Abstract
Background
Long non-coding RNA (lncRNA) is implicated in the progression of multiple cancers. This study aimed to explore the expression characteristics, biological function and molecular mechanism of lncRNA HAGLROS expression in NSCLC.
Methods
Quantitative real-time polymerase chain reaction (RT-PCR) was adopted to detect HAGLROS expression in NSCLC tissues and normal lung tissues. Survival curve was plotted by Kaplan–Meier method. Gain-of-function and loss-of-function models were respectively established to investigate the biological functions of HAGLROS, miR-100 and SMARCA5. MTT and Transwell assays were carried out to monitor the changes in proliferation, migration and invasion of NSCLC cells. Bioinformatics analysis and dual-luciferase reporter assay were used to verify the binding sites between HAGLROS and miR-100. Western blot was performed to determine the regulatory effects of HAGLROS and miR-100 on SMARCA5 protein expression.
Results
Up-regulated HAGLROS expression was observed in NSCLC tissues and cell lines. Over-expressed HAGLROS promoted the malignant phenotypes of NSCLC cells; conversely, HAGLROS knockdown repressed the malignant phenotypes of NSCLC cells. HAGLROS repressed miR-100 expression to promote SMARCA5 expression in NSCLC cells, and miR-100 overexpression or SMARCA5 knockdown counteracted the oncogenic functions of HAGLROS.
Conclusions
These results conclude that HAGLROS is a tumor promoter in NSCLC, and it regulates the malignant phenotypes of NSCLC cells via miR-100/SMARCA5 axis.
Keywords: NSCLC, HAGLROS, miR-100, SMARCA5
At a glance of commentary
Scientific background on the subject
Long non-coding RNA (lncRNA) HAGLROs have been reported to be associated with the progression of a variety of human cancers, including non-small cell lung cancer. This study aimed to further explore the function and molecular mechanism of HAGLROS in non-small cell lung cancer.
What this study adds to the field
This study found that HAGLROS was up-regulated in NSCLC tissues and cells. Over-expressed HAGLROS promoted the proliferation, migration and invasion of NSCLC cells. In contrast, HAGLROS knockdown had the opposite effect. In addition, HAGLROS promoted the progression of NSCLC by up regulating SMARCA5 through miR-100.
Non-small cell lung cancer (NSCLC) is the most common pathological subtype of lung cancer [1]. The early symptoms of NSCLC are insidious, and more than 40% of the patients are in advanced stage when they are diagnosed with NSCLC, and the prognosis of patients with metastasis or relapse is extremely adverse [2,3]. It's imperative to find novel targets to diagnose and treat NSCLC.
Non-coding RNAs (ncRNAs) have been regarded as potential markers or targets for multiple cancers [[4], [5], [6]]. Long non-coding RNA (lncRNA) is a type of ncRNA, defined as transcripts with lengths exceeding 200 nucleotides which could not be translated into proteins. LncRNAs play crucial roles in a wide range of biological processes [[7], [8], [9], [10]]. For example, lncRNA cancer susceptibility 2 (CASC2) exerts an inhibitory effect on hepatocellular carcinoma via regulating miR-367/F-box and WD repeat domain containing 7 (FBXW7) axis [5]. Up-regulation of lncRNA small nucleolar RNA host gene 12 (SNHG12) promotes the growth and invasion of cervical cancer cells by acting as a molecular sponge for miR-424-5p [11]. Recently, it is reported that HAGLR opposite strand lncRNA (HAGLROS) is highly expressed in NSCLC and it promotes the migration and invasion of NSCLC cells [12]. However, the role of HAGLROS in NSCLC has not been fully clarified.
MicroRNAs (miRNAs) contain about 22 nucleotides, and they regulate gene expression at post-transcriptional level [[13], [14], [15]]. MiR-100 belongs to miR-99 family, which is abnormally expressed in multiple cancers. Reportedly, miR-100 inhibits the growth of glioblastoma cells by targeting SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 (SMARCA5) and erb-b2 receptor tyrosine kinase 3 (ErbB3) [16]. Another study reports that miR-100 regulates epithelial-mesenchymal transition and Wnt/β-catenin signaling by targeting HOXA1 and functions as a tumor suppressor in NSCLC [17]. Moreover, HAGLROS can target miR-100 to regulate apoptosis and autophagy of WI-38 cells [18]. However, the specific role of the HAGLROS/miR-100/SMARCA5 axis in NSCLC progression remains unclear.
This study focused on the expression, function, mechanism and clinical implication of HAGLROS in NSCLC. We demonstrated that HAGLROS promoted the malignant phenotypes of NSCLC cells via repressing miR-100 and up-regulating SMARCA5.
Materials and methods
Sample collection
54 NSCLC patients in Xiangyang Central Hospital from 2014 to 2017 were enrolled in this research. Before the collection of the samples, written consents were obtained from the patients. The NSCLC tissues and corresponding adjacent tissues were collected during surgery, and the tissues were frozen and stored in liquid nitrogen at −196 °C before the extraction of total RNA.
Cell cultrue
Normal lung epithelial cells 16HBE, NSCLC cell lines A549, H5N1, H1299, SK-MES-1, H460 and NCI–H23 were purchased from Yaji Biological Co,. Ltd (Shanghai, China). The cells were incubated in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) and 100 U/mL penicillin, and 100 U/mL streptomycin (Hyclone, Logan, UT, USA) at 37 °C in 5% CO2.
Quantitative real-time polymerase chain reaction (RT-PCR)
Total RNA was extracted from cancer cells using RNAiso Plus reagent (Takara, Dalian, China). The total RNA was reversely transcribed with cDNA synthesis kit (Takara, Dalian, China). Afterwards, cDNA obtained was considered as a template, and RT-PCR was conducted with SYBR®Premix-Ex-Taq™ (Takara, Tokyo, Japan). The cycle threshold (Ct) values were used to quantify the relative gene expression with 2-△△Ct formula. U6 and GAPDH were used as the endogenous controls. The specific primer sequences were available in Table 1.
Table 1.
RT-PCR primer sequences.
| Name | Primer sequences |
|---|---|
| HAGLROS | Forward: 5′-TGTCACCCTTAAATACCGCTCT-3′ |
| Reverse: 5′-CTTCCTCCCACACAAATACTCC-3′ | |
| SMARCA5 | Forward: 5′-TCTGTTGCCAGATGTGTTTAATTCA-3′ |
| Reverse: 5′-CCCAAGGCAGTTGTTTGTATCA-3′ | |
| GAPDH | Forward: 5′-AGCCACATCGCTCAGACAC-3′ |
| Reverse: 5′-GCCCAATACGACCAAATCC-3′ | |
| miR-100 | Forward: 5′-GCGGCAACCCGTAGATCCGAA-3′ |
| Reverse: 5′-GTGCAGGGTCCGAGGT-3′ | |
| U6 | Forward: 5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| Reverse: 5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
Establishment of cell lines
The full sequence of HAGLROS lacking poly-A tail was synthesized and subcloned into pcDNA3.1 (GeneChem, Shanghai, China) to construct HAGLROS overexpression plasmid. SMARCA5 siRNA (si-SMARCA5), miR-100 mimics and inhibitors were purchased from GenePharma Co. Ltd. (Shanghai, China). Lipofecmine 2000 (Invitrogen, Carlsbad, CA, USA) was used as the transfection reagent. 48 h after transfection, NSCLC cells were harvested for the validation of overexpression or knockdown, and functional experiments.
MTT assay
NSCLC cells were inoculated in 96-well plate (103 cells in each well), and cultured for 12 h. After that, the medium was replaced by a 100 μl of complete medium containing 10 μl of MTT reagent (Beyotime, Shanghai, China) at different time points (12 h, 24 h, 48 h, 72 h and 96 h). Following that, the cells were cultured at 37 °C for 4 h. Following that, dimethyl sulfoxide (DMSO; Sigma, Shanghai, China) was added to resolve the formazan crystal. Finally, a microspectrophotometer was used to measure the absorbance of the cells at 570 nm.
Transwell assay
In the migration assay, NSCLC cells were suspended with serum-free medium, and inoculated in the upper chamber of the Transwell system (8 μm pore size; BD Biosciences, San Jose, CA, USA) (about 105 cells/well). The lower chamber was full of 500 μl of medium containing 10% FBS. 24 h later, the cells remaining on the upper surface of the filter were gently wiped off with cotton swabs, and cells passing through the filter were fixed with methanol, and then stained with crystal violet solution. Under an inverted microscope, five fields (including the center and periphery of the membrane) were randomly selected to count the number of cells. Matrigel (BD Biosciences, San Jose, CA, USA) was used to coat the filter of the Transwell system in the invasion assay, and the other procedures were the same as the migration assay.
Dual luciferase reporter gene assay
Briefly, DNA oligonucleotides and pMiR-Reporter vectors (Promega, Madison, WI, USA) were used to construct luciferase reporter vectors (pMiR-HAGLROS-wt/pMiR-HAGLROS-mut and pMiR-SMARCA5-wt/pMiR-SMARCA5-mut). pMiR-HAGLROS-wt or pMiR-HAGLROS-mut and miR-100 mimetics or negative controls (NC) were co-transfected into HEK293 cells. The luciferase activity was determined 48 h after the transfection using Dual Luciferase Reporter Assay Kit (Promega, Madison, WI, USA).
Northern blotting
20 μg of total RNA was analyzed on a 15% denaturing polyacrylamide-urea gel. miR-100 and HAGLROS were analysed on a 1.2% agarose gel in 1 × MOPS solution containing 1% formaldehyde. RNAs were separated by electrophoresis and transferred onto a Hyoid-N+ nylon membrane (GE healthcare, Shanghai, China). The membranes were incubated with hydration buffer containing probes in the labelling reaction system (20 mL) containing 2 mL of 10 × T4 PNK ligase buffer and 1 mL of T4 poly nucleotide kinase (NEB, Ipswich, MA, USA) and 2.5 mL of γ-[32 P]-ATP. Then the membranes were pre-hybridized in the hybridization buffer at 65 °C for 1 h and hybridized overnight at 65 °C. The membranes were then exposed to the phosphor screen for at least 12 h to show the bands.
Western blot
RIPA buffer (Beyotime, Shanghai, China) containing protease inhibitor cocktail (Roche, Nutley, NJ, USA) was employed to extract the total protein from NSCLC cells. Protein samples were subjected to SDS-PAGE and then transferred to nitrocellulose (NC) membrane (Millipore, Bedford, MA, USA). After the NC membranes were blocked with 5% fat-free milk, anti-SMARCA5 (Abcam, 1:1000), anti-proliferating cell nuclear antigen (PCNA) (Abcam, 1:1000), anti-matrix metallopeptidase 9 (MMP9) (Abcam, 1:1000) and anti-GAPDH (Santa Cruz, 1: 2000) antibodies were employed to incubate the membrane at 4 °C for 8 h. After the membranes were washed by TBST, the membranes were then incubated with horseradish peroxidase conjugated secondary antibody (1: 2000, Santa Cruz) at room temperature for 1 h. Next, the NC membranes were washed with TBST again, and ECL kit (Amersham Pharmacia Biotech, Little Chalfont, UK) was used to cover the membranes. Ultimately, ChemiDocXRS imaging system was adopted for detecting the protein bands.
Statistical analysis
All the assays were repeated for three times. SPSS13.0 statistical software (SPSS Inc., Chicago, IL, USA) was adopted to carry out statistical analyses. The Student's t test, one-way ANOVA, Fisher's Exact test or Chi-square test were used to evaluate the differences between different groups. Survival curves were plotted based on the Kaplan–Meier curves and log-rank tests. Correlations among HAGLROS expression, miR-100, and SMARCA5 were analyzed with Pearson's correlation analysis. Differences with p < 0.05 were considered to be statistically significant.
Results
HAGLROS expression was up-regulated in NSCLC tissues and cells
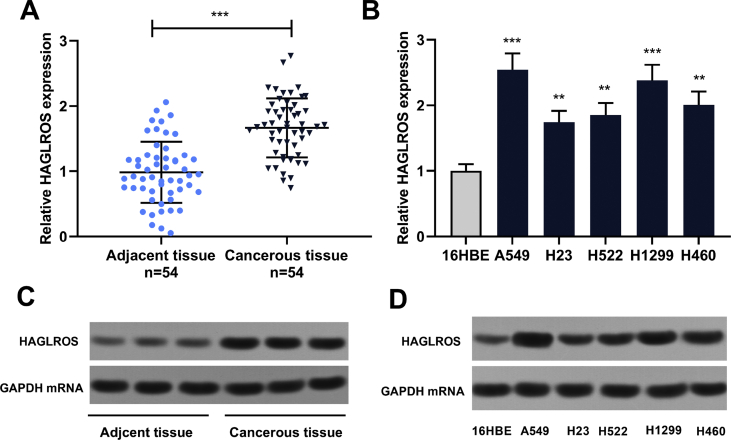
The expressions of HAGLROS in 54 pairs of NSCLC samples and adjacent tissues were detected. HAGLROS expression was observed to be notably up-regulated in NSCLC tissues compared with normal tissues [Fig. 1A]. Furthermore, HAGLROS in NSCLC cell lines including A549 and H1299 cells was be highly expressed [Fig. 1B]. Similarly, the Northern blot analysis demonstrated that the HAGLROS hybridization signal was stronger in NSCLC tissues and cell lines than that in adjacent tissues and 16HBE cells [Fig. 1C and D].
Fig. 1.
HAGLROS was up-regulated in NSCLC tissues and cell lines. (A) RT-PCR was performed to detect the expression of HAGLROS in 54 pairs of NSCLC tissues and matched adjacent normal tissues. (B) RT-PCR was adopted to detect HAGLROS expression in normal lung epithelial cells and NSCLC cell lines. (C and D) Northern blot was used to detect the expression of HAGLROS in 3 pairs of NSCLC tissues and 6 cell lines. ∗∗p < 0.01, ∗∗∗p < 0.001.
High HGALROS expression was correlated with poor prognosis of NSCLC patients
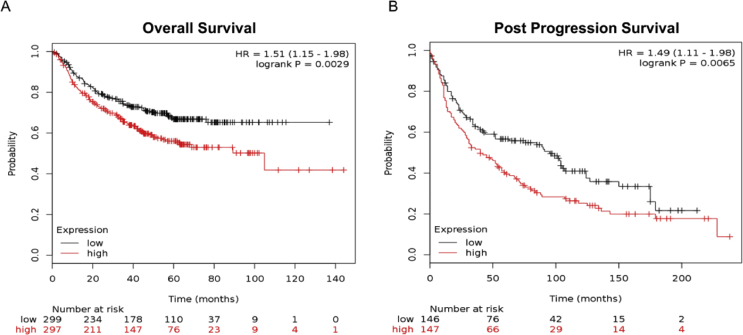
To explore whether HAGLROS expression was associated with the cliniopathological characteristics of NSCLC patients, the NSCLC samples with HGALROS expression higher than twice of the average HGALROS expression in adjacent tissues, were defined as “high expression”, and the other samples were defined as “low expression”. Chi-square test indicated that highly-expressed lncRNA HAGLROS in tumor tissues was correlated with T stage (p = 0.0219) and local lymph node metastasis (p = 0.0037) in NSCLC patients, but had no association with age, sex, smoking history and tumor type (p > 0.05) [Table 2]. Additionally, we used Kaplan–Meier plotter database (kmplot.com) to explore the correlation of HAGLROS with the prognosis of NSCLC patients. The results suggested that NSCLC patients with highly expressed HAGLROS had shorter relapse-free survival and overall survival than those with the low expression of HAGLROS [Fig. 2A and B]. The findings above informed us that HAGLROS could probably be used as a biomarker for NSCLC.
Table 2.
The correlation between lncRNA HAGLROS expression and clinicopathological features (n = 54).
| Clinical features | N | lncRNA HAGLROS expression |
Chi-square value | p value | |
|---|---|---|---|---|---|
| High expression | Low expression | ||||
| Age | |||||
| >60 | 22 | 14 | 8 | 0.1534 | 0.6953 |
| ≤60 | 32 | 22 | 10 | ||
| Gender | |||||
| Male | 31 | 21 | 10 | 0.0379 | 0.8457 |
| Female | 23 | 15 | 8 | ||
| Smoking history | |||||
| Smoker | 29 | 19 | 10 | 0.0372 | 0.8470 |
| Non-smoker | 25 | 17 | 8 | ||
| T stage | |||||
| T1-T2 | 7 | 2 | 5 | 0.034 | |
| T3-T4 | 47 | 34 | 13 | ||
| Lymph invasion | |||||
| N0 | 24 | 11 | 13 | 8.4375 | 0.0037 |
| N1–N2 | 30 | 25 | 5 | ||
| Histology | |||||
| Squamous cancer | 16 | 10 | 6 | 1.0000 | 0.6065 |
| Adenocarcinoma | 20 | 15 | 5 | ||
| Large cell carcinoma | 18 | 11 | 7 | ||
Fig. 2.
Patients with highly expressed HAGLROS in NSCLC have worse prognosis. (A and B) Kaplan–Meier analysis was carried out to analyze the differences of relapse survival time and overall survival time between NSCLC patients with high expression of HAGLROS and with low expression of HAGLROS.
HAGLROS facilitated the proliferation, migration and invasion of NSCLC cells
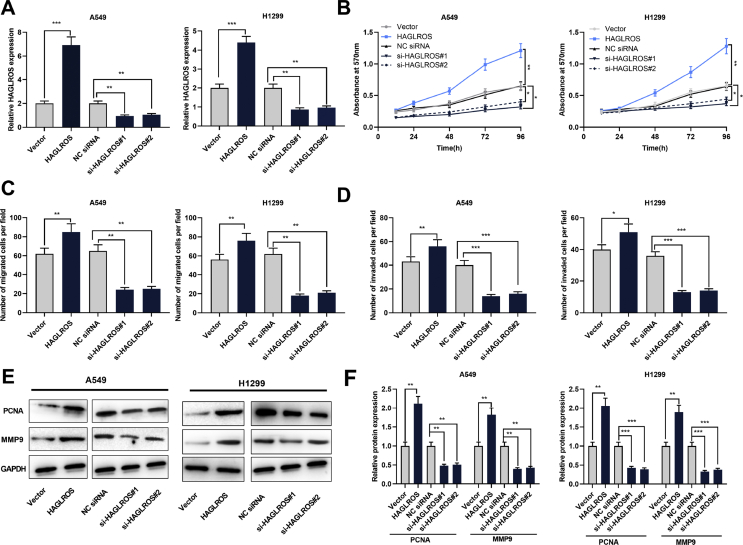
To probe into the impact of HAGLROS on NSCLC, we successfully established HAGLROS over-expression and knockdown cell lines with A549 and H1299, respectively [Fig. 3A]. MTT assay, subsequently, was carried out to monitor the proliferation of NSCLC cell lines A549 and H1299. The results demonstrated that over-expressed HAGLROS could promote the proliferation of NSCLC cells, while knockdown of HAGLROS showed the opposite effect [Fig. 3B]. Transwell assay confirmed the migration and invasion of NSCLC cells were facilitated by over-expressed HAGLROS, but restrained by knockdown of HAGLROS [Fig. 3C and D]. In addition, through Western blot analysis, it was found that overexpression of HAGLROS enhanced the expressions of PCNA and MMP9, and knockdown of HAGLROS inhibited the expressions of PCNA and MMP9 [Fig. 3E and F], which further indicated that HAGLROS regulated the proliferation, migration and invasion of NSCLC cells.
Fig. 3.
HAGLROS could promote the proliferation, migration and invasion of NSCLC cells. (A) Overexpression and knock-down of HAGLROS in A549 and H1299 cells were detected by RT-PCR. (B) MTT assay was used to monitor the proliferation of A549 and H1299 cells with HAGLROS overexpression or knockdown. (C and D) Transwell assay was conducted to determine the migration and invasion of A549 and H1299 cells with HAGLROS overexpression or knockdown. (E and F) Western blot was used to detect the expressions of PCNA and MMP9 proteins in A549 and H1299 cells with HAGLROS overexpression or knockdown. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
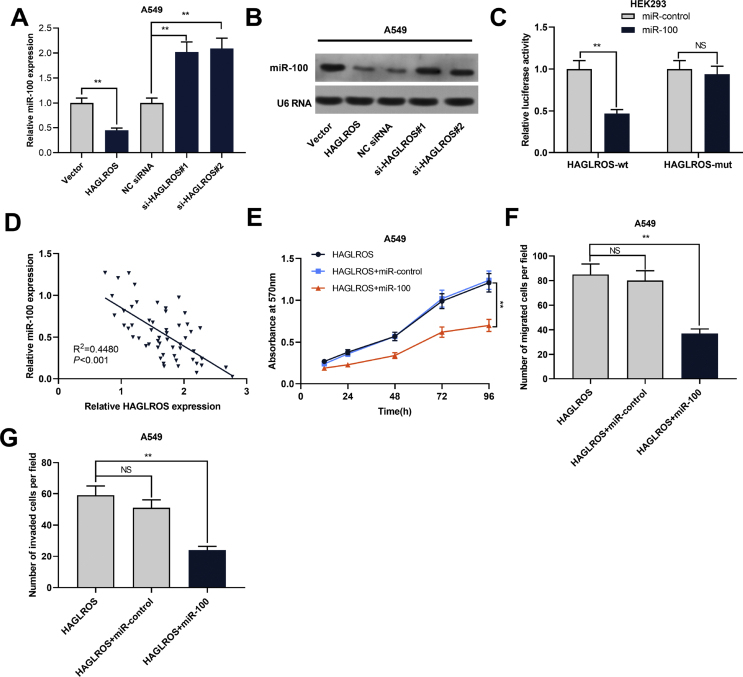
miR-100 interacted with HAGLROS in NSCLC cells
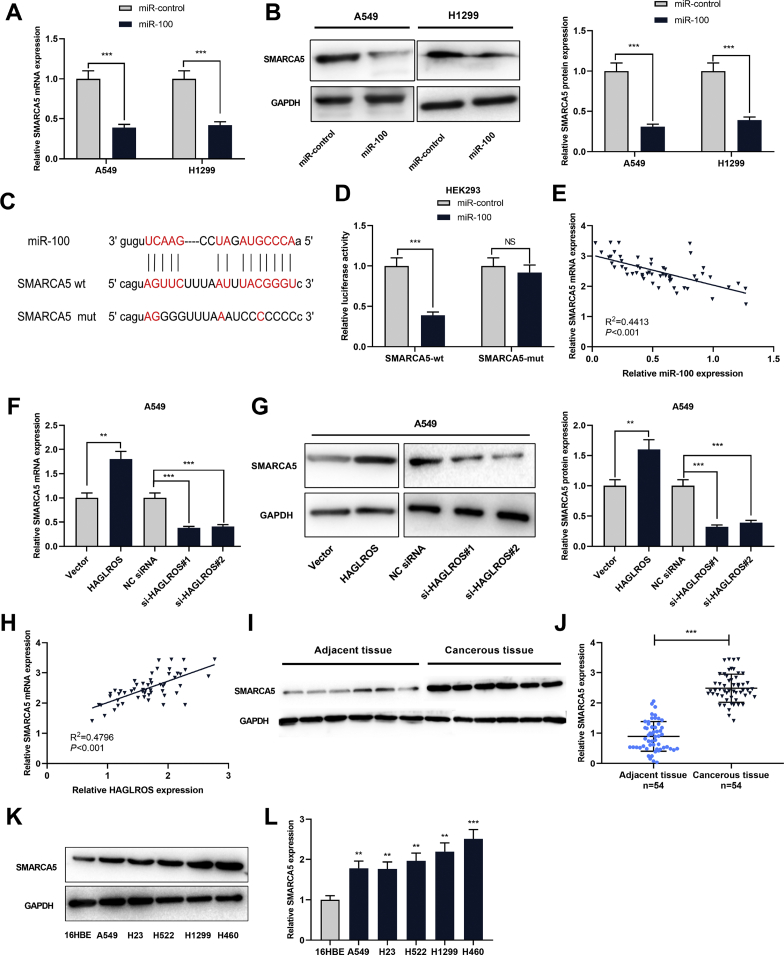
A recent study reported that HAGLROS could regulate the apoptosis and autophagy of WI-38 cells via regulating miR-100/NF-κB axis [18]. What's more, HAGLROS can modulate the apoptosis and autophagy of colorectal cancer cells via regulating miR-100/ATG5 axis [19]. To confirm whether HAGLROS could regulate the expression of miR-100 in NSCLC, we examined miR-100 expression in NSCLC cell lines after HAGLROS was knocked down or overexpressed with RT-PCR and Northern blot. It was observed that miR-100 expression was decreased in NSCLC cells with HAGLROS overexpression, while increased in NSCLC cells with HAGLROS knockdown [Fig. 4A]. Luciferase reporter gene assay indicated that miR-100 could directly bind to HAGLROS [Fig. 4B]. The results from RT-PCR further revealed that the expression level of HAGLROS in NSCLC samples was negatively correlated with miR-100 expression [Fig. 4C]. These results supported that miR-100 was a target of HAGLROS, and was negatively regulated by HAGLROS in NSCLC tissues and cells. Next, through MTT and Transwell assays, we found that compared with the cells with HAGLROS overexpression, the proliferation, migration and invasion of the cells in HAGLROS overexpression + miR-100 overexpression group were significantly reduced [Fig. 4D,E,F]. These data suggested that HAGLROS could probably regulate the malignant phenotypes of NSCLC cells via repressing miR-100.
Fig. 4.
MiR-100 interacted with HAGLROS in NSCLC. (A and B) The changes of miR-100 expression were examined by RT-PCR and Northern blot when HAGLROS was overexpressed or knocked down in NSCLC cells. (C) Dual-luciferase reporter assay was done to check the luciferase activity of cells to verify the binding relationship between HAGLROS and miR-100. (D) RT-PCR was carried out to explore the correlation between HAGLROS expression and miR-100 expression in 54 cases of NSCLC samples. (E) MTT assay was used to monitor the proliferation of A549 cells co-transfected with HAGLROS overexpression plasmids and miR-100 mimics. (F and G) Transwell assay was performed to monitor the migration and invasion of A549 cells co-transfected with HAGLROS overexpression plasmids and miR-100 mimics. ∗∗p < 0.01, NS: p > 0.05.
MiR-100 repressed SMARCA5 expression and HAGLROS induced SMARCA5 expression
We demonstrated that the expression of SMARCA5 was significantly down-regulated after miR-100 mimics were transfected into NSCLC cells [Fig. 5A and B]. By searching TargetScan database (http://www.targetscan.org/vert_72/), we found SMARCA5 was a potential direct target of miR-100 [Fig. 5C]. Luciferase reporter gene assay validated that miR-100 directly targeted SMARCA5 mRNA 3′-UTR [Fig. 5D]. We also demonstrated that the expression level of SMARCA5 in NSCLC samples was negatively correlated with miR-100 expression [Fig. 5E]. RT-PCR and Western blot revealed that HAGLROS could positively regulate SMARCA5 on mRNA and protein levels [Fig. 5F and G]. Furthermore, SMARCA5 in NSCLC samples was positively correlated with HAGLROS expression [Fig. 5H]. In addition, the results of Western blot and RT-PCR analysis showed that SMARCA5 protein and mRNA were significantly highly expressed in NSCLC tissues and cells [Fig. 5I–L]. Collectively, we concluded that HAGLROS/miR-100 axis regulated SMARCA5 expression in NSCLC.
Fig. 5.
In NSCLC, the expression of SMARCA5 can be inhibited by miR-100 and promoted by LncRNA HAGLROS. (A and B) RT-PCR and Western blot were used to detect SMARCA5 expression in A549 and H1299 cells with overexpressed miR-100. (C) The binding targets of miR-100 and SMARCA5 were predicted according to TargetScan database. (D) Dual-luciferase reporter assay was conducted to measure the luciferase activity to verify the targeting relationship between miR-100 and SMARCA5. (E) RT-PCR was employed to analyze the expression correlation between miR-100 and SMARCA5 in 54 cases of NSCLC samples. (F and G) RT-PCR and Western blot were done to assess the expression level of SMARCA5 mRNA and protein in A549 cells with HAGLROS overexpression or knockdown. (H) The correlation between HAGLROS and SMARCA5 expression in 54 cases of NSCLC samples was determined by RT-PCR. (I and J) Western blot and RT-PCR were used to detect the expression level of SMARCA5 protein and mRNA in NSCLC tissues and adjacent tissues. (K and L) Western blot and RT-PCR were used to detect the expression level of SMARCA5 protein and mRNA in NSCLC cells and 16HBE cells. ∗∗p < 0.01, ∗∗∗p < 0.001, NS: p > 0.05.
Knockdown of SMARCA5 attenuated the effect of overexpression of HAGLROS on the progression of NSCLC
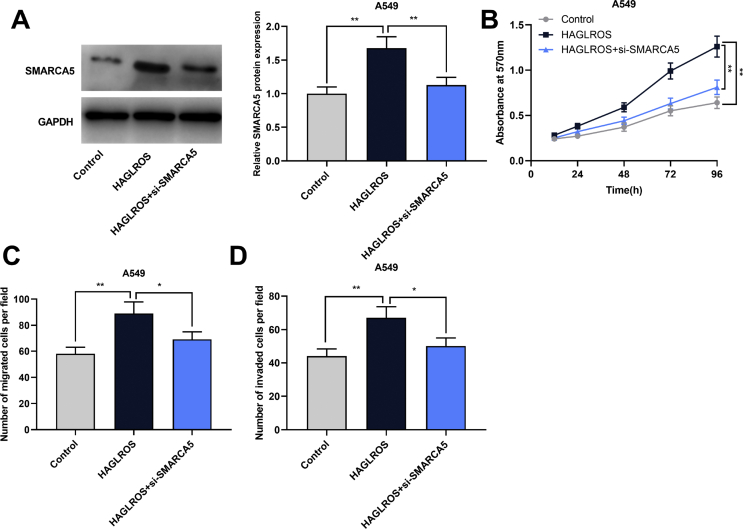
To explore whether SMARCA5 was a downstream effector of HAGLROS, si-SMARCA5 was transfected into A549 cells overexpressing HAGLROS. Western blot showed that the expression level of SMARCA5 in HAGLROS + si-SMARCA5 group was significantly lower than that in HAGLROS group [Fig. 6A]. Then the proliferation, migration and invasion of NSCLC cells were detected by CCK-8 and Transwell assays, and the results suggested that knockdown of SMARCA5 attenuated the promoting effects of HAGLROS overexpression on proliferation, migration, invasion [Fig. 6B–D].
Fig. 6.
The knockdown of SMARCA5 attenuated the effect of overexpression of HAGLROS on the malignant phenotypes of NSCLC cells. (A) Western blot was used to detect the expression level of SMARCA5 protein in A549 cells after co-transfection. (B–D) CCK-8 and Transwell assay were used to detect the proliferation, migration and invasion of A549 cells, respectively. ∗p < 0.05, ∗∗p < 0.01.
Discussion
Previously, lncRNA was considered as the “garbage” formed in the transcription, however, recent researches have revealed that lncRNAs features predominantly in regulating multiple biological processes, including tumorigenesis and cancer progression [[20], [21], [22], [23]]. For instance, lncRNA small nucleolar RNA host gene 15 (SNHG15) promotes the proliferation, migration and invasion of breast cancer cells [24]. LncRNA colorectal neoplasia differentially expressed (CRNDE) facilitates the proliferation of gastric cancer cells [25]. LncRNA urothelial cancer associated 1 (UCA1) meditates the growth and metastasis of pancreatic cancer cells [26]. In the present study, we demonstrated the up-regulation of HAGLROS in NSCLC, which is consistent with the previous report [12]. What's more, it was revealed that over-expressed HAGLROS was associated with unfavorable pathological characteristics and shorter survival time of NSCLC patients. Additionally, HAGLROS overexpression promoted the malignant phenotypes of NSCLC cells. These data suggest that HAGLROS is a promising biomarker and therapy target for NSCLC.
The role of miRNAs in tumors has been extensively studied. For instance, miR-494 inhibits the proliferation of oral cancer cells via regulating homeobox A10 (HOXA10) expression [27]. MiR-506 impedes the proliferation of esophageal cancer cells via targeting cAMP responsive element binding protein 1 (CREB1) [28]. Besides, miR-124 restrains the proliferation, migration and invasion of lung adenocarcinoma cells by directly targeting SRY-box transcription factor 9 (SOX9) [29]. MiR-100 is a potential molecular marker of NSCLC, which acts as a tumor suppressor by targeting polo-like kinase 1 (PLK1) [30]. Accumulating studies report that lncRNAs, as competitive endogenous RNA, can modulate the expression of miRNA. For instance, lncRNA colon cancer associated transcript 1 (CCAT1) promotes the proliferation, migration and invasion of thyroid cancer cell line FTC-133 by downregulating miR-143 [31]. LncRNA maternally expressed 3 (MEG3) suppresses the proliferation and invasion of melanoma cells by regulating miR-499-5p [32]. In NSCLC, lncRNA growth arrest specific 5 (GAS5) inhibits tumorigenesis by suppressing the expression of miR-23a, and lncRNA HOXA11 antisense RNA (HOXA11-AS) promotes epithelial-mesenchymal transformation of cancer cells via suppressing miR-200 b [33,34]. In this study, we found that HAGLROS could target miR-100 and repress its expression, and this regulatory relationship is similar with the previous reports [18,19].
SMARCA5 is a component of SWI/SNF chromatin-remodeling complex. It possesses a DNA-stimulated ATPase activity [35]. SMARCA5 is a crucial regulator in gene transcription, DNA repair and DNA replication, which is associated with malignant transformation [36,37]. Previously, some studies have reported abnormally high expression of SMARCA5 in breast cancer, gastric cancer, ovarian cancer and acute leukemia [[38], [39], [40]]. In the present work, we demonstrated that in NSCLC, SMARCA5 expression was significantly up-regulated, which was negative correlated with miR-100 expression and positively correlated with HAGLORS expression. Additionally, we validated that SMARCA5 was a target gene of miR-100 in NSCLC, which is also reported in glioblastoma and prostate cancer [16,41]. Importantly, we proved that SMARCA5 expression was up-regulated by HAGLROS overexpression in NSCLC cells, and SMARCA5 knockdown counteracted the biological effects of HAGLROS overexpression on NSCLC cells. These results suggest that HAGLROS exerts its oncogenic functions via regulating SMARCA5.
Conclusion
To conclude, the present study demonstrates that HAGLORS/miR-100/SMARCA5 axis is a novel molecular mechanism involved in NSCLC progression. Our work provides novel targets for the molecular treatment of NSCLC.
Ethics statement
Our study was approved by Medical College Review Board of the Xiangyang Central Hospital.
Funding
This research is supported by Natural Science Foundation of Xiangyang Central Hospital.
Conflicts of interest
The authors declare that they have no competing interest.
Acknowledgements
We thank all the laboratory members for continuous technical advice and helpful discussion.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Goldstraw P., Chansky K., Crowley J., Rami-Porta R., Asamura H., Eberhardt W.E., et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11:39–51. doi: 10.1016/j.jtho.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Torre L.A., Siegel R.L., Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 4.Liu Z., Dou C., Yao B., Xu M., Ding L., Wang Y., et al. Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. Oncotarget. 2016;7:25350–25365. doi: 10.18632/oncotarget.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Liu Z., Yao B., Li Q., Wang L., Wang C., et al. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer. 2017;16:123. doi: 10.1186/s12943-017-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y., Cheng X., Liang H., Jin Z. Long non-coding RNA HOTAIR and STAT3 synergistically regulate the cervical cancer cell migration and invasion. Chem Biol Interact. 2018;286:106–110. doi: 10.1016/j.cbi.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Schmitz S.U., Grote P., Herrmann B.G. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Lun L., Hou H., Tian R., Zhang H., Zhang Y. The value of lncRNA HULC as a prognostic factor for survival of cancer outcome: a meta-analysis. Cell Physiol Biochem. 2017;41:1424–1434. doi: 10.1159/000468005. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y.Y., Chen Z.H., Peng J.J., Wu J.L., Yuan Y.J., Zhai E.T., et al. Up-regulation of long non-coding RNA XLOC_010235 regulates epithelial-to-mesenchymal transition to promote metastasis by associating with Snail1 in gastric cancer. Sci Rep. 2017;7:2461. doi: 10.1038/s41598-017-02254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu X., Zhou C., Li R., Deng Y., Zhao L., Zhai W. Long noncoding RNA AFAP1-AS1 promoted tumor growth and invasion in cholangiocarcinoma. Cell Physiol Biochem. 2017;42:222–230. doi: 10.1159/000477319. [DOI] [PubMed] [Google Scholar]
- 11.Dong J., Wang Q., Li L., Xiao-Jin Z. Upregulation of long non-coding RNA small nucleolar RNA host gene 12 contributes to cell growth and invasion in cervical cancer by acting as a sponge for MiR-424-5p. Cell Physiol Biochem. 2018;45:2086–2094. doi: 10.1159/000488045. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y., Shen T., Ding X., Cheng L., Sheng L., Du X. HAGLROS is overexpressed and promotes non-small cell lung cancer migration and invasion. Jpn J Clin Oncol. 2020;50:1058–1067. doi: 10.1093/jjco/hyaa075. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 15.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 16.Alrfaei B.M., Clark P., Vemuganti R., Kuo J.S. MicroRNA miR-100 decreases glioblastoma growth by targeting SMARCA5 and ErbB3 in tumor-initiating cells. Technol Cancer Res Treat. 2020;19 doi: 10.1177/1533033820960748. 1533033820960748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han W., Ren X., Yang Y., Li H., Zhao L., Lin Z. microRNA-100 functions as a tumor suppressor in non-small cell lung cancer via regulating epithelial-mesenchymal transition and Wnt/β-catenin by targeting HOXA1. Thorac Cancer. 2020;11:1679–1688. doi: 10.1111/1759-7714.13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu M., Han T., Shi S., Chen E. Long noncoding RNA HAGLROS regulates cell apoptosis and autophagy in lipopolysaccharides-induced WI-38 cells via modulating miR-100/NF-κB axis. Biochem Biophys Res Commun. 2018;500:589–596. doi: 10.1016/j.bbrc.2018.04.109. [DOI] [PubMed] [Google Scholar]
- 19.Zheng Y., Tan K., Huang H. Long noncoding RNA HAGLROS regulates apoptosis and autophagy in colorectal cancer cells via sponging miR-100 to target ATG5 expression. J Cell Biochem. 2019;120:3922–3933. doi: 10.1002/jcb.27676. Retraction in: J Cell Biochem. 2021 Jun: 34101247. [DOI] [PubMed] [Google Scholar]
- 20.Chen Q.N., Wei C.C., Wang Z.X., Sun M. Long non-coding RNAs in anti-cancer drug resistance. Oncotarget. 2017;8:1925–1936. doi: 10.18632/oncotarget.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheetham S.W., Gruhl F., Mattick J.S., Dinger M.E. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricciuti B., Mencaroni C., Paglialunga L., Paciullo F., Crinò L., Chiari R., et al. Long noncoding RNAs: new insights into non-small cell lung cancer biology, diagnosis and therapy. Med Oncol. 2016;33:18. doi: 10.1007/s12032-016-0731-2. [DOI] [PubMed] [Google Scholar]
- 23.Chen J., Wang R., Zhang K., Chen L.B. Long non-coding RNAs in non-small cell lung cancer as biomarkers and therapeutic targets. J Cell Mol Med. 2014;18:2425–2436. doi: 10.1111/jcmm.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong Q., Qiu M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem Biophys Res Commun. 2018;495:1594–1600. doi: 10.1016/j.bbrc.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 25.Hu C.E., Du P.Z., Zhang H.D., Huang G.J. Long noncoding RNA CRNDE promotes proliferation of gastric cancer cells by targeting miR-145. Cell Physiol Biochem. 2017;42:13–21. doi: 10.1159/000477107. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X., Gao F., Zhou L., Wang H., Shi G., Tan X. UCA1 regulates the growth and metastasis of pancreatic cancer by sponging miR-135a. Oncol Res. 2017;25:1529–1541. doi: 10.3727/096504017X14888987683152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Libório-Kimura T.N., Jung H.M., Chan E.K. miR-494 represses HOXA10 expression and inhibits cell proliferation in oral cancer. Oral Oncol. 2015;51:151–157. doi: 10.1016/j.oraloncology.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Yao W.J., Wang Y.L., Lu J.G., Guo L., Qi B., Chen Z.J. MicroRNA-506 inhibits esophageal cancer cell proliferation via targeting CREB1. Int J Clin Exp Pathol. 2015;8:10868–10874. [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X., Liu Y., Liu X., Yang J., Teng G., Zhang L., et al. MiR-124 inhibits cell proliferation, migration and invasion by directly targeting SOX9 in lung adenocarcinoma. Oncol Rep. 2016;35:3115–3121. doi: 10.3892/or.2016.4648. [DOI] [PubMed] [Google Scholar]
- 30.Liu J., Lu K.H., Liu Z.L., Sun M., De W., Wang Z.X. MicroRNA-100 is a potential molecular marker of non-small cell lung cancer and functions as a tumor suppressor by targeting polo-like kinase 1. BMC Cancer. 2012;12:519. doi: 10.1186/1471-2407-12-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang T., Zhai H., Yan R., Zhou Z., Gao L., Wang L. lncRNA CCAT1 promotes cell proliferation, migration, and invasion by down-regulation of miR-143 in FTC-133 thyroid carcinoma cell line. Braz J Med Biol Res. 2018;51 doi: 10.1590/1414-431X20187046. Retraction in: Yang T, Zhai H, Yan R, Zhou Z, Gao L, Wang L. Braz J Med Biol Res 2021 14;54:e7046retraction. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Long J., Pi X. lncRNA-MEG3 suppresses the proliferation and invasion of melanoma by regulating CYLD expression mediated by sponging miR-499-5p. BioMed Res Int. 2018;2018:2086564. doi: 10.1155/2018/2086564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mei Y., Si J., Wang Y., Huang Z., Zhu H., Feng S., et al. Long noncoding RNA GAS5 suppresses tumorigenesis by inhibiting miR-23a expression in non-small cell lung cancer. Oncol Res. 2017;25:1027–1037. doi: 10.3727/096504016X14822800040451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J.H., Zhou L.Y., Xu S., Zheng Y.L., Wan Y.F., Hu C.P. Overexpression of lncRNA HOXA11-AS promotes cell epithelial-mesenchymal transition by repressing miR-200b in non-small cell lung cancer. Cancer Cell Int. 2017;17:64. doi: 10.1186/s12935-017-0433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohamed M.A., Greif P.A., Diamond J., Sharaf O., Maxwell P., Montironi R., et al. Epigenetic events, remodelling enzymes and their relationship to chromatin organization in prostatic intraepithelial neoplasia and prostatic adenocarcinoma. BJU Int. 2007;99:908–915. doi: 10.1111/j.1464-410X.2006.06704.x. [DOI] [PubMed] [Google Scholar]
- 36.Stopka T., Skoultchi A.I. The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci U S A. 2003;100:14097–14102. doi: 10.1073/pnas.2336105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reisman D., Glaros S., Thompson E.A. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 38.Jin Q., Mao X., Li B., Guan S., Yao F., Jin F. Overexpression of SMARCA5 correlates with cell proliferation and migration in breast cancer. Tumour Biol. 2015;36:1895–1902. doi: 10.1007/s13277-014-2791-2. [DOI] [PubMed] [Google Scholar]
- 39.Stopka T., Zakova D., Fuchs O., Kubrova O., Blafkova J., Jelinek J., et al. Chromatin remodeling gene SMARCA5 is dysregulated in primitive hematopoietic cells of acute leukemia. Leukemia. 2000;14:1247–1252. doi: 10.1038/sj.leu.2401807. [DOI] [PubMed] [Google Scholar]
- 40.Gigek C.O., Lisboa L.C., Leal M.F., Silva P.N., Lima E.M., Khayat A.S., et al. SMARCA5 methylation and expression in gastric cancer. Cancer Invest. 2011;29:162–166. doi: 10.3109/07357907.2010.543365. [DOI] [PubMed] [Google Scholar]
- 41.Sun D., Lee Y.S., Malhotra A., Kim H.K., Matecic M., Evans C., et al. miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res. 2011;71:1313–1324. doi: 10.1158/0008-5472.CAN-10-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]