Abstract
The possible neurodevelopmental consequences of SARS-CoV-2 infection are presently unknown. In utero exposure to SARS-CoV-2 has been hypothesized to affect the developing brain, possibly disrupting neurodevelopment of children. Spike protein interactors, such as ACE2, have been found expressed in the fetal brain, and could play a role in potential SARS-CoV-2 fetal brain pathogenesis. Apart from the possible direct involvement of SARS-CoV-2 or its specific viral components in the occurrence of neurological and neurodevelopmental manifestations, we recently reported the presence of toxin-like peptides in plasma, urine and fecal samples specifically from COVID-19 patients. In this study, we investigated the possible neurotoxic effects elicited upon 72-hour exposure to human relevant levels of recombinant spike protein, toxin-like peptides found in COVID-19 patients, as well as a combination of both in 3D human iPSC-derived neural stem cells differentiated for either 2 weeks (short-term) or 8 weeks (long-term, 2 weeks in suspension + 6 weeks on MEA) towards neurons/glia. Whole transcriptome and qPCR analysis revealed that spike protein and toxin-like peptides at non-cytotoxic concentrations differentially perturb the expression of SPHK1, ELN, GASK1B, HEY1, UTS2, ACE2 and some neuronal-, glia- and NSC-related genes critical during brain development. Additionally, exposure to spike protein caused a decrease of spontaneous electrical activity after two days in long-term differentiated cultures. The perturbations of these neurodevelopmental endpoints are discussed in the context of recent knowledge about the key events described in Adverse Outcome Pathways relevant to COVID-19, gathered in the context of the CIAO project (https://www.ciao-covid.net/).
Keywords: Spike protein, Toxin-like peptides, 3D neurospheres, Electrical activity, RNA-Seq, AOP, CIAO Project, brain development
1. Introduction
Coronavirus disease (COVID-19) resulting from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is still a public health issue. The effect of SARS-CoV-2 infection in pregnant women is of particular concern as studies suggest that they are at increased risk for severe COVID-19 associated with adverse fetal outcomes [1].
It is well documented that infections during pregnancy can increase the risk for the offspring to develop neurodevelopmental disorders [2], and in utero exposure to SARS-CoV-2 has been hypothesized to affect the developing brain [3], [4], possibly disrupting neurodevelopment [5], [6]. However, the mechanisms underlying these poor outcomes are still unknown, and may be due to the exacerbated pro-inflammatory environment of the pregnant mother, or to vertical transmission of SARS-CoV-2. SARS-CoV-2 has been suggested to cross the placenta and to infect syncytiotrophoblast of the placental barrier, as shown by molecular and immunohistochemical analyses and electron microscopy, with an antibody-dependent transcytosis mediated by FcRn (Neonatal Fc receptor) hypothesized as a potential mechanism underlying placental invasion [7], [8].
Moreover, anti-SARS-CoV-2 IgM antibodies have been detected in the blood of new-borns delivered by caesarean from mothers found positive for SARS-CoV-2, which suggests SARS-CoV-2 in utero exposure, or impaired placenta barrier with placental immune responses due to maternal respiratory SARS-CoV-2 infection [9], [10]. Since IgM antibodies do not cross the placenta except if disruption occurs and IgM represent the first immune response component against SARS-CoV-2, IgM antibodies in newborns may also indicate the possibility of recent exposure to SARS-CoV-2 of the fetus in the womb [11].
Additionally, transcytosis of opsonized or free viruses or viral particle transfer by infected blood cells could also cause placental infection and vertical transmission of the virus [12], [13]. Case reports documented placental SARS-CoV-2 infection [14], [15]. Although detection of RNA is considered not to be enough to conclude vertical transmission, a systematic review of the current literature aimed to evaluate the possibility of vertical transmission based on early RNA detection of SARS-CoV-2 after birth, and concluded that vertical transmission of the virus is very rare as observed in a minority of pregnant women tested positive for SARS-CoV-2 during the third trimester, with rates of infection similar to those of other pathogens causing congenital infections [16], [17]. Other recent systematic review studies suggest either that vertical transmission of the virus is not strongly supported by clinical evidence, e.g., [18], or that vertical transmission may be possible, although the likelihood of its occurrence is generally low, e.g., [19], [20].
In addition to the presence of viral particles, spike proteins (S1 and S2) have also been found immunolocalized in cytotrophoblast and syncytiotrophoblast cells of the placental villi, in one asymptomatic pregnant woman tested positive for SARS-CoV-2 [21]. In this study, viral RNA was detected in the amniotic fluid and S proteins were detected in the fetal membrane at 8–13 gestational weeks [21]. This study provides evidence of persistent placental infection by SARS-CoV-2, which could be causative of hydrops fetalis and intrauterine fetal demise during the first trimester of pregnancy [21]. In another study, placental vasculopathy (which could lead to fetal growth reduction and other complications) and the presence of SARS-CoV-2 across the placenta have been reported in a pauci-symptomatic pregnant woman [22], suggesting the possibility of viral vertical transmission during early pregnancy. Transplacental transmission of SARS-CoV-2 has been described also at later gestational weeks, e.g., in a 34-week pregnant woman, where the virus was found in the placenta as well as in several tissues of the fetus, who died as a consequence of severe placental thromboembolism [23]. Along the same line, transplacental viral transmission has been also detected in a woman at 35 weeks of gestation found positive for SARS-CoV-2, who showed neurological issues and delivered a baby presenting irritability, axial hypertonia, poor feeding and opisthotonos [24]. On the contrary, Garcia-Flores et al. showed that SARS-CoV-2 infection during pregnancy was associated with humoral and cellular immune responses in the maternal blood, as well as with altered cytokine profile in umbilical cord blood, although in the absence of placental infection [25].
Besides, the potential mechanisms of SARS-CoV-2 entry remain unclear in both placenta and fetal brain with variable findings reported [11], [26], [27]. In the placenta, the canonical ACE2 receptor was shown highly expressed during early gestation, then at negligible mRNA levels at full term, although term placentas from COVID-19 affected women showed increased ACE2 expression compared to healthy term placentas [17]. The study also showed that ACE2 protein is present in placenta despite low transcript levels. Noteworhty, by using an in vitro placental model, it has been shown that SARS-CoV-2 can infect the human placenta, and that ACE2 expression levels are directly associated with the release of SARS-CoV-2 [28].
In the human fetal brain, spike protein interactors, i.e., ACE2, TMPRSS2, FURIN and the recently discovered ZDHHC5, GOLGA7, and ATP1A1 have been found expressed. In particular, ACE2 and classical TMPRSS2 co-factor have been found expressed, although at a low level (being undetectable in some brain regions). Besides, the alternative receptors and co-factors FURIN, ZDHHC5, GOLGA7 and ATP1A1 are expressed at a high level in the fetal brain, and could play a direct or indirect role in potential SARS-CoV-2 fetal brain pathogenesis, especially during the 2nd and 3rd trimesters of pregnancy [29].
It is still unclear whether human placenta is susceptible to SARS-CoV-2 infection under normal physiological conditions; however, under conditions of systemic inflammation and of impaired placental barrier, which may occur in pregnant women with severe COVID-19, placental pathology and the possibility of vertical transmission should be further carefully investigated.
In a previous study [30], we reported the presence of toxin-like peptides in plasma, urine and fecal samples specifically and exclusively from symptomatic COVID-19 patients. In particular, the sequences of these (oligo)peptides (70–115, depending on the analysed samples) map with the sequences of known neurotoxic substances, i.e., conotoxins, metalloproteinases, prothrombin activators, phospholipases A2, and coagulation factors, which can be found in animal venoms and that are characterized by a high specificity and affinity towards human receptors, ion channels, and transporters of the CNS, like the nicotinic acetylcholine receptor [30]. We speculated that these toxin-like peptides could be involved in the reported COVID-19 neurological clinical manifestations. In addition, it is presently unclear whether these toxin-like peptides can cross the placenta. While the origin of these peptides is presently unknown, several hypotheses can be formulated, e.g.: (i) these peptides may be coded by specific SARS-CoV-2 RNA regions [31]; (ii) SARS-CoV-2 may be able to replicate in bacteria in a ‘bacteriophage-like’ manner [32]; (iii) bacteria, may produce and secrete these peptides in reaction to the presence of the virus through not fully defined mechanisms, including the involvement of rRNA [33], or small bacterial non-coding RNAs [34]; or (iv) a combination of the aforementioned mechanisms [30]. Noteworthy, while animal toxins have been discussed as potential drug candidates for the treatment of human diseases, including neurodegenerative diseases, cardiovascular diseases, cancer, neuropathic pain, and autoimmune diseases [35], [36], their possible detrimental effects on the developing brain have not been fully explored.
Several in vivo and in vitro models have been used to study SARS-CoV-2 mediated neurological and neuropathological modes of action. For instance, human cerebral organoids have been proven suitable to investigate SARS-CoV-2 infective mechanisms [37], [38], [39], [40], [41]. Human induced pluripotent stem cell (iPSC)-derived 3D models (e.g., neurospheres, brain spheres, organoids) can mimic key features of human fetal brain development [42], [43], [44], [45], [46], [47], and therefore may be used to assess the neurodevelopmental effects of SARS-CoV-2 and its components. For instance, by using human brain organoids, it has been shown that SARS-CoV-2 can cause impairment of excitatory synaptogenesis, affecting astrocytes’ synaptogenic functions [48].
While the possible neurodevelopmental consequences of SARS-CoV-2 infection are still not fully understood, the Adverse Outcome Pathway (AOP) framework could help improve interpretation and application of scientific understanding of COVID-19 pathological mechanisms [49]. In particular, the investigation of the molecular and cellular mechanisms underlying SARS-CoV-2 effects could be linked to the key events (KEs) described in COVID-19-relevant AOPs, which have been recently developed or are still under development in the context of the so called ‘CIAO’ project [49], [50], [51]. The project aims at modelling the pathogenesis of COVID-19 by exploiting the AOP framework approach. A similar strategy has been adopted also in the context of developmental neurotoxicity (DNT) testing, where mechanistic understanding of the molecular/cellular effects triggered by developmental neurotoxicants (e.g., deregulation of neural progenitor cell proliferation, neuronal and glial differentiation, migration, neurite outgrowth, synaptogenesis and neuronal network formation and function) could be anchored to KEs identified in the existing DNT relevant AOPs, as recently described [52], [53], [54]. Human 3D neuronal/glial models combined with emerging knowledge described in AOPs relevant for COVID-19 could enable mechanistic understanding of SARS-CoV-2 and viral components’ effects.
In this study, we investigated the possible neurotoxic effects elicited by 72 h exposure to spike protein (recombinant S1 + S2), toxin-like peptides found in symptomatic COVID-19 patients, and a combination of both on short- and long-term differentiated cultures of human iPSC-derived neural stem cells (NSCs) differentiated towards a mixed culture of neurons/glia as 3D neurospheres, as previously described [55]. Whole transcriptome (by RNA-seq) was assessed after 72 h exposure in short-term differentiated cultures (2-week old neurospheres), impact on viability and selected gene expression by qPCR analysis were evaluated in both short- and long-term cultures, while effects on the generation of spontaneous electrical activity by microelectrode array (MEA) were analysed in long-term differentiated cultures (2-weeks + 6-weeks on MEA). The perturbations of neurodevelopmental endpoints described in this in vitro study are discussed in the context of recent knowledge about the molecular and cellular KEs described in AOPs relevant to COVID-19-associated brain disorders, gathered in the context of the CIAO project and under development in the online platform AOP-Wiki (https://aopwiki.org/).
2. Materials and methods
2.1. Differentiation of human induced pluripotent stem cell (hiPSC)-derived neural stem cells (NSCs) into 3D neurospheres
The IMR90 cell line, a female human fetal lung fibroblast line obtained from a clinically normal 16 week old fetus, was originally developed at Coriell [56] with karyotype of a normal diploid female (46,XX). IMR90 fibroblasts were directly reprogrammed at I-Stem (Evry, France https://www.istem.eu/en/) by retroviral transduction of OCT4 and SOX2 using pMIG vectors (Addgene). Frozen colony fragments of IMR90-hiPSCs were kindly provided by Prof. Marc Peschanski (I-Stem). HiPSC colonies were phenotypically characterized by analysis of colony/cell morphology, analysis of PSC-specific markers (by immoncytochemistry and flow cytometry), as well as gene expression analysis of pluripotency-related genes and alkaline phosphate activity, as detailed in [57]. Neural stem cells (NSCs) were derived from IMR90-hiPSCs [57], [58], and differentiated as 3D neurospheres towards a mixed culture of neurons and astrocytes, in the presence of neuronal differentiation (ND) medium (i.e., Neurobasal Medium, N2 Supplements, B-27 Supplements, Penicillin/Streptomycin (50 U/mL), L-Glutamine (2 mM), laminin (mouse protein, 1 µg/mL), BDNF (2.5 ng/mL) and GDNF (1 ng/mL), all from ThermoFisher Scientific), as previously described [55]. Briefly, to generate neurospheres, NSCs were passaged using trypsin/EDTA and plated onto ultra-low adherent 6-well plates, at a density of 1 × 106/mL (2 mL/well) in the presence of ND medium. Already after 24 h, neurospheres were visible, and reached an average diameter of 200–250 µm after 1 week. If present, large, necrotic spheres were manually discarded, and medium was refreshed twice/week. Neurospheres were differentiated for minimum 2 weeks (up to 6 weeks), and plates were kept at 37 °C, 5% CO2 under constant gyratory shaking (86 rpm) on a plate shaker. Two-week old neurospheres were characterized by the presence of 31% β-III-tubulin+ neurons, 31% GFAP+ astrocytes, 18% CNPase+ oligodendrocytes, while 41% β-III-tubulin+, 39% GFAP+ and 11% CNPase+ cells were found in 6-week old cultures, as shown by flow cytometry analysis described in [55]. The percentage of remaining nestin+ NSCs was 48% and 8% in 2-week and 6-week old neurospheres, respectively [55].
Additionally, neurosphere cultures were characterized by the presence of a mixed culture of VGlut1+ glutamatergic, GABRE+ GABAergegic and TH+ dopaminergic neurons (see representative 40x confocal images of 4-week old neurospheres in Supplementary Figure 1). Electrical activity analysis by multi-well microelectrode array (MEA) showed that this culture is mainly responsive to the modulatory effects of CNQX, an AMPA receptor antagonist, indicating the presence of functional excitatory glutamatergic neurons, whilst Gabazine (a GABAA receptor antagonist) and Muscimol (a selective GABAA receptor agonist) did non cause significant changes of spontaneous electrical activity [55]. Further details about the protocol and the characterization of the model are provided in [55].
2.2. Exposure of 3D neurosphere to toxin-like peptides and recombinant spike protein
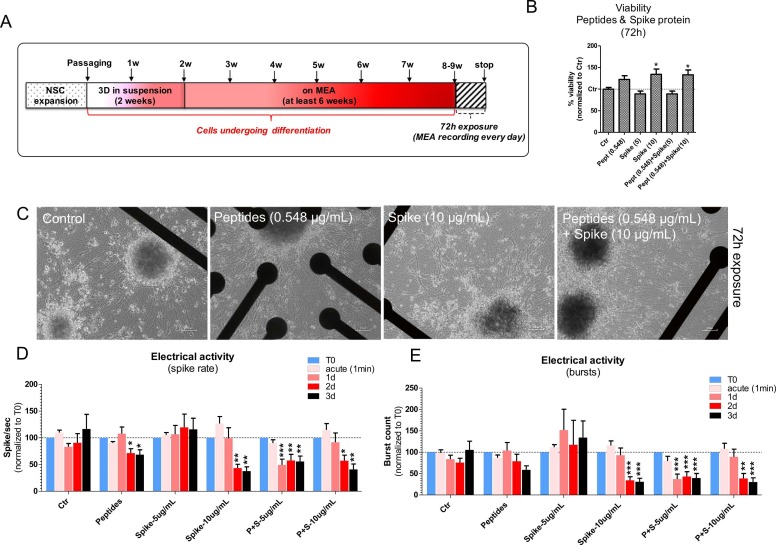
Two-week old neurospheres were manually transferred onto ultra-low adherence 24-well plates using a stereomicroscope (30 neurospheres/1 mL/well) and exposed for 72 h to recombinant 2019-nCoV S1 +S2 ECD protein with His tag (hereafter called ‘Spike’ or ‘S’) (Sigma-Merck, cat. SAB5700592, purity > 97% by SDS-PAGE) (stock concentration of 1000 µg/mL) at the concentrations of 0.3, 0.6, 1.3, 2.5, 5.0, 10.0 and 20.0 µg/mL, and a mix of toxin-like peptides (hereafter called ‘Peptides’ or ‘P’) derived from the supernatants of an in vitro faecal microbiota culture at 30 days (details on the used protocol are described in [30], [32] and reported below), obtained from a fecal sample (stool) of a subject positive for SARS-CoV-2 at the concentrations of 0.009, 0.017, 0.034, 0.069, 0.137, 0.548 and 1.096 µg/mL. Transmission electron microscopy (TEM) analysis of fecal matter samples contaminated with toxin-like peptides was carried out to assess the presence of the virus (Supplementary Figure 2), as described below. The stock concentration of the Peptide solution was 140 µg/mL (tested using Bradford approach) in ~ 50 µL volume, which corresponded to 10X, 50X and 100X the average level of toxin-like peptides found in faecal, blood and urine samples, respectively, of COVID-19 patients described in [30]. The stock solution was thus diluted 10 times with sterile PBS 1X (without calcium and magnesium) and filtered with a 0.22 µm pore filter to prevent bacterial contamination of cell cultures. After 72 h exposure to Spike and Peptides, cell viability was analysed as described below (compounds were not refreshed during the 72 h). In parallel experiments, 3D neurospheres were exposed to Spike (10 µg/mL), Peptides (0.548 µg/mL) and a combination of both (‘P + S’), and after 72 h neurospheres were collected for whole transcriptome analysis by RNA-seq (Bioclavis) as described below. To measure electrical activity, 2-week old 3D neurospheres were manually transferred using a stereomicroscope onto 24-well MEA plates and further differentiated for at least 6 additional weeks (total of 8–9 weeks in differentiation); then, cultures were exposed for 72 h to non-cytotoxic concentrations of Spike (5 and 10 µg/mL), Peptides (0.548 µg/mL) or a combination of both (‘P + S’). Electrical activity was recorded every day up to 72 h as detailed below.
2.3. Peptides mixture preparation
The mixture of Peptides was obtained from bacterial samples, grown as detailed in [30]. Briefly, bacterial cells were grown in NutriSelect™ Plus nutrient broth (Merck). Following the protocol recommended by the supplier, the medium was prepared as follows: 25 g were dissolved in 1 L of double distilled water and dispensed into tubes, which were sterilized by 15 min autoclave at 121 °C. All steps were conducted at temperature lower than 8 °C, protected from direct light. Final composition of the medium was Peptone (15 g/L), Yeast extract (3.0 g/L), Sodium chloride (6.0 g/L). D(+)-Glucose (1.0 g/L), pH 7.5 at 25 °C. Tubes/flasks with growth broth and bacteria were placed in an orbital shaker at 37 °C, and the liquid culture was left to grow for 30 days, monitoring bacterial growth by optical densities (OD) analysis using a (spectro)photometer absorbance microplate reader. After 30 days, the Peptides mixture was extracted and purified as follows: after a centrifugation at 13,000 g for 10 min, 700 µL were collected from each mL of supernatant, and filtered with a 0.2 µm filter. The obtained solution was checked by means of mass spectrometry (peptide proton rearrangement were considered during data acquisition and elaboration) to confirm the presence of toxin-like peptides. The total peptide and protein concentration were analysed using the Bradford approach. The identification of toxin-like peptides was obtained by using Liquid Chromatography-Surface Activated Chemical Ionization – Cloud Ion Mobility Mass Spectrometry (LC-SACI-CIMS), as described in [30]. The full set of manually reviewed venom proteins and toxins from UniprotKB database, mixed with a subset of non-venom proteins and toxins, was used as reference protein dataset in order to give statistical significance to the results. The identified toxin-like peptides include those reported in the list of representative toxin-like peptides mapped on 36 candidate protein sequences belonging to Chordata, Echinodermata and Mollusca, is detailed in Table 1 in [30], with information retrieved from UniprotKB and NCBI Taxonomy databases. This list is not expected to be exhaustive: in fact, several (oligo-) peptides (between 70 and 115, depending on the analysed sample [30]) matched with different animal venom proteins and toxins like conotoxins, phospholipases A2, and metalloproteinases (86% of assignments have a -log(e) > 25). Apart from these peptides mapped on toxins of animal origin, no signal with statistical significance and attributed to any known bacterial toxin was observed.
2.4. Transmission electron microscopy (TEM) analysis of fecal matter samples contaminated with toxin-like peptides
Gut bacteria derived from healthy donors were infected with the supernatant derived from COVID-19 affected individuals, containing bacteria and SARS-CoV-2 particles. Biological sample was directly deposited in a 3 µL drop on Formvar Carbon coated 200 mesh copper grids (Agar Scientific, USA), let to dry overnight in a desiccator, and the day after the sample was washed with ultrapure water and again let to dry overnight before analysis by JEOL JEM-2100 HR-transmission electron microscope at 120 kV (JEOL, Italy). TEM analysis conducted on unfiltered stock supernatants of Peptide samples demonstrated the presence of SARS-CoV-2 particles on the surface and inside gut bacteria (Supplementary Figure 2).
2.5. Analysis of cell viability with CellTiter-Blue®
After 72 h exposure to different concentrations of Spike or Peptides, cell viability was measured by incubating 3D neurospheres with CellTiter-Blue® Reagent (final 1:6 dilution in cell culture medium) at 37 °C and 5% CO2 for 4 h. After incubation, 100 µL medium/reagent were transferred into new 96-well plates accounting also for wells containing blanc solution (ND medium with CellTiter Blue reagent), and fluorescence was measured at 530–560 nm-/590 nm (excitation/emission) in a multiwell fluorimetric reader (Tecan). After blanc subtraction, data were normalised to the mean of control cells (i.e., cells in ND medium).
2.6. Immunocytochemistry of 3D neurospheres and confocal imaging
Three-week old neurospheres were fixed with 4% formaldehyde for 25 min, washed twice with PBS 1X (w/o calcium and magnesium) for 7 min, and stored in PBS 1X at 4 °C prior to staining. Neurospheres were incubated in PBS 1X containing 0.1% Triton-X-100% and 3.5% bovine serum albumin (BSA) (permeabilizing/blocking solution) for 30 min at room temperature under constant gyratory shaking. Neurospheres were incubated overnight (about 16 h) at 4 °C under constant gyratory shaking (50 rpm) with the following primary antibodies: β-III-tubulin (mouse, 1:500, Abcam Cat# ab41489, RRID:AB_727049, and chicken, 1:300, Abcam Cat# ab41489, RRID:AB_727049), glial fibrillary acidic protein (GFAP) (chicken, 1:500, Thermo Fisher Scientific Cat# 14–9892–82, RRID:AB_10598206), ACE2 (rabbit, 1:250, Sigma-Merck, Cat# SAB3500978), VGlut1 (rabbit, 1:250, Abcam Cat# ab72311, RRID:AB_1271456), GABRE (mouse 1:100, Thermo Fisher Scientific Cat# MA5–27696, RRID:AB_2735197), and Tyrosine hydroxylase (TH) (rabbit, 1:200, Thermo Fisher Scientific Cat# PA5–85167, RRID:AB_2792314), diluted in permeabilizing/blocking solution. The day after, neurospheres were washed twice with PBS 1X and further incubated for 1 h with Dy-Light -conjugated secondary antibodies (1:500, all from Abcam), and DAPI (1 µg/mL, ThermoFisher) in blocking solution (3.5% BSA in PBS 1X) under constant gyratory shaking. Neurospheres were transferred onto CytoVista™ Tissue Imaging Chamber (0.75 mm deep) on glass slides and mounted with ProLong™ Glass Antifade mounting medium (ThermoFisher). Pictures at 40x were taken using a Leica confocal microscope (Stellaris), and 3D reconstruction was done using Leica Application Suite X (LAS X) software (Version 4.1.0), considering taking images with 27–30 µm thickness and 0.8–1 µm z step size. Three biological replicates were considered, and at least 5 neurospheres were imaged for each experimental replicate.
2.7. Electrophysiological measurements using multi-well microelectrode array (MEA)
To assess the effects of Spike (5 and 10 µg/mL), Peptides (0.548 µg/mL) and a combination of both (‘P + S’) (vs control culture) on electrical activity, 2-week old neurospheres were manually transferred onto sterile 24-well microelectrode array (MEA) plates (24-well glass MEA plate (24W300/30 G-288) V.232) (30 neurospheres/500 µL/well) coated with polyethylenimine (PEI)- and mouse laminin. Cell cultures were further differentiated for at least 6 additional weeks in the presence of ND medium. Spontaneous electrical activity was recorded (for 5 min) starting 1 min after exposure and then every day up to 72 h (compounds were not refreshed during the 72 h). Electrical activity was recorded using the Multi-well MEA-System (Multi Channel Systems MCS GmbH), considering a Sampling Rate of 20000 Hz, a Low-Pass Filter Cutoff Frequency of 3500 Hz, and a High-Pass Filter Cutoff Frequency of 1 Hz. Spike detection was based on an automatic threshold estimation considering the following parameters: 20 individual segments, baseline duration (duration of each segment) of 100 ms, a rising edge of 5 St. Dev. and a falling edge of − 5 St. Dev, timing (dead time) of 3000 µs, cutouts pre trigger 1000 µs and post trigger 2000 µs, estimated for all wells. Unblinded data analyses of spike rate (number of spikes/sec) and burst count (considering a burst as a train of at least 4 spikes occurring within 50 ms, with maximum interval to start burst of 50 ms, maximum interval to end burst of 50 ms, minimum interval between bursts of 100 ms) were done using the "Multiwell-Analyzer" software, analysing the full recording and considering only active wells (i.e., wells characterized by at least 3 active channels, each active channel with minimum 10 spikes/min, and a minimum amplitude of 10 µV) (manual instructions are available at https://www.multichannelsystems.com/sites/multichannelsystems.com/files/documents/manuals/Multiwell-MEA-System_Manual.pdf) [59]. The average of spikes number and bursts number of selected active electrodes within each well were normalized to their respective T0 (i.e., cells not yet exposed to compounds). Four independent biological replicates were done, with 3–4 internal replicates per condition.
2.8. Quantitative PCR (qPCR) analysis of selected gene expression
Analysis of ACE2, MAP2 and GFAP gene expression by qPCR was performed in NSCs undergoing differentiation as 3D neurospheres (control culture) collecting samples for RNA isolation after 1, 2, 3, 4, 5 and 6 weeks of differentiation. Gene expression analysis of all other genes indicated in Table 1 was carried out in short-term and long-term differentiated cultures after 72 h exposure to Peptides 0.548 µg/mL, Spike 10 µg/mL, and P + S.
Table 1.
Genes and probes ID used for qPCR analysis (all from Thermo-Fisher).
| Gene name | Gene symbol | Assay ID |
|---|---|---|
| Angiotensin-Converting Enzyme 2 | ACE2 | Hs01085333_m1 |
| Sphingosine kinase 1 | SPHK1 | Hs00184211_m1 |
| Elastin | E LN | Hs00355783_m1 |
| Golgi associated kinase 1B | GASK1B (FAM198B) | Hs00930738_m1 |
| Hes Related Family BHLH Transcription Factor With YRPW Motif 1 | HEY1 | Hs00232618_m1 |
| Urotensin-II | UTS2 | Hs00922170_m1 |
| Microtubule Associated Protein 2 | MAP2 | Hs00258900_m1 |
| Paired Box 6 | PAX6 | Hs01088112_m1 |
| Nestin | NES | Hs04187831_g1 |
| Nuclear Receptor Subfamily 4 Group A Member 2 | NR4A2 | Hs00428691_m1 |
| Tyrosine Hydroxylase | TH | Hs00165941_m1 |
| Growth Associated Protein 43 | GAP43 | Hs00967138_m1 |
| Glutamate Ionotropic Receptor AMPA Type Subunit 1 | GRIA1 | Hs00181348_m1 |
| Glutamate Ionotropic Receptor AMPA Type Subunit 2 | GRIA2 | Hs00181331_m1 |
| Glutamate Ionotropic Receptor AMPA Type Subunit 3 | GRIA3 | Hs01557466_m1 |
| Choline O-Acetyltransferase | CHAT | Hs00252848_m1 |
| Solute Carrier Family 18 Member A3 | SLC18A3 | Hs00268179_s1 |
| Solute Carrier Family 5 Member 7 | SLC5A7 | Hs00222367_m1 |
| Gamma-Aminobutyric Acid Type A Receptor Subunit Alpha3 | GABRA3 | Hs00968132_m1 |
| Gamma-Aminobutyric Acid Type A Receptor Subunit Beta3 | GABRB3 | Hs00241459_m1 |
| Glial Fibrillary Acidic Protein | GFAP | Hs00909233_m1 |
| Bone Morphogenetic Protein Receptor Type 2 | BMPR2 | Hs00176148_m1 |
| Oligodendrocyte Transcription Factor 1 | OLIG1 | Hs00744293_s1 |
| Myelin Basic Protein | MBP | Hs00921945_m1 |
| Actin Beta | ACTB | Hs99999903_m1 |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | Hs02758991_g1 |
RNA was isolated using the RNAqueous®-Micro Kit (ThermoFisher) according to manufacturer's instructions, and 500 ng of total RNA were reverse transcribed by using the High Capacity cDNA Reverse Transcription Kit (as directed, ThermoFisher). qPCR reactions were run in duplicate using TaqMan® Gene Expression Master Mix (ThermoFisher) and the TaqMan gene expression assays indicated in Table 1. Amplification efficiencies of primers/probes were directly verified by the manufacturer (Thermo-Fisher) and were in the range of 100% (+/−10%) when measured over a 6-log dilution range (additional information are available at https://assets.thermofisher.com/TFS-Assets/LSG/Application-Notes/cms_040377.pdf) [60].
Fluorescent emission was recorded in real-time using the ABI PRISM Sequence Detection System 7900HT (ThermoFisher). PCR amplification conditions consisted of 45 cycles with primers annealing at 60 °C. Relative RNA quantities were normalized to the reference genes GAPDH and ACTB, and undifferentiated NSCs (for ACE2, MAP2 and GFAP expression in Fig. 1B-D) or Ctr (for all genes shown in Fig. 4, Fig. 5) were used to normalize the data (ΔΔCt Method). Three independent biological replicates were performed.
Fig. 1.
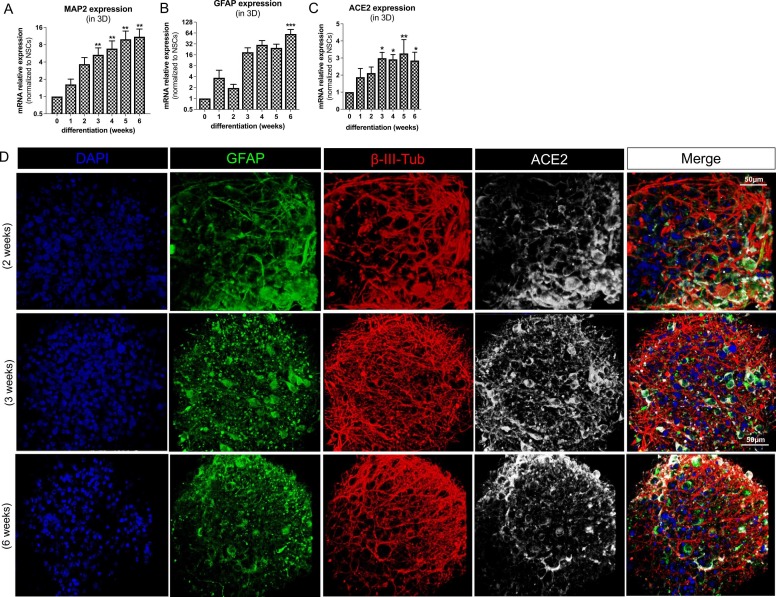
ACE2 expression in 3D neuronal/glial models undergoing differentiation. (A-C) Bar graphs showing MAP2, GFAP and ACE2 gene expression in 3D neurospheres differentiated for 0 (NSCs), 1, 2, 3, 4, 5 or 6 weeks; data were normalized to reference genes ACTB and GAPDH, and further normalized on NSCs (undifferentiated cells) (D) Representative fluorescent images (40x magnification) of 3D neurospheres differentiated for 2-, 3- and 6-weeks towards a mixed culture of neurons and glia; neurospheres were stained for GFAP (green), β-III-tubulin (red), ACE2 (white) and nuclei counterstained with DAPI (blue).
(A and B graphs are adapted from [55]).
Fig. 4.
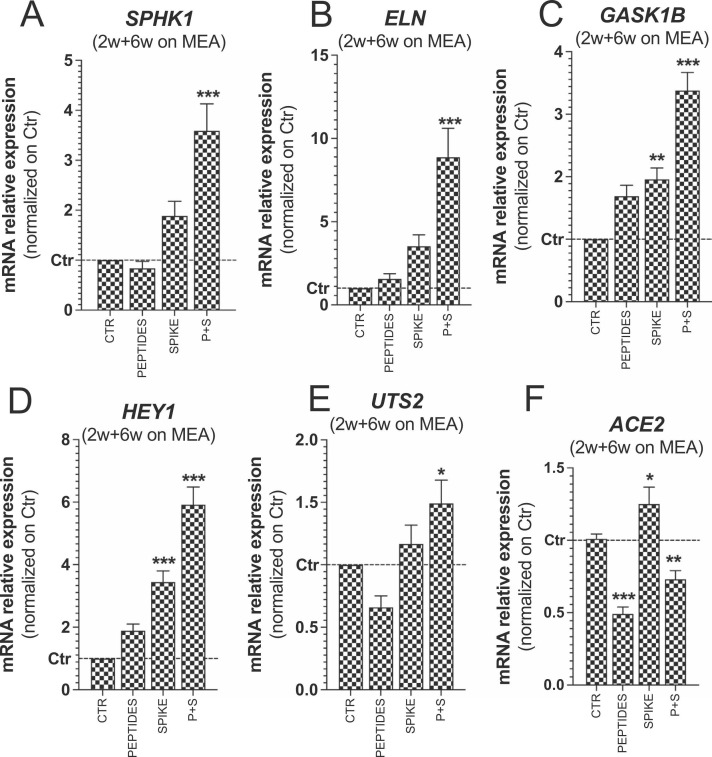
Effects of spike protein and toxin-like peptides on SPHK1, ELN, GASK1B, HEY1, UTS2 and ACE2 expression in long-term differentiated cultures. (A-F) Bar graphs showing expression of SPHK1, ELN, GASK1B, HEY1, UTS2 and ACE2 in long-term differentiated cultures exposed for 72 h to toxin-like peptides alone (0.548 µg/mL), spike protein alone (10 µg/mL) and a combination of both (P + S) vs Control. Data were normalized to reference genes ACTB and GAPDH, and further normalized to Ctr (mean ± S.E.M. of 3 biological replicates).
Fig. 5.
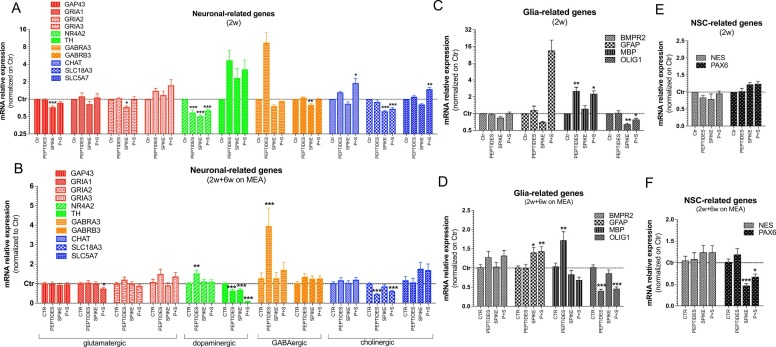
qPCR analyses of neuronal, glia and NSC-related genes in short and long-term differentiated cultures exposed for 72 h to peptides, spike protein and P + S. (A-F) Bar graphs showing expression of neuronal subtypes specific genes (A, B), glia-related genes (C, D), and NSC-related genes (E, F) in 2-week old neurosphere cultures (A, C, E) and 2-week old neurospheres further differentiated on MEA for 6 additional weeks (B, D, F). Both culture types were exposed at the end of differentiation period to toxin-like Peptides alone (0.548 µg/mL), Spike protein alone (10 µg/mL) and both compounds (P + S) vs Control for 72 h. Data were normalized to reference genes ACTB and GAPDH, and further normalized on Ctr (unexposed cells). For all analyses, mean ± S.E.M. of 3 biological replicates.
2.9. Whole transcriptome analysis by RNA sequencing (RNA-seq)
Analysis of gene expression by RNA-seq was performed in 2-week old neurospheres exposed for 72 h to Spike (10 µg/mL), Peptides (0.548 µg/mL) and a combination of both (‘P + S’) (vs Control) as briefly described. Neurospheres were decanted in 1.5 mL tubes, washed once with PBS 1X, lysed in 1X TempO-Seq Enhanced Lysis Buffer, and stored at − 80 °C prior to shipping. Samples (3 biological replicates) were supplied to BioClavis (BioClavis, ltd, Glasgow UK) (samples were received on 26/04/2021) for TempO-Seq analyses. The resulting FASTQ files were aligned using the STAR algorithm to the Human Whole Transcriptome v2.0 panel by BioClavis. BioClavis’ internal process control data confirmed the success of the sequencing (Supplementary Table 1). Next, a data matrix of gene expression level (raw counts) with sample names as column headers and gene names as rows was generated by BioClavis using the HTSEQ-count software. Further quality metrics were generated with the R package pcaExplorer (version 2.16.0) [61] . Expression analyses were performed using the DESeq2 (version 1.30.1) [62] and EdgeR (version 3.32.1) [63] R packages, relying on BioClavis’ data matrix filtered by low expressed genes (i.e., sum of raw counts < 10, considering all samples). Differentially expressed genes with FDR corrected p value < 0.1 were considered statistically significant. P value equal to 0.1 was set a priori before the data was analysed; a cut-off at 0.05 was also verified for comparative purposes.
2.10. Statistical analysis
Statistical significance of viability, MEA and qPCR data was assessed by one-way ANOVA with Dunnett's Multiple Comparison Test, comparing different conditions vs undifferentiated (NSC) or unexposed cells (Control or T0). Whole transcriptomic data were analysed by Wald Test. GraphPad Prism 9 software was used to compile and analyse data, which represent the average of at least three biological replicates ± standard error mean (S.E.M.). Data normality was visually assessed by Q-Q plot and statistically analysed using Shapiro-Wilk test and Kolmogorov-Smirnov test in GraphPad Prism 9. For all graphs, an asterisk over a data point indicates a significant difference with the control group or as indicated (* p < 0.05, ** p < 0.01, *** p < 0.001).
3. Results
3.1. Effects of spike protein and toxin-like peptides on cell viability and whole transcriptome analysis in short-term differentiated cultures
As expression of angiotensin-converting enzyme 2 (ACE2) receptor is critical to initiate viral entry into the cells through spike protein [64], [65], we verified the expression of ACE2 in hiPSC-derived NSCs undergoing differentiation towards a mixed culture of neuronal and glial cells as 3D neurospheres over 6 weeks (Fig. 1). Cultures underwent a progressive increase of MAP2 and GFAP gene expression over time (Fig. 1A and B, adapted from [55]); ACE2 appeared twice more expressed already after 1 week of differentiation (albeit not significant), and its expression progressively increased during differentiation (Fig. 1C).
ACE2 was found more expressed in astrocytes (positive for glial fibrillary acidic protein, GFAP, green), and at a lower level in neurons (stained with β-III-tubulin, red), as revealed by immunocytochemistry and confocal imaging analysis of 2-, 3- and 6-week old neurospheres (Fig. 1D).
Toxin-like peptides (Peptides) and spike protein (Spike) were first evaluated for their possible impact on cell viability in short-term differentiated (2-week old) cultures ( Fig. 2A-C), at a stage of increasing ACE2 expression. Two-week old neurospheres were exposed to different concentrations of Peptides (0.009, 0.017, 0.034, 0.069, 0.137, 0.548 and 1.096 µg/mL) and Spike (0.3, 0.6, 1.3, 2.5, 5.0, 10.0 and 20.0 µg/mL). Tested Spike levels are in line with spike proteins detected in the serum of some COVID-19 patients (between 2.5 and 17.5 µg/mL) [66], while tested levels of Peptides were lower than those found in blood of COVID-19 patients (on average 2–3 µg/mL) (Brogna and Cristoni, personal communication).
Fig. 2.
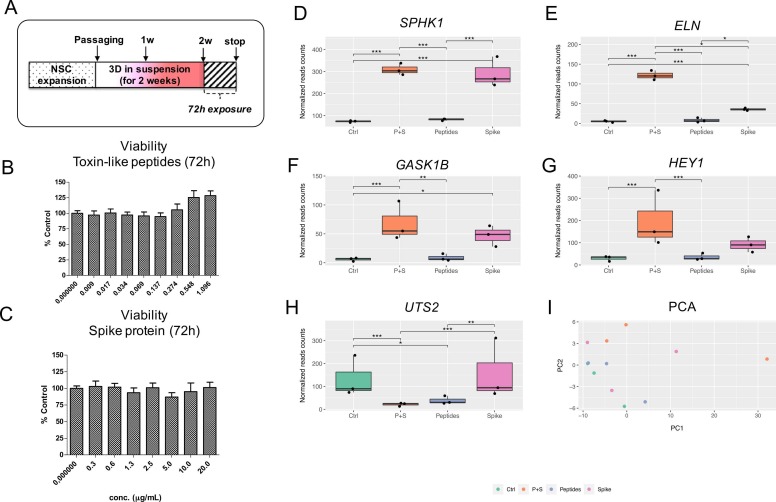
Effects of spike protein and toxin-like peptides on viability and whole transcriptome in 2-week old 3D neuronal/glial models after 72 h exposure. (A) NSCs were expanded, passaged and differentiated for 2 weeks as 3D neurospheres in suspension before being exposed for 72 h to different concentrations of toxin-like peptides or spike protein. (B, C) Cell viability analyses of 3D neurospheres after 72 h exposure to different concentrations of toxin-like peptides or spike protein; values were normalised to control (Ctr). (D-H) Differentially expressed genes with FDR corrected p value < 0.1; graphs show normalized reads counts for: Sphingosine kinase 1 (SPHK1) (D), Elastin (ELN) (E), Golgi associated kinase (GASK1B) (F), HEY1 (G), and Urotensin-II (UTS2) (H). (I) Principal component analysis (PCA). Data are representative of three biological replicates.
After 72 h, no impact on cell viability was observed in 3D neurospheres exposed to either different Peptides (Fig. 2B) or Spike (Fig. 2C), and no changes of neurosphere size could be observed (not shown).
We further assessed the effects of 72 h exposure to non-cytotoxic and physiologically relevant concentrations of Spike alone (10 µg/mL), Peptides alone (0.548 µg/mL) or a combination of both (vs Control) on overall transcriptome in 3D neurospheres pre-differentiated for 2 weeks (Fig. 2D-I). RNA-seq data showed that a small set of genes was significantly deregulated 72 h upon exposure to these compounds. In particular, by using DESeq2 (p value = 0.1), Sphingosine kinase 1 (SPHK1) (Fig. 2D), Elastin (ELN ) (Fig. 2E), and Golgi associated kinase (GASK1B) (Fig. 2F) were found upregulated by Spike and by the combination of Peptides+Spike (P + S). The use of EdgeR, confirmed the upregulation under the same conditions of SPHK1 and ELN , as well as the Notch3 effector HEY1 (Fig. 2G). By imposing a cut-off at 0.05, three genes, i.e., SPHK1, ELN and GASK1B showed to be differentially expressed, and with this cut-off no differences were found with respect to data reported in Fig. 2D-I.
Notably, while SPHK1, GASK1B and HEY1 upregulation was similar comparing samples exposed to Spike and P + S, the increase in ELN gene expression was significantly greater in cells exposed to P + S compared to Spike only (p < 0.05). On the contrary, Urotensin-II (UTS2) gene was down-regulated by both Peptides and P + S in a similar manner (Fig. 2H). Samples were characterized by homogeneous reads counts per sample and low variability, as shown by principal component analysis (PCA) (Fig. 2I).
3.2. Effects of spike protein and toxin-like peptides on spontaneous electrical activity in long-term differentiated cultures
We further explored the effects of Spike and Peptides on the generation of spontaneous electrical activity in long-term differentiated cultures by using the MEA technology. To this aim, neurospheres were pre-differentiated for 2 weeks in suspension, before being transferred onto MEA plates for recording of spontaneous electrical activity. Cultures were differentiated for at least 6 additional weeks on MEA before exposing them to Spike alone (5 and 10 µg/mL), Peptides alone (0.548 µg/mL) or a combination of both (P + S) vs Control cultures. Electrical activity was recorded 1 min after adding compounds (acute effects) and after 1-, 2- and 3-day exposure ( Fig. 3A). Under these exposure conditions, no cytotoxic effects were observed, and about 34% increase of cell viability was recorded upon exposure to Spike (10 µg/mL) alone and in combination with Peptides (P + S) (Fig. 3B).
Fig. 3.
Effects of spike protein and toxin-like peptides on electrical activity in long-term differentiated 3D neuronal/glial models. (A) NSCs were expanded, passaged and differentiated for 2 weeks as 3D neurospheres in suspension before being transferred to 24-well MEA plates for at least 6 additional weeks (total of 8–9 weeks in differentiation). Cultures were exposed for 72 h to Spike alone (5 and 10 µg/mL), Peptides alone (0.548 µg/mL) or a combination of both, and electrical activity was recorded for 5 min before (T0), 1 min after adding compounds (acute), and after 1-, 2- and 3-day exposure. (B) Cell viability analyses of cultures after 72 h exposure as indicated in A (values were normalised to Ctr). (C) Representative phase contrast images of cell cultures on 24-well MEA plates exposed to compounds as described in A. (D, E) Bar graphs showing spike rate (i.e., number of spikes/sec) (D), and number of bursts (E) of cultures exposed to compounds as described in A (data were normalized to T0). For all analyses, mean ± S.E.M. of 4 biological replicates.
MEA data showed that Peptides induced a decrease of spontaneous electrical activity (~30% decrease of spike rate) after 2d, whilst Spike at the concentration of 5 µg/mL had no significant impact on electrical activity (Fig. 3C, D, E). Spike protein at the higher tested concentration (10 µg/mL) caused a decrease of both spike rate (by ~57%) and overall number of bursts (by ~66%) after 2d (Fig. 3D, E). Combined exposure to both P + S-5 µg/mL caused a significant decrease of both spike rate (by ~50%) and bursts (by ~63%) after 1d, and a similar decrease was observed after 2d exposure to P + S-10 µg/mL (spike rate by ~43% and bursts by ~61%) (Fig. 3D, E). The effects elicited by Spike-10 alone were similar to those elicited by the combined exposure to P + S-10 µg/mL; on the other hand, the combined exposure to P + S-5 µg/mL had a greater impact on spontaneous electrical activity formation than either Spike-5 alone or Peptides alone, which may suggest a potentiated effect.
3.3. Effects of spike protein and toxin-like peptides on SPHK1, ELN, GASK1B, HEY1, UTS2 and ACE2 expression in long-term differentiated cultures
Moreover, we assessed by qPCR analysis the expression of SPHK1, ELN, GASK1B, HEY1 and UTS2 in long-term differentiated cultures upon exposure to tested compounds ( Fig. 4A-E). Similar to RNA-seq data on short-term differentiated cultures, P + S elicited a significant increase of SPHK1, ELN, GASK1B and HEY1; Spike was the main trigger of these effects, causing a significant increase of GASK1B and HEY1 expression (Fig. 4A-D). The expression of UTS2 was very modestly modulated under all conditions, showing a tendency towards a downregulation upon Peptides exposure, and a slight upregulation upon P + S exposure (Fig. 4E).
Notably, the expression of ACE2 did not significantly change in short-term differentiated cultures (not shown). However, in long-term cultures, ACE2 expression was found downregulated upon exposure to Peptides, while this downregulation was milder upon exposure to P + S; on the contrary, Spike protein caused a very modest but significant upregulation of ACE2 (Fig. 4F).
3.4. Effects of spike protein and toxin-like peptides on selected neuronal-, glia- and NSC-related gene expression in short- and long-term differentiated cultures
Finally, we analysed gene expression of a set of genes expressed by different neuronal subtypes and glial cells in both short- (2-weeks) and long-term differentiated cultures (2-weeks + 6-weeks on MEA) comparing the effects of Peptides alone (0.548 µg/mL), Spike alone (10 µg/mL), and P + S vs Ctr cultures. These data showed that Spike caused a slight decrease of GAP43 and GRIA2 in 2-week old neurospheres, while a very modest decrease of GRIA1 was seen after exposure to P + S in long-term differentiated cultures ( Fig. 5A, B).
The GABAergic gene GABRA3 was upregulated in both culture types after Peptide exposure (although not significantly in short-term differentiated culture), while GABRB3 resulted slightly downregulated after Spike exposure in short-term differentiated culture (Fig. 5A, B).
The dopaminergic genes NR4A2 and TH were found differentially regulated in short- vs long-term differentiated cultures, with NR4A2 undergoing downregulation in short-term differentiated cultures and found upregulated in long-term differentiated cultures exposed to Peptides, and TH showing a tendency towards an increase in short-term differentiated culture, and a significant decrease in long-term differentiated cultures at all tested conditions (Fig. 5A, B).
Morever, the cholinergic gene SLC18A3 resulted downregulated upon exposure to Spike and P + S (in short-term differentiated), and Peptides and P + S (in long-term differentiated cultures), whilst CHAT and SLC5A7 were found upregulated in short-term differentiated cultures upon exposure to P + S (Fig. 5A, B).
We also looked at the expression of some glia-related genes. While expression of BMPR2 did not change at either conditions, GFAP expression showed a tendency toward an increase in short-term differentiated culture exposed to P + S (not significant), whilst long-term differentiated cultures underwent a significant increase of GFAP expression upon exposure to both Spike and P + S (Fig. 5C, D). Noteworthy, MBP expression increased by about 3-fold in short-term differentiated cultures exposed to Peptides and P + S, while its expression was modestly upregulated by Peptides in long-term differentiated cultures. The oligodendrocyte marker OLIG1 was found significantly downregulated in short-term differentiated cells exposed to Spike and P + S, and resulted downregulated by Peptides and P + S in long-term differentiated cultures (Fig. 5C, D).
The increase in cell viability relative to control cultures observed in neurospheres exposed to Spike proteins (Fig. 3B) may reflect possible differences in cell proliferation. In line with this hypothesis, we assessed in both short- and long-term differentiated neurosphere cultures the expression of NES and PAX6, which are expressed in dividing NSCs [67], [68]. While the expression of both genes did not change in 2-week old neurospheres at any tested condition, PAX6 was found significantly downregulated by both Spike and P + S in long-term cultures, whilst NES expression did not significantly change (Fig. 5E, F).
4. Discussion
In a previous study [30], we reported the presence of toxin-like peptides in plasma, urine and fecal samples exclusively from COVID-19 patients. At present, the origin of these Peptides, whether they can cross the placenta, and what possible detrimental effects they may have on human perinatal development and the developing brain are unknown. In this study, we investigated the neurotoxic effects elicited upon 72 h exposure to Spike, Peptides mixture found in COVID-19 patients, and a combination of both (P + S) on human iPSC-derived NSCs differentiated towards a mixed culture of neurons/glia as 3D neurospheres at different differentiation stages. Expression of both glia and neuronal-related genes increased during differentiation in this 3D model, whose characterization is described in [55]. Notably, glia-related genes and proteins were observed as early as 2 weeks of differentiation [55], whilst other studies on hiPSC-derived neuronal and glial cell derivatives cultured as 3D organoids or neurospheres have shown that several weeks in differentiation were needed to obtain mature glia (e.g., [46], [69], [70]). Differences in temporal occurrence of gene expression and protein level changes may be linked to genetic, epigenetic and phenotypic differences in test systems, as well as differences in media formulation and experimental design. Notably, expression of ACE2 was found upregulated during differentiation in a time dependent manner, with ACE2 protein observed mainly in astrocytes, which suggests that 3D neurospheres may plausibly be infected by SARS-CoV-2 virus via Spike-ACE2 interaction. Our data show that Spike protein at 10 µg/mL caused a decrease of spontaneous electrical activity after 2d exposure in long-term differentiated cultures, and when combined with Peptides a similar decrease was observed. Notably, while 5 µg/mL Spike did not cause significant perturbations of electrical activity, when combined with Peptides, potentiated effects could be hypothesized, with a decrease of spike rate and bursts observed after 1d. Noteworthy, network connectivity perturbation observed in long-term differentiated cultures may be associated with the observed dysregulation of some critical neuronal-, glia- and NSC-related genes.
4.1. Neurodevelopment-related genes found dysregulated upon exposure to Peptides and Spike protein
GABRA3 gene expression (which resulted upregulated in both short- and long-term cultures upon exposure to Peptides) constitutes the dominant subunit in the forebrain tissue at birth [71]. GABRA3 upregulation has been associated with testosterone-mediated impulsive behaviour in rats [72], and was observed upon exposure to the psychostimulant methamphetamine [73]. On the other hand, glutamatergic gene expression (GAP43, GRIA1, GRIA2, GRIA3) was minimally modulated in both culture types at either conditions.
TH is known to catalyse the conversion of L-tyrosine to L-3,4-dihydroxyphenylalanine (L-DOPA), which is the initial and rate-limiting step in the biosynthesis of dopamine, noradrenaline, and adrenaline. TH resulted strongly downregulated particulalry in long-term cultures exposed to Peptides and Spike together. Noteworthy, TH downregulation has been observed in Parkinson’s disease (PD) [74] and in early parkinsonism [75].
Also downregulation of NR4A2 (Nurr1, a nuclear receptor essential for the differentiation, survival and maintenance of midbrain dopaminergic neurons), which was observed in short-term differentiated cultures upon exposure to all tested compounds, is associated with PD as observed in both PD patients and animal models [76], as well as neuroinflammation and neuronal cell death [77].
SLC18A3 (which was found downregulated mainly by Spike in short-term cultures, and Peptides in long-term cultures) encodes the Vesicular acetylcholine transporter (VAChT), which transports acetylcholine into secretory vesicles for release into the extracellular space [78]. A deficit of VAChT, induced by SLC18A3 variants, are associated with the presynaptic congenital myasthenic syndrome, which is functionally characterized by electrodecrement on low-frequency repetitive stimulation and a prolonged period of postactivation exhaustion, as shown by electrophysiology studies conducted on congenital myasthenic syndrome affected patients [79].
The observed OLIG1 downregulation induced by Spike in short-term differentiated cultures and by Peptides in long-term differentiated cultures, as well as the upregulation of MBP mainly elicited by Peptides in both short- and long-term differentiated cultures, suggest a dysregulation of oligodendrocyte development and maturation. Indeed, OLIG1 controls oligodendrocyte precursor differentiation into myelin-forming oligodendrocytes during development, and together with SOX10, activates MBP transcription [80].
Additionally, the tendency towards an upregulation of GFAP expression induced by P + S (not significant) in short-term cultures and the upregulation mainly induced by Spike in long-term cultures, could be associated with astrogliosis and microglial activation. In line with this, transgenic mice overexpressing wild type GFAP that develop encephalopathy showed upregulation of genes involved in glutathione metabolism, peroxide detoxification, and iron homeostasis at 3 months of age [81], as well as increased activation of cytokine, cytokine receptor genes, and complement components. With ageing, these transcripts resulted further elevated, with additional induction of macrophage-specific markers, indicating activation of microglia [81], [82], [83].
Additionally, PAX6 (which was found downregulated by Spike in long-term cultures), is an important transcription factor that controls NSC proliferation, multipotency, neurogenesis and cortical development [68], and its mutation or deletion has been shown to cause major brain defects and several neurodevelopmental disorders in the developing embryo [84]. Exposure to Spike protein may therefore have an impact on NSC self-renewal/proliferation.
Altogether these data suggest that Peptides and Spike protein in short- and long-term differentiated cultures, differentially affect genes involved in NSC self-renewal/proliferation, neuronal and glial differentiation, which may ultimately be associated with the observed perturbation of electrical activity.
4.2. Possible effects associated with SPHK1, ELN, GASK1B, HEY1, UTS2 and ACE2 dysregulation
Whole transcriptome analysis in short-term differentiated cultures revealed the upregulation of SPHK1, ELN, GASK1B, and HEY1 upon exposure to Spike or P + S, as well as the downregulation of UTS2 upon exposure to Peptides and P + S. qPCR analysis of these five genes showed that these genes were deregulated by tested compounds in a similar manner also in long-term differentiated cultures.
Upregulation of SPHK1 expression may be linked to induction of a pro-survival, pro-inflammatory mechanism in short-term and long-term cultures upon exposure to Spike and P + S. SPHK1 is known to control cell survival, migration and inflammation [85], [86], and its deregulation has been observed in various inflammatory and immune related-diseases, such as hypertension, atherosclerosis, Alzheimer's disease, inflammatory bowel disease, rheumatoid arthritis, and asthma [87], [88], [89]. It is also known to regulate microglial phagocytosis [88], and to modulate inflammation during cerebral ischemia/reperfusion injury [90], [91]. Moreover, SPHK1/S1PR2 signaling axis is closely associated with the course of Temporal Lobe Epilepsy [92]. The sphingolipid rheostat has been shown to play a role in viral replication, immune response modulation, and the maintenance of blood vessel integrity [93], [94].
The gene ELN (found upregulated especially by P + S in short- and long-term cultures) codes for elastin, which, together with elastases, play an important role in the aging of the arterial wall, skin and other connective tissues [95], [96], and elastin-derived peptides have been shown to potentiate atherosclerotic plaque formation [97]. ELN is also responsible for the vascular and connective tissue features of Williams syndrome, a relatively rare microdeletion disorder, characterized by cardiovascular disease, distinctive craniofacial appearance, intellectual disability and hypersociability [98]. Notably, ELN was found to play a neuroprotective role in response to preterm ischemia-hypoxia brain damage in a rat model [99]. To our knowledge, no studies have to date investigated the possible role of ELN in relation to SARS-CoV-2 associated brain (neurodevelopmental) sequelae.
Another gene found upregulated upon exposure to Spike and P + S in short- and long-term differentiated cultures is GASK1B (also known as FAM198A or C3orf41), which is expressed in nerve and epithelium during development [100], and is an integral active component of the Golgi apparatus, known to play a fundamental role in SARS-CoV-2 virion assembly [101]. Additionally, GASK1B is one of the most important caveolae-associated proteins, whose secretion plays an important role in the caveolae biogenesis pathway [102]. Notably, caveolae are involved in numerous membrane functions, including membrane trafficking and lipid metabolism, cell motility, and viral infection [103], [104]. Contrary to other coronaviridae, which use caveolae for internalization, SARS-CoV-2 seems to preferentially undergo clathrin-mediated endocytosis; however, the role of caveolae in SARS-CoV-2 remains disputed [105], with different plausible modes of entry depending also on the cell type [106].
HEY1 (found upregulated mainly upon exposure to P + S in short-term cultures) is a major Notch3 effector controlling NSC stemness in the vertebrate adult brain [107]. Overexpression of HEY1 has been shown to promote astrocyte differentiation and to inhibit neuronal differentiation in murine neural progenitor cells [108]. HEY1 resulted upregulated in long-term cultures exposed to Spike and more prominently P + S; together with PAX6 down-regulation observed under the same conditions, this suggests dysregulation of neuronal/glial differentiation processes in our test system mainly occurring as a consequence of Spike exposure.
Urotensin-II (UTS2) was found downregulated by Peptides and P + S in short-term cultures; in long-term cultures a tendency towards UTS2 downregulation was observed upon Peptides exposure, resulting very slightly upregulated upon P + S exposure. UTS2 is an 11-aminoacid neuropeptide that interacts with the urotensin receptor (UT), a specific G-protein coupled receptor [109]. UTS2 has both vasoconstrictor and vasodilatory actions, modulates cell proliferation, pro-fibrosis, neuroendocrine activity, controls insulin resistance, and has carcinogenic and inflammatory effects, playing a role in the onset and development of inflammatory diseases [110]. The ‘UTS2–UT’ system is widely distributed in cardiovascular tissue, the nervous system (especially cardiovascular control centres), the kidney, and the respiratory tract, and its downregulation has been linked to numerous pathophysiological conditions [109]. Interestingly, UTS2 has been found as an ACE2 and TMPRSS2 correlated gene by KEGG pathways analysis in several brain regions, including hypothalamus, insula, amygdala, myelencephalon, and parabrachial nuclei of pons, as shown by human brain gene-expression analyses and immunohistochemistry [111]. Additionally, UTS2 receptors have been found expressed in presynaptic cholinergic terminals in a subset of motor neuronal and non-motor neuronal perikarya, as well as in non-cholinergic nerve terminals, as observed in the ventral horn of the adult mouse cervical spinal cord [112]. In our study, downregulation of UTS2 was mainly triggered by Peptides, which could be linked to Peptides’ affinity towards nicotinic acetylcholine receptor [30].
In both short- and long-term cultures, apart from the decrease of UTS2 (triggered by Peptides), upregulation of SPHK1, ELN, GASK1B and HEY1 were mainly triggered by Spike, and these effects were slightly greater in cell cultures exposed to both Spike and Peptides (P + S), suggesting a possible exacerbating role of Peptides.
Noteworthy, expression of ACE2 resulted downregulated in long-term differentiated cultures exposed to Peptides. ACE2 expression starts to increase already after 1–2 weeks of differentiation in our neuronal/glial model; along the same line, ACE2 expression has been shown in neurons and glial cells in the brain (in particular in the brain stem and cardiovascular regulatory areas) [113], and may play a key role in the neural invasion of SARS-CoV-2 [114]. Downregulation of ACE2 has been shown to occur in response to SARS-CoV-2 cellular invasion [115]. Notably, ACE2 downregulation plays an important role in COVID-19 severity, with an imbalance of the renin-angiotensin system and consequential increase in the levels of substrates (e.g., angiotensin II, apelin-13, dynorphin-13), and a decrease of products (e.g., angiotensin (1−7), angiotensin (1−9), apelin-12, and dynorphin-12) in the human body. Substrates accumulation can cause inflammation, angiogenesis, thrombosis, neuronal and tissue damage; on the other hand, ACE2 products’ depletion can reduce the anti-inflammatory, anti-thrombotic and anti-angiogenic responses [116]. In particular, angiotensin (1−7) has a neuroprotective role, and its decrease may be associated with oxidative stress and neuronal cell death [117]. Moreover, ACE2 downregulation has been associated with glutamate-induced excitotoxicity in primary mouse cortical neurons [118].
4.3. Possible link between genes found dysregulated and key events described in neuro-related AOPs
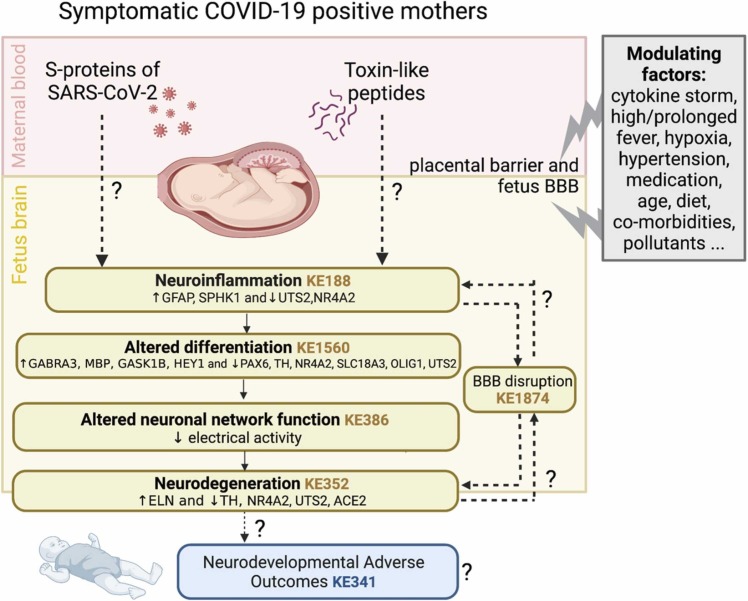
The observed transcriptional and functional developmental neurotoxic effects described in this study are in line with mechanistic knowledge described in COVID-19 relevant AOPs under development in the context of the CIAO project [119], [120], [121] and in other neuro-related AOPs. Potential penetration of SARS-CoV-2 and of contaminating toxin-like peptides present in maternal blood through placenta and blood-brain barriers may trigger neuroinflammation in the developing fetus, as suggested by increase in GFAP expression and SPHK1, along with the decrease of NR4A2 and UTS2. Neuroinflammation (described in KE188) may lead to alteration of differentiation (partially described in KE1560), which could be linked to dysregulation of critical neuronal-, glia- and NSC-related gene expression, as suggested by downregulation of PAX6, TH, NR4A2, SLC18A3, OLIG1 and UTS2, and the upregulation of GABRA3, MBP, GASK1B and HEY1. This may have detrimental effects on the modulation of neuronal as well as oligodendroglia differentiation, which may lead to alteration of neuronal network functions (KE386) as measured by the decrease of spontaneous electrical activity. Moreover, alteration of neuronal functionality may cause neurodegeneration (KE352), which may be characterized also by dysregulation of critical genes, such as NR4A2, TH, UTS2, ELN , and ACE2 as observed in our study. Noteworthy, both neuroinflammation and neurodegeneration may cause BBB disruption (KE1874). Ultimately, this sequence of KEs may lead to neurodevelopmental adverse outcomes, such as a decrease of learning and memory in children (KE341), which could be potentially monitored only in the next years in children born from infected mothers. Altogether, the emerging evidence prompts to implement AOPs with additional KEs and AOs relevant to the developing brain/fetus as this mechanistic knowledge may help clarify the mechanisms underlying possible SARS-CoV-2 impact on the developing brain.
The crucial question is still the possibility for SARS-CoV-2 to penetrate the developing fetus. Two studies have described placental viral invasion shown by immunohistochemistry and electron microscopy analyses [7], [8]. On the contrary, other studies have reported negative results for the presence of the virus in the neonates and placenta of pregnant women affected by SARS-CoV-2 [17]. Additionally, other studies reported about the presence of SARS-CoV-2 in one infant and one fetus affected from COVID-19 [10], [21].
In general, recent systematic reviews suggest that the likelihood of vertical transmission of SARS-CoV-2, which is only known for late third trimester infections, may be low [16], [18], [19], [20], [122]. However, the cytokine storm and hyperinflammation observed in pregnant women severely affected by COVID-19 [17], [25], [123] may cause perturbation of placenta integrity; prolonged fever, hypoxia, hypertension and medication side effects may exacerbate this phenomenon ( Fig. 6). It is presently unclear whether vertical transmission might also vary with different SARS-CoV-2 variants, which remains an open question for the future.
Fig. 6.
Summary of the effects possibly triggered by SARS-CoV-2 Spike protein and toxin-like Peptides, along with Key Events (KEs) describing these mechanisms, and additional modulating factors. Under each KE, the endpoints (genes and electrical activity) that have been found deregulated in this study are reported. Dashed lines and question marks indicate still unknown (not yet verified) processes
(image created with BioRender.com).
In the context of the CIAO project, several modulating factors, such as age, diet, gut microbiota, pre-existing comorbidities, environmental pollutants, etc., were investigated as factors modulating COVID-19 symptomatology [50]. Toxin-like peptides may represent additional exacerbating/detrimental modulatory factors possibly influencing SARS-CoV-2 impact on brain development.
4.4. Effects induced by SARS-CoV-2 Spike protein
In this study, we could not test the direct effects of SARS-CoV-2 viral particles for biosafety related reasons, thus limited our investigation to the effects of Spike protein, a critical component of viral structure.
Spike protein alone has been shown in one study to induce several damages associated with COVID-19, including damage to the lungs and arteries, inflammation of endothelial cells lining the pulmonary artery walls, together with impairment of mitochondrial function, decrease of ACE2 expression and eNOS activity, and increase of glycolysis as observed in vitro (on endothelial cells treated for 24 h with 4 μg/mL Spike) and in vivo (upon intratracheal administration of a pseudovirus expressing Spike protein to Syrian hamsters) [124]. Spike proteins (S1 and active trimer, 15 and 30 nM) have been found to induce mitochondrial damage in brain endothelial cells [125], and spike protein epitopes have been shown to interact with human toll-like receptor 8 (TLR 8), brain targeted Vascular Cell adhesion Molecules (VCAM1) proteins, Zonula Occludens (ZO), and some glia specific proteins (i.e., NDRG2 and Apo- S100B), which can lead to neuroinflammation [126]. Moreover, 10 nM SARS-CoV-2 viral spike proteins have been shown to alter BBB functions and induce pro-inflammatory response after 24 h in primary human brain microvascular endothelial cells (hBMVECs) cultured in 2D and 3D [127]. Radioiodinated spike S1 (12.5 ng per 300,000 c.p.m. BSA) has been found to cross the BBB and enter the parenchymal brain space in intravenously injected mice [128].
The concentrations of Spike protein tested in this study (5 and 10 µg/mL), are similar to what has been observed in the plasma of some COVID-19 patients (2.5–17.5 µg/mL) [66]. To the best of our knowledge, there are no published studies reporting the levels of spike protein in pregnant women affected by COVID-19. With regards to levels of spike protein found upon vaccination with mRNA-vaccines, very few studies are available. Ogata et al., by using a Quanterix assay, found that spike protein levels in the blood of people vaccinated with mRNA-1273 was < 50 pg/mL after vaccination [129], i.e., several orders of magnitude lower than the levels of spike tested in our study. On the other hand, Cognetti and Miller, by using a Disposable Photonics platform, found that 1–3 days after injection of mRNA vaccine BNT162b2, spike levels underwent an average shift of 15 pm, with a maximum concentration of 14.6 μg/mL, returning to a baseline level in less than a month [130]. The discrepancy between the two studies is high and not yet validated. Besides and again, the important missing data is the concentration of Spike proteins in cord blood or placenta.
Results reported in the present study are preliminary and would require further investigations, such as to assess the molecular and cellular effects induced by repeated dose exposure to Spike protein and Peptides, as well as the kinetics of spike protein entry into the cells. However, emerging evidence prompts the importance to further investigate and monitor the presently unknown long-term effects elicited by SARS-CoV-2 viral components on brain development. Use of S proteins, non-replicating SARS-CoV2 pseudoviruses and SARS-CoV-2 viruses might be informative as well to discriminate the impact of S protein binding on the receptor compared to viral entry and the infectious process.
Additionally, in this study we only assessed the expression of ACE2. Future analyses should aim to investigate the expression of other spike protein interactors (i.e., FURIN, ZDHHC5, GOLGA7 and ATP1A1) that are expressed at a high level in the fetal brain [29], in order to understand their role as alternative brain entry factors for SARS-CoV-2.
5. Conclusions
The functional and transcriptional perturbations described in the present study could contribute understanding some of the mechanisms underlying the neurodevelopmental manifestations that could be possibly associated with severe COVID-19. Our approach shows that the use of human in vitro models is crucial to gather insights about spike protein and SARS-CoV-2 effects and the role played by contaminating toxin-like peptides that have been exclusively found in biological samples of COVID-19 patients. Integrating emerging knowledge in AOPs could help improve interpretation of scientific understanding of COVID-19 pathological mechanisms in the fetal brain, and clarify the possible effects of SARS-CoV-2 viral components on the developing fetus.
Declaration of Competing Interest
The authors have no conflict of interests to declare.
Acknowledgements
The authors would like to thank Dr. Marc Peschanski (I-Stem, Évry, France) for providing IMR90-hiPSCs, and Dr. Anna Navarro Cuenca for providing the license for the use of BioRender.com.
Editor: Anna Price
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.reprotox.2022.04.011.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- 1.Zambrano L.D., Ellington S., Strid P., Galang R.R., Oduyebo T., Tong V.T., Woodworth K.R., Nahabedian J.F., 3rd, Azziz-Baumgartner E., Gilboa S.M., Meaney-Delman D., Pregnancy C.C.-R., Infant Linked Outcomes T. Update: characteristics of symptomatic women of reproductive age with laboratory-confirmed SARS-CoV-2 Infection by pregnancy status - United States, January 22-October 3, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69(44)):1641–1647. doi: 10.15585/mmwr.mm6944e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar-Valles A., Rodrigue B., Matta-Camacho E. Maternal immune activation and the development of dopaminergic neurotransmission of the offspring: relevance for schizophrenia and other psychoses. Front. Psychiat. 2020;11:852. doi: 10.3389/fpsyt.2020.00852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Figueiredo C.P., Fontes-Dantas F.L., da Poian A.T., Clarke J.R. SARS-CoV-2-associated cytokine storm during pregnancy as a possible risk factor for neuropsychiatric disorder development in post-pandemic infants. Neuropharmacology. 2021;201 doi: 10.1016/j.neuropharm.2021.108841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A.L. Düppers, B. Bohnhorst, E. Bültmann, T. Schulz, L. Higgins-Wood, C.S. von Kaisenberg, Severe fetal brain damage subsequent to acute maternal hypoxemic deterioration in COVID-19 Ultrasound in obstetrics & gynecology: the official journal of the International Society of Ultrasound in Obstetrics and Gynecology 58 3 2021 490 491. [DOI] [PMC free article] [PubMed]
- 5.G.M. Fernandes, F. Motta, L.M.P. Sasaki, P.D. Silva, Â A.M. Miranda, A.O. Carvalho, A.P.M. Gomides, A. Soares, A. Santos Jr., C.O. Alves, C.M. Gomes, C.C. Siracusa, D.A. Araújo Jr., D.L. Mendonça-Silva, J.A.L. Jesus, K.N. Costa, M.E.C. Castro, P.S. Kurizky, P.S. França, R. Tristão, Y.R. Pereira, L.C.G. Castro, A.M. Zaconeta, C.P. Albuquerque, L. Mota, Pregnancy outcomes and child development effects of SARS-CoV-2 Infection (PROUDEST Trial): Protocol for a Multicenter, Prospective Cohort Study, JMIR Research Protocols 10 4 2021 e26477. [DOI] [PMC free article] [PubMed]
- 6.Martins-Filho P.R., Tanajura D.M., Santos H.P., Jr., Santos V.S. COVID-19 during pregnancy: Potential risk for neurodevelopmental disorders in neonates? Eur. J. Obs. Gynecol. Reprod.Biol. 2020;250:255–256. doi: 10.1016/j.ejogrb.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosier H., Farhadian S.F., Morotti R.A., Deshmukh U., Lu-Culligan A., Campbell K.H., Yasumoto Y., Vogels C.B., Casanovas-Massana A., Vijayakumar P., Geng B., Odio C.D., Fournier J., Brito A.F., Fauver J.R., Liu F., Alpert T., Tal R., Szigeti-Buck K., Perincheri S., Larsen C., Gariepy A.M., Aguilar G., Fardelmann K.L., Harigopal M., Taylor H.S., Pettker C.M., Wyllie A.L., Cruz C.D., Ring A.M., Grubaugh N.D., Ko A.I., Horvath T.L., Iwasaki A., Reddy U.M., Lipkind H.S. SARS-CoV-2 infection of the placenta. J. Clinical Invest. 2020;130(9):4947–4953. doi: 10.1172/JCI139569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Algarroba G.N., Rekawek P., Vahanian S.A., Khullar P., Palaia T., Peltier M.R., Chavez M.R., Vintzileos A.M. Visualization of severe acute respiratory syndrome coronavirus 2 invading the human placenta using electron microscopy. Am. J. Obs. Gynecol. 2020;223(2):275–278. doi: 10.1016/j.ajog.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng H., Xu C., Fan J., Tang Y., Deng Q., Zhang W., Long X. Antibodies in infants born to mothers with COVID-19 pneumonia. Jama. 2020;323(18):1848–1849. doi: 10.1001/jama.2020.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong L., Tian J., He S., Zhu C., Wang J., Liu C., Yang J. Possible vertical transmission of SARS-CoV-2 from an infected mother to her newborn. Jama. 2020;323(18):1846–1848. doi: 10.1001/jama.2020.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pique-Regi R., Romero R., Tarca A.L., Luca F., Xu Y., Alazizi A., Leng Y., Hsu C.D., Gomez-Lopez N. Does the human placenta express the canonical cell entry mediators for SARS-CoV-2? Elife. 2020;9 doi: 10.7554/eLife.58716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egloff C., Vauloup-Fellous C., Picone O., Mandelbrot L., Roques P. Evidence and possible mechanisms of rare maternal-fetal transmission of SARS-CoV-2. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knyazev E., Nersisyan S., Tonevitsky A. Endocytosis and transcytosis of SARS-CoV-2 across the intestinal epithelium and other tissue barriers. Front. immunol. 2021;12 doi: 10.3389/fimmu.2021.636966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baud D., Greub G., Favre G., Gengler C., Jaton K., Dubruc E., Pomar L. Second-trimester miscarriage in a pregnant woman with SARS-CoV-2 infection. Jama. 2020;323(21):2198–2200. doi: 10.1001/jama.2020.7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecht J.L., Quade B., Deshpande V., Mino-Kenudson M., Ting D.T., Desai N., Dygulska B., Heyman T., Salafia C., Shen D., Bates S.V., Roberts D.J. SARS-CoV-2 can infect the placenta and is not associated with specific placental histopathology: a series of 19 placentas from COVID-19-positive mothers. Mod. Pathol. 2020;33(11):2092–2103. doi: 10.1038/s41379-020-0639-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotlyar A.M., Grechukhina O., Chen A., Popkhadze S., Grimshaw A., Tal O., Taylor H.S., Tal R. Vertical transmission of coronavirus disease 2019: a systematic review and meta-analysis. Am. J. Obs. Gynecol. 2021;224(1):35–53. doi: 10.1016/j.ajog.2020.07.049. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu-Culligan A., Chavan A.R., Vijayakumar P., Irshaid L., Courchaine E.M., Milano K.M., Tang Z., Pope S.D., Song E., Vogels C.B.F., Lu-Culligan W.J., Campbell K.H., Casanovas-Massana A., Bermejo S., Toothaker J.M., Lee H.J., Liu F., Schulz W., Fournier J., Muenker M.C., Moore A.J., Yale I.T., Konnikova L., Neugebauer K.M., Ring A., Grubaugh N.D., Ko A.I., Morotti R., Guller S., Kliman H.J., Iwasaki A., Farhadian S.F. Maternal respiratory SARS-CoV-2 infection in pregnancy is associated with a robust inflammatory response at the maternal-fetal interface. Medicine. 2021;2(5):591–610. doi: 10.1016/j.medj.2021.04.016. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Maiahy T.J., Al-Kuraishy H.M., Al-Gareeb A.I. Pregnancy and risk of vertical transmission in Covid-19. J. Pak. Med. Assoc. 2021;71(Suppl 8):S137–S143. [PubMed] [Google Scholar]
- 19.Ashraf P.K.M.A., Hosseinpour P., Erfani A., Roshanshad A., Pourdast A., Nowrouzi-Sohrabi P., Chaichian S., Poordast T. Coronavirus disease 2019 (COVID-19): a systematic review of pregnancy and the possibility of vertical transmission. J. Reprod. Infert. 2020;3:157–168. [PMC free article] [PubMed] [Google Scholar]
- 20.Rodrigues C., Baia I., Domingues R., Barros H. Pregnancy and breastfeeding during COVID-19 pandemic: a systematic review of published pregnancy cases. Front. in Pub. Health. 2020;8 doi: 10.3389/fpubh.2020.558144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shende P., Gaikwad P., Gandhewar M., Ukey P., Bhide A., Patel V., Bhagat S., Bhor V., Mahale S., Gajbhiye R., Modi D. Persistence of SARS-CoV-2 in the first trimester placenta leading to transplacental transmission and fetal demise from an asymptomatic mother. Human Reprod. 2021;36(4):899–906. doi: 10.1093/humrep/deaa367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu A.L., Guan M., Johannesen E., Stephens A.J., Khaleel N., Kagan N., Tuhlei B.C., Wan X.F. Placental SARS-CoV-2 in a pregnant woman with mild COVID-19 disease. J. Med. Virol. 2021;93(2):1038–1044. doi: 10.1002/jmv.26386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marinho P.S., da Cunha A., Chimelli L., Avvad-Portari E., Andreiuolo F.D.M., De Oliveira-Szejnfeld P.S., Mendes M.A., Gomes I.C., Souza L.R.Q., Guimarães M.Z., Goldman S.M., de Oliveira M.B.G., Rehen S., Amim J., Jr., Tovar-Moll F., Prata-Barbosa A. Case Report: SARS-CoV-2 mother-to-child transmission and fetal death associated with severe placental thromboembolism. Front. Med. 2021;8 doi: 10.3389/fmed.2021.677001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vivanti A.J., Vauloup-Fellous C., Prevot S., Zupan V., Suffee C., Do Cao J., Benachi A., De Luca D. Transplacental transmission of SARS-CoV-2 infection. Nat. Commun. 2020;11(1):3572. doi: 10.1038/s41467-020-17436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Flores V., Romero R., Xu Y., Theis K.R., Arenas-Hernandez M., Miller D., Peyvandipour A., Bhatti G., Galaz J., Gershater M., Levenson D., Pusod E., Tao L., Kracht D., Florova V., Leng Y., Motomura K., Para R., Faucett M., Hsu C.D., Zhang G., Tarca A.L., Pique-Regi R., Gomez-Lopez N. Maternal-fetal immune responses in pregnant women infected with SARS-CoV-2. Nat. Commun. 2022;13(1):320. doi: 10.1038/s41467-021-27745-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashary N., Bhide A., Chakraborty P., Colaco S., Mishra A., Chhabria K., Jolly M.K., Modi D. Single-Cell RNA-seq Identifies Cell Subsets in Human Placenta That Highly Expresses Factors Driving Pathogenesis of SARS-CoV-2. Front. Cell Dev. Biol. 2020;8:783. doi: 10.3389/fcell.2020.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M., Chen L., Zhang J., Xiong C., Li X. The SARS-CoV-2 receptor ACE2 expression of maternal-fetal interface and fetal organs by single-cell transcriptome study. PloS one. 2020;15(4) doi: 10.1371/journal.pone.0230295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fahmi A., Brugger M., Demoulins T., Zumkehr B., Oliveira Esteves B.I., Bracher L., Wotzkow C., Blank F., Thiel V., Baud D., Alves M.P. SARS-CoV-2 can infect and propagate in human placenta explants. Cell Rep. Med. 2021;2(12) doi: 10.1016/j.xcrm.2021.100456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varma P., Lybrand Z.R., Antopia M.C., Hsieh J. Novel Targets of SARS-CoV-2 spike protein in human fetal brain development suggest early pregnancy vulnerability. Front. Neurosci. 2020;14 doi: 10.3389/fnins.2020.614680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.C. Brogna, S. Cristoni, M. Petrillo, M. Querci, O. Piazza, G. Van den Eede, Toxin-like peptides in plasma, urine and faecal samples from COVID-19 patients, F1000Research 2021. [DOI] [PMC free article] [PubMed]
- 31.Brogna C. The Covid-19 virus double pathogenic mechanism. New Perspect. Preprints. 2020 2020040165. [Google Scholar]
- 32.Petrillo M., Brogna C., Cristoni S., Querci M., Piazza O., Van G. den Eede, Increase of SARS-CoV-2 RNA load in faecal samples prompts for rethinking of SARS-CoV-2 biology and COVID-19 epidemiology. F1000Res. 2021;10:370. doi: 10.12688/f1000research.52540.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Y.X. Chen, Z.Y. Xu, X. Ge, S. Sanyal, Z.J. Lu, B. Javid, Selective translation by alternative bacterial ribosomes Proceedings of the National Academy of Sciences of the United States of America 117 32 2020 19487 19496. [DOI] [PMC free article] [PubMed]
- 34.Coleman J., Green P.J., Inouye M. The use of RNAs complementary to specific mRNAs to regulate the expression of individual bacterial genes. Cell. 1984;37(2):429–436. doi: 10.1016/0092-8674(84)90373-8. [DOI] [PubMed] [Google Scholar]
- 35.Chen N., Xu S., Zhang Y., Wang F. Animal protein toxins: origins and therapeutic applications. Biophy. Rep. 2018;4(5):233–242. doi: 10.1007/s41048-018-0067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bordon K.C.F., Cologna C.T., Fornari-Baldo E.C., Pinheiro-Júnior E.L., Cerni F.A., Amorim F.G., Anjolette F.A.P., Cordeiro F.A., Wiezel G.A., Cardoso I.A., Ferreira I.G., de Oliveira I.S., Boldrini-França J., Pucca M.B., Baldo M.A., Arantes E.C. From animal poisons and venoms to medicines: achievements, challenges and perspectives in drug discovery. Front. Pharmacol. 2020;11:1132. doi: 10.3389/fphar.2020.01132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.F. Jacob, S.R. Pather, W.K. Huang, S.Z.H. Wong, H. Zhou, F. Zhang, B. Cubitt, C.Z. Chen, M. Xu, M. Pradhan, D.Y. Zhang, W. Zheng, A.G. Bang, H. Song, A.T.J.C. de, G.L. Ming, Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism, bioRxiv: the preprint server for biology (2020). [DOI] [PMC free article] [PubMed]
- 38.Pellegrini L., Albecka A., Mallery D.L., Kellner M.J., Paul D., Carter A.P., James L.C., Lancaster M.A. SARS-CoV-2 Infects the Brain Choroid Plexus and Disrupts the Blood-CSF Barrier in Human Brain Organoids. Cell Stem Cell. 2020;27(6):951–961. doi: 10.1016/j.stem.2020.10.001. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramani A., Müller L., Ostermann P.N., Gabriel E., Abida-Islam P., Müller-Schiffmann A., Mariappan A., Goureau O., Gruell H., Walker A., Andrée M., Hauka S., Houwaart T., Dilthey A., Wohlgemuth K., Omran H., Klein F., Wieczorek D., Adams O., Timm J., Korth C., Schaal H., Gopalakrishnan J. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020;39(20) doi: 10.15252/embj.2020106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.E. Song, C. Zhang, B. Israelow, P. Lu, O.E. Weizman, F. Liu, Neuroinvasive potential of SARS-CoV-2 revealed in a human brain organoid model, bioRxiv: the preprint server for biology (2020).
- 41.Zhang B.Z., Chu H., Han S., Shuai H., Deng J., Hu Y.F., Gong H.R., Lee A.C., Zou Z., Yau T., Wu W., Hung I.F., Chan J.F., Yuen K.Y., Huang J.D. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020;30(10):928–931. doi: 10.1038/s41422-020-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chesnut M., Hartung T., Hogberg H., Pamies D. Human Oligodendrocytes and Myelin In Vitro to Evaluate Developmental Neurotoxicity. Int. J. Mol. Sci. 2021;22(15) doi: 10.3390/ijms22157929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chesnut M., Paschoud H., Repond C., Smirnova L., Hartung T., Zurich M.G., Hogberg H.T., Pamies D. Human IPSC-derived model to study myelin disruption. Int. J. Mol. Sci. 2021;22(17) doi: 10.3390/ijms22179473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.T. Kadoshima, H. Sakaguchi, T. Nakano, M. Soen, S. Ando, M. Eiraku, Y. Sasai, Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex Proceedings of the National Academy of Sciences of the United States of America 110 50 2013 20284 20289. [DOI] [PMC free article] [PubMed]
- 45.Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pamies D., Barreras P., Block K., Makri G., Kumar A., Wiersma D., Smirnova L., Zang C., Bressler J., Christian K.M., Harris G., Ming G.L., Berlinicke C.J., Kyro K., Song H., Pardo C.A., Hartung T., Hogberg H.T. A human brain microphysiological system derived from induced pluripotent stem cells to study neurological diseases and toxicity. Altex. 2017;34(3):362–376. doi: 10.14573/altex.1609122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong X., Harris G., Smirnova L., Zufferey V., Sá R., Baldino Russo F., Baleeiro Beltrao Braga P.C., Chesnut M., Zurich M.G., Hogberg H.T., Hartung T., Pamies D. Antidepressant paroxetine exerts developmental neurotoxicity in an iPSC-Derived 3D human brain model. Front. Cell. Neurosci. 2020;14:25. doi: 10.3389/fncel.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.I. Mesci, A. Macia, A. Saleh, L. Martin-Sancho, X. Yin, C. Snethlage, S. Avansini, S.K. Chanda, A. Muotri, Sofosbuvir protects human brain organoids against SARS-CoV-2, bioRxiv: the preprint server for biology, 2020.
- 49.Nymark P., Sachana M., Leite S.B., Sund J., Krebs C.E., Sullivan K., Edwards S., Viviani L., Willett C., Landesmann B., Wittwehr C. Systematic organization of COVID-19 data supported by the adverse outcome pathway framework. Front. Pub. Health. 2021;9 doi: 10.3389/fpubh.2021.638605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clerbaux L.A., Amigo N., Amorim M.J., Bal-Price A., Batista Leite S., Beronius A., Bezemer G.F.G., Bostroem A.C., Carusi A., Coecke S., Concha R., Daskalopoulos E.P., Debernardi F., Edrosa E., Edwards S.W., Filipovska J., Garcia-Reyero N., Gavins F.N.E., Halappanavar S., Hargreaves A.J., Hogberg H.T., Huynh M.T., Jacobson D., Josephs-Spaulding J., Kim Y.J., Kong H.J., Krebs C.E., Lam A., Landesmann B., Layton A., Lee Y.O., Macmillan D.S., Mantovani A., Margiotta-Casaluci L., Martens M., Masereeuw R., Mayasich S.A., Mei L.M., Mortensen H., Munoz Pineiro A., Nymark P., Ohayon E., Ojasi J., Paini A., Parissis N., Parvatam S., Pistollato F., Sachana M., Sorli J.B., Sullivan K.M., Sund J., Tanabe S., Tsaioun K., Vinken M., Viviani L., Waspe J., Willett C., Wittwehr C. COVID-19 through adverse outcome pathways: building networks to better understand the disease - 3rd CIAO AOP Design Workshop. Altex. 2022 doi: 10.14573/altex.2112161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wittwehr C., Amorim M.J., Clerbaux L.A., Krebs C., Landesmann B., Macmillan D.S., Nymark P., Ram R., Garcia-Reyero N., Sachana M., Sullivan K., Sund J., Willett C. Understanding COVID-19 through adverse outcome pathways - 2nd CIAO AOP design workshop. Altex. 2021;38(2):351–357. doi: 10.14573/altex.2102221. [DOI] [PubMed] [Google Scholar]
- 52.Sachana M., Willett C., Pistollato F., Bal-Price A. The potential of mechanistic information organised within the AOP framework to increase regulatory uptake of the developmental neurotoxicity (DNT) in vitro battery of assays. Reprod.Toxicol. 2021;103:159–170. doi: 10.1016/j.reprotox.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pistollato F., Carpi D., Gyves E.M., Paini A., Bopp S.K., Worth A., Bal-Price A. Combining in vitro assays and mathematical modelling to study developmental neurotoxicity induced by chemical mixtures. Reprod Toxicol. 2021;105:101–119. doi: 10.1016/j.reprotox.2021.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pistollato F., de Gyves E.M., Carpi D., Bopp S.K., Nunes C., Worth A., Bal-Price A. Assessment of developmental neurotoxicity induced by chemical mixtures using an adverse outcome pathway concept. Environ. Health. 2020;19(1):23. doi: 10.1186/s12940-020-00578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nunes C., Gorczyca G., Mendoza-de Gyves E., Ponti J., Bogni A., Price A., Pistollato F. Upscaling biological complexity to boost neuronal/glial maturation and improve in vitro developmental neurotoxicity (DNT) evaluation (under review) Reprod Toxicol. 2022;110:124–140. doi: 10.1016/j.reprotox.2022.03.017. [DOI] [PubMed] [Google Scholar]
- 56.Coriell, NIA Aging Cell Repository at Coriell Institute for Medical Research.
- 57.Pistollato F., Louisse J., Scelfo B., Mennecozzi M., Accordi B., Basso G., Gaspar J.A., Zagoura D., Barilari M., Palosaari T., Sachinidis A., Bremer-Hoffmann S. Development of a pluripotent stem cell derived neuronal model to identify chemically induced pathway perturbations in relation to neurotoxicity: effects of CREB pathway inhibition. Toxicol. Appl. Pharmacol. 2014;280(2):378–388. doi: 10.1016/j.taap.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 58.Pistollato F., Canovas-Jorda D., Zagoura D., Price A. Protocol for the Differentiation of Human Induced Pluripotent Stem Cells into Mixed Cultures of Neurons and Glia for Neurotoxicity Testing. J. Vis. Exp. 2017;124 doi: 10.3791/55702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Multiwell-Mea-System, Version 2, 2021.
- 60.Amplification Effi ciency of TaqMan® Gene Expression Assays, in: A.A. Biosystems (Ed.).
- 61.Marini F., Binder H. pcaExplorer: an R/Bioconductor package for interacting with RNA-seq principal components. BMC Bioinform. 2019;20(1):331. doi: 10.1186/s12859-019-2879-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu W., Zhang Y., Fei P., Zhang T., Yao D., Gao Y., Liu J., Chen H., Lu Q., Mudianto T., Zhang X., Xiao C., Ye Y., Sun Q., Zhang J., Xie Q., Wang P.H., Wang J., Li Z., Lou J., Chen W. Mechanical activation of spike fosters SARS-CoV-2 viral infection. Cell Res. 2021;31(10):1047–1060. doi: 10.1038/s41422-021-00558-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qiao J., Li W., Bao J., Peng Q., Wen D., Wang J., Sun B. The expression of SARS-CoV-2 receptor ACE2 and CD147, and protease TMPRSS2 in human and mouse brain cells and mouse brain tissues. Biochem. Biophys. Res. Commun. 2020;533(4):867–871. doi: 10.1016/j.bbrc.2020.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.S. George, A. Chattopadhyay, Pal J. Gagnon, S. Timalsina, P. Singh, P. Vydyam, M. Munshi, J.E. Chiu, I. Renard, C.A. Harden, I.M. Ott, A.E. Watkins, C.B.F. Vogels, P. Lu, M. Tokuyama, A. Venkataraman, A. Casanovas-Massana, A.L. Wyllie, V. Rao, M. Campbell, S.F. Farhadian, N.D. Grubaugh, C.S. Dela Cruz, A.I. Ko, Evidence for SARS-CoV-2 Spike Protein in the Urine of COVID-19 patients Kidney360 2 6 2021 924 936. [DOI] [PMC free article] [PubMed]
- 67.Wiese C., Rolletschek A., Kania G., Blyszczuk P., Tarasov K.V., Tarasova Y., Wersto R.P., Boheler K.R., Wobus A.M. Nestin expression--a property of multi-lineage progenitor cells? Cell Mol. Life Sci. 2004;61(19–20):2510–2522. doi: 10.1007/s00018-004-4144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sansom S.N., Griffiths D.S., Faedo A., Kleinjan D.J., Ruan Y., Smith J., van Heyningen V., Rubenstein J.L., Livesey F.J. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5(6) doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koroleva A., Deiwick A., El-Tamer A., Koch L., Shi Y., Estevez-Priego E., Ludl A.A., Soriano J., Guseva D., Ponimaskin E., Chichkov B. In Vitro Development of Human iPSC-Derived Functional Neuronal Networks on Laser-Fabricated 3D Scaffolds. ACS Appl. Mater. Interfaces. 2021;13(7):7839–7853. doi: 10.1021/acsami.0c16616. [DOI] [PubMed] [Google Scholar]
- 70.Marton R.M., Miura Y., Sloan S.A., Li Q., Revah O., Levy R.J., Huguenard J.R., Pasca S.P. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat. Neurosci. 2019;22(3):484–491. doi: 10.1038/s41593-018-0316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ohlson J., Pedersen J.S., Haussler D., Ohman M. Editing modifies the GABA(A) receptor subunit alpha3. RNA. 2007;13(5):698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Agrawal J., Dwivedi Y. GABAA receptor subunit transcriptional regulation, expression organization, and mediated calmodulin signaling in prefrontal cortex of rats showing testosterone-mediated impulsive behavior. Front. Neurosci. 2020;14 doi: 10.3389/fnins.2020.600099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wearne T.A., Parker L.M., Franklin J.L., Goodchild A.K., Cornish J.L. GABAergic mRNA expression is upregulated in the prefrontal cortex of rats sensitized to methamphetamine. Behav. Brain Res. 2016;297:224–230. doi: 10.1016/j.bbr.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 74.Nagatsu T., Nakashima A., Ichinose H., Kobayashi K. Human tyrosine hydroxylase in Parkinson’s disease and in related disorders. J. Neural. Transm.) 2019;126(4):397–409. doi: 10.1007/s00702-018-1903-3. [DOI] [PubMed] [Google Scholar]
- 75.van Bloemendaal L., Ijzerman R.G., Ten Kulve J.S., Barkhof F., Diamant M., Veltman D.J., van Duinkerken E. Alterations in white matter volume and integrity in obesity and type 2 diabetes. Metab. Brain Dis. 2016;31(3):621–629. doi: 10.1007/s11011-016-9792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu X., He X., Ma S., Li M., Huang Q. Nurr1 downregulation is caused by CREB inactivation in a Parkinson’s disease mouse model. Neurosci. lett. 2021;759 doi: 10.1016/j.neulet.2021.136045. [DOI] [PubMed] [Google Scholar]
- 77.Jakaria M., Haque M.E., Cho D.Y., Azam S., Kim I.S., Choi D.K. Molecular insights into NR4A2(Nurr1): an emerging target for neuroprotective therapy against neuroinflammation and neuronal cell death. Mol. Neurobiol. 2019;56(8):5799–5814. doi: 10.1007/s12035-019-1487-4. [DOI] [PubMed] [Google Scholar]
- 78.SLC18A3 Gene - Solute Carrier Family 18 Member A3. 〈https://www.genecards.org/cgi-bin/carddisp.pl?gene=SLC18A3〉.
- 79.O'Grady G.L., Verschuuren C., Yuen M., Webster R., Menezes M., Fock J.M., Pride N., Best H.A., Benavides Damm T., Turner C., Lek M., Engel A.G., North K.N., Clarke N.F., MacArthur D.G., Kamsteeg E.J., Cooper S.T. Variants in SLC18A3, vesicular acetylcholine transporter, cause congenital myasthenic syndrome. Neurology. 2016;87(14):1442–1448. doi: 10.1212/WNL.0000000000003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wong C.K.H., Xiong X., Lau K.T.K., Chui C.S.L., Lai F.T.T., Li X., Chan E.W.Y., Wan E.Y.F., Au I.C.H., Cowling B.J., Lee C.K., Wong I.C.K. Impact of a delayed second dose of mRNA vaccine (BNT162b2) and inactivated SARS-CoV-2 vaccine (CoronaVac) on risks of all-cause mortality, emergency department visit, and unscheduled hospitalization. BMC Med. 2022;20(1):119. doi: 10.1186/s12916-022-02321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hagemann T.L., Gaeta S.A., Smith M.A., Johnson D.A., Johnson J.A., Messing A. Gene expression analysis in mice with elevated glial fibrillary acidic protein and Rosenthal fibers reveals a stress response followed by glial activation and neuronal dysfunction. Hum. Mol. Genet. 2005;14(16):2443–2458. doi: 10.1093/hmg/ddi248. [DOI] [PubMed] [Google Scholar]
- 82.Quinlan R.A., Brenner M., Goldman J.E., Messing A. GFAP and its role in Alexander disease. Exp. Cell Res. 2007;313(10):2077–2087. doi: 10.1016/j.yexcr.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schitine C., Nogaroli L., Costa M.R., Hedin-Pereira C. Astrocyte heterogeneity in the brain: from development to disease. Front. Cell. Neurosci. 2015;9:76. doi: 10.3389/fncel.2015.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Manuel M.N., Mi D., Mason J.O., Price D.J. Regulation of cerebral cortical neurogenesis by the Pax6 transcription factor. Front. Cell. Neurosci. 2015;9:70. doi: 10.3389/fncel.2015.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Z., Min X., Xiao S.H., Johnstone S., Romanow W., Meininger D., Xu H., Liu J., Dai J., An S., Thibault S., Walker N. Molecular basis of sphingosine kinase 1 substrate recognition and catalysis. Structure. 2013;21(5):798–809. doi: 10.1016/j.str.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 86.Kharel Y., Mathews T.P., Gellett A.M., Tomsig J.L., Kennedy P.C., Moyer M.L., Macdonald T.L., Lynch K.R. Sphingosine kinase type 1 inhibition reveals rapid turnover of circulating sphingosine 1-phosphate. Biochem. J. 2011;440(3):345–353. doi: 10.1042/BJ20110817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bu Y., Wu H., Deng R., Wang Y. Therapeutic Potential of SphK1 Inhibitors Based on Abnormal Expression of SphK1 in Inflammatory Immune Related-Diseases. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.733387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee J.Y., Han S.H., Park M.H., Baek B., Song I.S., Choi M.K., Takuwa Y., Ryu H., Kim S.H., He X., Schuchman E.H., Bae J.S., Jin H.K. Neuronal SphK1 acetylates COX2 and contributes to pathogenesis in a model of Alzheimer’s Disease. Nat. Commun. 2018;9(1):1479. doi: 10.1038/s41467-018-03674-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Giovagnoni C., Ali M., Eijssen L.M.T., Maes R., Choe K., Mulder M., Kleinjans J., Del Sol A., Glaab E., Mastroeni D., Delvaux E., Coleman P., Losen M., Pishva E., Martinez-Martinez P., van den Hove D.L.A. Altered sphingolipid function in Alzheimer’s disease; a gene regulatory network approach. Neurobiol. Aging. 2021;102:178–187. doi: 10.1016/j.neurobiolaging.2021.02.001. [DOI] [PubMed] [Google Scholar]
- 90.Zhou F., Wang Y.K., Zhang C.G., Wu B.Y. miR-19a/b-3p promotes inflammation during cerebral ischemia/reperfusion injury via SIRT1/FoxO3/SPHK1 pathway. J. Neuroinflamm. 2021;18(1):122. doi: 10.1186/s12974-021-02172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang M., Zhou D., Ouyang Z., Yu M., Jiang Y. Sphingosine kinase 1 promotes cerebral ischemia-reperfusion injury through inducing ER stress and activating the NF-κB signaling pathway. J. Cell. Physiol. 2020;235(10):6605–6614. doi: 10.1002/jcp.29546. [DOI] [PubMed] [Google Scholar]
- 92.Dong Y.Y., Xia M., Wang L., Cui S., Li Q.B., Zhang J.C., Meng S.S., Zhang Y.K., Kong Q.X. Spatiotemporal Expression of SphK1 and S1PR2 in the Hippocampus of Pilocarpine Rat Model and the Epileptic Foci of Temporal Lobe Epilepsy. Front. Cell Dev. Biol. 2020;8:800. doi: 10.3389/fcell.2020.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xiong Y., Hla T. S1P control of endothelial integrity. Curr. Topics Microbiol. Immunol. 2014;378:85–105. doi: 10.1007/978-3-319-05879-5_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Spiegel S., Milstien S. The outs and the ins of sphingosine-1-phosphate in immunity. Nat. Rev. Immunol. 2011;11(6):403–415. doi: 10.1038/nri2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Robert L., Jacob M.P., Frances C., Godeau G., Hornebeck W. Interaction between elastin and elastases and its role in the aging of the arterial wall, skin and other connective tissues. A review. Mechan. Ageing Dev. 1984;28(2–3):155–166. doi: 10.1016/0047-6374(84)90015-0. [DOI] [PubMed] [Google Scholar]
- 96.Scandolera A., Rabenoelina F., Chaintreuil C., Rusciani A., Maurice P., Blaise S., Romier-Crouzet B., El Btaouri H., Martiny L., Debelle L., Duca L. Uncoupling of elastin complex receptor during in vitro aging is related to modifications in its intrinsic sialidase activity and the subsequent lactosylceramide production. PloS one. 2015;10(6) doi: 10.1371/journal.pone.0129994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gayral S., Garnotel R., Castaing-Berthou A., Blaise S., Fougerat A., Berge E., Montheil A., Malet N., Wymann M.P., Maurice P., Debelle L., Martiny L., Martinez L.O., Pshezhetsky A.V., Duca L., Laffargue M. Elastin-derived peptides potentiate atherosclerosis through the immune Neu1-PI3Kγ pathway. Cardiovasc. Res. 2014;102(1):118–127. doi: 10.1093/cvr/cvt336. [DOI] [PubMed] [Google Scholar]
- 98.Kozel B.A., Barak B., Kim C.A., Mervis C.B., Osborne L.R., Porter M., Pober B.R. Williams syndrome. Nat. Rev. Dis. Primers. 2021;7(1):42. doi: 10.1038/s41572-021-00276-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Feng E., Jiang L. Peptidomic analysis of hippocampal tissue for explore leptin neuroprotective effect on the preterm ischemia-hypoxia brain damage model rats. Neuropharmacology. 2020;162 doi: 10.1016/j.neuropharm.2019.107803. [DOI] [PubMed] [Google Scholar]
- 100.nextprot, GASK1B - Function, 2021. 〈https://www.nextprot.org/entry/NX_Q6UWH4/〉. (Accessed 15 Nov 2021.
- 101.Tiwari R., Mishra A.R., Gupta A., Nayak D. Structural similarity-based prediction of host factors associated with SARS-CoV-2 infection and pathogenesis. J. Biomol. Struct. Dynam. 2021:1–12. doi: 10.1080/07391102.2021.1874532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wei Z., Liu T., Lei J., Wu Y., Wang S., Liao K. Fam198a, a member of secreted kinase, secrets through caveolae biogenesis pathway. Acta Biochimica Et Biophysica Sinica. 2018;50(10):968–975. doi: 10.1093/abbs/gmy105. [DOI] [PubMed] [Google Scholar]
- 103.Thompson M.A., Prakash Y.S., Pabelick C.M. The role of caveolae in the pathophysiology of lung diseases. Expert Rev. Resp. Med. 2014;8(1):111–122. doi: 10.1586/17476348.2014.855610. [DOI] [PubMed] [Google Scholar]
- 104.Nassar Z.D., Parat M.O. Cavin family: new players in the biology of caveolae. International Rev. Cell Mol. Biol. 2015;320:235–305. doi: 10.1016/bs.ircmb.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 105.Filippini A., D’Alessio A. Caveolae and lipid rafts in endothelium: valuable organelles for multiple functions. Biomolecules. 2020;10(9) doi: 10.3390/biom10091218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Glebov O.O. Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing, The. FEBS journal. 2020;287(17):3664–3671. doi: 10.1111/febs.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Than-Trong E., Ortica-Gatti S., Mella S., Nepal C., Alunni A., Bally-Cuif L. Neural stem cell quiescence and stemness are molecularly distinct outputs of the Notch3 signalling cascade in the vertebrate adult brain. Development. 2018;145(10) doi: 10.1242/dev.161034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jalali A., Bassuk A.G., Kan L., Israsena N., Mukhopadhyay A., McGuire T., Kessler J.A. HeyL promotes neuronal differentiation of neural progenitor cells. J. Neurosci. Res. 2011;89(3):299–309. doi: 10.1002/jnr.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Castel H., Desrues L., Joubert J.E., Tonon M.C., Prézeau L., Chabbert M., Morin F., Gandolfo P. The G Protein-Coupled Receptor UT of the Neuropeptide Urotensin II Displays Structural and Functional Chemokine Features. Front. Endocrinol. 2017;8:76. doi: 10.3389/fendo.2017.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.S.L. Sun, L.M. Liu, Urotensin II: an inflammatory cytokine, The Journal of endocrinology 2019. [DOI] [PubMed]
- 111.S. Nampoothiri, F. Sauve, G. Ternier, D. Fernandois, C. Coelho, M. Imbernon, E. Deligia, R. Perbet, V. Florent, M. Baroncini, F. Pasquier, F. Trottein, C.A. Maurage, V. Mattot, P. Giacobini, S. Rasika, V. Prevot, The hypothalamus as a hub for SARS-CoV-2 brain infection and pathogenesis, bioRxiv: the preprint server for biology 2020.
- 112.Bruzzone F., Cervetto C., Mazzotta M.C., Bianchini P., Ronzitti E., Leprince J., Diaspro A., Maura G., Vallarino M., Vaudry H., Marcoli M. Urotensin II receptor and acetylcholine release from mouse cervical spinal cord nerve terminals. Neuroscience. 2010;170(1):67–77. doi: 10.1016/j.neuroscience.2010.06.070. [DOI] [PubMed] [Google Scholar]
- 113.Bourgonje A.R., Abdulle A.E., Timens W., Hillebrands J.L., Navis G.J., Gordijn S.J., Bolling M.C., Dijkstra G., Voors A.A., Osterhaus A.D., van der Voort P.H., Mulder D.J., van Goor H. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19) J. Pathol. 2020;251(3):228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tang X., Zheng F. A review of ischemic stroke in COVID-19: currently known pathophysiological mechanisms. Neurol. Sci. 2022;43(1):67–79. doi: 10.1007/s10072-021-05679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mehrabadi M.E., Hemmati R., Tashakor A., Homaei A., Yousefzadeh M., Hemati K., Hosseinkhani S. Induced dysregulation of ACE2 by SARS-CoV-2 plays a key role in COVID-19 severity. Biomed. Pharmacother. 2021;137 doi: 10.1016/j.biopha.2021.111363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li T., Huang H.Y., Wang H.D., Gao C.C., Liang H., Deng C.L., Zhao X., Han Y.L., Zhou M.L. Restoration of brain angiotensin-converting enzyme 2 alleviates neurological deficits after severe traumatic brain injury via mitigation of pyroptosis and apoptosis. J. Neurotrauma. 2022;39(5–6):423–434. doi: 10.1089/neu.2021.0382. [DOI] [PubMed] [Google Scholar]
- 118.Xu J., Sriramula S., Lazartigues E. Excessive glutamate stimulation impairs ACE2 activity through ADAM17-mediated shedding in cultured cortical neurons. Cell. Mol. Neurobiol. 2018;38(6):1235–1243. doi: 10.1007/s10571-018-0591-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.A. Price, F. Pistollato, AOP 374: Binding of Sars-CoV-2 spike protein to ACE 2 receptors expressed on brain cells (neuronal and non-neuronal) leads to neuroinflammation resulting in encephalitis, 2021. 〈https://aopwiki.org/aops/374〉. (Accessed 15 Nov 2021).
- 120.F. De Bernardi, S. Saravan, A. Munoz, N. Sachana, S. Coecke, AOP 394: Sars-cov-2 infection leading to impaired olfactory function (anosmia), 2021. (Accessed Nov 15, 2021).
- 121.M. Sachana, AOP 395: Binding of Sars-CoV-2 spike protein to ACE 2 receptors expressed on pericytes leads to disseminated intravascular coagulation resulting in cerebrovascular disease (stroke), 2021. 〈https://aopwiki.org/aops/395〉. (Accessed 15 Nov 2021.
- 122.Novoa R.H., Quintana W., Llancari P., Urbina-Quispe K., Guevara-Rios E., Ventura W. Maternal clinical characteristics and perinatal outcomes among pregnant women with coronavirus disease 2019. A systematic review. Travel Med. Infect Dis. 2021;39 doi: 10.1016/j.tmaid.2020.101919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gee S., Chandiramani M., Seow J., Pollock E., Modestini C., Das A., Tree T., Doores K.J., Tribe R.M., Gibbons D.L. The legacy of maternal SARS-CoV-2 infection on the immunology of the neonate. Nat. Immunol. 2021;22(12):1490–1502. doi: 10.1038/s41590-021-01049-2. [DOI] [PubMed] [Google Scholar]
- 124.Lei Y., Zhang J., Schiavon C.R., He M., Chen L., Shen H., Zhang Y., Yin Q., Cho Y., Andrade L., Shadel G.S., Hepokoski M., Lei T., Wang H., Zhang J., Yuan J.X., Malhotra A., Manor U., Wang S., Yuan Z.Y., Shyy J.Y. SARS-CoV-2 Spike protein impairs endothelial function via downregulation of ACE 2. Circulat. Res. 2021;128(9):1323–1326. doi: 10.1161/CIRCRESAHA.121.318902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim E.S., Jeon M.T., Kim K.S., Lee S., Kim S., Kim D.G. Spike Proteins of SARS-CoV-2 induce pathological changes in molecular delivery and metabolic function in the brain endothelial cells. Viruses. 2021;13(10) doi: 10.3390/v13102021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dasgupta S., Bandyopadhyay M. Molecular docking of SARS-COV-2 Spike epitope sequences identifies heterodimeric peptide-protein complex formation with human Zo-1, TLR8 and brain specific glial proteins. Med. Hypothes. 2021;157 doi: 10.1016/j.mehy.2021.110706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Buzhdygan T.P., DeOre B.J., Baldwin-Leclair A., Bullock T.A., McGary H.M., Khan J.A., Razmpour R., Hale J.F., Galie P.A., Potula R., Andrews A.M., Ramirez S.H. The SARS-CoV-2 spike protein alters barrier function in 2D static and 3D microfluidic in-vitro models of the human blood-brain barrier. Neurobiol. Dis. 2020;146 doi: 10.1016/j.nbd.2020.105131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rhea E.M., Logsdon A.F., Hansen K.M., Williams L.M., Reed M.J., Baumann K.K., Holden S.J., Raber J., Banks W.A., Erickson M.A. The S1 protein of SARS-CoV-2 crosses the blood-brain barrier in mice. Nat. Neurosci. 2021;24(3):368–378. doi: 10.1038/s41593-020-00771-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Polykretis P. Role of the antigen presentation process in the immunization mechanism of the genetic vaccines against COVID-19 and the need for biodistribution evaluations. Scand. J. Immunol. 2022 doi: 10.1111/sji.13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cognetti J.S., Miller B.L. Monitoring Serum Spike Protein with Disposable Photonic Biosensors Following SARS-CoV-2 Vaccination. Sensors. 2021;21(17) doi: 10.3390/s21175857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material