Abstract
Purpose
Emerging evidence suggest that infection-dependent hyperactivation of complement system (CS) may worsen COVID-19 outcome. We investigated the role of predicted high impact rare variants — referred as qualifying variants (QVs) — of CS genes in predisposing asymptomatic COVID-19 in elderly individuals, known to be more susceptible to severe disease.
Methods
Exploiting exome sequencing data and 56 CS genes, we performed a gene-based collapsing test between 164 asymptomatic subjects (aged ≥60 years) and 56,885 European individuals from the Genome Aggregation Database. We replicated this test comparing the same asymptomatic individuals with 147 hospitalized patients with COVID-19.
Results
We found an enrichment of QVs in 3 genes (MASP1, COLEC11, and COLEC10), which belong to the lectin pathway, in the asymptomatic cohort. Analyses of complement activity in serum showed decreased activity of lectin pathway in asymptomatic individuals with QVs. Finally, we found allelic variants associated with asymptomatic COVID-19 phenotype and with a decreased expression of MASP1, COLEC11, and COLEC10 in lung tissue.
Conclusion
This study suggests that genetic rare variants can protect from severe COVID-19 by mitigating the activity of lectin pathway and prothrombin. The genetic data obtained through ES of 786 asymptomatic and 147 hospitalized individuals are publicly available at http://espocovid.ceinge.unina.it/
Keywords: Complement system, COVID-19, SARS-CoV-2, ES, Lectin pathway
Introduction
The spreading of SARS-CoV-2 has emerged as a social and health issue.1 Till February 2022, more than 370 million people have an active infection and more than 5.5 million deaths have been caused by SARS-CoV-2 (https://covid19.who.int/). COVID-19, in which SARS-CoV-2 is the etiological agent, is a respiratory syndrome characterized by a broad spectrum of clinical outcomes, ranging from an asymptomatic infection to a severe respiratory disease that results in lung failure and eventually death.2 Clinical heterogeneity of COVID-19 depends on both host’s and pathogen’s characteristics. Presence of chronic comorbidities, such as diabetes, hypertension, and advanced age are among the main risk factors for severe disease progression,3 rendering elderly individuals more vulnerable to SARS-CoV-2 infection.4
Studies on large cohorts highlighted that genetic variants affecting genes involved in host–pathogen interaction can influence COVID-19 outcome. In particular, genome-wide association studies (GWAS) have identified several loci (3p21.31, 19p13.3, 12q24.13, 21q22.1) associated with severe forms of COVID-19 disease that harbor genes involved in SARS-CoV-2 response.5, 6, 7 Recently, we also showed that common genetic variants predispose to an increased risk of hospitalization by affecting the expression of CCR5, TMPRSS2, and MX1, genes known to play a key role in SARS-CoV-2 pathogenesis.8, 9, 10
Regarding the effect of rare genetic variants on the SARS-CoV-2 pathogenesis, the first large study, including both exome sequencing (ES) and genome sequencing data identified rare deleterious variants affecting interferon pathway genes in individuals with severe COVID-19,11 but subsequent exome-wide association analyses in a very large population found no significant genetic association.11 Our targeted analysis of ES data showed that rare pathogenic variants of common variable immunodeficiency-associated genes are more frequent in severe COVID-19 patients than in nonsevere ones.9 Moreover, severely affected subjects showed a recurrent rare variant, p.His159Tyr (H159Y), in the TNFRSF13C gene, encoding the B cell-activating factor receptor. Additional exome-wide and targeted gene sequencing studies are needed to confirm these studies and establish if rare genetic variation can significantly affect different COVID-19 manifestations and outcomes.
Among the immune system factors modulating SARS-CoV-2 infection, complement system (CS) plays a pivotal role.12 CS is a first-line defense of mammals, consisting of a group of noncellular mediators that stimulate pathogen clearance both directly—by forming a pore on the microbe’s surface—and indirectly—by promoting inflammation and immune system stimulation.12 CS activation is triggered by several viruses, and it is also known to be implicated in pathogenesis and severity of SARS-CoV-2 and other coronaviruses.12 Hyperstimulation of CS by such viruses can indeed promote both cytokine storm—by increasing the concentration of C5a and C3a anaphylatoxins13—and activation of the coagulation cascade,14 2 events directly associated to severe COVID-19 manifestation. Moreover, both CS and coagulation cascade genes are upregulated in the more severe forms of COVID-19 and single-nucleotide variation (SNV) mapping within CS genes are associated with poor prognosis and increased expression of complement factors.14
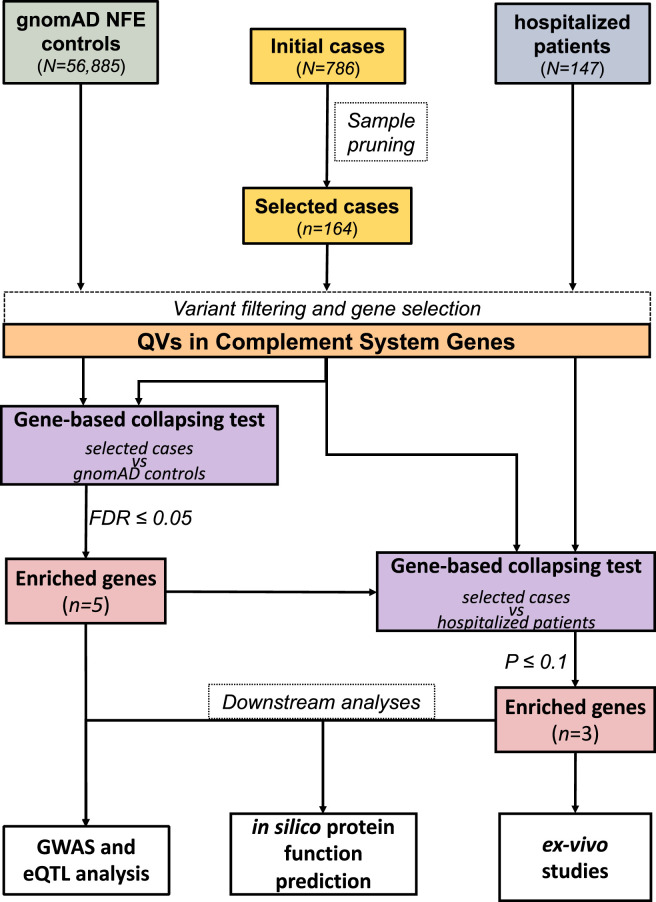
We hypothesized that individuals at high risk of developing a severe COVID-19, such as elderly people, without clinically relevant symptoms of SARS-CoV-2, are more likely to carry deleterious variants in genes involved in CS activation. In this study, we investigated the contribution of rare germline variants in asymptomatic COVID-19 by comparing a group of 164 elderly asymptomatic subjects (age ≥60 years old) from Campania (Italy) with 56,885 non-Finnish European (NFE) individuals from the Genome Aggregation Database (gnomAD) (https://gnomad.broadinstitute.org). We carried out a targeted analysis of ES data and highlighted an enrichment of predicted high impact rare variants—defined as qualifying variants (QVs)—in 3 CS activators (MASP1, COLEC11, and COLEC10) and in 2 CS regulator genes (CD55 and CFHR2) in asymptomatic subjects. Comparable enrichments were obtained for 3 of these genes (MASP1, COLEC11, and COLEC10) when comparing the same asymptomatic individuals against an independent cohort of 147 hospitalized SARS-CoV-2 infected subjects. A schematic representation of the study is depicted in the Figure 1 .
Figure 1.
Schematic representation of the study design. Exome sequencing (ES) data from 786 asymptomatic COVID-19 individuals (initial cases, top center) were pruned on the basis of age, relatedness, genetic ancestry, and heterozygosity (sample pruning); afterward, 164 subjects were retained (selected cases). ES data of 56,885 NFE individuals from gnomAD database (top left) were used as controls. First, we applied variant quality control filters on both gnomAD controls and asymptomatic subjects — coverage harmonization, minor allele depth threshold, and quality by depth (QD) — and selected only coding variants falling in 56 complement system genes (variant filtering and gene selection). Subsequently, we counted the number of QVs (QVs in complement system genes) for each one of the 56 genes. A detailed definition of QVs is provided in Materials and Methods and Supplemental Methods sections. On the basis of the count of QVs, we performed a gene-based collapsing test between the 2 cohorts to assess the enrichment in the asymptomatic individuals; we found a significant enrichment (FDR ≤ 0.05) in 5 genes (MASP1, COLEC11, COLEC10, CFHR, and CD55). We replicated the same test comparing the asymptomatic subjects with 147 COVID-19 hospitalized patients (top right). After applying variant quality filters, we selected QVs falling in the 5 significantly enriched genes, and performed a second gene-based collapsing test between the 2 cohorts; 3 of 5 genes (MASP1, COLEC11, and COLEC10) resulted significantly enriched of QVs in the asymptomatic cohort. Integrated GWAS and eQTL analyses were preformed to test if common sequence variants regulating MASP1, COLEC11, COLEC10, CFHR, and CD55 expression may be associated with asymptomatic phenotype. Finally, we performed further analysis on MASP1, COLEC11, and COLEC10, such as functional and structural protein product predictions and functional ex vivo analyses to test complement and coagulation activity in asymptomatic cases with QVs in MASP1, COLEC11, and COLEC10, as detailly described in the text. FDR, False Discovery Rate; eQTL, expression quantitative loci traits; gnomAD, Genome Aggregation Database; GWAS, genome-wide association study; NEF, non-Finnish European; QV, qualifying variant.
Materials and Methods
Individuals recruitment and DNA extraction
The COVID-19 asymptomatic cohort (N = 786) (Supplemental Table 1) were selected as previously described.15 The cohort included individuals from Campania (Italy) screened in May 2020, for SARS-CoV-2 and resulting positive for SARS-CoV-2 antibodies and without any of COVID-19 symptoms in the 3 previous months, including hospitalization requirement, fever, cough, or at least 2 symptoms among sore throat, headache, diarrhea, vomit, asthenia, muscle pain, joint pain, smell or taste loss, and shortness of breath.16 The COVID-19 hospitalized cohort (N = 147) were selected as previously described.9 Clinical features of 147 COVID-19 hospitalized patients are summarized in Supplemental Table 2. Germline DNA of both asymptomatic individuals and hospitalized patients was extracted using a Maxwell RSC Blood DNA Kit (Promega). Publicly available exome data of 56,885 NFE individuals were retrieved from gnomAD and used as controls. Figure 1 shows a schematic representation of the study design.
Bioinformatic analysis of ES
Paired end sequencing reads were aligned on GRCh37/hg19 reference genome. Subsequently, SNVs and insertions and deletions (INDELs) were detected using HaplotypeCaller (genomic analysis toolkit - GATK - suite) and the resulting variant call format files were annotated using ANNOVAR. Further details on DNA sequencing and downstream bioinformatic pipeline are provided in Supplementary Methods.
Sample pruning and data harmonization
Elderly individuals are more likely to suffer from severe COVID-19;4 therefore, to facilitate the identification of genetic heritable variants of SARS-CoV-2 host response, we first selected only 192 out of 786 SARS-CoV-2 infected and asymptomatic subjects analyzed using ES, aged ≥60 years (n = 192, 24.4%). Subsequently, because sequencing-based cohort comparisons can be inflated using statistical artifacts, we applied sample-level quality control filters to our data sets (Supplemental Table 1). To this end, variant data was converted into PLINK format to perform individual-level quality control; a subset of common (minor allele frequency ≥ 0.05) and linkage disequilibrium pruned variants was used to identify and remove related individuals and PCA outliers. Specifically, we removed the following: (1) samples with heterozygosity rate deviating >3 SDs from the mean (Supplemental Figure 1A), (2) one of each pair of related individuals with PI-HAT > 0.1875 (in between third and second degree related17) (Supplemental Figure 1B), and (3) individuals scoring >6 SDs away from the mean of the top 10 principal components (Supplemental Figure 1C). All filtering steps was performed in PLINK v2.0 (https://www.cog-genomics.org/plink/2.0/), and principal components calculated using FlashPCA v2.0. Upon sample pruning, 21.0% of the initial cohort was retained for further analyses (n = 164).
To harmonize gnomAD and asymptomatic individuals data, we only retained bases at ≥10× coverage in >90% of samples in both cohorts using precalculated data for gnomAD or calculated using samtools depth for the COVID-19 asymptomatic cohorts. Furthermore, in all the data sets, bases with a quality by depth <5 and variants with minor allele depth <8 were further discarded from downstream analyses.
Definition of QVs
The Human Genome Organization Gene Nomenclature Committee website was interrogated to select CS genes (N = 56): 27 were indicated as activators whereas 29 as regulators of the CS (Supplemental Table 3). We defined all missense variants as QVs that (1) had a minor allele frequency ≤1% in NFE population and (2) were predicted as pathogenic by Combined Annotation Dependent Depletion, Mendelian Clinically Applicable Pathogenicity, and MutationTaster algorithms (see Supplementary Methods). The combination of the earlier-described predictor tools has been reported as the most powerful in detecting rare pathogenic variants.18 A variant was also considered as qualifying when rare and annotated as frameshift, start-loss, stop-gain, stop-loss, or splicing, regardless of its pathogenicity scores. Rare variants (minor allele frequency ≤1% in NFE population) affecting CS genes in the asymptomatic data set along with pathogenicity scores are reported in Supplemental Table 4.
Gene-based collapsing test and functional predictions of nonsynonymous variants
For each CS gene, we counted the number of individuals carrying at least 1 QV in each gene of interest as described by Guo et al.19 In brief, for each gene, the total number of heterozygous subjects was equal to the sum of the subjects who harbored at least 1 QV in heterozygosis. Similarly, the total number of homozygous subjects was equal to the sum of the homozygosis subjects for at least 1 QV.
We then compared the number of heterozygous and homozygous subjects for each gene between the asymptomatic and the gnomAD cohorts and the asymptomatic and the hospitalized cohort according to a dominant model. We performed a 2-tailed Fisher exact test to assess the enrichment of QVs burden in each gene between the asymptomatic and gnomAD data sets, and adjusted resulting P values with Benjamini–Hochberg method to account for type I errors. To specifically evaluate the enrichment of QVs in asymptomatic individuals, we performed a 1-tailed Fisher exact test when comparing the latter with the hospitalized cohort.
DUET20 and Screening for Non-Acceptable Polymorphisms 2 (SNAP2)21 web-based tools were used to predict structural and functional effects of nonsynonymous QVs in MASP1, COLEC11, and COLEC10 protein products, respectively.
C3, CH50, and AH50 serum levels and prothrombin time assessment
Serum concentrations of complement C3 were evaluated using Cobas 8000 fully automated platform by Roche Diagnostics. This instrument is based on an immunoturbidimetric principle. The reference ranges include 90 to 180 mg/dL for human C3. Coagulation tests, which included activated partial thromboplastin time, antithrombin, fibrin/fibrinogen degradation products, fibrinogen, prothrombin time, international normalized ratio (INR), prothrombin time activity, and thrombin time, were performed using an ACL TOP (Instrumentation Laboratory) automatic coagulation analyzer. The CH50 serum levels were measured using the commercially available Optilite assay on the fully automated Optilite turbidimetric analyzer (The Binding Site Group, Ltd), according to the manufacturer’s instructions. AH50 serum levels were assessed using enzyme-linked immunosorbent assay kit (Sunlong Biotech Co, LtD), according to the manufacturer’s instructions.
Integrative analysis of GWAS and expression quantitative loci traits data sets
Summary statistics of a GWAS, including 4829 laboratory-confirmed SARS-CoV-2 infections and hospitalized for COVID-19 patients and 11,816 not-hospitalized individuals with European genetic ancestry, were retrieved from the COVID-19 Host Genetics Initiative portal (data set COVID19_HGI_B1_ALL_eur_leave_23andme_20210107.b37, www.COVID19hg.org).22 SNVs with a P-value less or equal than 0.05 mapping to MASP1, COLEC11, COLEC10, CD55, and CFHR2 and expression quantitative loci traits (eQTLs) data from the lung tissue (N = 515) from GTEx database v8 (https://gtexportal.org/home, accessed on June 2021) were obtained.
Results
ES quality control and statistics
ES of 786 asymptomatic COVID-19 individuals produced, on average, 6.5×107 raw reads per sample (Supplemental Figure 2A), of which 0.2% were discarded as sequencing artifacts. The percentage of bases with a phred-scale quality score of ≥20 (Q20) and ≥30 (Q30) was 97.8% and 93.9%, respectively (Supplemental Figure 2B). On average, a 99.6% of target region coverage was achieved (Supplemental Figure 2C), and the mean sequencing depth was 92.3× (Supplemental Figure 2D). Among all the target regions, 94.6% had at least 20× coverage (Supplemental Figure 2E) that suffices for reliable variant calling. The average number of raw SNVs and small INDELs were 84,278 and 11,956, respectively (Supplemental Figure 2F-G).
A total of 147 hospitalized COVID-19 subjects, already included in other studies,9 , 23 were also used for comparison purposes. In brief, the sequencing process returned an average of 4.3×107 raw reads per sample (Supplemental Figure 3A), of which 98.4% was retained after removing sequencing artifacts. The percentage of bases at Q20 and Q30 was 97.7% and 93.8%, respectively (Supplemental Figure 3B). The percentage of the coverage of target regions was 99.63%, the average sequencing depth was 149.1×, and the percentage of those covered by at least 20 reads was 95.1% (Supplemental Figure 3C-D). The number of raw SNVs and INDELs was 189,582 and 31,676 (Supplemental Figure 3F-G), respectively. Full list of the identified genetic variants is publicly available at http://espocovid.ceinge.unina.it/.
Enrichment of QVs in MASP1, COLEC11, and COLEC10 in elderly asymptomatic individuals
To test the hypothesis that asymptomatic SARS-CoV-2 infected elderly individuals can harbor functional protective variants in complement genes, we performed a gene-based collapsing test of QVs mapping within 27 complement activators and 29 complement regulators genes (Supplemental Table 3) among 164 asymptomatic elderly (aged ≥60 years), carefully selected from our case cohort (Supplemental Table 1), and 56,885 gnomAD individuals. Our strategy, depicted in Figure 1, consisted of first exploiting the large gnomAD database used as control data set to yield enough power and identify genes enriched in QVs and then to test the subset of identified enriched genes in a relatively small but carefully selected cohort of 147 hospitalized patients (Supplemental Table 2).
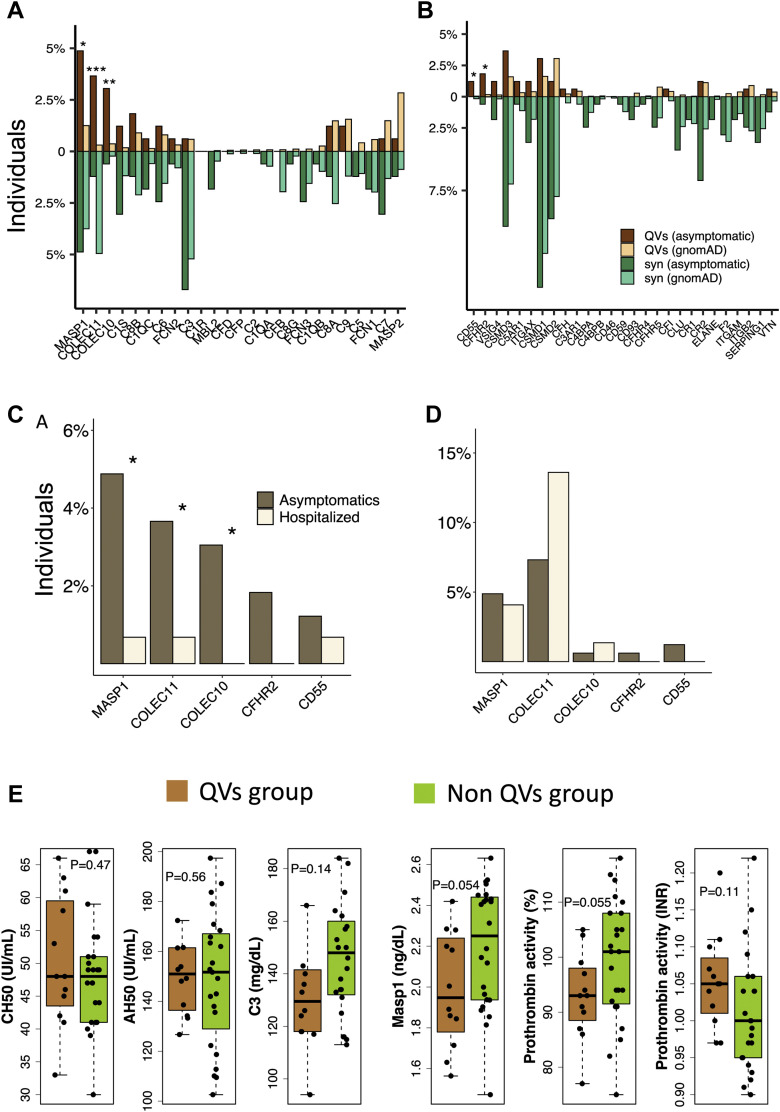
We found an enrichment of QVs in 3 complement activators genes—MASP1 (odds ratio [OR] = 4.05, False Discovery Rate [FDR] = 9.6 × 10–3), COLEC11 (OR = 12.4, FDR = 3.5 × 10–4), and COLEC10 (OR = 8.57, FDR = 4.8 × 10–3) (Figure 2 A)—and in 2 complement regulator genes—CD55 (OR = 27.0, FDR = 0.040) and CFHR2 (OR = 11.4, FDR = 0.040) (Figure 2B)—when comparing COVID-19 asymptomatic individuals and gnomAD controls. Table 1 includes all QVs in these 5 genes in the asymptomatic data set. No significant enrichment of synonymous variants (mostly benign) was found in complement genes (Figure 2A and B), suggesting that no underlying bias influenced the test statistics and that the cohorts were relatively well-harmonized.
Figure 2.
Percentage of QVs in complement genes and C3 levels and INR of asymptomatic subjects harboring QVs in significant genes. Percentage-expressed frequency of QVs (dark and light brown) and synonymous rare variants (minor allele frequency ≤ 0.01) (syn, dark and light green) of the asymptomatic and gnomAD cohorts in complement activator (A) and regulator (B) genes. Stars above the bars denote significantly QVs-enriched genes (∗ indicates FDR < 0.05; ∗∗ indicates FDR < 0.005; ∗∗∗ indicates FDR < 0.0005); percentage-expressed frequency of QVs (C) and synonymous rare variants (D) in the asymptomatic and hospitalized cohorts (∗P < .1). E. (from left to right) Box plots showing the distribution of CH50, AH50, C3, and MASP1 serum levels, along with prothrombin time, expressed either as percentage activity or international normalized ratio (INR), in a set of asymptomatic individuals harboring heterozygous QVs in MASP1 (n = 5), COLEC11 (n = 3), and COLEC10 (n = 4) (QVs group, light brown) and without any QV in these genes (n = 24) (non-QVs group, green). gnomAD, Genome Aggregation Database; INR, international normalized ratio; QV, qualifying variant; syn, synonymous.
Table 1.
Qualifying variants in the top 5 significant genes in the asymptomatic cohort
| Variant | Gene | rsID | Variant Effect | MAF (NFE Population)a | CADD Score | MutationTaster | M-CAP score | No of Heterozygotes |
|---|---|---|---|---|---|---|---|---|
| p.(Ser109Cys) | MASP1 | NA | Missense SNV | 0 | 27.2 | Disease | 0.03738 | 1 |
| p.(Arg677Cys) | MASP1 | rs368168610 | Missense SNV | 0.00003434 | 29 | Disease | 0.231827 | 1 |
| p.(Tyr244Cys) | MASP1 | rs28945071 | Missense SNV | 0.0005 | 27.4 | Disease | 0.138472 | 1 |
| p.(Val666Leu) | MASP1 | NA | Missense SNV | 0.000001804 | 33 | Disease | 0.172555 | 2 |
| p.(Lys591Alafs∗14) | MASP1 | rs533160857 | Frameshift deletion | 0.00013334 | 39 | NA | NA | 3 |
| p.(Ser174Leu) | COLEC11 | rs140226372 | Missense SNV | 0 | 32 | Disease | 0.070292 | 2 |
| p.(Glu34Gly) | COLEC11 | NA | Missense SNV | 0 | 25.3 | Disease | 0.620558 | 4 |
| p.(Asp81Gly) | COLEC10 | rs770053333 | Missense SNV | 0.000001796 | 17.21 | Disease | 0.05912 | 5 |
| p.(Arg130∗) | CFHR2 | rs41313888 | Stop gain | 0.000575 | 34 | Disease | NA | 1 |
| p.(Thr67Asnfs∗3) | CFHR2 | rs779257906 | Frameshift insertion | 0.000008975 | NA | NA | NA | 1 |
| p.(Pro166Ser) | CD55 | rs924315999 | Missense SNV | 0.000002245 | 24.9 | Disease | 0.201836 | 1 |
| p.(Cys225Arg) | CD55 | NA | Missense SNV | 0 | 23.5 | Disease | 0.645858 | 1 |
CADD, Combined Annotation Dependent Depletion; ExAC, Exome Aggregation Consortium; gnomAD, Genome Aggregation Database; MAF, minor allele frequency; M-CAP, Mendelian Clinically Applicable Pathogenicity; NA, not applicable; NFE, non-Finnish European; SNV, single-nucleotide variation.
Mean of MAF frequency in NFE population according to gnomAD, ExAC, and 1000 Genomes projects.
To confirm our finding, we next performed a gene-based collapsing test on the top 5 significantly enriched genes in the asymptomatic cohort against 147 hospitalized COVID-19 patients, who we refer to as the COVID-19 hospitalized cohort. The mean age (hospitalized patients = 61.4, asymptomatic subjects = 69.4) and sex proportion (male = 59% and 50%, female = 41% and 50%, respectively in hospitalized and asymptomatic cohorts) were comparable between the 2 groups. We confirmed an enrichment of QVs in MASP1 (P = .027), COLEC11 (P = .080), and COLEC10 (P = .040) in the asymptomatic cases (Figure 2C), whereas no significant difference in the tally of synonymous variants was observed (Figure 2D).
Functional and structural prediction of QVs
The 3 genes (MASP1, COLEC11, and COLEC10) found significantly enriched in QVs in asymptomatic individuals belong to lectin pathway of complement and are known to be biologically and functionally connected to each other.24 , 25
To assess the effect of MASP1, COLEC11, and COLEC10 variants on their protein products, we focused on the QVs of these genes in asymptomatic subjects (Supplemental Figure 4, Table 2 ). QVs in MASP1 were found in 8 asymptomatic subjects: 3 individuals were heterozygous for the frameshift deletion p.(Lys591Alafs∗14) in the catalytic domain predicted as loss-of-function by gnomAD,26 whereas 5 individuals hosted 4 different heterozygous missense variants in both the catalytic and the interaction domains (Supplemental Figure 4A), and 6 and 5 subjects were heterozygous for missense variants in COLEC11 and COLEC10, respectively (Supplemental Figure 4B and C). Prediction of the protein-level effect of QVs indicated that all the missense variants of MASP1 were likely to hamper both protein function and structure. The missense variant S248L, falling within the lectin domain of COLEC11 and found in 2 individuals, was predicted to have an effect on the function of the gene product, but was the only one predicted as stabilizing. We were not able to establish a structural effect for QVs in COLEC10 and the interaction domain of COLEC11, although all of them were predicted to have an effect at the protein level (Table 2).
Table 2.
Structural and functional prediction of missense QVs in MASP1, COLEC11, and COLEC10 in the asymptomatic cohort
| Gene | Amino Acid Change | No of Heterozygotes | Protein Domain (Function) | DUET ΔΔG (Kcal/mol) | SNAP2 Score |
|---|---|---|---|---|---|
| MASP1 | p.(Ser109Cys)a,b | 1 | CUB1 (interaction) | –0.337 | 82 |
| MASP1 | p.(Arg677Cys)a,b | 1 | Serine protease (catalytic) | –1.106 | 37 |
| MASP1 | p.(Tyr244Cys)a,b | 1 | CUB2 (interaction) | –0.058 | 88 |
| MASP1 | p.(Val666Leu)a | 2 | Serine protease (catalytic) | –0.233 | 63 |
| COLEC11 | p.(Ser174Leu) | 2 | Lectin (carbohydrate recognition) | 0.228 | 30 |
| COLEC11 | p.(Glu108Gly) | 4 | Collagen-like (interaction) | NA | 29 |
| COLEC10 | p.(Asp81Gly) | 5 | Collagen-like (interaction) | NA | 14 |
QV, qualifying variant; NA, not applicable.
Variants affecting MASP1 isoform.
Variants affecting MASP3 isoform.
To evaluate if genetic alterations of MASP1, COLEC11, and COLEC10 may influence the complement activation, we measured the C3, CH50, and AH50 levels in the blood of 2 groups of asymptomatic subjects who either harbored a QV in 1 of these genes (MASP1, n = 5; COLEC11, n = 3; COLEC10, n = 4) or not (n = 24), hereby referred as the QVs group and non-QVs group, respectively. We observed no significant difference in CH50 (P = .47) and AH50 (P = .56) levels (Figure 2E), suggesting no alterations of classical and alternative complement pathways. Only a trend toward the association for lower C3 levels in non-QVs group was observed (Figure 2E, P = .14). We thus tested the MASP1 serum levels to verify a potential alteration of lectin pathway and found that QVs group had lower levels of MASP1 (Figure 2E, P = .054). We then sought to see if these variants could affect the coagulation processes by testing the levels and prothrombin time expressed as percentage activity and INR. These assays revealed a decreased prothrombin activity in the QVs group (Figure 2E, percentage prothrombin activity: P = .055 and INR: P = .11).
Common alleles are associated with asymptomatic or paucisymptomatic phenotype and reduced expression of lectin pathway genes
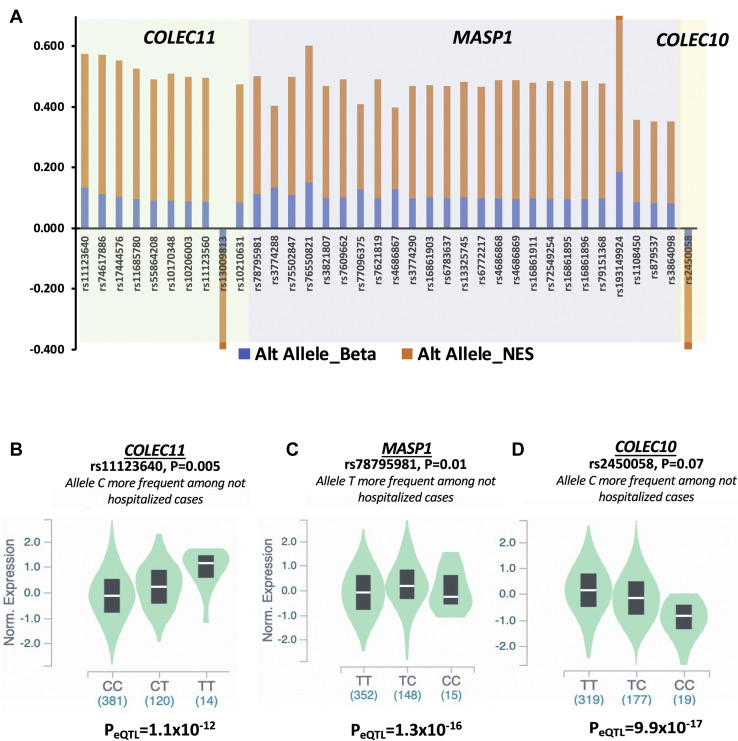
On the basis of the hypothesis that an attenuated reaction of CS may protect against the more severe forms of SARS-CoV-2 infection, we verified if common variants associated with asymptomatic individuals are also correlated with reduced expression of QVs-enriched complement genes. We integrated the summary statistics of a large GWAS data set including 4829 hospitalized cases and 11,816 not-hospitalized cases with eQTLs of lung tissues (see Materials and Methods). The phenotypes definition used in the GWAS were comparable to those adopted in our study; the not-hospitalized cases can be considered with good confidence as asymptomatic or paucisymptomatic SARS-CoV-2 infected individuals. We thus applied an integrative analysis strategy selecting SNVs that were (1) nominally associated (P ≤ .05) with not-hospitalized cases; (2) located within and ±500 kilobases surrounding the genes MASP1, COLEC11, COLEC10, CD55, and CFHR2; and (3) eQTLs for same genes in the lung tissue. Although no significant eQTLs were found for CD55 and CFHR2 (data not shown), we found a total of 35 significant eQTLs for MASP1 and COLEC11 (Supplemental Table 5). We also found 1 eQTL for COLEC10, although slightly above the nominal significance threshold (P = .079) (Supplemental Table 5). In line with our hypothesis, all alleles of selected SNVs with a protective effect against the severe COVID-19 correlated with lower expression of their respective target gene (ie, same direction of association for the genetic association and normalized effect size for eQTL results) (Figure 3 A and Supplemental Table 5). In Figure 3, we show, as representative example, the expression of COLEC11 (Figure 3B), MASP1 (Figure 3C), and COLEC10 ( Figure 3D) in the lung tissue, stratified according to the most significant SNVs. These results further support the hypothesis that genetic variation that mitigates an excessive action of the lectin pathway genes can protect from developing a severe form of COVID-19.
Figure 3.
Common variants are associated with not-hospitalized COVID-19 phenotype and low expression of COLEC11, MASP1, and COLEC10 in the lung tissue. A. The SNVs associated with favorable outcome of COVID-19 and eQTLs for COLEC11, MASP1, and COLEC10 are plotted and ordered according to the P values for each target gene. The effect size (Beta) and NES of altered alleles from the genetic association and eQTL analyses are reported for each target gene. The violin plots show the median expressions of the target genes (B) COLEC11, (C) MASP1, and (D) COLEC10 stratified per genotypes of the most significant SNVs associated with not hospitalized phenotype. eQTL, expression quantitative loci traits; NES, normalized effect size.
Discussion
On the basis of the assumption that genetic variation causing a reduced functionality of CS can contribute to asymptomatic SARS-CoV-2 infection in elderly individuals, known to be at high-risk for developing COVID-19 with rapidly progressive clinical deterioration, we performed a gene-based collapsing test comparing exome data of 164 asymptomatic ≥60 years old subjects with 56,885 NFE individuals from gnomAD database. We found an enrichment of rare germline QVs in 5 CS genes (MASP1, COLEC11, COLEC10, CD55, and CFHR2). We replicated the enrichment in 3 of these genes (MASP1, COLEC11, and COLEC10) when testing the same data set against an independent cohort of 147 hospitalized COVID-19 patients.
Our results are supported by several lines of evidence. First, we used a cohort of individuals with specific features that emphasize the genetic effect on the asymptomatic phenotype for the following reasons: (1) all subjects got infected in a small window of time around March and April 2020, at the beginning of the virus spread in Italy and thus likely with the same viral strain and (2) our cohort included extreme phenotype samples: individuals who developed no severe clinical manifestation even if infected with SARS-CoV-2 at advanced age (average age = 69.4 and 43%, 71/164 with age 70 years or above). Second, the identified mutated genes are functionally closely related to each other, because their respective proteins are implicated in the lectin pathway of the CS. In this pathway, proteins like CL-L1 and CL-K1 (ie, the protein product of COLEC10 and COLEC11, respectively)—known as collectins—recognize specific carbohydrate patterns on pathogen surface and, upon homo- and hetero-multimerization, interact with downstream serine-proteases,—such as MASP1 and MASP3 (ie, the main isoforms produced by MASP1)—to converge toward the activation of the common pathway of CS via cleavage of the C3 fragments.27 The functional convergence of MASP1, COLEC11, and COLEC10 is further supported by the fact that germline variants affecting these genes are associated with 4 autosomal recessive syndromes with overlapping phenotypes—collectively known as the 3MC syndrome.25 Third, the QVs falling in these 3 genes were predicted to be deleterious for the protein functions and recur in different cases. In MASP1, most QVs occurred in the serine-protease domain (6 out of 8), putatively affecting the catalytic function; the other QVs (2 out of 8) fell in interaction domains, probably hampering the ability of interaction of Masp1/3 proteins with collectins. Of note, the same frameshift deletion p.(Lys591Alafs∗14) recurred in 3 cases. In COLEC11, we found 6 QVs: the p.(Glu108Gly), observed in 4 cases, occurred in the collagen-like domain—which allows multimerization—whereas the p.(Ser248Leu) variant, found in other 2 different cases, is located in the carbohydrate recognition domain. In COLEC10, the aminoacidic substitution, p.(Asp81Gly), recurred in 5 cases and is located in the collagen-like domain.
Moreover, we found no statistically significant difference in CH50 and AH50 levels between asymptomatic subjects who either harbored QV (QVs group) or not (non-QVs group) in MASP1, COLEC11, and COLEC10. These results suggested that the classical and alternative complement pathways—represented by CH50 and AH50 markers, respectively—were not affected by these variants. Thus, to assess if the lectin pathway was altered in subjects who harbor QVs in MASP1, COLEC11, and COLEC10, we assessed the serum levels of MASP1 and found a statistically significant reduction of MASP1 levels in the QVs group compared with non-QVs group. We performed latter analysis for the following reasons: (1) because both COLEC10 and COLEC11 interact with Masp1-3 in plasma,28 , 29 a reduction in Masp1 is reflective of an impairment of the lectin pathway owing to the genetic alterations affecting these 3 proteins, (2) Masp1 is crucial for lectin pathway activation,30 and (3) Masp1 binds and activates the other mannan-binding lectin-associated serine protease, Masp2,31 which is known to be required for a proper activation of the lectin pathway.32 Thus, these results collectively support the evidence that the found genetic rare variants in MASP1, COLEC11, and COLEC10 can mitigate the overall activity of lectin pathway. We additionally found that these variants affect the coagulation processes; indeed, QVs group showed an overall lower prothrombin activity than the non-QVs group. Although above the significance threshold, we found reduced serum C3 levels in the QVs group. However, on the basis of the results obtained on other CS components (Masp1, CH50, and AH50 serum levels), we ruled out that this phenomenon could be due to a hyperactivation of one of the CS pathways. Interestingly, this trend is in line with a study conducted on White population which correlated the serum levels of CS components with age, sex, and other CS factors, in which a positive correlation between Masp1 and C3 levels was reported but above the significance threshold.33 Thus, additional functional studies are needed to investigate the possible causes of Masp1 and C3 correlation in serum.
Finally, our integrative genomic analysis of GWAS and eQTL data showed that common alleles that are more frequent in asymptomatic and paucisymptomatic phenotypes also correlated with a decreased expression of MASP1, COLEC11, and COLEC10 genes in the lung tissues. It is worth to note that, in agreement with the results from gene-set collapsing test, only 3 genes, MASP1, COLEC11, and COLEC10, and not CD55 and CFHR2, resulted significant in this different approach. One possible limitation of our analysis is that the GWAS SNVs mapping to the selected genes did not yield genome-wide significance levels. However, this could be just the result of the relatively low statistical power to detect such a small effect size of common variants compared with effect of rare variants on the phenotype.
Taken together, these results suggest that rare genetic variants may attenuate lectin pathway activation upon SARS-CoV-2 infection, predisposing infected individuals—especially those who present risk factors, such as advanced age—to an asymptomatic form of COVID-19.
The involvement of lectin pathway in the SARS-CoV-2 response is supported by growing evidence.34 It is well-known that the role of the lectin pathway in the defense against enveloped viruses, such as influenza A virus and SARS-CoV35 , 36—the latter evolutionarily related to SARS-CoV-2.37 Moreover, the interaction between nucleocapsid (N) protein of SARS-CoV-2 and proteins belonging to the MASP family can enhance aberrant lectin pathway hyperactivation (Gao T, Hu M, Zhang X, et al. Highly pathogenic coronavirus N protein aggravates lung injury by MASP2-mediated complement over-activation. 2020). In addition, a study that integrated the effects of both common and rare germline variants in COVID-19 reported a correlation between MASP1 rare variants and a mild phenotype,38 whereas gene candidate-driven study found SNVs that was associated with severe COVID-19 risk and high expression of COLEC11 and CD55 genes.14
In conclusion, consistent with previous published data, our results suggest an involvement of the lectin pathway in COVID-19 pathogenesis. We showed that deleterious germline QVs in MASP1, COLEC11, and COLEC10 are associated with an asymptomatic phenotype of the disease and reduced activity of lectin pathway and prothrombin. Our results provide further insights on COVID-19 clinical heterogeneity and contribute to clarify some pathogenicity mechanisms of SARS-CoV-2 infection.
Data Availability
The genetic data obtained through exome sequencing of the 786 asymptomatic subjects and 147 hospitalized patients are publicly available at http://espocovid.ceinge.unina.it/.
Acknowledgments
This research was funded by the project “CEINGE Task-Force Covid-19,” grant number D64I200003800, by Regione Campania for the fight against Covid-19 (DGR n. 140 del 17 Marzo 2020). We would also like to acknowledge the support of Macrogen Europe in carrying out the library preparation and sequencing of DNA samples.
Author Information
Conceptualization, Supervision, and Project Administration: M.C., M.Z., A.I.
Investigation, Methodology, and Formal Analysis: M.C., G.D., V.A.L., F.B., U.E.
Resources:G.F. (Giulia Frisso), B.E.R., P.A., G.M.C., G.S., I.G., M.D.M, G.F. (Giuseppe Fiorentino), C.P., C.B., R.S., F.A., B.P.
Data curation: G.D., V.A.L., U.E., A.B., G.P., P.d.A., S.C., B.E.R., R.R., I.A., V.F.
Writing-original draft: M.C., G.D., F.B. All authors have read and agreed to the published version of the manuscript.
Ethics Declaration
This study was reviewed and accepted by the University of Naples Federico II Research Ethics Committee, protocol number 180/20. Informed consent was obtained from all the subjects involved.
Conflict of Interest
The authors declare no conflicts of interest.
Footnotes
Additional Information
The online version of this article (https://doi.org/10.1016/j.gim.2022.04.007) contains supplementary material, which is available to authorized users.
Supplementary Material
References
- 1.Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. Published correction appears in Lancet. 2020;395(10223):496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou F., Yu T., Du R., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. Published correction appears in Lancet. 2020;395(10229):1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C., Chen X., Cai Y., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. Published correction appears in JAMA Intern Med. 2020;180(7):1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Severe Covid-19 GWAS Group. Ellinghaus D., Degenhardt F., et al. Genomewide Association Study of severe covid-19 with respiratory failure. N Engl J Med. 2020;383(16):1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pairo-Castineira E., Clohisey S., Klaric L., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591(7848):92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 7.Shelton J.F., Shastri A.J., Ye C., et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat Genet. 2021;53(6):801–808. doi: 10.1038/s41588-021-00854-7. [DOI] [PubMed] [Google Scholar]
- 8.Andolfo I., Russo R., Lasorsa V.A., et al. Common variants at 21q22.3 locus influence MX1 and TMPRSS2 gene expression and susceptibility to severe COVID-19. iScience. 2021;24(4):102322. doi: 10.1016/j.isci.2021.102322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantalupo S., Lasorsa V.A., Russo R., et al. Regulatory noncoding and predicted pathogenic coding variants of CCR5 predispose to severe COVID-19. Int J Mol Sci. 2021;22(10):5372. doi: 10.3390/ijms22105372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russo R., Andolfo I., Lasorsa V.A., Iolascon A., Capasso M. Genetic analysis of the coronavirus SARS-CoV-2 host protease TMPRSS2 in different populations. Front Genet. 2020;11:872. doi: 10.3389/fgene.2020.00872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q., Bastard P., COVID Human Genetic Effort. Cobat A., Casanova J.L. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature. 2022;603(7902):587–598. doi: 10.1038/s41586-022-04447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Java A., Apicelli A.J., Liszewski M.K., et al. The complement system in COVID-19: friend and foe? JCI Insight. 2020;5(15):e140711. doi: 10.1172/jci.insight.140711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahmudpour M., Roozbeh J., Keshavarz M., Farrokhi S., Nabipour I. Vol. 133. Cytokine; 2020. p. 155151. (COVID-19 cytokine storm: the anger of inflammation). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramlall V., Thangaraj P.M., Meydan C., et al. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat Med. 2020;26(10):1609–1615. doi: 10.1038/s41591-020-1021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerino P., Coppola A., Volzone P., et al. Seroprevalence of SARS-CoV-2-specific antibodies in the town of Ariano Irpino (Avellino, Campania, Italy): a population-based study. Future Sci OA. 2021;7(4):FSO673. doi: 10.2144/fsoa-2020-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavezzo E., Franchin E., Ciavarella C., et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo. Nature. 2020;584(7821):425–429. doi: 10.1038/s41586-020-2488-1. Published correction appears in Nature. 2021;590(7844):E11. [DOI] [PubMed] [Google Scholar]
- 17.Anderson C.A., Pettersson F.H., Clarke G.M., Cardon L.R., Morris A.P., Zondervan K.T. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5(9):1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh R., Oak N., Plon S.E. Evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. Genome Biol. 2017;18(1):225. doi: 10.1186/s13059-017-1353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo M.H., Plummer L., Chan Y.M., Hirschhorn J.N., Lippincott M.F. Burden testing of rare variants identified through exome sequencing via publicly available control data. Am J Hum Genet. 2018;103(4):522–534. doi: 10.1016/j.ajhg.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pires D.E.V., Ascher D.B., Blundell T.L. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014;42(Web Server issue):W314–W319. doi: 10.1093/nar/gku411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bromberg Y., Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35(11):3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.COVID-19 Host Genetics Initiative The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet. 2020;28(6):715–718. doi: 10.1038/s41431-020-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russo R., Andolfo I., Lasorsa V.A., et al. The TNFRSF13C H159Y variant is associated with severe COVID-19: a retrospective study of 500 patients from Southern Italy. Genes (Basel) 2021;12(6):881. doi: 10.3390/genes12060881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasta G.R., Quesenberry M., Ahmed H., O’Leary N. C-type lectins and galectins mediate innate and adaptive immune functions: their roles in the complement activation pathway. Dev Comp Immunol. 1999;23(4-5):401–420. doi: 10.1016/s0145-305x(99)00020-8. [DOI] [PubMed] [Google Scholar]
- 25.Rooryck C., Diaz-Font A., Osborn D.P.S., et al. Mutations in lectin complement pathway genes COLEC11 and MASP1 cause 3MC syndrome. Nat Genet. 2011;43(3):197–203. doi: 10.1038/ng.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacArthur D.G., Balasubramanian S., Frankish A., et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335(6070):823–828. doi: 10.1126/science.1215040. Published correction appears in Science. 2012;336(6079):296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsushita M., Fujita T. Activation of the classical complement pathway by mannose-binding protein in association with a novel C1s-like serine protease. J Exp Med. 1992;176(6):1497–1502. doi: 10.1084/jem.176.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen S., Selman L., Palaniyar N., et al. Collectin 11 (CL-11, CL-K1) is a MASP-1/3-associated plasma collectin with microbial-binding activity. J Immunol. 2010;185(10):6096–6104. doi: 10.4049/jimmunol.1002185. [DOI] [PubMed] [Google Scholar]
- 29.Henriksen M.L., Brandt J., Andrieu J.P., et al. Heteromeric complexes of native collectin kidney 1 and collectin liver 1 are found in the circulation with MASPs and activate the complement system. J Immunol. 2013;191(12):6117–6127. doi: 10.4049/jimmunol.1302121. [DOI] [PubMed] [Google Scholar]
- 30.Degn S.E., Jensen L., Hansen A.G., et al. Mannan-binding lectin-associated serine protease (MASP)-1 is crucial for lectin pathway activation in human serum, whereas neither MASP-1 nor MASP-3 is required for alternative pathway function. J Immunol. 2012;189(8):3957–3969. doi: 10.4049/jimmunol.1201736. [DOI] [PubMed] [Google Scholar]
- 31.Héja D., Kocsis A., Dobó J., et al. Revised mechanism of complement lectin-pathway activation revealing the role of serine protease MASP-1 as the exclusive activator of MASP-2. Proc Natl Acad Sci U S A. 2012;109(26):10498–10503. doi: 10.1073/pnas.1202588109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiel S., Vorup-Jensen T., Stover C.M., et al. A second serine protease associated with mannan-binding lectin that activates complement. Nature. 1997;386(6624):506–510. doi: 10.1038/386506a0. [DOI] [PubMed] [Google Scholar]
- 33.Gaya da Costa M., Poppelaars F., van Kooten C., et al. Age and sex-associated changes of complement activity and complement levels in a healthy Caucasian population. Front Immunol. 2018;9:2664. doi: 10.3389/fimmu.2018.02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polycarpou A., Howard M., Farrar C.A., et al. Rationale for targeting complement in COVID-19. EMBO Mol Med. 2020;12(8) doi: 10.15252/emmm.202012642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gralinski L.E., Sheahan T.P., Morrison T.E., et al. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio. 2018;9(5) doi: 10.1128/mBio.01753-18. e01753-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Brien K.B., Morrison T.E., Dundore D.Y., Heise M.T., Schultz-Cherry S. A protective role for complement C3 protein during pandemic 2009 H1N1 and H5N1 influenza A virus infection. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0017377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X., Zai J., Zhao Q., et al. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J Med Virol. 2020;92(6):602–611. doi: 10.1002/jmv.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picchiotti N., Benetti E., Fallerini C., et al. Post-Mendelian genetic model in COVID-19. Cardiol Cardiovasc Med. 2021;5(6):673–694. doi: 10.26502/fccm.92920232. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genetic data obtained through exome sequencing of the 786 asymptomatic subjects and 147 hospitalized patients are publicly available at http://espocovid.ceinge.unina.it/.