Abstract
The transition to agriculture occurred relatively late in Eastern Europe, leading researchers to debate whether it was a gradual, interactive process or a colonisation event. In the forest and forest-steppe regions of Ukraine, farming appeared during the fifth millennium BCE, associated with the Cucuteni-Trypillia cultural complex (CTCC, ~ 5000–3000 BCE). Across Europe, the Neolithisation process was highly variable across space and over time. Here, we investigate the population dynamics of early agriculturalists from the eastern forest-steppe region based on the analyses of 20 ancient genomes from the site of Verteba Cave (3935–825 cal BCE). Results reveal that the CTCC individuals’ ancestry is related to both western hunter-gatherers and Near Eastern farmers, has no local ancestry associated with Ukrainian Neolithic hunter-gatherers and has steppe ancestry. An Early Bronze Age individual has an ancestry profile related to the Yamnaya expansions but with 20% of ancestry related to the other Trypillian individuals, which suggests admixture between the Trypillians and the incoming populations carrying steppe-related ancestry. A Late Bronze Age individual dated to 980–825 cal BCE has a genetic profile indicating affinity to Beaker-related populations, detected close to 1000 years after the end of the Bell Beaker phenomenon during the third millennium BCE.
Subject terms: Anthropology, Archaeology, Population genetics
Introduction
The Neolithisation process in Europe resulted in dramatic technological and cultural shifts, which included novel subsistence practices1. There are two major groups of models that explain the Neolithisation process: demic diffusion models describe Neolithisation as a colonisation process by farmers which is propelled by exponential population growth characteristic of the Neolithic, whereas acculturation models outline the process as one in which at least some of the transition entails indigenous hunting-foraging groups that adopt farming following periods of variable length during which they interact with neighbouring exogenous farmers2. Across most of Europe, the Neolithic transition was genetically defined by a profound population replacement, consistent with the demic diffusion of peoples from Anatolia3–5. The Anatolian farmers reached the Balkans and other regions of Southeast Europe in the seventh millennium BCE6,7 and subsequently spread further via the Mediterranean and later through the Danube, substantially replacing indigenous Mesolithic European populations8,9.
In contrast to Central Europe10,11, areas of Eastern Europe including Ukraine, Moldova, Western Russia, and Romania did not adopt agriculture until the Late Neolithic (~ 4500 BCE), although various sedentary and semi-sedentary hunter-gatherers Mesolithic groups in these regions began using pottery as early as 8500 BCE12,13.
The Cucuteni-Trypillia cultural complex (CTCC) is a grouping of several interrelated Middle Neolithic/Eneolithic archaeological cultures in parts of Ukraine, Moldova, and Romania14,15. This complex stretches from the Transylvanian Alps to the Dnieper River and is named for the type-sites of Cucuteni in Iași County, Romania and Trypillia (also known as Tripolye, in Russian) in Kyiv oblast, Ukraine. The Cucuteni and Trypillia cultures have common roots in the Precucuteni culture; the earliest CTCC sites are found in the piedmont of the Carpathian Mountains and the earliest radiocarbon dates (from the Precucuteni 2 period) date to around 4800 BCE16,17. The CTCC originated from the interaction of several Danubian Neolithic groups, with evidence for similarities in house construction, ceramic styles, and lithic artifact production10,16,18,19.
Following the origin of this cultural complex in the Carpathian piedmont, the CTCC eventually occupied a territory covering much of the modern territories of Ukraine, Moldova, and Romania. The first diagnostically Early Trypillia (Trypillia A) sites diverged from the Precucuteni culture ~ 4500 BCE in the Dniester River valley. Later population movements, occurring from the middle period (Trypillia BI) onward, saw the Trypillia culture expand to Volhynia in the west and the Dnieper River in the east. This territorial expansion is believed to have resulted primarily from demographic increases associated with a successful agropastoral subsistence strategy, and the search for new arable land for cultivation18. However, some population growth may have been the product of Trypillian populations incorporating indigenous hunting and gathering (HG) groups, such as members of the Bug-Dneister culture. Another mode of population increase could have been the acculturation of refugees following the collapse of the Neolithic in Romania, Hungary, and Bulgaria. During the middle-to-late periods of the Trypillia culture (Trypillia BII to CI; 4100–3400 BCE), some CTCC groups established extremely large settlements in Central Ukraine, often referred to as “giant-settlements'' or “megasites,” which attained sizes of 100–320 ha20. Rapid demographic growth within the CTCC around the turn of the fourth millennium BCE necessitated the exploitation of new territories, precipitating migrations to previously peripheral areas21.
Hypotheses for the rise of the megasites in particular are varied; it has been suggested that they may have been a defensive response to threats posed by steppe pastoralists or competing sub-groups within the CTCC22,23, or that they simply represent ephemeral episodes of population agglomeration due to large-scale migration from the Dniester region24.
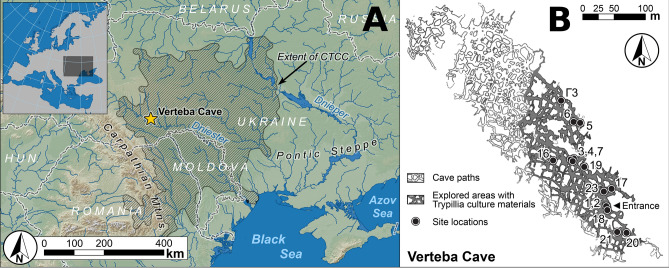
Even though Trypillian populations established a high density of settlements in Western and Central Ukraine25, very few burials have been located. Only a handful of cemeteries dating to the Late Trypillia period were excavated during the 1960s and 1970s, such as Chapaievka in Ukraine26 and Vykhvatyntsi in Moldova27. While these sites give some glimpse into Trypillian mortuary behavior, they are limited in their temporal scope and have not been subjected to modern laboratory analyses. To better understand the origins, connections, and diversity of the CTCC, we collected human remains from three chambers at the site of Verteba Cave (VC) in Ternopil oblast, Ukraine, one of the few sites that contain human remains associated with the CTCC (Fig. 1). Accelerator mass spectrometer (AMS) 14C dates of human and faunal remains place the Trypillian occupation of VC to between 3950 and 3520 cal BCE (2σ)28,29. On the basis of the ceramic assemblages present in the cave and a sample of lower-resolution liquid scintillation 14C dates, it is probable that occupation continued for some time into the Late Trypillia (CII) period and Early Bronze Age transition30. More recently, AMS radiocarbon dating has also identified deposits at different locations throughout the cave dating to the Mesolithic (7950–7490 cal BCE [2σ]), Bronze Age, Iron Age and Medieval period31. Skeletal assemblages were taken from three separate chambers (Site 7, Site 17, and Site 20) of the cave (Table S2, Fig. 1). Each of these chambers contain CTCC material culture; however, burials in the cave are secondary in nature and disturbance through human activity during antiquity and bioturbation complicate reconstruction of the cave’s use and chronology. Most individuals in this study come from Site 7, which has been extensively documented through ceramic analysis and radiocarbon dating31,32, with peak occupation dated to periods CI and early CII of the Trypillia periodization (~ 3900–3350 BCE). Interpretations regarding the use of the cave are varied, including its use as a temporary shelter, ritual site or mortuary location29,33,34. There is additional evidence to support the idea that the burials in the cave, which are largely commingled and secondary in nature, are representative of victims of warfare or sacrifice35.
Figure 1.
(A) Location of Verteba cave in Ternopil Oblast, Ukraine, plotted against the overall distribution of CTCC sites. (B) Map of sites within Verteba cave; individuals included in this publication were found at sites 7, 17 and 20, Adapted from Ledogar et al.31.
The paleogenetics of the Trypillian population is limited to the analysis of uniparental markers (mtDNA) and genome-wide analysis of 8 individuals. Mitochondrial haplogroups typical of ancient Eurasian farming groups (H, HV, T, K, J) have been observed for these individuals scattered throughout the cave30,32,36. Schmidt et al. found within a single chamber evidence for Haplogroup W, which has been observed for steppe populations associated with the Corded Ware and Unetice cultures of the middle Volga region8. Genome-wide analyses of CTCC individuals have shown ancestral components predominately of Early Neolithic farming groups (estimated to be 60–80%), confirming that the early farmers who settled Western and Central Ukraine were largely derived from the same source population as the farmers of Anatolia and Western Europe13,37. The remaining 20 percent of their ancestry is less certain. Mathieson et al. (2018) found that this ancestral component was a mix of western (WHG) and eastern (EHG) hunter-gatherers (HG) found in HG groups inhabiting the region during the Neolithic. Immel et al. recovered genome-wide data from four individuals buried at two separate sites in northern Moldova that date to 3500–3100 cal BCE (during the Late Trypillia period), five centuries younger than the radiocarbon dates from Verteba cave, and found a larger degree of steppe-related ancestry (albeit in varying proportions among the sampled individuals). This observation may be explained by the gradual assimilation of local Mesolithic and Neolithic HG groups into the Trypillian population, at least for groups who settled in Moldova.
The settlement systems of the Trypillia culture interacted with both Central European and the steppe populations. Archaeological evidence for steppe interaction is found in shell-tempered pottery, which are similar to steppe-style wares35,38. Some of these look nearly identical to pottery found at steppe sites, while others combine shell tempering with CTCC decorative motifs35. Symbolic objects influenced by—or directly imported from—steppe communities, such as stone mace heads, are found at some Middle-to-Late Trypillia sites35, and exchanges of pottery are evident as early as Trypillia BII39. There was undoubtedly some degree of interaction between Trypillian populations and the Dnieper-Donets culture, while any synchrony between the Trypillia culture and the following Yamnaya horizon was likely very brief. Regardless, however, some Trypillian populations were likely in permanent contact with steppe populations40. Interestingly, after around 3400 BCE, the Trypillian mega-sites were largely abandoned. The cause of this abandonment has been widely debated, one hypothesis is an increase in conflict due to the westward expansion of steppe populations. Such a hypothesis may find corroboration in the frequent evidence of violent death discovered in Verteba cave23,35.
Here, we recovered genome-wide sequence data from 20 individuals buried in VC, eight of which are directly dated by AMS 14C to the interval of 3790 to 825 cal BCE (Table S1). We use this data to specifically test several questions: (1) is there evidence for admixture with local HGs, as has been suggested by Rascovan et al.29; (2) using our expanded dataset with higher coverage than in Mathieson et al.13, can we clarify the Neolithic ancestral component of the Trypillian population, i.e., can we show that they are more similar to early farmers from Anatolia, the LBK (Linearbandkeramik), or elsewhere; (3) since the CTCC individuals lived in close proximity to steppe populations, is there evidence for genetic admixture with the Yamnaya or earlier steppe populations; and (4) do later Bronze Age populations that settled in the region share genetic affinities with the CTCC group from Verteba Cave?
Results
DNA was extracted from 20 petrous bones. Eight of the samples were directly radiocarbon dated and we determined that six (VERT-035, VERT-106, VERT-031, VERT-100, VERT-104 and VERT-015) date to 3790–3535 cal BCE (2σ; Late Eneolithic), one individual (VERT-113) from Site 7 dates to 1960–1770 cal BCE (Middle Bronze Age; MBA) and one from Site 17 (VERT-114) dates to 980–825 cal BCE (Late Bronze Age; LBA) (Table S2). Endogenous content of the sequenced samples ranged from 59 to 82% and yielded genomic coverages between 0.2X and 2.2X (Table S2). Aligned reads were authenticated using dedicated ancient DNA pipelines (Supplementary methods). All analyzed sequences showed the typical pattern of aDNA: an excess of C>T transition in the 5’ end and G>A transitions in the 3’ end, consistent with the age of the samples; additionally, no signs of contamination were found in the sequences41. Further details of the contamination assessment is provided in Supplementary information. We were able to assign molecular sex to all the individuals, from which 8 are female and 12 are male (Table S2). In all cases the molecular sex is concordant with morphological sex. No familiar relationships have been identified in the analyzed data.
Uniparental markers
The analyzed individuals that are generally thought to come from the Eneolithic Period have maternal haplogroups T2b, H, HV, K1, N1, J1, U5 and T2c (Table S2). The MBA sample shows haplogroup HV, typical from several Neolithic cultures such as the ALPC5 as well as European Bronze Age individuals42,43. The LBA individual shows haplogroup T2, also associated with multiple BA individuals and cultures43–45. These haplogroups are typically found in European Neolithic and Bronze Age populations5,13,43. Male individuals exhibit Y-chromosome haplogroups G, I and C, which have also been previously reported in Neolithic and Bronze Age populations of Europe13. Both the mtDNA and Y-chr haplogroups of all individuals are fully concordant with the previously reported data. (Table S2).
Population genetics
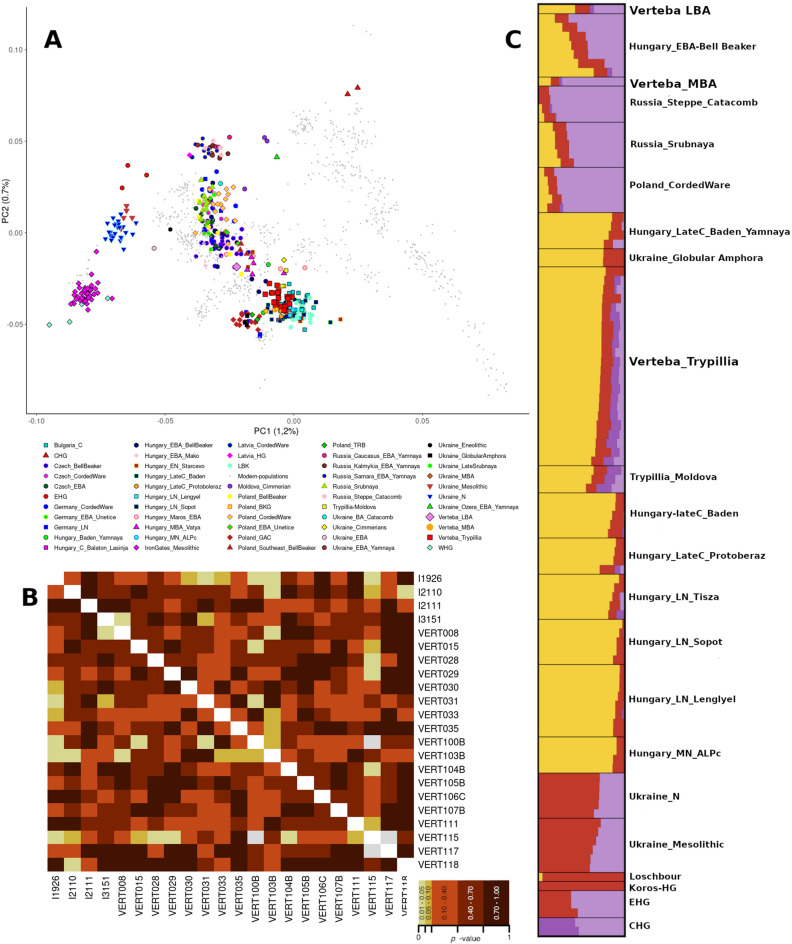
To place VC individuals within present-day and archaeological Eurasian populations, we used a principal component analysis (PCA)46 built with 729 modern individuals from Europe and the Mediterranean Basin3. Together with the 20 Verteba genomes, 478 additional ancient genomes (Table S3) were projected onto the PCA. 18 out of the 20 VC individuals are placed closely to Neolithic and Eneolithic European populations such as LBK, Central European Middle and Late Neolithic samples and the Moldova Trypillian individuals37 (Fig. 2A). The PCA also evidenced the extreme similarity between the 18 newly reported Trypillians and the other four Trypillians from Verteba Cave previously sequenced13, therefore all these 22 individuals were labeled together as Verteba_Trypillia and further analyzed together. The two Bronze Age individuals are clear outliers. Individual VERT-114 falls within the Bell-Beaker diversity and appears to have a position close to the Czech, Hungarian and Polish Bell-Beaker groups. Individual VERT-113 appears close to European Corded-Ware and Srubnaya populations, showing a strong affinity to steppe samples. We have then explored the presence of structure in the Trypillian population (only using the 22 samples from the main cluster) using qpWave. The results have shown the absence of population structure, therefore all samples have been analyzed together, as no individual showed statistically significant pairwise differences to the rest using a threshold of 0.05 (Fig. 2B).
Figure 2.
Population genetics: (A) PCA built with modern European populations in which Neolithic and Bronze Age populations of Eastern Europe have been projected. It is observed that the Verteba_Trypillia individuals are located within the European Neolithic populations genetic diversity while the Verteba_MBA and Verteba_LBA individuals are located close to other Bronze Age individuals suggesting genetic similarity. (B) ADMIXTURE analysis of the most representative populations included in the analysis (K = 4). The different colors represent the
source ancestries of the studied individuals: Yellow represents Anatolia_N related ancestry, Red represents WHG related ancestry and the purple colors represent Steppe related ancestries, each individual is represented by the proportions of these ancestries. (C) Heatmap built with 1 × 1 qpWave results of the Verteba_Trypillia individuals, where no individuals show a sign to be clustered in a different population suggesting that these individuals belong to the same genetic population.
We then explored the genetic diversity of the VC individuals using ADMIXTURE47. The 22 individuals (labeled ‘Verteba_Trypillia’) in the PCA that showed affinity with Eneolithic samples are mostly defined by the ancestral component dominant in Anatolia-Neolithic individuals, which suggests a strong relationship with European Neolithic populations, similar to previous studies13,37. However, these samples also show the presence of EHG, CHG, and WHG components as described in Mathiesson et al.13, with the exception of one individual (I3151), who seems to be absent of any EHG/CHG ancestry. Individual VERT-114 (LBA) shows a predominant Anatolia Neolithic component and a great presence of an EHG component. The MBA individual (VERT-113) exhibits a high degree of similarity with Corded Ware and Yamnaya steppe populations (Fig. 2C, Fig. S1).
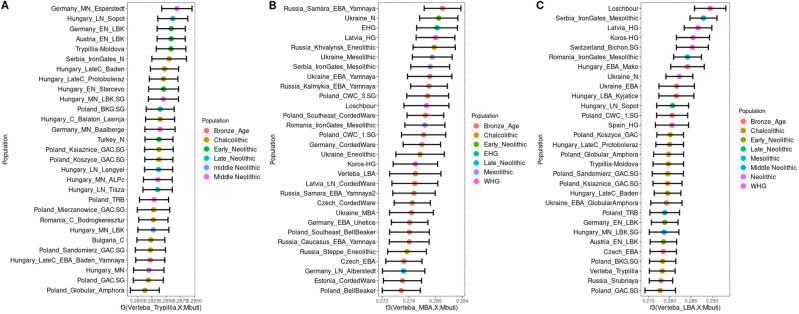
Next, we investigated the genetic affinities of the VC individuals using f-statistics. We used f3-outgroup statistics to quantify the amount of shared genetic drift of Verteba_Trypillia, VERT-114, and VERT-113, tested against other ancient European populations. Overall, the Verteba_Trypillia individuals share more derived SNPs with Neolithic European populations (Fig. 3). Individual VERT-114 shows a high level of derived SNPs with HG populations as well as with Late Neolithic and Bronze Age populations. In turn, individual VERT-113 shares derived SNPs with HG populations and some Steppe-related populations such as Central European Corded-Ware.
Figure 3.
Outgroup-f3 statistics, the samples have been clustered between: Verteba individuals, individual VERT-114 (Verteba_LBA) and individual VERT-113 (Verteba_MBA). We have plotted the 20 populations with the highest values.
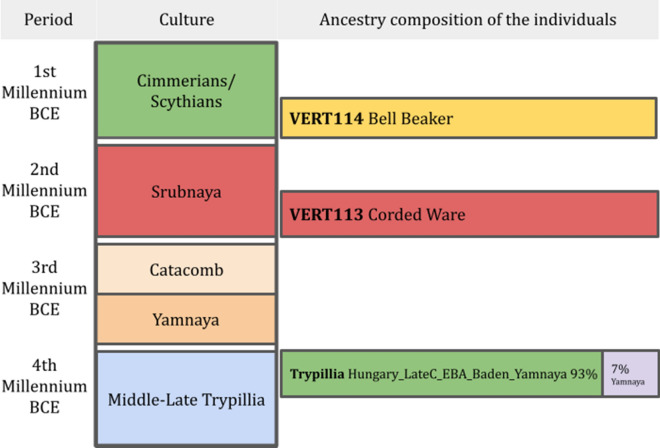
We ran f4 statistics and qpAdm46 to quantify ancestry components as well as to estimate the direction of gene flow (Table S4, Fig. S2). We ran several tests to understand the genetic composition of Verteba_Trypillia (excluding the outliers) and the possible sources of genetic admixture for this population. We first ran qpAdm using populations chronologically close to the CTCC individuals. five models worked (Table S5), with the simplest ones involving about 93% of Hungary_LateC_EBA_Baden_Yamnaya ancestry plus 7% from Yamnaya-related populations, evidencing the connection between Trypillians and steppe populations as Hungary_LateC_EBA_Baden_Yamnaya also has Steppe ancestry (Fig. 4). We then tested possible connections with specific steppe-related populations using f4 statistical test in the form f4(Mbuti,Verteba_Trypillia;Russia_Samara_EBA_Yamnaya,Ukraine_EBA_Yamnaya) = − 0.000525, Z score = − 1.843, which does not statistically connects the Trypillians with the Ukrainian or Russian Yamnaya populations.
Figure 4.
Ancestry and culture summary: Chronology of the different cultures that settled in Ukraine between the 4th and the 1st Millenium BCE, on the right the ancestry components of the analyzed individuals from Verteba cave according to the qpAdm analyses. The colors on the period column represent the different historical periods in the west of Ukraine. The Ancestry composition column colors graphically show the results of qpAdm. Green corresponds to Hungary_LateC_EBA_Baden_Yamnaya, Violet to Yamnaya, Red to Corded_Ware and Yellow to Bell_beaker.
We then explored the same individuals, but this time using populations representing different basal ancestries that might have contributed to the CTCC genetic pool (EHG, CHG, Anatolia_N, WHG and Ukranie_N) (Table S6). Only one model worked, which involved 40% of Anatolia Neolithic-related ancestry, 20% of WHG and 40% of CHG. These results were also observed in Mathiesson et al. with very similar proportions, but were not feasible in the Moldovan Trypillians from Immel et al. To investigate the possible sources of the HG component of the 22 Verteba_Trypillia individuals we ran statistics in the form of: f4(Mbuti,Verteba_Trypillia;Ukraine_N,Serbia_IronGates_Mesolithic) = 0.001603, Z score = 12.173, f4(Mbuti, Verteba_Trypillia;Ukraine_N,Loschbour) = 0.001730, Z score = 6.380, f4(Mbuti, Verteba_Trypillia;Ukraine_N,Koros-HG) = 0.002037, Z score = 7.573, f4(Mbuti, Verteba_Trypillia;Koros-HG,Serbia_IronGates_Mesolithic) = − 0.00047, Z score = − 1.879. Showing that Koros-HG is the WHG source with the highest genetic affinity to Ukraine Trypillians. When compared to Central European Eneolithic populations, the Eneolithic Verteba individuals do not seem to share a statistically significant affinity with the CTCC population of Moldova, as shown in the form of f4(Mbuti, Verteba_Trypillia;Hungary_LateC_EBA_Baden_Yamnaya, Trypillia_Moldova):,-0.000304 Z score = − 0.936. qpAdm also indicates that Ukraine_N and WHG are two likely sources of HG-related ancestry to Verteba_Trypillia, in addition to Hungary_LateC_EBA_Baden_Yamnaya (Table S5). The statistic f4(Mbuti, Verteba_Trypillia; Ukraine_N, WHG) shows a clear tendency towards WHG: 0.001192, Z score = 6.26, suggesting very little presence of ancestry from the local Hunther_Gatherers in the Verteba individuals.
To detect different individual ancestry compositions, we ran qpAdm tests individually on the 22 Verteba_Trypillian individuals showing that most of them can be modeled with a single source using: Trypillia_Moldova, Hungary_LateC_EBA_Baden_Yamnaya or Hungary_LN_Tisza, indicating a clear affinity for Late_Neolithic populations with steppe ancestry (Table S7) as all these populations show the presence of steppe component. Surprisingly, only four of the Verteba_Trypillia individuals can be modeled using Trypillia-Moldova as a single source. To investigate if there are statistically significant differences between the possible sources for these individuals we ran the tests f4(Mbuti, VERTXXX; Hungary_LateC_EBA_Baden_Yamnaya, Trypillia-Moldova) individually (Table S8). The results show that only one individual (VERT-035) is statistically more related to Hungary_LateC_EBA_Baden_Yamnaya than to Trypillian-Moldova, pointing towards the existence of some variability within the Trypillians in Ukraine. As we did for the general population, we also performed the qpAdm analyses using distal sources (EHG, CHG, Anatolia_N, WHG and Ukranie_N). The results show that most of the individuals can be modeled using Anatolia_N plus a ~ 20% of WHG/Ukranie_N. When the f4(Mbuti, VERTXXX; Ukraine_N, WHG) is tested individually the statistics show no differences (Table S9), however the affinity to WHG at a population level is clear revealing the importance of having big sample sizes to perform f-statistics based assessments (Table S4).
VERT-113, dated to the MBA, shows a clear signal of steppe-related ancestry, and is the only individual in the dataset that shows a strong influx of this ancestry: f4(Mbuti, Verteba_MBA; Russia_Samara_EBA_Yamnaya, LBK): -0.00398 Z score = − 7.848. The same test with individual VERT-114 was not statistically significant (Z = 1.382). Relevantly, we observe a major affinity to Russia_Yamnaya over Ukraine_Yamnaya using f4 statistics (Table S5). Furthermore, this is the only individual that shows a major affinity to Ukranian_N over WHG as the source of HG-related ancestry, as shown by the statistic f4(Mbuti, VERT-113; Ukraine_N, WHG): − 0.001276, Z score (Z = − 4.202). The distal models of qpAdm using basal ancestries reveal that this individual exhibits up to 33% of Ukraine_N and 66% of CHG, supporting high amounts of steppe-related ancestry. When modeled with close chronology populations the individual requires a single source related to the Corded_Ware (Table S5). We tried to assess if the signal could, however, correspond to similar genetic populations but more contemporary and geographically closer to VERT-113, such as Srubnaya, using the statistic f4(Mbuti, VERT-113; Poland_Southeast_CordedWare, Russia_Srubnaya), but the results (f4 = 0.00003, Z = 0.11) show that there is no statistical relationship, which indicates no evidence supporting the Srubnaya origin.
Individual VERT-114, dated around the LBA, showed a genetic position close to Bell-Beaker populations in PCA and ADMIXTURE. This individual shows a higher influx of ancestry from WHG than from EHG populations f4(Mbuti, VERT-114; WHG, EHG,) − 0.002, Z score = − 8,64), similar to the results obtained for the aggregate group of 22 Verteba individuals. qpAdm results for this individual show that a single model with a Bell_Beaker population as a single source works. Many of the two-way models involve populations related to Ukraine_Globular_Amphora and to Steppe populations, with about 60% of ancestry from the former and the rest from the latter. We also explored the possible connections between this individual and the Cimmerians, who established themselves in present-day Ukraine around the year 1000 BCE, being contemporary to VERT-114. Both f4(Mbuti, VERT-114; Moldova_Cimmerian.SG, Poland_Southeast_BellBeaker) f4 = 0.00428 Zscore = 7.171) and f4(Mbuti, VERT-114; Moldova_Cimmerian.SG, Hungary_EBA_BellBeaker) f4 = 0.003301, Zscore = 4.043) showed a clear affinity of VERT-114 to the Bell_Beaker individuals over the Cimmerians. This individual can also be modeled as an admixture between Turkey_N (67%) and Ukraine_N (33%) (Table S6).
We used the approach presented in Ringbauer et al.48, to explore the presence of runs of homozygosity (ROH) in the sample. We observed that the samples tested present very few parts of the genome under ROH (Fig. S3), meaning that the individuals were part of large populations. An exception was VERT-100 who showed long ROH segments, suggesting that this individual was an offspring of related individuals.
Phenotypic positions
We genotyped 105 SNPs linked with metabolic, pigmentation and pathogen resistance phenotypic traits. Pseudohaploid genotypes are shown in Table S10. From these genotypes it is evident that none of the tested individuals from Verteba cave was lactose tolerant as all are homozygous for the non-tolerant variants for SNPs rs498823549 and rs18254950. It is also interesting to remark that, except for two individuals, the majority of individuals from Verteba cave have the variant of SNP rs12913832 associated with blue eyes and the other two associated with dark ones51.
Discussion
The CTCC is an important archaeological complex that brought farming to Eastern Europe16,18. Prior to our research, the CTCC genomic record consisted of only four individuals from Verteba Cave and four individuals from Moldova, nevertheless, the previously reported diversity within the CTCC37 showed that more research on the genetic diversity of this culture is needed to understand its origin, dynamics, and collapse. Recent publications have revealed the utility and the relevance of large-scale projects focused on specific sites43,52. Here, we have presented genomic data from 20 individuals buried in Verteba Cave that are dated to the fourth, second and first millennia BCE. The genetic analysis of these individuals has revealed important genetic turnovers both in the Early Bronze Age and during the Late Bronze Age. In the future, more individuals should be sequenced to clarify these observations, in particular to obtain more individuals from the third millennium BCE onward, as the genomic record in Ukraine from the Bronze Age is limited to six individuals from the second and third millennia BCE. Importantly, we also provide eight new radiocarbon dates, which are extremely relevant, as previous studies have demonstrated the presence of diverse material in Verteba Cave’s, caused by repeated use of the site from at least the Mesolithic up until modern times28.
Previous analyses of CTCC individuals’ mitochondrial DNA HVRI indicated their close maternal ancestry with early Neolithic groups, with lineages that are representative of the Neolithic ‘package’, including haplogroups H, HV, T, V, J, and K53,54. With the exception of two individuals with haplogroup U5a, all the other 18 individuals that were included in our analyses have haplogroups that are similar to Central European Neolithic groups13. This diversity is in stark contrast to individuals from earlier non-agricultural Neolithic sites from the Ukraine that have only haplogroup U, likely the result of continuity with previous Mesolithic hunter-gatherers (HG). The mtDNA haplogroup diversity suggests that local populations were largely replaced by those associated with the Trypillian culture. The majority of VC individuals exhibit the G2a2 Y haplogroup, which is widely present in Anatolia-related Neolithic European individuals5,13. The other identified haplogroups, C1 and I2, have been also reported among European Neolithic populations, pointing to an origin of the CTCC individuals without a sex-biased migratory past, which contrasts with the steppe migrations during the Bronze Age55.
Population genetic analyses indicate that the individuals buried at Verteba Cave during the Late Eneolithic (3790–3535 cal BCE) genetically resemble other previously published CTCC individuals, and are closely related to other published CTCC individuals in Moldova37. These observations broadly suggest that Eneolithic CTCC individuals descended from the same, or closely related, population that spread the Neolithic across most of Europe and without little or no sign of admixture with earlier Ukrainian Mesolithic or Neolithic groups composed of hunter-gatherer-related ancestry and specifically pointing towards the Baden individuals from Hungary. In fact, most of the Trypillian individuals can be modeled by Eneolithic populations from Europe that have steppe ancestry, however four out of the 20 individuals could be modeled as Moldovan Trypillians. These results in the qpAdm modeling suggest that there were differences in the ancestry composition of the Trypillians of Verteba Cave, which could be linked to the proportion of HG in the individuals, although this variability is not substantial enough to differentiate the individuals into different populations.
Previous studies of CTCC individuals could not provide a clear origin for the HG component of CTCC-associated groups. Here, despite observing that models including Ukranian_N individuals and WHG seem to work, the f4-statistics suggest that the source of the HG component would be mainly WHG. In addition, not a single qpAdm model using EHG as a source works, which supports that observation. The significant proportion of WHG ancestry found in the Trypillians (up to 18%) might be related to the hunter gatherer resurgence seen in other Middle Neolithic populations of Central Europe, likely due to admixture with groups in the west who already had a higher WHG component derived from Anatolia-related Neolithic groups prior to the origin of the CTCC8,13,56. This would also indicate that the HG Neolithic populations from Ukraine did not contribute much ancestry to the Trypillians. In addition, we also observe the presence of steppe-related ancestry in these individuals, as was revealed in Moldova37, although the proportion in the Verteba individuals is lower, which could correlate with the age of the individuals suggesting a continuous pulse from the East to the West gradually increasing the Yamnaya-related ancestry during the fourth millenium BCE.
Individual VERT-113, dated to the Middle Bronze Age (1960–1770 cal BCE [2σ]), has an ancestry profile that is quite different from the earlier CTCC individuals. There is significantly more Caucasus HG/Yamnaya and EHG ancestry, and thus this individual was related to the Yamnaya expansions. qpAdm results suggest a link between VERT-113 and Corded Ware populations from Poland, pointing to a similarity between this individual and these populations. Also, this is the only individual with a higher genetic affinity to the Ukraine_N than to the WHG, suggesting that the population that originated MBA in the second millenium BCE may have had shared affinities with the Ukraine_N populations.
Interestingly, VERT-114 (Late Bronze Age) does not show many genetic connections with MBA VERT-113 according to the f3 values, who is clearly associated with the Yamnaya pastoralists. The genomic composition of VERT-114 suggests a relationship with Beaker-related populations, despite being almost 1000 years younger than the end of the Bell Beaker phenomenon57, and with a date that would be more coincident with the Cimmerians or Scythians58. However, no qpAdm models with these cultures work and the f4 results do seem to confirm the similarity with the Bell Beakers over the Cimmerians. The genetic background of this individual, with its strong western affinities, supports the evidence shown in Narasimhan et al.59 of a western influx into the Steppe during the Late Bronze Age. Further sequencing and analysis of individuals from the cave and the area surrounding VC dating from the third millennium BCE will be critical for exploring cave use after abandonment by the CTCC-related peoples.
The results of our paleogenomic analysis have important implications for understanding the Neolithisation process in far eastern Europe. As the populations of the CTCC expanded from Romania and Moldova into the forest-steppe areas of western and central Ukraine, they would have come into contact with populations associated with the indigenous Bug-Dniester culture, a group whose subsistence system focused primarily on foraging60. This group was likely descendant from Mesolithic hunter-gatherers. The paleogenomics of the Verteba Cave individuals suggest that local Mesolithic hunter-gatherers did not contribute significantly to later Trypillian ancestry, indicating that the process to Neolithisation in western Ukraine was the product of substantial migration rather than indigenous adoption of agricultural practices.
Our results also provide support to the idea that a long-lasting frontier existed between the sedentary agriculturalists of the forest-steppe ecozone and the neighboring nomadic pastoralists from the Pontic Steppe. This frontier is characterized by drastic contrasts in material culture and subsistence regimes, and was likely maintained in prehistory due to these factors as well as by major linguistic differences60. Documenting the lack of admixture on this cultural frontier is key to understanding the context from which the Yamnaya migration occurred8.
In conclusion, the results show that Verteba Cave represents a significant mortuary site that connects East and West. The genetic structure of the CTCC peoples includes ancestry related to both earlier hunter-gatherers from the west and farmers from the Near East, and one that is genetically distinct from those of Moldovan CTCC peoples. The lack of local ancestry associated with Ukrainian Neolithic hunter-gatherers suggests that these farmers mostly replaced local foragers and did not mix with the neighbouring steppe populations. Additionally, during the Bronze Age, Verteba Cave was used by successive waves of nomadic pastoralists from the east that eventually brought significant genetic and cultural changes to Europe that eventually mixed with the local descendants of Trypillia-culture population. Additional genomic sampling from these later time periods will help to answer questions of site chronology and possibly indicate how the Trypliian culture eventually collapsed.
Materials and methods
To perform the present study, 23 samples were collected from Verteba cave (Ukraine). Due to low coverage, two samples were not included in the final analyses. The complete description of the methods can be found in the Supplementary Information section.
AMS radiocarbon analysis
The ages of individuals from this study were determined using AMS 14C dating; here we report eight new dates run at the Pennsylvania State University Accelerator Mass Spectrometry Laboratory (lab code: PSUAMS) and the Oxford Radiocarbon Accelerator Unit (lab code: OxA) (see Table S1).
Bone collagen extraction for 14C and stable isotope analysis was extracted and purified at the Pennsylvania State University using a modified Longin method with ultrafiltration61. In cases where this method returned an unacceptably low gelatin yield, samples were processed according to the XAD amino acid purification method62. Samples were prepared first by manual cleaning adhering sediment and removing exposed surfaces with an X-acto blade. This was followed by demineralization for 24–36 h in 0.5 N HCl at 5 °C. The pseudomorph was then rinsed to neutrality in multiple changes of Nanopure H2O, before being gelatinized for 10 h at 60 °C in 0.01 N HCl. The resulting gelatin was lyophilized, visually inspected and then weighed to assess bone collagen preservation. Rehydrated gelatin solution was pipetted into pre-cleaned Centriprep63 ultrafilters (retaining 30 kDa molecular weight gelatin) and centrifuged 3 times for 20 min, diluted with Nanopure H2O, and centrifuged 3 more times for 20 min to desalt the solution. Carbon and nitrogen concentrations and stable isotope ratios were measured at the Yale Analytical and Stable Isotope Center with a Costech elemental analyzer (ECS 4010) and Thermo DeltaPlus analyzer. Sample quality was evaluated by examining the % crude gelatin yield, %C, %N and C:N ratios before AMS 14C dating. C:N ratios for the dated samples fell between 3.15 and 3.42, indicating acceptable collagen preservation64. Collagen samples were combusted for three hours at 900 °C in vacuum-sealed quartz tubes with CuO and Ag wires. Sample CO2 was reduced to graphite at 550 °C using H2 and a Fe catalyst, with reaction water drawn off with Mg(ClO4)265. Graphite samples were pressed into targets in Al cathodes and loaded on the target wheel for AMS analysis. The 14C ages were corrected for mass-dependent fractionation with measured δ13C values66 and compared with samples of Pleistocene whale bone (backgrounds, 48,000 14C BP), late Holocene bison bone (~ 1850 14C BP), late AD 1800s cow bone and OX-2 oxalic acid standards for calibration. Sample preparation protocols at OxA follow similar standards and have been published in depth elsewhere (Brock et al. 2010).
Dates were calibrated according to the IntCal20 Northern Hemisphere calibration curve67 using OxCal 4.468. Six of the eight dates display a tight distribution ranging from 3790 to 3535 cal BCE (2σ), with mean results spread across a single ~ 80-year period from 3720–3640 cal BCE. These results are comparable with dates from other sites in Romania, Moldova and Ukraine dating to the latter part of period Trypillia CI, and are consistent with the late Trypillia CI (Shypynetska local group) occupation identified at the site (Nikitin et al. 2010). A single date corresponds with the Middle Bronze Age (PSUAMS-3153; VERT-113), dating to the range of 1960–1770 cal BCE (2σ).
DNA extraction and library preparation
All laboratory work was performed in dedicated ancient DNA laboratories at University College, Dublin. These facilities are physically located from other molecular biology laboratories, and measures are taken to minimize contamination of ancient individuals, including head-to-toe suits, face masks, hair nets, multiple layers of gloves, bleaching of all surfaces and UV decontamination of all (non-sensitive) reagents. All laboratory tools used to process samples were decontaminated using bleach (1:5 concentration) and UV irradiated in a cross-linker. The final step of library preparation (amplification) was performed outside the ancient DNA laboratory. We included extraction negative controls (no powder), library and PCR negative controls (extract was supplemented by water) in every batch of samples processed and carried them through the entire wet laboratory processing to test for reagent contamination.
Samples were initially UV irradiated on both sides for ~ 10 min. We targeted the inner ear region of the petrous bone69,70 using a sandblaster (Renfert). Fragments of the cochlea were then powdered using a mixer mill (Retsch Mixer Mill 400). Twenty-three petrous bone samples were initially screened. Using ~ 75 mg of powder, DNA was extracted using an optimized DNA extraction protocol71. Illumina sequencing libraries were constructed using 12.5-25uL of extract, amplified using Accuprime Pfx Supermix (Thermo Fisher Scientific), following Gamba et al.72; a protocol adopted from73. Quality assessment of the amplified library was performed on an Agilent 2100 Bioanalyzer and a Qubit 2.0 Fluorometer. All amplified libraries were initially screened using an Illumina MiSeq. After screening, additional libraries were sequenced to ~ 1X on the NextSeq platform.
Bioinformatic analysis
Sequencing reads were trimmed using cutadapt (Version. 1.2.1)74 and aligned to the human reference genome (GRCh37), with the mitochondrial genome replaced by the revised Cambridge reference sequence (rCRS) using BWA75 (Version 0.7.5). Duplicate mapped reads were removed using Picard Tools76. Reads with mapping qualities below 30 were also removed. Unique and filtered reads were analyzed with qualimap-277 to assess the coverage of the genomes. MapDamage-278 was used to estimate the level of deamination and the authenticity of the data. We have clipped two bases per read end to minimize the effect of damage.
Pseudo-haplotypes were called using sequenceTools79 filtering the calls with mapping and base quality below 30. As a reference panel we used the positions of the 1240 k capture dataset8. The Verteba calls were merged with a panel of 750 modern individuals from 46 populations3 and 611 individuals from 67 ancient populations3–5,8,13,37,45,56,57,80–89. Molecular sex was determined using ry_compute.py90, we also explored the presence of familiar relationships with READ91.
Mitochondria aligned reads were processed with Schmutzi92 to generate a consensus sequence of the mitochondrial genomes using –uselength option. We determined the mitochondrial haplogroups of the mitochondrial consensus sequences with Haplogrep v2.093. Y chromosome haplogroups of male individuals were determined using Yleaf94.
Principal component analysis was built with 597,573 SNPs and 750 modern genomes using smartpca from Eigensoft package46. Resulting data was plotted using R95. Ancient samples were projected in the PCA built with the modern ones using the option lsqproject. Two rounds of outlier removal were used. Results were plotted with R.
An unsupervised ADMIXTURE analysis was performed with ADMIXTURE47. 611 ancient individuals, 2068 modern individuals together with the 20 Verteba individuals were used for the analysis. From the 597,573 SNPs of the Human Origins dataset was filtered removing SNPs with MAF below 0.05 and more than 5% of missing sites. Filtered SNPs were pruned by linkage-disequilibrium (LD) using PLINK 1.996 flag –indep-pairwise with a windows size of 200 SNPs, advanced by 50 SNPs and establishing an r2 threshold of 0.4. The ADMIXTURE analysis was performed with 417,913 SNP s, with K ranging from 2 to 15 and 10 bootstrap replications. Admixture was plotted with R.
F-statistics were run using admixtools46 in the form f3(Test, X; Mbuti) using all the ancient European populations available. D statistics were also run using the same package. We have used the form f4(Mbuti, Test; PopA, PopB) using a list of possible sources of Hunter-Gatherer, Neolithic and steppe components. In this analysis, we excluded results with less than 100,000 shared SNPs.
We performed qpAdm using the admixtools package46 In this analysis we have used the same proxies as the ones used for D statistic plus Verteba in case of modeling VERT-113 and VERT-114 individuals and setting allSNPs: NO. The list of samples included in each category is displayed in table S6. As outgroups we used: Russia_Kostenki14.SG, Italy_North_Villabruna_HG,Han.DG,Mbuti.DG,ONG.SG,Papuan.SDG,Russia_MA1_HG.SG,Ust_Ishim. In a second round we added the rest of tested populations to the right making the models compete between them as described in Harney et al.97. We also performed qpWave analysis to assess the presence of substructure in the Verteba_Trypillia population using the same software and right population set listed below, the threshold was set to a p-value of 0,05.
We calculated the ROH segment distribution following the protocol described in Ringbauer et al.48. The phenotypic positions analyzed were genotyped using the pseudo haploid calls. The frequencies of the present day populations were obtained from the 100 genomes data98.
Supplementary Information
Author contributions
R.W.S. and J.K.K. collected samples, R.W.S. performed wet laboratory work, P.G. and D.M.F. analyzed the data, T.K.H. and D.J.K. performed radiocarbon dating and isotope analysis, P.G., R.W.S., D.M.F., J.K.K., S.H.L., T.K.H., H.O. and R.P. wrote the paper with input from all co-authors.
Funding
Open access funding provided by University of Vienna. Funding was provided by the European Commission’s Marie Skłodowska-Curie Individual Fellowships (PACE #70245 to RWS and RP), Grand Valley State University Professional Development Grant (GDM), the University of Wisconsin-Oshkosh (JKK) and the National Science Foundation (BCS-1725067 to TKH and DJK).
Data availability
Sequencing reads have been deposited in the European Nucleotide Archive (ENA) with the accession code PRJEB38797.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Pere Gelabert and Ryan W. Schmidt.
Contributor Information
Pere Gelabert, Email: pere.gelabert@unive.ac.at.
Ryan W. Schmidt, Email: ryan.schmidt@cibio.up.pt
Ron Pinhasi, Email: ron.pinhasi@univie.ac.at.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-11117-8.
References
- 1.Cavalli-Sforza LL. Genes, peoples, and languages. Proc. Natl. Acad. Sci. U. S. A. 1997;94:7719–7724. doi: 10.1073/pnas.94.15.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shennan S. The First Farmers of Europe: An Evolutionary Perspective. Cambridge University Press; 2018. [Google Scholar]
- 3.Lazaridis I, et al. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofmanová Z, et al. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc. Natl. Acad. Sci. U. S. A. 2016;113:6886–6891. doi: 10.1073/pnas.1523951113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lipson M, et al. Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature. 2017;551:368–372. doi: 10.1038/nature24476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ancient Europe 8000 B.C.—A.D. 1000—(Encyclopedia of the Barbarian World, Vol. 1. (Charles Scribner & Sons, 2004).
- 7.Barton L. First farmers: The origins of agricultural societies by P. S. Bellwood, and: The peopling of East Asia: Putting together archaeology, linguistics and genetics ed. by L. Sagart, R. Blench, and A. Sanchez-Mazas, and: The Origins of Pottery and Agriculture ed. by Y. Yasuda (review) Asian Perspect. 2012;51:321–333. doi: 10.1353/asi.2014.0000. [DOI] [Google Scholar]
- 8.Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skoglund P, et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336:466–469. doi: 10.1126/science.1216304. [DOI] [PubMed] [Google Scholar]
- 10.Zvelebil M, Dolukhanov P. The transition to farming in Eastern and Northern Europe. J. World Prehist. 1991;5:233–278. doi: 10.1007/BF00974991. [DOI] [Google Scholar]
- 11.Zvelebil M, Rowley-Conwy P. Transition to farming in Northern Europe: A hunter-gatherer perspective. Nor. Archaeol. Rev. 1984;17:104–128. doi: 10.1080/00293652.1984.9965402. [DOI] [Google Scholar]
- 12.Jones ER, et al. The neolithic transition in the Baltic was not driven by admixture with early European farmers. Curr. Biol. 2017;27:576–582. doi: 10.1016/j.cub.2016.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathieson I, et al. The genomic history of southeastern Europe. Nature. 2018;555:197–203. doi: 10.1038/nature25778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis L. The Cucuteni-Tripolye Culture: A Study in Technology and the Origins of Complex Society. BAR Oxford; 1984. [Google Scholar]
- 15.Milisauskas S, Kruk J. Middle neolithic/early copper age, continuity, diversity, and greater complexity, 5500/5000–3500 BC. In: Milisauskas S, editor. European Prehistory: A Survey. Springer; 2011. pp. 223–291. [Google Scholar]
- 16.Zbenovich VG. The Tripolye culture: Centenary of research. J. World Prehist. 1996;10:199–241. doi: 10.1007/BF02221076. [DOI] [Google Scholar]
- 17.Rassamakin, Y., Menotti, F. & Korvin-Piotrovskiy, A. Absolute chronology of Ukrainian Tripolye settlements. The Tripolye Culture Giant-Settlements in Ukraine. Formation, Development and Decline. Oxford: Oxbow Books 19–69 (2012).
- 18.Menotti, F. & Korvin-Piotrovskiy, A. The Tripolye Culture Giant-Settlements in Ukraine: Formation, Development and Decline 264 (Oxbow Books, 2012).
- 19.Marinescu-Bîlcu, S. Cultura Precucuteni pe teritoriul României, Bucureşti: Ed. Academiei RSR (1974).
- 20.Rassmann K, et al. High precision Tripolye settlement plans, demographic estimations and settlement organization. J. Neolit. Archaeol. 2014;96:134. doi: 10.12766/jna.2014.3. [DOI] [Google Scholar]
- 21.Harper TK, Diachenko A, Rassamakin YY, Kennett DJ. Ecological dimensions of population dynamics and subsistence in Neo-Eneolithic Eastern Europe. J. Anthropol. Archaeol. 2019;53:92–101. doi: 10.1016/j.jaa.2018.11.006. [DOI] [Google Scholar]
- 22.Дepгaчeв, B. A. O cкипeтpax, o лoшaдяx, o вoйнe. (Hecтop-Иcтopия, 2007).
- 23.Videiko MY. Reasons of origins and development of the Tripolian proto-cities. Archaeology. 1998;4:145–151. [Google Scholar]
- 24.Diachenko A, Menotti F. The gravity model: Monitoring the formation and development of the Tripolye culture giant-settlements in Ukraine. J. Archaeol. Sci. 2012;39:2810–2817. doi: 10.1016/j.jas.2012.04.025. [DOI] [Google Scholar]
- 25.Müller J, Rassmann K, Videiko M. Trypillia Mega-Sites and European Prehistory: 4100–3400 BCE. Routledge; 2016. [Google Scholar]
- 26.Кpyц, B. A. Пoзднeтpипoльcкиe пaмятники Cpeднeгo Пoднeпpoвья. (Hayк. дyмкa, 1977).
- 27.Beликaнoвa, M. C. Пaлeoaнтpoпoлoгия Пpyтcкo-Днecтpoвcкoгo мeждypeчья. (нayкa, 1975).
- 28.Lillie MC, et al. First isotope analysis and new radiocarbon dating of Trypillia (Tripolye) farmers from Verteba Cave, Bilche Zolote, Ukraine. Doc. Praehist. 2017;44:306. doi: 10.4312/dp.44.18. [DOI] [Google Scholar]
- 29.Madden GD, Karsten JK, Ledogar SH, Schmidt R, Sokhatsky MP. Violence at verteba cave, Ukraine: New insights into the Late Neolithic intergroup conflict. Int. J. Osteoarchaeol. 2018;28:44–53. doi: 10.1002/oa.2633. [DOI] [Google Scholar]
- 30.Nikitin AG, Sokhatsky MP, Kovaliukh MM, Videiko MY. Comprehensive site chronology and ancient mitochondrial DNA analysis from verteba cave: A trypillian culture site of eneolithic Ukraine. Interdiscip. Archaeol. 2010;1:9–18. [Google Scholar]
- 31.Ledogar SH, et al. New AMS dates for verteba cave and stable isotope evidence of human diet in the holocene forest-steppe, Ukraine. Radiocarbon. 2019;61:141–158. doi: 10.1017/RDC.2018.52. [DOI] [Google Scholar]
- 32.Nikitin AG, Sokhatsky MP, Kovaliukh MM, Videiko MY. Comprehensive site chronology and ancient Mitochondrial dna analysis from Verteba cave: A trypillian culture site of eneolithic ukraine. PLoS ONE. 2017;1:9–18. [Google Scholar]
- 33.Kadrow S, Pokutta DA. The Verteba cave: A subterranean sanctuary of the Cucuteni-Trypillia culture in Western Ukraine. J. Neolit. Archaeol. 2016;18:1–21. [Google Scholar]
- 34.Ledogar SH. A Zooarchaeological and Geochemical Analysis of the Faunal Remains from the Tripolye site Verteba Cave Ukraine. State University of New York at Albany; 2017. [Google Scholar]
- 35.Anthony DW. The Horse, the Wheel, and Language: How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World. Princeton University Press; 2007. [Google Scholar]
- 36.Schmidt RW, et al. Analysis of ancient human mitochondrial DNA from Verteba Cave, Ukraine: Insights into the Late Neolithic-Chalcolithic Cucuteni-Tripolye culture. Anthropol. Sci. 2020;128:1–10. doi: 10.1537/ase.200205. [DOI] [Google Scholar]
- 37.Immel A, et al. Gene-flow from steppe individuals into Cucuteni-Trypillia associated populations indicates long-standing contacts and gradual admixture. Sci. Rep. 2020;10:4253. doi: 10.1038/s41598-020-61190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellis L. Analysis of Cucuteni-Tripolye and Kurgan pottery and the implications for ceramic technology in the transformation of European and Anatolian culture 4500–2500 BC and its legacy. J. Indo-European Stud. Wash. 1980;8:211–230. [Google Scholar]
- 39.Tsvek E, Rassamakin YY. The interactions between the Eastern Tripolye culture and the Pontic steppe area: Some aspects of the problem. Cucuteni. 2005;120:173–192. [Google Scholar]
- 40.Chapman, J. Expansion and social change at the time of Varna. Counterpoint: Essays in Archaeology and Heritage Studies in honour of Professor Kristian Kristiansen 301–308 (2013).
- 41.Sawyer S, Krause J, Guschanski K, Savolainen V, Pääbo S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS ONE. 2012;7:e34131. doi: 10.1371/journal.pone.0034131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olalde I, et al. The genomic history of the Iberian Peninsula over the past 8000 years. Science. 2019;363:1230–1234. doi: 10.1126/science.aav4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mittnik A, et al. Kinship-based social inequality in Bronze Age Europe. Science. 2019;366:731–734. doi: 10.1126/science.aax6219. [DOI] [PubMed] [Google Scholar]
- 44.Saag L, et al. The arrival of Siberian ancestry connecting the eastern Baltic to Uralic speakers further East. Curr. Biol. 2019;29:1701–1711.e16. doi: 10.1016/j.cub.2019.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang C-C, et al. Ancient human genome-wide data from a 3000-year interval in the Caucasus corresponds with eco-geographic regions. Nat. Commun. 2019;10:590. doi: 10.1038/s41467-018-08220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ringbauer H, Novembre J, Steinrücken M. Detecting runs of homozygosity from low-coverage ancient DNA. bioRxiv. 2020 doi: 10.1101/2020.05.31.126912. [DOI] [Google Scholar]
- 49.Enattah NS, et al. Identification of a variant associated with adult-type hypolactasia. Nat. Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 50.Bersaglieri T, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eiberg H, et al. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum. Genet. 2008;123:177–187. doi: 10.1007/s00439-007-0460-x. [DOI] [PubMed] [Google Scholar]
- 52.Amorim CEG, et al. Understanding 6th-century barbarian social organization and migration through paleogenomics. Nat. Commun. 2018;9:3547. doi: 10.1038/s41467-018-06024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brandt G, et al. Ancient DNA reveals key stages in the formation of central European mitochondrial genetic diversity. Science. 2013;342:257–261. doi: 10.1126/science.1241844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nikitin AG, et al. Mitochondrial DNA analysis of eneolithic trypillians from Ukraine reveals neolithic farming genetic roots. PLoS ONE. 2017;12:e0172952. doi: 10.1371/journal.pone.0172952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldberg A, Günther T, Rosenberg NA, Jakobsson M. Ancient X chromosomes reveal contrasting sex bias in Neolithic and Bronze Age Eurasian migrations. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2657–2662. doi: 10.1073/pnas.1616392114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathieson I, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olalde I, et al. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature. 2018;555:190–196. doi: 10.1038/nature25738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.MacKenzie D, Curran MW. A History of Russia, The Soviet Union, and Beyond. Wadsworth Publishing Company; 2002. [Google Scholar]
- 59.Narasimhan VM, et al. The formation of human populations in South and Central Asia. Science. 2019;365:eaat7487. doi: 10.1126/science.aat7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anthony DW, Brown DR. The secondary products revolution, horse-riding, and mounted warfare. J. World Prehist. 2011;24:131. doi: 10.1007/s10963-011-9051-9. [DOI] [Google Scholar]
- 61.Kennett DJ, et al. Archaeogenomic evidence reveals prehistoric matrilineal dynasty. Nat. Commun. 2017;8:14115. doi: 10.1038/ncomms14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lohse JC, Madsen DB, Culleton BJ, Kennett DJ. Isotope paleoecology of episodic mid-to-late Holocene bison population expansions in the Southern Plains, U.S.A. Quart. Sci. Rev. 2014;102:14–26. doi: 10.1016/j.quascirev.2014.07.021. [DOI] [Google Scholar]
- 63.McClure SB, Puchol OG, Culleton BJ. Ams dating of human bone from cova de la Pastora: New evidence of ritual continuity in the prehistory of Eastern Spain. Radiocarbon. 2010;52:25–32. doi: 10.1017/S0033822200045008. [DOI] [Google Scholar]
- 64.van Klinken GJ. Bone collagen quality indicators for palaeodietary and radiocarbon measurements. J. Archaeol. Sci. 1999;26:687–695. doi: 10.1006/jasc.1998.0385. [DOI] [Google Scholar]
- 65.Santos GM, Southon JR, Druffel-Rodriguez KC, Griffin S, Mazon M. Magnesium perchlorate as an alternative water trap in AMS graphite sample preparation: A report on sample preparation at Kccams at the University of California, Irvine. Radiocarbon. 2004;46:165–173. doi: 10.1017/S0033822200039485. [DOI] [Google Scholar]
- 66.Stuiver M, Polach HA. Discussion reporting of 14C data. Radiocarbon. 1977;19:355–363. doi: 10.1017/S0033822200003672. [DOI] [Google Scholar]
- 67.Reimer PJ, et al. The IntCal20 northern hemisphere radiocarbon age calibration curve (0–55 cal kBP) Radiocarbon. 2020;62:725–757. doi: 10.1017/RDC.2020.41. [DOI] [Google Scholar]
- 68.Ramsey CB. Bayesian analysis of radiocarbon dates. Radiocarbon. 2009;51:337–360. doi: 10.1017/S0033822200033865. [DOI] [Google Scholar]
- 69.Pinhasi R, Fernandes DM, Sirak K, Cheronet O. Isolating the human cochlea to generate bone powder for ancient DNA analysis. Nat. Protoc. 2019;14:1194–1205. doi: 10.1038/s41596-019-0137-7. [DOI] [PubMed] [Google Scholar]
- 70.Pinhasi R, et al. Optimal ancient DNA yields from the inner ear part of the human petrous bone. PLoS ONE. 2015;10:e0129102. doi: 10.1371/journal.pone.0129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dabney J, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U. S. A. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gamba C, et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 2014;5:5257. doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010;2010:db.prot.5448. doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 74.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 75.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Picard-tools. http://broadinstitute.github.io/picard.
- 77.Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schiffels S, et al. Iron Age and Anglo-Saxon genomes from East England reveal British migration history. Nat. Commun. 2016;7:10408. doi: 10.1038/ncomms10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fernandes DM, et al. A genomic Neolithic time transect of hunter-farmer admixture in central Poland. Sci. Rep. 2018;8:14879. doi: 10.1038/s41598-018-33067-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sikora M, et al. The population history of northeastern Siberia since the Pleistocene. Nature. 2019;570:182–188. doi: 10.1038/s41586-019-1279-z. [DOI] [PubMed] [Google Scholar]
- 82.Kılınç GM, et al. The demographic development of the first farmers in anatolia. Curr. Biol. 2016;26:2659–2666. doi: 10.1016/j.cub.2016.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fu Q, et al. The genetic history of Ice Age Europe. Nature. 2016;534:200–205. doi: 10.1038/nature17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Damgaard PB, et al. 137 ancient human genomes from across the Eurasian steppes. Nature. 2018;557:369–374. doi: 10.1038/s41586-018-0094-2. [DOI] [PubMed] [Google Scholar]
- 85.Lazaridis I, et al. Genetic origins of the Minoans and Mycenaeans. Nature. 2017;548:214–218. doi: 10.1038/nature23310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raghavan M, et al. The genetic prehistory of the New World Arctic. Science. 2014;345:1255832. doi: 10.1126/science.1255832. [DOI] [PubMed] [Google Scholar]
- 87.Olalde I, et al. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature. 2014;507:225–228. doi: 10.1038/nature12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Keller A, et al. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat. Commun. 2012;3:698. doi: 10.1038/ncomms1701. [DOI] [PubMed] [Google Scholar]
- 90.Skoglund P, Storå J, Götherström A, Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 2013;40:4477–4482. doi: 10.1016/j.jas.2013.07.004. [DOI] [Google Scholar]
- 91.Monroy Kuhn JM, Jakobsson M, Günther T. Estimating genetic kin relationships in prehistoric populations. PLoS ONE. 2018;13:e0195491. doi: 10.1371/journal.pone.0195491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Renaud G, Slon V, Duggan AT, Kelso J. Schmutzi: Estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA. Genome Biol. 2015;16:224. doi: 10.1186/s13059-015-0776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weissensteiner H, et al. HaploGrep 2: Mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44:W58–63. doi: 10.1093/nar/gkw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ralf A, Montiel González D, Zhong K, Kayser M. Yleaf: Software for human Y-chromosomal haplogroup inference from next-generation sequencing data. Mol. Biol. Evol. 2018;35:1291–1294. doi: 10.1093/molbev/msy032. [DOI] [PubMed] [Google Scholar]
- 95.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2013. [Google Scholar]
- 96.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harney É, Patterson N, Reich D, Wakeley J. Assessing the performance of qpAdm: A statistical tool for studying population admixture. Genetics. 2021;217:iyaa045. doi: 10.1093/genetics/iyaa045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing reads have been deposited in the European Nucleotide Archive (ENA) with the accession code PRJEB38797.