Abstract
As a critical member of the ubiquitin-specific proteolytic enzyme family, ubiquitin-specific peptidase 20 (USP20) regulates the stability of proteins via multiple signaling pathways. In addition, USP20 upregulation is associated with various cellular biological processes, such as cell cycle progression, proliferation, migration, and invasion. Emerging studies have revealed the pivotal role of USP20 in the tumorigenesis of various cancer types, such as breast cancer, colon cancer, lung cancer, gastric cancer and adult T cell leukemia. In our review, we highlight the different mechanisms of USP20 in various tumor types and demonstrate that USP20 regulates the stability of multiple proteins. Therefore, regulating the activity of USP20 is a novel tumor treatment. However, the clinical significance of USP20 in cancer treatment merits more evidence. Finally, different prospects exist for the continued research focus of USP20.
Subject terms: Targeted therapies, Ubiquitylation
Facts
USP20 is upregulated in multiple cancer types.
USP20 mainly exhibits a promotive effect in breast cancer, lung cancer, and colon cancer; however, it shows inhibitory effects in gastric cancer.
Regulating the expression of USP20 is a novel treatment for tumors.
Deletions of USP20 exhibit an inhibitory effect on the tumorigenesis of some cancer types.
Open questions
Does USP20 have a common mechanism in tumors?
Can these emerging inhibitors of USP20 be successfully applied soon in clinical practice?
Introduction
The ubiquitin–proteasome system (UPS) is the most important post-transcriptional modification in eukaryotic cells [1, 2]. The components of this system include ubiquitin (ub), E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, E3 ubiquitin ligase and 26S proteasome [3]. Ubiquitylation is a reversible posttranslational modification that plays a role in various biological processes, including protein degradation [4], DNA damage and repair [5], cell cycle progression [6], and immune response [7]. Ubiquitin, comprising 76 amino acids, is highly conserved among eukaryotes and contains seven lysine residues, K6, K11, K27, K29, K33, K48, and K63 [8]. Of these residues, K48 regulates the degradation of target proteins by linking polyubiquitin, K63 increases cell signal transduction and protein kinase activation by linking linear polyubiquitin chains, and the others are linked by mono- or polyubiquitin chains [9]. The UPS includes two steps. First, in ubiquitylation, one or more ubiquitin proteins are added to tag the substrate proteins. Second, the marked proteins will be identified by the 26S proteasome for cleavage, degradation, and recycling [10]. Deubiquitylation is the opposite process of ubiquitylation. Ubiquitination and deubiquitylation are always in a state of dynamic equilibrium [10]. Deubiquitinating enzymes (DUBs) are involved in deubiquitylation, which can rescue the marked substrate proteins by remodeling and removing conjugated ubiquitin chains [11] (Fig. 1A). The balance between ubiquitin enzymes and DUBs ultimately determines the ubiquitination status of a given target protein, making protein ubiquitination a multifunctional and dynamic posttranslational modification. Ubiquitination plays a key role in multiple cellular processes, including gene expression [12], cell cycle progression [13], DNA damage and repair [14], cell growth [15], and apoptosis [16]. These ubiquitination-regulated processes are critical to maintain cellular homeostasis, and abnormal regulation of these processes contributes to tumor development [17, 18]. The importance of ubiquitination in cancer-related cell function and successful use of the proteasome inhibitor bortezomib in multiple myeloma have attracted increased attention concerning the potential of ubiquitination/deubiquitination proteins in tumor therapy [19, 20].
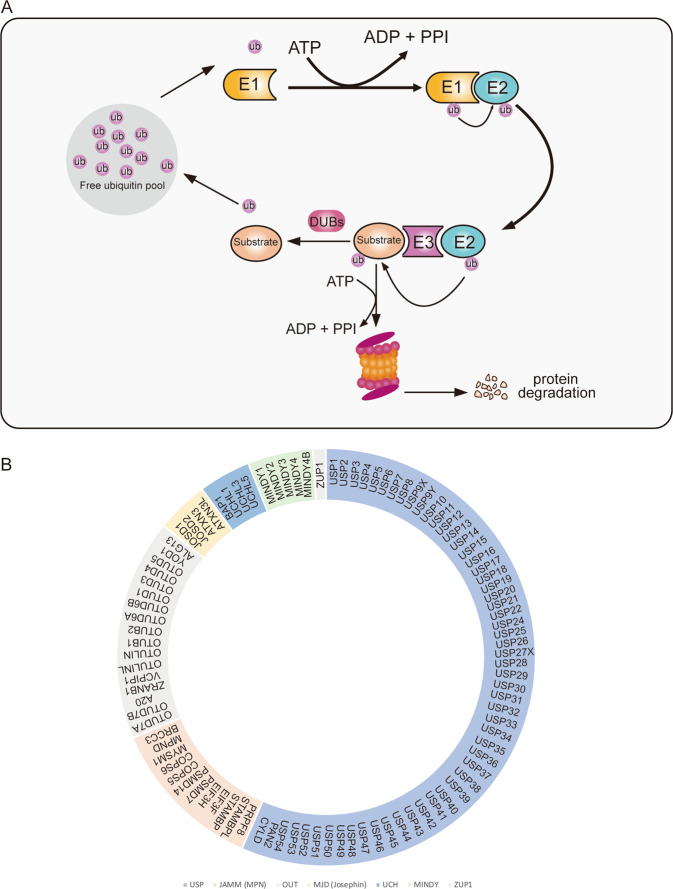
Fig. 1. The ubiquitin–proteasome system cascade and the classification of deubiquitylase family.
A Diagrammatic of key events in ubiquitylation and deubiquitylation. The E1 enzyme activates and combines with ubiquitin moiety in an ATP-dependent manner. Then the ubiquitin moiety is transferred to an E2 conjugating enzyme, Finally, ubiquitin is transferred directly from E2 enzyme to substrate protein by E3 ligase, on the one hand, the labeled protein is degraded by the 26s proteasome. Or the DUBs stabilize the targeted protein by removing the ubiquitin moiety, the ubiquitin becomes free ubiquitin to reuse. B The reported subclass of DUBs including ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), JAMMs (also known as MPN+), MJDs (also known as Josephins), and the two new families: MINDY family and the ZUP1 family.
Recently, an increasing number of studies have demonstrated that DUBs regulate various biological or pathological processes by stabilizing tumor or antitumor proteins [21–24]. As recently described, human DUBs are subdivided into seven families, ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), JAMMs (also known as MPN+ and hereafter referred to as JAMM/MPN+), MJDs (also known as Josephins), the MINDY family and the ZUP1 family (these two new families of DUBs were discovered recently) [25] (Fig. 1B). Among them, USPs are the largest subfamily of DUBs that participate in the progression of multiple tumors [26–29]. Therefore, studies on DUBs as a therapeutic target warrant further exploration. Table 1 shows detailed information about different types of DUBs and their roles in different cancers. Presently, many related inhibitors of DUBs have been used in tumor therapeutic models, further illustrating the potential of DUBs as therapeutic targets. Previous studies have described that ubiquitin-specific peptidase 20 (USP20) plays a critical role in tumorigenesis [30]. In this article, we first briefly discuss the function of USP20, focusing on its increasingly recognized potential as a target in cancer treatment. Finally, we summarize the alterations of USP20 in multiple human cancers and discuss novel findings regarding the potential of this enzyme as a tumor therapeutic.
Table 1.
Other DUBs and cancer.
| DUB | Cancer type | Tumorigenesis | Reference |
|---|---|---|---|
| USP1 | Osteosarcoma | Promotes the invasion of osteosarcoma cells | [84] |
| Glioma | USP1 stabilizes EZH2 to activate β-catenin to drive glioma tumorigenesis | [85] | |
| Breast cancer | Regulates metastasis | [86] | |
| USP2 | Breast cancer | Promotes cell migration and invasion | [87] |
| USP4 | Breast cancer | Promotes invasion | [88] |
| Melanoma | May be an oncogene | [89] | |
| Glioblastoma | Promotes glioblastoma multiforme | [90] | |
| Facilitates chemoresistance | [91] | ||
| USP5 | Pancreatic cancer | Promotes tumorigenesis and progression | [92] |
| Non-small cell lung cancer | Upregulation of USP5 contributes to tumorigenesis | [93] | |
| Colorectal cancer | Promotes cell growth and resistance to chemotherapeutics | [94] | |
| USP7 | Medulloblastoma | Promotes medulloblastoma cell survival and metastasis | [95] |
| Breast cancer | Promotes breast carcinogenesis via stabilizing PHF8 | [96] | |
| Osteosarcoma | Promotes metastasis by inducing EMT | [97] | |
| Lung cancer | Modulates the antitumor immune response | [98] | |
| USP8 | Cervical cancer | Associated with a poor prognosis in cervical squamous cell carcinoma patients | [99] |
| Suppresses apoptosis by stabilizing FLIPL | [100] | ||
| USP10 | Colon cancer | Promotes tumor proliferation | [101] |
| Lung cancer | Inhibits cell proliferation, invasion and cell growth | [102, 103] | |
| The USP10-HDAC6 confers cisplatin resistance | [104] | ||
| Liver cancer | Promotes cell proliferation by stabilizing YAP/TAZ | [105] | |
| Promotes metastasis by stabilizing Smad4 | [106] | ||
| Inhibits p53 Signaling and constricts poor outcome | [107] | ||
| Acute myeloid leukemia | Stabilizes oncogenic FLT3 | [108] | |
| USP11 | Colorectal cancer | Promotes growth and metastasis | [109] |
| Ovarian cancer | Promotes EMT by stabilizing Snail | [110] | |
| USP13 | Non-small cell lung cancer | Promotes tumor progression | [111] |
| USP14 | Breast cancer | Regulates cell cycle progression and cell proliferation and metastasis | [112–114] |
| Enhances sensitivity to enzalutamide | [115] | ||
| USP30 | Liver cancer | Stabilizes DRP1 to promote hepatocarcinogenesis | [116] |
| UCHL1 | Gastric cancer | Promotes metastasis | [117] |
| OTUB2 | Non-small cell lung cancer | Promotes tumorigenesis | [118] |
Structure and function of USP20
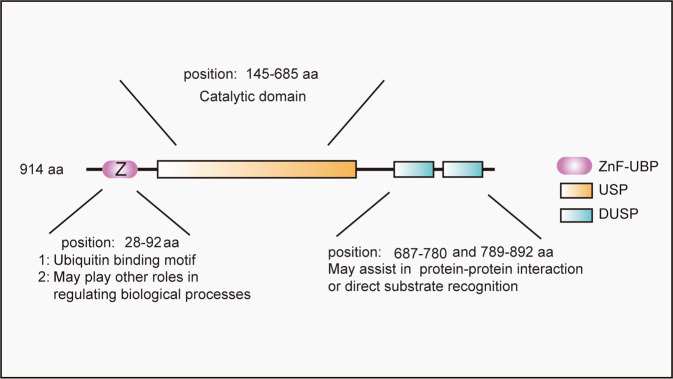
The ubiquitin-specific proteolytic enzyme family is the largest subtype of DUBs identified thus far. USP20, a specific member of this family, is also called pVHL-interacting deubiquitinating enzyme 2 (VDU2) [31, 32], and was first identified as a von Hippel–Lindau (VHL) syndrome-related deubiquitinase [33, 34]. Human USP20 comprises 914 amino acids with a predicted molecular weight of 102 kDa [34]. The primary structure of human USP20 includes an N-terminal zinc finger ubiquitin-specific protease (ZnF-UBP) domain, a USP catalytic domain and two tandem domain present in ubiquitin-specific proteases (DUSP) domains [35] (Fig. 2). The ZnF-UBP domain is also found in some other USPs and is the ubiquitin-binding motif [36]. However, the definite function of this domain of USP20 has not been demonstrated very clearly until now. Yang et al. demonstrated that the ZnF-UBP domain of USP20 presented weak binding capacity to monoubiquitin [34], however, this domain characteristically binds with K48-linked di-ubiquitin [37]. The USP catalytic domain, the structure of which is similar to other members of the USP family, is the most important functional area of USPs and exhibits strong homology in two regions that surround the catalytic Cys box and His box [38]. USP20 comprises three domains that form a shape that looks like the right hand extended. These three domains are similar to the “palm”, “finger” and “thumb” domains. This right-hand-like structure can form a ubiquitin-binding surface that is convenient for ubiquitin binding. The catalytic center is located between the “palm” and “thumb”, and the “hand” holds the ubiquitin molecule of the target proteins. Next, the ubiquitin molecules are removed from the labeled proteins so that the ubiquitin molecules and proteins are recycled and reused [39, 40]. The DUSP domain is a tripod structure similar to AB3 formed by three alpha-helices forming a bundle structure to support three strands of antiparallel beta folds [35]. Studies have reported that the DUSP domain in DUBs may play a crucial role in protein–protein interactions or direct substrate recognition [35]. Kommaddi et al. demonstrated that the phosphorylation status at serine 333 of USP20 is critical for deubiquitinase activity. Subsequently, they found that protein kinase A phosphorylates USP20 on serine 333, inhibits the trafficking of the substrate β2AR (β2 adrenergic receptors) and decreases degradation via autophagosomes [41]. Berthouze et al. also demonstrated that USP20 served as a novel regulator for β2AR recycling and resensitization [42]. Lu et al. demonstrated that mechanistic target of rapamycin complex 1 phosphorylated USP20 at serine 132 and serine 134 and then increased the stability of HMG-CoA reductase (HMGCR), the rate-limiting enzyme in the cholesterol biosynthetic pathway [43].
Fig. 2. The structure of USP20 and the function of each domain of USP20.
USP20 contains three different domains, including ZNF-UBP, USP, and DUSP domain. Those different domains of USP20 play crucial roles in different biological processes.
USP20 as a potential cancer target
As a member of the largest subfamily of DUBs, USP20 plays key roles in various tumors by stabilizing tumorigenic or antitumor proteins. USP20 was first identified as a substrate of the VHL tumor suppressor protein [32]. The pVHL protein is encoded by the VHL gene, which plays a key role in cellular oxygen sensing by ubiquitinating hypoxia-inducing factors and is then degraded by the proteasome [44]. The dysregulation of the pVHL protein is related to VHL (Hippel–Lindau syndrome) disease. This disease is a rare autosomal dominant hereditary tumor syndrome involving multiple systems, manifested as multiple familial benign and malignant tumors and cysts of the central nervous system and internal organs [45–47]. Considering that USP20 is a substrate of the pVHL protein, we speculate that pVHL protein inactivation induces decreased degradation of USP20, promoting the progression of VHL disease. An additional characterized substrate of the pVHL E3 ligase complex is the α-subunit of hypoxia-inducible factor 1 (HIF1α) [48]. Early studies have reported that HIF1α is overexpressed in various cancer types and regulates the expression of most genes involved in many essential biological and pathological processes [49–52]. Li et al. found that USP20 recognizes and binds to HIF1α, removes its ubiquitin chain, and maintains high expression levels of HIF1α, increasing the transcription of hypoxia-inducible element genes [48]. Considering that HIF1α activates the transcription of genes that are involved in crucial aspects of cancer biology, including angiogenesis [53], cell survival [54], glucose metabolism [55], and invasion [56], by triggering multiple signaling pathways, USP20 can regulate multiple biological processes by stabilizing tumorigenic or antitumor proteins. In addition, studies have demonstrated that USP20 triggers the activation of multiple pathways, including Wnt, MAPK, HIF1, NF-κB, cell cycle checkpoint and many other signaling pathways [33, 57–60], promoting the processes of multiple cancer types (Fig. 3). As a results, these findings prompt USP20 may be a potential cancer target.
Fig. 3. The mechanism of deubiquitinating enzyme USP20 in cancer.
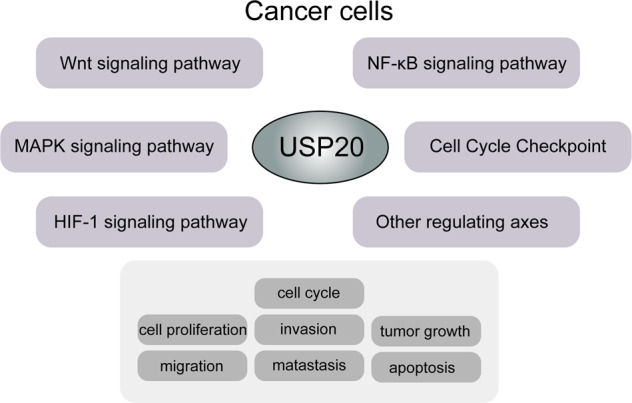
USP20 regulates cell proliferation, migration, invasion, metastasis, and tumor growth by participating in multiple signaling pathways in various cancer types.
Role of USP20 in several types of cancer
Colon cancer
Colon cancer is a leading cause of cancer-related death worldwide [61]. Wu et al. first identified β-catenin as a substrate of USP20, and their study also suggested that USP20 stabilizes β-catenin by decreasing the ubiquitination of β-catenin in vivo and in vitro [57]. To further investigate the role of the USP20-β-catenin axis in cancers, they detected the protein and mRNA levels of USP20 and β-catenin in colon cancer and other cancer cell lines, including osteosarcoma, cervical, breast, ovarian, and colorectal cancer cell lines [57]. They demonstrated that USP20 and β-catenin were upregulated and correlated in most of these cancer cell lines. And they also observed that USP20 overexpression markedly increased cell proliferation, migration and invasion in these cancer cell lines. They demonstrated that USP20 expression was positively correlated with the β-catenin levels in clinical colon cancer samples [57]. Previous publications have identified β-catenin as an oncogene [62], prompting us to speculate that USP20 promotes tumorigenesis by regulating β-catenin stability. Thus, the USP20-β-catenin axis may be an emerging therapeutic target in colon cancer.
Breast cancer
Breast cancer is the leading cause of cancer-related death among women [61]. Sowa et al. first identified USP20 as a candidate interacting with the extracellular signal-regulated kinase 3 (ERK3) protein via global proteomics analysis [63]. Mathien et al. found that USP20 was regulated in breast cancer and that the discovered USP20 played a crucial role in the migration of breast cancer lines [58]. Among the mechanisms by which USP20 affects the migration of breast cancer cell lines, they demonstrated that USP20 was correlated with ERK3, and the stability of ERK3 protein was increased by USP20 [58]. And they investigated the possible effect of USP20 on the contribution of ERK3. They demonstrated that USP20 overexpression could enhance HeLa cell migration, and they further examined the impact of the USP20-ERK3 axis in regulating the migration of breast cancer cell lines [58]. First, their studies showed that both USP20 and ERK3 are overexpressed in multiple breast cancer cell lines, including MCF7, T47D, and SKBR3. They observed a strong significant correlation between the USP20 and ERK3 protein levels. Second, they depleted USP20 in MCF7 cell lines, resulting in markedly decreased ERK3 protein levels and reduced migration of MCF7 cells [58]. These results indicate that the USP20-ERK3 axis can promote the migration of breast cancer cell lines, suggesting that USP20 may play an important role in the tumorigenesis of breast cancer.
Another important regulating mechanism in breast cancer is the USP20-SNAI2 axis. SNAI2 (also known as SLUG) is reported as a metastasis-related transcription factor [64]. Previous studies have shown that estrogen receptor α (ERα) repressed the expression of SNAI2 [65], and the protein level of ERα correlated inversely with SNAI2 in breast cancer cell lines and tissues [66]. Li et al. first identified SNAI2 as one substrate of USP20, they found that USP20 can increase the stability of SNAI2, and subsequently increase the migration and invasion of ER– breast cancer cell lines [67]. In an in vitro study, they found that knockdown of USP20 can suppress the lung colonization of breast cancer cell lines. Meanwhile, they detected USP20 and SNAI2 in ER– clinical breast cancer samples, and demonstrated that USP20 positively correlated with SNAI2. Higher protein level of USP20 and SNAI2 was also demonstrated to predict worse prognosis in ER– breast cancer patients [67]. Thus, their study suggested that USP20-SNAI2 axis may serve as a novel therapeutic target axis in breast cancer.
Cervical cancer
Cervical cancer has been a cause of cancer-related death in recent years among women [61]. Ha et al. first demonstrated that USP20 plays a critical role in regulating the stability of p62 in tumor necrosis factor (TNFα)-mediated nuclear factor kappa light chain enhancer of activated B cells (NF-κB) activation [59]. In their study, they found that USP20 increased the stability of p62 by deubiquitinating lysine 48 (K48)-linked polyubiquitination, promoting cell survival. They further demonstrated the depletion of UPS20 in HeLa cervical cancer cell lines, resulting in a reduced NF-κB-mediated pro-survival signal and increased receptor-interacting serine/threonine protein kinase 1 (RIPK1)-independent apoptosis [59]. Their data defined a novel mechanism by which USP20 regulates the protein level of p62. They found that a high level of p62 protein promoted cell survival and decreased the cell death of HeLa cell lines [59]. This report is the first to reveal the role of the USP20-p62 axis in NF-κB-mediated cell survival. This finding suggests that USP20 acts as an essential regulator in cancer progression and will be a novel therapeutic target for cancer.
Autophagy is critical in tumorigenesis in multiple cancer types [68–70]. Autophagy begins with the formation of autophagosomes, a process that depends on the activity of the serine/threonine kinase ULK1 (hATG1) [71]. A previous study reported that USP20 is localized to the endoplasmic reticulum (ER) [72, 73], and USP20 may play an important role in autophagy. Kim et al. demonstrated that USP20 acts as a positive regulator of the autophagy process by increasing the stability of unc51-like kinase 1 (ULK1) in the HeLa cell line [74]. They found that this regulation also existed in the colon cancer cell lines HCT116 and HT29, and the basal level of ULK1 was the critical factor in inducing autophagy initiation [74]. In their study, they also reported USP20 dissociated from ULK1 at a later time after autophagy induction and promoted the next step in autophagy. The dissociated ULK1 transited into lysosomes to degrade and maintain the basal level. They also found that USP20 is critical for cell survival under starvation [74]. Dynamic regulation of the USP20-ULK1 axis may act as a promising target to inhibit autophagy in certain human diseases.
Gastric cancer
Wang et al. verified that both USP20 and Claspin proteins are expressed at low levels in human gastric cancer tissues, and this phenomenon was also observed in gastric cancer cell lines (MGC-803, NCI-N87, MKN45, BGC-823, KATO III, SGC-7901, AGS, SNU-1, SNU-16 and MKN74) [60]. They also found that low USP20 expression levels correlated with a poor prognosis in patients [60]. In their study, they demonstrated USP20 regulated the stability of Claspin and thus modulating the activation of the cell cycle checkpoint. Low expression of USP20 significantly promoted cell proliferation and accelerated the transition from G1 to S phase of the cell cycle [60]. This finding suggested that USP20 could inhibit cell proliferation of gastric cancer cells by regulating Claspin. It prompted us to speculate that USP20 is a promising new molecular target to design new therapeutic modalities to control the development and progression of gastric cancer.
Adult T cell leukemia
Adult T cell leukemia (ATL) is a fatal hematopoietic malignant tumor caused by type 1 human T cell leukemia virus (HTLV-1) infection [75–77]. Yasunaga et al. first identified USP20 as the first DUB shown to deubiquitinate Tax, and their findings also suggested that ubiquitinated Tax was necessary for activation of the NF-κB signaling pathway [78]. Tax is expressed in many ATL cell lines [77]. Interestingly, Yasunaga et al. found that USP20 expression was significantly low in these cell lines [78]. Because NF-κB is a pro-survival and pro-proliferative factor in HTLV-1 cells, they discovered that the upregulation of USP20 protein negatively regulated NF-κB signal transduction and inhibited cell proliferation of the ATL2 cell line [78]. A previous study showed that ubiquitinated TNF receptor associated factor 6 (TRAF6) is a key regulator of the activation of the NF-κB signaling pathway [79]. Yasunaga et al. also found that Tax overexpression increases TRAF6 ubiquitination, activating NF-κB. TRAF6 ubiquitination induced by Tax was also sensitive to USP20 deubiquitination [78]. In summary, their findings demonstrated that USP20 acted as a DUB for Tax and TRAF6 and as a negative regulator of NF-κB signal transduction. These results prompted us to speculate that USP20 may be a potential new therapeutic target for ATL.
Challenges and prospects
In the past few years, researchers have overcome many difficulties and have screened many small-molecule compounds, including inhibitors and activators. Table 2 presents many preclinical inhibitors of DUBs. However, only one small-molecule compound exists for USP20. GSK2643943A, a small-molecule inhibitor targeting USP20/Ub-Rho, was first identified by GlaxoSmithKline (GSK) from a screen involving compounds. The structure of this USP20 inhibitor is shown in Fig. 4, but this inhibitor is still in the preclinical stage [80, 81]. Lu et al. demonstrated that mammalian target of rapamycin complex 1 phosphorylates USP20 at S132 and S134 and then stabilizes HMGCR, increasing cholesterol biosynthesis in the liver [43]. And their study also showed genetic deletion or pharmacological inhibition of USP20 markedly decreased diet-induced body weight gain and reduced lipid levels in the serum and liver [43]. Although no DUB inhibitors are in ongoing clinical trials, with the rapid development of inhibitors, DUBs are likely increasingly attractive as drug targets. USP20, as a member of the largest deubiquitinase family, was first identified as an oncogene [30]. Although USP20 attracts our attention as a potential therapeutic target, it is associated with challenges. The first challenge is to develop DUB inhibitors or activators, including the development of USP20 inhibitors or activators. On the one hand, the mechanisms of function of DUB enzymes are usually complicated and involve regulating enzyme activity through allosteric and/or substrate-mediated catalysis. Many DUBs dynamically change between active and inactive conformations [81, 82]. On the other hand, because most DUBs perform ubiquitin transfer via reactive thiol groups, most standard assays used to identify inhibitors are nonselective redox or alkylation false-positives [83]. Second, to date, only one USP20 inhibitor has been identified. Although Lu et al. found that this USP20 inhibitor, GSK2643943A, inhibits the function of USP20 to improve metabolic-related diseases [43], no data are available in tumor-related studies. The lack of preclinical research data will be one of the biggest challenges for the transfer of USP20 inhibitors to clinical applications. Third, the mechanism by which USP20 promotes tumor progression in this review has been elucidated in a few types of cancer cells, but the mechanism is unclear in most cancer cell lines that highly express USP20. However, a common mechanism in different types of tumors should also be a challenge for transferring the inhibitor to clinical applications. We require more evidence to support USP20 as a potential tumor therapeutic target or combine USP20 inhibitors with other drugs in cancer treatment. Fourth, presently, the impact of USP20 on tumor development occurs only in cells or animal models, and clinical samples must be further verified. Fifth, through searching currently available literature, we have summarized the role of USP20 in the regulation of cellular proliferation, migration, tumor growth, and metastasis. Although USP20 serves as an oncogene in varied cancer types, it also acts as a tumor suppressor in a few cancer types, including ATL and gastric cancer. The different roles of USP20 may attribute to the heterogeneity of different tumor types. Specifically, different from the observation in cervical cancer that USP20 regulates the stability of p62 in TNFα mediated NF-κB activation, USP20 act as a DUB that negatively regulates NF-κB signal transduction in ATL. Moreover, it regulates the cell cycle checkpoint activation in gastric cancer. In this respect, more studies are needed to further elucidate the function of USP20 and the underlying mechanism in diverse malignancies. Screening activators for USP20 as a therapeutic alternative is also promising for malignancies with low USP20 levels. However, USP20 as a tumor therapeutic target has its limitations. We believe that USP20 as a tumor therapy target is expected to become a reality in the clinic with the in-depth exploration of the role of USP20 in tumor progression.
Table 2.
Other representative inhibitors of DUBs.
| DUB inhibitor | Target | Structure | Stage of development | IC50 | Reference |
|---|---|---|---|---|---|
| SJB3-019A | USP1 |  |
Preclinical | 78.1 nM | [119] |
| ML323 | USP1 | 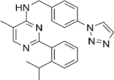 |
Preclinical | 76 nM | [120] |
| ML364 | USP2 | 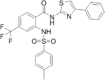 |
Preclinical | 1.1 μM | [121] |
| P5091 | USP7 | 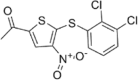 |
Preclinical | 4.2 μM | [122, 123] |
| FT671 | USP7 | 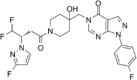 |
Preclinical | 52 nM | [124] |
| GNE-6776 | USP7 | 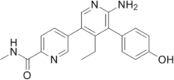 |
Preclinical | N.A. | [125] |
| DUBs-IN-1 | USP8 | 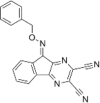 |
Preclinical | 0.24 μM | [126] |
| Spautin1 | USP13 and USP10 | 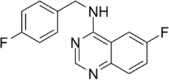 |
Preclinical | 0.6–0.7 μM | [127, 128] |
| Mitoxantrone | USP11 | 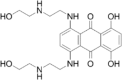 |
Preclinical | 8.5 μM | [129] |
| IU1 | USP14 | 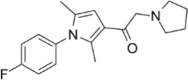 |
Preclinical | 4–5 μM | [130] |
| IU1-47 | USP14 | 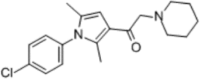 |
Preclinical | 0.6 μM | [131] |
| IU1-248 | USP14 | 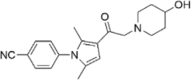 |
Preclinical | 0.83 μM | [132] |
| PR619 | Broad range DUB inhibitor | 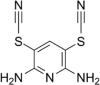 |
Preclinical | [133, 134] | |
| 1,10-Phenanthroline | JAMM type Isopeptidase | 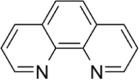 |
Preclinical | [135] | |
| VLX1570 | USP14 and UCHL5 | 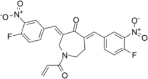 |
Clinical trial phase (now suspended) | 10 μM | [136] |
| GSK2643943A | USP20/Ub-Rho | 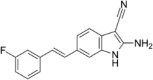 |
Preclinical | 160 nM | [43] |
Fig. 4. The structure of GSK2643943A.
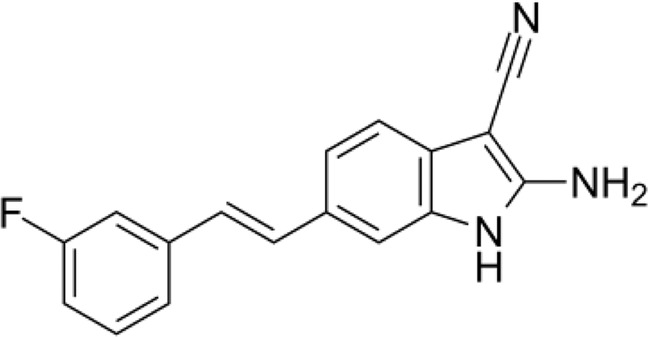
The structure of GSK2643943A, one small molecule inhibitor for USP20/Ub-Rho.
Conclusion
In this summary, USP20, a DUB belonging to USPs, is responsible for removing ubiquitin moieties from ubiquitin-labeled proteins. An increasing number of researchers have focused on exploring the function of USP20 in regulating tumorigenesis. Recently, many breakthroughs have been made in clarifying the role of USP20 in regulating cell proliferation, migration, tumor growth, and glucose metabolism by regulating different signaling pathways. These results also provide evidence for our speculation that USP20 is a target for tumor therapy. More importantly, GSK screened one inhibitor of USP20 and showed that this inhibitor affects the expression of USP33, which shares approximately 59% identity with USP20 and has strong homology at the amino and carboxy termini. Therefore, novel USP20 inhibitors may provide potential treatment options for USP20-overexpressing cancer types. Further analysis of the molecular signaling pathway of USP20 can offer new insights into its tumorigenesis or antitumor mechanisms. The specific regulation of tumorigenesis by USP20 may be a hot research topic in the future. The development of clinical drugs for USP20 will also provide a new opportunity for tumor treatment.
Acknowledgements
We would like to thank the authors of the primary studies. We used Adobe Photoshop, Adobe Illustrator to create the figures in this article.
Author contributions
QL collected related literature. QL and CY wrote the manuscript. JR, JS, FL, TT, and PZ provided guidance and revised the manuscript. All authors read and approved the final manuscript.
Funding
This project was funded by the National Natural Science Foundation of China (Grant Nos. 81874173, 81472346), the Natural Science Foundation of Zhejiang Province (Grant Nos. LY20H160033,2017205840, 2018274816, LGJ18H160001). There was no participation of the funding bodies in the study design, and lyses, collection, data interpretation and manuscript writing.
Competing interests
The authors declare no competing interests.
Footnotes
Edited by Dr. Francesca Bernassola
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Qiong Li, Chanqi Ye.
References
- 1.Park J, Cho J, Song EJ. Ubiquitin-proteasome system (UPS) as a target for anticancer treatment. Arch Pharmacal Res. 2020;43:1144–61. doi: 10.1007/s12272-020-01281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nandi D, Tahiliani P, Kumar A, Chandu D. The ubiquitin-proteasome system. J Biosci. 2006;31:137–55. doi: 10.1007/BF02705243. [DOI] [PubMed] [Google Scholar]
- 3.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol cell Biol. 2008;9:679–90. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.AC AH. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Sato K, Sundaramoorthy E, Rajendra E, Hattori H, Jeyasekharan AD, Ayoub N, et al. A DNA-damage selective role for BRCA1 E3 ligase in claspin ubiquitylation, CHK1 activation, and DNA repair. Curr Biol. 2012;22:1659–66. doi: 10.1016/j.cub.2012.07.034. [DOI] [PubMed] [Google Scholar]
- 6.Gennaro VJ, Stanek TJ, Peck AR, Sun Y, Wang F, Qie S, et al. Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells. Proc Natl Acad Sci USA. 2018;115:E9298–E9307. doi: 10.1073/pnas.1807704115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palazon-Riquelme P, Worboys JD, Green J, Valera A, Martin-Sanchez F, Pellegrini C, et al. USP7 and USP47 deubiquitinases regulate NLRP3 inflammasome activation. EMBO Rep. 2018;19:e44766.. doi: 10.15252/embr.201744766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–6. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 9.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol cell Biol. 2009;10:550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 10.CA. GM. The ubiquitin-proteasome proteolytic pathway destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 11.Wolberger C. Mechanisms for regulating deubiquitinating enzymes. Protein Sci. 2014;23:344–53. doi: 10.1002/pro.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi M, Oshima S, Maeyashiki C, Nibe Y, Otsubo K, Matsuzawa Y, et al. The ubiquitin hybrid gene UBA52 regulates ubiquitination of ribosome and sustains embryonic development. Sci Rep. 2016;6:36780. doi: 10.1038/srep36780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choudhury AD, Xu H, Baer R. Ubiquitination and proteasomal degradation of the BRCA1 tumor suppressor is regulated during cell cycle progression. J Biol Chem. 2004;279:33909–18. doi: 10.1074/jbc.M403646200. [DOI] [PubMed] [Google Scholar]
- 14.Ginjala V, Nacerddine K, Kulkarni A, Oza J, Hill SJ, Yao M, et al. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol. 2011;31:1972–82. doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu L, Dong L, Li H, Liu Z, Luo Z, Duan G, et al. Ubiquitination-mediated degradation of SIRT1 by SMURF2 suppresses CRC cell proliferation and tumorigenesis. Oncogene. 2020;39:4450–64. doi: 10.1038/s41388-020-1298-0. [DOI] [PubMed] [Google Scholar]
- 16.C W. Regulation of apoptosis by the ubiquitin and proteasome pathway. J Cell Mol Med. 2002;6:25–48. doi: 10.1111/j.1582-4934.2002.tb00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Du L, Ren Y, Liu X, Jiao Q, Cui D, et al. SKP2 promotes breast cancer tumorigenesis and radiation tolerance through PDCD4 ubiquitination. J Exp Clin Cancer Res. 2019;38:76. doi: 10.1186/s13046-019-1069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Li B, Rezaeian AH, Xu X, Chou PC, Jin G, et al. H3 ubiquitination by NEDD4 regulates H3 acetylation and tumorigenesis. Nat Commun. 2017;8:14799. doi: 10.1038/ncomms14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahlis NJSH, White D, Sebag M, Lentzsch S, Kotb R, Venner CP, et al. Selinexor plus low-dose bortezomib and dexamethasone for patients with relapsed or refractory multiple myeloma. Blood. 2018;132:2546–54. doi: 10.1182/blood-2018-06-858852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott K, Hayden PJ, Will A, Wheatley K, Coyne I. Bortezomib for the treatment of multiple myeloma. Cochrane Database Syst Rev. 2016;4:CD010816. doi: 10.1002/14651858.CD010816.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin J, Zhou Z, Chen W, Wang C, Zhang H, Ge G, et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun. 2015;6:8471. doi: 10.1038/ncomms9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu P, Xiao H, Yang Q, Hu R, Jiang L, Bi R, et al. The USP21/YY1/SNHG16 axis contributes to tumor proliferation, migration, and invasion of non-small-cell lung cancer. Exp Mol Med. 2020;52:41–55. doi: 10.1038/s12276-019-0356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S, Yuan J, Zheng R. Suppression of ubiquitin-specific peptidase 17 (USP17) inhibits tumorigenesis and invasion in non-small cell lung cancer cells. Oncol Res. 2016;24:263–9. doi: 10.3727/096504016X14666990347392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S, Juan J, Zhang Z, Du Y, Xu Y, Tong J, et al. Inhibition of the deubiquitinase USP5 leads to c-Maf protein degradation and myeloma cell apoptosis. Cell Death Dis. 2017;8:e3058. doi: 10.1038/cddis.2017.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clague MJ, Urbe S, Komander D. Breaking the chains: deubiquitylating enzyme specificity begets function. Nat Rev Mol Cell Biol. 2019;20:338–52. doi: 10.1038/s41580-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 26.Shan J, Zhao W, Gu W. Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol Cell. 2009;36:469–76. doi: 10.1016/j.molcel.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji AL, Li T, Zu G, Feng DC, Li Y, Wang GZ, et al. Ubiquitin-specific protease 22 enhances intestinal cell proliferation and tissue regeneration after intestinal ischemia reperfusion injury. World J Gastroenterol. 2019;25:824–36. doi: 10.3748/wjg.v25.i7.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gersch M, Wagstaff JL, Toms AV, Graves B, Freund SMV, Komander D. Distinct USP25 and USP28 oligomerization states regulate deubiquitinating activity. Mol Cell. 2019;74:436–51.e437. doi: 10.1016/j.molcel.2019.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Fan Y, Xie F, Zhou H, Jin K, Shao L, et al. Breast cancer metastasis suppressor OTUD1 deubiquitinates SMAD7. Nat Commun. 2017;8:2116. doi: 10.1038/s41467-017-02029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholson BMJ, Butt TR, Mattern MR. Deubiquitinating enzymes as novel anticancer targets. Future Oncol. 2007;3:191–9. doi: 10.2217/14796694.3.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park JJ, Yun JH, Baek KH. Polyclonal and monoclonal antibodies specific for ubiquitin-specific protease 20. Monoclon Antib Immunodiagn Immunother. 2013;32:193–9. doi: 10.1089/mab.2012.0120. [DOI] [PubMed] [Google Scholar]
- 32.Curcio-Morelli C, Zavacki AM, Christofollete M, Gereben B, de Freitas BCG, Harney JW, et al. Deubiquitination of type 2 iodothyronine deiodinase by von Hippel–Lindau protein–interacting deubiquitinating enzymes regulates thyroid hormone activation. J Clin Investig. 2003;112:189–96. doi: 10.1172/JCI18348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li ZWD, Na X, Schoen SR, Messing EM, Wu G. Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel-Lindau tumor suppressor. Biochem Biophys Res Commun. 2002;294:700–9. doi: 10.1016/S0006-291X(02)00534-X. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Wen Y, Zhang N. (1)H, (13)C and (15)N backbone and side-chain resonance assignments of the ZnF-UBP domain of USP20/VDU2. Biomolecular NMR Assign. 2017;11:91–93. doi: 10.1007/s12104-017-9726-y. [DOI] [PubMed] [Google Scholar]
- 35.de Jong RN, Ab E, Diercks T, Truffault V, Daniels M, Kaptein R, et al. Solution structure of the human ubiquitin-specific protease 15 DUSP domain. J Biol Chem. 2006;281:5026–31. doi: 10.1074/jbc.M510993200. [DOI] [PubMed] [Google Scholar]
- 36.Allen MD, Bycroft M. The solution structure of the ZnF UBP domain of USP33/VDU1. Protein Sci. 2007;16:2072–5. doi: 10.1110/ps.072967807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, Ding Y, Zhou C, Wen Y, Zhang N. Structural and functional studies of USP20 ZnF-UBP domain by NMR. Protein Sci. 2019;28:1606–19. doi: 10.1002/pro.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quesada V, Diaz-Perales A, Gutierrez-Fernandez A, Garabaya C, Cal S, Lopez-Otin C. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem Biophys Res Commun. 2004;314:54–62. doi: 10.1016/j.bbrc.2003.12.050. [DOI] [PubMed] [Google Scholar]
- 39.Minervini G, Quaglia F, Tabaro F, Tosatto SCE. Insights into the molecular features of the von Hippel-Lindau-like protein. Amino Acids. 2019;51:1461–74. doi: 10.1007/s00726-019-02781-8. [DOI] [PubMed] [Google Scholar]
- 40.Clerici M, Luna-Vargas MP, Faesen AC, Sixma TK. The DUSP-Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nat Commun. 2014;5:5399. doi: 10.1038/ncomms6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kommaddi RP, Jean-Charles PY, Shenoy SK. Phosphorylation of the deubiquitinase USP20 by protein kinase A regulates post-endocytic trafficking of beta2 adrenergic receptors to autophagosomes during physiological stress. J Biol Chem. 2015;290:8888–903. doi: 10.1074/jbc.M114.630541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berthouze M, Venkataramanan V, Li Y, Shenoy SK. The deubiquitinases USP33 and USP20 coordinate beta2 adrenergic receptor recycling and resensitization. EMBO J. 2009;28:1684–96. doi: 10.1038/emboj.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu XY, Shi XJ, Hu A, Wang JQ, Ding Y, Jiang W, et al. Feeding induces cholesterol biosynthesis via the mTORC1-USP20-HMGCR axis. Nature. 2020;588:479–84. doi: 10.1038/s41586-020-2928-y. [DOI] [PubMed] [Google Scholar]
- 44.Maxwell PHWM, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–5. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 45.Lonser RR, Glenn GM, Walther M, Chew EY, Libutti SK, Linehan WM, et al. von Hippel-Lindau disease. Lancet. 2003;361:2059–67. doi: 10.1016/S0140-6736(03)13643-4. [DOI] [PubMed] [Google Scholar]
- 46.Chittiboina P, Lonser RR. Von Hippel-Lindau disease. Handb Clin Neurol. 2015;132:139–56. doi: 10.1016/B978-0-444-62702-5.00010-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ben-Skowronek I, Kozaczuk S. Von Hippel-Lindau syndrome. Horm Res Paediatr. 2015;84:145–52. doi: 10.1159/000431323. [DOI] [PubMed] [Google Scholar]
- 48.Li Z, Wang D, Messing EM, Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep. 2005;6:373–8. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shao M, Hepler C, Zhang Q, Shan B, Vishvanath L, Henry GH, et al. Pathologic HIF1alpha signaling drives adipose progenitor dysfunction in obesity. Cell Stem Cell. 2021;28:685–701.e687. doi: 10.1016/j.stem.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia Z, Wu S, Wei X, Liao Y, Yi P, Liu Y, et al. Hypoxic ER stress suppresses beta-catenin expression and promotes cooperation between the transcription factors XBP1 and HIF1alpha for cell survival. J Biol Chem. 2019;294:13811–21. doi: 10.1074/jbc.RA119.008353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, et al. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joseph JV, Conroy S, Pavlov K, Sontakke P, Tomar T, Eggens-Meijer E, et al. Hypoxia enhances migration and invasion in glioblastoma by promoting a mesenchymal shift mediated by the HIF1alpha-ZEB1 axis. Cancer Lett. 2015;359:107–16. doi: 10.1016/j.canlet.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 53.Lyu X, Wang J, Guo X, Wu G, Jiao Y, Faleti OD, et al. EBV-miR-BART1-5P activates AMPK/mTOR/HIF1 pathway via a PTEN independent manner to promote glycolysis and angiogenesis in nasopharyngeal carcinoma. PLoS Pathog. 2018;14:e1007484. doi: 10.1371/journal.ppat.1007484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Logsdon DP, Shah F, Carta F, Supuran CT, Kamocka M, Jacobsen MH, et al. Blocking HIF signaling via novel inhibitors of CA9 and APE1/Ref-1 dramatically affects pancreatic cancer cell survival. Sci Rep. 2018;8:13759. doi: 10.1038/s41598-018-32034-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moriyama H, Moriyama M, Ozawa T, Tsuruta D, Iguchi T, Tamada S, et al. Notch signaling enhances stemness by regulating metabolic pathways through modifying p53, NF-kappaB, and HIF-1alpha. Stem Cells Dev. 2018;27:935–47. doi: 10.1089/scd.2017.0260. [DOI] [PubMed] [Google Scholar]
- 56.Chen WJ, Huang RS. Low-folate stress reprograms cancer stem cell-like potentials and bioenergetics metabolism through activation of mTOR signaling pathway to promote in vitro invasion and in vivo tumorigenicity of lung cancers. J Nutritional Biochem. 2018;53:28–38. doi: 10.1016/j.jnutbio.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 57.Wu C, Luo K, Zhao F, Yin P, Song Y, Deng M, et al. USP20 positively regulates tumorigenesis and chemoresistance through beta-catenin stabilization. Cell Death Differ. 2018;25:1855–69. doi: 10.1038/s41418-018-0138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mathien SDP, Soulez M, Voisin L, Meloche S. Deubiquitinating enzyme USP20 regulates extracellular signal-regulated kinase 3 stability and biological activity. Mol Cell Biol. 2017;37:e00432–00416.. doi: 10.1128/MCB.00432-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ha J, Kim M, Seo D, Park JS, Lee J, Lee J, et al. The deubiquitinating enzyme USP20 regulates the TNFalpha-induced NF-kappaB signaling pathway through stabilization of p62. Int J Mol Sci. 2020;21:3116.. doi: 10.3390/ijms21093116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang C, Yang C, Ji J, Jiang J, Shi M, Cai Q, et al. Deubiquitinating enzyme USP20 is a positive regulator of Claspin and suppresses the malignant characteristics of gastric cancer cells. Int J Oncol. 2017;50:1136–46. doi: 10.3892/ijo.2017.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 62.Liang Y, Feng Y, Zong M, Wei XF, Lee J, Feng Y, et al. beta-catenin deficiency in hepatocytes aggravates hepatocarcinogenesis driven by oncogenic beta-catenin and MET. Hepatology. 2018;67:1807–22. doi: 10.1002/hep.29661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shih JY, Yang PC. The EMT regulator slug and lung carcinogenesis. Carcinogenesis. 2011;32:1299–304. doi: 10.1093/carcin/bgr110. [DOI] [PubMed] [Google Scholar]
- 65.Ye Y, Xiao Y, Wang W, Yearsley K, Gao JX, Shetuni B, et al. ERalpha signaling through slug regulates E-cadherin and EMT. Oncogene. 2010;29:1451–62. doi: 10.1038/onc.2009.433. [DOI] [PubMed] [Google Scholar]
- 66.Bai JW, Chen MN, Wei XL, Li YC, Lin HY, Chen M, et al. The zinc-finger transcriptional factor Slug transcriptionally downregulates ERalpha by recruiting lysine-specific demethylase 1 in human breast cancer. Oncogenesis. 2017;6:e330. doi: 10.1038/oncsis.2017.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W, Shen M, Jiang YZ, Zhang R, Zheng H, Wei Y, et al. Deubiquitinase USP20 promotes breast cancer metastasis by stabilizing SNAI2. Genes Dev. 2020;34:1310–5. doi: 10.1101/gad.339804.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marsh T, Debnath J. Autophagy suppresses breast cancer metastasis by degrading NBR1. Autophagy. 2020;16:1164–5. doi: 10.1080/15548627.2020.1753001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang X, Ding H, Liang M, Chen X, Yan Y, Wan N, et al. Curcumin induces ferroptosis in non-small-cell lung cancer via activating autophagy. Thorac Cancer. 2021;12:1219–30. doi: 10.1111/1759-7714.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z, Sun Q, Guo J, Wang S, Song G, Liu W, et al. GRSF1-mediated MIR-G-1 promotes malignant behavior and nuclear autophagy by directly upregulating TMED5 and LMNB1 in cervical cancer cells. Autophagy. 2019;15:668–85. doi: 10.1080/15548627.2018.1539590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chan EY, Tooze SA. Evolution of Atg1 function and regulation. Autophagy. 2009;5:758–65. doi: 10.4161/auto.8709. [DOI] [PubMed] [Google Scholar]
- 72.Thorne C, Eccles RL, Coulson JM, Urbe S, Clague MJ. Isoform-specific localization of the deubiquitinase USP33 to the Golgi apparatus. Traffic. 2011;12:1563–74. doi: 10.1111/j.1600-0854.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- 73.Culver JA, Mariappan M. Deubiquitinases USP20/33 promote the biogenesis of tail-anchored membrane proteins. J Cell Biol. 2021;220:e202004086. doi: 10.1083/jcb.202004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim JH, Seo D, Kim SJ, Choi DW, Park JS, Ha J, et al. The deubiquitinating enzyme USP20 stabilizes ULK1 and promotes autophagy initiation. EMBO Rep. 2018;19:e44378.. doi: 10.15252/embr.201744378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taylor GP, Matsuoka M. Natural history of adult T-cell leukemia/lymphoma and approaches to therapy. Oncogene. 2005;24:6047–57. doi: 10.1038/sj.onc.1208979. [DOI] [PubMed] [Google Scholar]
- 76.Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7:270–80. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- 77.Pujari R, Hunte R, Thomas R, van der Weyden L, Rauch D, Ratner L, et al. Human T-cell leukemia virus type 1 (HTLV-1) tax requires CADM1/TSLC1 for inactivation of the NF-kappaB inhibitor A20 and constitutive NF-kappaB signaling. PLoS Pathog. 2015;11:e1004721. doi: 10.1371/journal.ppat.1004721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yasunaga J, Lin FC, Lu X, Jeang KT. Ubiquitin-specific peptidase 20 targets TRAF6 and human T cell leukemia virus type 1 tax to negatively regulate NF-kappaB signaling. J Virol. 2011;85:6212–9. doi: 10.1128/JVI.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Min Y, Kim MJ, Lee S, Chun E, Lee KY. Inhibition of TRAF6 ubiquitin-ligase activity by PRDX1 leads to inhibition of NFKB activation and autophagy activation. Autophagy. 2018;14:1347–58. doi: 10.1080/15548627.2018.1474995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kemp M. Recent advances in the discovery of deubiquitinating enzyme inhibitors. Prog Med Chem. 2016;55:149–92. doi: 10.1016/bs.pmch.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sahtoe DD, Sixma TK. Layers of DUB regulation. Trends Biochem Sci. 2015;40:456–67. doi: 10.1016/j.tibs.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 82.Mevissen TET, Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem. 2017;86:159–92. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 83.Wrigley JD, Eckersley K, Hardern IM, Millard L, Walters M, Peters SW, et al. Enzymatic characterisation of USP7 deubiquitinating activity and inhibition. Cell Biochem Biophys. 2011;60:99–111. doi: 10.1007/s12013-011-9186-4. [DOI] [PubMed] [Google Scholar]
- 84.Liu J, Zhu H, Zhong N, Jiang Z, Xu L, Deng Y, et al. Gene silencing of USP1 by lentivirus effectively inhibits proliferation and invasion of human osteosarcoma cells. Int J Oncol. 2016;49:2549–57. doi: 10.3892/ijo.2016.3752. [DOI] [PubMed] [Google Scholar]
- 85.Ma L, Lin K, Chang G, Chen Y, Yue C, Guo Q, et al. Aberrant activation of beta-catenin signaling drives glioma tumorigenesis via USP1-mediated stabilization of EZH2. Cancer Res. 2019;79:72–85. doi: 10.1158/0008-5472.CAN-18-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma A, Tang M, Zhang L, Wang B, Yang Z, Liu Y, et al. USP1 inhibition destabilizes KPNA2 and suppresses breast cancer metastasis. Oncogene. 2019;38:2405–19. doi: 10.1038/s41388-018-0590-8. [DOI] [PubMed] [Google Scholar]
- 87.Qu Q, Mao Y, Xiao G, Fei X, Wang J, Zhang Y, et al. USP2 promotes cell migration and invasion in triple negative breast cancer cell lines. Tumour Biol. 2015;36:5415–23. doi: 10.1007/s13277-015-3207-7. [DOI] [PubMed] [Google Scholar]
- 88.Cao WHLX, Meng SL, Gao YW, Wang Y, Ma ZL, Wang XG, et al. USP4 promotes invasion of breast cancer cells via Relaxin/TGF-beta1/Smad2/MMP-9 signal. Eur Rev Med Pharm Sci. 2016;20:1115–22. [PubMed] [Google Scholar]
- 89.Guo W, Ma J, Pei T, Zhao T, Guo S, Yi X, et al. Up-regulated deubiquitinase USP4 plays an oncogenic role in melanoma. J Cell Mol Med. 2018;22:2944–54. doi: 10.1111/jcmm.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou Y, Liang P, Ji W, Yu Z, Chen H, Jiang L. Ubiquitin-specific protease 4 promotes glioblastoma multiforme via activating ERK pathway. OncoTargets Ther. 2019;12:1825–39. doi: 10.2147/OTT.S176582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qin N, Han F, Li L, Ge Y, Lin W, Wang J, et al. Deubiquitinating enzyme 4 facilitates chemoresistance in glioblastoma by inhibiting P53 activity. Oncol Lett. 2019;17:958–64. doi: 10.3892/ol.2018.9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li XY, Wu HY, Mao XF, Jiang LX, Wang YX. USP5 promotes tumorigenesis and progression of pancreatic cancer by stabilizing FoxM1 protein. Biochem Biophys Res Commun. 2017;492:48–54. doi: 10.1016/j.bbrc.2017.08.040. [DOI] [PubMed] [Google Scholar]
- 93.Ma XQW, Pan H, Yang F, Deng J. Overexpression of USP5 contributes to tumorigenesis in non-small cell lung cancer via the stabilization of beta-catenin protein. Am J Cancer Res. 2017;8:2284–95. [PMC free article] [PubMed] [Google Scholar]
- 94.Xu X, Huang A, Cui X, Han K, Hou X, Wang Q, et al. Ubiquitin specific peptidase 5 regulates colorectal cancer cell growth by stabilizing Tu translation elongation factor. Theranostics. 2019;9:4208–20. doi: 10.7150/thno.33803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhan M, Sun X, Liu J, Li Y, Li Y, He X, et al. Usp7 promotes medulloblastoma cell survival and metastasis by activating Shh pathway. Biochem Biophys Res Commun. 2017;484:429–34. doi: 10.1016/j.bbrc.2017.01.144. [DOI] [PubMed] [Google Scholar]
- 96.Wang QMS, Song N, Li X, Liu L, Yang S, Ding X, et al. Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. J Clin Invest. 2016;126:2205–20. doi: 10.1172/JCI85747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zeng Q, Li Z, Zhao X, Guo L, Yu C, Qin J, et al. Ubiquitinspecific protease 7 promotes osteosarcoma cell metastasis by inducing epithelialmesenchymal transition. Oncol Rep. 2019;41:543–51. doi: 10.3892/or.2018.6835. [DOI] [PubMed] [Google Scholar]
- 98.Dai X, Lu L, Deng S, Meng J, Wan C, Huang J, et al. USP7 targeting modulates anti-tumor immune response by reprogramming tumor-associated macrophages in lung cancer. Theranostics. 2020;10:9332–47. doi: 10.7150/thno.47137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yan M, Zhao C, Wei N, Wu X, Cui J, Xing Y. High expression of ubiquitin-specific protease 8 (USP8) is associated with poor prognosis in patients with cervical squamous cell carcinoma. Med Sci Monit. 2018;24:4934–43. doi: 10.12659/MSM.909235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jeong M, Lee EW, Seong D, Seo J, Kim JH, Grootjans S, et al. USP8 suppresses death receptor-mediated apoptosis by enhancing FLIPL stability. Oncogene. 2017;36:458–70. doi: 10.1038/onc.2016.215. [DOI] [PubMed] [Google Scholar]
- 101.Ouyang SW, Liu TT, Liu XS, Zhu FX, Zhu FM, Liu XN, et al. USP10 regulates Musashi-2 stability via deubiquitination and promotes tumour proliferation in colon cancer. FEBS Lett. 2019;593:406–13. doi: 10.1002/1873-3468.13323. [DOI] [PubMed] [Google Scholar]
- 102.He Y, Jiang S, Mao C, Zheng H, Cao B, Zhang Z, et al. The deubiquitinase USP10 restores PTEN activity and inhibits non-small cell lung cancer cell proliferation. J Biol Chem. 2021;297:101088. doi: 10.1016/j.jbc.2021.101088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sun J, Li T, Zhao Y, Huang L, Sun H, Wu H, et al. USP10 inhibits lung cancer cell growth and invasion by upregulating PTEN. Mol Cell Biochem. 2018;441:1–7. doi: 10.1007/s11010-017-3170-2. [DOI] [PubMed] [Google Scholar]
- 104.Hu C, Zhang M, Moses N, Hu CL, Polin L, Chen W, et al. The USP10-HDAC6 axis confers cisplatin resistance in non-small cell lung cancer lacking wild-type p53. Cell Death Dis. 2020;11:328. doi: 10.1038/s41419-020-2519-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhu H, Yan F, Yuan T, Qian M, Zhou T, Dai X, et al. USP10 promotes proliferation of hepatocellular carcinoma by deubiquitinating and stabilizing YAP/TAZ. Cancer Res. 2020;80:2204–16. doi: 10.1158/0008-5472.CAN-19-2388. [DOI] [PubMed] [Google Scholar]
- 106.Yuan T, Chen Z, Yan F, Qian M, Luo H, Ye S, et al. Deubiquitinating enzyme USP10 promotes hepatocellular carcinoma metastasis through deubiquitinating and stabilizing Smad4 protein. Mol Oncol. 2020;14:197–210. doi: 10.1002/1878-0261.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Takayama KI, Suzuki T, Fujimura T, Takahashi S, Inoue S. Association of USP10 with G3BP2 inhibits p53 signaling and contributes to poor outcome in prostate cancer. Mol Cancer Res. 2018;16:846–56. doi: 10.1158/1541-7786.MCR-17-0471. [DOI] [PubMed] [Google Scholar]
- 108.Weisberg EL, Schauer NJ, Yang J, Lamberto I, Doherty L, Bhatt S, et al. Inhibition of USP10 induces degradation of oncogenic FLT3. Nat Chem Biol. 2017;13:1207–15. doi: 10.1038/nchembio.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sun H, Ou B, Zhao S, Liu X, Song L, Liu X, et al. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. EBioMedicine. 2019;48:236–47. doi: 10.1016/j.ebiom.2019.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang W, Wang J, Yan H, Zhang K, Liu Y. Upregulation of USP11 promotes epithelialtomesenchymal transition by deubiquitinating Snail in ovarian cancer. Oncol Rep. 2019;41:1739–48. doi: 10.3892/or.2018.6924. [DOI] [PubMed] [Google Scholar]
- 111.Wu Y, Zhang Y, Liu C, Zhang Y, Wang D, Wang S, et al. Amplification of USP13 drives non-small cell lung cancer progression mediated by AKT/MAPK signaling. Biomed Pharmacother. 2019;114:108831. doi: 10.1016/j.biopha.2019.108831. [DOI] [PubMed] [Google Scholar]
- 112.Liu B, Liu Y, Wang Y, Xie C, Gan M, Han T, et al. CyclinB1 deubiquitination by USP14 regulates cell cycle progression in breast cancer. Pathol Res Pract. 2019;215:152592. doi: 10.1016/j.prp.2019.152592. [DOI] [PubMed] [Google Scholar]
- 113.Li Y, Huang J, Zeng B, Yang D, Sun J, Yin X, et al. PSMD2 regulates breast cancer cell proliferation and cell cycle progression by modulating p21 and p27 proteasomal degradation. Cancer Lett. 2018;430:109–22. doi: 10.1016/j.canlet.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 114.Zhu L, Yang S, He S, Qiang F, Cai J, Liu R, et al. Downregulation of ubiquitin-specific protease 14 (USP14) inhibits breast cancer cell proliferation and metastasis, but promotes apoptosis. J Mol Histol. 2016;47:69–80. doi: 10.1007/s10735-015-9650-3. [DOI] [PubMed] [Google Scholar]
- 115.Xia X, Huang C, Liao Y, Liu Y, He J, Guo Z, et al. Inhibition of USP14 enhances the sensitivity of breast cancer to enzalutamide. J Exp Clin Cancer Res. 2019;38:220. doi: 10.1186/s13046-019-1227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gu L, Zhu Y, Lin X, Li Y, Cui K, Prochownik EV, et al. Amplification of glyceronephosphate O-acyltransferase and recruitment of USP30 stabilize DRP1 to promote hepatocarcinogenesis. Cancer Res. 2018;78:5808–19. doi: 10.1158/0008-5472.CAN-18-0340. [DOI] [PubMed] [Google Scholar]
- 117.Gu YY, Yang M, Zhao M, Luo Q, Yang L, Peng H, et al. The de-ubiquitinase UCHL1 promotes gastric cancer metastasis via the Akt and Erk1/2 pathways. Tumour Biol. 2015;36:8379–87. doi: 10.1007/s13277-015-3566-0. [DOI] [PubMed] [Google Scholar]
- 118.Li J, Cheng D, Zhu M, Yu H, Pan Z, Liu L, et al. OTUB2 stabilizes U2AF2 to promote the Warburg effect and tumorigenesis via the AKT/mTOR signaling pathway in non-small cell lung cancer. Theranostics. 2019;9:179–95. doi: 10.7150/thno.29545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Das DS, Das A, Ray A, Song Y, Samur MK, Munshi NC, et al. Blockade of deubiquitylating enzyme USP1 inhibits DNA repair and triggers apoptosis in multiple myeloma cells. Clin Cancer Res. 2017;23:4280–9. doi: 10.1158/1078-0432.CCR-16-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liang Q, Dexheimer TS, Zhang P, Rosenthal AS, Villamil MA, You C, et al. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat Chem Biol. 2014;10:298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Davis MI, Pragani R, Fox JT, Shen M, Parmar K, Gaudiano EF, et al. Small molecule inhibition of the ubiquitin-specific protease USP2 accelerates cyclin D1 degradation and leads to cell cycle arrest in colorectal cancer and mantle cell lymphoma models. J Biol Chem. 2016;291:24628–40. doi: 10.1074/jbc.M116.738567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22:345–58. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.An T, Gong Y, Li X, Kong L, Ma P, Gong L, et al. USP7 inhibitor P5091 inhibits Wnt signaling and colorectal tumor growth. Biochemical Pharmacol. 2017;131:29–39. doi: 10.1016/j.bcp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 124.Turnbull AP, Ioannidis S, Krajewski WW, Pinto-Fernandez A, Heride C, Martin ACL, et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature. 2017;550:481–6. doi: 10.1038/nature24451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kategaya L, Di Lello P, Rouge L, Pastor R, Clark KR, Drummond J, et al. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature. 2017;550:534–8. doi: 10.1038/nature24006. [DOI] [PubMed] [Google Scholar]
- 126.Colombo M, Vallese S, Peretto I, Jacq X, Rain JC, Colland F, et al. Synthesis and biological evaluation of 9-oxo-9H-indeno[1,2-b]pyrazine-2,3-dicarbonitrile analogues as potential inhibitors of deubiquitinating enzymes. ChemMedChem. 2010;5:552–8. doi: 10.1002/cmdc.200900409. [DOI] [PubMed] [Google Scholar]
- 127.Shao S, Li S, Qin Y, Wang X, Yang Y, Bai H, et al. Spautin-1, a novel autophagy inhibitor, enhances imatinib-induced apoptosis in chronic myeloid leukemia. Int J Oncol. 2014;44:1661–8. doi: 10.3892/ijo.2014.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–34. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jacko AM, Nan L, Li S, Tan J, Zhao J, Kass DJ, et al. De-ubiquitinating enzyme, USP11, promotes transforming growth factor beta-1 signaling through stabilization of transforming growth factor beta receptor II. Cell Death Dis. 2016;7:e2474. doi: 10.1038/cddis.2016.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–84. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Boselli M, Lee BH, Robert J, Prado MA, Min SW, Cheng C, et al. An inhibitor of the proteasomal deubiquitinating enzyme USP14 induces tau elimination in cultured neurons. J Biol Chem. 2017;292:19209–25. doi: 10.1074/jbc.M117.815126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang Y, Jiang Y, Ding S, Li J, Song N, Ren Y, et al. Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade. Cell Res. 2018;28:1186–94. doi: 10.1038/s41422-018-0091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kuo KL, Liu SH, Lin WC, Chow PM, Chang YW, Yang SP, et al. The deubiquitinating enzyme inhibitor PR-619 enhances the cytotoxicity of cisplatin via the suppression of anti-apoptotic Bcl-2 protein: in vitro and in vivo study. Cells. 2019;8:1268.. doi: 10.3390/cells8101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cowell IG, Ling EM, Swan RL, Brooks MLW, Austin CA. The deubiquitinating enzyme inhibitor PR-619 is a potent DNA topoisomerase II poison. Mol Pharmacol. 2019;96:562–72. doi: 10.1124/mol.119.117390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Altun M, Kramer HB, Willems LI, McDermott JL, Leach CA, Goldenberg SJ, et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem Biol. 2011;18:1401–12. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 136.Wang X, Mazurkiewicz M, Hillert EK, Olofsson MH, Pierrou S, Hillertz P, et al. The proteasome deubiquitinase inhibitor VLX1570 shows selectivity for ubiquitin-specific protease-14 and induces apoptosis of multiple myeloma cells. Sci Rep. 2016;6:26979. doi: 10.1038/srep26979. [DOI] [PMC free article] [PubMed] [Google Scholar]