Abstract
Background:
Clinical exome sequencing typically achieves diagnostic yields of 30–57.5% in individuals with monogenic rare diseases. Undiagnosed Disease Programs (UDPs) implement strategies to improve diagnostic outcomes for these individuals.
Aim:
We share the lessons learned from the first three years of UDP-Vic, an Australian program embedded within a clinical genetics service in the state of Victoria with a focus on paediatric rare disease.
Methods:
We enrolled families who remained without a diagnosis after clinical genomic (panel, exome or genome) sequencing between 2016 and 2018. We utilized family-based exome sequencing (family ES), family-based genome sequencing (family GS), RNA sequencing (RNA-seq), and high-resolution chromosomal microarray (CMA) with research-based analysis.
Results:
In 150 families, we achieved a diagnosis or strong candidate in 64 (42.7%) families (37 in known genes with a consistent phenotype, three in known genes with a novel phenotype, and 24 in novel disease-genes). Fifty-four diagnoses or strong candidates were made by family ES, six by family GS with RNA-seq, two by high-resolution CMA, and two by data reanalysis.
Conclusion:
We share our lessons learned from the program. Flexible implementation of multiple strategies allowed for scalability and response to the availability of new technologies. Broad implementation of family ES with research-based analysis showed promising yields post a negative clinical singleton ES. RNA-seq offered multiple benefits in family ES-negative populations. International data-sharing strategies were critical in facilitating collaborations to establish novel disease-gene associations. And finally, the integrated approach of a multi-skilled, multidisciplinary team was fundamental in having diverse perspectives and strategic decision-making.
Keywords: Whole Exome Sequencing, Rare Diseases/diagnosis*, Rare Diseases/genetics* Genetic Techniques, Genetic Diseases, Inborn, Exome, Undiagnosed
INTRODUCTION
Collaborative diagnostic research initiatives are instrumental in the systematic approach to the diagnosis of rare genetic disease. Pioneered in 2008 with the National Institute of Health’s (NIH) Undiagnosed Diseases Program (UDP),1 which has since evolved into the NIH’s Undiagnosed Diseases Network (UDN),2 diagnostic programs have successfully utilized a range of technologies to investigate rare genetic disease. The National Genomic Research Institute launched the Centers for Mendelian Genomics in 2012 to discover the genetic basis underlying Mendelian traits and accelerate discoveries by disseminating the obtained knowledge and effective approaches through international collaborations.3 These programs are driven by experts in rare disease, have bridged research and clinical spaces to identify novel disease-genes, broadened access to novel technologies, and improved integration within the health system to develop appropriate referral pathways. Collectively, they serve as a crucial tool to meet the International Rare Diseases Research Consortium’s (IRDiRC) ambitious goals to improve diagnostic rates and reduce delay in the diagnosis of rare disease by 2027.4
Despite heterogeneity in approaches between programs, the most widely utilized genomic technology is exome sequencing (ES), which has allowed for diagnostic rates of 25–57.5% across a range of heterogeneous rare disease cohorts,5–8 and has accelerated novel disease-gene discovery.9 Singleton ES – that is, sequencing of the affected proband only – is commonly utilized in the clinical setting due to its lower cost compared to family-based ES – that is, sequencing of the affected proband with additional family members (typically both biological parents and the proband as a trio). While clinical ES only examines genes already associated with disease, research-based ES analyses in UDPs take a broader approach to identify pathogenic variant(s) in genes not previously known to be associated with human disease. Together with the use of global data-sharing platforms10 and pathways to laboratory functional studies and animal models, these are key components of undiagnosed disease programs that connect researchers and facilitate novel gene discovery.11 There is significant heterogeneity between programs in their eligibility criteria and diagnostic approaches (Table 1). Some centres recruit individuals who have never had next-generation sequencing (NGS) investigations (panel, exome or genome sequencing (GS)) but have had extensive other investigations.12–14 Other programs recruit individuals that may have already had NGS and outline approaches after a negative result, and some take a mixed approach by also accepting sequencing-naïve individuals.15–17 Phenotypic data collection also varies and may be obtained through a proband’s standard clinical course of care or require specific study visits to complete a detailed protocol.16 17
Table 1.
Global diagnostic programs for rare genetic disease
| Program (Clinical site if applicable) | Launch year | Population characteristics | Prior genomic sequencing | Analytic strategies utilized |
|---|---|---|---|---|
|
| ||||
| Undiagnosed Diseases Program 1 | 2008 | Heterogenous phenotypes | Nil prior genomic sequencing | Traditional genetic and biochemical investigations, ES, GS |
| Finding Of Rare disease GEnes (FORGE) Canada Project 55 56 | 2010 | Paediatric, heterogeneous phenotypes | Nil prior genomic sequencing | Traditional genetic and biochemical investigations, ES |
| Deciphering Developmental Disorders study 57 58 | 2011 | Paediatric, primarily neurodevelopmental phenotypes | Nil prior genomic sequencing | Traditional genetic and biochemical investigations, ES |
| Centers for Mendelian Genomics 3 | 2012 | Heterogenous phenotypes | Various genomic sequencing | ES, GS, RNA-seq, methylation, long read sequencing |
| Undiagnosed Diseases Network 2 | 2014 | Heterogenous phenotypes | Various genomic sequencing (32% ES-negative) | Clinical review, targeted genetic and biochemical investigations, ES, GS |
| Undiagnosed Diseases Network (Dukes/Columbia Clinical Site) 16 | 2014 | Heterogenous phenotypes, predominantly paediatric | Various genomic sequencing (60% ES-negative) | Re-phenotyping, targeting genetic and biochemical investigations, and GS. |
| Undiagnosed Diseases Network (University of California-Los Angeles Clinical Site) 15 | 2014 | Heterogenous phenotypes, 79% paediatric | Various genomic sequencing | ES, GS, reanalysis of prior genomic sequencing, RNA-seq |
| Care4Rare Canadian Consortium 11 59 | 2014 | Heterozygous phenotypes | Nil previous genomic sequencing | ES, GS, reanalysis of sequencing data, multi-omics |
| SpainUDP 12 | 2015 | Heterogenous phenotypes | Nil previous genomic sequencing | Phenotyping, ES |
| Italian undiagnosed Rare diseases network 60 | 2016 | Heterogenous phenotypes | Nil previous genomic sequencing | Phenotyping, ES |
| Undiagnosed Diseases Program - Western Australia 14 | 2016 | Heterogenous phenotypes | Nil previous genomic sequencing | Traditional genetic and biochemical investigations, ES, GS |
| Japan’s initiative on rare and undiagnosed diseases 61 | 2017 | Heterogenous phenotypes | Undiagnosed post six months of ‘standard’ investigations | NGS technologies |
| Korean Undiagnosed Diseases Program 13 | 2017 | Primarily paediatric, neurological phenotypes | Nil previous genomic testing | Traditional genetic and biochemical investigations, ES |
Table 1 highlights other published international undiagnosed disease programs. These are primarily ongoing diagnostic programs, therefore the data summarised in this table reflect the state of the program at the time of publication and do not summarise the true total number of participants or tests in each program. Generally, there are two main study designs. Either the clinical visit/evaluation with genomic analysis is performed involving those doing the clinical evaluation, or the sequencing is completed independently, with phenotype data provided by the referring physician. All programs allow for pathways to functional studies for novel disease-gene discoveries. Entry into each program required an extensive clinical workup, though we have highlighted if the program reports previous genomic sequence before entry, such as ES or GS. The Undiagnosed Diseases Network (UDN) in the United States has multiple clinical sites. We have only included two of these listed in separate rows in the table to underline different methods reported. Abbreviations – ES exome sequencing; GS – genome sequencing; RNA-seq – RNA sequencing
In our centre, while clinical singleton ES achieves a diagnostic rate of 52–57.5% across a range of paediatric phenotypes,7 8 this still leaves many affected individuals who undergo NGS without a diagnosis. These patients are offered streamlined recruitment to the Murdoch Children’s Research Institute’s Undiagnosed Diseases Program Victoria Australia (UDP-Vic), initially funded by philanthropic donation and established in collaboration with The Broad Center for Mendelian Genomics (Broad CMG) at The Broad Institute of MIT and Harvard, USA. The UDP-Vic recruits patients after a negative clinical NGS (panel, ES or GS) result and incorporates multifaceted case-specific analytic strategies including family ES, GS, RNA-sequencing (RNA-seq), and high-resolution chromosomal microarray (CMA). The UDP-Vic team is multidisciplinary with analysis of each case led by the recruiting clinical geneticist and integrated with the proband’s standard clinical care. Here we share our experiences and lessons learned over the first three years of the program.
METHODS
Study design and population
We prospectively recruited individuals undiagnosed after a negative NGS (panel, ES or GS) result from a single centre, Victorian Clinical Genetics Services (VCGS) in Melbourne, Australia from March 2016 to June 2018. The recruiting clinical geneticist proposed each individual at a clinical review panel at which time the following criteria were assessed: (i) their phenotype was likely to be monogenic; (ii) appropriate testing had been undertaken, including standard resolution CMA (CytoSNP300K, Core Exome, or GSA; Illumina, San Diego USA) and singleton ES; (iii) phenotypically relevant genomic lesions not tractable by ES had been excluded, e.g. FMR1 triplet repeat analysis or methylation studies for imprinting disorders; and (iv) additional family members for sequencing were available if appropriate. Ethics approval was granted by the Royal Children’s Hospital Ethics Committee (HREC 36291A). Written informed consent was obtained from each family.
Sequencing
The program applied family ES as the primary investigation to all families, with the deployment of adjunct tests in a case-specific manner. Reanalysis of previously generated ES data was performed iteratively by analysts and the recruiting clinical geneticists. Family-based GS with accompanying fibroblast or muscle RNA-seq of the affected proband was performed in a subset of families who had already undergone non-diagnostic family ES. Individuals were selected for family-based GS and RNA-seq if there was a high ongoing suspicion of a monogenic condition, the family provided consent, and appropriate samples were able to be obtained during the study period. Aberrant gene expression or mRNA splicing events identified from RNA-seq were correlated with nearby rare GS variants that may be causative of the change in gene expression or splicing.
All research-based sequencing – ES, GS, and RNA-seq – and data processing were performed by the Genomics Platform at the Broad Institute of MIT and Harvard University. Additional bioinformatic analysis was performed at MCRI for RNA-seq analysis and mitochondrial genome variant calling. Further details are included in the supplementary methods. The ES, GS, and ES-based copy number variant call-set data were uploaded to an open-source web interface (seqr) for collaborative analysis between the Broad CMG and local investigators. Following analysis, each case was discussed in a multidisciplinary team teleconference comprising individuals from VCGS and Broad CMG including bioinformaticians, genomic analysts, clinical geneticists, genetic counsellors, and other disciplines such as cytogeneticists where this expertise was required.
We performed high-resolution CMAs utilising the Illumina Omni 5M SNP platform (Illumina, San Diego USA) in cases where there was clinical suspicion of an intragenic CNV not detected by standard resolution CMA and ES. This array is validated to detect heterozygous copy number changes of 20 consecutive probes, giving an effective resolution of 1–10 kilobases. The high-resolution Omni arrays allowed for the detection of smaller structural variants and INDELS not detected by ES or the standard-resolution CMA required to enter the program.18 Further details are included in the supplementary methods.
Data collection
We collected data relating to phenotype, demographics, clinical investigations, and outcomes from February to July 2019, with a further update in January 2021. Data were extracted manually from electronic medical records and genetic files and stored in Phenotips19 and REDCap (Research Electronic Data Capture)20 – electronic data capture tools hosted at the Murdoch Children’s Research Institute.
A molecular diagnosis, or strong candidate for primarily novel gene discoveries, was considered the endpoint of an individual’s diagnostic trajectory. To reach a molecular diagnosis in an established disease gene, the variant was required to reach the criteria for the American College of Medical Genetics’ and Genomics’ classification of ‘pathogenic’ or ‘likely pathogenic.’21 For novel disease-gene variants and newly characterized (novel) but unpublished phenotypes of known disease genes, a ‘strong candidate diagnosis’ was proposed. We required a strong candidate diagnosis to meet the following three criteria: (i) phenotypically similar unrelated individuals (matched through data sharing platforms) with a variant in the same gene and population allele frequencies compatible with disease penetrance and inheritance pattern; (ii) in vitro or in vivo functional validation, either planned or underway via local or external research collaborators; and (iii) multidisciplinary agreement that variants in the proposed gene(s) were likely causative for the phenotype(s) and recommended for functional confirmation.
We also report families with a gene of uncertain significance (GUS) if a potential novel disease-gene or known disease gene with a novel phenotype has been proposed, but with insufficient or conflicting evidence regarding a gene-disease association. Similar to strong candidates, a GUS required multidisciplinary agreement that variants in the proposed gene(s) are a plausible cause for the phenotype and warrant functional confirmation. However, the additional evidence of phenotypically similar unrelated individuals with a variant in the same gene was lacking. We considered all probands with a variant in a GUS as undiagnosed.
RESULTS
Diagnoses
From 2016–2018, 150 families with a suspected rare genetic disease were recruited to UDP-Vic. Of these, 144 (96.0%) probands had a negative ES result prior to enrolment, of which 17 had a negative family ES result (one duo; 15 trios; one quad). Of the remaining six probands, four were enrolled after an appropriate clinical gene panel showed no pathogenic variant(s), one proband was enrolled after a negative clinical GS result, and one proband was enrolled after reportedly negative research GS, but on further inquiry, prior research sequencing had not actually occurred. In total, 144/150 (96.0%) probands were paediatric (less than 18 years of age) at the time of enrolment and 115 (76.7%) had neurodevelopmental phenotypes (Table 2).
Table 2.
Characteristics of UDP-Vic
| Total Cohort | 150 |
|---|---|
| Sex of proband | |
| Male | 87 (58.0) |
| Female | 63 (42.0) |
| Consanguineous family | |
| Yes | 10 (6.7) |
| No | 140 (93.3) |
| Affected relatives | |
| Affected first-degree relatives | 16 (10.7) |
| Affected second-degree relatives | 3 (2.0) |
| Phenotype category | |
| Syndromic neurodevelopmental | 93 (62.0) |
| Non-syndromic neurodevelopmental | 22 (14.7) |
| MCA / Dysmorphism (non-neurodevelopmental) | 19 (12.7) |
| Neuromuscular | 5 (3.3) |
| Craniofacial | 2 (1.3) |
| Musculoskeletal | 4 (2.7) |
| Dermatological | 2 (1.3) |
| Haematological | 2 (1.3) |
| Ocular | 1 (1.1) |
| Deceased | |
| Total deceased | 10 (6.7) |
| Deceased at the time of diagnosis | 2 (1.3) |
Table 2 shows the phenotype characteristics of the UDP-Vic. n(%) Abbreviations: MCA - multiple congenital anomalies
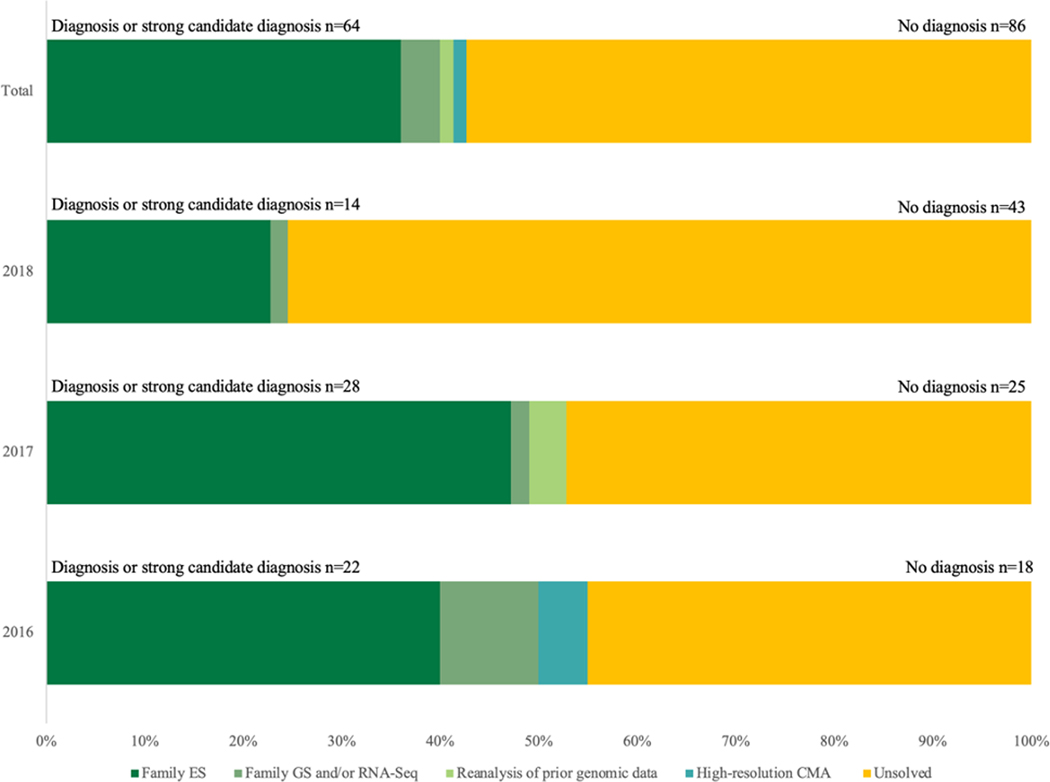
Overall, 64/150 (42.7%) families achieved a molecular diagnosis or strong candidate diagnosis (Figure 1). Of these, 37 were in known disease-genes with a phenotype consistent with others with pathogenic variant(s) in that gene. For the remaining 27 with a diagnosis or strong candidate diagnosis, three were in known genes with novel phenotypes, and 24 were in novel disease genes. If we considered only the 144 families that had undergone ES prior to recruitment, rather than a panel (n=4), GS (n=1), or no previous sequencing (n=1), 62 (43.1%) achieved a molecular diagnosis or strong candidate diagnosis.
Figure 1:
Diagnoses or strong candidate diagnoses by intake year and analytic approach
Figure 1 shows the diagnoses or strong candidate diagnoses by intake year and diagnostic test. The total number of diagnoses or strong candidate diagnoses is also included by intake year.
Abbreviations: family ES – family-based exome sequencing; family GS – family-based genome sequencing; RNA-seq – ribonucleic acid sequencing
As part of the UDP-Vic workflow, 15 disease gene manuscripts have been published (Supp Table 1). Of the 24 novel gene discoveries, nine have been published.22–30 A further two of our novel disease-gene strong candidates matched with multiple research groups via Matchmaker Exchange and these manuscripts have been published.31 32 At the time of matching, these external groups’ manuscripts had sufficiently progressed such that the UDP-Vic was not part of their publication or discoveries. However, we annotate these discoveries as novel (see Supp Table 2) as they were unpublished at the time of discovery within the UDP-Vic. The remaining 13 candidate novel genes are yet unpublished but are considered strong candidates with further functional studies underway. Of the three novel phenotype discoveries in known disease genes, two have been published33 34 with one strong candidate awaiting publication. Finally, of the 37 diagnoses in known genes with known phenotypes, four have been published as part of descriptive cohorts.35–38
Of the remaining 86 families without a diagnosis or strong candidate diagnosis, three families have variants in GUS (GTF3C1, FZD8, PSMD6). These variants have insufficient evidence to be a strong candidate as, to our knowledge, there are no matches with phenotypically similar unrelated individuals and considered undiagnosed.
Sequencing of other affected and unaffected family members (Family ES)
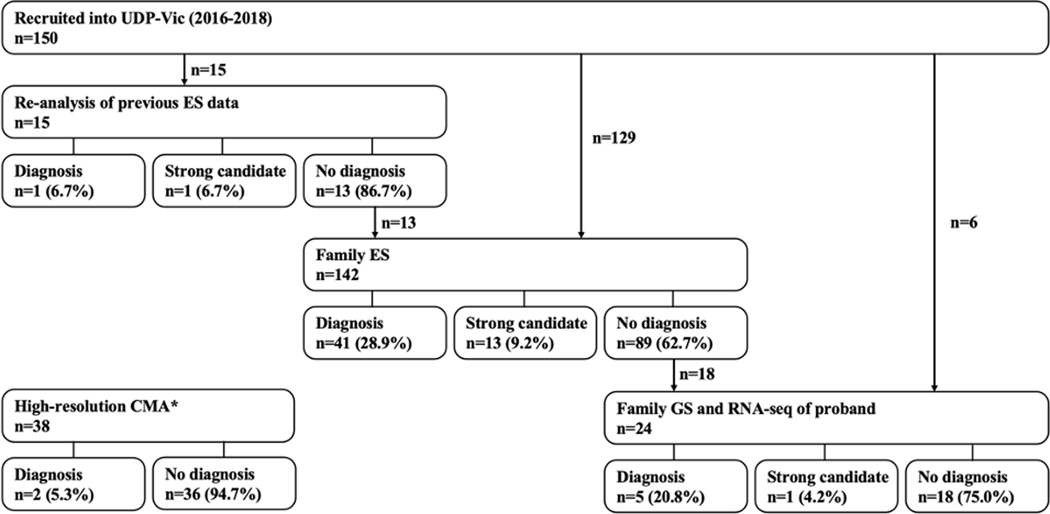
Family-based ES made the majority of diagnoses in the cohort (54/64 diagnoses or strong candidate diagnoses; Figure 2). In total, 142 families underwent family ES (128 were trios, 11 quads, and three quints) with a diagnostic rate of 38.0%. These 54 families achieved a molecular diagnosis or strong candidate diagnosis despite 52 having a negative ES result prior to recruitment. Of these 52, 22 were novel discoveries (19 novel disease genes and three known disease genes with novel phenotypes). The remaining 30/52 diagnoses were achieved by a variety of methods including: new literature confirming novel gene-disease associations after the initial negative clinical sES or family ES (six families); small structural variants detected via CNV calling as part of the family ES pipeline (five families including one with new gene-disease association); improvements in coverage and updated bioinformatic pipeline on our ES platform compared to the initial ES prior to entry to the UDP-Vic (three families, one with a mosaic variant, one with a variant in a repetitive genomic region, and one with a variant in the mitochondrial genome); broader research-based analysis of family ES wherein genes that were initially masked on clinical analysis were reviewed (one family); finally, application of family ES over singleton ES allowed improved curation of causative variants (15 families). Such was the case for the diagnosis of an in-frame deletion in USP9X in a female in middle childhood with syndromic intellectual disability (FAM16, Supp Table 2).38 The variant was present in the singleton ES data but not selected for curation as it was not recognized at the time that in-frame deletions in USP9X might be a causative variant for this phenotype. The addition of parental data with family ES led to the recognition of this variant as de novo which prompted further analysis and established the diagnosis.
Figure 2.
Pathway to diagnoses or strong candidate diagnoses after entry into the UDP-Vic
Figure 2 shows the investigations that reached a diagnosis or strong candidate diagnosis in the cohort of 150 families enrolled in the UDP-Vic. Note that six families who had undergone family ES prior to enrolment in the UDP underwent family GS and RNA-seq directly, rather than repeated family ES within the UDP-Vic. Research-based sequencing was performed at the Broad CMG. *High-resolution CMAs were used before, alongside, or after investigations at The Broad CMG, therefore this testing option is not placed in connection with other testing options. However, the two families that reached a molecular diagnosis via high-resolution CMA both occur after a negative family ES result prior to the implementation of CNV calling within the family ES pipeline. Abbreviations: UDP-Vic – Undiagnosed Diseases Program-Victoria; ES – exome sequencing; GS – genome sequencing; RNA-seq – Ribonucleic acid sequencing.
Family-based GS and RNA-seq
Of the 64 diagnoses or strong candidate diagnoses, six were made by family GS and proband RNA-seq (Figure 2). In total, 24 families underwent family GS (21 trios, three quads) and proband RNA-seq leading to a diagnostic rate of 25.0% (6/24). In two families, the superior coverage of GS compared to ES led to the identification of a pathogenic variant. In a child with hypertelorism, congenital glaucoma, anterior segment dysgenesis, and tetralogy of Fallot, a pathogenic variant in FOXC1 was identified after negative sES and family ES in two centres (FAM11, Supp Table 2). The causative FOXC1 variant was not found on review of the ES data likely because the high guanine-cytosine (G-C) content of the region makes it challenging to detect with ES. This was the only family in our cohort with an affected parent, which modified our variant search strategy with family sequencing data. In a child with severe global developmental delay, cortical visual impairment, and infantile spasms, a de novo missense variant in the mitochondrial gene MT-ND1 was identified by updated bioinformatic analysis using family GS data (FAM149, Supp Table 2).
RNA-seq was instrumental in resolving synonymous variants or deep intronic variants that led to aberrant splicing in the remaining four diagnoses or strong candidate diagnoses. Firstly, in a child with severe global developmental delay, microcephaly, diaphragmatic hernia, and talipes equinovarus born to non-consanguineous parents, RNA-seq on fibroblasts identified aberrant splicing and reduced expression of TRAPPC4 (FAM27, Supp Table 2).37 A homozygous intronic variant resulted in either exon 3 skipping or a 40 nucleotide extension of exon 3; both aberrant transcripts were predicted to undergo nonsense-mediated decay. Secondly, in a child with optic atrophy, ophthalmoplegia, global developmental delay, episodic ataxia, and symmetrical lesions in the brainstem and thalami on brain MRI and normal respiratory chain enzyme analysis on muscle, a missense variant and a synonymous splice site variant in NDUFV1 were detected in a compound heterozygous state by family GS (FAM42, Supp Table 2). RNA-seq on fibroblasts demonstrated aberrant splicing with skipping of exon 8 leading to frameshift. This, combined with fibroblast respiratory chain enzyme analysis confirmed the diagnosis of mitochondrial complex 1 deficiency [MIM:618225]. Thirdly, in siblings with global developmental delay, microcephaly, sensorineural hearing impairment, radial ray defect, and abnormal signal intensities on brain MRI born of fourth-degree consanguineous parents, RNA-seq detected an aberrant splicing event implicating a homozygous deep intronic variant in NDUFB10 (FAM4, Supp Table 2) leading to inclusion of a cryptic exon and frameshift resulting in nonsense-mediated decay.27 Finally, in siblings with microcephaly, severe intellectual disability, and ataxia born to healthy unaffected parents, compound heterozygous variants (c.6625C>T; p.(Arg2209Ter) and c.2610+5G>A) were identified in a strong candidate novel disease-gene (TPR, FAM29). Both variants were initially identified on family ES and family GS, however RNA-seq was able to show that the c.2610+5G>A variant disrupted splicing and shortening of exon 20, suggesting that the phenotype might be related to loss-of-function (van Bergen et al, manuscript in preparation).
High-resolution CMA
Of the 64 diagnoses or strong candidate diagnoses, two molecular diagnoses were made by high-resolution CMA, neither of which were detected on standard CMA (Figure 2). High-resolution CMA was performed in 38 families, yielding a diagnostic rate of 5.2% (2/38). Firstly, a deletion in exon 10 of MSL3 (FAM18, Supp Table 2) identified on systematic review of X chromosome copy number losses in a male, in conjunction with global data sharing, led to collaboration and identification of a novel disease-gene association.24 Secondly, in a male in late adolescence with clinical features suggestive of Rubenstein-Taybi syndrome [MIM #180849], we identified a de novo chromosome 16p13.3 intragenic deletion of exon 2 of CREBBP. Both diagnoses had been missed by ES; however, in both cases, the family ES was conducted prior to the implementation of our CNV-calling pipeline. Retrospective application of CNV-calling confirmed the deletion was visible in the family ES data of both families.
ES reanalysis
Finally, of the 64 diagnoses or strong candidate diagnoses, two were made only by reanalysis of previously generated singleton ES data (Figure 2). Singleton ES reanalysis was undertaken in an ad hoc manner in our cohort, usually by their recruiting clinical geneticist, and only performed in 15 probands, leading to a diagnostic rate of 13.3%. The primary ES data were reanalysed one to 17 months after the initial negative ES result. Firstly, one diagnostic variant in SLC2A1 (FAM78, Supp Table 2) was identified after updating the phenotypic data with the primary clinician. Secondly, in a female toddler with global developmental delay, microcephaly, spasticity, and abnormality of the cerebral white matter, a strong candidate variant in PYCR3 was identified after an analysis of clinical singleton ES data was expanded to investigate potential novel disease-genes utilizing a research-based approach (FAM81, Supp Table 2). The variant was established to be de novo by Sanger sequencing.
Diagnostic odyssey
We sought to understand the diagnostic trajectories of families recruited to UDP-Vic in order to inform our counselling and management of expectations. The median time from the negative NGS result before UDP-Vic recruitment to the establishment of a diagnosis or strong candidate diagnosis was 1.42 years (IQR 1.03–2.48 years). The median time from first genetics consultation to diagnosis or strong candidate diagnosis was 5.14 years (IQR 2.71–7.31 years). As families recruited early in the research program have been studied for longer periods, each intake year has reached a higher diagnostic yield than the following (55.0% for 2016, 52.8% for 2017, 24.6% for 2018) (Figure 1).
Data sharing
Data sharing, primarily through Matchmaker Exchange,10 was undertaken for variants in 72 (48.0%) families identified by research-based sequencing, of which 51 (70.8%) families reached at least one match. Of all 26 novel diagnoses and strong candidate diagnoses (variants in novel disease-genes and known genes with novel phenotypes), 19 (73.1%) potentially causative variants were matched and pathogenicity confirmed in unrelated individuals using Matchmaker Exchange. Matches through international data sharing networks, have led to 10 publications to date.22–24 26 28–30 33 34 38 The remaining have functional studies underway or are the subject of manuscripts in preparation.
DISCUSSION
We report here on the first three years, 150 families enrolled, and 64 (42.7%) diagnoses or strong candidate diagnoses in our Undiagnosed Diseases Program, considering both our successes and potential areas for improvement. By analysing each of our testing strategies, our organizational and personnel structure, and the timelines involved in testing, we share the lessons we have learned from the program.
Lesson 1: Flexible implementation of testing strategies allowed for scalability and response to the availability of new technologies
The stepwise approach utilizing multiple testing methodologies developed iteratively as the program grew in scope and provided multiple benefits. Firstly, it allowed for improvements in the scalability of the process. The UDP-Vic requires considerable resources as clinicians, analysts, and other researchers are extensively involved in all stages from recruitment to diagnosis. By careful selection of a subset of the cohort to access more resource-intensive testing, such as family GS and RNA-seq, we were able to scale the program quickly without overwhelming our systems. Secondly, it allowed the program to respond as different technologies emerged or became achievable in our setting. This shift was clearly observed when high-resolution CMAs became available in our centre. We successfully demonstrated their utility in detecting smaller CNVs missed by ES in two probands in our first intake cohort, such that by the third intake year, high-resolution CMA analysis was frequently applied within our centre earlier in the diagnostic workup before family ES. This iterative and adaptive approach was also found to be successful in the study of the Duke/Columbia site of the UDN.16 Looking towards the future, as family ES becomes more widely available as a first-line clinical test in suspected rare genetic disease,39 we anticipate a shift of the UDP-Vic to perform family GS with RNA-seq after negative family ES and copy number analysis.
Lesson 2: Family ES with research-based analysis provided the highest diagnostic yield
We found family ES to achieve a high diagnostic yield (38.0%) in our primarily singleton ES-negative population. While this study was not designed to directly compare rates between testing methodologies, family ES was effective at prioritising de novo variants and thus allowed for diagnostic improvements that justified its use instead of solely employing ES-reanalysis or moving directly to family GS. Such was the case in the diagnosis of a de novo variant in USP9X (FAM16, Supp Table 2), a diagnosis where the use of family ES added additional diagnostic evidence of a variant that is an in-frame deletion in an X-chromosome gene in a female. Family ES was especially helpful in identifying novel disease-gene candidates that could be considered for submission to data sharing services. ES-reanalysis also proved useful with our diagnostic rate of 13.3% similar to the median rate of 15% reported by others and reviewed previously.40 As seven diagnoses made by family ES were aided by new information published following an initial negative clinical exome, reanalysis of singleton ES data after 18 months could be an alternative strategy to family ES in cost-constrained settings.
Whilst consensus exists that ES is a first line diagnostic tool for certain phenotypes,39 the choice of either singleton or family-based sequencing strategy is likely to be context-dependent.41 In our centre clinical singleton ES achieves a reasonably high diagnostic rate (52–57.5%)7 8 and family ES is less frequently deployed as first-line due to cost considerations. However, with the lessons learned in the UDP-Vic and incremental gains in diagnostic yield from family ES,42 our practice continues to evolve. Where funding allows for family ES as a diagnostic test, data from this study and others would support its use, especially for undifferentiated and complex phenotypes.
After a negative family ES result, GS was most useful in detecting causative variants when coupled with RNA-seq. This combined approach allowed the correlation of aberrant splicing events with deep intronic variants not detectable by family ES. Only two of the six diagnoses made by family GS did not require RNA-seq. We were surprised to not identify more individuals with pathogenic CNVs on GS data but this may change as we increasingly utilise this strategy. The incremental diagnostic potential of GS over ES43 comes at a cost, both financially and in data processing resources.44 As GS reference libraries and pipelines continue to improve, family GS with RNA-seq may become the research test of choice, particularly as family ES is increasingly used in clinical settings.
Lesson 3: RNA-seq offers utility in family ES-negative populations
RNA-seq played two important roles in advancing our path towards diagnosis in some individuals. Firstly, similar to the University of California-Los Angeles clinical site of the UDN,15 it was useful in investigating variants of uncertain significance from ES or GS by assisting in the interpretation and validation of abnormal splicing due to intronic or splice site variants. Secondly, RNA-seq was able to act as an a priori diagnostic tool when no candidate had previously been proposed by ES or GS, as was the case for the deep intronic variant in NDUFB10 (FAM4, Supp Table 2). RNA-seq has a limitation of often requiring biologically relevant tissues to be sampled, and in our cohort introduced delays and invasive biopsies in acquiring fibroblast samples from probands. Given our predominantly neurodevelopmental cohort, we may not be interrogating the most appropriate tissue (nervous tissue), and we may increase the diagnostic yield of RNA-seq if this tissue was accessible. Regardless, we highlight the benefits of utilizing RNA-seq in family ES negative populations within a UDP.
Lesson 4: Analysis informed by deep phenotyping and knowledge of disease mechanisms underpinned many of our novel disease-gene discoveries
Many of our novel discoveries were clinically driven and supported by expertise in disease mechanisms. Such was the case in our diagnosis of biallelic variants in ADARB1 (FAM8, Supp Table 2). The proband’s phenotype of microcephaly, severe global developmental delay, and seizures had sufficient overlap with other RNA editing syndromes,45 leading to the recognition of ADARB1, which has a role in RNA editing, as a clear candidate. Data sharing and international collaboration led to a novel disease-gene discovery.28 Similarly, in another proband whose phenotypic features were strongly suggestive of a ciliopathy, the homozygous variant in SMO, a gene implicated in SHH-GLI signalling, was considered a novel candidate to pursue even though mosaic mono-allelic variants in SMO had previously been associated with Curry-Jones syndrome [MIM #601707]. This again led to international collaboration and the discovery of a novel phenotype associated with biallelic variants in SMO (FAM7, Supp Table 2).33
Lesson 5: Data sharing was critical in facilitating rapid collaborations to establish novel disease-gene associations
Research-based sequencing with the use of data-sharing tools and functional work was able to accelerate novel discoveries. In particular, we found that international collaboration via Matchmaker Exchange contributed to 19 diagnoses or strong candidate diagnoses, similar to other heterogeneous populations with suspected Mendelian disease.46 For the three GUS and other potential variants in undiagnosed families, data sharing strategies will be a critical tool in order to reach a diagnosis. Integrating these translational tools into a UDP workflow is critical to the acceleration of novel discoveries, an essential component in the diagnosis of rare diseases.
Lesson 6: Allowing for the passage of time, and the rapid change in rare-disease literature, led to additional diagnoses in families previously without a diagnosis
The diagnosis of rare diseases is a rapidly moving field, with ongoing novel disease-gene discoveries, disease-gene phenotype expansions, improvements in referral pathways and systems, and implementation of novel technologies. Seven of our diagnoses in known genes were made in genes associated with human disease after the previous negative exome and before resequencing within the UDP-Vic. The cutting-edge nature of rare disease research is such that novel discoveries are continually being made.47 Two of our diagnoses with novel disease genes were concurrently being published by external groups at the time of matching via data sharing networks, and these manuscripts had sufficiently progressed such that the UDP-Vic was not part of their publication or discoveries. While we did not contribute to these novel manuscripts, we mention these diagnoses to highlight the timely nature of novel discoveries and the rate of change within the field.
The time that passed from initial clinical exome to research-based analysis allowed for this new literature to be published, and additionally allowed new clinical information to manifest, potential candidates to be analysed over multiple multidisciplinary meetings, matching on data-sharing platforms, exchange of information between multiple international research groups, and functional studies to be performed. Our diagnostic rate of probands in the first intake year continued to increase in year three and will likely continue to increase.46 While our time to diagnosis falls short of the IRDiRC’s goal of within one year if their disorder is present in the medical literature,4 programs such as the UDP-Vic pave the way for clinical translation of multifaceted analytic strategies in rare disease diagnostics. Improvements in diagnostic timelines may now lie at two bottlenecks – recognition of rare disease by general clinicians and thus early referral to genetics centres, a timeline which has not been measured in this study, and efficiently improving diagnostic pathways within such programs.
Lesson 7: Multidisciplinary expertise was fundamental in having diverse perspectives and strategic decision-making
Recruitment, data analysis, and testing strategy decision-making occurred within a multidisciplinary team. Such a process allowed for a wide range of specialist perspectives and different approaches tailored to the undiagnosed case. Inclusion of the treating clinician in variant curation increases the diagnostic yield owing to a deep knowledge of the phenotype, as well as centralised analysis with access to phenotypic data via medical records.48 49 Expanding this to subspecialist analysis by bioinformaticians and cytogeneticists enriched our diagnostic capacities. An example of this was FAM18, who remained undiagnosed after singleton and family ES. Analysis by high-resolution CMA and manual analysis by a cytogeneticist on our team led to the recognition of a novel X chromosome CNV in MSL3, a novel gene at the time. Our multidisciplinary international team met monthly, and all processes were embedded within our clinical service, facilitating communication and rapid translation of findings. This also led to upskilling of all team members, including trainees, in the different strategies required for successful outcomes.
Limitations
We have reported our experiences with our UDP and acknowledge our approach involves a relatively small number of primarily paediatric patients with syndromic neurodevelopmental phenotypes. While the aim was to select patients most likely to benefit from such a program, diagnostic yields may not be replicable at other services with phenotypically different patients. Our cohort was already extensively investigated with the majority (96%) having already received a negative singleton ES result, a point of difference compared to many UDPs. Our genetic investigations occurred over an extended time course and were resource-intensive, a process that requires substantial investment for any clinical service. Additionally, comparisons of diagnostic yields per genetic testing strategy are difficult given different subsets of the cohort had undergone each test. In its current format, scalability is a major hurdle to overcome as our clinical and research teams are small, and few steps in our process are automated.
Ongoing diagnostic odyssey
There is little consensus on the optimal strategies for the remaining 86 families of our cohort who remain without a diagnosis.50 For some, continued ES-reanalysis with research-based strategies and data-sharing networks may reach a molecular diagnosis.51 For others, many potential strategies may be on the horizon. While RNA-seq is particularly suited to gene expression analysis, other ‘omics’ analytic modalities could complement genomic testing where gene expression is not affected. Proteomics, metabolomics, and epigenomics could all play adjunctive roles in improving the likelihood of elucidating the diagnostic cause.52 53 Alternatively, long-read genome sequencing (LRS) promises to address the shortcomings of short-read sequencing technologies.54
CONCLUSION
UDPs provide a concentration of expertise – structural, technological, and workforce - in improving outcomes in the complex task of diagnosing rare diseases. Our experience in the first three years of the UDP-Vic suggest that (i) stepwise approaches are useful in the flexibility and scalability of a UDP allowing for the incorporation of new testing modalities over time; (ii) family ES is highly effective in achieving a diagnosis following a negative singleton ES result; (iii) RNA-seq in conjunction with GS offers significant benefit after a negative family ES result, but the incremental diagnostic gain of family GS alone requires further investigation; (iv) in-built adjunctive research-based methods such as the use of international data-sharing strategies and confirmatory functional studies are critical to novel diagnoses; (v) extended timelines may be necessary for some novel diagnoses; and finally, (vi) diverse multidisciplinary perspectives were fundamental to strategic decision-making about diagnostic approaches to each case. For those who remain undiagnosed even after the above methodologies, future technology such as LRS and the promise of ‘omics’ technologies provide significant hope. Further research is necessary on the impacts of a diagnosis on medical management from such programs, the psychosocial impacts such programs have on their participants, and economic analysis comparing our approach of implementing a UDP fully integrated with an outpatient genetics service compared to necessitating travel to dedicated centers such as those of the UDN for completion of a detailed research protocol.16 17 Our findings provide additional perspectives to implementing a program in the diagnosis of rare genetic disease, emphasizes the high diagnostic rate after negative singleton ES achievable in a UDP, and highlights the value of a multidisciplinary team utilizing different diagnostic technologies whilst fully interdigitated with the clinical service.
Supplementary Material
ACKNOWLEDGMENTS
Additionally, GATK-gCNV calls on ES data were kindly generated by Isaac Wong, Jack Fu, Harrison Brand, and Michael Talkowski.
FUNDING
Funding for sequencing and analysis was provided by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung and Blood Institute Center for Mendelian Genomics grant (UM1 HG008900) and by National Human Genome Research Institute grant R01 HG009141.
We acknowledge financial support from the Murdoch Children’s Research Institute and the Harbig Foundation. Research conducted at the Murdoch Children’s Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program.
The Illumina Omni 5M SNP platform was donated at no cost by Illumina Australia.
This work was completed as fulfillment of TC’s University of Melbourne MD Research Project.
Footnotes
ETHICS DECLIRATION
Ethics approval was granted by the Royal Children’s Hospital Ethics Committee (HREC 36291A). Written informed consent was obtained from each family.
DATA SHARING
Data are available upon reasonable request. Please contact the corresponding author for data sharing requests.
COMPETING INTERESTS
None declared
References
- 1.Gahl WA, Mulvihill JJ, Toro C, Markello TC, Wise AL, Ramoni RB, Adams DR, Tifft CJ. The NIH Undiagnosed Diseases Program and Network: Applications to modern medicine. Mol Genet Metab 2016;117(4):393–400 doi: 10.1016/j.ymgme.2016.01.007published Online First: 2016/02/06]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Splinter K, Adams DR, Bacino CA, Bellen HJ, Bernstein JA, Cheatle-Jarvela AM, Eng CM, Esteves C, Gahl WA, Hamid R, Jacob HJ, Kikani B, Koeller DM, Kohane IS, Lee BH, Loscalzo J, Luo X, McCray AT, Metz TO, Mulvihill JJ, Nelson SF, Palmer CGS, Phillips JA, Pick L, Postlethwait JH, Reuter C, Shashi V, Sweetser DA, Tifft CJ, Walley NM, Wangler MF, Westerfield M, Wheeler MT, Wise AL, Worthey EA, Yamamoto S, Ashley EA. Effect of genetic diagnosis on patients with previously undiagnosed disease. N Engl J Med 2018;379(22):2131–39 doi: 10.1056/NEJMoa1714458published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Posey JE, O’Donnell-Luria AH, Chong JX, Harel T, Jhangiani SN, Coban Akdemir ZH, Buyske S, Pehlivan D, Carvalho CMB, Baxter S, Sobreira N, Liu P, Wu N, Rosenfeld JA, Kumar S, Avramopoulos D, White JJ, Doheny KF, Witmer PD, Boehm C, Sutton VR, Muzny DM, Boerwinkle E, Gunel M, Nickerson DA, Mane S, MacArthur DG, Gibbs RA, Hamosh A, Lifton RP, Matise TC, Rehm HL, Gerstein M, Bamshad MJ, Valle D, Lupski JR, Centers for Mendelian G. Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet Med 2019;21(4):798–812 doi: 10.1038/s41436-018-0408-7published Online First: 2019/01/19]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Austin CP, Cutillo CM, Lau LPL, Jonker AH, Rath A, Julkowska D, Thomson D, Terry SF, de Montleau B, Ardigo D, Hivert V, Boycott KM, Baynam G, Kaufmann P, Taruscio D, Lochmuller H, Suematsu M, Incerti C, Draghia-Akli R, Norstedt I, Wang L, Dawkins HJS, International Rare Diseases Research C. Future of Rare Diseases Research 2017–2027: An IRDiRC Perspective. Clin Transl Sci 2018;11(1):21–27 doi: 10.1111/cts.12500published Online First: 2017/08/11]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark MM, Stark Z, Farnaes L, Tan TY, White SM, Dimmock D, Kingsmore SF. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom Med 2018;3:16 Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith HS, Swint JM, Lalani SR, Yamal JM, de Oliveira Otto MC, Castellanos S, Taylor A, Lee BH, Russell HV. Clinical Application of Genome and Exome Sequencing as a Diagnostic Tool for Pediatric Patients: a Scoping Review of the Literature. Genet Med 2019;21(1):3–16 doi: 10.1038/s41436-018-0024-6published Online First. [DOI] [PubMed] [Google Scholar]
- 7.Stark Z, Tan TY, Chong B, Brett GR, Yap P, Walsh M, Yeung A, Peters H, Mordaunt D, Cowie S, Amor DJ, Savarirayan R, McGillivray G, Downie L, Ekert PG, Theda C, James PA, Yaplito-Lee J, Ryan MM, Leventer RJ, Creed E, Macciocca I, Bell KM, Oshlack A, Sadedin S, Georgeson P, Anderson C, Thorne N, Melbourne Genomics Health A, Gaff C, White SM. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med 2016;18(11):1090–96 Online First. [DOI] [PubMed] [Google Scholar]
- 8.Tan TY, Dillon OJ, Stark Z, Schofield D, Alam K, Shrestha R, Chong B, Phelan D, Brett GR, Creed E, Jarmolowicz A, Yap P, Walsh M, Downie L, Amor DJ, Savarirayan R, McGillivray G, Yeung A, Peters H, Robertson SJ, Robinson AJ, Macciocca I, Sadedin S, Bell K, Oshlack A, Georgeson P, Thorne N, Gaff C, White SM. Diagnostic Impact and Cost-effectiveness of Whole-Exome Sequencing for Ambulant Children With Suspected Monogenic Conditions. JAMA Pediatr 2017;171(9):855–62 doi: 10.1001/jamapediatrics.2017.1755published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boycott KM, Rath A, Chong JX, Hartley T, Alkuraya FS, Baynam G, Brookes AJ, Brudno M, Carracedo A, den Dunnen JT, Dyke SOM, Estivill X, Goldblatt J, Gonthier C, Groft SC, Gut I, Hamosh A, Hieter P, Hohn S, Hurles ME, Kaufmann P, Knoppers BM, Krischer JP, Macek M Jr., Matthijs G, Olry A, Parker S, Paschall J, Philippakis AA, Rehm HL, Robinson PN, Sham PC, Stefanov R, Taruscio D, Unni D, Vanstone MR, Zhang F, Brunner H, Bamshad MJ, Lochmuller H. International Cooperation to Enable the Diagnosis of All Rare Genetic Diseases. Am J Hum Genet 2017;100(5):695–705 doi: 10.1016/j.ajhg.2017.04.003published Online First: 2017/05/06]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, Brunner HG, Buske OJ, Carey K, Doll C. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum Mutat 2015;36(10):915–21 Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boycott KM, Campeau PM, Howley HE, Pavlidis P, Rogic S, Oriel C, Berman JN, Hamilton RM, Hicks GG, Lipshitz HD, Masson JY, Shoubridge EA, Junker A, Leroux MR, McMaster CR, Michaud JL, Turvey SE, Dyment D, Innes AM, van Karnebeek CD, Lehman A, Cohn RD, MacDonald IM, Rachubinski RA, Frosk P, Vandersteen A, Wozniak RW, Pena IA, Wen XY, Lacaze-Masmonteil T, Rankin C, Hieter P. The Canadian Rare Diseases Models and Mechanisms (RDMM) Network: Connecting Understudied Genes to Model Organisms. Am J Hum Genet 2020;106(2):143–52 doi: 10.1016/j.ajhg.2020.01.009published Online First: 2020/02/08]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Martin E, Martinez-Delgado B, Bermejo-Sanchez E, Alonso J, Spain UDPN, Posada M. SpainUDP: The Spanish Undiagnosed Rare Diseases Program. Int J Environ Res Public Health 2018;15(8) doi: 10.3390/ijerph15081746published Online First: 2018/08/17]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SY, Lim BC, Lee JS, Kim WJ, Kim H, Ko JM, Kim KJ, Choi SA, Kim H, Hwang H, Choi JE, Cho A, Moon J, Seong MW, Park SS, Lee YJ, Kim YO, Kim JS, Kim WS, Kwon YS, Park JD, Ahn Y, Hwang JY, Park HY, Lee Y, Choi M, Chae JH. The Korean undiagnosed diseases program: lessons from a one-year pilot project. Orphanet J Rare Dis 2019;14(1):68 doi: 10.1186/s13023-019-1041-5published Online First: 2019/03/22]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baynam G, Broley S, Bauskis A, Pachter N, McKenzie F, Townshend S, Slee J, Kiraly-Borri C, Vasudevan A, Hawkins A, Schofield L, Helmholz P, Palmer R, Kung S, Walker CE, Molster C, Lewis B, Mina K, Beilby J, Pathak G, Poulton C, Groza T, Zankl A, Roscioli T, Dinger ME, Mattick JS, Gahl W, Groft S, Tifft C, Taruscio D, Lasko P, Kosaki K, Wilhelm H, Melegh B, Carapetis J, Jana S, Chaney G, Johns A, Owen PW, Daly F, Weeramanthri T, Dawkins H, Goldblatt J. Initiating an undiagnosed diseases program in the Western Australian public health system. Orphanet J Rare Dis 2017;12(1):83 doi: 10.1186/s13023-017-0619-zpublished Online First: 2017/05/05]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H, Huang AY, Wang LK, Yoon AJ, Renteria G, Eskin A, Signer RH, Dorrani N, Nieves-Rodriguez S, Wan J, Douine ED, Woods JD, Dell’Angelica EC, Fogel BL, Martin MG, Butte MJ, Parker NH, Wang RT, Shieh PB, Wong DA, Gallant N, Singh KE, Tavyev Asher YJ, Sinsheimer JS, Krakow D, Loo SK, Allard P, Papp JC, Undiagnosed Diseases N, Palmer CGS, Martinez-Agosto JA, Nelson SF. Diagnostic utility of transcriptome sequencing for rare Mendelian diseases. Genet Med 2020;22(3):490–99 doi: 10.1038/s41436-019-0672-1published Online First: 2019/10/15]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shashi V, Schoch K, Spillmann R, Cope H, Tan QK, Walley N, Pena L, McConkie-Rosell A, Jiang YH, Stong N, Need AC, Goldstein DB, Undiagnosed Diseases N. A comprehensive iterative approach is highly effective in diagnosing individuals who are exome negative. Genet Med 2019;21(1):161–72 doi: 10.1038/s41436-018-0044-2published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoch K, Esteves C, Bican A, Spillmann R, Cope H, McConkie-Rosell A, Walley N, Fernandez L, Kohler JN, Bonner D, Reuter C, Stong N, Mulvihill JJ, Novacic D, Wolfe L, Abdelbaki A, Toro C, Tifft C, Malicdan M, Gahl W, Liu P, Newman J, Goldstein DB, Hom J, Sampson J, Wheeler MT, Undiagnosed Diseases N, Cogan J, Bernstein JA, Adams DR, McCray AT, Shashi V. Clinical sites of the Undiagnosed Diseases Network: unique contributions to genomic medicine and science. Genet Med 2021;23(2):259–71 doi: 10.1038/s41436-020-00984-zpublished Online First: 2020/10/24]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haraksingh RR, Abyzov A, Urban AE. Comprehensive performance comparison of high-resolution array platforms for genome-wide Copy Number Variation (CNV) analysis in humans. BMC Genomics 2017;18(1):321 doi: 10.1186/s12864-017-3658-xpublished Online First: 2017/04/26]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girdea M, Dumitriu S, Fiume M, Bowdin S, Boycott KM, Chenier S, Chitayat D, Faghfoury H, Meyn MS, Ray PN, So J, Stavropoulos DJ, Brudno M. PhenoTips: patient phenotyping software for clinical and research use. Hum Mutat 2013;34(8):1057–65 doi: 10.1002/humu.22347published Online First: 2013/05/03]. [DOI] [PubMed] [Google Scholar]
- 20.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42(2):377–81 Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17(5):405–23 doi: 10.1038/gim.2015.30published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yates TM, Vasudevan PC, Chandler KE, Donnelly DE, Stark Z, Sadedin S, Willoughby J, Broad Center for Mendelian G, study DDD, Balasubramanian M. De novo mutations in HNRNPU result in a neurodevelopmental syndrome. Am J Med Genet A 2017;173(11):3003–12 doi: 10.1002/ajmg.a.38492published Online First: 2017/09/26]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker K, Gordon SL, Melland H, Bumbak F, Scott DJ, Jiang TJ, Owen D, Turner BJ, Boyd SG, Rossi M, Al-Raqad M, Elpeleg O, Peck D, Mancini GMS, Wilke M, Zollino M, Marangi G, Weigand H, Borggraefe I, Haack T, Stark Z, Sadedin S, Broad Center for Mendelian G, Tan TY, Jiang Y, Gibbs RA, Ellingwood S, Amaral M, Kelley W, Kurian MA, Cousin MA, Raymond FL SYT1-associated neurodevelopmental disorder: a case series. Brain 2018;141(9):2576–91 doi: 10.1093/brain/awy209published Online First: 2018/08/15]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basilicata MF, Bruel AL, Semplicio G, Valsecchi CIK, Aktas T, Duffourd Y, Rumpf T, Morton J, Bache I, Szymanski WG, Gilissen C, Vanakker O, Ounap K, Mittler G, van der Burgt I, El Chehadeh S, Cho MT, Pfundt R, Tan TY, Kirchhoff M, Menten B, Vergult S, Lindstrom K, Reis A, Johnson DS, Fryer A, McKay V, Study DDD, Fisher RB, Thauvin-Robinet C, Francis D, Roscioli T, Pajusalu S, Radtke K, Ganesh J, Brunner HG, Wilson M, Faivre L, Kalscheuer VM, Thevenon J, Akhtar A. De novo mutations in MSL3 cause an X-linked syndrome marked by impaired histone H4 lysine 16 acetylation. Nat Genet 2018;50(10):1442–51 doi: 10.1038/s41588-018-0220-ypublished Online First: 2018/09/19]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruszka P, Hu T, Hong S, Signer R, Cogne B, Isidor B, Mazzola SE, Giltay JC, van Gassen KLI, England EM, Pais L, Ockeloen CW, Sanchez-Lara PA, Kinning E, Adams DJ, Treat K, Torres-Martinez W, Bedeschi MF, Iascone M, Blaney S, Bell O, Tan TY, Delrue MA, Jurgens J, Barry BJ, Engle EC, Savage SK, Fleischer N, Martinez-Agosto JA, Boycott K, Zackai EH, Muenke M. Phenotype delineation of ZNF462 related syndrome. Am J Med Genet A 2019;179(10):2075–82 doi: 10.1002/ajmg.a.61306published Online First: 2019/07/31]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castilla-Vallmanya L, Selmer KK, Dimartino C, Rabionet R, Blanco-Sanchez B, Yang S, Reijnders MRF, van Essen AJ, Oufadem M, Vigeland MD, Stadheim B, Houge G, Cox H, Kingston H, Clayton-Smith J, Innis JW, Iascone M, Cereda A, Gabbiadini S, Chung WK, Sanders V, Charrow J, Bryant E, Millichap J, Vitobello A, Thauvin C, Mau-Them FT, Faivre L, Lesca G, Labalme A, Rougeot C, Chatron N, Sanlaville D, Christensen KM, Kirby A, Lewandowski R, Gannaway R, Aly M, Lehman A, Clarke L, Graul-Neumann L, Zweier C, Lessel D, Lozic B, Aukrust I, Peretz R, Stratton R, Smol T, Dieux-Coeslier A, Meira J, Wohler E, Sobreira N, Beaver EM, Heeley J, Briere LC, High FA, Sweetser DA, Walker MA, Keegan CE, Jayakar P, Shinawi M, Kerstjens-Frederikse WS, Earl DL, Siu VM, Reesor E, Yao T, Hegele RA, Vaske OM, Rego S, Undiagnosed Diseases Network CRCC, Shapiro KA, Wong B, Gambello MJ, McDonald M, Karlowicz D, Colombo R, Serretti A, Pais L, O’Donnell-Luria A, Wray A, Sadedin S, Chong B, Tan TY, Christodoulou J, White SM, Slavotinek A, Barbouth D, Morel Swols D, Parisot M, Bole-Feysot C, Nitschke P, Pingault V, Munnich A, Cho MT, Cormier-Daire V, Balcells S, Lyonnet S, Grinberg D, Amiel J, Urreizti R, Gordon CT. Phenotypic spectrum and transcriptomic profile associated with germline variants in TRAF7. Genet Med 2020;22(7):1215–26 doi: 10.1038/s41436-020-0792-7published Online First: 2020/05/08]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helman G, Compton AG, Hock DH, Walkiewicz M, Brett GR, Pais L, Tan TY, De Paoli-Iseppi R, Clark MB, Christodoulou J, White SM, Thorburn DR, Stroud DA, Stark Z, Simons C. Multiomic analysis elucidates Complex I deficiency caused by a deep intronic variant in NDUFB10. Hum Mutat 2020. doi: 10.1002/humu.24135published Online First: 2020/11/11]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan TY, Sedmik J, Fitzgerald MP, Halevy RS, Keegan LP, Helbig I, Basel-Salmon L, Cohen L, Straussberg R, Chung WK, Helal M, Maroofian R, Houlden H, Juusola J, Sadedin S, Pais L, Howell KB, White SM, Christodoulou J, O’Connell MA. Bi-allelic ADARB1 Variants Associated with Microcephaly, Intellectual Disability, and Seizures. Am J Hum Genet 2020;106(4):467–83 doi: 10.1016/j.ajhg.2020.02.015published Online First: 2020/03/30]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen AW, Arora P, Khayat MM, Smith LJ, Lewis AM, Rossetti LZ, Jayaseelan J, Cristian I, Haynes D, DiTroia S, Meeks N, Delgado MR, Rosenfeld JA, Pais L, White SM, Meng Q, Pehlivan D, Liu P, Gingras MC, Wangler MF, Muzny DM, Lupski JR, Kaplan CD, Gibbs RA. Germline mutation in POLR2A: a heterogeneous, multi-systemic developmental disorder characterized by transcriptional dysregulation. HGG Adv 2021;2(1) doi: 10.1016/j.xhgg.2020.100014published Online First: 2021/03/06]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radio FC, Pang K, Ciolfi A, Levy MA, Hernández-García A, Pedace L, Pantaleoni F, Liu Z, de Boer E, Jackson A, Bruselles A, McConkey H, Stellacci E, Lo Cicero S, Motta M, Carrozzo R, Dentici ML, McWalter K, Desai M, Monaghan KG, Telegrafi A, Philippe C, Vitobello A, Au M, Grand K, Sanchez-Lara PA, Baez J, Lindstrom K, Kulch P, Sebastian J, Madan-Khetarpal S, Roadhouse C, MacKenzie JJ, Monteleone B, Saunders CJ, Jean Cuevas JK, Cross L, Zhou D, Hartley T, Sawyer SL, Monteiro FP, Secches TV, Kok F, Schultz-Rogers LE, Macke EL, Morava E, Klee EW, Kemppainen J, Iascone M, Selicorni A, Tenconi R, Amor DJ, Pais L, Gallacher L, Turnpenny PD, Stals K, Ellard S, Cabet S, Lesca G, Pascal J, Steindl K, Ravid S, Weiss K, Castle AMR, Carter MT, Kalsner L, de Vries BBA, van Bon BW, Wevers MR, Pfundt R, Stegmann APA, Kerr B, Kingston HM, Chandler KE, Sheehan W, Elias AF, Shinde DN, Towne MC, Robin NH, Goodloe D, Vanderver A, Sherbini O, Bluske K, Hagelstrom RT, Zanus C, Faletra F, Musante L, Kurtz-Nelson EC, Earl RK, Anderlid BM, Morin G, van Slegtenhorst M, Diderich KEM, Brooks AS, Gribnau J, Boers RG, Finestra TR, Carter LB, Rauch A, Gasparini P, Boycott KM, Barakat TS, Graham JM Jr., Faivre L, Banka S, Wang T, Eichler EE, Priolo M, Dallapiccola B, Vissers L, Sadikovic B, Scott DA, Holder JL Jr., Tartaglia M. SPEN haploinsufficiency causes a neurodevelopmental disorder overlapping proximal 1p36 deletion syndrome with an episignature of X chromosomes in females. Am J Hum Genet 2021. doi: 10.1016/j.ajhg.2021.01.015published Online First: 2021/02/18]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nahorski MS, Maddirevula S, Ishimura R, Alsahli S, Brady AF, Begemann A, Mizushima T, Guzman-Vega FJ, Obata M, Ichimura Y, Alsaif HS, Anazi S, Ibrahim N, Abdulwahab F, Hashem M, Monies D, Abouelhoda M, Meyer BF, Alfadhel M, Eyaid W, Zweier M, Steindl K, Rauch A, Arold ST, Woods CG, Komatsu M, Alkuraya FS. Biallelic UFM1 and UFC1 mutations expand the essential role of ufmylation in brain development. Brain 2018;141(7):1934–45 doi: 10.1093/brain/awy135published Online First: 2018/06/06]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Straub J, Konrad EDH, Gruner J, Toutain A, Bok LA, Cho MT, Crawford HP, Dubbs H, Douglas G, Jobling R, Johnson D, Krock B, Mikati MA, Nesbitt A, Nicolai J, Phillips M, Poduri A, Ortiz-Gonzalez XR, Powis Z, Santani A, Smith L, Stegmann APA, Stumpel C, Vreeburg M, Deciphering Developmental Disorders S, Fliedner A, Gregor A, Sticht H, Zweier C. Missense Variants in RHOBTB2 Cause a Developmental and Epileptic Encephalopathy in Humans, and Altered Levels Cause Neurological Defects in Drosophila. Am J Hum Genet 2018;102(1):44–57 doi: 10.1016/j.ajhg.2017.11.008published Online First: 2017/12/26]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le TL, Sribudiani Y, Dong X, Huber C, Kois C, Baujat G, Gordon CT, Mayne V, Galmiche L, Serre V, Goudin N, Zarhrate M, Bole-Feysot C, Masson C, Nitschke P, Verheijen FW, Pais L, Pelet A, Sadedin S, Pugh JA, Shur N, White SM, El Chehadeh S, Christodoulou J, Cormier-Daire V, Hofstra RMW, Lyonnet S, Tan TY, Attie-Bitach T, Kerstjens-Frederikse WS, Amiel J, Thomas S. Bi-allelic Variations of SMO in Humans Cause a Broad Spectrum of Developmental Anomalies Due to Abnormal Hedgehog Signaling. Am J Hum Genet 2020;106(6):779–92 doi: 10.1016/j.ajhg.2020.04.010published Online First: 2020/05/16]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenk GM, Berry IR, Stutterd CA, Blyth M, Green L, Vadlamani G, Warren D, Craven I, Fanjul-Fernandez M, Rodriguez-Casero V, Lockhart PJ, Vanderver A, Simons C, Gibb S, Sadedin S, Broad Center for Mendelian G, White SM, Christodoulou J, Skibina O, Ruddle J, Tan TY, Leventer RJ, Livingston JH, Meisler MH Cerebral hypomyelination associated with biallelic variants of FIG4. Hum Mutat 2019;40(5):619–30 doi: 10.1002/humu.23720published Online First: 2019/02/12]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar R, Gardner A, Homan CC, Douglas E, Mefford H, Wieczorek D, Ludecke HJ, Stark Z, Sadedin S, Broad CMG, Nowak CB, Douglas J, Parsons G, Mark P, Loidi L, Herman GE, Mihalic Mosher T, Gillespie MK, Brady L, Tarnopolsky M, Madrigal I, Eiris J, Domenech Salgado L, Rabionet R, Strom TM, Ishihara N, Inagaki H, Kurahashi H, Dudding-Byth T, Palmer EE, Field M, Gecz J. Severe neurocognitive and growth disorders due to variation in THOC2, an essential component of nuclear mRNA export machinery. Hum Mutat 2018;39(8):1126–38 doi: 10.1002/humu.23557published Online First: 2018/06/01]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rius R, Van Bergen NJ, Compton AG, Riley LG, Kava MP, Balasubramaniam S, Amor DJ, Fanjul-Fernandez M, Cowley MJ, Fahey MC, Koenig MK, Enns GM, Sadedin S, Wilson MJ, Tan TY, Thorburn DR, Christodoulou J. Clinical Spectrum and Functional Consequences Associated with Bi-Allelic Pathogenic PNPT1 Variants. J Clin Med 2019;8(11) doi: 10.3390/jcm8112020published Online First: 2019/11/23]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh SG, Scala M, Beetz C, Helman G, Stanley V, Yang X, Breuss MW, Mazaheri N, Selim L, Hadipour F, Pais L, Stutterd CA, Karageorgou V, Begtrup A, Crunk A, Juusola J, Willaert R, Flore LA, Kennelly K, Spencer C, Brown M, Trapane P, Hurst ACE, Lane Rutledge S, Goodloe DH, McDonald MT, Shashi V, Schoch K, Undiagnosed Diseases N, Tomoum H, Zaitoun R, Hadipour Z, Galehdari H, Pagnamenta AT, Mojarrad M, Sedaghat A, Dias P, Quintas S, Eslahi A, Shariati G, Bauer P, Simons C, Houlden H, Issa MY, Zaki MS, Maroofian R, Gleeson JG. A relatively common homozygous TRAPPC4 splicing variant is associated with an early-infantile neurodegenerative syndrome. Eur J Hum Genet 2020. doi: 10.1038/s41431-020-00717-5published Online First: 2020/09/10]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jolly LA, Parnell E, Gardner AE, Corbett MA, Perez-Jurado LA, Shaw M, Lesca G, Keegan C, Schneider MC, Griffin E, Maier F, Kiss C, Guerin A, Crosby K, Rosenbaum K, Tanpaiboon P, Whalen S, Keren B, McCarrier J, Basel D, Sadedin S, White SM, Delatycki MB, Kleefstra T, Kury S, Brusco A, Sukarova-Angelovska E, Trajkova S, Yoon S, Wood SA, Piper M, Penzes P, Gecz J. Missense variant contribution to USP9X-female syndrome. NPJ Genom Med 2020;5(1):53 doi: 10.1038/s41525-020-00162-9published Online First: 2020/12/11]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srivastava S, Love-Nichols JA, Dies KA, Ledbetter DH, Martin CL, Chung WK, Firth HV, Frazier T, Hansen RL, Prock L, Brunner H, Hoang N, Scherer SW, Sahin M, Miller DT, Group NDDESRW. Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet Med 2019. doi: 10.1038/s41436-019-0554-6published Online First: 2019/06/12]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan NB, Stapleton R, Stark Z, Delatycki MB, Yeung A, Hunter MF, Amor DJ, Brown NJ, Stutterd CA, McGillivray G, Yap P, Regan M, Chong B, Fanjul Fernandez M, Marum J, Phelan D, Pais LS, White SM, Lunke S, Tan TY. Evaluating systematic reanalysis of clinical genomic data in rare disease from single center experience and literature review. Mol Genet Genomic Med 2020;8(11):e1508 doi: 10.1002/mgg3.1508published Online First: 2020/09/25]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan TY, Lunke S, Chong B, Phelan D, Fanjul-Fernandez M, Marum JE, Kumar VS, Stark Z, Yeung A, Brown NJ, Stutterd C, Delatycki MB, Sadedin S, Martyn M, Goranitis I, Thorne N, Gaff CL, White SM. A head-to-head evaluation of the diagnostic efficacy and costs of trio versus singleton exome sequencing analysis. Eur J Hum Genet 2019;27(12):1791–99 doi: 10.1038/s41431-019-0471-9published Online First: 2019/07/20]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farwell KD, Shahmirzadi L, El-Khechen D, Powis Z, Chao EC, Tippin Davis B, Baxter RM, Zeng W, Mroske C, Parra MC, Gandomi SK, Lu I, Li X, Lu H, Lu HM, Salvador D, Ruble D, Lao M, Fischbach S, Wen J, Lee S, Elliott A, Dunlop CL, Tang S. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med 2015;17(7):578–86 doi: 10.1038/gim.2014.154published Online First: 2014/10/31]. [DOI] [PubMed] [Google Scholar]
- 43.Alfares A, Aloraini T, subaie LA, Alissa A, Qudsi AA, Alahmad A, Mutairi FA, Alswaid A, Alothaim A, Eyaid W, Albalwi M, Alturki S, Alfadhel M. Whole-genome sequencing offers additional but limited clinical utility compared with reanalysis of whole-exome sequencing. Genet Med 2018;20(11):1328–33 doi: 10.1038/gim.2018.41published Online First. [DOI] [PubMed] [Google Scholar]
- 44.Schwarze K, Buchanan J, Taylor JC, Wordsworth S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med 2018;20(10):1122–30 doi: 10.1038/gim.2017.247published Online First: 2018/02/16]. [DOI] [PubMed] [Google Scholar]
- 45.Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nature genetics 2012;44(11):1243–48 Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nambot S, Thevenon J, Kuentz P, Duffourd Y, Tisserant E, Bruel AL, Mosca-Boidron AL, Masurel-Paulet A, Lehalle D, Jean-Marcais N, Lefebvre M, Vabres P, El Chehadeh-Djebbar S, Philippe C, Tran Mau-Them F, St-Onge J, Jouan T, Chevarin M, Poe C, Carmignac V, Vitobello A, Callier P, Riviere JB, Faivre L, Thauvin-Robinet C. Clinical whole-exome sequencing for the diagnosis of rare disorders with congenital anomalies and/or intellectual disability: Substantial interest of prospective annual reanalysis. Genet Med 2018;20(6):645–54 doi: 10.1038/gim.2017.162published Online First. [DOI] [PubMed] [Google Scholar]
- 47.Bamshad MJ, Nickerson DA, Chong JX. Mendelian Gene Discovery: Fast and Furious with No End in Sight. Am J Hum Genet 2019;105(3):448–55 doi: 10.1016/j.ajhg.2019.07.011published Online First: 2019/09/07]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baldridge D, Heeley J, Vineyard M, Manwaring L, Toler TL, Fassi E, Fiala E, Brown S, Goss CW, Willing M, Grange DK, Kozel BA, Shinawi M. The Exome Clinic and the role of medical genetics expertise in the interpretation of exome sequencing results. Genet Med 2017;19(9):1040–48 doi: 10.1038/gim.2016.224published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salmon LB, Orenstein N, Markus-Bustani K, Ruhrman-Shahar N, Kilim Y, Magal N, Hubshman MW, Bazak L. Improved diagnostics by exome sequencing following raw data reevaluation by clinical geneticists involved in the medical care of the individuals tested. Genet Med 2018. doi: 10.1038/s41436-018-0343-7published Online First. [DOI] [PubMed] [Google Scholar]
- 50.Fresard L, Montgomery SB. Diagnosing rare diseases after the exome. Cold Spring Harb Mol Case Stud 2018;4(6) doi: 10.1101/mcs.a003392published Online First: 2018/12/19]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zastrow DB, Kohler JN, Bonner D, Reuter CM, Fernandez L, Grove ME, Fisk DG, Undiagnosed Diseases N, Yang Y, Eng CM, Ward PA, Bick D, Worthey EA, Fisher PG, Ashley EA, Bernstein JA, Wheeler MT. A toolkit for genetics providers in follow-up of patients with non-diagnostic exome sequencing. J Genet Couns 2019;28(2):213–28 doi: 10.1002/jgc4.1119published Online First: 2019/04/10]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aref-Eshghi E, Bend EG, Colaiacovo S, Caudle M, Chakrabarti R, Napier M, Brick L, Brady L, Carere DA, Levy MA, Kerkhof J, Stuart A, Saleh M, Beaudet AL, Li C, Kozenko M, Karp N, Prasad C, Siu VM, Tarnopolsky MA, Ainsworth PJ, Lin H, Rodenhiser DI, Krantz ID, Deardorff MA, Schwartz CE, Sadikovic B. Diagnostic Utility of Genome-wide DNA Methylation Testing in Genetically Unsolved Individuals with Suspected Hereditary Conditions. Am J Hum Genet 2019;104(4):685–700 doi: 10.1016/j.ajhg.2019.03.008published Online First: 2019/04/02]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crowther LM, Poms M, Plecko B. Multiomics tools for the diagnosis and treatment of rare neurological disease. J Inherit Metab Dis 2018;41(3):425–34 doi: 10.1007/s10545-018-0154-7published Online First: 2018/03/15]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merker JD, Wenger AM, Sneddon T, Grove M, Zappala Z, Fresard L, Waggott D, Utiramerur S, Hou Y, Smith KS, Montgomery SB, Wheeler M, Buchan JG, Lambert CC, Eng KS, Hickey L, Korlach J, Ford J, Ashley EA. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet Med 2018;20(1):159–63 doi: 10.1038/gim.2017.86published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beaulieu CL, Majewski J, Schwartzentruber J, Samuels ME, Fernandez BA, Bernier FP, Brudno M, Knoppers B, Marcadier J, Dyment D, Adam S, Bulman DE, Jones SJ, Avard D, Nguyen MT, Rousseau F, Marshall C, Wintle RF, Shen Y, Scherer SW, Consortium FC, Friedman JM, Michaud JL, Boycott KM. FORGE Canada Consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am J Hum Genet 2014;94(6):809–17 doi: 10.1016/j.ajhg.2014.05.003published Online First: 2014/06/07]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sawyer SL, Hartley T, Dyment DA, Beaulieu CL, Schwartzentruber J, Smith A, Bedford HM, Bernard G, Bernier FP, Brais B, Bulman DE, Warman Chardon J, Chitayat D, Deladoey J, Fernandez BA, Frosk P, Geraghty MT, Gerull B, Gibson W, Gow RM, Graham GE, Green JS, Heon E, Horvath G, Innes AM, Jabado N, Kim RH, Koenekoop RK, Khan A, Lehmann OJ, Mendoza-Londono R, Michaud JL, Nikkel SM, Penney LS, Polychronakos C, Richer J, Rouleau GA, Samuels ME, Siu VM, Suchowersky O, Tarnopolsky MA, Yoon G, Zahir FR, Majewski J, Boycott KM. Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: Time to address gaps in care. Clin Genet 2016;89(3):275–84 doi: 10.1111/cge.12654published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wright CF, McRae JF, Clayton S, Gallone G, Aitken S, FitzGerald TW, Jones P, Prigmore E, Rajan D, Lord J, Sifrim A, Kelsell R, Parker MJ, Barrett JC, Hurles ME, FitzPatrick DR, Firth HV. Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders. Genet Med 2018;20(10):1216–23 doi: 10.1038/gim.2017.246published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deciphering Developmental Disorders S. Large-scale discovery of novel genetic causes of developmental disorders. Nature 2015;519(7542):223–8 doi: 10.1038/nature14135published Online First: 2014/12/24]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dyment DA, Tetreault M, Beaulieu CL, Hartley T, Ferreira P, Chardon JW, Marcadier J, Sawyer SL, Mosca SJ, Innes AM, Parboosingh JS, Bulman DE, Schwartzentruber J, Majewski J, Tarnopolsky M, Boycott KM, Consortium FC, Care4Rare C. Whole-exome sequencing broadens the phenotypic spectrum of rare pediatric epilepsy: a retrospective study. Clin Genet 2015;88(1):34–40 doi: 10.1111/cge.12464published Online First: 2014/07/22]. [DOI] [PubMed] [Google Scholar]
- 60.Salvatore M, Polizzi A, De Stefano MC, Floridia G, Baldovino S, Roccatello D, Sciascia S, Menegatti E, Remuzzi G, Daina E, Iatropoulos P, Bembi B, Da Riol RM, Ferlini A, Neri M, Novelli G, Sangiuolo F, Brancati F, Taruscio D. Improving diagnosis for rare diseases: the experience of the Italian undiagnosed Rare diseases network. Ital J Pediatr 2020;46(1):130 doi: 10.1186/s13052-020-00883-8published Online First: 2020/09/16]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adachi T, Kawamura K, Furusawa Y, Nishizaki Y, Imanishi N, Umehara S, Izumi K, Suematsu M. Japan’s initiative on rare and undiagnosed diseases (IRUD): towards an end to the diagnostic odyssey. Eur J Hum Genet 2017;25(9):1025–28 doi: 10.1038/ejhg.2017.106published Online First: 2017/08/11]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.