Abstract
Single-gene mutations account for more than 6000 diseases, 10% of all pediatric hospital admissions, and 20% of infant deaths. Down syndrome and other aneuploidies occur in more than 0.2% of births worldwide and are on the rise because of advanced reproductive age. Birth defects of genetic origin can be diagnosed in utero after invasive extraction of fetal tissues. Noninvasive testing with circulating cell-free fetal DNA is limited by a low fetal DNA fraction. Both modalities are unavailable until the end of the first trimester. We have isolated intact trophoblast cells from Papanicolaou smears collected noninvasively at 5 to 19 weeks of gestation for next-generation sequencing of fetal DNA. Consecutive matched maternal, placental, and fetal samples (n = 20) were profiled by multiplex targeted DNA sequencing of 59 short tandem repeat and 94 single-nucleotide variant sites across all 24 chromosomes. The data revealed fetal DNA fractions of 85 to 99.9%, with 100% correct fetal haplotyping. This noninvasive platform has the potential to provide comprehensive fetal genomic profiling as early as 5 weeks of gestation.
INTRODUCTION
According to the World Health Organization, single-gene disorders, such as thalassemia (1:8000 pregnancies in some regions) and sickle cell anemia (1:500 pregnancies among African Americans), are a major health burden. Demographic shifts, including advanced maternal age (>35 years), increase the overall risk for fetal chromosomal abnormalities that cause congenital birth defects. Down syndrome is estimated to affect more than 250,000 families in the United States. Thousands of inherited genetic disorders and de novo mutations can now be diagnosed and screened to manage the health of affected individuals where testing is available. It is advantageous to identify inherited disease early in life, even before birth, and to provide immediate care in utero for metabolic disorders such as congenital adrenal hyperplasia, biotin-responsive multiple carboxylase deficiency, B12-responsive methylmalonic aciduria, and others.
Prenatal testing for genetic disorders can currently be performed invasively using amniocentesis (no earlier than 12 weeks) or chorionic villous sampling (CVS; no earlier than 9 weeks) (1, 2), and these procedures have a limited but definite risk of complications. In addition, preimplantation genetic screening is available as an adjunct to in vitro fertilization (3).
In the past five decades, tremendous efforts have been made to develop reliable, noninvasive, early, and safe prenatal testing techniques using intact fetal erythrocytes, cell-free fetal DNA, and endocervical trophoblast cells (4–6). These approaches have identified chromosomal abnormalities, sex-linked diseases, and Rhesus blood group D genotype in fetuses. Prenatal screening of cell-free fetal DNA in maternal blood (earliest at 8 weeks) has been implemented clinically with some restrictions. The technology relies on fetal DNA fragments that rapidly turn over in maternal blood, with analysis by massively parallel sequencing to distinguish fetal from maternal haplotypes (1, 4, 7). However, the small fetal fraction (4 to 10% at 10 weeks of gestation) and fragmentation of degrading DNA [146 base pair (bp)] in the circulation limit its reliability for probing a broad range of major genetic anomalies (7, 8). The fetal fraction of DNA rises steadily with gestational age but must exceed 4% for accurate and reliable assessment of specific trisomies (8). With a low fetal fraction, it is particularly challenging to detect a fetal haplotype that is overrepresented in the maternal fraction of DNA, such as a male fetus at risk for an X-linked disorder (7). Digital polymerase chain reaction (PCR) assays can detect autosomal recessive sickle cell anemia (9) and X-linked hemophilia (10) but rely on exact quantification of the fetal fraction, which can be problematic in the first trimester because of the overwhelming amount of copurifying maternal DNA. The American College of Obstetricians and Gynecologists recommends that positive cell-free DNA results should be verified by the invasive tests CVS and amniocentesis, which are considered diagnostic (11).
A simple, reliable, and safe approach to fully interrogate the intact fetal genome early in the first trimester of pregnancy at single-nucleotide resolution currently does not exist. To address and close this gap, we developed a straightforward alternative that uses a Papanicolaou (Pap) smear to capture intact fetal trophoblast cells in numbers sufficient for next-generation sequencing as early as 5 weeks of gestation. Fetal cells with a placenta (trophoblast)–like cellular phenotype naturally migrate from the conceptus into the reproductive tract by a poorly understood mechanism (12–14). To date, their presence offered little benefit for non-invasive testing due to low cell recovery and a 2000-fold excess of maternal cells and DNA in endocervical specimens (15). We first reported that fetal trophoblast cells can be purified to near homogeneity by trophoblast retrieval isolation from the cervix (TRIC) using Pap smears safely collected during early pregnancy with a nylon cytobrush from the lower segment of the endocervical canal (16–18). In contrast to cell-free fetal DNA (8), we showed that isolation of fetal trophoblast cells by TRIC is unaffected by gestational age between 5 and 20 weeks, or by maternal obesity (19), thus providing a robust source of fetal DNA during a period of pregnancy when it is most useful clinically.
Here, we report the development of a DNA isolation and genotyping protocol that provides a high fetal fraction from trophoblast cells obtained by TRIC during ongoing pregnancies at 5 to 19 weeks of gestation (Fig. 1). Fetal DNA was assessed in 20 specimens using multiplex targeted sequencing to distinguish it from maternal DNA, based on the profiling of 153 short tandem repeat (STR) and single-nucleotide variant (SNV) sites across the entire genome.
Fig. 1. Trophoblast isolation, assessment of cell purity, and fetal DNA isolation for sequencing.
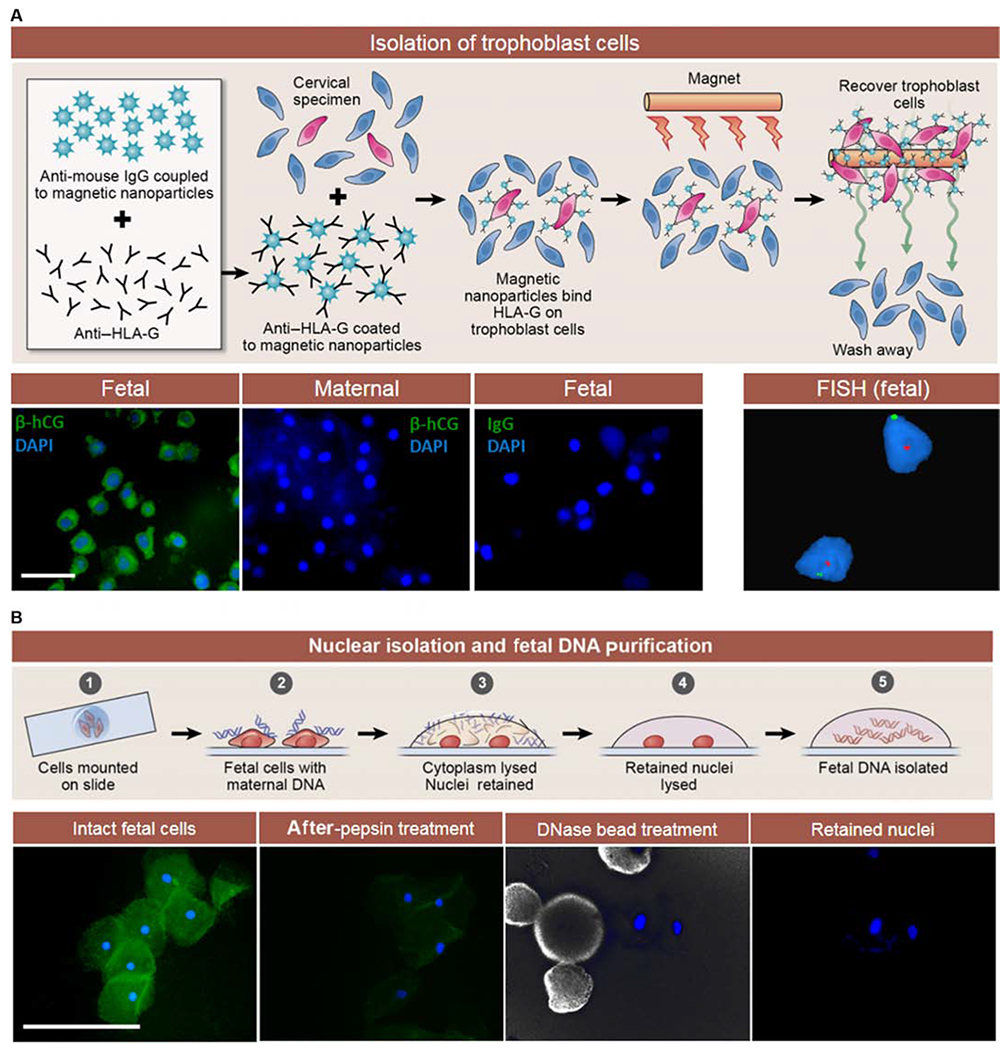
(A) Endocervical specimen collection and immunomagnetic separation of trophoblast cells (red) using anti–human leukocyte antigen–G (anti–HLA-G)–coated magnetic nanoparticles. Bottom: Trophoblast purity assessment by immunofluorescence staining for β-hCG (green) showing negative results for maternal cells and immunoglobulin G (IgG) control (first three panels). Screening for gender was performed by FISH of the X and Y chromosomes (right panel, male sample). (B) Top: Nuclear DNA isolation from the fetal trophoblast cells mounted on microscope slides (step 1). Maternal DNA is blue, and intact fetal trophoblast cells are red (step 2). Fetal nuclei remain adherent to slides after limited cell proteolysis (step 3). Nuclei are washed and lysed (step 4), providing fetal DNA (step 5). The lower panels show immunofluorescence microscopy images of fetal trophoblast cells during the DNA isolation procedure. F-actin is labeled with fluorescein isothiocyanate–phalloidin (green), and nuclei are labeled with 4’,6’-diamidino-2-phenylindole (DAPI) (blue). Pepsin digestion removed F-actin. Deoxyribonuclease (DNase) immobilized on beads was used to degrade residual maternal DNA. Nuclei were then available for fetal DNA purification. Scale bars, 100 μm.
RESULTS
Isolation and characterization of trophoblast cells
By TRIC (16), maternal and fetal trophoblast cells were isolated from consecutive endocervical specimens (n = 20), as illustrated in Fig. 1A, at gestational ages ranging from 5 to 19 weeks (8.3 ± 3.6 weeks; Table 1). Matched maternal cells and placental tissues were used as references for analysis. An average of 282 trophoblast cells (range, 170 to 410) was separated from maternal cells that were in excess of 200,000 cells. Purity of fetal cells was assessed by staining for expression of the trophoblast-specific protein, β-subunit human chorionic gonadotropin (β-hCG). On average, 89.2% (±5.0%) of the isolated fetal trophoblast cells stained positive for β-hCG, which was completely absent in maternal cells (Table 1 and Fig. 1A, lower panel). The gender of the isolated cells was determined by fluorescence in situ hybridization (FISH) for the X and Y chromosomes. In pregnancies with a male fetus, the fetal trophoblast cells showed positive probe binding for both X and Y chromosomes, as expected (Fig. 1A, lower panel). Fetal and maternal DNA were obtained from 234 (±75) and 335 (±104) cells, respectively, using endocervical specimens (n = 20) and small pieces of placental tissue (Table 1). Initially, fetal DNA obtained from pooled trophoblast cells contained large amounts of maternal DNA (fig. S1). For fetal trophoblast cells, nuclear isolation combined with DNase treatment after TRIC was necessary (fig. S1) to ensure high-quality fetal DNA samples (Fig. 1).
Table 1.
Overview of samples and results for targeted sequencing.
| Sample ID | Gestational age (weeks) | Trophoblast cell purity (% β-hCG) | Fetal gender | Autosomal STR alleles in TRIC | % Fetal fraction (% maternal fraction) | Number of SNVs in fetal trophoblast cells | |||
|---|---|---|---|---|---|---|---|---|---|
| Median | Range | After threshold (% of total number of SNVs) | Number of discriminatory SNV (% of total number of SNV after threshold) | Incorrectly called in fetal trophoblast cells after threshold | |||||
| SA | 5 | 86 | Male | 94.8 | 85.6–100 | 97.6 (2.4) | 93 (98.9) | 41 (44.0) | 0 |
| SB | 5 | 89 | Female | 85.5 | 75.1–100 | 92.2 (7.8) | 77 (81.9) | 31 (40.2) | 0 |
| SC | 5 | 91 | Female | 89.1 | 73.5–100 | 87.2 (12.8) | 82 (87.2) | 39 (47.5) | 0 |
| SD | 6 | 85 | Female | 91.9 | 80.8–98.7 | 95.2 (4.8) | 89 (94.7) | 40 (44.9) | 0 |
| SE | 6 | 92 | Male | 86.4 | 85.7–96.9 | 85.6 (14.4) | 62 (66.0) | 25 (40.3) | 0 |
| SF | 6 | 93 | Male | 88.1 | 73.2–99.4 | 75.6 (24.4) | 55 (58.5) | 11 (20.0) | 0 |
| SG | 6 | 89 | Male | 93.3 | 84.9–100 | 93.9 (6.1) | 91 (96.8) | 45 (49.4) | 0 |
| SH | 6 | 86 | Female | 92.7 | 77.8–98.5 | 98.6 (1.4) | 89 (94.7) | 47 (52.8) | 0 |
| SI | 6 | 89 | Male | 95 | 88.9–100 | 100 (0.0) | 90 (95.7) | 35 (38.8) | 0 |
| SJ | 7 | 92 | Female | 89 | 77.7–96.6 | 82.1 (17.9) | 68 (72.3) | 19 (27.9) | 0 |
| SK | 8 | 80 | Female | 89 | 72.3–100 | 82.4 (17.6) | 74 (78.7) | 22 (29.7) | 0 |
| SL | 8 | 96 | Female | 92 | 79.6–99.4 | 97.2 (2.8) | 87 (92.6) | 38 (43.6) | 0 |
| SM | 8 | 96 | Female | 90.2 | 78.3–100 | 90.8 (9.2) | 91 (96.8) | 44 (48.3) | 0 |
| SN | 8 | 92 | Female | 93.3 | 76.5–99.2 | 92.8 (7.2) | 90 (95.7) | 42 (46.6) | 0 |
| SO | 9 | 85 | Male | 93.6 | 79.8–100 | 92.9 (7.1) | 90 (95.7) | 46 (51.1) | 0 |
| SP | 9 | 94 | Male | 91.9 | 81.9–100 | 100 (0.0) | 92 (97.9) | 37 (40.2) | 0 |
| SQ | 11 | 85 | Female | 91.9 | 73.8–97.6 | 95.9 (4.1) | 89 (94.7) | 39 (43.8) | 0 |
| SR | 13 | 91 | Male | 87.4 | 77.7–100 | 91.9 (8.1) | 77 (81.9) | 41 (53.2) | 0 |
| SS | 14 | 90 | Male | 91.7 | 81.8–100 | 92.6 (7.4) | 93 (98.9) | 44 (47.3) | 0 |
| ST | 19 | 80 | Female | 93.2 | 85.3–99.5 | 98.7 (1.3) | 92 (97.9) | 48 (52.1) | 0 |
Targeted multiplex next-generation sequencing
We determined whether fetal DNA obtained by TRIC had the fidelity for reliable detection of single-nucleotide mutations, as well as small DNA insertions and deletions. To do this, we used the ForenSeq platform to genotype and distinguish the fetus and the mother, using small quantities of DNA (20–22) to target 59 STRs (27 autosomal, 7 X, and 24 Y haplotype markers) and 94 SNVs on all chromosomes (table S1). On average, 1.9 ± 0.9 ng of DNA from fetal trophoblast cells and matched placental and maternal cells (n = 20) was sequenced. All 153 loci (table S1) were sequenced at a mean sequencing depth of 5268 ± 1714 per STR and 760 ± 218 per SNV. The fetal nuclear isolation strategy developed here generated high-resolution STR profiles, detailing multiple shared and discriminatory alleles between mother and fetus. Detailed analysis revealed concordance between allelic profiles of fetal DNA obtained by TRIC and reference placental DNA, which were both distinct from maternal DNA, and provided 100% correct fetal gender identification in all samples compared to fetal reference DNA (Fig. 2A and Table 1).
Fig. 2. Targeted sequencing of fetal trophoblast cells obtained by TRIC.
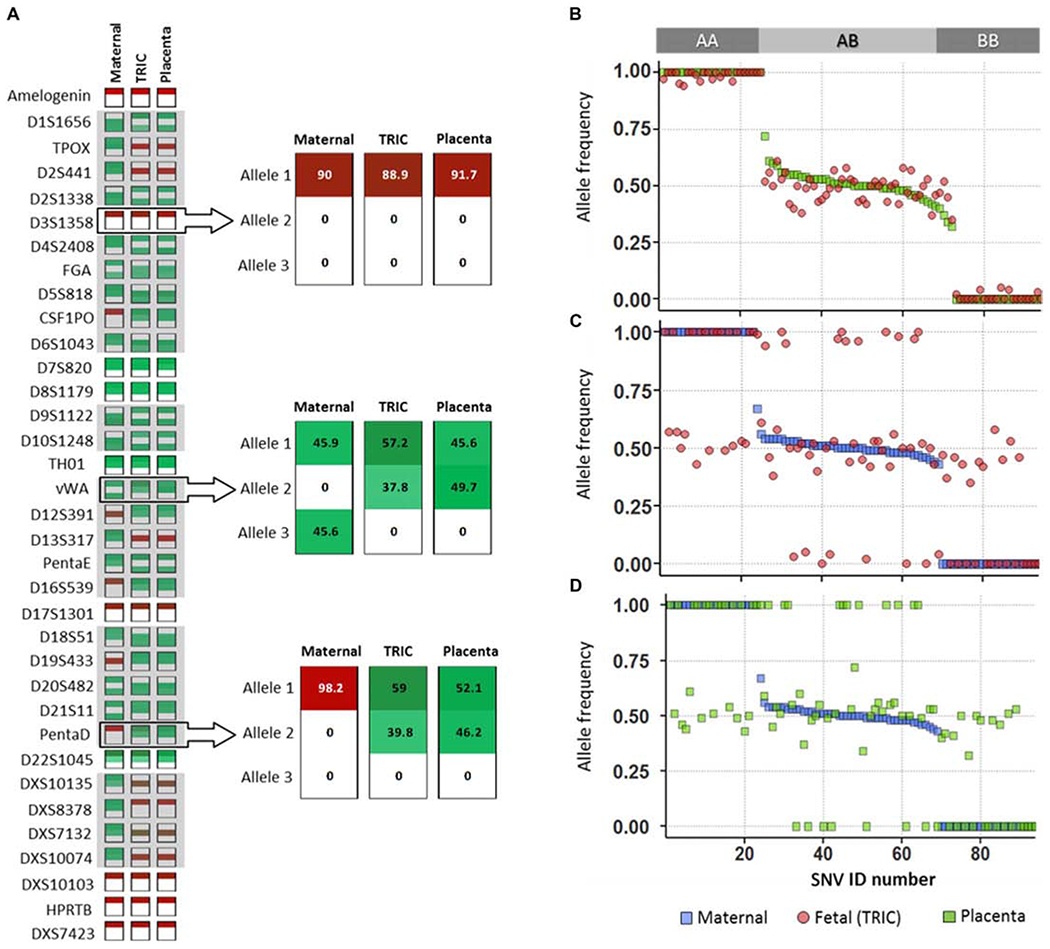
(A) Graphical representation of STR profiles of DNA triads composed of maternal cells (Maternal), fetal trophoblast cells (TRIC), and placental tissue (Placenta) from sample SN (Table 1). For eachDNA sample, the two dominant alleles with the highest relative peaks (green), or single peak when homozygous (red), were determined for each STR and compared among the triad. The adjacent miniplots show distribution of shared STRs (upper miniplot) and discriminatory STRs (middle and lower miniplots) with the percentage of reads indicated numerically and by color intensity for each dominant allele. (B to D) Comparisons of allelic frequencies for the 94 sequenced SNVs in sample SD (Table 1). (B) Fetal trophoblast and maternal cells, (C) fetal trophoblast cells and placenta, and (D) maternal cells and placenta.
We observed that all 94 SNVs were detected in the fetal samples (Fig. 2, B to D) and matched the placental reference DNA haplotypes (Fig. 2C). Discriminatory SNV analysis, comparing the maternal haplotype to the reference placental DNA and fetal haplotype (Fig. 2, B and D), revealed an average fetal DNA fraction of 92.2 ± 6.5%, indicative of low amounts of maternal DNA in the fetal samples (Table 1). Additionally, all of the maternal and fetal haplotype pairs lacked forbidden combinations (AA/BB) at any allele (Fig. 2B), consistent with unique individuals, related as parent and offspring.
Accuracy of haplotype determinations
Using a threshold filter derived from reference placental DNA to account for variant reads resulting from maternal DNA that copurified with fetal DNA, homozygous SNVs were only called when the variant allele reads were in the range of 0 to 10% or 90 to 100%, whereas heterozygous SNVs were only called when between 40 and 60%. We observed that even after the threshold was applied, we were able to call 89 ± 12% of the 94 SNVs in fetal samples (Table 1). The haplotype of any SNV that passed the filter always matched the placental reference DNA and was, therefore, correctly determined. The number of SNVs recovered after filtering inversely correlated with the amount of copurifying maternal DNA present. Stratifying the 20 samples, we found that the presence of less than 5% maternal DNA (n = 8) resulted in 95.4 ± 2.5% recovery of the SNVs available before filtering; for 6.1 to 9.2% maternal DNA (n = 7), average recovery was 92.5 ± 7.3% of SNVs, whereas higher contamination (12.8 to 24.4%, n = 5) reduced recovery of SNVs to 62.5 ± 15%.
The data were further analyzed to determine when the purity of the fetal fraction was sufficient to call the correct fetal haplotypes in the absence of reference (placental) DNA. An internal quality control was established using the median summed percent reads of the autosomal STRs. A 100% correct fetal haplotyping is needed for clinically acceptable testing. The cutoff STR allele percentages were based on the 20 samples presented in this study and 4 samples analyzed in preliminary experiments before full optimization of the DNA isolation protocol (n = 24; fig. S2). With median summed percent reads above 82.5%, the fetal haplotype was always correctly called.
DISCUSSION
Genetic disorders pose a health risk for the newborn. After 45 years of exploring the diagnostic utility of fetal trophoblast cells residing in the female reproductive tract (12, 14), we report that sequences across the fetal genome can be analyzed accurately and in detail with a safe and standardized procedure, beginning at 3 weeks after conception (5 weeks after the previous menses). The TRIC protocol, followed by nuclear isolation, provided access to the full, unfragmented (16) fetal genome earlier in gestation compared to other procedures currently in practice (fig. S3).
Previous studies have suggested genetic testing protocols using fetal trophoblast cells captured from the cervix by size selection and laser microdissection (23–27). A persistent obstacle was the inability to generate pure, well-characterized fetal cell populations. The lack of sufficient quantities of fetal DNA for reliable molecular analysis limited DNA characterization and reproducibility. As a result, those approaches were not adapted for clinical use. To overcome these limitations, we have developed a rapid isolation protocol that provides fetal trophoblast cells within 1 day using immunomagnetic isolation with anti–HLA-G (16). Although trophoblast HLA-G expression decreases in women with placental disorders, such as preeclampsia (28), fetal trophoblast cell yield is not significantly hindered in those pregnancies (19). As a result, the number of dropouts due to placental pathologies when using TRIC is minimal, reducing the need for resampling. Initial experiments that isolated DNA from fetal trophoblast cell extracts (fig. S1) failed to reliably provide a high fetal DNA fraction that was adequate for clinical analysis, suggesting that maternal DNA fragments bound to the fetal trophoblast cells had copurified. This complication has likely hindered comprehensive analysis of the fetal genome by other groups working with specimens collected from the reproductive tract (14). Nuclear isolation eliminated the maternal DNA, resulting in adequate fetal DNA purity from endocervical specimens. An average fetal fraction of 91.5% was obtained before 10 weeks of gestation, with a fetal fraction of up to 99.9% as early as 6 weeks of gestation.
Next-generation sequencing is a powerful tool for both global analysis of the genome and targeted sequencing of selected loci. Whole-genome and exome sequencing has the advantage of identifying chromosome structural anomalies, as well as de novo mutations. However, those approaches require larger quantities of DNA for complete coverage and adequate read depth (>30×) to obtain clinically relevant single-nucleotide resolution. The small fraction of fetal DNA available in cell-free DNA limits its usefulness to chromosome copy number variance testing as a noninvasive perinatal screen. However, about 90% of congenital defects are not caused by aneuploidy (29). By accessing intact fetal trophoblast cells with entire genomes, the TRIC protocol reliably provides fetal genetic analysis as demonstrated here using targeted sequencing. Targeted sequencing without whole-genome amplification is the method of choice for diagnosing specific gene mutations when DNA is limited, and a high read depth is desired. It requires less DNA, provides greater depth (>300×), and is a reliable, inexpensive approach. Additionally, targeted sequencing is easier to analyze than whole-genome sequencing and avoids the potential of uncovering maternal disorders, including cancers, that can be detected with the current shotgun sequencing approaches (30).
The fetal genotype was determined reliably with 125 isolated fetal trophoblast cells by targeted multiplex sequencing of 157 loci across all chromosomes. SNVs are naturally occurring single-nucleotide variations in the genome that are technically comparable to pathogenic point mutations. Similarly, STRs mimic small insertions and deletions in the genome that distinguish some clinically relevant mutations. Targeted sequencing of SNVs has been extended on a large scale for aneuploidy analysis of cell-free fetal DNA (7). The ability to detect both SNVs and STRs is evidence that prenatal genotyping by TRIC is capable of precisely haplotyping fetal genetic loci. However, in our study, some SNVs did not pass the filtering criterion, mainly because of their lower sequencing depth, leading to more imbalanced SNV percentages. This was likely due to less efficient PCR amplification compared to other loci in this highly multiplexed primer panel. Use of specific and highly efficient primers for multiplexed PCR could help to eliminate the potential amplification bias.
Achieving a high fetal fraction through TRIC and nuclear isolation provides a solution for the detection of gene mutations in the first trimester of pregnancy. By including discriminatory SNVs or STRs in the targeted sequencing strategy, interfering maternal DNA can be quantified and subtracted to determine the fetal haplotype for precise prenatal genotyping. With a fetal fraction of close to 100%, the DNA quality is comparable to that obtained by CVS or amniocentesis and would be compatible with established commercial pipelines. Because we were able to analyze STR loci on the X chromosome, it should be possible to detect recessive, autosomal, single-nucleotide disorders in male fetuses without the need of paternal haplotyping. With advances in targeted multiplex sequencing approaches, Nicolaides et al. have successfully used 19,488 parallel reactions to detect aneuploidies in cell-free DNA (31), which should be feasible when using fetal trophoblast cells from the cervix to screen for aneuploidies, as well as multiple genetic disorders.
The targeted sequencing approach used in this study can be adapted to test for genetic disorders or variations. Genomic regions that are difficult to amplify would require appropriate PCR primer design and optimization of PCR strategies to ensure sufficient sequencing depth. Other limitations of the technology have to be considered before clinical application. Reduced fetal trophoblast cell recovery due to improper cervical sampling requires resampling, which is feasible in this noninvasive approach. Fetal genotyping discrepancies due to confined placental mosaicism and multiparity need to be evaluated, which would require a large longitudinal study.
TRIC fills a technical gap in current noninvasive prenatal testing by potentially offering the precision of current invasive technology without being hampered by limitations caused by a large maternal DNA fraction, rapid fetal DNA catabolism, early gestational age, or maternal obesity. Because of its simplicity, TRIC should enable prenatal genetic analysis in a diagnostic clinical setting without the necessity of complex bioinformatics for interpretation of massively parallel sequencing. Isolation of fetal trophoblast cells by TRIC holds promise not only for prenatal genetic testing but also for investigation of trophoblast biology and pathophysiology in ongoing pregnancies (14, 32, 33). The findings of this study establish that TRIC isolates cells with a fetal genotype and introduce a practical approach for precise interrogation of the fetal genome at the outset of pregnancy.
MATERIALS AND METHODS
Study design
The objective of this study was to develop an approach for single-nucleotide resolution genotyping of fetal DNA from cells obtained at 5 to 20 weeks of gestation using a noninvasive Pap smear. Endocervical specimens and a portion of the placenta were obtained from 20 enrolled patients with both male and female singleton conceptuses. Fetal trophoblast and maternal cells were isolated by TRIC from endocervical specimens. We developed a nuclear isolation protocol to remove maternal DNA from fetal trophoblast cells before DNA purification. DNA isolated from fetal trophoblast cells, maternal cells, and matched placentas was analyzed by targeted sequencing of 59 STRs and 94 SNVs on a MiSeq FGx platform. STR and SNV haplotypes within each triad were identified using the ForenSeq analytic pipeline. We assessed the purity of the fetal DNA fraction and genotyped the fetus by comparing SNV read depths to those of corresponding SNVs in the maternal DNA.
Patient selection
The institutional review board of Wayne State University approved this study, and each participating patient provided written informed consent. For this study, pregnant women (n = 20) between 5 and 19 weeks of gestational age and undergoing voluntary interruption of pregnancy were consecutively recruited at the Safe and Sound for Women clinic in Las Vegas, NV, and the Northland Family Planning Centers of Michigan. The patients were counseled for collection of endocervical specimens. Endocervical sampling was performed before placement of a laminaria. Women experiencing active vaginal bleeding or with multiple gestations were excluded from the study. All samples used for the study were collected consecutively.
Isolation of endocervical fetal trophoblast cells
Endocervical sampling was performed, as described previously (15). Endocervical cells were immediately fixed after the Pap smear procedure using a ThinPrep kit (Hologic) to preserve cells, proteins, and nucleic acids. Trophoblast cells were isolated from each specimen using TRIC (16). Briefly, cells were centrifuged and resuspended in 10 ml of phosphate-buffered saline (PBS) and washed three times. After a final resuspension in 1.5 ml of PBS, anti–HLA-G–coated magnetic nanoparticles were added and incubated overnight at 4°C with mixing. The nonbound (maternal) cells were collected after magnetic immobilization of HLA-G–positive cells, which were recovered after three washings in PBS. The HLA-G–positive (fetal) cells were treated with DNase I, immobilized on F7M matrix (MoBiTec) for 30 min at room temperature in solution (1× digestion buffer), and the matrix removed by washing three times with PBS. This was followed by immunofluorescence microscopy for the expression of β-hCG (Fig. 1A), and the percentage of the labeled cells was determined.
Fluorescence in situ hybridization
Isolated fetal trophoblast cells were mounted on slides and probed for X and Y chromosomes by FISH at the Detroit Medical Center Cytogenetics Laboratory using the DYZ1 satellite III on the Y chromosome and the DXZ1 α satellite on the X chromosome as fluorescently labeled probes (Abbott Molecular). Nuclei were counterstained with DAPI and scored for each chromosome to quantify cells that were XX or XY. Nuclei were first isolated from fetal trophoblast cells before next-generation sequencing by ForenSeq.
Targeted sequencing
DNA was obtained from fetal trophoblast and maternal cells isolated by TRIC, and tissue was obtained from the corresponding placenta. Targeted sequencing was performed using pregnancies with both male and female fetuses (n = 20).
Nuclear isolation and DNA extraction
Targeted sequencing was performed after digestion of exogenous DNA. Fetal and maternal cells obtained by TRIC were suspended in PBS, deposited on microscope slides, and dried on a slide warmer at 40°C. Before DNA extraction, the slides were immersed in pepsin solution (0.01 g in 100 ml of 0.1 N HCl) for 11 min, followed by a PBS wash for 5 min to remove cell membranes and contaminating maternal DNA fragments. Exogenous DNA was further eliminated from the glass-bound nuclei by adding 10 μl of washed, immobilized DNase (DNase I, immobilized on Matrix F7M, MoBiTec) onto the pepsin-treated slides and incubating for 5 min at room temperature. The slides were washed with PBS to remove the beads and terminate DNA digestion. The nuclei were lysed by overnight incubation at 65°C with 0.5 μl per cell of Arcturus PicoPure DNA lysis buffer (Applied Biosystems) and then inactivated at 95°C for 30 min.
Placental villi (20 to 25 mg) were dissected and suspended in 100 μl of PBS. Immobilized DNase (10 μl) was added and shaken for 10 min at room temperature to ensure efficient extracellular maternal and fetal DNA digestion. DNase beads were removed by washing in PBS. DNA was extracted from the tissue using the Qiagen DNeasy Blood and Tissue Kit. All DNA was purified using the Qiagen MinElute PCR Purification Kit, eluted in a final volume of 20 μl, and quantified using the PicoGreen assay.
Library preparation for ForenSeq
DNA libraries were prepared with the ForenSeq DNA Signature Prep Kit (Illumina) using primer mix A, as instructed by the manufacturer. Individual samples were quantified by Qubit and analyzed using Agilent high-sensitivity DNA chips before library normalization. The percentage of libraries between 200 and 1000 bp was calculated on the basis of the library traces and was mixed accordingly at the genomic core facility at the University of Illinois. A pool size of 200 bp to 1 kb was selected, and the size-selected negative controls were mixed into the pool. The samples were sequenced on a MiSeq FGx system, including positive and negative controls (Illumina).
ForenSeq data analysis
ForenSeq with primer mix A analyzed 94 SNV loci and 59 STR loci on all human chromosomes (table S1). The ForenSeq Universal Analysis software provides a run quality report and detailed genotype. From this report, data were extracted, and allele percentages were determined using the read depth of each STR and SNV. For STR data, the two dominant alleles with the highest read depths were analyzed to establish sample quality.
To determine the purity of fetal DNA (fetal fraction), the extent of contamination with maternal DNA was calculated from the SNV data. For each sample triad (fetal, maternal, and placental), the homozygosity or heterozygosity of each SNV was determined (first in the placenta, then comparing to fetal), and the percentage deviation of the fetal DNA was calculated. Deviations from homozygosity or heterozygosity were also calculated in the SNV profile of maternal DNA to determine a threshold of technical and/or biological variation. Maternal and placental SNV profiles were then compared, and the nondiscriminatory SNVs (identical maternal and placental alleles) were filtered out. For the discriminatory SNVs, the deviations obtained from the maternal sample were subtracted from the fetal sample and multiplied by two (to account for two maternal alleles contributing to the fetal sample), yielding the percentage of maternal contamination.
To investigate whether the fetal genotype could be accurately determined in the absence of reference DNA, autosomal STRs were used. A cutoff for STR allele percentages was established on the basis of the samples presented in this study (n = 20), as well as earlier analyzed samples (n = 4) that were examined before full optimization of the DNA isolation protocol (fig. S2). The dominant alleles with the highest read depth were summed to establish the median and range, which discriminated the high- from the lower-quality samples. Good samples had median STR alleles of >85%. A cutoff of 82.5% for discriminating good from lower-quality samples was chosen on the basis of receiver operating characteristic analysis. As a result, the determined threshold for autosomal STR alleles then provided 100% accuracy in calling the fetal haplotype in further SNV analysis.
For SNV analysis, a maximal SNV-specific maternal contamination of 20% was allowed in the fetal samples. To achieve this, a homozygous SNV only passed this SNV threshold when the variant allele represented either 0 to 10% or 90 to 100% of the read depth, whereas heterozygous SNVs only passed when the variant allele read depth was between 40 and 60%.
Supplementary Material
Fig. S1. Comparison of fetal fractions obtained with and without nuclear isolation.
Fig. S2. Cutoff determination for STR allele percentages.
Fig. S3. Comparison of approaches to obtain fetal DNA in ongoing pregnancies.
Table S1. List of SNVs and STRs targeted by ForenSeq.
Acknowledgments:
We thank the Safe and Sound for Women clinic of Las Vegas, NV, the Northland Family Planning Centers of Michigan, and their patients for participating in this research study. We also thank D. Schloff of the Detroit Medical Center Cytogenetics Laboratory and A. G. Hernandez of the DNA Services at the University of Illinois for technical assistance.
Funding:
This research was supported by NIH grants HL128628 (S.D.), HD071408 (D.R.A)., and K12HD001254 (R.F.), the W.K. Kellogg Foundation (D.R.A. and S.D.), and the March of Dimes (S.D.).
Footnotes
Competing interests:
D.R.A. and S.D. have pending patents and received payment for intellectual property that has been licensed on their behalf by Wayne State University to PerkinElmer Inc. and are inventors on patent applications (PCT/US2015/055126 and PCT/US2013/065570) held and/or submitted by Wayne State University that covers the isolation and use of endocervical trophoblast cells for fetal diagnosis.
Data and materials availability:
Sequencing data are available from the Dryad Digital Repository at http://dx.doi.org/10.5061/dryad.43n2j.
REFERENCES AND NOTES
- 1.Wapner RJ, Invasive prenatal diagnostic techniques. Semin. Perinatol 29, 401–404 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Mujezinovic F, Alfirevic Z, Procedure-related complications of amniocentesis and chorionic villous sampling: A systematic review. Obstet. Gynecol 110, 687–694 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Sermon K, Van Steirteghem A, Liebaers I, Preimplantation genetic diagnosis. Lancet 363, 1633–1641 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Gil MM, Quezada MS, Revello R, Akolekar R, Nicolaides KH, Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: Updated meta-analysis. Ultrasound Obstet. Gynecol 45, 249–266 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Rodeck C, Tutschek B, Sherlock J, Kingdom J, Methods for the transcervical collection of fetal cells during the first trimester of pregnancy. Prenat. Diagn 15, 933–942 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Chitty LS, Bianchi DW, Noninvasive prenatal testing: The paradigm is shifting rapidly. Prenat. Diagn 33, 511–513 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Wong FCK, Lo YMD, Prenatal diagnosis innovation: Genome sequencing of maternal plasma. Annu. Rev. Med 67, 419–432 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Wang E, Batey A, Struble C, Musci T, Song K, Oliphant A, Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma. Prenat. Diagn 33, 662–666 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Barrett AN, McDonnell TCR, Chan KCA, Chitty LS, Digital PCR analysis of maternal plasma for noninvasive detection of sickle cell anemia. Clin. Chem 58, 1026–1032 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Tsui NBY, Kadir RA, Chan KCA, Chi C, Mellars G, Tuddenham EG, Leung TY, Lau TK, Chiu RWK, Lo YMD, Noninvasive prenatal diagnosis of hemophilia by microfluidics digital PCR analysis of maternal plasma DNA. Blood 117, 3684–3691 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Committee Opinion No. 640: Cell-free DNA screening for fetal aneuploidy. Obstet. Gynecol 126, e31–e37 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Shettles LB, Use of the Y chromosome in prenatal sex determination. Nature 230, 52–53 (1971). [DOI] [PubMed] [Google Scholar]
- 13.Adinolfi M, Sherlock J, First trimester prenatal diagnosis using transcervical cells: An evaluation. Hum. Reprod. Update 3, 383–392 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Imudia AN, Kumar S, Diamond MP, DeCherney AH, Armant DR, Transcervical retrieval of fetal cells in the practice of modern medicine: A review of the current literature and future direction. Fertil. Steril 93, 1725–1730 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imudia AN, Suzuki Y, Kilburn BA, Yelian FD, Diamond MP, Romero R, Armant DR, Retrieval of trophoblast cells from the cervical canal for prediction of abnormal pregnancy: A pilot study. Hum. Reprod 24, 2086–2092 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolnick JM, Kilburn BA, Bajpayee S, Reddy N, Jeelani R, Crone B, Simmerman N, Singh M, Diamond MP, Armant DR, Trophoblast retrieval and isolation from the cervix (TRIC) for noninvasive prenatal screening at 5 to 20 weeks of gestation. Fertil. Steril 102, 135–142.e6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holt J, Stiltner L, Jamieson B, Fashner J, Clinical inquiries. Should a nylon brush be used for Pap smears from pregnant women? J. Fam. Pract 54, 463–464 (2005). [PubMed] [Google Scholar]
- 18.Paraiso MF, Brady K, Helmchen R, Roat TW, Evaluation of the endocervical Cytobrush and Cervex-Brush in pregnant women. Obstet. Gynecol 84, 539–543 (1994). [PubMed] [Google Scholar]
- 19.Fritz R, Kohan-Ghadr HR, Sacher A, Bolnick AD, Kilburn BA, Bolnick JM, Diamond MP, Drewlo S, Armant DR, Trophoblast retrieval and isolation from the cervix (TRIC) is unaffected by early gestational age or maternal obesity. Prenat. Diagn 35, 1218–1222 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kidd KK, Pakstis AJ, Speed WC, Grigorenko EL, Kajuna SLB, Karoma NJ, Kungulilo S, Kim J-J, Lu R-B, Odunsi A, Okonofua F, Parnas J, Schulz LO, Zhukova OV, Kidd JR, Developing a SNP panel for forensic identification of individuals. Forensic Sci. Int 164, 20–32 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Sanchez JJ, Phillips C, Børsting C, Balogh K, Bogus M, Fondevila M, Harrison CD, Musgrave-Brown E, Salas A, Syndercombe-Court D, Schneider PM, Carracedo A, Morling N, A multiplex assay with 52 single nucleotide polymorphisms for human identification. Electrophoresis 27, 1713–1724 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Churchill JD, Schmedes SE, King JL, Budowle B, Evaluation of the Illumina® Beta Version ForenSeq™ DNA Signature Prep Kit for use in genetic profiling. Forensic Sci. Int. Genet 20, 20–29 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Pfeifer I, Benachi A, Saker A, Bonnefont JP, Mouawia H, Broncy L, Frydman R, Brival ML, Lacour B, Dachez R, Paterlini-Bréchot P, Cervical trophoblasts for noninvasive single-cell genotyping and prenatal diagnosis. Placenta 37, 56–60 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Bussani C, Cioni R, Mattei A, Fambrini M, Marchionni M, Scarselli G, Prenatal diagnosis of common aneuploidies in transcervical samples using quantitative fluorescent-PCR analysis. Mol. Diagn. Ther 11, 117–121 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Bussani C, Cioni R, Scarselli B, Barciulli F, Bucciantini S, Simi P, Fogli A, Scarselli G, Strategies for the isolation and detection of fetal cells in transcervical samples. Prenat. Diagn 22, 1098–1101 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Katz-Jaffe MG, Mantzaris D, Cram DS, DNA identification of fetal cells isolated fromcervical mucus: Potential for early non-invasive prenatal diagnosis. BJOG 112, 595–600 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Mantzaris D, Cram DS, Potential of syncytiotrophoblasts isolated from the cervical mucus for early non-invasive prenatal diagnosis: Evidence of a vanishing twin. Clin. Chim. Acta 438, 309–315 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Goldman-Wohl DS, Ariel I, Greenfield C, Hochner-Celnikier D, Cross J, Fisher S, Yagel S, Lack of human leukocyte antigen-G expression in extravillous trophoblasts is associated with pre-eclampsia. Mol. Hum. Reprod 6, 88–95 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Christianson A, Modell B, Medical genetics in developing countries. Annu. Rev. Genomics Hum. Genet 5, 219–265 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Bianchi DW, Chudova D, Sehnert AJ, Bhatt S, Murray K, Prosen TL, Garber JE, Wilkins-Haug L, Vora NL, Warsof S, Goldberg J, Ziainia T, Halks-Miller M, Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA 314, 162–169 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Nicolaides KH, Syngelaki A, Gil M, Atanasova V, Markova D, Validation of targeted sequencing of single-nucleotide polymorphisms for non-invasive prenatal detection of aneuploidy of chromosomes 13, 18, 21, X, and Y. Prenat. Diagn 33, 575–579 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Fritz R, Kohan-Ghadr H-R, Bolnick JM, Bolnick AD, Kilburn BA, Diamond MP, Drewlo S, Armant DR, Noninvasive detection of trophoblast protein signatures linked to early pregnancy loss using trophoblast retrieval and isolation from the cervix (TRIC). Fertil. Steril 104, 339–346.e4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolnick JM, Kohan-Ghadr H-R, Fritz R, Bolnick AD, Kilburn BA, Diamond MP, Armant DR, Drewlo S, Altered biomarkers in trophoblast cells obtained noninvasively prior to clinical manifestation of perinatal disease. Sci. Rep 6, 32382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Comparison of fetal fractions obtained with and without nuclear isolation.
Fig. S2. Cutoff determination for STR allele percentages.
Fig. S3. Comparison of approaches to obtain fetal DNA in ongoing pregnancies.
Table S1. List of SNVs and STRs targeted by ForenSeq.
Data Availability Statement
Sequencing data are available from the Dryad Digital Repository at http://dx.doi.org/10.5061/dryad.43n2j.


