purified using phenol chloroform extraction. Primers were designed between an anchor fragment located near the Arc promoter and other fragments both upstream and downstream of the gene (Fig. 2E and table S1). Standard quantitative real-time PCR method (qPCR) procedures (40 cycles using PowerUp SYBR) were used (see quantitative real-time PCR method) to calculate Cts, and interaction strength was calculated by controlling for random interactions (using Arc bacterial artificial chromosome clone, CH230-456-015; BACPAC Genomics Inc.), normalizing for loading using internal primers to Hprt1, normalizing to a control interaction with Arc SARE, and adjusting for primer efficiency using standard curve data. The resulting interaction is referred to as interaction strength, as used by other investigators (34).
Immunofluorescence staining
Animals were anesthetized and perfused with 4% paraformaldehyde, and then brains were processed for immunofluorescence staining (58). Brains were sliced to 20-μm sections using a cryostat and washed two times with 0.01 M PBS. Brain sections were blocked with 10% normal goat serum and then with 1% bovine serum albumin. After this, they were incubated overnight at 4°C with primary antibodies of NeuN and CRISPR-Cas9 (conjugated with Alexa Fluor 488). For NeuN staining, sections were incubated with Alexa Fluor 647–conjugated secondary antibody for 2 hours. Last, sections were washed several times with 0.01 M PBS and then mounted on slides and coverslipped using Fluoromount-G with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen 0-4959). sgRNAs were cloned into vector tagged with mCherry fluorescent protein (DsRed) and visualized in red color. All images were taken using a confocal microscope (Zeiss LSM 710). Information on antibodies is provided in table S2.
Statistics
Statistical analyses were performed using SigmaStat, and data were visualized using GraphPad Prism 8. Comparisons between two groups were performed with two-tailed Student’s t tests. Two-way analysis of variance (ANOVA) (2 × 2 factorial design) was used for comparison between four groups followed by Tukey’s post hoc test. If comparisons did not meet the assumptions for parametric tests, then Mann-Whitney rank sum tests (two sided) were performed. For 2BC experiments, repeated-measures two-way ANOVA mixed model followed by Tukey’s post hoc test was used to determine significance.
Acknowledgments
Some figures (Figs. 2D, 4E, and 5) were prepared using BioRender.com.
Funding: This work was supported by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) grants, U01AA019971, U24AA024605 [Neurobiology of Adolescent Drinking in Adulthood (NADIA) project], P50AA022538 (Center for Alcohol Research in Epigenetics), and by the Department of Veterans Affairs Senior Research Career Scientist Award to S.C.P. and F32AA027410 to J.P.B. The content is solely the responsibility of the authors and does not represent the official views of the NIH or U.S. Department of Veterans Affairs.
Author contributions: S.C.P. conceived the idea, received funding, and supervised the study. S.C.P. discussed the experimental plan with J.P.B. and designed the experiments. H.Z. and J.P.B. generated animals and performed the viral injections and behavioral experiments. J.P.B., H.Z., G.M.W., D.H., and E.J.K. performed the biochemical and histochemical experiments. J.P.B., D.H., and A.W.L. generated viral vectors, designed gRNAs, and assembled CRISPR tools. All data were analyzed by J.P.B., and figures were prepared by J.P.B. and H.Z. and finalized by S.C.P. and G.M.W. S.C.P. and J.P.B. interpreted the data and wrote the manuscript that is edited and approved by all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. As described in Materials and Methods, plasmids related to dCas9-P300, dCas9-KRAB, and gRNA were obtained from Addgene with a material transfer agreement and were from C. Gersbach.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 to S6
REFERENCES AND NOTES
- 1.Witkiewitz K., Litten R. Z., Leggio L., Advances in the science and treatment of alcohol use disorder. Sci. Adv. 5, eaax4043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koob G. F., Drug addiction: Hyperkatifeia/negative reinforcement as a framework for medications development. Pharmacol. Rev. 73, 163–201 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sacks J. J., Gonzales K. R., Bouchery E. E., Tomedi L. E., Brewer R. D., 2010 national and state costs of excessive alcohol consumption. Am. J. Prev. Med. 49, e73–e79 (2015). [DOI] [PubMed] [Google Scholar]
- 4.DeWit D. J., Adlaf E. M., Offord D. R., Ogborne A. C., Age at first alcohol use: A risk factor for the development of alcohol disorders. Am. J. Psychiatry 157, 745–750 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Grant B. F., Stinson F. S., Harford T. C., Age at onset of alcohol use and DSM-IV alcohol abuse and dependence: A 12-year follow-up. J. Subst. Abuse 13, 493–504 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Hingson R. W., Heeren T., Winter M. R., Age at drinking onset and alcohol dependence, age at onset, duration, and severity. Arch. Pediatr. Adolesc. Med. 160, 739–746 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Mustonen A., Alakokkare A. E., Salom C., Hurtig T., Levola J., Scott J. G., Miettunen J., Niemelä S., Age of first alcohol intoxication and psychiatric disorders in young adulthood–A prospective birth cohort study. Addict. Behav. 118, 106910 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Pacek L. R., Storr C. L., Mojtabai R., Green K. M., La Flair L. N., Alvanzo A. A. H., Cullen B. A., Crum R. M., Comorbid alcohol dependence and anxiety disorders: A national survey. J. Dual Diagn. 9, 271–280 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kyzar E. J., Floreani C., Teppen T. L., Pandey S. C., Adolescent alcohol exposure: Burden of epigenetic reprogramming, synaptic remodeling, and adult psychopathology. Front. Neurosci. 10, 222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spear L. P., Effects of adolescent alcohol consumption on the brain and behaviour. Nat. Rev. Neurosci. 19, 197–214 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Kyzar E. J., Zhang H., Pandey S. C., Adolescent alcohol exposure epigenetically suppresses amygdala Arc enhancer RNA expression to confer adult anxiety susceptibility. Biol. Psychiatry 85, 904–914 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bohnsack J. P., Teppen T., Kyzar E. J., Dzitoyeva S., Pandey S. C., The lncRNA BDNF-AS is an epigenetic regulator in the human amygdala in early onset alcohol use disorders. Transl. Psychiatry 9, 34 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilpin N. W., Herman M. A., Roberto M., The central amygdala as an integrative hub for anxiety and alcohol use disorders. Biol. Psychiatry 77, 859–869 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wall M. J., Collins D. R., Chery S. L., Allen Z. D., Pastuzyn E. D., George A. J., Nikolova V. D., Moy S. S., Philpot B. D., Shepherd J. D., Müller J., Ehlers M. D., Mabb A. M., Corrêa S. A. L., The temporal dynamics of Arc expression regulate cognitive flexibility. Neuron 98, 1124–1132.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim T. K., Hemberg M., Gray J. M., Costa A. M., Bear D. M., Wu J., Harmin D. A., Laptewicz M., Barbara-Haley K., Kuersten S., Markenscoff-Papadimitriou E., Kuhl D., Bito H., Worley P. F., Kreiman G., Greenberg M. E., Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawashima T., Okuno H., Nonaka M., Adachi-Morishima A., Kyo N., Okamura M., Takemoto-Kimura S., Worley P. F., Bito H., Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proc. Natl. Acad. Sci. U.S.A. 106, 316–321 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez-Albert J., Lipinski M., Lopez-Cascales M. T., Rowley M. J., Martin-Gonzalez A. M., del Blanco B., Corces V. G., Barco A., Immediate and deferred epigenomic signatures of in vivo neuronal activation in mouse hippocampus. Nat. Neurosci. 22, 1718–1730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korb E., Finkbeiner S., Arc in synaptic plasticity: From gene to behavior. Trends Neurosci. 34, 591–598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohnsack J. P., Pandey S. C., Histone modifications, DNA methylation, and the epigenetic code of alcohol use disorder. Int. Rev. Neurobiol. 156, 1–62 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peña C. J., Bagot R. C., Labonté B., Nestler E. J., Epigenetic signaling in psychiatric disorders. J. Mol. Biol. 426, 3389–3412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sweatt J. D., The emerging field of neuroepigenetics. Neuron 80, 624–632 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yim Y. Y., Teague C. D., Nestler E. J., In vivo locus-specific editing of the neuroepigenome. Nat. Rev. Neurosci. 21, 471–484 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Creyghton M. P., Cheng A. W., Welstead G. G., Kooistra T., Carey B. W., Steine E. J., Hanna J., Lodato M. A., Frampton G. M., Sharp P. A., Boyer L. A., Young R. A., Jaenisch R., Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A., Wysocka J., A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klann T. S., Black J. B., Chellappan M., Safi A., Song L., Hilton I. B., Crawford G. E., Reddy T. E., Gersbach C. A., CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol. 35, 561–568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilton I. B., D’Ippolito A. M., Vockley C. M., Thakore P. I., Crawford G. E., Reddy T. E., Gersbach C. A., Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33, 510–517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura M., Gao Y., Dominguez A. A., Qi L. S., CRISPR technologies for precise epigenome editing. Nat. Cell Biol. 23, 11–22 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Pandey S. C., Sakharkar A. J., Tang L., Zhang H., Potential role of adolescent alcohol exposure-induced amygdaloid histone modifications in anxiety and alcohol intake during adulthood. Neurobiol. Dis. 82, 607–619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spear L. P., Adolescent alcohol exposure: Are there separable vulnerable periods within adolescence? Physiol. Behav. 148, 122–130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyzar E. J., Bohnsack J. P., Zhang H., Pandey S. C., MicroRNA-137 drives epigenetic reprogramming in the adult amygdala and behavioral changes after adolescent alcohol exposure. eNeuro 6, ENEURO.0401-19.2019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Q., Yu L. R., Wang L., Zhang Z., Kasper L. H., Lee J. E., Wang C., Brindle P. K., Dent S. Y. R., Ge K., Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H., Kyzar E. J., Bohnsack J. P., Kokare D. M., Teppen T., Pandey S. C., Adolescent alcohol exposure epigenetically regulates CREB signaling in the adult amygdala. Sci. Rep. 8, 10376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth S. Y., Denu J. M., Allis C. D., Histone acetyltransferases. Annu. Rev. Biochem. 70, 81–120 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Schaukowitch K., Joo J. Y., Liu X., Watts J. K., Martinez C., Kim T. K., Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 56, 29–42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Y., Sun L., Chen Z., Whitaker J. W., Wang T., Wang W., Predicting enhancer transcription and activity from chromatin modifications. Nucleic Acids Res. 41, 10032–10043 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagège H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forné T., Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722–1733 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Alerasool N., Segal D., Lee H., Taipale M., An efficient KRAB domain for CRISPRi applications in human cells. Nat. Methods 17, 1093–1096 (2020). [DOI] [PubMed] [Google Scholar]
- 38.O’Geen H., Bates S. L., Carter S. S., Nisson K. A., Halmai J., Fink K. D., Rhie S. K., Farnham P. J., Segal D. J., Ezh2-dCas9 and KRAB-dCas9 enable engineering of epigenetic memory in a context-dependent manner. Epigenetics Chromatin 12, 26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oudelaar A. M., Higgs D. R., The relationship between genome structure and function. Nat. Rev. Genet. 22, 154–168 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Fitz J., Neumann T., Steininger M., Wiedemann E.-M., Garcia A. C., Athanasiadis A., Schoeberl U. E., Pavri R., Spt5-mediated enhancer transcription directly couples enhancer activation with physical promoter interaction. Nat. Genet. 52, 505–515 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Groner A. C., Meylan S., Ciuffi A., Zangger N., Ambrosini G., Dénervaud N., Bucher P., Trono D., KRAB–Zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLOS Genet. 6, e1000869 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thakore P. I., D’Ippolito A. M., Song L., Safi A., Shivakumar N. K., Kabadi A. M., Reddy T. E., Crawford G. E., Gersbach C. A., Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crews F. T., Robinson D. L., Chandler L. J., Ehlers C. L., Mulholland P. J., Pandey S. C., Rodd Z. A., Spear L. P., Swartzwelder H. S., Vetreno R. P., Mechanisms of persistent neurobiological changes following adolescent alcohol exposure: NADIA consortium findings. Alcohol. Clin. Exp. Res. 43, 1806–1822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakharkar A. J., Kyzar E. J., Gavin D. P., Zhang H., Chen Y., Krishnan H. R., Grayson D. R., Pandey S. C., Altered amygdala DNA methylation mechanisms after adolescent alcohol exposure contribute to adult anxiety and alcohol drinking. Neuropharmacology 157, 107679 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moonat S., Sakharkar A. J., Zhang H., Tang L., Pandey S. C., Aberrant histone deacetylase2-mediated histone modifications and synaptic plasticity in the amygdala predisposes to anxiety and alcoholism. Biol. Psychiatry 73, 763–773 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barbier E., Tapocik J. D., Juergens N., Pitcairn C., Borich A., Schank J. R., Sun H., Schuebel K., Zhou Z., Yuan Q., Vendruscolo L. F., Goldman D., Heilig M., DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. J. Neurosci. 35, 6153–6164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warnault V., Darcq E., Levine A., Barak S., Ron D., Chromatin remodeling—A novel strategy to control excessive alcohol drinking. Transl. Psychiatry 3, e231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon-O’Brien E., Alaux-Cantin S., Warnault V., Buttolo R., Naassila M., Vilpoux C., The histone deacetylase inhibitor sodium butyrate decreases excessive ethanol intake in dependent animals. Addict. Biol. 20, 676–689 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Becker J. B., Koob G. F., Sex differences in animal models: Focus on addiction. Pharmacol. Rev. 68, 242–263 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koob G. F., Volkow N. D., Neurobiology of addiction: A neurocircuitry analysis. Lancet Psychiatry 3, 760–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lorsch Z. S., Hamilton P. J., Ramakrishnan A., Parise E. M., Salery M., Wright W. J., Lepack A. E., Mews P., Issler O., McKenzie A., Zhou X., Parise L. F., Pirpinias S. T., Torres I. O., Kronman H. G., Montgomery S. E., Eddie Loh Y. H., Labonté B., Conkey A., Symonds A. E., Neve R. L., Turecki G., Maze I., Dong Y., Zhang B., Shen L., Bagot R. C., Nestler E. J., Stress resilience is promoted by a Zfp189-driven transcriptional network in prefrontal cortex. Nat. Neurosci. 22, 1413–1423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matharu N., Rattanasopha S., Tamura S., Maliskova L., Wang Y., Bernard A., Hardin A., Eckalbar W. L., Vaisse C., Ahituv N., CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science 363, eaau0629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heller E. A., Cates H. M., Peña C. J., Sun H., Shao N., Feng J., Golden S. A., Herman J. P., Walsh J. J., Mazei-Robison M., Ferguson D., Knight S., Gerber M. A., Nievera C., Han M. H., Russo S. J., Tamminga C. S., Neve R. L., Shen L., Zhang H. S., Zhang F., Nestler E. J., Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat. Neurosci. 17, 1720–1727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morgan S. L., Mariano N. C., Bermudez A., Arruda N. L., Wu F., Luo Y., Shankar G., Jia L., Chen H., Hu J. F., Hoffman A. R., Huang C. C., Pitteri S. J., Wang K. C., Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat. Commun. 8, 15993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., Agarwala V., Li Y., Fine E. J., Wu X., Shalem O., Cradick T. J., Marraffini L. A., Bao G., Zhang F., DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leibowitz M. L., Papathanasiou S., Doerfler P. A., Blaine L. J., Sun L., Yao Y., Zhang C.-Z., Weiss M. J., Pellman D., Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nat. Genet. 53, 895–905 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lasek A. W., Janak P. H., He L., Whistler J. L., Heberlein U., Downregulation of mu opioid receptor by RNA interference in the ventral tegmental area reduces ethanol consumption in mice. Genes Brain Behav. 6, 728–735 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Zhang H., Sakharkar A. J., Shi G., Ugale R., Prakash A., Pandey S. C., Neuropeptide Y signaling in the central nucleus of amygdala regulates alcohol-drinking and anxiety-like behaviors of alcohol-preferring rats. Alcohol. Clin. Exp. Res. 34, 451–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nelson J. D., Denisenko O., Bomsztyk K., Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1, 179–185 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S6