Abstract
A physical unclonable function (PUF) is a physical entity that provides a measurable output that can be used as a unique and irreproducible identifier for the artifact wherein it is embedded. Popularized by the electronics industry, silicon PUFs leverage the inherent physical variations of semiconductor manufacturing to establish intrinsic security primitives for attesting integrated circuits. Owing to the stochastic nature of these variations, photolithographically manufactured silicon PUFs are impossible to reproduce (thus unclonable). Inspired by the success of silicon PUFs, we sought to create the first generation of genetic PUFs in human cells. We demonstrate that these PUFs are robust (i.e., they repeatedly produce the same output), unique (i.e., they do not coincide with any other identically produced PUF), and unclonable (i.e., they are virtually impossible to replicate). Furthermore, we demonstrate that CRISPR-engineered PUFs (CRISPR-PUFs) can serve as a foundational principle for establishing provenance attestation protocols.
CRISPR-engineered physical unclonable functions establish a foundational security technology for provenance attestation protocols.
INTRODUCTION
A physical unclonable function (PUF) is a hardware security primitive that exploits the inherent randomness of its manufacturing process to enable attestation of the entity wherein it is embodied (1–4). A PUF is typically modeled as a mapping between input stimuli (challenges) and output values (responses), which is established stochastically among a vast array of options and is, therefore, unique and irreproducible. Upon manufacturing, a PUF is interrogated and a database comprising valid challenge-response pairs (CRPs) produced by this PUF is populated. Attestation can, thus, be achieved by issuing a challenge to the holder of the physical entity embodying the PUF, receiving the response and comparing against the golden references stored in the database. Accordingly, typical quality metrics for evaluating a PUF include robustness (i.e., the probability that, given the same challenge, it will consistently produce the same response) and uniqueness (i.e., the probability that its mapping does not coincide with the mapping of any other identically manufactured PUF).
While PUF-like concepts were proposed earlier in the literature, their popularity soared after their first implementation in silicon, as part of electronic integrated circuits (5). By exploiting the inherent variation of advanced semiconductor manufacturing processes, silicon PUFs became a commercial success, serving as the foundation of many security protocols implemented both in software and in hardware. While this success stimulated similar efforts in various other domains, to date, PUFs have yet to be adopted in the context of biological sciences, wherein they could find numerous applications.
Similar to the use of silicon PUFs (in their simplest form) as unique IDs for verifying genuineness of electronic circuits, genetic PUFs could be embedded in cell lines to attest their provenance. More specifically, genetic PUFs could enable the producer of a valuable cell line to insert a unique, robust, and unclonable signature in each legitimately produced copy of this cell line. Upon thawing of a frozen sample and before its initial use, a customer who purchased a copy of the cell line can obtain this signature and communicate it to the producer who compares it against the signature database of legitimately produced copies of this cell line and, thereby, attests its provenance (fig. S1). Through this protocol, the producer of the cell line can ensure that anyone publicly claiming ownership of a copy of this cell line has acquired it legitimately. At the same time, the customer can be assured of the source and quality of the procured cell line, as the producer explicitly confirms its origin and assumes responsibility for its production.
Toward developing PUFs, we hypothesized that a process that combines molecular barcoding with nonhomologous end joining (NHEJ) repair and exploits the inherent stochasticity of the latter (Fig. 1A) yields measurable genetic changes that satisfy all PUF conditions. More specifically, a two-dimensional mapping between barcodes and indels resulting from this process, which can be obtained by sequencing a genetic locus of the cell line, is a robust yet unique and unclonable signature.
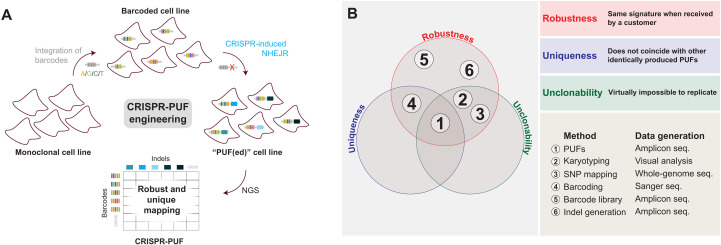
Fig. 1. Overview of the CRISPR-PUF generation process.
(A) Schematic illustration of design of CRISPR-PUFs. Barcodes were stably integrated into cell lines of interest, which were subsequently subjected to CRISPR-SpCas9 treatment to induce nonhomologous end joining repair (NHEJR). The resulting two-dimensional mapping between barcodes and indels is evaluated for robustness and uniqueness. (B) Venn diagram comparing PUFs to other methods/technologies.
As visualized in a Venn diagram (Fig. 1B), clustered regularly interspaced short palindromic repeats (CRISPR)–engineered PUF (CRISPR-PUF) is the only known methodology that satisfies all three PUF criteria. Barcodes and indels alone are not PUFs and cannot be used for provenance attestation. Indels are not PUFs because they are not unique (6, 7) and are clonable (thus violating two of the three PUF conditions). Barcodes are also not PUFs, as they violate the uniqueness criterion. As shown later here, when we integrated a 5-nucleotide (nt) barcode library into the AAVS1 locus of human embryonic kidney (HEK) 293 cells via CRISPR-SpCas9 in six parallel replicates, we observed that the uniqueness criterion is not satisfied. We emphasize that increasing the size of the barcode would not resolve the uniqueness criterion but would merely increase complexity. In contrast, the uniqueness of our PUF design is not based on a scalar property, such as the complexity or entropy of barcodes or indels, but rather on the joint probability distributions of both barcodes and indels in the cell population. Last, natural genetic variations such as short nucleotide polymorphisms or short tandem repeats can be used for cell line authentication but not for provenance attestation, because they are, generally, not unique or unclonable (Fig. 1B). As an example, all cell lines derived from a single monoclonal source share the same SNP mapping or karyotyping information and thus violate the uniqueness requirement.
RESULTS
Implementation of the first-generation CRISPR-PUF
To validate our hypothesis, and toward implementing the first generation of genetic PUFs, we carried out a pilot study where we leverage genome engineering using CRISPR (8–11). CRISPR is an immune response mechanism against bacteriophage infections in bacteria and archaea that has revolutionized the field of genome editing and spurred myriads of applications critically relevant to agriculture, biomanufacturing, and human health (9, 12–16). Critically, Cas9 can be programmed to bind to a specific region of DNA and generate a double-stranded break, which, in turn, initiates the error-prone DNA repair pathway NHEJ. Our method involved the following steps.
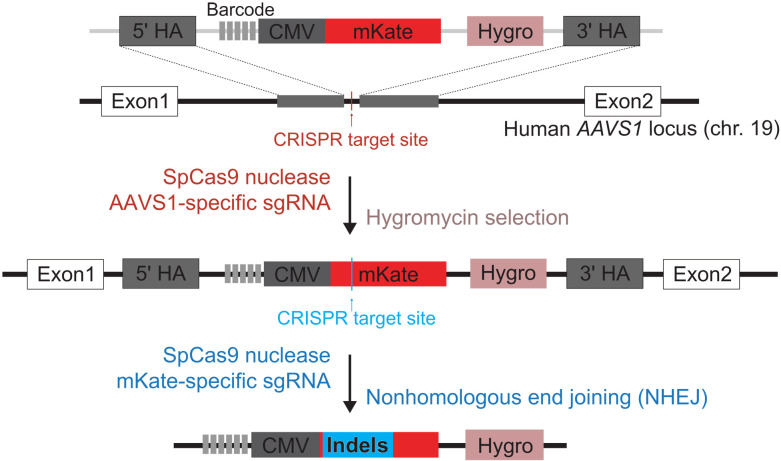
First, we stably integrated a 5-nt barcode library into the AAVS1 locus of human HEK293 cells via CRISPR-SpCas9–mediated homologous recombination. Specifically, as shown in Fig. 2, a 5-nt barcode (5′-NNNNN-3′; complexity: 45 = 1024) was placed immediately upstream of a truncated cytomegalovirus [CMV; 225 base pairs (bp) versus 612 bp of the full-length CMV promoter] mKate construct and a phosphoglycerate kinase 1 promoter (PGK-1)-hygromycin resistance gene for drug selection. The safe harbor AAVS1 locus was chosen as the integration site to minimize potential disruption of normal cellular functions upon the stable integration of the transgenes (17). Subsequently, the genomic DNA from the resulting stable cells was collected and used as the polymerase chain reaction (PCR) template (table S1, primers P1 and P2) to isolate the complementary DNA (cDNA) transcript harboring the barcode, which were subsequently subjected to NGS (next-generation sequencing)–based amplicon sequencing. In total, 805 distinct barcodes were detected (Fig. 3 and table S2).
Fig. 2. Schematic illustration of implementation of CRISPR-PUFs.
Using CRISPR-Cas9 system, a set of synthetic constructs containing an array of 5–base pair (bp) barcodes (5′-NNNNN-3′), constitutive fluorescent reporter, and hygromycin resistance gene was stably integrated into the human AAVS1 safe harbor locus. Next, the cells were transiently transfected with CRISPR to induce NHEJ.
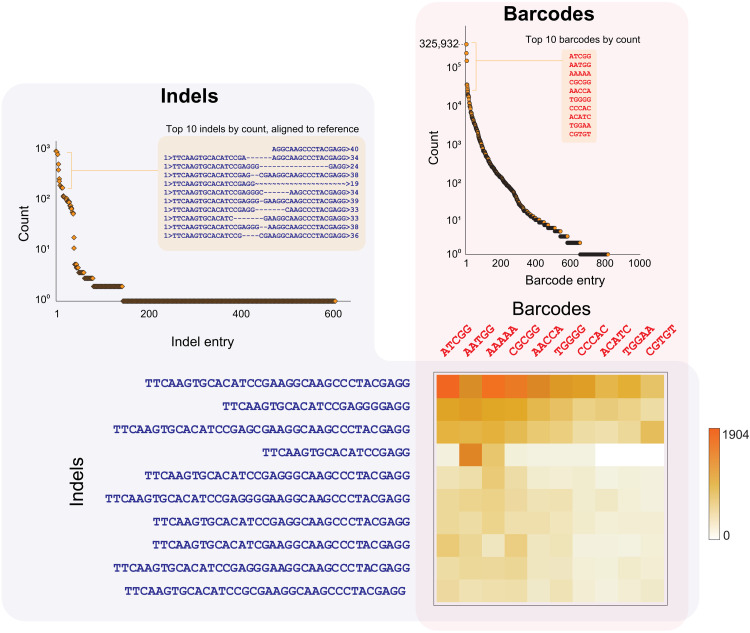
Fig. 3. Distribution of indels and barcodes that makeup a CRISPR-PUF.
(Right) Frequency of detected barcodes. In total, 805 unique barcodes were observed. (Left) Frequency of detected indels. In total, 569 unique indels were observed. (Bottom) Barcode/indel matrix. Heatmap presentation of the pilot CRISPR-PUF matrix consisting of the 10 most frequently occurring barcodes and indels. The color of each tile in the heatmap indicates the number of cells with the corresponding combination of indel and barcode.
Next, we aimed to combine the randomness of transfection into the barcoded cells and the inherent stochasticity of the cellular DNA error-repair processes to create a unique two-dimensional mapping between the barcodes and the indels. To this end, we initially screened five single-guide RNAs (sgRNAs; fig. S2) (18) for targeting efficiency by designing the sgRNA to target the open reading frame of the fluorescence reporter mKate. As shown in fig. S2, when cotransfected with SpCas9, all five sgRNAs efficiently suppressed the expression of mKate (sgRNA-5 was used for all subsequent experiments). We, therefore, proceeded by transiently transfecting the barcoded cell line with a sgRNA that targets adjacent to the integrated barcode to induce NHEJ repair.
Subsequently, the genomic DNA from the CRISPR-treated barcoded cell line was extracted, and the amplicons containing both the barcodes and the expected indel sequences were prepared using PCR (primers P1 and P2). This was followed by NGS sequencing (100-bp paired-end reads), which provided both the barcode sequence (forward end) and the indel sequence (reverse end). As shown in Fig. 3, in total, 569 distinct indels were observed (table S3), and the most frequently occurring indels demonstrated deletions of 1 to 16 nt flanking the predicted SpCas9 cutting site (cutting site: between 5′-CGAGGG-3′ and 5′-CGAAGG-3′; protospacer adjacent motif (PAM): AGG) (7).
The detected indels were associated with their corresponding barcodes from the same reads, and the resulting two-dimensional matrix was sorted by the frequencies of barcoded indels (table S4). As expected, CRISPR-mediated editing occurred in a subpopulation of a nonuniformly distributed barcoded cell population (table S2), resulting in 218 of the total 805 barcodes being present in the barcode and indels matrix. We provide the cropped matrix for the most frequently detected barcode and indel sequences in Fig. 3. By simple inspection, the utility of this matrix as a PUF to support authentication of mammalian cells becomes apparent: Using silicon PUF terminology, a vector of (barcode, indel) elements in this matrix can be used as a challenge, while the corresponding vector of frequencies can be used as the response.
Qualitative analysis of CRISPR-PUFs for uniqueness and robustness
Before relying on CRISPR-PUFs for attesting provenance of a cell line, we sought to evaluate their aptitude as PUFs (Fig. 1B). To this end, building upon our experience with the initial pilot experiments, we thoroughly assessed CRISPR-PUFs using the strategy illustrated in Fig. 4. With numerous PUFs constructed across various human cell lines, we performed individual PUF and pairwise comparisons to establish their robustness (i.e., their ability to produce matching signatures when a cell line is sequenced multiple times, e.g., at the vendor and at the customer site) and uniqueness (i.e., their ability to produce distinct signatures when multiple, identically produced copies of the same cell line are sequenced).
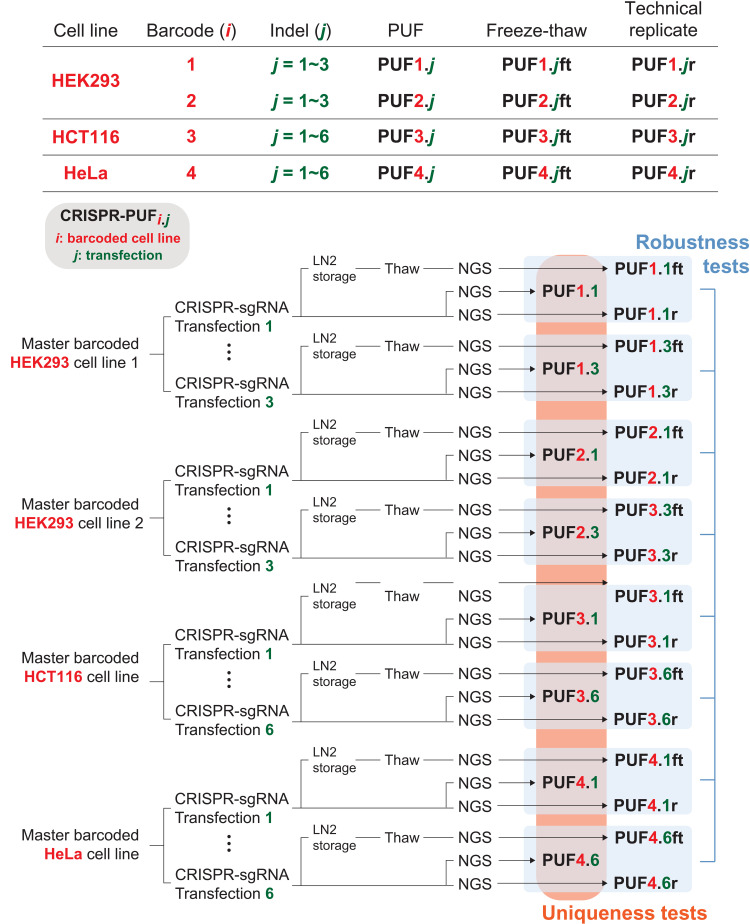
Fig. 4. List of all CRISPR-PUFs generated for this study.
In this study, two independently barcoded HEK293 cell lines were each transfected with identical sgRNA in three separate instances, resulting in a total of six unique PUFs. In addition, a HCT116 cell line and a HeLa cell line, each with a distinct set of barcodes, were each transfected six times to generated six unique PUFs for each cell line. A portion of each PUF was subjected to one cycle of freeze-thaw (denoted as ft) before proceeding with next-generation sequencing (NGS). To account for sequencing errors, PUFs from each barcoded cell line were also sequenced twice (denoted as r). Therefore, in total, 50 samples were sequenced via NGS, each resulting in a unique matrix of barcode/indel frequencies. To assess robustness to NGS measurement error, the matrix for each PUFi.j is compared against the matrix of its technical replicate PUFi.jr (i = {1, 2, 3, 4}). Similarly, to assess robustness to the freeze-thaw process, the matrix for each PUFi.j is compared against the matrix of its freeze-thaw counterpart PUFi.jft (i = {1, 2, 3, 4}). To assess uniqueness, the matrices of all PUFi.j (i = {1, 2, 3, 4}) are compared pairwise.
To facilitate such comparisons, two independently engineered, barcoded cell lines (barcoded cell line #1 and barcoded cell line #2) were prepared for HEK293 cells. In parallel, two additional barcoded cell lines were also generated for HCT116 (barcoded cell line #3) and HeLa (barcodes cell line #4) cells, respectively. Next, for each of the two cell lines derived from HEK293, we transfected the barcoded cells with the same sgRNA (sgRNA-5; fig. S2) three times (independent experiments), producing a total of six CRISPR-PUFs (PUF1.1, PUF1.2, and PUF1.3 from barcoded cell line #1 and PUF2.1, PUF2.2, and PUF2.3 from barcoded cell line #2). We also subjected all engineered cells to one cycle of freezing and thawing, resulting in PUF1.1ft, PUF1.2ft, and PUF1.3ft for barcoded cell line #1 and PUF2.1ft, PUF2.2ft, and PUF2.3ft for barcoded cell line #2. These CRISPR-PUFs were subjected to NGS analysis to produce the previously described barcode-indel matrix for each one of them. To incorporate and account for measurement errors introduced at the NGS step, PUF1.1 and PUF2.1 were sequenced twice, with the repeat results named PUF1.1r and PUF2.1r, respectively. Similarly, the two cell lines derived from HCT116 and HeLa were each subjected to six independent CRISPR-sgRNA treatments, and the resulting cells (PUF3.j and PUF4.j, respectively) were subjected to one cycle of freezing and thawing (PUFi.jft, i = {3, 4}, j = {1–6}), as well as repeated NGS sequencing (PUFi.jr, i = {3, 4}, j = {1–6}). All the CRISPR-PUFs produced by our experiments and used in our evaluation are summarized in Fig. 4.
To evaluate robustness, we compare the NGS-generated barcode/indel matrix of PUFi.j to those of PUFi.jr and PUFi.jft (i = {1, 2, 3, 4}), anticipating that they match (Fig. 4, robustness tests). Similarly, to evaluate CRISPR-PUF uniqueness that stems from the stochastic nature of NHEJ repair and the random association with the barcodes, we compare the NGS-generated barcode/indel matrix across all PUFs (Fig. 4, uniqueness tests), anticipating that they are distinct.
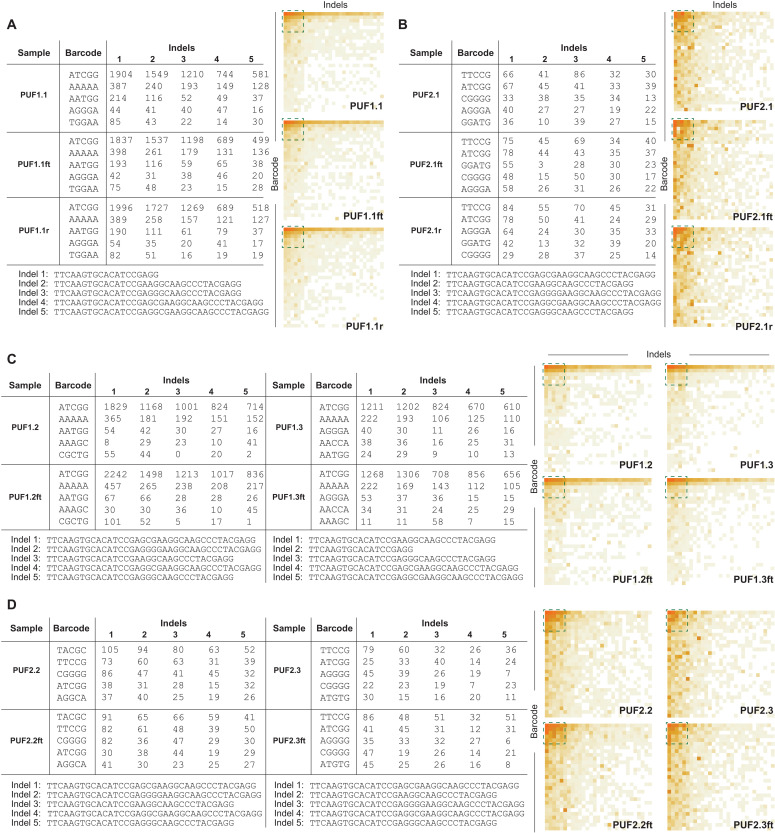
For a qualitative assessment, we focus on the most densely populated area of the barcode/indel matrix. As an example, in Fig. 5 (A and B), we provide the frequencies and sequences of the five most frequently observed barcodes and indels for PUFi.1, PUFi.1r, and PUFi.1ft (i = {1, 2}) from HEK293 cells. We also provide heatmaps of the 30 most frequently observed barcodes and indels (complete sequence data in tables S5 to S8). These remain qualitatively the same and suggest a high level of robustness among these samples (which will be quantified in the following sections).
Fig. 5. Qualitative assessment of CRISPR-PUFs generated using HEK293.
(A to D) Frequencies of barcode-indel addresses consisting of the five most commonly observed barcodes and indels (left) and heatmap based on the same data but expanded to the top 30 most commonly observed barcodes and indels (right) for a given PUF and its freeze-thaw counterparts and technical replicates (if applicable). The green dashed square on the heatmap represents the data shown on the table. Data shown in (A) and (B) are barcode-indel addresses for PUF1.1 (A) and PUF2.1 (B) with their respective freeze-thaw counterpart and technical replicate. Data shown in (C) and (D) are barcode-indel addresses for PUF1.2 and PUF1.3 (C) and PUF2.2 and PUF2.3 (D) with their respective freeze-thaw counterpart.
In contrast, different PUFs exhibit dissimilar patterns of the cropped CRISPR-PUF matrices (e.g., PUF1.2 and PUF1.3 in Fig. 5C) and different representation in the most frequently observed barcodes and indels. As an example, the third and fourth most frequently observed barcodes for PUF1.2 were 5′-AATGG-3′ and 5′-AAAGC-3′, while for PUF1.3, they were 5′-AGGGA-3′ and 5′-AACCA-3′, respectively. Similarly, the most frequent indel from PUF1.2 was 5′-TTCAAGTGCACATCCGAGG-3′, while for PUF1.3, it was 5′-TTCAAGTGCACATCCGAAGGCAAGCCCTACGAGG-3′ (table S5). These results suggest that a CRISPR-PUF identifier based on a combination of the barcode/indel sequences and their respective counts can satisfy both robustness and uniqueness.
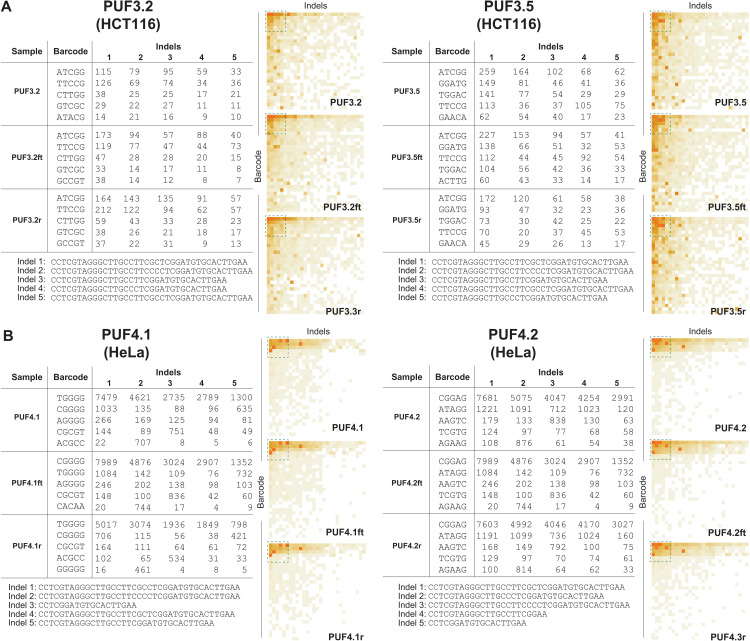
As mentioned earlier, we introduced six PUFs in each of two additional human cell lines (HCT116 and HeLa). Our sequencing results (all PUFs in figs. S3 and S4) show that, qualitatively, these PUFs also satisfy both robustness and uniqueness. We provide the frequencies and sequences of the five most frequently observed barcodes and indels for representative PUFs for both cell lines (Fig. 6). For example, for HCT116 cells, the heatmaps were visually similar among PUF3.2, PUF3.2ft, and PUF3.2r, while being distinct between PUF3.2 and the rest of the PUFs (Fig. 6A and fig. S3). Similarly, for HeLa cells, while the fifth most frequently observed indels from PUF4.2, PUF4.2ft, and PUF4.2r remained as 5′-CCTCGGATGTGCACTTGAA-3′, this sequence was not observed in the most frequent indel list (top five) from PUF4.1 sample (Fig. 6B and fig. S4). All barcode and indel sequences from HCT116 and HeLa are included in tables S9 and S10.
Fig. 6. Qualitative assessment of CRISPR-PUFs generated using HCT116 and HeLa.
Qualitative analysis of PUFs is shown, with HCT116 (A) and HeLa (B). In both cell types, heatmaps of barcode-indel addresses from intra-PUFs were visually similar, via different between inter-PUFs.
Quantitative analysis of CRISPR-PUFs for uniqueness and robustness
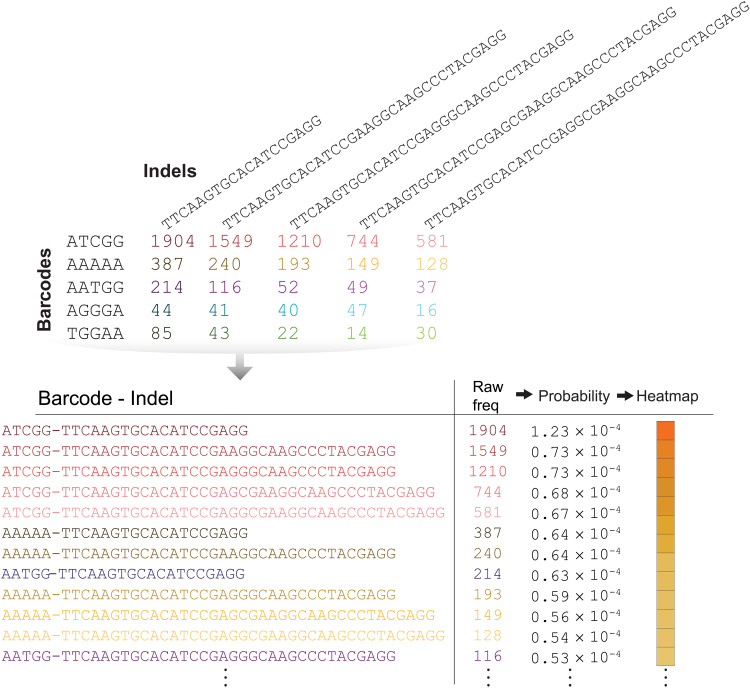
For provenance attestation, the end user of a CRISPR-PUF(ed) cell line must provide the NGS data (i.e., barcode/indel matrix), which is then compared against the values stored in a database to determine whether there is a match. To facilitate quantitative evaluation of the similarity between CRISPR-PUF matrices, we first concatenate the barcode and indel sequences to generate unique addresses (Fig. 7). This allows us to express each CRISPR-PUF as a probability distribution (table S11), based on the frequency of occurrence for each unique barcode-indel address.
Fig. 7. Quantitative assessment of CRISPR-PUFs.
To quantitatively assess the effectiveness of CRISPR-PUFs, the NGS result was converted to a frequency-based array of barcode-indel combinations. The corresponding probability density functions were then calculated to enable comparison between samples.
To perform a pairwise comparison between CRISPR-PUF derived from each cell line, we use a standard metric for computing distance between probability distributions, the total variation distance. The results (figs. S5 to S7) reveal that intra-PUF distances (defined as the variation between a specific CRISPR-PUFi.j and its corresponding repeat or freeze-thaw counterparts) are significantly smaller than inter-PUF distances (defined as the variation between two different CRISPR-PUFs) (table S12) in all three cell lines. As an example, in HEK293 cells, for each of the two PUFi families (i = {1, 2}), a threshold on total variation distance can be selected (i.e., 0.007 and 0.019) such that all intra-PUF distances are below threshold (indicating a match) and all inter-PUF distances are above threshold (indicating a no match). Similarly, such thresholds can also be established in PUFs derived from HCT116 and HeLa cells (0.037 for HCT116 and 0.013 for HeLa). This can also be visually confirmed by contrasting intra-PUF color intensity (i.e., inside the red boxes of figs. S5 to S7) to inter-PUF color intensity (i.e., outside the red boxes) for each of the four PUFi families (i = {1, 2, 3, 4}).
In practice, provenance attestation can be performed quantitatively by using the Bray-Curtis dissimilarity (19) between the end user’s CRISPR-PUF and the values stored in a database. The Bray-Curtis dissimilarity quantifies the compositional dissimilarity between two samples based on count of each species that make up the sample. The computation involves summing the absolute differences between the counts and dividing it by the sum of the abundances in the two samples (see Materials and Methods). Two samples of identical composition will yield Bray-Curtis dissimilarity value of 0, while two samples with no overlap between their composition will yield Bray-Curtis dissimilarity value of 1.
To demonstrate the use of the Bray-Curtis similarity in this context, we compute the intra-PUF and inter-PUF dissimilarities using the rank-ordered N most-frequent barcode-indel addresses of PUF1.1 as the reference (fig. S8A). As the number of used addresses increases toward the full list (N = 3478), we observe that the Bray-Curtis value between the reference (PUF1.1) and the CRISPR-PUFs originating from the same barcoded cell line #1 (i.e., PUF1.1r, PUF1.1ft, and PUF1.j, where j = {2, 3}) also increases (fig. S8B). On the other hand, the Bray-Curtis dissimilarity value(s) from the CRISPR-PUFs originating from barcoded cell line #2 (i.e., PUF2.j, where j = {1, 2, 3}) remains close to the maximum (fig. S8B). We also observe that it is possible to obtain appreciably different intra-PUF and inter-PUF values by using as few as N = 10 addresses (fig. S8C). We find it unnecessary to use the complete list, because the contribution of additional barcode-indel addresses to the difference between intra-PUF and inter-PUF Bray-Curtis dissimilarities diminishes as N increases. Overall, we observe that Bray-Curtis dissimilarity calculation using approximately 15% of the barcode-indel addresses (for all cell lines) results in lists that can provide an indisputable identification signature (figs. S8 to S13) while being sufficiently large to prevent unauthorized reproduction, as discussed later.
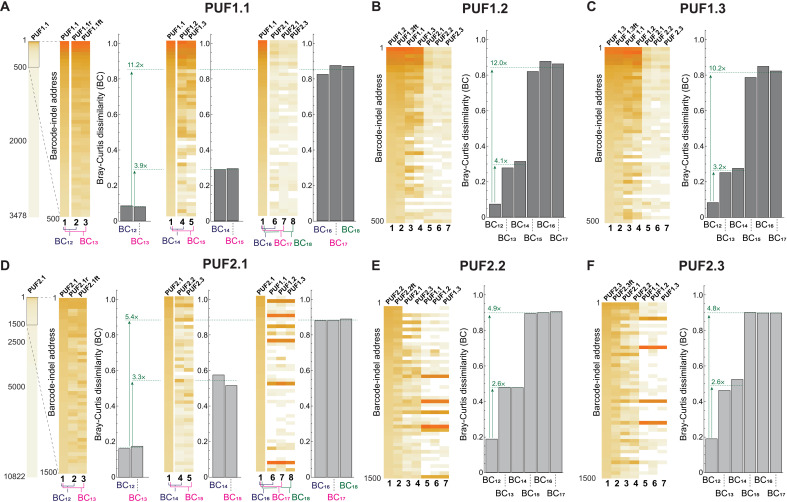
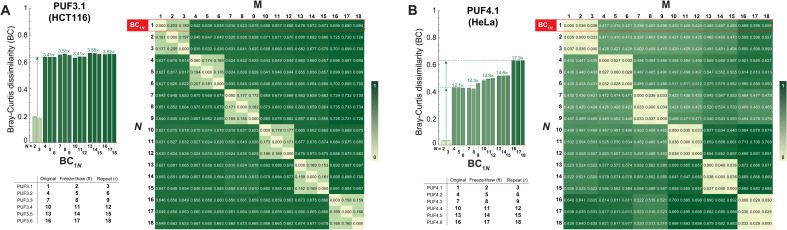
On the basis of the above observations, we calculated the Bray-Curtis dissimilarities between all the CRISPR-PUF in each of the three cell lines, each time using PUFi.j as a reference and comparing to its repeat and freeze-thaw versions, as well as to all other CRISPR-PUFs. As shown therein, a Bray-Curtis distance of 0.2 is an appropriate threshold for matching a CRISPR-PUF to its repeat and freeze-thaw counterparts in HEK293-derived PUFs (Fig. 8) while ensuring a no-match outcome when comparing to any other CRISPR-PUF. For any given PUFs generated in a HEK293 cell line, the intra-PUF Bray-Curtis dissimilarity value is never higher than 0.2, and the inter-PUF Bray-Curtis dissimilarity value of these PUFs against those generated using the same set of barcodes (e.g., PUF1.2 versus PUF1.3) is at least 2.6-fold higher than the corresponding intra-PUF Bray-Curtis dissimilarity value. When compared against PUFs generated from a different set of barcodes (e.g., PUF1.2 versus PUF2.2), the difference rises to a minimum of 4.8-fold and a maximum of 12-fold increase in Bray-Curtis dissimilarity. We observe that PUFs generated using HCT116 and HeLa cells show a similar trend (Fig. 9, A and B, respectively). As an example, using PUF3.1 as the reference, the inter-PUF Bray-Curtis dissimilarities were at least 3.4-fold higher than the corresponding intra-PUF dissimilarities (Fig. 9A and fig. S14), a pattern that was even more pronounced in HeLa-derived PUF4.1 (>12-fold differences between inter- and intra-PUF dissimilarities; Fig. 9B and fig. S15).
Fig. 8. Quantitative assessment of HEK293-derived CRISPR-PUFs using Bray-Curtis dissimilarity.
(A) The difference between each pair of barcode-indel arrays is quantified using the Bray-Curtis dissimilarity method. Before calculating the Bray-Curtis value, the barcode-indel arrays are trimmed down to approximately 15% of the total dataset (left). Then, the Bray-Curtis dissimilarity calculations against the reference PUF (i.e., PUF1.1) are made for three groups: (i) technical replicates (left), (ii) PUFs originating from the same barcoded cell line (center), and (iii) PUFs originating from a different barcoded cell line (right). Bray-Curtis values shown in (B) and (C) are results of an identical analysis as in (A) but using PUF1.2 and PUF1.3 as the reference, respectively. Bray-Curtis values shown in (D), (E), and (F) are analogous results to (A), (B), and (C), respectively, using PUF2.1, PUF2.2, and PUF.2.3 as the reference, respectively. Again, approximately 15% of the total dataset is used.
Fig. 9. Quantitative assessment of HCT116- and HeLa-derived CRISPR-PUFs using Bray-Curtis dissimilarity.
(A) Left: Comparison of Bray-Curtis dissimilarities for a single PUF (PUF3.1) generated in HCT116 against 17 other PUFs generated in the same cell line. The barcode-indel arrays are trimmed down as described previously in Fig. 8. Right: Matrix of pair-wise Bray-Curtis dissimilarity for all 18 PUFs generated in HCT116. Results shown in (B) are same analysis as before, with PUFs generated in HeLa cell line (PUF4.1).
We point out that a universal threshold is unnecessary, even if possible. In provenance attestation, it is sufficient to set an individual threshold for each cell line wherein a PUF has been introduced. Given a metric (e.g., Bray-Curtis dissimilarity), this threshold should be chosen to accept the signatures of all legitimately produced copies of the cell line, which the vendor stores in the CRP database, allowing a small margin to account for expected signature variation due to the freeze-thaw process or due to sequencing error, as further explained below. By individually setting this threshold for each cell line, we can optimize its ability to differentiate between PUF signatures of legitimately produced copies and illegitimate clones of a cell line.
In a noise-free case, the Bray-Curtis dissimilarity value would be 0 for valid PUFs. In reality, this is not the case. An important consideration here is that the Bray-Curtis values depend on the quality of the sequencing data. NGS is known to have a substitution error rate of 0.1 to 1% per base (20). Therefore, in addition to our repeated sequencing experiments (i.e., PUFi.jr) and to determine the worst-case Bray-Curtis dissimilarity values originating strictly from sequencing errors, for each of the reference PUFs derived from HEK293 cells, we generated 100 (artificially) mutated sequences using an error rate of 1% per base. Subsequently, the Bray-Curtis values between these mutated sequences and their PUF references were calculated using the rank-ordered barcode-indel addresses of the reference. Using these simulations, we calculated the upper bound for the Bray-Curtis dissimilarity for “valid” PUFs (fig. S16). As shown in table S13, the simulated worst-case dissimilarity values accurately match a CRISPR-PUF to its repeat and freeze-thaw counterparts while ensuring a no-match outcome when comparing to any other CRISPR-PUF. We note that the simulated worst-case dissimilarity values are different among PUF samples. This is expected because the underlying barcode distributions before applying the CRISPR-induced NHEJ are different and the absolute Bray-Curtis value depends on the average length of the sequencing reads (see “Bray-Curtis and sequencing reads” in the Supplementary Text section of the Supplementary Materials).
CRISPR-PUFs generate complexity necessary for function as PUFs
As described earlier in Fig. 1B, barcodes alone do not satisfy the properties required to qualify as a PUF. To validate this claim, we stably integrated a 5-nt barcode library into the AAVS1 locus of HEK293 cells in six parallel trials (BARCODE1 to BARCODE6, table S14) and subjected the samples to the two independent NGS-based amplicon sequencings. The overall barcode distribution patterns were notably similar among the repeats (fig. S17A). Next, the Bray-Curtis dissimilarities between a BARCODEi and its sequencing repeat (BARCODEir), as well as between two distinct samples, were calculated as before (fig. S17B). As shown in table S15, the intra-PUF dissimilarities generally overlapped with those of inter-PUFs (as an example, the Bray-Curtis dissimilarity between BARCODE2 and BARCODE2r was 0.013, which was higher than the Bray-Curtis dissimilarity between BARCODE3 and BARCODE4, which was 0.011). These results confirmed our conjecture that barcodes alone do not satisfy the uniqueness requirement and therefore are not suitable to be used as PUFs.
To further investigate the uniqueness of our generated PUFs, we performed additional computational analysis. Specifically, we tested whether the observed distribution of the barcode-indel addresses represents a unique combination of barcodes and indels that cannot be replicated. To achieve this, we randomly sampled a barcode sequence and an indel sequence from each of the reference HEK293-derived PUFs’ probability distribution functions and subsequently concatenated these two sequences to generate artificial combinations of barcode-indel addresses (fig. S18). The same number of concatenated addresses as in the original PUF was simulated to form a novel “resampled” PUF. Specifically, for each reference PUF, 100 resampled PUFs were generated. Next, the Bray-Curtis values between these simulated sequences and their PUF references were calculated (table S16). As shown in fig. S19, for all reference CRISPR-PUFs, the simulated inter-PUF dissimilarities (i.e., Bray-Curtis values between a reference and its reshuffled samples) are between 2.8× and 3.7× larger than intra-PUF dissimilarities (i.e., Bray-Curtis values between a reference and its repeat or freeze-thaw counterparts), and, additionally, are all larger than the worst-case dissimilarity values identified in our earlier analysis.
Collectively, these additional computational and experimental results confirm that CRISPR-PUFs satisfy both the robustness and the uniqueness criteria required for serving as a cell-line provenance attestation mechanism. We further posit that CRISPR-PUFs are also virtually impossible to replicate, thus unclonable. In the electronics industry, uniqueness and unclonability go hand-in-hand because silicon PUFs are inherent by-products of the randomness of semiconductor manufacturing. Even if the PUF function is known, manufacturing an exact clone is impossible. In biology, counterfeiting a CRISPR-PUF whose barcode-indel matrix is known would require DNA synthesis and integration of each individual sequence into a target cell line, followed by mixing the monoclonal cell populations to achieve the desired CRISPR-PUF frequencies. While gene synthesis is becoming cheaper and synthesizing each individual fragment is feasible, integration, single-cell isolation, mixing at desired proportions and, lastly, validation require prohibitive resource and time investment (see the “Reverse Engineering a CRISPR-PUF” section in the Supplementary Materials). Notably, the key determinants of synthesis costs and complexity (i.e., distance between the barcode and indel location and the number of barcode/indel combinations) are dictated by the CRISPR-PUF owner.
We note that other safe harbor sites, such as ROSA26 or CCR5, can be used to introduce PUFs. In addition, the human AAVS1 locus (GenBank, AC010327.8) is relatively large (146 kb), and, in theory, could accommodate multiple transgene integration events, especially when specialized genetic elements, such as insulators, are used to minimize the cross-talk between integrated cassettes.
DISCUSSION
Here, we exploit the complexity of barcode libraries and the inherent stochasticity of DNA error-repair induced via genome editing to engineer the first genetic PUFs in human cells. CRISPR-PUFs constitute a novel technology that can be used to establish security and trust in human cell engineering and synthetic biology applications. We demonstrate the use of the technology for provenance attestation in cell line distribution networks, but successful proliferation of genetic PUFs can be transformative in a wide range of applications.
Before silicon PUFs, the lack of provenance attestation methods fueled a counterfeiting industry (intellectual property theft through reverse engineering, illicit overproduction, integrated circuit recycling, remarking, etc.), resulting in an estimated (21) annual loss of $100 billion by legitimate semiconductor companies. The invention of silicon PUFs not only has significantly curtailed the problem but also has particularly succeeded in preventing counterfeiting of the latest cutting-edge products. Silicon PUFs were introduced for the purpose of providing a unique, robust, and unclonable digital fingerprint in each copy of a legitimately produced fabricated integrated circuit. While this digital fingerprint can be used as a key to support cryptographic algorithms, its main intent is provenance attestation of the integrated circuit.
We believe that the first application of CRISPR-PUFs will be for provenance attestation in cell line distribution networks. Recent advances in synthetic biology and genome editing (22–31) have enabled development of a broad range of engineered cells and have fueled emergence of a novel industry that seeks to produce specialized cell lines (32–35) and monetize them through commercial distribution networks. Many such highly customized proprietary cell lines are the result of extensive and expensive research and development efforts and come with price tags in the tens of thousands of dollars. Therefore, the legitimate producers of these valuable cell lines have a vested interest to protect their intellectual property and recover their investment by ensuring that their proprietary cell line does not get illicitly copied and distributed. At the same time, customers who acquire such expensive cell lines also have a vested interest in being assured of the origin (and, thereby, the quality) of their purchase, as well as holding proof of legitimate ownership of the cell line. In short, this emerging industry is in need of novel protocols for formally verifying the sale transaction of proprietary cell lines. As valuable cell lines continue to emerge, provenance attestation to protect the investment and intellectual property of the producing company from illegal replication and to authenticate each clients’ legitimate ownership of the purchased product is bound to become essential.
Moreover, cross-contamination or misidentification of cell lines due to poor handling, mislabeling, or procurement from dubious or undocumented sources is a rampant problem, resulting in innumerable financial and time losses (36–41). For example, a major German cell repository has reported that 20% of its human cell line stocks were cross-contaminated with other cell lines (36), and the China Center for Type Culture Collection demonstrated that 85% of cell lines in their repository, supposedly established from primary isolates, were actually HeLa cells (42). Such issues undermine quality, repeatability, and, ultimately, overall efficiency of medical research. Therefore, quality control and source verification provisions are paramount toward safeguarding against working with unsuitable cell line models and producing false data.
As provenance attestation takes place only once following thawing of the frozen sample and before initial use of the population, temporal stability is not necessary for the intended application. Once a customer has attested the provenance of a cell line after thawing, the option of subculturing and freezing the cell line again is available. Because temporal stability is maintained while frozen, any future use can again be attested using the CRISPR-PUF. This is analogous to the use of a silicon PUF for attesting provenance of an electronic chip every time the power is turned on. Inorganic physical PUFs also change over time due to silicon aging, but the time scales are, of course, different; silicon chips remain functional for a decade or more, while the cell cultures are usually propagated for a few months. Even so, silicon PUFs often include provisions (i.e., error correcting codes) for dealing with the degradation of the PUF responses over time, and one can implement a similar strategy in biology.
To explore the temporal stability of our current PUF designs, we created an additional HCT116-based PUF (PUF3.7) following the same protocol and subsequently passed this polyclonal cell line for 11 passages (~4 days per passage). Next, genomic DNA was extracted from cells collected at each passage number, and the amplicons containing barcodes and indels were PCR-amplified (protocol as described earlier here). Last, using a sample from passage 0 as the reference, the Bray-Curtis values were calculated for samples collected from each of the 11 passages. As shown in fig. S20 and table S17, the Bray-Curtis dissimilarity values increase along with the passage number, indicating that, as expected, PUF signatures change with cell propagation. When we compare this trend against the validation results obtained using the same HCT116-based PUFs (PUF3.1 to 3.6), we find that the Bray-Curtis dissimilarity due to temporal instability crosses the minimum observed inter-PUF dissimilarity of 0.6 at passage 6. Thus, our design can tolerate up to six rounds of passage or 20 days continuous cell culturing. This result provides a comfortable margin for ensuring robustness of CRISPR-PUFs for provenance at the point of sale, which is the application targeted by this manuscript.
From the customer perspective, authentication of ownership via CRISPR-PUF can assure them that they have a unique copy of a cell line, a subculture whose origin from a desired cell line has been attested. Therefore, successful proliferation of such genetic PUFs can be transformative for intellectual property protection of engineered cell lines. Companies can introduce CRISPR-PUFs to their cells to enable unique authorization and validation (fig. S21), laboratories across the world may use this technology as a starting point for validating point of source, and funding agencies and journals may require CRISPR-PUFs in published documents and reports for quality control and for ensuring reproducibility.
MATERIALS AND METHODS
Cell culture and transient transfection
The HEK293 cells (catalog number CRL-1573), HCT116 cells (catalog number CCL-247), and HeLa cells (catalog number CCL-2) were acquired from the American Type Culture Collection and maintained at 37°C, 100% humidity, and 5% CO2. The cells were grown in Dulbecco’s modified Eagle’s medium (Invitrogen, catalog number 11965-1181) supplemented with 10% fetal bovine serum (Invitrogen, catalog number 26140), 0.1 mM minimal essential medium nonessential amino acids (Invitrogen, catalog number 11140-050), and penicillin (0.045 U/ml) and streptomycin (0.045 U/ml) (penicillin-streptomycin liquid; Invitrogen, catalog number 15140). To pass the cells, the adherent culture was first washed with phosphate-buffered saline (PBS; Dulbecco’s PBS; MediaTech, catalog number 21-030-CM), then trypsinized with trypsin-EDTA (0.25% trypsin with EDTAX4Na; Invitrogen, catalog number 25200), and lastly diluted in fresh medium. For transient transfection, ~300,000 cells in 1 ml of complete medium were plated into each well of 12-well culture–treated plastic plates (Greiner Bio-One, catalog number 665180) and grown for 16 to 20 hours. All transfections were then performed using 1.75 μl of jetPRIME (Polyplus Transfection) and 75 μl of jetPRIME buffer. The transfection mixture was then applied to the cells and mixed with the medium by gentle shaking.
Flow cytometry
Forty-eight to 72 hours after transfection, cells from each well of the 12-well plates were trypsinized with 0.1 ml of 0.25% trypsin-EDTA at 37°C for 3 min. Trypsin-EDTA was then neutralized by adding 0.9 ml of complete medium. The cell suspension was centrifuged at 1000 rpm for 5 min, and, after removal of supernatants, the cell pellets were resuspended in 0.5 ml of PBS buffer. The cells were analyzed on a BD LSRFortessa flow analyzer. Cyan fluorescent protein (CFP) was measured with a 445-nm laser and a 515/20-nm band-pass filter, and mKate was measured with a 561-nm laser, 610-nm emission filter, and 610/20-nm band-pass filter. For data analysis, 100,000 events were collected. A forward scatter/side scatter gate was generated using an untransfected negative sample and applied to all cell samples. The mKate and CFP readings from untransfected HEK293 cells were set as baseline values and were subtracted from all other experimental samples. The normalized mKate values (mKate/CFP) were then collected and processed by FlowJo. All experiments were performed in triplicates.
Generation of barcoded stable cells
To generate the barcoded stable cells, ~10 million of the cells were seeded onto a 10-cm petri dish. Sixteen hours later, the cells were transiently transfected with 1 μg of the donor plasmid (barcode-truncated CMV-mKate-PGK1-hygromycin resistance gene) and 9 μg of CMV-SpCas9-U6-AAVS1/sgRNA plasmid using the jetPRIME reagent (Polyplus Transfection). Forty-eight hours later, hygromycin B (Thermo Fisher Scientific, catalog number 10687010) was added at the final concentration of 200 μg/ml. The selection lasted ~2 weeks, after which the surviving clones were pooled to generate the polyclonal stable cells. The barcoded stable cells were further expanded and maintained in the complete growth medium containing hygromycin (200 μg/ml).
NGS-based amplicon sequencing
To determine the abundance of the barcode and indel sequences, total genomic DNA was isolated from CRISPR-PUF cells transfected with CMV-SpCas9-U6-sgRNA5 using the DNeasy Blood & Tissue Kit (QIAGEN, catalog number 69504). cDNA fragments harboring both barcode and expected indel sequences were PCR-amplified by using ~100 ng of the genomic DNA and primers P1 and P2, which added the 5′-overhang adapter sequence P12 and the 3′-overhang adapter sequence P13 for subsequent Illumina NGS amplicon sequencing. The PCR conditions were as follows: first one cycle of 30 s at 98°C, followed by 40 cycles of 10 s at 98°C, 30 s at 60°C, and 1 min at 72°C. The purified PCR products were then subjected to NGS-based amplicon sequencing (Illumina 100-bp paired-end sequencing), which was performed at the Genome Sequencing Facility at The University of Texas Health Science Center at San Antonio. One million individual reads were generated for each sample.
Total variation distance
The total variation distance, δTVD, between two probability measures P and Q for a countable sample space Ω is equal to the half of the L1 norm of these distributions or equivalently, half of the elementwise sum of the absolute difference of P and Q, as defined in Eq. 1
| (1) |
In addition, the total variation distance is the area between the two probability distribution curves defined as CP ≝ {(ω, P(ω)}ω∈Ω and CQ ≝ {(ω, Q(ω)}ω∈Ω. It can be shown that for a finite set Ω, the total variation distance is equal to the largest difference in probability, taken over all subsets of Ω, i.e., all possible events.
Bray-Curtis dissimilarity
The Bray-Curtis dissimilarity δBC between two vectors u and v of same length n is defined in Eq. 2
| (2) |
The Bray-Curtis dissimilarity has values between 0 and 1 when all coordinates are positive.
Acknowledgments
Funding: This work was funded by the University of Texas at Dallas SPIRE mechanism and partially by U.S. National Science Foundation (NSF) CAREER grant 1351354 and NSF 1361355, a Cecil H. and Ida Green Endowment, and the University of Texas at Dallas.
Author contributions: Y.L. and M.M.B. performed research. Y.L., L.B., M.M.B, and T.K. processed the data. All authors wrote the manuscript. L.B., Y.L., and Y.M. designed the experiments and analysis. L.B. supervised the project.
Competing interests: L.B., Y.L., and Y.M. are named as inventors on pending patent, serial no. PCT/US2021/033108 published on 25 November 2021 as WO 2021-236740 A2. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S21
Supplementary Text
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S17
Scripts S1 and S2
REFERENCES AND NOTES
- 1.Rührmair U., Sölter J., Sehnke F., On the foundations of physical unclonable functions. Cryptol. ePrint Arch. , 1–20 (2009). [Google Scholar]
- 2.Herder C., Yu M. D., Koushanfar F., Devadas S., Physical unclonable functions and applications: A tutorial. Proc. IEEE 102, 1126–1141 (2014). [Google Scholar]
- 3.McGrath T., Bagci I. E., Wang Z. M., Roedig U., Young R. J., A PUF taxonomy. Appl. Phys. Rev. 6, 011303 (2019). [Google Scholar]
- 4.Gao Y., Al-Sarawi S. F., Abbott D., Physical unclonable functions. Nat. Electron. 3, 81–91 (2020). [Google Scholar]
- 5.B. Gassend, D. Clarke, M. van Dijk, S. Devadas, in Proceedings of the 9th ACM Conference on Computer and Communications Security - CCS ‘02 (ACM Press, New York, New York, USA, 2002), pp. 148–160. [Google Scholar]
- 6.van Overbeek M., Capurso D., Carter M. M., Thompson M. S., Frias E., Russ C., Reece-Hoyes J. S., Nye C., Gradia S., Vidal B., Zheng J., Hoffman G. R., Fuller C. K., May A. P., DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell 63, 633–646 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Chen W., McKenna A., Schreiber J., Haeussler M., Yin Y., Agarwal V., Noble W. S., Shendure J., Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Res. 47, 7989–8003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelson T., Heckl D., Ebert B. L., Root D. E., Doench J. G., Zhang F., Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E., A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E., Church G. M., RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y. Y., Nowak C. M. C. M., Withers D., Pertsemlidis A., Bleris L., CRISPR-based editing reveals edge-specific effects in biological networks. Cris. J. 1, 286–293 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Hsu P. D., Lander E. S., Zhang F., Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ran F. A., Cong L., Yan W. X., Scott D. A., Gootenberg J. S., Kriz A. J., Zetsche B., Shalem O., Wu X., Makarova K. S., Koonin E. V., Sharp P. A., Zhang F., In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert L. A. A., Larson M. H. H., Morsut L., Liu Z., Brar G. A. A., Torres S. E. E., Stern-Ginossar N., Brandman O., Whitehead E. H. H., Doudna J. A. A., Lim W. A., Weissman J. S., Qi L. S., CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., Lim W. A., Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang L., Mali P., Kim-Kiselak C., Church G., CRISPR-Cas-mediated targeted genome editing in human cells. Methods Mol. Biol. 1114, 245–267 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Sadelain M., Papapetrou E. P., Bushman F. D., Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer 12, 51–58 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Nowak C. M., Lawson S., Zerez M., Bleris L., Guide RNA engineering for versatile Cas9 functionality. Nucleic Acids Res. 44, 9555–9564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bray J. R., Curtis J. T., An ordination of the upland forest communities of southern wisconsin. Ecol. Monogr. 27, 325–349 (1957). [Google Scholar]
- 20.Petrackova A., Vasinek M., Sedlarikova L., Dyskova T., Schneiderova P., Novosad T., Papajik T., Kriegova E., Standardization of sequencing coverage depth in NGS: Recommendation for detection of clonal and subclonal mutations in cancer diagnostics. Front. Oncol. 9, 851 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guin U., Huang K., DiMase D., Carulli J. M., Tehranipoor M., Makris Y., Counterfeit integrated circuits: A rising threat in the global semiconductor supply chain. Proc. IEEE 102, 1207–1228 (2014). [Google Scholar]
- 22.Rinaudo K., Bleris L., Maddamsetti R., Subramanian S., Weiss R., Benenson Y., A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 25, 795–801 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Moore R., Spinhirne A., Lai M. J., Preisser S., Li Y., Kang T., Bleris L., CRISPR-based self-cleaving mechanism for controllable gene delivery in human cells. Nucleic Acids Res. 43, 1297–1303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinberg B. H., Pham N. T. H., Caraballo L. D., Lozanoski T., Engel A., Bhatia S., Wong W. W., Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells. Nat. Biotechnol. 35, 453–462 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim T., Lu T. K., CRISPR/Cas-based devices for mammalian synthetic biology. Curr. Opin. Chem. Biol. 52, 23–30 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Chavez A., Scheiman J., Vora S., Pruitt B. W., Tuttle M., Iyer E. P. R., Lin S., Kiani S., Guzman C. D., Wiegand D. J., Ter-Ovanesyan D., Braff J. L., Davidsohn N., Housden B. E., Perrimon N., Weiss R., Aach J., Collins J. J., Church G. M., Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., Zhang F., Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leisner M., Bleris L., Lohmueller J., Xie Z., Benenson Y., Rationally designed logic integration of regulatory signals in mammalian cells. Nat. Nanotechnol. 5, 666–670 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lapique N., Benenson Y., Genetic programs can be compressed and autonomously decompressed in live cells. Nat. Nanotechnol. 13, 309–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao X. J., Chong L. S., Kim M. S., Elowitz M. B., Programmable protein circuits in living cells. Science 361, 1252–1258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krzysztoń R., Wan Y., Petreczky J., Balázsi G., Gene-circuit therapy on the horizon: Synthetic biology tools for engineered therapeutics*. Acta Biochim. Pol. 68, 377–383 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aijaz A., Li M., Smith D., Khong D., LeBlon C., Fenton O. S., Olabisi R. M., Libutti S., Tischfield J., Maus M. V., Deans R., Barcia R. N., Anderson D. G., Ritz J., Preti R., Parekkadan B., Biomanufacturing for clinically advanced cell therapies. Nat. Biomed. Eng. 2, 362–376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J. S., Grav L. M., Lewis N. E., Faustrup Kildegaard H., CRISPR/Cas9-mediated genome engineering of CHO cell factories: Application and perspectives. Biotechnol. J. 10, 979–994 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Donohoue P. D., Barrangou R., May A. P., Advances in industrial biotechnology using CRISPR-cas systems. Trends Biotechnol. 36, 134–146 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Quarton T., Kang T., Papakis V., Nguyen K., Nowak C., Li Y., Bleris L., Uncoupling gene expression noise along the central dogma using genome engineered human cell lines. Nucleic Acids Res. 48, 9406–9413 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Capes-Davis A., Theodosopoulos G., Atkin I., Drexler H. G., Kohara A., MacLeod R. A. F., Masters J. R., Nakamura Y., Reid Y. A., Reddel R. R., Freshney R. I., Check your cultures! A list of cross-contaminated or misidentified cell lines. Int. J. Cancer 127, 1–8 (2010). [DOI] [PubMed] [Google Scholar]
- 37.MacLeod R. A. F., Dirks W. G., Matsuo Y., Kaufmann M., Milch H., Drexler H. G., Widespread intraspecies cross-contamination of human tumor cell lines arising at source. Int. J. Cancer 83, 555–563 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Dirks W. G., MacLeod R. A. F., Nakamura Y., Kohara A., Reid Y., Milch H., Drexler H. G., Mizusawa H., Cell line cross-contamination initiative: An interactive reference database of STR profiles covering common cancer cell lines. Int. J. Cancer 126, 303–304 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Lichter P., Allgayer H., Bartsch H., Fusenig N., Hemminki K., Doeberitz M. V. K., Kyewski B., Miller A. B., Zur Hausen H., Obligation for cell line authentication: Appeal for concerted action. Int. J. Cancer 126, 1 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Freshney R. I., Database of misidentified cell lines. Int. J. Cancer 126, 302 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Cheung S. T., Chan S. L., Lo K. W., Contaminated and misidentified cell lines commonly use in cancer research. Mol. Carcinog. 59, 573–574 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Ye F., Chen C., Qin J., Liu J., Zheng C., Genetic profiling reveals an alarming rate of cross-contamination among human cell lines used in China. FASEB J. 29, 4268–4272 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S21
Supplementary Text
Tables S1 to S17
Scripts S1 and S2