Abstract
Background
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a long non-coding RNA (lncRNA) that regulates disease progression in various types of cancers. The aim of this study was to explore the role of MALAT1 in breast cancer (BC) progression and doxorubicin resistance.
Methods
Real-time polymerase chain reaction (RT-PCR) was used to determine the expression of MALAT1 in BC tissues and cells; MTT and Transwell assay were used to detect the proliferation, migration and invasion of BC cells, respectively; drug resistance test was performed to assess the sensitivity of BC cells to doxorubicin; dual-luciferase reporter gene assay was conducted to verify the interaction between MALAT1 and miR-570–3p.
Results
MALAT1 was highly expressed in BC tissues compared with normal tissues adjacent to cancer as well as in BC cells. In addition, inhibition the expression of MALAT1 could significantly suppress the proliferation, migration and invasion of BC cells. Meanwhile, down-regulation of MALAT1 sensitized BC cells to doxorubicin. Moreover, bioinformatics analysis suggested that miR-570–3p was the potential downstream target of MALAT1. Dual-luciferase reporter gene assay confirmed that MALAT1 could directly target miR-570–3p. Additionally, miR-570–3p was lowly expressed in BC tissues and cells. Up-regulation of miR-570–3p not only significantly inhibited the proliferation, metastasis, and invasion of BC cells, but also increased the sensitivity of BC cells to doxorubicin.
Conclusion
MALAT1 functions as a novel oncogenic lncRNA in regulating the progression and doxorubicin resistance of BC by targeting miR-570–3p.
Keywords: Breast cancer, Long non-coding RNA, Metastasis-associated lung adenocarcinoma transcript 1, miR-570–3p
At a glance commentary
Scientific background on the subject
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a long non-coding RNA (lncRNA) that regulates disease progression in various types of cancers. However, the role and mechanism of MALAT1 in doxorubicin resistance of breast cancer (BC) cells has not been elucidated.
What this study adds to the field
This study found that MALAT1 promoted BC cell proliferation, migration and invasion by targeting miR-570-3p and promoted doxorubicin resistance of BC cells. This study would be expected to enrich the understanding of doxorubicin resistance of BC and provide new ideas for the treatment strategies of BC.
Presently, breast cancer (BC) has become one of the most common malignant tumors, which endangers the health of women all over the world with an increasing morbidity every year [1]. Surgery, radiotherapy and chemotherapy are routine treatment strategies for BC, however a lot of patients have a postoperative recurrence and poor prognosis; additionally, chemoresistance contributes to the progression of the disease [2]. Doxorubicin is a first-line chemotherapeutic drug for adjuvant chemotherapy and palliative chemotherapy for BC. However, chemoresistance is the most important factor limiting the clinical application of doxorubicin [3]. It remains important to understand the mechanism of tumorigenesis and progression of BC so as to provide new targets for improving the prognosis of the patients [4].
Long non-coding RNA (lncRNAs) and microRNAs (miRNAs), both belonging to non-coding RNA, are closely related to cancers [5,6]. An increasing number of studies have shown that lncRNAs act as key regulators in tumor progressions, regulating cell proliferation, migration, invasion, differentiation, apoptosis and so on [7]. LncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a widely investigated lncRNA in cancer biology. For example, it can promote the proliferation and metastasis of epithelial ovarian cancer cells through regulating PI3K/AKT pathway [8]. Silencing of MALAT1 promotes apoptosis and inhibit cell proliferation by inhibiting Wnt/β-catenin signaling pathway in colonic cancer [9]. On the other hand, miRNAs, a class of endogenous, single-stranded non-coding RNAs with a length about 20–25 nucleotides, also play a critical role in various biological processer [10]. For example, miR-96 inhibits the development of pancreatic cancer as a tumor suppressor [11]; miR-26b can inhibit the proliferation of BC cells [12].
LncRNA can function as a molecular sponge (also called competitive endogenous RNA, ceRNA) to adsorb miRNA to repress its expression [13]. This mechanism has been proved to play an important role in cancer progression. For instance, it has been reported that lncRNA HNF1A-AS1 promoted the progression of esophageal squamous cell carcinoma by targeting and down-regulating the expression of miR-214 [14]. MALAT1 regulates the expression of Rac 1 by sponging miR-101b [15]. Moreover, MALAT1 can regulate the expression of TGF-α by targeting miR-376a to promote the development of osteosarcoma [16]. However, the interactions between MALAT1 and miRNA in BC have not been fully clarified. In this study, we examined the expression, function and potential mechanism of MALAT1 in BC. We demonstrated that MALAT1 promoted the progression and doxorubicin resistance of BC by targeting miR-570–3p.
Materials and methods
Tissue samples
BC specimens from 81 patients were obtained in Liaoning Cancer Hospital during 2012–2017, and the written informed consent was obtained from each participant. Cancer tissues and the adjacent normal tissues were collected and stored in liquid nitrogen after removal immediately during the surgery. The study obtained the approval of the Ethics Review Board of Liaoning Cancer Hospital.
Cell culture
Human BC cell lines (MCF-7, SK-BR-3, MDA-MB-468, MDA-MB-231, T-47d and MDA-MB-453) and normal breast epithelial cell line (MCF-10A) were obtained from Zeye Biotechnology Co., LTD (Shanghai, China). The cells were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) supplemented 10% fetal bovine serum (FBS) (Gbico, Detroit, MI, USA) and 100 U/mL penicillin, and 100 μg/mL streptomycin (Hyclone, Logan, UT, USA) in a humidified atmosphere of 5% CO2 at 37 °C.
Cell transfection
miR-570–3p mimics and negative control (miR-control) were obtained from RiBoBio (Guangzhou, China). pcDNA3.1, pcDNA3.1-MALAT1, MALAT1-specific short hairpin RNA (sh-MATAL1) and negative control shRNA were synthesized and provided by GenPharma (Shanghai, China). BC cells were transfected with 100 mM of the indicated oligonucleotide or 50 ng of plasmid using Lipofectamine® 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the protocols provided by the manufacturer.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from BC cells using RNAiso Plus reagent (Takara, Dalian, China). To detect the expression of miR-570–3p, total RNA was reversely transcribed using MMLV reverse transcriptase (Takara, Dalian, China). The obtained cDNA was then used as the template for RT-PCR with SYBR Prime Script RT-PCR Kits (Takara, Dalian, China). The quantitative results were calculated after the reaction with 2 −ΔΔCt method, and the detailed information of the primer sequences was shown in Table 1.
Table 1.
qRT-PCR Primer Sequence (5′–3′).
| Name | Primer Sequences |
|---|---|
| lncRNA MALAT1 | Forward: 5′-GAAGATAGGCATTTGAGTGGCT-3′ |
| Reverse: 5′-CTGAAGAGCATTGGAGATCAGC-3′ | |
| miR-570–3p | Forward: 5′-ACACTCCAGCTGGGCGAAAACAGCA-3′ |
| Reverse: 5′-CGGCAATTCAGTTGAGGCAAAGGT-3′ | |
| U6 | Forward: 5′-GCTTCGGCAGCACATATACT-3′ |
| Reverse: 5′-ACGCTTCACGAATTTGCGTG-3′ | |
| β-actin | Forward: 5′-ACTCGTCATACTCCTGCT-3′ |
| Reverse: 5′-GAAACTACCTTCAACTCC-3′ |
MTT assay
In cell proliferation assay, cells in logarithmic growth phase were inoculated in 96-well plates with 5000 cells per well (100 μL medium). One each time point, 20 μl MTT solution (Sigma, St.Louis, MO, USA; 5 mg/mL) was then added to each well. After 4 h of incubation and at 37 °C, he solution of each well was discarded, and 150 μL of dimethyl sulfoxide (DMSO, Beyotime Biotechnology, Shanghai, China) was supplemented to each well. After the precipitate was completely dissolved by shaking, a microplate reader (Bio-Rad, Hercules, CA, USA) was applied to measure the optical density (OD) values of the wells at 570 nm. The OD values were detected for 4 consecutive days. In the drug resistance experiment, doxorubicin with different concentrations (0.0001–100 μM) was added when cells were cultured for 24 h. After the cell culture was continued for 48 h at 37 °C, MTT assay was performed. The procedures were the same as that of the cell proliferation assay, and then the IC50 was calculated.
Transwell assay
In the migration assay, the transfected BC cells were inoculated into Transwell chambers (BD Biosciences, San Jose, CA, USA, pore size: 8 μm) placed on a 24-well plate, and 2 duplicated wells were set in each group. 105 BC cells were suspended in the upper chamber containg 200 μL serum-free medium. The wells of the 24-well plate contained 500 μL DMEM medium containing 10% FBS. 24 h after incubation, the cells on the upper surface of the membrane were gently wiped off with a cotton swab, whereas the migrated cells on the underside of the membrane were fixed with methanol and stained with crystal violet solution to count the number of cells. As for the invasion assay, the experiment was conducted with the Transwell chambers coated with Matrigel. The other steps are the same as that of the migration assay.
Dual-luciferase reporter gene assay
The target sequence of MALAT1 for miR-570–3p was predicted by StarBase. Wild type (WT) or mutant type (MUT) MALAT1 was subcloned into the pmirGLO vector (Promega Cooperation, Madison, WI, USA). MiR-570–3p mimics or negative control miRNA (miR-control), and the reporter vectors were co-transfected into HEK293T cells with Lipofectamine® 2000 (Invitrogen, Carlsbad, CA, USA). After 48 h of transfection, the luciferase activity was measured by Dual-luciferase® Reporter Assay System (Promega Corp., Madison, WI, USA).
Statistical analysis
All experiments were performed for at least three times and the data were presented as mean ± SD. To examine whether the data were normally distributed, One-Sample Kolmogorov–Smirnov test was performed. For normally distributed data, to compare the data between two groups, Student's t test was performed. One-way ANOVA was performed to compare the data among three or more groups, and if the data showed significant difference, Tukey's post hoc test was performed to compare the difference between two groups. For data that were skewed distributed, Wilcoxon signed-rank test was performed to examine the difference between two groups. The Pearson's correlation coefficient was used to calculate the correlation between MALAT1 and miR-570–3p expression in BC samples. All statistical analyses were carried out using SPSS software (SPSS Inc., Chicago, IL, USA). A p value of less than 0.05 (p < 0.05) was considered to be statistically significant.
Results
MALAT1 was up-regulated in BC tissues and cell lines
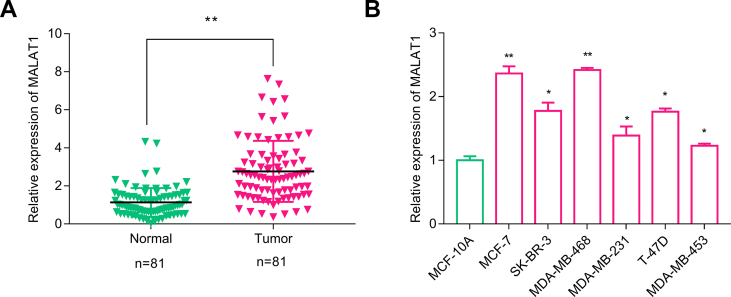
To investigate the relationship between lncRNA MALAT1 and BC progression, qRT-PCR was performed to detect MALAT1 expression in 81 cases of BC tissues and paired adjacent normal breast tissues. It was found that MALAT1 expression in BC tissues was significantly higher than that in normal breast tissues (p < 0.05) [Fig. 1A]. What's more, the expression of MALAT1 in BC cell lines (MCF-7, SK-BR-3, MDA-MB-468, MDA-MB-231, T-47d and MDA-MB-453) was significantly higher than that in the normal breast epithelial cell line (MCF-10A) [Fig. 1B], suggesting that MALAT1 may be involved in the regulation of BC development.
Fig. 1.
Relative expression of MALAT1 was detected by qRT-PCR in (A) BC tissues and adjacent normal breast tissues; (b) normal breast epithelial cell line (MCF-10A) and BC cell lines (MCF-7, SK-BR-3, MDA-MB-468, MDA-MB-231, T-47D and MDA-MB-453). All experiments were performed in triplicate. In (A), Wilcoxon signed-rank test was performed; in (B) One-way ANOVA and Tukey's post hoc test were performed. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
MALAT1 contributed to the malignant phenotypes of BC as an oncogenic lncRNA
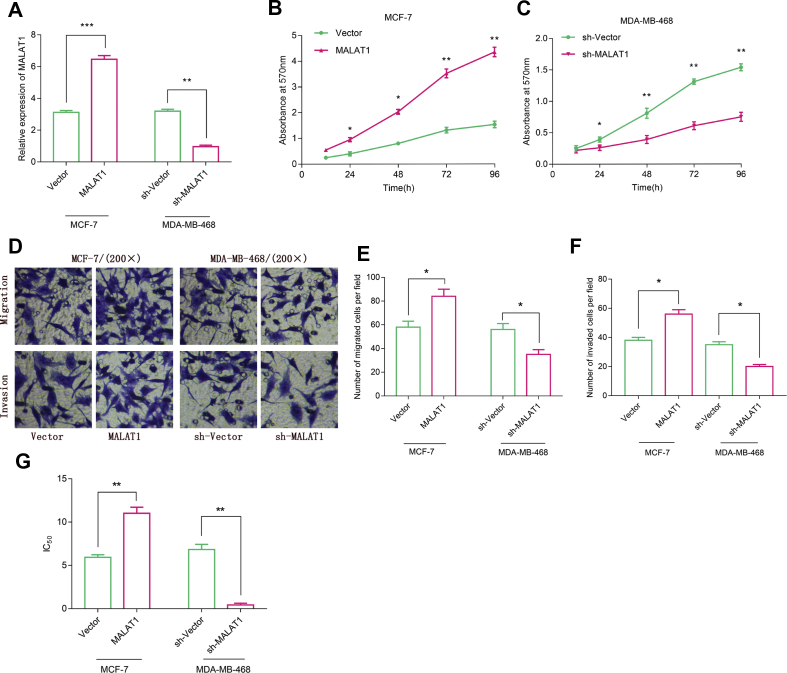
To explore the function of MALAT1 in BC, we constructed over-expressed MALAT1 and low-expressed MALAT1 cell models in MCF-7 and MDA-MB-468, respectively [Fig. 2A]. After that, MTT assay was conducted and we found that over-expression of MALAT1 could facilitate cancer cell proliferation, while low-expression of MALAT1 could arrest cell proliferation [Fig. 2B and C]. Furthermore, Transwell assay revealed that over-expression of MALAT1 accelerated the migration and invasion of MCF-7 cells, whereas knockdown of MALAT1 suppressed the migration and invasion of MDA-MB-468 cells [Fig. 2D–F]. Additionally, MTT assay showed that over-expression of MALAT1 significantly enhanced the doxorubicin resistance of BC cells (p < 0.05); conversely, inhibiting MALAT1 weakened BC cells’ doxorubicin resistance [Fig. 2G].
Fig. 2.
Function of MALAT1 in BC cells. (A) MALAT1 expression in MCF-7 and MDA-MB-468 cells was analyzed by qRT-PCR after transfection (B, C) The proliferation of BC cells was detected by MTT assay (D, E, F) The numbers of migrated and invaded BC cells were measured by Transwell assay. (G) The IC50 value of doxorubicin of BC cells was determined by MTT assay. All experiments were performed in triplicate. Student's t test was performed. ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001.
miR-570–3p is a target of MALAT1 in BC
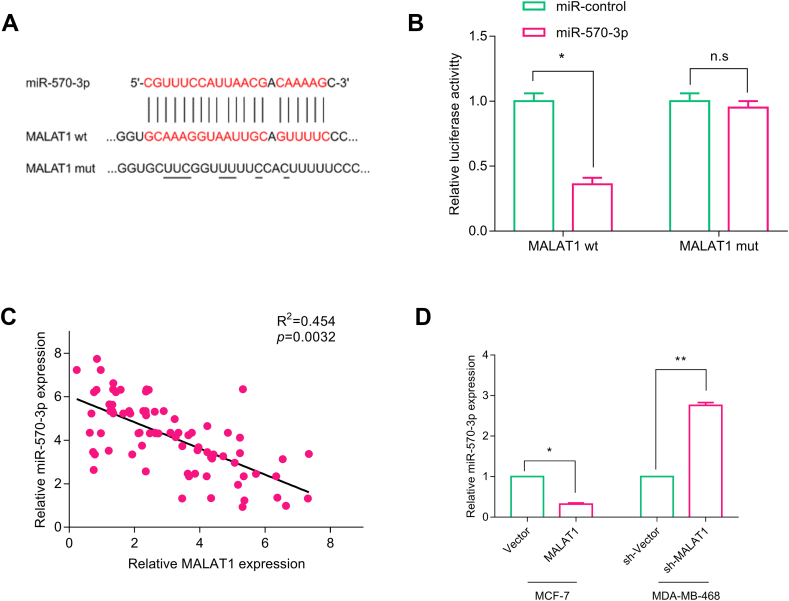
We found that miR-570–3p was a potential target of MALAT1 and there existed a presumed binding site between miR-570–3p and MALAT1 by referring to Starbase database (available at: http://starbase.sysu.edu.cn) [Fig. 3A]. To determine whether miR-570–3p was sponged by MALAT1, dual-luciferase reporter gene assay was performed. The results suggested that miR-570–3p significantly inhibited the luciferase activity of MALAT1 WT reporter (p < 0.05), but it didn't change the luciferase activity of MALA1 MUT reporter [Fig. 3B]. To investigate whether MALAT1 could regulate the expression of miR-570–3p in BC, the expressions of MALAT1 and miR-570–3p in clinical samples were assessed by qRT-PCR, and then Pearson's correlation coefficient was calculated. It showed that the MALAT1 expression in BC samples was negatively correlated with miR-570–3p expression [Fig. 3C]. To further verify whether miR-570–3p was a target of MALAT1, we detected the expression of miR-570–3p in BC cell lines with over-expressed and low-expressed MALAT1 by qRT-PCR. As expected, miR-570–3p expression in over-expressed MALAT1 cells were decreased while miR-570–3p expression in low-expressed MALAT1 cells were increased [Fig. 3D]. In summary, these results supported the conclusion that miR-570–3p was an target for MALAT1 in BC.
Fig. 3.
miR-570–3p was a target miRNA of MALAT1 in BC. (A) Schematic diagram of the presumed binding site between MALAT1 and miR-570–3p as well as the sequence of MALAT1 mutant. (B) Luciferase reporter gene assay was used to verify the predicted binding site. (C) The correlation between MALAT1 expression and miR-570–3p expression in BC tissues. (D) miR-570–3p expressions in MALAT1 over-expression and low-expression groups were measured by qRT-PCR. All experiments were performed in triplicate. In (B) and (D), Student's t test was performed; in (C) Pearson's correlation analysis was performed. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
miR-570–3p expression was down-regulated in BC tissues and cell lines
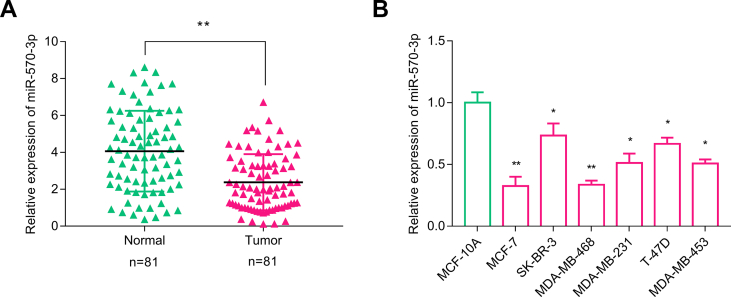
To explore the relationship between miR-570–3p and BC progression, we performed qRT-PCR to detect miR-570–3p expression in BC tissues and paired adjacent normal tissues from 81 cases. It was observed miR-570–3p expression in BC tissues was significantly lower than that in normal breast tissues (p < 0.05) [Fig. 4A]. It is also down-regulated in BC cell lines (p < 0.05) [Fig. 4B].
Fig. 4.
Relative expression of miR-570–3p was detected by qRT-PCR in (A) BC tissues and adjacent normal breast tissues; (B) normal breast epithelial cell line (MCF-10A) and BC cell lines (MCF-7, SK-BR-3, MDA-MB-468, MDA-MB-231, T-47D and MDA-MB-453). All experiments were performed in triplicate. In (A), Wilcoxon signed-rank test was performed; in (B) One-way ANOVA and Tukey's post hoc test were performed. ∗p < 0.05; ∗∗p < 0.01.
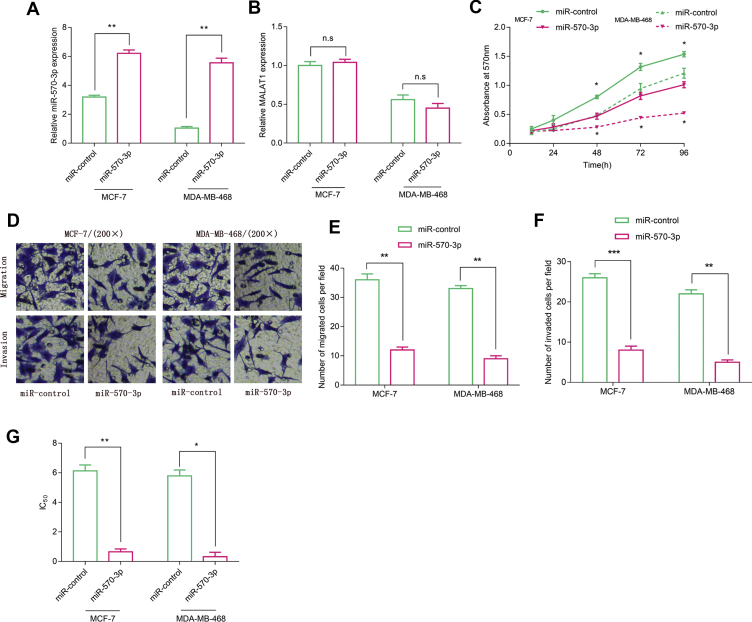
MiR-570–3p acted as a tumor suppressor in BC to inhibit the malignant phenotypes
To further investigate the biological function of miR-570–3p on BC cells, we successfully constructed miR-570–3p over-expressed cell lines [Fig. 5A]. The over-expression of miR-570–3p exerted no significant effect on the expression of MALAT1 [Fig. 5B]. Furthermore, MTT assay showed that miR-570–3p over-expression repressed the proliferation of BC cells (p < 0.05) [Fig. 5C]. Besides, the results from Transwell assay indicated that miR-570–3p over-expression could not only significantly inhibit the migration and invasion of BC cells (p < 0.05) [Fig. 5D–F] but also dramatically reduce the IC50 value of doxorubicin (p < 0.05) [Fig. 5F], indicating that over-expression of miR-570–3p could increase doxorubicin sensitivity in BC cells.
Fig. 5.
Functions of miR-570–3p in BC cells. qRT-PCR was used to detect the expressions of (A) miR-570–3p, (B)MALAT1 after transfection. (C) The proliferation of BC cells was tested by MTT assay (D-F) The numbers of migrated and invaded BC cells were measured Transwell assay. (G) The IC50 value of doxorubicin in BC cells was determined by MTT assay. All experiments were performed in triplicate. Student's t test was performed. ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001.
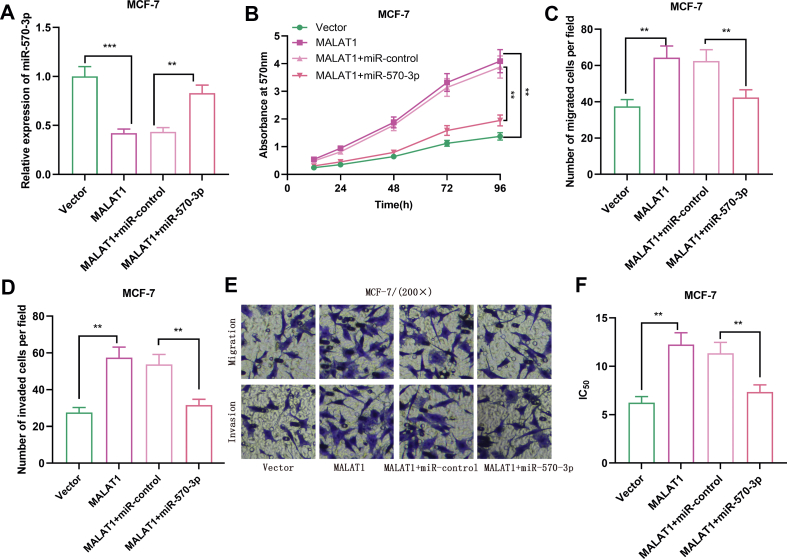
miR-570–3p over-expression partly reversed the cancer-promoting effect of MALAT1 on BC cells
Next, we constructed BC cell lines with over-expression of MALAT1 and miR-570–3p [Fig. 6A]. The results from MTT assay informed us that compared with the MALAT1 over-expression group, the proliferation and metastasis of cells in the MALAT1+miR-570–3p over-expression group were significantly increased [Fig. 6B–E], and the drug sensitivity to doxorubicin was also enhanced [Fig. 6F]. Collectively, we concluded that the cancer-promoting effect of MALAT1 in BC was partly mediated by its regulation on miR-570–3p.
Fig. 6.
MALAT1 regulated the malignant phenotype of BC cells by interacting with miR-570–3p. (A) qRT-PCR was used to detect the expression level of miR-570–3p in MCF-7 cells transfected with MALAT1 overexpression plasmid or co-transfected with miR-570–3p mimic. (B) The proliferation of BC cells was detected by MTT assay (C-E) The numbers of migrated and invaded BC cells were measured by Transwell assay. (F) The IC50 value of doxorubicin in BC cells was tested by MTT assay. All experiments were performed in triplicate. Student's t test was performed. ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001.
Discussion
There are accumulating studies indicating that lncRNAs are key regulators in cancer biology [17]. The abnormal expressions of lncRNAs have been prevalently found in malignant tumors.For example, lncRNA RMRP acted as an oncogenic lncRNA in lung cancer [18]; lncRNA SNHG1 promoted the proliferation of gastric cancer cells [19]; lncRNA UCA1 contributes to the epithelial–mesenchymal transition of BC cells [20]; lncRNA ANCR facilitate the metastasis of BC cells [21]; furthermore, the high-expression of lncRNA p10247 in BC was proven to be associated with cancer metastasis [22]. MALAT1 was firstly identified in non-small cell lung cancer and immediately raised great concern [23]. MALAT1 has been confirmed to be dysregulated in a variety of malignancies. For instance, MALAT1 is significantly up-regulated in osteosarcoma, which can promote osteosarcoma cell metastasis by activating PI3K/Akt signaling pathway [24]. Highly expressed MALAT1 is significantly related to the adverse prognosis of cervical cancer patients [25]. Down-regulation of MALAT1 inhibits the proliferation, invasion and migration of colorectal cancer cells by regulating SOX9 [26]. In BC, it is reported that MALAT1 inhibits the metastasis but facilitates the angiogenesis of BC [27]. However, another study demonstrates that MALAT1 induces the migration and invasion of BC cells [28]. Therefore, it is still controversial on the role of MALAT1 in tumorigenesis, progression and metastasis of BC. In this study, we found that MALAT1 was highly expressed in BC tissues and cell lines. Additionally, in vitro experiments showed that the cell proliferation, migration, invasion and drug resistance were significantly decreased after MALAT1 was knocked down, while increased after MALAT1 was overexpressed. These data supported that MALAT1 could contribute to the progression of BC as a cancer-promoting lncRNA.
In recent years, extensive studies on the relationship between miRNA and cancer biology have been reported [29]. For instance, miR-148b/miR-152 could mediate the proliferation and invasion of pituitary adenomas cells [30]; miR-1290 could enhance the proliferation, migration and invasion of glioma cells [31]. Moreover, studies have confirmed that the dysregulated expression of miR-570–3p is related to tumorigenesis. For example, miR-570–3p is up-regulated in bladder cancer and it promotes cancer cell migration and invasion by targeting KLF10 [32]. Another study demonstrates that up-regulation of miR-570–3p inhibits metformin-induced osteosarcoma cell metastasis by targeting LCMR1 and ATG12 [33]. However, the specific role of miR-570–3p in BC remains largely unknown. In the present study, we discovered that the expression of miR-570–3p was down-regulated in BC tissues and cells. Importantly, we found that increasing the expression of miR-570–3p could restrain the proliferation and metastasis, and improve the sensitivity of BC cells to doxorubicin. These results validated that miR-570–3p functioned as a tumor suppressor to suppress the progression of BC.
Accumulating studies have discovered that lncRNAs, as a ceRNA, could regulate the expression level of genes by targeting miRNA [[34], [35], [36], [37]]. For example, LINC00511 regulates E2F1/Nanog by competitively binding with miR-185–3p to promote the progression of BC [38]. LncRNA NEAT1 promotes the metastasis of BC cells by regulating the miR-133b/TIMM17A axis [39]. LINC01116 can act as a molecular sponge for miR-145 and up-regulate the expression of ESR1 in BC cells [40]. In this study, we identified a binding site between MALAT1 and miR-570–3p. Our further experiments confirmed that the over-expression of MALAT1 notably inhibited the expression of miR-570–3p. Moreover, over-expressed miR-570–3p could not only significantly impede the proliferation, migration and doxorubicin resistance of BC cells, but also offset the promotive effect induced by MALAT1. The findings above validated that miR-570–3p exerted cancer-suppressive functions in BC, and MALAT1 exerted cancer-promoting effects by down-regulating the expression of miR-570–3p.
This study has several limitations. Firstly, the expression pattern of MALAT1 and miR-570–3p remains to be further verified in a larger population since the samples in the current work were from a single institution. Additionally, we only performed in vitro studies in this work, and the biological functions of MALAT1 and miR-570–3p require validation with in vivo studies. Furthermore, the downstream targets of miR-570–3p are not screened and verified. Last but not least, whether MALAT1/miR-570–3p axis regulates the sensitivity of BC cells to other chemotherapeutics is worth studying.
Conclusion
In summary, MALAT1 is up-regulated in BC and it targets miR-570–3p. MALAT1/miR-570–3p axis regulates the malignant phenotypes of BC cells and contributes to the resistance to doxorubicin. Our work helps clarify the mechanism of BC progression, and provides novel insights to the diagnosis and treatment of BC.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Dent R., Trudeau M., Pritchard K.I., Hanna W.M., Kahn H.K., Sawka C.A., et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 3.Kartal-Yandim M., Adan-Gokbulut A., Baran Y. Molecular mechanisms of drug resistance and its reversal in cancer. Crit Rev Biotechnol. 2016;36:716–726. doi: 10.3109/07388551.2015.1015957. [DOI] [PubMed] [Google Scholar]
- 4.Jin S., Ye K. Targeted drug delivery for breast cancer treatment. Recent Pat Anti-Cancer Drug Discov. 2013;8:143–153. [PubMed] [Google Scholar]
- 5.Roberts A., Pimentel H., Trapnell C., Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 6.1000 Genomes Project Consortium, Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Y., Feng S.J., Qiu S., Shao N., Zheng J.H. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 2017;21:3176–3184. [PubMed] [Google Scholar]
- 9.Zhang J., Li Q., Xue B., He R. MALAT1 inhibits the Wnt/β-catenin signaling pathway in colon cancer cells and affects cell proliferation and apoptosis. Bosn J Basic Med Sci. 2020;20:357–364. doi: 10.17305/bjbms.2019.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yates L.A., Norbury C.J., Gilbert R.J. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Huang X., Lv W., Zhang J.H., Lu D.L. miR-96 functions as a tumor suppressor gene by targeting NUAK1 in pancreatic cancer. Int J Mol Med. 2014;34:1599–1605. doi: 10.3892/ijmm.2014.1940. [DOI] [PubMed] [Google Scholar]
- 12.Tormo E., Adam-Artigues A., Ballester S., Pineda B., Zazo S., González-Alonso P., et al. The role of miR-26a and miR-30b in HER2+ breast cancer trastuzumab resistance and regulation of the CCNE2 gene. Sci Rep. 2017;7:41309. doi: 10.1038/srep41309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anastasiadou E., Jacob L.S., Slack F.J. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G., Zhao W., Gao X., Zhang D., Li Y., Zhang Y., et al. HNF1A-AS1 promotes growth and metastasis of esophageal squamous cell carcinoma by sponging miR-214 to upregulate the expression of SOX-4. Int J Oncol. 2017;51:657–667. doi: 10.3892/ijo.2017.4034. [DOI] [PubMed] [Google Scholar]
- 15.Yu F., Lu Z., Cai J., Huang K., Chen B., Li G., et al. MALAT1 functions as a competing endogenous RNA to mediate Rac 1 expression by sequestering miR-101b in liver fibrosis. Cell Cycle. 2015;14:3885–3896. doi: 10.1080/15384101.2015.1120917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo W., He H., Xiao W., Liu Q., Deng Z., Lu Y., et al. MALAT1 promotes osteosarcoma development by targeting TGFA via MIR376A. Oncotarget. 2016;7:54733–54743. doi: 10.18632/oncotarget.10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 18.Meng Q., Ren M., Li Y., Song X. LncRNA-RMRP acts as an oncogene in lung cancer. PloS One. 2016;11 doi: 10.1371/journal.pone.0164845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y., Ma Z., He Y., Liu W., Su Y., Tang Z. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochem Biophys Res Commun. 2017;491:926–931. doi: 10.1016/j.bbrc.2017.07.137. [DOI] [PubMed] [Google Scholar]
- 20.Huang J., Zhou N., Watabe K., Lu Z., Wu F., Xu M., et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip 1) Cell Death Dis. 2014;5 doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z., Dong M., Fan D., Hou P., Li H., Liu L., et al. LncRNA ANCR down-regulation promotes TGF-β-induced EMT and metastasis in breast cancer. Oncotarget. 2017;8:67329–67343. doi: 10.18632/oncotarget.18622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang YX, Li W, Zhang YU, Hayano T, Li Q, Barragan I, et al. Long non-coding RNA p10247, high expressed in breast cancer (lncRNA-BCHE), is correlated with metastasis. Clin Exp Metastasis. 2018;35:109–121. doi: 10.1007/s10585-018-9901-2. [DOI] [PubMed] [Google Scholar]
- 23.Gutschner T., Hämmerle M., Diederichs S. MALAT1--a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y., Huang W., Sun W., Zheng B., Wang C., Luo Z., et al. LncRNA MALAT1 promotes cancer metastasis in osteosarcoma via activation of the PI3K-akt signaling pathway. Cell Physiol Biochem. 2018;51:1313-1326. doi: 10.1159/000495550. [DOI] [PubMed] [Google Scholar]
- 25.Yang L., Bai H.S., Deng Y., Fan L. High MALAT1 expression predicts a poor prognosis of cervical cancer and promotes cancer cell growth and invasion. Eur Rev Med Pharmacol Sci. 2015;19:3187–3193. [PubMed] [Google Scholar]
- 26.Xu Y., Zhang X., Hu X., Zhou W., Zhang P., Zhang J., et al. The effects of lncRNA MALAT1 on proliferation, invasion and migration in colorectal cancer through regulating SOX9. Mol Med. 2018;24:52. doi: 10.1186/s10020-018-0050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang X.J., Xia Y., He G.F., Zheng L.L., Cai Y.P., Yin Y., et al. MALAT1 promotes angiogenesis of breast cancer. Oncol Rep. 2018;40:2683–2689. doi: 10.3892/or.2018.6705. [DOI] [PubMed] [Google Scholar]
- 28.Chou J., Wang B., Zheng T., Li X., Zheng L., Hu J., et al. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc 42. Biochem Biophys Res Commun. 2016 Mar 25;472:262–269. doi: 10.1016/j.bbrc.2016.02.102. [DOI] [PubMed] [Google Scholar]
- 29.Kontomanolis E.N., Koukouli A., Liberis G., Stanulov H., Achouhan A., Pagkalos A. MiRNAs: regulators of human disease. Eur J Gynaecol Oncol. 2016;37:759–765. [PubMed] [Google Scholar]
- 30.He W., Huang L., Li M., Yang Y., Chen Z., Shen X. MiR-148b, MiR-152/ALCAM Axis regulates the proliferation and invasion of pituitary adenomas cells. Cell Physiol Biochem. 2017;44:792–803. doi: 10.1159/000485342. [DOI] [PubMed] [Google Scholar]
- 31.Yan L., Cai K., Sun K., Gui J., Liang J. MiR-1290 promotes proliferation, migration, and invasion of glioma cells by targeting LHX6. J Cell Physiol. 2018;233:6621–6629. doi: 10.1002/jcp.26381. [DOI] [PubMed] [Google Scholar]
- 32.He Q., Yan D., Dong W., Bi J., Huang L., Yang M., et al. circRNA circFUT8 upregulates krüpple-like factor 10 to inhibit the metastasis of bladder cancer via sponging miR-570-3p. Mol Ther Oncolytics. 2020;16:172–187. doi: 10.1016/j.omto.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bao X., Zhao L., Guan H., Li F. Inhibition of LCMR1 and ATG12 by demethylation-activated miR-570-3p is involved in the anti-metastasis effects of metformin on human osteosarcoma. Cell Death Dis. 2018;9:611. doi: 10.1038/s41419-018-0620-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong Q., Li O., Zheng W., Xiao W.Z., Zhang L., Wu D., et al. LncRNA HOTAIR regulates HIF-1α/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Tan J.Y., Sirey T., Honti F., Graham B., Piovesan A., Merkenschlager M., et al. Corrigendum: extensive microRNA-mediated crosstalk between lncRNAs and mRNAs in mouse embryonic stem cells. Genome Res. 2015;25:1410–1411. [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K., Liu C.Y., Zhou L.Y., Wang J.X., Wang M., Zhao B., et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun. 2015;6:6779. doi: 10.1038/ncomms7779. [DOI] [PubMed] [Google Scholar]
- 37.Thum T., Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res. 2015;116:751–762. doi: 10.1161/CIRCRESAHA.116.303549. [DOI] [PubMed] [Google Scholar]
- 38.Lu G., Li Y., Ma Y., Lu J., Chen Y., Jiang Q., et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Canc Res. 2018;37:289. doi: 10.1186/s13046-018-0945-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X., Deng S., Pang X., Song Y., Luo S., Jin L., et al. LncRNA NEAT1 silenced miR-133b promotes migration and invasion of breast cancer cells. Int J Mol Sci. 2019;20:3616. doi: 10.3390/ijms20153616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu H.B., Chen Q., Ding S.Q. LncRNA LINC01116 competes with miR-145 for the regulation of ESR1 expression in breast cancer. Eur Rev Med Pharmacol Sci. 2018;22:1987–1993. doi: 10.26355/eurrev_201804_14726. [DOI] [PubMed] [Google Scholar]