Summary
Chromatin is essentially an array of nucleosomes, each of which consists of the DNA double-stranded fiber wrapped around a histone octamer. This organization supports cellular processes such as DNA replication, DNA transcription, and DNA repair in all eukaryotes. Human histone H4 is encoded by fourteen canonical histone H4 genes, all differing at the nucleotide level but encoding an invariant protein. Here, we present a cohort of 29 subjects with de novo missense variants in six H4 genes (H4C3, H4C4, H4C5, H4C6, H4C9, and H4C11) identified by whole-exome sequencing and matchmaking. All individuals present with neurodevelopmental features of intellectual disability and motor and/or gross developmental delay, while non-neurological features are more variable. Ten amino acids are affected, six recurrently, and are all located within the H4 core or C-terminal tail. These variants cluster to specific regions of the core H4 globular domain, where protein-protein interactions occur with either other histone subunits or histone chaperones. Functional consequences of the identified variants were evaluated in zebrafish embryos, which displayed abnormal general development, defective head organs, and reduced body axis length, providing compelling evidence for the causality of the reported disorder(s). While multiple developmental syndromes have been linked to chromatin-associated factors, missense-bearing histone variants (e.g., H3 oncohistones) are only recently emerging as a major cause of pathogenicity. Our findings establish a broader involvement of H4 variants in developmental syndromes.
Keywords: histone H4, nucleosome, neurodevelopmental disorder, intellectual disability, microcephaly, zebrafish
Main text
Histones are among the most slowly evolving genes in eukaryotes.1,2 Histone H4 acts as a functional unit by forming a dimer with H3, but unlike H3, there are no variant isoforms linked to specific cellular processes. We previously reported four individuals with a primordial dwarfism phenotype with de novo missense variants affecting Lys91 in H4C3 (HIST1H4C; MIM: 602827) and H4C11 (HIST1H4J; MIM: 602826), a critical residue near the C terminus of the protein prone to post-translational modifications (PTMs) such as acetylation3 or, more important here, ubiquitination.4, 5, 6 Somatic variation in H4 is potentially relevant in a cancer setting,7,8 although described variation impacts multiple residues in contrast to the highly recurrent pathogenic variants observed for the established oncohistone, H3.8 This could be because while the functional importance of PTMs on N-tails of H3 is linked directly to the chromatin effectors targeting these sites,9, 10, 11 variants affecting the histone core (H4 in this case) have more direct and global consequences on chromatin organization, as they impact nucleosome structure and dynamics.12
We have identified an additional cohort of 29 individuals with de novo missense variants in six different histone H4 genes: H4C3 (HIST1H4C), H4C4 (HIST1H4D; MIM: 602823), H4C5 (HIST1H4E; MIM: 602830), H4C6 (HIST1H4F; MIM: 602824), H4C9 (HIST1H4I; MIM: 602833), and H4C11 (HIST1H4J) (Figure 1A, Table 1). The cohort was collected with trio-based whole-exome sequencing in combination with data sharing via Genematcher13 and DECIPHER.14 This study received ethical approval from the New Zealand Health and Disability Ethics Committee (16/STH/3) and London–Riverside REC (09/H0706/20). All families provided consent to be involved in this project and separate consent was obtained for the use of photos.
Figure 1.
H4 variants identified in the cohort
(A) Highly recurrent variants were found in six different H4 genes (H4C3, H4C4, H4C5, H4C6, H4C9, and H4C11), which all encode an identical protein. Aggregate prevalence of disease-causing amino acid changes is also shown. The N-terminal methionine is cleaved from histone H4, and therefore all numbering is relative to the mature polypeptide, in keeping with the protein literature.
(B) The affected residues of H4 (orange ribbon) either cluster to the N-terminal α-helix facing toward DNA (cluster 1, purple spheres) or are located in regions buried within the nucleosome core (cluster 2, orange spheres). Size of sphere indicates the relative prevalence of substitutions affecting that residue.
Table 1.
Variants identified in H4 genes
| Number of individuals | Gene | Chromosomal position (hg38) | cDNA | Protein | H4anumbering | CADD PHRED | REVEL score |
|---|---|---|---|---|---|---|---|
| 1 | H4C3 | 6: 26,104,044 | c.97C>G | p.Pro33Ala | Pro32Ala | 23.1 | 0.526 |
| 2 | H4C3 | 6: 26,104,045 | c.98C>T | p.Pro33Leu | Pro32Leu | 25.7 | 0.438 |
| 3 | H4C3 | 6: 26,104,221 | c.274A>C | p.Lys92Gln | Lys91Gln | 26.6 | 0.706 |
| 1 | H4C4 | 6: 26,188,955 | c.122G>A | p.Arg41His | Arg40His | 23.9 | 0.459 |
| 1 | H4C5 | 6: 26,204,739 | c.95A>C | p.Lys32Thr | Lys31Thr | 26.6 | 0.523 |
| 1 | H4C5 | 6: 26,204,742 | c.98C>G | p.Pro33Arg | Pro32Arg | 25.3 | 0.541 |
| 1 | H4C5 | 6: 26,204,750 | c.106C>T | p.Arg36Trp | Arg35Trp | 26.6 | 0.494 |
| 1 | H4C5 | 6: 26,204,757 | c.113T>C | p.Leu38Pro | Leu37Pro | 27.5 | 0.659 |
| 4 | H4C5 | 6: 26,204,765 | c.121C>T | p.Arg41Cys | Arg40Cys | 26.6 | 0.446 |
| 7 | H4C5 | 6: 26,204,780 | c.136C>T | p.Arg46Cys | Arg45Cys | 26.3 | 0.589 |
| 2 | H4C5 | 6: 26,204,939 | c.295T>C | p.Tyr99His | Tyr98His | 26.9 | 0.317 |
| 1 | H4C6 | 6: 26,240,708 | c.283G>A | p.Gly95Arg | Gly94Arg | 24.5 | 0.677 |
| 1 | H4C9 | 6: 27,139,430 | c.122G>T | p.Arg41Leu | Arg40Leu | 26.7 | 0.628 |
| 2 | H4C9 | 6: 27,139,535 | c.227A>G | p.His76Arg | His75Arg | 25.3 | 0.754 |
| 1 | H4C11 | 6: 27,824,245 | c.121C>T | p.Arg41Cys | Arg40Cys | 25.8 | 0.541 |
RefSeq IDs: GenBank: NM_003542.4 (H4C3), GenBank: NM_003539.4 (H4C4), GenBank: NM_003545.3 (H4C5), GenBank: NM_003540.4 (H4C6), GenBank: NM_003495.2 (H4C9), GenBank: NM_021968.4 (H4C11).
Note on nomenclature: to refer to the residues belonging to this study, HGVS Variant nomenclature would include Methionine-1 (Met1) at the translation initiating site, e.g., H4C3 Pro33Ala (c.97C>G [p.Pro33Ala]). However, as the research field of epigenetics and oncohistones typically drops this first post-translationally removed methionine, we have also done so. Therefore, the above-mentioned example (included in this study) is referred to as H4C3 Pro32Ala.
There are fourteen canonical histone H4 genes in the human genome, clustering in three genomic loci. At a nucleotide level, all genes are different, but together they encode an identical protein. Transcription of these genes is independently regulated, and differing expression levels are observed during brain development15,16 and in human tissues.4 The genes harboring variants identified in our cohort are among the more highly expressed, however, as H4 transcripts are not polyadenylated and therefore missed in most RNA sequencing (RNA-seq) protocols, limited expression data are available.
All variants observed were absent from control databases (1KG, gnomAD v2.1.1).17 Furthermore, there were no missense variants affecting Lys91 in any of the 14 canonical histone H4 genes in gnomAD, and only extremely rarely were substitutions observed for the other positions in different H4 genes. The absence of any substitutions at Lys91 could reflect a stronger requirement for fidelity at this position, especially given the post-translation modifications of Lys91.18
The genetic findings of the cohort are striking, especially given that histones are some of the slowest evolving eukaryotic proteins and human H4 is 92% conserved with the yeast ortholog.1,2 In the human population, the histone H4 genes are tolerant to both loss-of-function and missense variation (gnomAD). We identify nine sites across the 103 amino acid protein with a variant, six of which were found recurrently (Pro32, Arg40, Arg45, His75, Lys91, and Tyr98), including two where the same site (Pro32 and Arg40) is altered in multiple different H4 genes. All sites are conserved through to Saccharomyces cerevisiae. The altered residues cluster in two main regions of histone H4 (Figure 1B); one cluster centers on the first α-helix of H4 (Figure 1B; purple spheres), a region important for DNA contacts and protein interactions with H3 and histone chaperones.18 Arg45 is positioned in the loop following the first α-helix and forms one of the sprockets of the nucleosome that contacts the minor groove of DNA. Substitutions at Arg45 in S. cerevisiae have proven to be deleterious to growth and fitness with altered chromatin remodeling.7,19,20 The second cluster is within the core of the nucleosome (Figure 1B, orange spheres), where important structural contacts exist between the H3-H4 dimer and with histone chaperones.18 Suggestive evidence for these other variants also originates from S. cerevisiae, where a Gly94 mutant (a key residue for H4 C-terminal flexibility) confers reduced viability,21 whereas a His75 mutant disrupts DNA damage repair processes.22 Other studied variants in similar regions (either somatic “oncohistone” variants in H4,7,8 or recently described germline variants in H323) are predicted to perturb either nucleosome stability or interaction with histone chaperones, suggesting the H4 variants described in our cohort most likely cause similar effects. These complementary observations, alongside the significant recurrence of specific variants, provide strong evidence for pathogenicity.
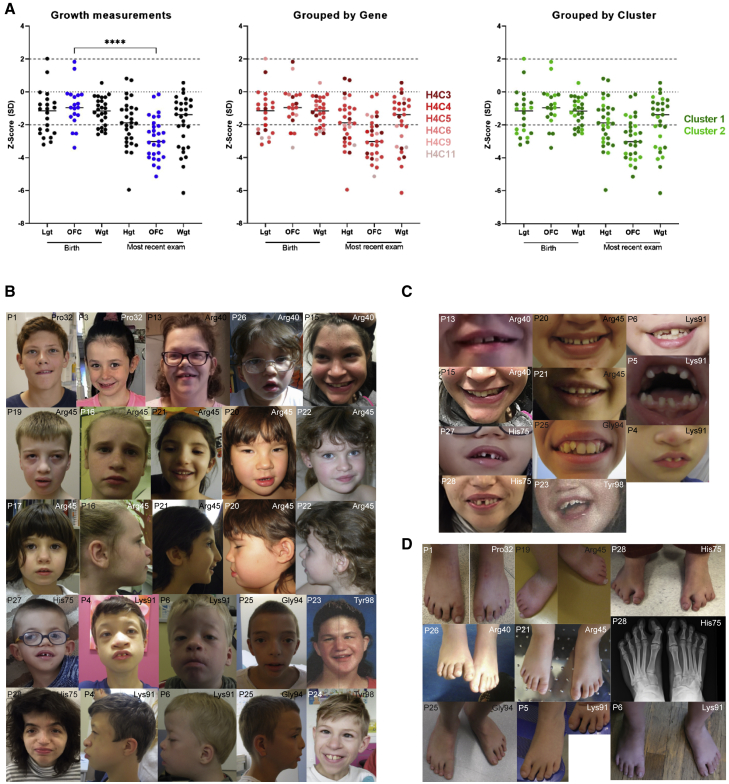
All individuals displayed intellectual disability (ID) and the majority demonstrated global and/or gross motor developmental delay (Table 2, Table S1). Other neurodevelopmental features such as hypotonia (34%), seizures (17%), or autism (17%) were present in some individuals but less common. Brain MRI was generally normal except for two individuals with delayed myelination. Microcephaly was commonly observed (Figure 2A); an occipitofrontal circumference Z score smaller than −2 SD was evident in 16% of individuals at birth and became progressively more severe with age (76% at the most recent exam). Short stature (defined as Z score for height smaller than −2 SD) and failure to thrive were common features (38%), however without significant change over time (Figure 2A). Clustering of anthropometric data by H4 gene or protein region revealed no obvious genotype-phenotype patterns (Figure 2A). The age range of the cohort is between 10 months and 52 years. Interestingly, the oldest individual shows signs of premature aging with greyed, thinning hair and wrinkly skin, looking at least two decades beyond his biological age, which did not occur in his parents. Premature aging has also been observed in Rahman syndrome (MIM: 617537), caused by pathogenic variants in H1-4 (HIST1H1E; MIM: 142220).24 This phenotype appears to be milder in our H4 cohort, however the majority of the individuals are still quite young. One individual died from leukemia stemming from myelodysplasia (P28), but no other individuals were reported to have bone marrow abnormalities.
Table 2.
Clinical features of individuals in the H4 cohort
| Clinical feature | Proportion (percentage) |
|---|---|
| Neurodevelopment | |
| Intellectual disability | 29/29 (100%) |
| Developmental delay | 29/29 (100%) |
| Hypotonia | 10/29 (34%) |
| Seizures | 5/29 (17%) |
| Autism | 5/29 (17%) |
| Ataxia | 4/29 (14%) |
| Growth | |
| Microcephaly—prenatal onset | 2/19 (11%) |
| Microcephaly—postnatal | 20/29 (69%) |
| Short stature | 11/29 (38%) |
| Failure to thrive | 11/29 (38%) |
| Skeletal features | |
| Craniosynostosis | 2/29 (7%) |
| Digit anomalies | 4/29 (14%) |
| Vertebral anomalies | 4/27 (15%) |
| Facial features | |
| Hypertelorism | 5/29 (17%) |
| Upslanting palpebral fissures | 3/29 (10%) |
| Broad nasal tip | 11/29 (38%) |
| Thin upper lip/vermillion | 4/29 (14%) |
| Teeth anomalies | 6/29 (21%) |
| Other features | |
| Recurrent infections | 4/29 (14%) |
| Visual impairment | 17/28 (61%) |
| Hearing impairment | 7/29 (24%) |
| Age range | 10 months – 52 years (median 10 years 11 months) |
Figure 2.
Clinical characteristics of individuals with histone H4 gene variants
(A) Individuals with variants in histone H4 genes demonstrate a reduction in height, weight, and brain growth (OFC, occipitofrontal circumference); the latter significantly progresses as the individuals age. There are no detectable genotype-phenotype patterns separating by the specific histone H4 gene or variant cluster. ∗∗∗∗p < 0.0001.
(B) Facial dysmorphism affecting midline structures is noticeable among the cohort, but highly variable, with no obvious genotype-phenotype correlation.
(C) Individuals can present with abnormalities in the appearance and position of teeth (for example, P5, P25). A recurring feature present in several individuals is a noticeable gap between the upper central incisors.
(D) Individuals with variants in histone H4 genes also show a spectrum of toe anomalies, ranging from no anomalies present (for example, P21) through to severe 2–3 toe (P1, P28) or 3–4 toe (P25) syndactyly, which can be bilateral. Toes can also be short (P19, P28).
Non-neurological features were variable across the cohort. Facial features comprised a wide spectrum (Figure 2B). While some individuals were relatively non-dysmorphic, others had a common presentation affecting the facial midline, with hypertelorism (17%), a high nasal bridge (or conversely very low nasal bridge) with a broad nasal base (38%) and narrow nares, wide mouth with a gap between central incisors or other tooth anomalies (21%) (Figure 2C), and moderately pointy chin. Visual impairment such as strabismus, astigmatism, or myopia were reasonably common (61% individuals), and 24% individuals demonstrated hearing impairment. Skeletal development was normal for most individuals, however recurring features such as vertebral or digit abnormalities were present in several individuals (Figure 2D), and particularly severe in individual P28. Variability in clinical features and growth was noted even among the seven individuals harboring the same de novo variant in H4C5 encoding Arg45Cys (Figure 2B, Figure S1).
To validate the pathogenic effect of these variants, we expressed wild-type human histone H4 and the histone H4 variants pertaining to this study in zebrafish by means of mRNA microinjection (Figure 3). The complete conservation of the zebrafish and human histone H4 proteins at the amino acid sequence level and the early, mRNA-mediated ectopic overexpression make this an optimal set-up for assaying the variants’ effect on early development. Early embryonic effects were evaluated at 28 hours post fertilization (hpf) as fundamental developmental processes such as gastrulation and primary organogenesis are completed by this time point. All variants tested displayed significant visible developmental effects compared to microinjection of the corresponding wild-type H4 gene, except the Pro32Ala and Arg40Cys variants. Specifically, classification was based on cephalic development, anterior-posterior axis establishment, and tissue necrosis during early embryonic development, as these parameters captured the major defects and the cellular toxicity (Figures 3A–3C) observed across all variants analyzed. Phenotypic observation revealed that variants affecting Lys91 and His75 caused the strongest effects. Interestingly, these two variants both cluster at the core of the nucleosome, and His75 plays a role in DNA damage repair,22 a process previously suggested to be involved in the syndromic features of Lys91 individuals.4 His75 is at the interface between two histone molecules within the octamer (in this case, H4 and H2B) and it is located in the lrs (loss-of-ribosomal DNA silencing) domain of H4,22 a nucleosomal surface structure reported to have gene-specific silencing function in yeast.19 Additionally, we observed a dosage-dependent effect for Arg40His and Arg45Cys (Figures 3D and 3E), which has been noted for other sprocket arginine substitutions and may relate to possible roles in higher-order organization of chromatin.25 Interestingly, such sprocket function has been reported for both the sin (switch-independent) and lrs H4 domains containing Arg45 and His75, respectively.26 These regions have an almost identical three-dimensional structure26 and play a role in gene regulation (repression), the perturbation of which most likely results in pathogenicity and was detected in our functional assay. The variability in frequency of phenotype occurrence across variants may reflect the importance of the affected residues in cellular processes crucial to active cell proliferation, as the early zebrafish embryo is a system in which cells have a relatively short cell-cycle time. The milder effect observed in our functional assay for variants affecting Pro32 and Arg40 may point to a moderate requirement, however the strong recurrence of variants affecting these residues in our cohort provides alternative evidence to support their pathogenicity. Altogether, these results provide a first picture of the variety of phenotypes caused by the assayed variants, supporting our genetic data. However, as their interpretation is limited by their transience and the inherent variability of our current mRNA assay, further testing in a stable model is required to obtain more insight in the molecular mechanisms linking genetic variants and phenotype.
Figure 3.
H4 variants induce developmental defects in zebrafish embryos
(A) Phenotypical characterization in 28 hpf embryos. Representative images of observed phenotypes in zebrafish embryos 28 hpf microinjected with mRNA encoding either wild-type or identified variants at the one-cell stage. The different classes are defined on general development and necrosis. Arrowhead, cephalic necrosis; arrow, curved tail.
(B and C) High magnification examples of cephalic necrosis (B) and curved tail (C) phenotypes.
(D and E) Quantification of the phenotypical classification as described in (A). Variants reported in (D) were microinjected with 50 pg/embryo, and additional testing with 100 pg/embryo is reported in (E). Data marked with a hash symbol was previously published in Tessadori et al.4 Fisher’s exact test: ns, not significant; p > 0.05; ∗p < 0.05; ∗∗∗∗p < 0.0001. Scale bars: 100 μm (A); 50 μm (B and C).
Through genetic and developmental findings, we have identified pathogenic substitutions in six genes encoding histone H4 in a large cohort of individuals with a neurodevelopmental syndrome. Despite well-established cellular requirements for post-translational modification of the N-terminal tail of H4,27, 28, 29 it is notable that no de novo variants were identified in this region.
Recently, a large cohort of individuals with a neurodegenerative and developmental disorder were reported harboring de novo missense variants in the histone H3 replication-independent genes H3F3A (MIM: 601128) and H3F3B (MIM: 601058).23 While there are phenotypic commonalities between the H3.3 cohort and individuals presented here, the presentations are distinct. The H3F3A/H3F3B cohort appears to have a more expansive neurological dysfunction with anomalies noted on imaging, accompanied by septal or genital abnormalities and craniosynostosis. In contrast, our H4 cohort has ID/DD and microcephaly as presenting features, but only more rarely are there other neurodevelopmental abnormalities. The pathogenic variants identified in H3F3A and H3F3B are located throughout the protein, with a smaller number of recurrently occurring variants. In comparison, for histone H4, the high level of recurrent variants observed is significant, along with the clear clustering of the variants in two regions of the H4 protein.
The redundancy of the H4 genes in the human genome is remarkable. Loss-of-function variants in H4 genes are present in the healthy population17 and are even present in homozygous form,30 supporting our hypothesis that the variants identified here act through a dominant effect. This disease mechanism, combined with the paralogous landscape of H4 genes, presents opportunities for future treatment strategies through targeted knockdown of specific H4 gene products.
Acknowledgments
The authors thank all individuals and their families who were involved in this study; Marine Tessarech for molecular diagnostic assistance; Anisha Chopra for experimental work; and Jacques Giltay, Meriel McEntagart, and Cynthia Morton for advice. F.T. is supported by NWO grant NWO/OCENW.GROOT.2019.029, A.O.M.W. was supported by the NIHR Oxford Biomedical Research Centre, K.K. and L.S.B. were supported by the Marsden Fund, and L.S.B. was supported by a Rutherford Discovery Fellowship, both administered by the Royal Society of New Zealand. A.O.D.L. is supported by a Manton Endowed Scholar award. Project support was provided by Fondazione Bambino Gesù (Vite Coraggiose), Italian Ministry of Health (5x1000, CCR-2017-23669081 and RCR-2020-23670068_001), Italian Ministry of Research (FOE 2019), and the Cliff Broad Family Trust, administered by the Neurological Foundation of New Zealand. The opinions expressed here are those of the authors and do not reflect those of the Navy, the Department of Defense, or the United States government. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant HICF-1009-003), a parallel funding partnership between the Wellcome Trust with the Department of Health and the Wellcome Trust Sanger Institute (grant WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12, granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research through the Comprehensive Clinical Research Network.
Declaration of interests
The authors declare no competing interests.
Published: February 23, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.02.003.
Contributor Information
Louise S. Bicknell, Email: louise.bicknell@otago.ac.nz.
Gijs van Haaften, Email: g.vanhaaften@umcutrecht.nl.
Data and code availability
Full genetic data are not available due to privacy regulations.
Web resources
Online Mendelian Inheritance in Man, https://omim.org/
Supplemental information
References
- 1.Kamakaka R.T., Biggins S. Histone variants: deviants? Genes Dev. 2005;19:295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- 2.Pusarla R.H., Bhargava P. Histones in functional diversification. Core histone variants. FEBS J. 2005;272:5149–5168. doi: 10.1111/j.1742-4658.2005.04930.x. [DOI] [PubMed] [Google Scholar]
- 3.Ye J., Ai X., Eugeni E.E., Zhang L., Carpenter L.R., Jelinek M.A., Freitas M.A., Parthun M.R. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol. Cell. 2005;18:123–130. doi: 10.1016/j.molcel.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tessadori F., Giltay J.C., Hurst J.A., Massink M.P., Duran K., Vos H.R., van Es R.M., Scott R.H., van Gassen K.L.I., Bakkers J., van Haaften G., Deciphering Developmental Disorders Study Germline mutations affecting the histone H4 core cause a developmental syndrome by altering DNA damage response and cell cycle control. Nat. Genet. 2017;49:1642–1646. doi: 10.1038/ng.3956. [DOI] [PubMed] [Google Scholar]
- 5.Tessadori F., Rehman A.U., Giltay J.C., Xia F., Streff H., Duran K., Bakkers J., Lalani S.R., van Haaften G. A de novo variant in the human HIST1H4J gene causes a syndrome analogous to the HIST1H4C-associated neurodevelopmental disorder. Eur. J. Hum. Genet. 2020;28:674–678. doi: 10.1038/s41431-019-0552-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan Q., Dutt S., Xu R., Graves K., Juszczynski P., Manis J.P., Shipp M.A. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol. Cell. 2009;36:110–120. doi: 10.1016/j.molcel.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bagert J.D., Mitchener M.M., Patriotis A.L., Dul B.E., Wojcik F., Nacev B.A., Feng L., Allis C.D., Muir T.W. Oncohistone mutations enhance chromatin remodeling and alter cell fates. Nat. Chem. Biol. 2021;17:403–411. doi: 10.1038/s41589-021-00738-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nacev B.A., Feng L., Bagert J.D., Lemiesz A.E., Gao J., Soshnev A.A., Kundra R., Schultz N., Muir T.W., Allis C.D. The expanding landscape of ‘oncohistone’ mutations in human cancers. Nature. 2019;567:473–478. doi: 10.1038/s41586-019-1038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis P.W., Müller M.M., Koletsky M.S., Cordero F., Lin S., Banaszynski L.A., Garcia B.A., Muir T.W., Becher O.J., Allis C.D. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu C., Jain S.U., Hoelper D., Bechet D., Molden R.C., Ran L., Murphy D., Venneti S., Hameed M., Pawel B.R., et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science. 2016;352:844–849. doi: 10.1126/science.aac7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papillon-Cavanagh S., Lu C., Gayden T., Mikael L.G., Bechet D., Karamboulas C., Ailles L., Karamchandani J., Marchione D.M., Garcia B.A., et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat. Genet. 2017;49:180–185. doi: 10.1038/ng.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tropberger P., Schneider R. Scratching the (lateral) surface of chromatin regulation by histone modifications. Nat. Struct. Mol. Biol. 2013;20:657–661. doi: 10.1038/nsmb.2581. [DOI] [PubMed] [Google Scholar]
- 13.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J.A., Ding S.L., Sunkin S.M., Smith K.A., Ng L., Szafer A., Ebbert A., Riley Z.L., Royall J.J., Aiona K., et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollen A.A., Nowakowski T.J., Chen J., Retallack H., Sandoval-Espinosa C., Nicholas C.R., Shuga J., Liu S.J., Oldham M.C., Diaz A., et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tessarz P., Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 19.Kruger W., Peterson C.L., Sil A., Coburn C., Arents G., Moudrianakis E.N., Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 20.Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 21.Chavez M.S., Scorgie J.K., Dennehey B.K., Noone S., Tyler J.K., Churchill M.E. The conformational flexibility of the C-terminus of histone H4 promotes histone octamer and nucleosome stability and yeast viability. Epigenetics Chromatin. 2012;5:5. doi: 10.1186/1756-8935-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selvam K., Rahman S.A., Li S. Histone H4 H75E mutation attenuates global genomic and Rad26-independent transcription-coupled nucleotide excision repair. Nucleic Acids Res. 2019;47:7392–7401. doi: 10.1093/nar/gkz453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bryant L., Li D., Cox S.G., Marchione D., Joiner E.F., Wilson K., Janssen K., Lee P., March M.E., Nair D., et al. Histone H3.3 beyond cancer: Germline mutations in Histone 3 Family 3A and 3B cause a previously unidentified neurodegenerative disorder in 46 patients. Sci. Adv. 2020;6:eabc9207. doi: 10.1126/sciadv.abc9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flex E., Martinelli S., Van Dijck A., Ciolfi A., Cecchetti S., Coluzzi E., Pannone L., Andreoli C., Radio F.C., Pizzi S., et al. Aberrant Function of the C-Terminal Tail of HIST1H1E Accelerates Cellular Senescence and Causes Premature Aging. Am. J. Hum. Genet. 2019;105:493–508. doi: 10.1016/j.ajhg.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodges A.J., Gallegos I.J., Laughery M.F., Meas R., Tran L., Wyrick J.J. Histone Sprocket Arginine Residues Are Important for Gene Expression, DNA Repair, and Cell Viability in Saccharomyces cerevisiae. Genetics. 2015;200:795–806. doi: 10.1534/genetics.115.175885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J.H., Cosgrove M.S., Youngman E., Wolberger C., Boeke J.D. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 2002;32:273–279. doi: 10.1038/ng982. [DOI] [PubMed] [Google Scholar]
- 27.Sanders S.L., Portoso M., Mata J., Bähler J., Allshire R.C., Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Shogren-Knaak M., Ishii H., Sun J.M., Pazin M.J., Davie J.R., Peterson C.L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 29.Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D., Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeBoever C., Tanigawa Y., Lindholm M.E., McInnes G., Lavertu A., Ingelsson E., Chang C., Ashley E.A., Bustamante C.D., Daly M.J., Rivas M.A. Medical relevance of protein-truncating variants across 337,205 individuals in the UK Biobank study. Nat. Commun. 2018;9:1612. doi: 10.1038/s41467-018-03910-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Full genetic data are not available due to privacy regulations.