Abstract
Broadly HIV-1–neutralizing VRC01-class antibodies bind the CD4-binding site of Env and contain VH1-2*02–derived heavy chains paired with light chains expressing five–amino acid–long CDRL3s. Their unmutated germline forms do not recognize HIV-1 Env, and their lack of elicitation in human clinical trials could be due to the absence of activation of the corresponding naïve B cells by the vaccine immunogens. To address this point, we examined Env-specific B cell receptor sequences from participants in the HVTN 100 clinical trial. Of all the sequences analyzed, only one displayed homology to VRC01-class antibodies, but the corresponding antibody (FH1) recognized the C1C2 gp120 domain. For FH1 to switch epitope recognition to the CD4-binding site, alterations in the CDRH3 and CDRL3 were necessary. Only germ line–targeting Env immunogens efficiently activated VRC01 B cells, even in the presence of FH1 B cells. Our findings support the use of these immunogens to activate VRC01 B cells in humans.
Abs with VRC01-class features can bind C1C2.
INTRODUCTION
HIV-1 broadly neutralizing antibodies (bnAbs) target diverse areas of the viral Env spike, including the CD4-binding site (CD4-BS), the apex region, the N332 glycan patch, the interface between the gp120 and gp41 subunits, and the gp41 subunit itself (1–3). bnAbs that bind the same Env region and share common genetic and/or structural features are grouped into “classes” (1). The anti–CD4-BS VRC01-class antibodies are among the most potent bnAbs known (4); they prevent SHIV infection of rhesus macaques and HIV-1 infection of humanized mice (5, 6), and monoclonal Ab (mAb) VRC01 was recently shown to protect from infection by susceptible circulating primary viruses in large clinical trials (7). They are therefore logical targets for HIV-1 vaccine development.
VRC01-class antibodies have been isolated from several HIV-1–infected subjects, but their heavy chain (HC) variable domains are derived exclusively from the VH1-2*02 gene allele and are paired with light chains (LCs) expressing five–amino acid–long CDRL3s (8–15). Their variable region of immunoglobulin heavy chain (VH) and light chain (VL) genes are extensively mutated (9, 12, 14, 16). However, despite extensive amino acid sequence diversity among them, VRC01-class antibodies adopt similar overall structures and engage the CD4-BS with similar angles of approach (9, 13).
In contrast to their mature mutated forms, recombinant (rec) antibodies generated from their inferred unmutated antibody sequences (“germ line”, gl) do not bind recEnv or neutralize HIV-1 (17–20). These observations lead to the proposal that Envs used as immunogens in human clinical trials would not have activated naïve B cells expressing glVRC01-class B cell receptors (BCRs) (17, 19, 21). If true, this would, in part, explain the absence of VRC01-like neutralizing activities in sera collected from vaccinees. However, experimental evidence for this is presently lacking.
Participants of the HVTN 100 phase 1 and 2 clinical trial were immunized with ALVAC vectors (vCP2438), expressing clade C ZM96 gp120 with the gp41 transmembrane sequence of the subtype B LAI strain and the gag and protease of LAI, at months 0 and 2 and then coadministered with recombinant clade C Env gp120 proteins (TV1.C and 1086.c) formulated in squalene-based adjuvant MF59 at 4, 6, and 12 months (22, 23). Known glVRC01-class antibodies do not bind these Env proteins, and thus, they were not expected to have activated B cells expressing VRC01-class BCRs in vivo. Following an in-depth analysis of BCRs expressed by vaccine-specific gp120+ memory B cells isolated at the end of the immunization regimen from a subset of vaccinated participants, 339 BCRs were analyzed. Only one, FH1, displayed classic VRC01-like HC and LC characteristics, i.e., an HC derived from VH1-2*02 paired with a k3-20 LC expressing a five–amino acid–long CDRL3. The amino acid sequence homology between FH1 and glVRC01 (outside the CDRH3 and CDRL3) is 95% (95 of 100) in the HC and 98% (92 of 94) in the LC. The isolation of FH1 potentially suggests that Env-based immunogens that are not specifically designed to activate VRC01-class B cells may do so, albeit infrequently.
Here, we characterized the structural, antigenic, and functional properties of FH1 and determined that, in contrast to VRC01-class antibodies, it recognizes the C1C2 domain of gp120, where non-neutralizing antibodies with antibody-dependent cellular cytotoxicity (ADCC) activities also bind (24, 25). We also report that for FH1 to switch its epitope specificity toward the CD4-BS, both CDRH3 and CDRL3 domains had to be modified to those of VRC01. By engineering B cells to express either FH1 or glVRC01 BCRs, we were able to confirm that the latter can only be activated by specifically designed germ line–targeting immunogens but not by the Env used in the HVTN100 trial. Hence, our study provides evidence in support of the clinical evaluation of Env immunogens that have been specifically designed to target VRC01-expressing naïve B cells.
RESULTS
Isolation of an antibody with VRC01-like genetic characteristics
HVTN 100 was a phase 1/2 clinical trial of an RV144-like vaccine regimen, which had been modified with subtype C–specific immunogens for the South African epidemic (fig. S1) (22). Two weeks after the fifth immunization, vaccine-specific 1086.C gp120+ immunoglobulin G (IgG)+ and vaccine-nonspecific 1086.C gp120− IgG+ B cell populations were isolated and their VH/VL genes sequenced from 14 vaccine recipients.
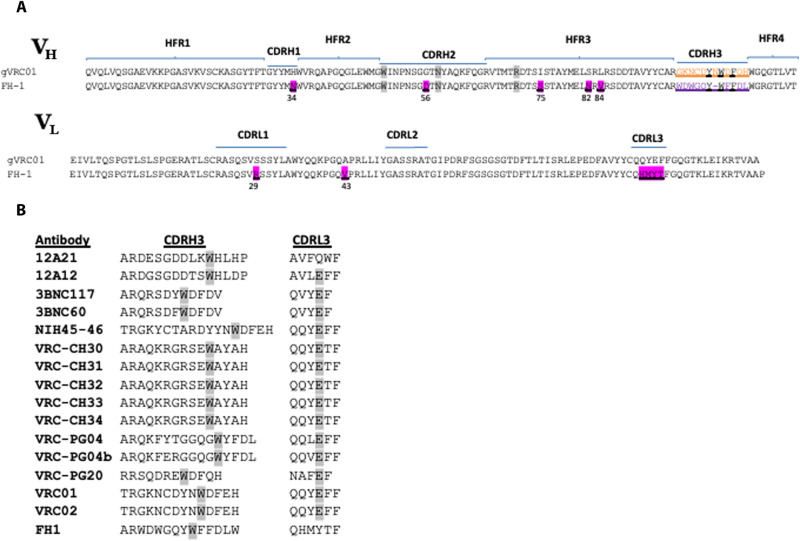
Env-specific B cells were single-cell–sorted from the vaccine recipients with the highest proportion of VH1-2*02 gene usage. Of the Env-specific B cells from which we recovered paired VH and VL genes, we identified that one VH1-2*02 HC was paired with a Vκ3-20 with a five–amino acid–long CDRL3 (“FH1”; Fig. 1A).
Fig. 1. Amino acid sequence of FH1 and VRC01-class antibodies.
(A) Alignment of the VH and VL amino acid sequences of FH1 and glVRC01. Amino acids that differ between the two antibody sequences are highlighted in ink and are underlined. The three gene-encoded amino acids in the VH1-2*02 domains (Trp50HC, Asn58HC, and Arg71HC, Kabat numbering) that play key roles in the interaction between VRC01-class antibodies and the CD4-BS of Env are highlighted in gray. (B) Sequence alignment of the CDRH3 and CDRL3 of FH1 and of known VRC01-class antibodies. Tryptophan residues located exactly five amino acids away from the C terminus of CDRH3 are highlighted in gray. Glutamic acid residues located at position 96 in the CDRL3 of most VRC01-class antibodies are highlighted in gray.
Both VH and VL antibody domains were minimally mutated with five amino acid changes in VH1-2*02 (H34N in CDRH1, G56D in CDRH2, and I75S, S82N, and L84V in FRH3) and only two changes in κ3-20 (S29R in CDRL1, and A43V). Overall, there was 92% amino acid homology between FH1 and glVRC01 in both their VH and VL domains. The CDRH3 of FH1 is 11 amino acids long, which is within the range of CDRH3s of VRC01-class antibodies (Fig. 1B) and, like most VRC01-class antibodies, expresses a Trp exactly five amino acids away from the C-terminal domain of CDRH3 (14). The CDRL3 sequence (QHMYT) of FH1 differs from the prevalent κ3-20 CDRL3 sequence motifs (QQY/LEF) present in known VRC01-class antibodies (Fig. 1B) (14, 26, 27). Thus, FH1 displays VH and VL sequence similarities to known VRC01-class antibodies.
FH1 binds to the C1C2 gp120 domain
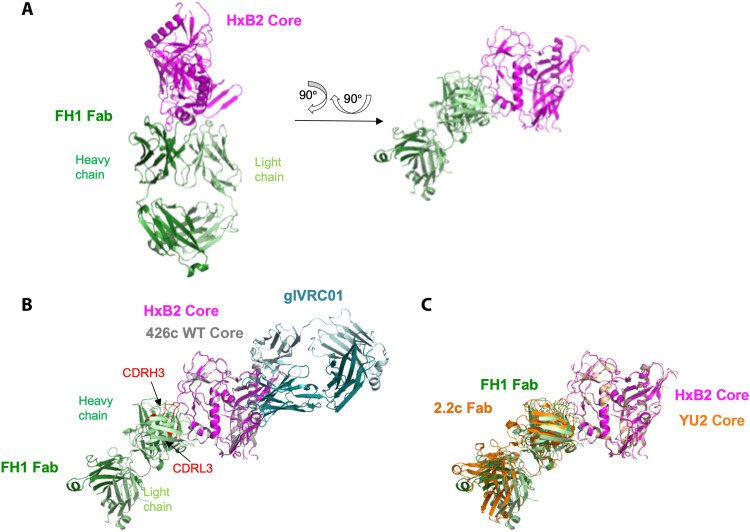
We generated and analyzed a 3.5-Å structure of FH1 Fab complexed with the core gp120 domain of the HxB2 Env expressed in GnTI−/− cells (Fig. 2A and table S1). The structure revealed that FH1 binds primarily to the C1 region of the gp120 Env subunit [95% of the total buried surface area (BSA)] with some contacts in the C2 region (5% of the total BSA) (table S2), both in the inner domain of gp120. FH1 binds Env with a total BSA of ~1067 Å2, 62% contributed from the HC [CDRH1 (8%), CDRH2 (22%), CDRH3 (28%), and the rest from FR regions] and 38% from the LC [CDRL1 (19%), CDRL3 (15%), and the rest from FR1] (table S2). In comparison, glVRC01 binds Env with a total BSA of ~950 Å2, 75% contributed from the HC [CDRH2 (52%), CDRH3 (11%), and the rest from FR regions] and 25% from the LC [CDRL1 (1%), CDRL3 (17%), and the rest from FR1].
Fig. 2. Crystal structure of FH1 Fab bound to HxB2 gp120 Core indicates mode of binding.
(A) Cartoon representation of the overall complex structure with HxB2 Core shown in magenta and FH1 Fab shown in green (HC in dark green and LC in light green). Two views are shown. (B) Superposition of structure of WT 426c Core (gray) bound to glVRC01 (marine) onto that of HxB2 Core bound to FH1. The FH1 CDRH3 and CDRL3 domains are highlighted in red. (C) Superposition of YU2 Core bound to 2.2c Fab structure (orange) onto that of HxB2 Core bound to FH1 indicates a similar epitope.
Three gene-encoded amino acids in the VH1-2*02 domains of VRC01-class antibodies (Trp50HC, Asn58HC, and Arg71HC; Kabat numbering) play key roles in the antibody interactions with the CD4-BS of Env. Trp50HC (in FWR2) interacts with Asn280gp120 in loop D, Asn58HC (in FWR3) interacts with Arg456gp120 in V5, and Arg71HC (in FWR3) interacts with Asp368 gp120 within the CD4-binding loop (26, 28). These three amino acids remain unaltered during the maturation of VRC01-class antibodies. In addition, Trp100BHC in CDRH3 interacts with Asn/Asp279 in loop D. In contrast, in the case of FH1, Trp50, Asn58, and Trp100A interact with the C1 and C2 domains, while Arg71 does not contact Env (table S2). Thus, while VRC01-class antibodies recognize the CD4-BS of Env, FH1 recognizes elements of the C1/C2 domains of gp120 (Fig. 2B). The overall angle of approach of FH1 to gp120, however, is similar to that of the weakly ADCC-inducing A32-like antibody, 2.2c (Fig. 2C) (25).
Env cross-recognition properties of FH1
FH1 recognizes both the 1086.C and TV1 Envs used in the HVTN 100 vaccine (Fig. 3A). So far, only a handful of Env-derived recombinant proteins that bind germline forms of VRC01-class antibodies (“germ line–targeting” proteins) have been designed: 426c Core, 426c OD, eOD-CT6/GT8, and BG505 GT1.1 and were tested as immunogens in knock-in mice (29–38). While 426c Core expresses both the inner and outer gp120 domains of the clade C 426c Env, eOD-GT8 and 426c OD express the outer gp120 domain of the clade B HxB2– and 426c-derived Envs, respectively.
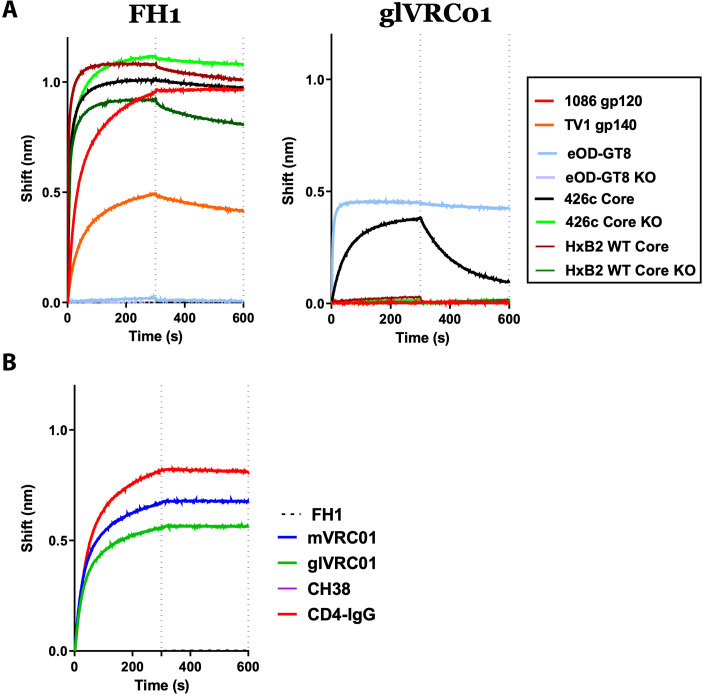
Fig. 3. Antibody-Env interactions.
(A) The interactions between FH1 or glVRC01 and the indicated recombinant Envs were determined by biolayer interferometry. KO indicates the presence of amino acid mutations that abrogate the binding of CD4-BS antibodies (D368R, E370A, and D279A in the case of 426c Core and HxB2 Core and D368R and D279A in the case of eOD-GT8). (B) The binding of FH1 to 426c Core monomer in the absence or presence of indicated antibodies and IgG-CD4 was monitored by biolayer interferometry.
FH1 did not bind eOD-GT8 (which lacks C1 and C2) but bound 426c Core (which expresses both C1 and C2) (Fig. 3A). As expected, FH1 bound equally well to a variant of 426c Core expressing the CD4-BS knockout (KO) mutations (D368R, E370A, and D279A) (426c Core CD4-BS KO) and recognized the HxB2 wild-type (WT) Core protein in a CD4-BS–independent manner [i.e., it bound the HxB2 WT Core even in the presence of the abovementioned CD4-BS KO mutations, HxB2 WT Core KO]. In contrast, and as expected, glVRC01 mAb bound eOD-GT8 and 426c Core in a CD4-BS–dependent manner, as previously discussed (31, 32, 39) but not the HxB2 WT Core, 1086.C, or TV1 Envs.
Antibody competition experiments performed between FH1 and mVRC01 (mature VRC01), glVRC01, CD4-IgG, or CH38 (an anti-C1 MAb) (Fig. 3B) indicated that FH1 does not compete with the anti–CD4-BS antibodies or IgG-CD4 but competes with CH38 (and with itself). Collectively, the above results confirm that FH1 recognizes a known target of vaccine-elicited antibodies with ADCC activities (24, 40, 41).
The affinity of FH1 to 426c Core is about two orders of magnitude greater than that of glVRC01 (fig. S2). As expected, the affinity of FH1 for 1086.C gp120, contained in the vaccine boost, is approximately 12-fold greater than for the 426 Core. In summary, although the VH and VL regions of FH1 and VRC01-class antibodies share very similar amino acid sequences, the two antibodies recognize different Env domains.
Both CDRH3 and CDRL3 dictate epitope specificity of FH1
As discussed above, the gene-encoded portions of the VH and VL domains of glVRC01-class antibodies and those of FH1 differ in only seven amino acid positions (five in VH and two in VL). The major sequence differences are in CDRH3 and CDRL3. We therefore examined whether the differences in epitope-recognition of FH1 and glVRC01 are due to sequence variations in either CDRH3, CDRL3, or both (Fig. 4). For this analysis, we used eOD-GT8 as the target Env as it is recognized by glVRC01 but not FH1 (Fig. 3A).
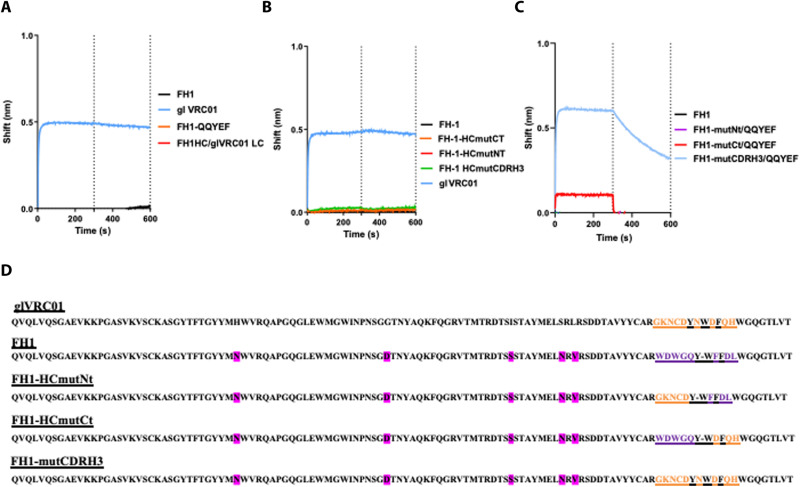
Fig. 4. Role of CDRH3 and CDRL3 is defining the epitope specificity of FH1.
The interactions between FH1 and the various FH1/VRC01 chimeric antibodies with eOD-GT8 (monomer) were determined by biolayer interferometry. (A) Effect of substitutions in CDRL3, (B) effect of substitutions in CDRH3, and (C) effect of substitutions in both the CDRL3 and CDRH3. (D) The amino acid sequences in HC of all antibodies examined are presented. Amino acids that differ between the HC sequences of FH1 and glVRC01 are highlighted in pink.
We first replaced the CDRL3 amino acid sequence of FH1 (QHMYT) with that of VRC01 (QQYEF) (Fig. 4A). That antibody, FH1-QQYEF, like FH1, did not bind eOD-GT8. We then paired the FH1 HC with the entire κ3-20 LC of glVRC01, but that antibody (FH1 HC/glVRC01 LC) did not bind eOD-GT8 either (Fig. 4A).
Next, we examined whether amino acid differences in CDRH3 were responsible for the differential epitope specificities of FH1 and VRC01. To this end, we replaced the C-terminal half of the CDRH3 of FH1 with that of VRC01 (FH1-HCmutCT), the N-terminal half of the CDRH3 of FH1 with that of VRC01 (FH1-HCmutNT), or the entire CDRH3 of FH1 with that of VRC01 (FH1-HCmutCDRH3) and examined the effect of these alterations on antibody binding to eOD-GT8. None of these antibodies bound eOD-GT8 (Fig. 4B). Collectively, these results indicate that the amino acid differences in the individual CDRH3 and CDRL3 domains between FH1 and VRC01 are not, by themselves, responsible for the different epitope specificities of these antibodies.
We thus examined whether the different epitope specificities were due to differences in both the CDRH3 and CDRL3 domains. To this end, we engineered chimeric antibodies expressing the HCs of the abovementioned FH1-HCmutNT, FH1-HCmutCT, and FH1-HCmutCDRH3 antibodies with the LC of FH1-QQYEF and examined their binding to eOD-GT8 (Fig. 4C). No binding was observed with the FH1-HCmutNt/QQYEF antibody, weak binding was observed with the FH1-HCmutCt/QQYEF antibody, but robust binding was observed with the FH1-HCmutCDRH3/QQYEF antibody. We conclude, therefore, that for FH1 to change its epitope specificity from the C1/C2 domain to the CD4-BS of gp120, extensive amino acid changes in both the CDRH3 and CDRL3 domains would be required.
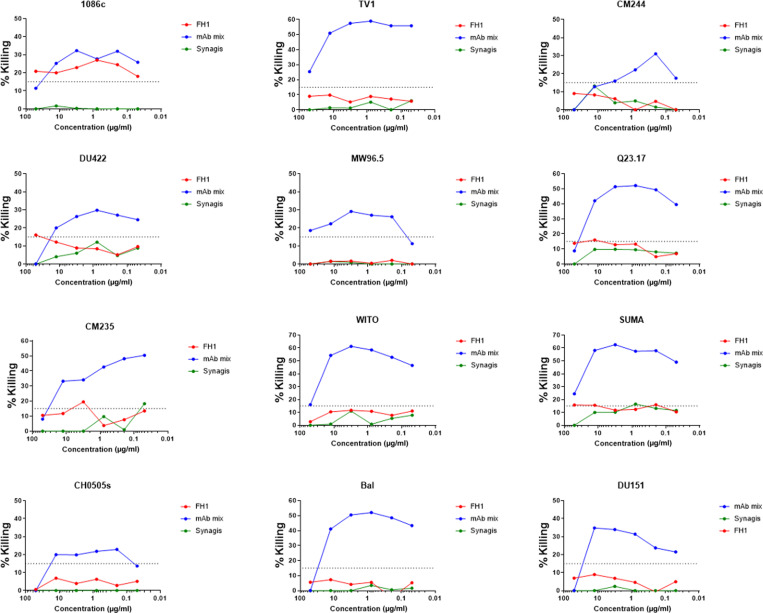
FH1 has ADCC but not neutralizing potential
We evaluated the ability of FH1 to neutralize the vaccine-matched 1086c strain and the panel of 426c viral mutants that have been engineered to be sensitive to neutralization by the glVRC01 mAb (42). There was no neutralization of the viruses detected at up to 50 mg/ml. We also evaluated the ability of FH1 to mediate ADCC to a panel of infectious molecular clones. FH1 was able to mediate ADCC against the vaccine-matched strain 1086c, even at very low concentrations (>0.05 mg/ml), which is the same strain that was used as a gp120 to isolate this antibody (Fig. 5). However, FH1 was unable to mediate ADCC against any other viruses tested, including TV1, which is a subtype C vaccine–matched virus, or others from a global panel of 10 heterologous viruses tested.
Fig. 5. ADCC activities.
The ADCC activities of FH1, a cocktail of anti–HIV-1 mAbs (A32, 2G12, CH44, and 7B2) and of mAb palivizumab (Synagis) were determined against the indicated viruses. The ADCC-positive cutoff was 15% specific killing based on our previous reports.
Competition between FH1- and VRC01-expressing B cells for Env
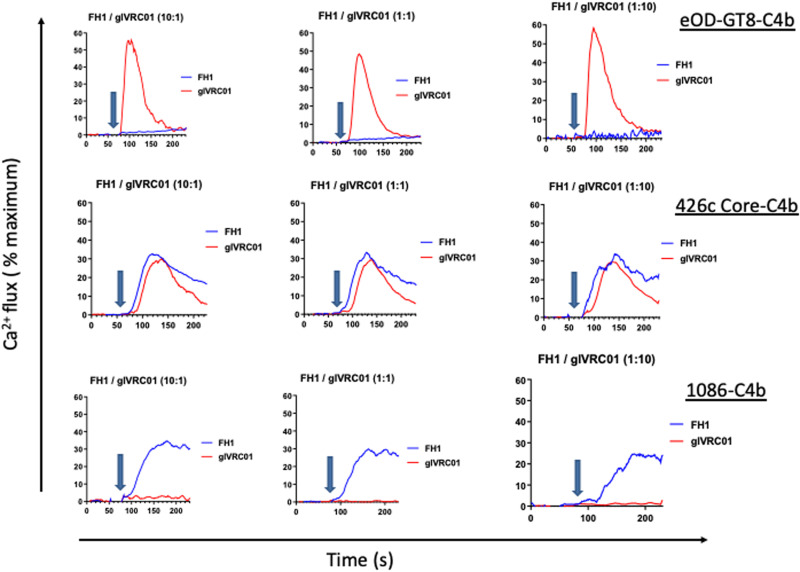
The results presented above suggested that the 426c Core immunogen, which is designed to activate B cells expressing glVRC01-class Abs (32, 34), will also activate B cells expressing FH1-like BCRs, which can give rise to antibodies with anti–HIV-1 ADCC activities. Potentially, because the relative affinity of the FH1-like BCRs for 426c Core are higher than those of glVRC01 B cells, if the frequency of FH1-like B cells is high enough, then the 426c Core may not be able to activate the latter cells. In contrast, the expectation is that the 1086 Env would only activate FH1-expressing B cells, while eOD-GT8 would activate VRC01-expressing B cells but not FH1-expressing B cells. To investigate this directly, we engineered B cells expressing FH1 and glVRC01 BCRs and examined their activation by 1086-C4b, 426c Core-C4b, and eOD-GT8-C4b self-assembling nanoparticles (7meric) at three different FH1/glVRC01 B cell ratios (10:1, 1:1, and 1:10) (Fig. 6). eOD-GT8-C4b activated glVRC01 B cells but not FH1 cells irrespective of the relative FH/glVRC01 B cell ratios (Fig. 6, top row), 426c Core-C4b activated both B cells irrespective of the relative B cells ratios (Fig. 6, middle row), while 1086-C4b only activated FH1 B cells irrespective of the relative B cell ratios (Fig. 6, bottom row). These data suggest that eOD-GT8 and 426c Core, both germ line–targeting immunogens, are therefore more likely to activate glVRC01-class B cells rather than non–germ line–targeting Env immunogens. In addition, the 426c Core may activate anti-C1C2 gp120 domain B cells that are linked with anti–HIV-1 ADCC activities.
Fig. 6. Activation of B cells expressing FH1 or glVRC01 BCRs.
B cells engineered to express either FH1 or glVRC01 BCRs were mixed at the indicated ratios and incubated with eOD-GT8-C4b (top), 426c Core-C4b (middle), or 1086-C4b (bottom). The relative activation of FH1 (blue traces) or glVRC01 (red traces) was evaluated by monitoring changes in intracellular Ca2+ concentrations over time. Arrows indicate when the indicated Envs were added.
DISCUSSION
The VH and VL genes of FH1 are minimally mutated. This implies that the corresponding B cell did not undergo extensive somatic hypermutation and may have been activated late during the HVTN 100 immunization series. Because 1086 and TV1 were coadministered, and both bind FH1, it is presently unknown which of the two immunogens initiated the activation of the corresponding naïve B cell.
Still, the possibility exists that during a more prolonged germinal center reaction, the FH1 BCR may accumulate somatic mutations that would lead to a change in its epitope specificity from the C1/C2 domain to the CD4-BS. However, here, we demonstrate that such a dramatic epitope switch would require extensive and simultaneous changes in both the CDRH3 and CDRL3 domains. Thus, it is unlikely that FH1-like–expressing B cells, which become activated by non–germ line–targeting Envs, will become VRC01-like through the accumulation of somatic hypermutations in these two domains. It is, however, conceivable that during affinity maturation, some somatic mutations within and/or outside the CDRH3/CDRL3 alter the relative conformation of these two regions so that the antibody changes its epitope specificity. Our findings also support observations that despite the predominance of V gene–encoded antibody domain CDRH2 in the interaction of VRC01-class antibodies with the CD4-BS, their CDRH3 domains influence their epitope specificity (39, 43).
Efforts to elicit VRC01-like antibodies have focused on designing recombinant Env immunogens capable of binding with high affinity to the unmutated forms of these antibodies, as expressed on naïve human B cells (17, 19, 32–34, 37, 44). These germ line–targeting immunogens present not only the epitope of interest (the VRC01 epitope) but also other epitopes recognized by non–VRC01-like antibodies, which are non-neutralizing (16, 45). Depending on the relative affinities and frequencies of the on-target and off-target B cells, the former B cells may not be efficiently activated by germ line–targeting immunogens (46, 47). Here, we show that even if the off-target B cells (non–VRC01-like and non-neutralizing) are in large excess over the on-target B cells (VRC01-like), germ line–targeting immunogens can still activate the on-target B cells. In contrast, even if the on-target B cells are in large excess over the off-target B cells, a non–germ line–targeting immunogen will not activate the former cells but will readily activate the latter cells. The observation that FH1 and glVRC01-like antibodies display high degrees of amino acid sequence homology, but different epitope specificities, highlights the importance of combining paired VH/VL gene sequence analysis with actual antibody-binding and structural analysis to properly assess the success of upcoming human immunizations aiming at activating VRC01-class BCRs.
In summary, our study highly suggests that Env immunogens that are not designed to engage the unmutated forms of VRC01-class antibodies are unlikely to activate the corresponding B cells in humans. Our study supports the proposal that the in vivo activation of B cells expressing the precursor BCR forms of VRC01-class bnAbs will require specifically designed protein immunogens.
MATERIALS AND METHODS
HVTN 100 clinical trial
HVTN 100 was a randomized, controlled, double-blind study phase 1 and 2 clinical trial, which enrolled 252 healthy HIV-uninfected 18- to 40-year-old participants at six sites in South Africa. The relevant research ethics committees approved the study. All participants gave written informed consent in English or their local language. Additional details about the HVTN 100 study design, eligibility criteria, participants and their baseline characteristics, randomization, blinding, and study products are available in previous reports (22). The trial was registered with the South African National Clinical Trials Registry (DOH-27-0215-4796) and ClinicalTrials.gov (NCT02404311), and all approved Institutional Review Board protocols were followed with human participants.
Recombinant envelope protein and antibody expression and purification
Details on the expression and purification of recombinant HIV-1 envelope proteins in human embryonic kidney (HEK) 293 EBNA-6E cells (National Council Canada) and of mAbs have been extensively presented (34, 37, 48, 49). The Envs used during the Ca2+ flux assays were multimerized using the C4b-binding protein oligomerization motif, as we previously described in detail (32, 34, 37).
Complex crystallization and X-ray data collection
HxB2 was transfected in GNTI−/− cells (provided by P. Bjorkman at CalTech) and purified with GNA lectin and size exclusion chromatography (SEC). FH1 Fab was obtained by digesting FH1 IgG with Lys C, collecting the flow through from a protein A column and purifying it by SEC. HxB2 and FH1 Fab were incubated at a 1.1 molar excess of Fab and treated with EndoH for 1 hour at room temperature, and the complex was purified over SEC. The complex was screened against the Hampton Crystal HT, ProPlex HT-96, and Wizard Precipitant Synergy no. 2 crystallization screens. The NT8 robotic system (FORMULATRIX) was used to set initial sitting drop crystallization trials. Following initial hits, crystallization conditions were optimized using hanging-drop vapor diffusion. Crystals were grown in 0.1 M NH4SO4, 20% polyethylene glycol, molecular weight 1500 (PEG 1500), and 0.1 M tris (pH 7.5, 10 mg/ml). Crystals were flash-frozen in 0.15 M (NH4)2SO4, 30% PEG 1500, and 0.15 M tris (pH 7.5) supplemented with 20% ethylene glycol. Datasets were processed using HKL2000 (50), and initial models were generated using molecular replacement in Phenix (51). Following molecular replacement, iterative model building and refinement were achieved using COOT (52) and Phenix, respectively.
Biolayer interferometry
Biolayer interferometry experiments were performed as previously described (34, 37). Briefly, kinetic analyses were performed using recombinant Fabs loaded onto FAB2G biosensors (ForteBio, catalog no. 18-5126) (at 40 ug in 1× phosphate-buffered saline) and twofold dilutions of Env monomers. The assay parameters are the same as for measuring the binding of IgG but with an extended dissociation phase of 600 s. Curve fitting used to determine relative apparent antibody affinities for envelope was performed using a 1:1 binding model and the data analysis software (ForteBio). Mean kon, koff, and dissociation constant (KD) values were determined by averaging all binding curves within a dilution series having coefficient of determination values of greater than 95% confidence level.
HIV-1 neutralization and ADCC assays
All neutralization experiments were performed as previously described (42). ADCC was determined by a luciferase-based assay as previously described (53, 54). Briefly, CEM.NKRCCR5 cells [National Institutes of Health (NIH) AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH from A. Trkola] (PMC112928) were used as targets after infection with the HIV-1 IMCs. Recombinant FH1 mAb was tested across a range of concentrations using fivefold serial dilutions starting at 50 μg/ml. The final readout was the luminescence intensity [relative light units (RLU)] generated by the presence of residual intact target cells that have not been lysed by the effector population in the presence of ADCC-mediating mAb (ViviRen substrate, Promega, Madison, WI). The percentage of specific killing was calculated using the formula: percent specific killing = [(number of RLU of target and effector well − number of RLU of test well)/number of RLU of target and effector well] × 100. In this analysis, the RLU of the target plus effector wells represents spontaneous lysis in absence of any source of Ab. The ADCC-positive cutoff was 15% specific killing based on our previous reports. The mAb palivizumab (Synagis), which mediates ADCC (55) but is specific for respiratory syncytial virus, and a cocktail of HIV-1 mAbs (HIV-1 mAb mix) demonstrated to mediate ADCC [A32 (24), 2G12 (56), CH44 (57), and 7B2 (58)] were used as negative and positive controls, respectively.
Generation of FH1- and glVRC01-A20 cell lines
FH1- and glVRC01-A20 cell lines [American Type Culture Collection (ATCC), catalog no. ATCC TIB-208] were generated using CRISPR technique as described previously (59). The single-guide RNA (sgRNA) muIgH367 sequence is TTATACAGTATCCGATGCATAGG (PAM site in bold) targeting the region between the last JH gene and the Eu enhancer, which is upstream of endogenous IgH constant region. Cas9 proteins (18 μmol; Invitrogen) and sgRNA (54 μmol; Synthego) were precomplexed in 5-μl Neon Buffer R at room temperature for 20 min. A total of 250,000 A20 cells were resuspended in 7-μl R buffer, added to the Cas9/sgRNA complex, and electroporated using 20-ms pulse at 1725 V using the Neon Transfection System (Invitrogen). The cleavage efficiency of sgRNA was about 60% as assessed by Synthego ICE CRISPR software after polymerase chain reaction amplification and sequencing of the targeted genomic DNA. After electroporation, cells were recovered in prewarmed RPMI 1640 complete medium without antibiotics for 30 min and then transduced by FH1 or glVRC01 adeno-associated virus (AAV) viruses. After expanding for 3 days, cells were stained with Q3W24 BGF BITH BUV395 anti-CD19 (BD Biosciences), AF647-conjugated 426cDMRScore-c4b, and near-infrared live dead dye (Thermo Fisher Scientific) to sort out the live, CD19-positive, Env-binding population on FACSAria (BD Biosciences).
AAV viruses were produced by cotransfection of FH1- or glVRC01-AAV plasmids, serotype 6 capsid, and adenoviral helper plasmids into HEK 293 cells using polyethylenimine. Viruses were purified over iodixanol gradient. Their titers were approximately 1 × 109 per microliter. FH1 and glVRC01 AAV plasmids were constructed by In-Fusion (Takara) recombination of ECoRV-linearized AAV backbone plasmid PCH19-mb3AAV-HR (gift from J. Taylor’s laboratory) and FH1 or glVRC01 gBlocks (IDT). The FH1 and glVRC01 gBlocks contain the full-length LC, the 57–amino acid glycine-serine linker, and the HC VDJ, plus In-Fusion homology sequences to the backbone plasmid.
Calcium influx
Calcium influx experiments were performed as previously described (18, 45). Briefly, FH1- or glVRC01-expressing A20 cells were resuspended in RPMI 1640 complete medium (1.5 × 106/ml), mixed (1:1 in volume) with Fluo 4 dye (Fluo 4 Direct Calcium Assay Kit, Thermo Fisher Scientific), and incubated at 37°C for 30 min. Fluorescence was recorded for 1 min (baseline), and 0.25 μM Env was added, and the fluorescence was recorded for 3 min, at which point, ionomycin was added (2 μg/ml). The Env proteins tested were eOD-GT8-c4b, 426c Core-c4b, and 1086-C4b. Data were analyzed using the Kinetics function in FlowJo software. Baseline fluorescence was subtracted from fluorescence intensity at all time points using Prism 7 software. Calcium influx in mixed FH1- and glVRC01-expressing A20 cells was performed as previously described (45). Briefly, FH1- and glVRC01-A20 cells (1.0 × 106) were labeled with 1 μM CellTrace Violet and 1 μM CellTrace Yellow (Thermo Fisher Scientific), respectively, according to the manufacturer’s protocol. The cells were washed, resuspended in RPMI 1640 complete medium, and stained with Fluo 4 dye. FH1- and glVRC01-expressing A20 cells were then mixed at 1:10, 1:1, or 10:1 ratios and incubated with eOD-GT8-c4b, 426c Core-c4b, or 1086-c4b, as described above. Analysis was performed in FlowJo; FH1 and glVRC01 cells were gated on CellTrace Violet and CellTrace Yellow positive separately and graphed for Fluo 4 fluorescence intensity over time.
Acknowledgments
We appreciate the efforts of the HVTN 100 protocol team, including protocol chairs, L. G. Bekker and F. Laher, and the study participants in the conduct of that trial. We thank the Fred Hutchinson Cancer Research Center’s Vector Core for generating AAV viruses. Beamline 5.0.2 of the Advanced Light Source, a U.S. DOE Office of Science User Facility under contract no. DE-AC02-05CH11231, is supported, in part, by the ALS-ENABLE program funded by the NIH, National Institute of General Medical Sciences, grant P30 GM124169-01.
Funding: This study was funded by NIH grants R01AI081625 (L.S.), R01AI104384 (L.S.), P01 AI138212 (L.S.), U19 AI128914 (L.S.), UM1AI144462 (subaward M.J.M.), and UM1AI068618 (M.J.M.).
Author contributions: Conceptualization: L.S., M.J.M., and M.P. Investigation: M.D.G., J.F., C.E.W., L.B.-F., A.J.M., C.N.H., J.J.T., D.M., and G.F. Supervision: K.W.C., M.P., M.J.M., and L.S. Writing—original draft: L.S. Writing—review and editing: all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Atomic coordinates generated in this study have been deposited in the PDB under accession number 7RDW.
Supplementary Materials
This PDF file includes:
Figs. S1 and S2
Tables S1 and S2
REFERENCES AND NOTES
- 1.Kwong P. D., Mascola J. R., Human antibodies that neutralize HIV-1: Identification, structures, and B cell ontogenies. Immunity 37, 412–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burton D. R., Hangartner L., Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol. 34, 635–659 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCoy L. E., Burton D. R., Identification and specificity of broadly neutralizing antibodies against HIV. Immunol. Rev. 275, 11–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sok D., Burton D. R., HIV broadly neutralizing antibodies: Taking good care of the 98. Immunity 45, 958–960 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balazs A. B., Chen J., Hong C. M., Rao D. S., Yang L., Baltimore D., Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 481, 81–84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shingai M., Donau O. K., Plishka R. J., Buckler-White A., Mascola J. R., Nabel G. J., Nason M. C., Montefiori D., Moldt B., Poignard P., Diskin R., Bjorkman P. J., Eckhaus M. A., Klein F., Mouquet H., Cetrulo Lorenzi J. C., Gazumyan A., Burton D. R., Nussenzweig M. C., Martin M. A., Nishimura Y., Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J. Exp. Med. 211, 2061–2074 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corey L., Gilbert P. B., Juraska M., Montefiori D. C., Morris L., Karuna S. T., Edupuganti S., Mgodi N. M., deCamp A., Rudnicki E., Huang Y., Gonzales P., Cabello R., Orrell C., Lama J. R., Laher F., Lazarus E. M., Sanchez J., Frank I., Hinojosa J., Sobieszczyk M. E., Marshall K. E., Mukwekwerere P. G., Makhema J., Baden L. R., Mullins J. I., Williamson C., Hural J., McElrath M., Bentley C., Takuva S., Gomez Lorenzo M. M., Burns D. N., Espy N., Randhawa A. K., Kochar N., Piwowar-Manning E., Donnell D. J., Sista N., Andrew P., Kublin J. G., Gray G., Ledgerwood J. E., Mascola J. R., Cohen M. S.; HVTN 704/HPTN 085 and HVTN 703/HPTN 081 Study Teams , Two randomized trials of neutralizing antibodies to prevent HIV-1 acquisition. N. Engl. J. Med. 384, 1003–1014 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sajadi M. M., Dashti A., Tehrani Z. R., Tolbert W. D., Seaman M. S., Ouyang X., Gohain N., Pazgier M., Kim D., Cavet G., Yared J., Redfield R. R., Lewis G. K., De Vico A. L., Identification of near-pan-neutralizing antibodies against HIV-1 by deconvolution of plasma humoral responses. Cell 173, 1783–1795.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheid J. F., Mouquet H., Ueberheide B., Diskin R., Klein F., Oliveira T. Y. K., Pietzsch J., Fenyo D., Abadir A., Velinzon K., Hurley A., Myung S., Boulad F., Poignard P., Burton D. R., Pereyra F., Ho D. D., Walker B. D., Seaman M. S., Bjorkman P. J., Chait B. T., Nussenzweig M. C., Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Umotoy J., Bagaya B. S., Joyce C., Schiffner T., Menis S., Saye-Francisco K. L., Biddle T., Mohan S., Vollbrecht T., Kalyuzhniy O., Madzorera S., Kitchin D., Lambson B., Nonyane M., Kilembe W.; IAVI Protocol C Investigators; IAVI African HIV Research Network, Poignard P., Schief W. R., Burton D. R., Murrell B., Moore P. L., Briney B., Sok D., Landais E., Rapid and focused maturation of a VRC01-class HIV broadly neutralizing antibody lineage involves both binding and accommodation of the N276-glycan. Immunity 51, 141–154.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X., Zhang Z., Schramm C. A., Joyce M. G., Do Kwon Y., Zhou T., Sheng Z., Zhang B., O’Dell S., McKee K., Georgiev I. S., Chuang G. Y., Longo N. S., Lynch R. M., Saunders K. O., Soto C., Srivatsan S., Yang Y., Bailer R. T., Louder M. K., Mullikin J. C., Connors M., Kwong P. D., Mascola J. R., Shapiro L., Benjamin B., Blakesley R., Bouffard G., Brooks S., Coleman H., Dekhtyar M., Gregory M., Guan X., Gupta J., Han J., Hargrove A., Ho S. L., Legaspi R., Maduro Q., Masiello C., Maskeri B., McDowell J., Montemayor C., Park M., Riebow N., Schandler K., Schmidt B., Sison C., Stantripop M., Thomas J., Thomas P., Vemulapalli M., Young A., Maturation and diversity of the VRC01-antibody lineage over 15 years of chronic HIV-1 infection. Cell 161, 470–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu X., Zhou T., Zhu J., Zhang B., Georgiev I., Wang C., Chen X., Longo N. S., Louder M., McKee K., O’Dell S., Perfetto S., Schmidt S. D., Shi W., Wu L., Yang Y., Yang Z. Y., Yang Z., Zhang Z., Bonsignori M., Crump J. A., Kapiga S. H., Sam N. E., Haynes B. F., Simek M., Burton D. R., Koff W. C., Doria-Rose N. A., Connors M.; NISC Comparative Sequencing Program, Mullikin J. C., Nabel G. J., Roederer M., Shapiro L., Kwong P. D., Mascola J. R., Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou T., Georgiev I., Wu X., Yang Z. Y., Dai K., Finzi A., Do Kwon Y., Scheid J. F., Shi W., Xu L., Yang Y., Zhu J., Nussenzweig M. C., Sodroski J., Shapiro L., Nabel G. J., Mascola J. R., Kwong P. D., Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou T., Zhu J., Wu X., Moquin S., Zhang B., Acharya P., Georgiev I. S., Altae-Tran H. R., Chuang G. Y., Joyce M. G., Kwon Y. D., Longo N. S., Louder M. K., Luongo T., McKee K., Schramm C. A., Skinner J., Yang Y., Yang Z., Zhang Z., Zheng A., Bonsignori M., Haynes B. F., Scheid J. F., Nussenzweig M. C., Simek M., Burton D. R., Koff W. C.; NISC Comparative Sequencing Program, Mullikin J. C., Connors M., Shapiro L., Nabel G. J., Mascola J. R., Kwong P. D., Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245–258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J., Kang B. H., Ishida E., Zhou T., Griesman T., Sheng Z., Wu F., Doria-Rose N. A., Zhang B., McKee K., O’Dell S., Chuang G. Y., Druz A., Georgiev I. S., Schramm C. A., Zheng A., Joyce M. G., Asokan M., Ransier A., Darko S., Migueles S. A., Bailer R. T., Louder M. K., Alam S. M., Parks R., Kelsoe G., von Holle T., Haynes B. F., Douek D. C., Hirsch V., Seaman M. S., Shapiro L., Mascola J. R., Kwong P. D., Connors M., Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity 45, 1108–1121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havenar-Daughton C., Sarkar A., Kulp D. W., Toy L., Hu X., Deresa I., Kalyuzhniy O., Kaushik K., Upadhyay A. A., Menis S., Landais E., Cao L., Diedrich J. K., Kumar S., Schiffner T., Reiss S. M., Seumois G., Yates J. R., Paulson J. C., Bosinger S. E., Wilson I. A., Schief W. R., Crotty S., The human naive B cell repertoire contains distinct subclasses for a germline-targeting HIV-1 vaccine immunogen. Sci. Transl. Med. 10, eaat0381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jardine J., Julien J. P., Menis S., Ota T., Kalyuzhniy O., McGuire A., Sok D., Huang P. S., MacPherson S., Jones M., Nieusma T., Mathison J., Baker D., Ward A. B., Burton D. R., Stamatatos L., Nemazee D., Wilson I. A., Schief W. R., Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGuire A. T., Glenn J. A., Lippy A., Stamatatos L., Diverse recombinant HIV-1 Envs fail to activate B cells expressing the germline B cell receptors of the broadly neutralizing anti-HIV-1 antibodies PG9 and 447-52D. J. Virol. 88, 2645–2657 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGuire A. T., Hoot S., Dreyer A. M., Lippy A., Stuart A., Cohen K. W., Jardine J., Menis S., Scheid J. F., West A. P., Schief W. R., Stamatatos L., Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J. Exp. Med. 210, 655–663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoot S., McGuire A. T., Cohen K. W., Strong R. K., Hangartner L., Klein F., Diskin R., Scheid J. F., Sather D. N., Burton D. R., Stamatatos L., Recombinant HIV envelope proteins fail to engage germline versions of Anti-CD4bs bNAbs. PLOS Pathog. 9, e1003106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamatatos L., Pancera M., McGuire A. T., Germline-targeting immunogens. Immunol. Rev. 275, 203–216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bekker L. G., Moodie Z., Grunenberg N., Laher F., Tomaras G. D., Cohen K. W., Allen M., Malahleha M., Mngadi K., Daniels B., Innes C., Bentley C., Frahm N., Morris D. E., Morris L., Mkhize N. N., Montefiori D. C., Sarzotti-Kelsoe M., Grant S., Yu C., Mehra V. L., Pensiero M. N., Phogat S., DiazGranados C., Barnett S. W., Kanesa-Thasan N., Koutsoukos M., Michael N. L., Robb M. L., Kublin J. G., Gilbert P. B., Corey L., Gray G. E., McElrath M.; HVTN 100 Protocol Team , Subtype C ALVAC-HIV and bivalent subtype C gp120/MF59 HIV-1 vaccine in low-risk, HIV-uninfected, South African adults: A phase 1/2 trial. Lancet HIV 5, e366–e378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen X., Laher F., Moodie Z., McMillan A. S., Spreng R. L., Gilbert P. B., Huang Y., Yates N. L., Grunenberg N., Juliana McElrath M., Allen M., Pensiero M., Mehra V. L., der Meeren O. V., Barnett S. W., Phogat S., Gray G. E., Bekker L. G., Corey L., Tomaras G. D., HIV-1 vaccine sequences impact V1V2 antibody responses: A comparison of two poxvirus prime gp120 boost vaccine regimens. Sci. Rep. 10, 2093 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrari G., Pollara J., Kozink D., Harms T., Drinker M., Freel S., Moody M. A., Alam S. M., Tomaras G. D., Ochsenbauer C., Kappes J. C., Shaw G. M., Hoxie J. A., Robinson J. E., Haynes B. F., An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J. Virol. 85, 7029–7036 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Acharya P., Tolbert W. D., Gohain N., Wu X., Yu L., Liu T., Huang W., Huang C. C., Kwon Y. D., Louder R. K., Luongo T. S., McLellan J. S., Pancera M., Yang Y., Zhang B., Flinko R., Foulke J. S. Jr., Sajadi M. M., Kamin-Lewis R., Robinson J. E., Martin L., Kwong P. D., Guan Y., DeVico A. L., Lewis G. K., Pazgier M., Structural definition of an antibody-dependent cellular cytotoxicity response implicated in reduced risk for HIV-1 infection. J. Virol. 88, 12895–12906 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.West A. P. Jr., Diskin R., Nussenzweig M. C., Bjorkman P. J., Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc. Natl. Acad. Sci. U.S.A. 109, E2083–E2090 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diskin R., Scheid J. F., Marcovecchio P. M., West A. P. Jr., Klein F., Gao H., Gnanapragasam P. N. P., Abadir A., Seaman M. S., Nussenzweig M. C., Bjorkman P. J., Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334, 1289–1293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scharf L., West A. P. Jr., Gao H., Lee T., Scheid J. F., Nussenzweig M. C., Bjorkman P. J., Diskin R., Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody. Proc. Natl. Acad. Sci. U.S.A. 110, 6049–6054 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briney B., Sok D., Jardine J. G., Kulp D. W., Skog P., Menis S., Jacak R., Kalyuzhniy O., de Val N., Sesterhenn F., Le K. M., Ramos A., Jones M., Saye-Francisco K. L., Blane T. R., Spencer S., Georgeson E., Hu X., Ozorowski G., Adachi Y., Kubitz M., Sarkar A., Wilson I. A., Ward A. B., Nemazee D., Burton D. R., Schief W. R., Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell 166, 1459–1470.e11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dosenovic P., von Boehmer L., Escolano A., Jardine J., Freund N. T., Gitlin A. D., McGuire A. T., Kulp D. W., Oliveira T., Scharf L., Pietzsch J., Gray M. D., Cupo A., van Gils M. J., Yao K. H., Liu C., Gazumyan A., Seaman M. S., Björkman P. J., Sanders R. W., Moore J. P., Stamatatos L., Schief W. R., Nussenzweig M. C., Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell 161, 1505–1515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jardine J. G., Ota T., Sok D., Pauthner M., Kulp D. W., Kalyuzhniy O., Skog P. D., Thinnes T. C., Bhullar D., Briney B., Menis S., Jones M., Kubitz M., Spencer S., Adachi Y., Burton D. R., Schief W. R., Nemazee D., Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349, 156–161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGuire A. T., Gray M. D., Dosenovic P., Gitlin A. D., Freund N. T., Petersen J., Correnti C., Johnsen W., Kegel R., Stuart A. B., Glenn J., Seaman M. S., Schief W. R., Strong R. K., Nussenzweig M. C., Stamatatos L., Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat. Commun. 7, 10618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medina-Ramirez M., Garces F., Escolano A., Skog P., de Taeye S. W., Del Moral-Sanchez I., Guire A. T. M., Yasmeen A., Behrens A.-J., Ozorowski G., van den Kerkhof T. L. G. M., Freund N. T., Dosenovic P., Hua Y., Gitlin A. D., Cupo A., van der Woude P., Golabek M., Sliepen K., Blane T., Kootstra N., van Breemen M. J., Pritchard L. K., Stanfield R. L., Crispin M., Ward A. B., Stamatatos L., Klasse P. J., Moore J. P., Nemazee D., Nussenzweig M. C., Wilson I. A., Sanders R. W., Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. J. Exp. Med. 214, 2573–2590 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parks K. R., MacCamy A. J., Trichka J., Gray M., Weidle C., Borst A. J., Khechaduri A., Takushi B., Agrawal P., Guenaga J., Wyatt R. T., Coler R., Seaman M., Branche C. L., Montefiori D. C., Veesler D., Pancera M., Guire A. M., Stamatatos L., Overcoming steric restrictions of VRC01 HIV-1 neutralizing antibodies through immunization. Cell Rep. 29, 3060–3072.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian M., Cheng C., Chen X., Duan H., Cheng H.-L., Dao M., Sheng Z., Kimble M., Wang L., Lin S., Schmidt S. D., Du Z., Joyce M. G., Chen Y., De Kosky B. J., Chen Y., Normandin E., Cantor E., Chen R. E., Doria-Rose N. A., Zhang Y., Shi W., Kong W.-P., Choe M., Henry A. R., Laboune F., Georgiev I. S., Huang P.-Y., Jain S., McGuire A. T., Georgeson E., Menis S., Douek D. C., Schief W. R., Stamatatos L., Kwong P. D., Shapiro L., Haynes B. F., Mascola J. R., Alt F. W., Induction of HIV neutralizing antibody lineages in mice with diverse precursor repertoires. Cell 166, 1471–1484.e18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duan H., Chen X., Boyington J. C., Cheng C., Zhang Y., Jafari A. J., Stephens T., Tsybovsky Y., Kalyuzhniy O., Zhao P., Menis S., Nason M. C., Normandin E., Mukhamedova M., De Kosky B. J., Wells L., Schief W. R., Tian M., Alt F. W., Kwong P. D., Mascola J. R., Glycan masking focuses immune responses to the HIV-1 CD4-binding site and enhances elicitation of VRC01-class precursor antibodies. Immunity 49, 301–311.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Y.-R., Parks K. R., Weidle C., Naidu A. S., Khechaduri A., Riker A. O., Takushi B., Chun J.-H., Borst A. J., Veesler D., Stuart A., Agrawal P., Gray M., Pancera M., Huang P.-S., Stamatatos L., HIV-1 VRC01 germline-targeting immunogens select distinct epitope-specific B cell receptors. Immunity 53, 840–851.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sok D., Briney B., Jardine J. G., Kulp D. W., Menis S., Pauthner M., Wood A., Lee E. C., le K. M., Jones M., Ramos A., Kalyuzhniy O., Adachi Y., Kubitz M., MacPherson S., Bradley A., Friedrich G. A., Schief W. R., Burton D. R., Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science 353, 1557–1560 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yacoob C., Pancera M., Vigdorovich V., Oliver B. G., Glenn J. A., Feng J., Sather D. N., McGuire A. T., Stamatatos L., Differences in allelic frequency and CDRH3 region limit the engagement of HIV Env immunogens by putative VRC01 neutralizing antibody precursors. Cell Rep. 17, 1560–1570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonsignori M., Pollara J., Moody M. A., Alpert M. D., Chen X., Hwang K. K., Gilbert P. B., Huang Y., Gurley T. C., Kozink D. M., Marshall D. J., Whitesides J. F., Tsao C. Y., Kaewkungwal J., Nitayaphan S., Pitisuttithum P., Rerks-Ngarm S., Kim J. H., Michael N. L., Tomaras G. D., Montefiori D. C., Lewis G. K., DeVico A., Evans D. T., Ferrari G., Liao H. X., Haynes B. F., Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J. Virol. 86, 11521–11532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Easterhoff D., Moody M. A., Fera D., Cheng H., Ackerman M., Wiehe K., Saunders K. O., Pollara J., Vandergrift N., Parks R., Kim J., Michael N. L., O’Connell R. J., Excler J. L., Robb M. L., Vasan S., Rerks-Ngarm S., Kaewkungwal J., Pitisuttithum P., Nitayaphan S., Sinangil F., Tartaglia J., Phogat S., Kepler T. B., Alam S. M., Liao H. X., Ferrari G., Seaman M. S., Montefiori D. C., Tomaras G. D., Harrison S. C., Haynes B. F., Boosting of HIV envelope CD4 binding site antibodies with long variable heavy third complementarity determining region in the randomized double blind RV305 HIV-1 vaccine trial. PLOS Pathog. 13, e1006182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LaBranche C. C., Guire A. T. M., Gray M. D., Behrens S., Kwong P. D., Chen X., Zhou T., Sattentau Q. J., Peacock J., Eaton A., Greene K., Gao H., Tang H., Perez L. G., Chen X., Saunders K. O., Kwong P. D., Mascola J. R., Haynes B. F., Stamatatos L., Montefiori D. C., HIV-1 envelope glycan modifications that permit neutralization by germline-reverted VRC01-class broadly neutralizing antibodies. PLOS Pathog. 14, e1007431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonsignori M., Hwang K. K., Chen X., Tsao C. Y., Morris L., Gray E., Marshall D. J., Crump J. A., Kapiga S. H., Sam N. E., Sinangil F., Pancera M., Yongping Y., Zhang B., Zhu J., Kwong P. D., O’Dell S., Mascola J. R., Wu L., Nabel G. J., Phogat S., Seaman M. S., Whitesides J. F., Moody M. A., Kelsoe G., Yang X., Sodroski J., Shaw G. M., Montefiori D. C., Kepler T. B., Tomaras G. D., Alam S. M., Liao H. X., Haynes B. F., Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 85, 9998–10009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jardine J. G., Kulp D. W., Havenar-Daughton C., Sarkar A., Briney B., Sok D., Sesterhenn F., Ereño-Orbea J., Kalyuzhniy O., Deresa I., Hu X., Spencer S., Jones M., Georgeson E., Adachi Y., Kubitz M., deCamp A. C., Julien J. P., Wilson I. A., Burton D. R., Crotty S., Schief W. R., HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 351, 1458–1463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGuire A. T., Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies. Science 346, 1380–1383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abbott R. K., Lee J. H., Menis S., Skog P., Rossi M., Ota T., Kulp D. W., Bhullar D., Kalyuzhniy O., Havenar-Daughton C., Schief W. R., Nemazee D., Crotty S., Precursor frequency and affinity determine B cell competitive fitness in germinal centers, tested with germline-targeting HIV vaccine immunogens. Immunity 48, 133–146.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dosenovic P., Kara E. E., Pettersson A. K., McGuire A. T., Gray M., Hartweger H., Thientosapol E. S., Stamatatos L., Nussenzweig M. C., Anti-HIV-1 B cell responses are dependent on B cell precursor frequency and antigen-binding affinity. Proc. Natl. Acad. Sci. U.S.A. 115, 4743–4748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sellhorn G., Caldwell Z., Mineart C., Stamatatos L., Improving the expression of recombinant soluble HIV Envelope glycoproteins using pseudo-stable transient transfection. Vaccine 28, 430–436 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Sellhorn G., Kraft Z., Caldwell Z., Ellingson K., Mineart C., Seaman M. S., Montefiori D. C., Lagerquist E., Stamatatos L., Engineering, expression, purification, and characterization of stable clade A/B recombinant soluble heterotrimeric gp140 proteins. J. Virol. 86, 128–142 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 51.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Liao H. X., Bonsignori M., Alam S. M., McLellan J. S., Tomaras G. D., Moody M. A., Kozink D. M., Hwang K. K., Chen X., Tsao C. Y., Liu P., Lu X., Parks R. J., Montefiori D. C., Ferrari G., Pollara J., Rao M., Peachman K. K., Santra S., Letvin N. L., Karasavvas N., Yang Z. Y., Dai K., Pancera M., Gorman J., Wiehe K., Nicely N. I., Rerks-Ngarm S., Nitayaphan S., Kaewkungwal J., Pitisuttithum P., Tartaglia J., Sinangil F., Kim J. H., Michael N. L., Kepler T. B., Kwong P. D., Mascola J. R., Nabel G. J., Pinter A., Zolla-Pazner S., Haynes B. F., Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity 38, 176–186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chua J. V., Davis C., Husson J. S., Nelson A., Prado I., Flinko R., Lam K. W. J., Mutumbi L., Mayer B. T., Dong D., Fulp W., Mahoney C., Gerber M., Gottardo R., Gilliam B. L., Greene K., Gao H., Yates N., Ferrari G., Tomaras G., Montefiori D., Schwartz J. A., Fouts T., DeVico A. L., Lewis G. K., Gallo R. C., Sajadi M. M., Safety and immunogenicity of an HIV-1 gp120-CD4 chimeric subunit vaccine in a phase 1a randomized controlled trial. Vaccine 39, 3879–3891 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hiatt A., Bohorova N., Bohorov O., Goodman C., Kim D., Pauly M. H., Velasco J., Whaley K. J., Piedra P. A., Gilbert B. E., Zeitlin L., Glycan variants of a respiratory syncytial virus antibody with enhanced effector function and in vivo efficacy. Proc. Natl. Acad. Sci. U.S.A. 111, 5992–5997 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trkola A., Purtscher M., Muster T., Ballaun C., Buchacher A., Sullivan N., Srinivasan K., Sodroski J., Moore J. P., Katinger H., Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70, 1100–1108 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moody M. A., Gao F., Gurley T. C., Amos J. D., Kumar A., Hora B., Marshall D. J., Whitesides J. F., Xia S. M., Parks R., Lloyd K. E., Hwang K. K., Lu X., Bonsignori M., Finzi A., Vandergrift N. A., Alam S. M., Ferrari G., Shen X., Tomaras G. D., Kamanga G., Cohen M. S., Sam N. E., Kapiga S., Gray E. S., Tumba N. L., Morris L., Zolla-Pazner S., Gorny M. K., Mascola J. R., Hahn B. H., Shaw G. M., Sodroski J. G., Liao H. X., Montefiori D. C., Hraber P. T., Korber B. T., Haynes B. F., Strain-specific V3 and CD4 binding site autologous HIV-1 neutralizing antibodies select neutralization-resistant viruses. Cell Host Microbe 18, 354–362 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sadraeian M., Guimarães F. E. G., Araújo A. P. U., Worthylake D. K., LeCour L., Pincus S. H., Selective cytotoxicity of a novel immunotoxin based on pulchellin A chain for cells expressing HIV envelope. Sci. Rep. 7, 7579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moffett H. F., Harms C. K., Fitzpatrick K. S., Tooley M. R., Boonyaratanakornkit J., Taylor J. J., B cells engineered to express pathogen-specific antibodies protect against infection. Sci. Immunol. 4, eaax0644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 and S2
Tables S1 and S2