Abstract
Prolonged treatment of chronic hepatitis B virus (HBV) infection with lamivudine ([−]-β-l-2′,3′-dideoxy-3′ thiacytidine) or famciclovir may select for viral mutants that are drug resistant due to point mutations in the polymerase gene. Determining whether such HBV mutants are sensitive to new antiviral agents is therefore important. We used a transient transfection system to compare the sensitivities of wild-type HBV and four lamivudine- and/or famciclovir-resistant HBV mutants to adefovir [9-(2-phosphonyl-methoxyethyl)-adenine; PMEA] and the nucleoside analogues (−)-β-d-2, 6-diaminopurine dioxolane (DAPD) and 2′-fluoro-5-methyl-β-l-arabinofuranosyluracil (l-FMAU). The drug-resistant mutants contained amino acid substitutions in the polymerase protein. We found that the M550I and M550V plus L526M substitutions, which confer lamivudine resistance, did not confer cross-resistance to adefovir or DAPD, but conferred cross-resistance to l-FMAU. The M550V substitution in isolation conferred a similar phenotype to M550I, except that it did not confer significant resistance to l-FMAU. The L526M substitution, which is associated with famciclovir resistance, conferred cross-resistance to l-FMAU but not to adefovir or DAPD. Inhibition of HBV secretion by DAPD, l-FMAU, and adefovir did not always correlate with inhibition of the generation of intracellular HBV replicative intermediates, suggesting that these analogs may preferentially inhibit specific stages of the viral replication cycle.
Chronic infection with hepatitis B virus (HBV) creates significant economic and social burdens globally. Although both the spread of HBV-related infection and development of hepatocellular carcinoma can be effectively checked by vaccination, treatment with antiviral agents remains the only therapeutic option available to individuals chronically infected with HBV (27). The World Health Organization estimates that the number of chronically infected individuals, who constitute a large reservoir of potentially infectious virus, will be close to 400 million by the end of this year (27). Alpha-interferon and lamivudine are the only two drugs currently licensed for the treatment of chronic hepatitis B in most countries. Clinicians can now choose to treat with lamivudine, a well-tolerated, orally available drug that has minimal side effects (20); with alpha-interferon, which must be administered by subcutaneous injection and may cause some adverse events (44); or with the combination of lamivudine and interferon, which appears to act additively (36).
Successful response is usually defined as seroconversion from hepatitis B antigen (HbeAg) positive to anti-HBe-positive status together with a permanent reduction in serum HBV DNA to levels that are undetectable by DNA liquid hybridization assay and normalization of serum alanine aminotransferase (ALT) activity (27). Responses may be confirmed histologically by liver biopsy. Sixteen to 17% of patients treated with lamivudine respond favorably within 12 months (15, 22), and the response frequency increases to about 40% after 24 months of continuous treatment, which is 6 to 10 times greater than the spontaneous HBeAg seroconversion frequency (T.-T. Chang, C. L. Lai, Y.-F. Liaw, N. Y. Leung, R. Guan, S. G. Lim, C. M. Lee, K. Y. Ng, S. Edmundson, C. Stevenson, and J. C. Dent, Abstr. Hepatol. 30:420A, abstr. 1038, 1999). Prolonged monotherapy with lamivudine or other antiviral agents may select for HBV mutants that are drug resistant. Lamivudine resistance has been reported in about 15 to 25% of patients after 12 months of treatment, increasing with treatment duration (15, 22, 24). Development of resistance to lamivudine and other nucleoside analogues is usually associated with mutations in the HBV DNA polymerase gene (reviewed in reference 13). One of the common mutations associated with lamivudine resistance results in a methionine-to-isoleucine substitution (M550I) in the YMDD (tyrosine-methionine-aspartate-aspartate) nucleotide-binding motif in the catalytic (C) domain of the polymerase. Substitution with valine (M550V) has also been described, almost invariably in conjunction with an upstream leucine-to-methionine (L526M) change in the polymerase B domain (1, 3, 24, 40, 43, 47). Selection for HBV mutants that contain the L526M change without associated changes in the C domain may occur during treatment with famciclovir (14), the oral prodrug for the deoxyguanosine analogue penciclovir (19, 39, 42). The emergence of HBV drug resistance highlights the need for development of more anti-HBV drugs and alternative therapeutic strategies such as combination chemotherapy (7, 13, 41). Additional anti-HBV drugs, some of which have already entered clinical trials, will be required in future. They may include, among others, (−)-β-d-2, 6-diaminopurine dioxolane (DAPD), a prodrug for the deoxyguanosine analogue dioxolane guanine (DXG) [10; Schinazi, R. F., H. M. McClure, F. D. Boudinot, Y. Jxiang, and C. K. Chu., Abstr. Antivir. Res. 23(Suppl):81, 1994], clevudine (2′-fluoro-5-methyl-β-L-arabinofuranosyluracil [l-FMAU]), a thymidine analogue (2, 6) and adefovir [9-(2-phosphonylmethoxyethyl)-adenine; PMEA], a 5′-dAMP analogue, a prodrug of which (adefovir dipivoxil) is currently undergoing phase III clinical trials against HBV (17).
It is important to determine whether new drugs are active against existing drug-resistant HBV mutants and, if possible, to determine their mechanism(s) of action, since these parameters will determine their suitability for use in specific circumstances such as preexisting drug resistance (7, 13, 41). Accordingly, the present study was designed to compare the activities of DAPD and l-FMAU with those of lamivudine and adefovir in parallel antiviral assays in hepatoma cells transiently transfected with either wild-type HBV or drug-resistant mutants which contained the L526M, M550I, M550V, or L526M and M550V (L526M+M550V) substitutions in the viral polymerase.
MATERIALS AND METHODS
Chemicals and reagents.
DAPD, l-FMAU, and lamivudine were provided by Triangle Pharmaceuticals (Triangle Park, N.C.). Adefovir was provided by Gilead Sciences (Foster City, Calif.). Stock solutions (50 mM) of l-FMAU, lamivudine, and adefovir were prepared in distilled water, DAPD was dissolved in dimethyl sulfoxide (DMSO). All other chemicals and reagents were purchased from local suppliers and were analytical grade.
Site-directed mutagenesis.
Point mutations were introduced by site-directed mutagenesis into a 1.5-times genome-length wild-type HBV genotype A, subtype adw2, which was inserted into the plasmid pBluescript KS+ (5). Mutagenesis was carried out according to instructions provided with the Quickchange mutagenesis kit, which was supplied by Stratagene (La Jolla, Calif.). Primers (Table 1) for the mutagenesis reaction were synthesized by GeneWorks (Adelaide, SA, Australia) and purified to 99.9% homogeneity by high-performance liquid chromatography. Successive mutagenesis reactions were performed to generate the double mutant that contained the L526M+M550V changes. Automated DNA sequencing in both directions (BigDye Terminator Cycle Sequencing; PE Applied Biosystems, Foster City, Calif.) verified the sequences of the mutants.
TABLE 1.
Primer sequences used to generate polymerase gene mutations
| Primer | Sequence |
|---|---|
| L526M forward | 5′CCT CAG TCC GTT TCT CAT GGC TCA GTT TAC TAG 3′ |
| L526M reverse | 3′GGA GTC AGG CAA AGA GTA CCG CGT CAA ATG ATC 5′ |
| M550I forward | 5′GGC TTT CAG CTA TAT CGA TGA TGT GGT ATT GGG GG 3′ |
| M550I reverse | 3′CCG AAA GTC GAT ATA GCT ACT ACA CCA TAA CCC CC 5′ |
| M550V forward | 5′GGC TTT CAG CTA TGT GGA TGA TGT GGT ATT GGG 3′ |
| M550V reverse | 3′CCG AAA GTC GAT ACA CCT ACT ACA CCA TAA CCC 5′ |
Cell culture and transfection.
HepG2 cells were grown in minimum essential medium (Gibco BRL) supplemented with 10% (vol/vol) fetal bovine serum at 37°C and 5% CO2. For transfections, cells were seeded to semiconfluence in 60-mm-diameter tissue culture dishes (Greiner, Frickenheim, Germany) and allowed to adhere overnight. On the following day (day 1), cells were pretreated with lamivudine, adefovir, DAPD, or l-FMAU for 4 h prior to the transfection. The culture medium was changed on day 3. Final drug concentrations were 0, 5, 10, 50, 100, or 200 μM for DAPD; 0, 0.05, 0.1, 0.5, 1, 5, 10, or 50 μM for lamivudine and l-FMAU; and 0, 0.05, 0.1, 0.2, 0.5, 1, 5, or 10 μM for adefovir. Transient transfections were achieved with Fugene6 transfection reagent (Roche Diagnostics, Mannheim, Germany) according to the instructions provided by the manufacturer. Media and cell lysates were harvested for analysis 5 days after transfection. Intracellular HBeAg expression, as measured by enzyme immunoassay (Abbott Laboratories, North Chicago, Ill.), was used as an indicator of transfection efficiency.
Assays for cytotoxic or cytostatic effects.
A colorimetric assay for cell viability was used to detect possible cytotoxic or cytostatic effects due to exposure to antiviral drugs. Triplicate sets of HepG2 cells were seeded into multiwell tissue culture plates at semiconfluency and allowed to adhere overnight. The cells were transfected as described above, except that the concentrations of plasmid and Fugene6 were reduced in proportion to well volume. Transfected cells were exposed continuously to test drugs at concentrations up to 1 mM. When cells in untreated control wells reached confluence, cell viability was assayed by MTT reduction (30).
Cell harvest and extraction of HBV DNA.
HBV genomic replication occurs within viral nucleocapsids in the cytoplasm of infected cells (37) and is protected from nuclease digestion. At harvest, cells were rinsed twice with cold phosphate-buffered saline and lysed with 800 μl of 0.5% (vol/vol) NP40 in 150 mM NaCl–50 mM Tris-HCl (pH 7.5). After clarification of cell lysates by centrifugation for 3 min, supernatants were transferred to fresh microcentrifuge tubes, and 1 M MgCl2 was added to give a final Mg2+ concentration of 10 mM. Contaminating input plasmid was then digested for 1 h at 37°C with 20 U of DNase I (Roche Diagnostics) before the reaction was terminated with 10 mM EDTA. After protein digestion with 0.5% sodium dodecyl sulfate (SDS)–0.5-mg/ml proteinase K (Roche Diagnostics) at 37°C for 2 h, DNA was extracted by sequential phenol-chloroform treatment and precipitated with isopropanol. Precipitated DNA was redissolved in 5 mM EDTA, and sample volumes were standardized based on HBeAg assay results to allow for differences in transfection efficiency. Aliquots of each sample were then electrophoresed through 1% agarose in Tris-Acetate-EDTA buffer. HBV replicative intermediates (RI) were detected by Southern hybridization after capillary transfer to positively charged nylon membranes. For analysis of extracellular virus, aliquots of culture medium collected on day 5 posttransfection were clarified by centrifugation before precipitation of virions at 4°C overnight with 26% polyethylene glycol 8000–1.4 M NaCl–25 mM EDTA. Precipitated virions were collected by centrifugation for 30 min at 10,000 × g and then resuspended in 200 μl of 5.5 mM MgCl2–10 mM Tris-HCl (pH 7.5) before extraction of HBV DNA and processing as described above.
Detection of HBV RI.
A 1.2-kb fragment of HBV DNA was labeled with [α-32P]dCTP (3,000 Ci/mmol) to a specific activity of 109 cpm/μg of HBV DNA by using a Random Primer Plus extension kit (Du Pont-NEN, Boston, Mass.) and used as a probe to detect both plus- and minus-strand HBV DNA RI. Blots were hybridized overnight at 65°C in 7% SDS–1 mM EDTA–0.25 M NaH2PO4, rinsed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS for 10 min at 65°C, washed twice with 0.1× SSC–0.1% SDS for 30 min at 65°C, then exposed to Super RX film (Fuji, Tokyo, Japan) at −70°C.
Antiviral effects and data analysis.
Image densities from suitably exposed autoradiographs of Southern blots were measured with a scanning densitometer (model GC-67 with Molecular Analyst software; Bio-Rad Laboratories, Hercules, Calif.). The densities of bands corresponding to intracellular single-stranded (minus strand) DNA (ssDNA) or extracellular relaxed circular DNA (rcDNA) were used to measure antiviral effect. The amount of viral replication in drug-treated samples was expressed as a percentage of the amount of replication in drug-free controls. Data were fitted to logistic dose-response curves, and curve parameters were estimated with the aid of TableCurve2D, a curve-fitting software package from Jandel Scientific (San Rafael, Calif.) as described previously (8, 9, 12). Drug concentrations that reduced the replication or secretion of each mutant HBV to 50% of the average amount measured in the corresponding drug-free controls (50% inhibitory concentrations [IC50s]) were estimated from dose-response curves. The ratios of mutant to wild-type IC50S for each drug (arbitrarily named “resistance factors”) were determined (12). Drug sensitivity and resistance were defined on the basis of resistance factors: <1, hypersensitive; <5, sensitive; >5 < 10, equivocal; > 10, resistant. Similar criteria have frequently been adopted to define human immunodeficiency virus (HIV) drug resistance phenotypes. Where possible, Mann-Whitney (nonparametric, two tailed) t tests were used to establish statistical significance, which was defined as P < 0.05.
RESULTS
Toxicity.
All cell monolayers remained intact for the entire duration of the assays, and daily microscopic examination revealed no evidence of cytotoxicity. Neither l-FMAU nor DAPD was cytotoxic at concentrations up to 1 mM. At 1 mM lamivudine or PMEA, cell viability was 30 or 55%, respectively. The lamivudine concentration, which caused a 50% reduction in cell viability, was estimated to be 524 ± 151 μM. At the highest concentration present (2% [vol/vol]), the solvent DMSO had no effect on toxicity.
Replication efficiency of transfectants.
In the absence of antiviral drugs, the apparent replication efficiencies of the wild type and mutants differed substantially. After adjustment based on HBeAg production, the average amounts of intracellular ssDNA generated relative to the wild type (100%) were (in decreasing order) L526M (60%) > L526M+M550V (40%) > M5501 (20%) ≅ M550V (18%). The ranking was reproducible between experiments, regardless of absolute amounts of HBV ssDNA.
Relative antiviral efficacies.
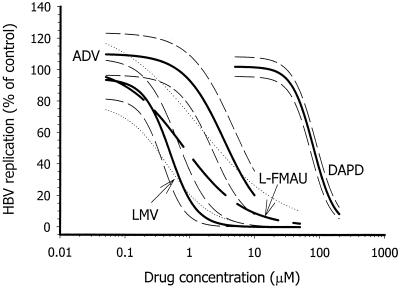
Intracellular HBV RI and genomic DNA were extracted from the cytoplasm of transfected cells and cell culture medium, respectively, after 5 days of continuous exposure to different antiviral drugs and drug concentrations. Analysis by Southern hybridization, autoradiography, and scanning densitometry generated matched sets of data (Fig. 1), to which logistic dose-response curves were fitted with the aid of TableCurve2D (Fig. 2). Where possible, the drug concentration that reduced HBV replication by 50% (the IC50) was estimated from the equation that described the appropriate dose-response plot. All analogs inhibited replication of wild-type HBV in a dose-dependent manner. From three independent experiments, the decreasing order of drug efficacy (against wild-type HBV) was found to be l-FMAU (0.44 ± 0.48 μM) ≅ lamivudine (0.53 ± 0.35 μM) > adefovir (3.5 ± 1.9 μM) ≫ DAPD (154 ± 73 μM). (Values in parentheses are mean IC50S ± standard deviations.) The ranking of drug efficacies was consistent, although there was considerable interexperimental variation in estimated IC50S for individual drugs. Despite interexperimental variation in IC50S for individual drugs, confidence intervals for dose-response curves (Fig. 2) and intraexperimental coefficients of variation for IC50 estimates were generally within ±25% (Table 2). Table 2 summarizes the results of a single representative set of experiments in which inhibition of wild-type HBV and all mutants was compared in parallel. (The <2-fold difference in efficacy between l-FMAU and lamivudine against wild-type HBV was not significant and did not affect the ranking.)
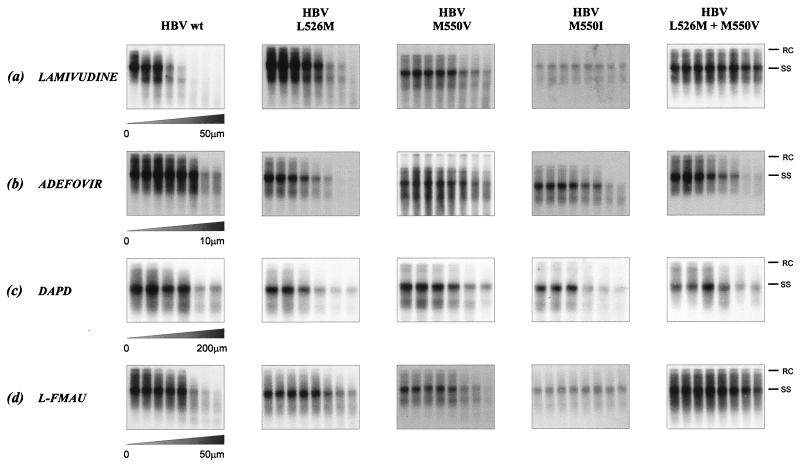
FIG. 1.
Southern hybridization of HBV RI extracted from intracellular core particles extracted from HepG2 cells transiently transfected with plasmid vectors which contained either wild-type (wt) HBV or L526M, M5501, or L526M + M550V mutants following treatment with (a) lamivudine, (b) adefovir, (c) DAPD, or (d) l-FMAU. Bands which correspond to the rcDNA (RC) and ssDNA (SS) species are indicated.
FIG. 2.
Graphical analysis of data obtained from autoradiographs shown in Fig. 1. Dose-response plots for inhibition of wild-type HBV replication by lamivudine (LMV), adefovir (ADV), l-FMAU, and DAPD are shown. The finer lines on either side of each plot represent 95% confidence intervals for each curve fit.
TABLE 2.
Sensitivity of replication of wild-type and mutant HBV to inhibition by lamivudine, adefovir, DAPD, and l-FMAUa
| HBV | Result for equation parameter
|
Correlation (r2) | Result for sensitivity parameter
|
||||
|---|---|---|---|---|---|---|---|
| a | b | c | IC50 (μM) | % Inhibition at wild-type IC50b | Resistance factorc | ||
| Lamivudine | |||||||
| Wild type | 94 ± 5.2 | 0.50 ± 0.09 | 2.1 ± 0.84 | 0.97 | 0.47 | 50 ± 16 | 1.0 |
| L526M | 98 ± 2.8 | 0.83 ± 0.09 | 1.2 ± 0.16 | 0.99 | 0.80 | 36 ± 6 | 1.7 (2.5) |
| M550V | 111 ± 6.1d | 7.0 ± 2.0 | 1.0 ± 0.29 | 0.94 | 8.5 | −4 ± 10f | 18 (22) |
| M550I | NDRe | >50 | 0 | >106 (>100) | |||
| Dual | NDRe | >50 | 0 | >106 (>100) | |||
| Adefovir | |||||||
| Wild type | 110 ± 5.4 | 3.4 ± 0.76 | 1.4 ± 0.35 | 0.97 | 3.9 | 50 ± 15 | 1.0 |
| L526M | 113 ± 12 | 1.5 ± 0.63 | 0.78 ± 0.05 | 0.97 | 2.0 | 54 ± 11 | 0.5 (0.4) |
| M550V | 102 ± 10 | 2.3 ± 1.3 | 1.0 ± 0.4 | 0.91 | 2.8 | 41 ± 23 | 0.7 (0.5) |
| M550I | 110 ± 17 | 1.9 ± 1.8 | 0.61 ± 0.28 | 0.81 | 2.7 | 44 ± 25 | 0.7 (0.7) |
| Dual | 119 ± 11 | 0.55 ± 0.12 | 2.5 ± 1.3 | 0.93 | 0.64 | 99 ± 7 | 0.2 (0.4) |
| l-FMAU | |||||||
| Wild type | 103 ± 7.8 | 0.79 ± 0.30 | 0.92 ± 0.27 | 0.98 | 0.83 | 50 ± 17 | 1.0 |
| L526M | 101 ± 1.0 | 59 ± 1.8 | 1.3 ± 0.05 | 1.0 | >100 | −5 ± 9f | >120 (>100) |
| M550V | 103 ± 10 | 1.3 ± 0.89 | 0.55 ± 0.17 | 0.96 | 1.5 | 42 ± 22 | 18 (2) |
| M550I | NDRe | >100 | 0 | >120 (>100) | |||
| Dual | NDRe | >100 | 0 | >120 (>100) | |||
| DAPD | |||||||
| Wild type | 100 ± 1.5 | 79 ± 3.2 | 2.0 ± 0.16 | 1.0 | 79 | 50 ± 5 | 1.0 |
| L526M | 101 ± 1.0 | 59 ± 1.8 | 1.3 ± 0.05 | 1.0 | 59 | 41 ± 3 | 0.8 (0.7) |
| M550V | 99 ± 8.7 | 37 ± 16.1 | 0.81 ± 0.25 | 0.97 | 36 | 35 ± 16 | 0.4 (0.5) |
| M550I | 98 ± 1.0 | 67 ± 3.6 | 1.7 ± 0.14 | 1.0 | 65 | 41 ± 8 | 0.8 (0.6) |
| Dual | 102 ± 7.0 | 60 ± 17.3 | 0.89 ± 0.24 | 0.97 | 62 | 45 ± 20 | 0.8 (0.4) |
The results presented here are derived from complete sets of representative experiments in which each analog was assayed as an inhibitor of all HBV strains (wild type and four drug-resistant variants) in parallel. The image densities of the HBV ssDNA bands in each autoradiograph were measured with a scanning densitometer. The results were expressed as percentages of the mean density of the corresponding untreated controls. Parallel inhibition of intracellular rcDNA and extracellular virion DNA was seen in all cases, except as noted in the Discussion section. The parameters a, b, and c define a logistic dose-response curve, which is described by the equation y = a/[1 + (x/b)c], where y = percentage of inhibition inhibition, a is the curve's amplitude, b is the x value at its transition center, and c is a parameter that defines the transition width (see references 8, 9, and 12 for further information).
The percentage of inhibition (given with 95% confidence interval) that occurred at the drug concentration required to reduce replication of wild-type HBV by 50% (the IC50) in the same experiment was estimated from each fitted plot. For example, the percentages tabulated for lamivudine are those that occur at 0.47 μM lamivudine, which inhibited wild-type HBV by 50% ± 16%.
The resistance factor is the factor (to the nearest decimal place) by which the IC50 estimated for the mutant differs from the corresponding estimate for the wild type (see reference 12). It was calculated by dividing the IC50 for the mutant by the IC50 for the wild type. The phenotype has arbitrarity been defined by values of <1 (hypersensitive), >1 <5 (sensitive), >5 <10 (equivocal) and >10 (resistant). In replicate experiments, the ranking of resistance to each drug was consistent with the ranking shown here. Values in parentheses are the averages of resistance factors from two or three experiments. As noted in the text, three independent experiments gave the following mean ± standard deviation IC50 for wild-type HBV: l-FMAU, 0.44± 0.48 μM; lamivudine, 0.53 ± 0.35 μM; adefovir 3.15 ± 1.9 μM; and DAPD 154 ± 73 μM. The reversal in ranking of wild-type IC50 for lamivudine and l-FMAU in this set of experiments is consistent with the previously noted ranking (in decreasing order of efficacy): l-FMAU ≅ lamivudine > adefovir >> DAPD, since the <2-fold difference in IC50 is not statistically significant.
In some cases, low concentrations of inhibitor stimulated HBV replication, reflected by a values of >100% (compare tabulated a values with the illustration in Fig. 1).
NDR, no dose-response relationship. Logistic dose-response equations (9, 12) did not accurately describe these data sets because the percentage inhibition even at the highest drug concentration was insufficient to establish any meaningful dose-response relationship.
Negative values represent stimulation of replication relative to untreated controls.
Relative sensitivities of wild-type and mutant HBV to lamivudine and adefovir.
The lamivudine sensitivity of wild-type HBV and its mutant derivatives was compared in order to confirm that the point mutations that created the M550I, M550V, or L526M + M550V polymerase substitutions conferred lamivudine resistance to the HBV clone used in our experiments. As expected, these three mutants were lamivudine resistant. It was not possible to estimate accurate IC50s or resistance factors for M550I or L526M + M550V, which showed the greatest resistance to lamivudine. The L526M substitution did not confer significant lamivudine resistance (resistance factor, <5). The ranking of lamivudine sensitivity was reproducible, showing sensitivity to lamivudine consistently decreasing in the order wild type > L526M > M550V >> M550I ≅ (L526M + M550V) by factors of <5 (L526M), <20 (M550V), and >100 (M550I and dual), respectively. No evidence for selective inhibition by lamivudine of any particular step in the HBV replication cycle was seen.
Exposure to adefovir caused dose-dependent inhibition of replication of both wild-type and mutant HBV. Dose-response curves and IC50 values derived from them (Table 2) indicated that adefovir was equally, if not more, effective as an inhibitor of replication of lamivudine-resistant HBV mutants than of wild-type HBV. Resistance factors were close to or less than 1.0, confirming that none of the mutants was cross-resistant to adefovir. Although resistance factors <1.0 suggest hypersensitivity, even the maximum difference (about fivefold decrease for the dual mutant) was not statistically significant. Differential effects of adefovir on generation of intracellular RI were not observed. The extracellular release of mutant virions with M550 changes appeared to be less sensitive to inhibition by DAPD than generation of intracellular RI, but the differences were not statistically significant.
Relative sensitivities of wild-type and mutant HBV to DAPD and l-FMAU.
DAPD caused dose-dependent inhibition of replication of wild-type and mutant HBV, although high concentrations (>35 μM) were required to produce 50% inhibition. Differential effects of DAPD on generation of intracellular RI were not observed, but as with adefovir, the mutants appeared to be more sensitive than the wild type, although the differences (a maximum decrease of 2.5-fold in the case of M550V) were not statistically significant. Similarly, the extracellular release of virions with M550 changes appeared to be less sensitive to inhibition by DAPD than generation of intracellular RI, but the differences were not statistically significant.
l-FMAU was as effective as lamivudine as an inhibitor of wild-type HBV replication in this assay system. Replication of all mutants except M550V was essentially l-FMAU resistant (resistance factors >100). Differential effects of l-FMAU on generation of intracellular RI were not observed, but in contrast to DAPD, the release of virions into the extracellular medium appeared to be relatively more sensitive to inhibition by l-FMAU. Again, differences were not statistically significant.
DISCUSSION
In this study, we evaluated the antiviral activity of adefovir and two novel antiviral nucleoside analogues, DAPD and l-FMAU, against wild-type HBV and HBV mutants that contained amino acid changes in the polymerase protein that confer phenotypic resistance to lamivudine and/or famciclovir. We found that the L526M substitution, which is associated with famciclovir resistance, conferred resistance to l-FMAU, both in isolation and in association with M550V, but it did not significantly affect sensitivity to lamivudine, adefovir, or DAPD. The M550I change conferred cross-resistance to l-FMAU, but did not significantly affect sensitivity to adefovir or DAPD (Fig. 1 and Table 2).
The pattern of lamivudine resistance that we observed is consistent with previous reports of experimental and clinical studies (1, 3, 15, 16, 22, 24–26, 31–33, 35, 40, 42, 43, 47, 48). Similarly, retention of adefovir sensitivity by lamivudine- and/or famciclovir-resistant mutants was expected on the basis of previous reports (34, 45, 48). One of the latter (45) showed that PMEApp (the active intracellular anabolite of adefovir) inhibited activity of mutant HBV polymerases more efficiently than the wild-type in cell-free assays, indicating increased, rather than decreased, sensitivity to adefovir. These observations are consistent with ours (Table 2) and with reports that lamivudine-resistant strains of HIV-1 that have analogous changes in the reverse transcriptase also exhibit adefovir hypersensitivity (29). For HIV, the increased sensitivity to adefovir is believed to be due to the increased affinity for PMEApp that occurs as a result of conformational changes in the mutant polymerases (29), and the same may be true for HBV, since the polymerases share substantial sequence and structural homology (13). The ranking of drug efficacy reported here (in decreasing order: l-FMAU ≅ lamivudine > adefovir > DAPD) is also consistent with most previous reports (16, 28, 32, 48) as is the ranking of relative replication competency of wild-type and mutant HBV (in decreasing order: wild type > L526M > L526M + M550V >> M550I ≅ M550V). The observed ranking of replication competence supports the notion that the L526M substitution partially restores replication competence to M550 mutants (32). Overall, the patterns of lamivudine resistance, drug efficacy, and replication competence confirm previous observations made with different assay systems and serve to validate the transient transfection assay described here.
In this system, l-FMAU showed potent activity against wild-type HBV, with an IC50 of ≅0.44 μM, comparable with IC50s found by using either stably or transiently HBV-transfected human (2.2.15 or HuH-7, respectively) cells (2, 32, 48). The observed l-FMAU resistance of the M550I and L526M+M550V mutants confirms recent observations by Fu and colleagues (16). Our observation that the M550V mutant is not significantly resistant to l-FMAU is also in accord with the report by Ying et al. (48), who used HepG2-derived HepAD cells in their assays. In these cells, the expression of stably transfected HBV genomes (wild type in HepAD38 and M550V in HepAD79, respectively) is controlled by a tetracycline-sensitive promoter (21).
In our assays, we found IC50s in the range 35 to 225 μM for inhibition of replication of wild-type HBV by DAPD, the actual value depending on which particular HBV RI was used to estimate this parameter (Table 2). We found IC50s about an order of magnitude lower for DAPD (in the range 15 to 50 μM) in HBV-transfected avian LMH (Leghorn male hepatoma cells, which phosphorylate deoxyguanosine analogs almost 100-fold more efficiently than human hepatoma cell lines (4; R. Chin, S. Locarnini, T. Shaw, and G. Civitico; unpublished data). DAPD and DXG were reported to have IC50s of 0.1 and 1.0 μM, respectively, in 2.2.15 cells [R. F. Schinazi, H. M. McClure, F. D. Boudinot, Y. Jxiang, and C. K. Chu, Antivir. Res. 23(Suppl):81, 1994]. Higher IC50s were reported by Ying and coworkers (48), who performed anti-HBV assays with both 2.2.15 cells (IC50s were 13.0 and 3.5 μM for DAPD and DXG, respectively) and HepAD38 cells (in which the corresponding IC50s were 14.0 and 16.0 μM). More recently, it was reported that DAPD was relatively ineffective (IC50, >>10 μM) as an inhibitor of HBV replication in transfected HuH-7 cells (32). Large differences in the sensitivity of HBV replication to inhibition by DAPD in different assay systems that use different cell lines or different subclones of the same cell line presumably reflect differences in cellular metabolism or differences in intracellular concentrations and accessibility of endogenous nucleotides. The common mutations that confer lamivudine and/or famciclovir resistance did not confer cross-resistance to DAPD, consistent with the report that for HIV, mutations associated with resistance to other analogs, including lamivudine and adefovir, do not confer cross-resistance to DAPD (18).
Each of the analogs tested in the present assay presumably acts mainly by termination of nascent DNA chains during either first (RNA dependent)- or second-strand (DNA dependent) viral DNA synthesis. Although l-FMAU possesses a 3′ hydroxyl equivalent, which might theoretically permit its internal incorporation into viral or cellular DNA, there is no evidence that this occurs (2, 6). However, it remains possible that some dNTP analogues may preferentially inhibit other specific catalytic or regulatory activities of the HBV DNA polymerase, which is a multifunctional protein that also functions as an RNase H, primes first-strand DNA synthesis, and coordinates virion assembly (37). In general, the analogs tested here appeared to inhibit first-strand HBV (ssDNA) synthesis less effectively than they inhibited synthesis of other DNA RI or production of extracellular virion DNA. Although we observed that in this assay system, the DNA-dependent activity of the HBV polymerase appears to be more susceptible to inhibition than is its RNA-dependent (reverse transcriptase) activity, the low efficiency of transient transfection, together with relatively large interassay variations in transfection efficiency, make it difficult to establish the significance of differences in antiviral efficacy unless they are relatively large. The approximately fivefold range of apparent replication competence exhibited by wild-type and mutant HBV compounds these problems. These assays may underestimate drug resistance of mutants which replicate poorly, because low drug concentrations are sufficient to reduce the autoradiographic signal to undetectable levels. This cannot be compensated for by longer exposure, because it results in unacceptable background and signal/noise ratios. For example, M550V in isolation was found not to significantly increase l-FMAU resistance (Table 2). However, if the sensitivity of detection were increased five-fold to compensate for the replication efficiency (about 20% of the wild type), the drug concentration required to decrease the autoradiographic signal to an undetectable level may increase correspondingly, which would imply significant resistance (with a resistance factor of about 10). In this case, the poor dynamic range afforded by transient transfection assays may hamper detection of drug resistance. Further studies with other in vitro assay systems that have a greater dynamic range and in which HBV replication is more efficient and controllable will probably be required to better quantify drug resistance and to elucidate mechanisms of action. The recently developed recombinant baculovirus transfection system (11, 12) as well as AD cell lines (21) are likely to prove useful for such studies. Because of cell-dependent differences in nucleotide metabolism, cell-free enzyme assays (such as those described by Seifer et al. [38] and Xiong et al. [45, 46]) may be required for precise quantitative studies.
The potential for selection of new drug-resistant HBV mutants highlights the need to develop additional anti-HBV drugs and therapeutic strategies (7, 13, 41). Reliable in vitro systems that are capable of detecting drug resistance as it arises are required, as are assays that are capable of predicting which drug combinations are optimal for delaying or preventing resistance and for controlling resistance if it develops (13). On the other hand, in vitro studies are not always accurate predictors of effectiveness in vivo, and clinical trials are the ultimate test of treatment efficacy.
In conclusion, this report confirms that the L526M, M550I, and M550V + L526M substitutions that confer lamivudine and/or famciclovir resistance also confer resistance to l-FMAU, but do not confer cross-resistance to adefovir or DAPD. The lack of cross-resistance to adefovir is consistent with recent clinical observations (34). Whether DAPD has potential for clinical use against the most common lamivudine-resistant HBV mutants will depend on whether the high concentrations of DAPD that are needed to inhibit HBV replication in vitro are also necessary in vivo and, if so, whether they can be achieved and sustained during treatment.
ACKNOWLEDGMENTS
R.C. was partly supported by a Postgraduate Fellowship from SmithKline Beecham Pharamceuticals (Australia).
We thank Graham Brown for his support and encouragement and Scott Bowden for reviewing the manuscript.
REFERENCES
- 1.Allen M I, Deslauriers M, Andrews C W, Tipples G A, Walters K A, Tyrrell D L, Brown N, Condreay L D. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology. 1998;27:1670–1677. doi: 10.1002/hep.510270628. [DOI] [PubMed] [Google Scholar]
- 2.Balakrishna Pai S, Liu S-H, Zhu Y-L, Chu C K, Cheng Y-C. Inhibition of hepatitis B virus by a novel l-nucleoside, 2′-fluoro-5-methyl-β-l-arabinofuranosyl uracil. Antimicrob Agents Chemother. 1996;40:380–386. doi: 10.1128/aac.40.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartholomew M M, Jansen R W, Jeffers L J, Reddy K R, Johnson L C, Bunzendahl H, Condreay L D, Tzakis A G, Schiff E R, Brown N A. Hepatitis-B-virus resistance to lamivudine given for recurrent infection after orthotopic liver transplantation. Lancet. 1997;349:20–22. doi: 10.1016/S0140-6736(96)02266-0. [DOI] [PubMed] [Google Scholar]
- 4.Bennett L L, Jr, Allan P W, Arnett G, Shealy Y F, Shewach D S, Mason W S, Fourel I, Parker W B. Metabolism in human cells of the d and l enantiomers of the carbocyclic analog of 2′-deoxyguanosine: substrate activity with deoxycytidine kinase, mitochondrial deoxyguanosine kinase, and 5′-nucleotidase. Antimicrob Agents Chemother. 1998;42:1045–1051. doi: 10.1128/aac.42.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bock C T, Tillmann H L, Maschek H J, Manns M P, Trautwein C. A preS mutation isolated from a patient with chronic hepatitis B infection leads to virus retention and misassembly. Gastroenterology. 1997;113:1976–1982. doi: 10.1016/s0016-5085(97)70018-0. [DOI] [PubMed] [Google Scholar]
- 6.Chu C K, Boudinot F D, Peek S F, Hong J H, Choi Y, Korba B E, Gerin J L, Cote P J, Tennant B C, Cheng Y C. Preclinical investigation of L-FMAU as an anti-hepatitis B virus agent. Antivir Ther. 1998;3:113–121. [PubMed] [Google Scholar]
- 7.Colacino J M, Staschke K A. The identification and development of antiviral agents for the treatment of chronic hepatitis B virus infection. Prog Drug Res. 1998;50:259–322. doi: 10.1007/978-3-0348-8833-2_6. [DOI] [PubMed] [Google Scholar]
- 8.Colledge D, Civitico G, Locarnini S, Shaw T. In vitro antihepadnaviral activities of combinations of penciclovir, lamivudine, and adefovir. Antimicrob Agents Chemother. 2000;44:551–560. doi: 10.1128/aac.44.3.551-560.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colledge D, Locamini S, Shaw T. Synergistic inhibition of hepadnaviral replication by lamivudine in combination with penciclovir in vitro. Hepatology. 1997;26:216–225. doi: 10.1053/jhep.1997.v26.pm0009214473. [DOI] [PubMed] [Google Scholar]
- 10.Cui L, Schinazi R F, Gosselin G, Imbach J L, Chu C K, Rando R F, Revankar G R, Sommadossi J P. Effect of beta-enantiomeric and racemic nucleoside analogues on mitochondrial functions in HepG2 cells. Implications for predicting drug hepatotoxicity. Biochem Pharmacol. 1996;52:1577–1584. doi: 10.1016/s0006-2952(96)00562-x. [DOI] [PubMed] [Google Scholar]
- 11.Delaney W E, Isom H C. Hepatitis B virus replication in human HepG2 cells mediated by hepatitis B virus recombinant baculovirus. Hepatology. 1998;28:1134–1146. doi: 10.1002/hep.510280432. [DOI] [PubMed] [Google Scholar]
- 12.Delaney W E, IV, Edwards R, Colledge D, Shaw T, Torresi J, Miller T G, Isom H C, Bock C T, Manns M P, Trautwein C, Locarnini S. Cross-resistance testing of antihepadnaviral compounds using novel recombinant baculoviruses which encode drug-resistant strains of hepatitis B virus. Antimicrob Agents Chemother. 2001;45:1705–1713. doi: 10.1128/AAC.45.6.1705-1713.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delaney W E, Locarnini S, Shaw T. Resistance of hepatitis B virus to antiviral drugs: current aspects and directions for future investigation. Antiviral Chem Chemother. 2001;12:41–75. doi: 10.1177/095632020101200101. [DOI] [PubMed] [Google Scholar]
- 14.de Man R A, Marcellin P, Habal F, Desmond P, Wright T, Rose T, Jurewicz R, Young C. A randomized, placebo-controlled study to evaluate the efficacy of 12-month famciclovir treatment in patients with chronic hepatitis B e antigen-positive hepatitis B. Hepatology. 2000;32:413–417. doi: 10.1053/jhep.2000.9407. [DOI] [PubMed] [Google Scholar]
- 15.Dienstag J L, Schiff E R, Wright T L, Perrillo R P, Hann H W, Goodman Z, Crowther L, Condreay L D, Woessner M, Rubin M, Brown N A. Lamivudine as initial treatment for chronic hepatitis B in the United States. N Engl J Med. 1999;341:1256–1263. doi: 10.1056/NEJM199910213411702. [DOI] [PubMed] [Google Scholar]
- 16.Fu L, Liu S H, Cheng Y C. Sensitivity of L-(−)2,3-dideoxythiacytidine resistant hepatitis B virus to other antiviral nucleoside analogues. Biochem Pharmacol. 1999;57:1351–1359. doi: 10.1016/s0006-2952(99)00073-8. [DOI] [PubMed] [Google Scholar]
- 17.Gilson R J, Chopra K B, Newell A M, Murray-Lyon I M, Nelson M R, Rice S J, Tedder R S, Toole J, Jaffe H S, Weller I V. A placebo-controlled phase I/II study of adefovir dipivoxil in patients with chronic hepatitis B virus infection. J Viral Hepatitis. 1999;6:387–395. doi: 10.1046/j.1365-2893.1999.00182.x. [DOI] [PubMed] [Google Scholar]
- 18.Gu Z, Wainberg M A, Nguyen-Ba N, L'Heureux L, de Muys J M, Bowlin T L, Rando R F. Mechanism of action and in vitro activity of 1′,3′-dioxolanylpurine nucleoside analogues against sensitive and drug-resistant human immunodeficiency virus type 1 variants. Antimicrob Agents Chemother. 1999;43:2376–2382. doi: 10.1128/aac.43.10.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunther S, von Breunig F, Santantonio T, Jung M C, Gaeta G B, Fischer L, Sterneck M, Will H. Absence of mutations in the YMDD motif/B region of the hepatitis B virus polymerase in famciclovir therapy failure. J Hepatol. 1999;30:749–754. doi: 10.1016/s0168-8278(99)80124-x. [DOI] [PubMed] [Google Scholar]
- 20.Jarvis B, Faulds D. Lamivudine. A review of its therapeutic potential in chronic hepatitis B. Drugs. 1999;58:101–141. doi: 10.2165/00003495-199958010-00015. [DOI] [PubMed] [Google Scholar]
- 21.Ladner S K, Otto M J, Barker C S, Zaifert K, Wang G-H, Guo J-T, Seeger C, King R W. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother. 1997;41:1715–1720. doi: 10.1128/aac.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai C L, Chien R N, Leung N W, Chang T T, Guan R, Tai D I, Ng K Y, Wu P C, Dent J C, Barber J, Stephenson S L, Gray D F. A one-year trial of lamivudine for chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. N Engl J Med. 1998;339:61–68. doi: 10.1056/NEJM199807093390201. [DOI] [PubMed] [Google Scholar]
- 23.Lau G K K, Tsiang M, Hou J, Yuen S, Carman W F, Zhang L, Gibbs C S, Lam S. Combination therapy with lamivudine and famciclovir for chronic hepatitis B infected chinese: a viral dynamics study. Hepatology. 2000;32:394–399. doi: 10.1053/jhep.2000.9143. [DOI] [PubMed] [Google Scholar]
- 24.Liaw Y F, Leung N W, Chang T T, Guan R, Tai D I, Ng K Y, Chien R N, Dent J, Roman L, Edmundson S, Lai C L. Effects of extended lamivudine therapy in Asian patients with chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. Gastroenterology. 2000;119:172–180. doi: 10.1053/gast.2000.8559. [DOI] [PubMed] [Google Scholar]
- 25.Liaw Y F, Chien R N, Yeh C T, Tsai S L, Chu C M. Acute exacerbation and hepatitis B virus clearance after emergence of YMDD motif mutation during lamivudine therapy. Hepatology. 1999;30:567–572. doi: 10.1002/hep.510300221. [DOI] [PubMed] [Google Scholar]
- 26.Ling R, Mutimer D, Ahmed M, Boxall E H, Elias E, Dusheiko G M, Harrison T J. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with lamivudine. Hepatology. 1996;24:711–713. doi: 10.1002/hep.510240339. [DOI] [PubMed] [Google Scholar]
- 27.Lok A S. Hepatitis B infection: pathogenesis and management. J Hepatol. 2000;32:89–97. doi: 10.1016/s0168-8278(00)80418-3. [DOI] [PubMed] [Google Scholar]
- 28.Melegari M, Scaglioni P P, Wands J R. Hepatitis B virus mutants associated with 3TC and famciclovir administration are replication defective. Hepatology. 1998;27:628–633. doi: 10.1002/hep.510270243. [DOI] [PubMed] [Google Scholar]
- 29.Miller M D, Anton K E, Mulato A S, Lamy P D, Cherrington J M. Human immunodeficiency virus type 1 expressing the lamivudine-associated M184V mutation in reverse transcriptase shows increased susceptibility to adefovir and decreased replication capability in vitro. J Infect Dis. 1999;179:92–100. doi: 10.1086/314560. [DOI] [PubMed] [Google Scholar]
- 30.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 31.Ogata N, Fujii K, Takigawa S, Nomoto M, Ichida T, Asakura H. Novel patterns of amino acid mutations in the hepatitis B virus polymerase in association with resistance to lamivudine therapy in Japanese patients with chronic hepatitis B. J Med Virol. 1999;59:270–276. [PubMed] [Google Scholar]
- 32.Ono S K, Kato N, Shiratori Y, Kato J, Goto T, Schinazi R F, Carrilho F J, Omata M. The polymerase L528M mutation cooperates with nucleotide binding-site mutations, increasing hepatitis B virus replication and drug resistance. J Clin Investig. 2001;107:449–455. doi: 10.1172/JCI11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono-Nita S K, Kato N, Shiratori Y, Lan K H, Yoshida H, Carrilho F J, Omata M. Susceptibility of lamivudine-resistant hepatitis B virus to other reverse transcriptase inhibitors. J Clin Investig. 1999;103:1635–1640. doi: 10.1172/JCI5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perrillo R, Schiff E, Yoshida E, Statler A, Hirsch K, Wright T, Gutfreund K, Lamy P, Murray A. Adefovir dipivoxil for the treatment of lamivudine-resistant hepatitis B mutants. Hepatology. 2000;32:129–134. doi: 10.1053/jhep.2000.8626. [DOI] [PubMed] [Google Scholar]
- 35.Santantonio T, Mazzola M, Iacovazzi T, Miglietta A, Guastadisegni A, Pastore G. Long-term follow-up of patients with anti-HBe/HBV DNA-positive chronic hepatitis B treated for 12 months with lamivudine. J Hepatol. 2000;32:300–306. doi: 10.1016/s0168-8278(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 36.Schalm S W, Heathcote J, Cianciara J, Farrell G, Sherman M, Willems B, Dhillon A, Moorat A, Barber J, Gray D F. Lamivudine and alpha interferon combination treatment of patients with chronic hepatitis B infection: a randomised trial. Gut. 2000;46:562–568. doi: 10.1136/gut.46.4.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seeger C, Mason W S. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seifer M, Hamatake R, Bifano M, Standring D N. Generation of replication-competent hepatitis B virus nucleocapsids in insect cells. J Virol. 1998;72:2765–2776. doi: 10.1128/jvi.72.4.2765-2776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seigneres B, Pichoud C, Ahmed S S, Hantz O, Trepo C, Zoulim F. Evolution of hepatitis B virus polymerase gene sequence during famciclovir therapy for chronic hepatitis B. J Infect Dis. 2000;181:1221–1233. doi: 10.1086/315368. [DOI] [PubMed] [Google Scholar]
- 40.Seta T, Yokosuka O, Imazeki F, Tagawa M, Saisho H. Emergence of YMDD motif mutants of hepatitis B virus during lamivudine treatment of immunocompetent type B hepatitis patients. J Med Virol. 2000;60:8–16. doi: 10.1002/(sici)1096-9071(200001)60:1<8::aid-jmv2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 41.Shaw T, Locarnini S. Combination therapy for chronic hepatitis B: the path forward? Drugs. 2000;60:517–531. doi: 10.2165/00003495-200060030-00001. [DOI] [PubMed] [Google Scholar]
- 42.Tillmann H L, Trautwein C, Bock T, Boker K H, Jackel E, Glowienka M, Oldhafer K, Bruns I, Gauthier J, Condreay L D, Raab H R, Manns M P. Mutational pattern of hepatitis B virus on sequential therapy with famciclovir and lamivudine in patients with hepatitis B virus reinfection occurring under HBIg immunoglobulin after liver transplantation. Hepatology. 1999;30:244–256. doi: 10.1002/hep.510300141. [DOI] [PubMed] [Google Scholar]
- 43.Tipples G A, Ma M M, Fischer K P, Bain V G, Kneteman N M, Tyrrell D L. Mutation in HBV RNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology. 1996;24:714–717. doi: 10.1002/hep.510240340. [DOI] [PubMed] [Google Scholar]
- 44.Wong D K, Cheung A M, O'Rourke K, Naylor C D, Detsky A S, Heathcote J. Effect of alpha-interferon treatment in patients with hepatitis B e antigen-positive chronic hepatitis B. A meta-analysis. Ann Intern Med. 1993;119:312–323. doi: 10.7326/0003-4819-119-4-199308150-00011. [DOI] [PubMed] [Google Scholar]
- 45.Xiong X, Flores C, Yang H, Toole J J, Gibbs C S. Mutations in hepatitis B DNA polymerase associated with resistance to lamivudine do not confer resistance to adefovir in vitro. Hepatology. 1998;28:1669–1673. doi: 10.1002/hep.510280629. [DOI] [PubMed] [Google Scholar]
- 46.Xiong X, Yang H, Westland C E, Zou R, Gibbs C S. In vitro evaluation of hepatitis B virus polymerase mutations associated with famciclovir resistance. Hepatology. 2000;31:219–224. doi: 10.1002/hep.510310132. [DOI] [PubMed] [Google Scholar]
- 47.Yeh C T, Chien R N, Chu C M, Liaw Y F. Clearance of the original hepatitis B virus YMDD-motif mutants with emergence of distinct lamivudine-resistant mutants during prolonged lamivudine therapy. Hepatology. 2000;31:1318–1326. doi: 10.1053/jhep.2000.7296. [DOI] [PubMed] [Google Scholar]
- 48.Ying C, De Clercq E, Nicholson W, Furman P, Neyts J. Inhibition of the replication of the DNA polymerase M550V mutation variant of human hepatitis B virus by adefovir, tenofovir, L-FMAU, DAPD, penciclovir and lobucavir. J Viral Hepatitis. 2000;7:161–165. doi: 10.1046/j.1365-2893.2000.00210.x. [DOI] [PubMed] [Google Scholar]