Summary
Neurodevelopmental disorders are highly heterogenous conditions resulting from abnormalities of brain architecture and/or function. FBXW7 (F-box and WD-repeat-domain-containing 7), a recognized developmental regulator and tumor suppressor, has been shown to regulate cell-cycle progression and cell growth and survival by targeting substrates including CYCLIN E1/2 and NOTCH for degradation via the ubiquitin proteasome system. We used a genotype-first approach and global data-sharing platforms to identify 35 individuals harboring de novo and inherited FBXW7 germline monoallelic chromosomal deletions and nonsense, frameshift, splice-site, and missense variants associated with a neurodevelopmental syndrome. The FBXW7 neurodevelopmental syndrome is distinguished by global developmental delay, borderline to severe intellectual disability, hypotonia, and gastrointestinal issues. Brain imaging detailed variable underlying structural abnormalities affecting the cerebellum, corpus collosum, and white matter. A crystal-structure model of FBXW7 predicted that missense variants were clustered at the substrate-binding surface of the WD40 domain and that these might reduce FBXW7 substrate binding affinity. Expression of recombinant FBXW7 missense variants in cultured cells demonstrated impaired CYCLIN E1 and CYCLIN E2 turnover. Pan-neuronal knockdown of the Drosophila ortholog, archipelago, impaired learning and neuronal function. Collectively, the data presented herein provide compelling evidence of an F-Box protein-related, phenotypically variable neurodevelopmental disorder associated with monoallelic variants in FBXW7.
Keywords: Neurodevelopment, global developmental delay, brain malformation, epilepsy, macrocephaly, intellectual disability, hypotonia, gastrointestinal issues, FBXW7, F-box protein
Introduction
Neurodevelopment is a complex spatiotemporal process requiring the coordinated action of genetic and environmental cues to regulate a multitude of developmental processes, including cellular proliferation, differentiation, migration, and formation of neural circuits. Neurodevelopmental disorders affect ∼2%–5% of children and result in variable neurocognitive symptoms.1, 2, 3 They are genetically and phenotypically heterogeneous and often require untargeted genomic analysis and a genotype-first approach for the discovery of novel phenotypes.4 Several neurodevelopmental disorders have been attributed to genes that regulate cell division, underscoring the importance of this process in the development of the central nervous system.5,6 F-box (FBX) proteins are essential for regulating the ubiquitination of proteins involved in the cell cycle. There are 69 human FBX proteins, which are classified into three subcategories on the basis of the structural class of their substrate-binding domains: FBXW proteins contain a tryptophan-aspartic acid 40 (WD40) repeat domain; FBXL proteins contain a leucine-rich repeat; and FBXO proteins contain other protein-interaction domains (reviewed in Nguyen et al. 7 and Zhang et al. 8). FBX proteins are incorporated as one subunit of a tetrameric SCF (SKP1-CUL1-FBX) ubiquitin ligase complex. First, the FBX protein aggregates the phosphorylated target protein independently of the other complex subunits, then it attaches to the adaptor protein S-phase kinase-associated protein 1 (SKP1), which links it to the major structural scaffold protein cullin 1 (CUL1). CUL1 links SKP1 to the ring-box 1 (RBX1) protein, which facilitates the transfer of a ubiquitin molecule to the protein target, now marked for degradation via the ubiquitin proteasome system (UPS).8
To date, germline variants in five genes encoding FBX proteins have been found to underlie neurodevelopmental disorders. De novo missense variants in FBXW11 (MIM: 605651) located in the encoded WD40 domain repeats have been associated with mild to severe neurodevelopmental disability, often accompanied by behavioral abnormalities and mandibular, ocular, and digital features.9 De novo frameshift, nonsense, splicing, and missense variants in FBXO11 (MIM: 607871) result in mild to severe intellectual disability with dysmorphic facies and behavioral abnormalities.10,11 De novo variants in FBXO28 (MIM: 609100) have been identified in individuals with severe to profound intellectual disability (ID) and epilepsy with various seizure types,12,13 confirming the initial suggestions that the gene was the primary phenotypic determinant in chromosome 1q41q42 microdeletion syndrome.14,15 Autosomal-recessive inheritance has also been observed in FBX-related phenotypes; biallelic variants in FBXL4 (MIM: 605654) cause mitochondrial DNA depletion syndrome with encephalomyopathy,16,17 and in FBXL3 they cause intellectual disability with dysmorphic features and short stature.18 Additionally, KDM2B (MIM: 609078), also known as FBXL10, is a candidate neurodevelopmental-disease-associated gene with a homozygous variant identified in two siblings with developmental delay, hypotonia, and infantile spasms;19 additionally, monoallelic single-nucleotide variants and chromosomal microdeletions involving this gene have also been identified in individuals with syndromic intellectual disability.20,21
F-box- and WD-repeat-domain-containing 7 (FBXW7; GenBank: NG_029466.2; MIM: 606278) has been extensively studied as a tumor suppressor (reviewed in Yeh et al.22 and Sailo et al.23). However, it has also been implicated in a variety of diverse biological processes, including the immune response,24,25 liver lipid metabolism,26 angiogenesis,27,28 cardiac hypertrophy,29 haemopoiesis,30 neurodevelopment31, 32, 33, 34, 35, 36 and excitotoxicity.37,38 Herein we provide a detailed characterization of 35 individuals from 32 families identified through global matchmaking databases and found to have 28 germline de novo and inherited monoallelic FBXW7 variants associated with neurodevelopmental disability and variable features. Evidence from in silico protein modeling, cell-based functional studies, and Drosophila neuronal knockdown converge to support the discovery that pathogenic variants in FBXW7 cause an FBX-related neurodevelopmental syndrome.
Subjects and methods
Subjects and FBXW7 variant analysis
All procedures were approved by institutional human research ethics committees, and informed consent was obtained for all individuals. Individuals were clinically evaluated in separate centers, and DNA samples were analyzed by chromosomal microarray or genomic sequencing (exome or genome, with singleton or trio analysis) on a clinical or research basis. Contact between researchers was facilitated with web-based tools Matchmaker Exchange39 and GeneMatcher.40 High-confidence candidate variants, categorized as either predicted LoF or damaging candidates, absent from gnomAD and classified as pathogenic according to the American College of Medical Genetics (ACMG) guidelines41 are reported. The functional outcome of splice-site variants was predicted with BDGP NNSPLICE 0.9,42 NetGene243,44 and Splice AI.45
In silico modeling of the impact of FBXW7 variant interaction with CYCLIN E1
The structure of CYCLIN E1 (amino acids [aa] 89–395) was built under the default parameters of the i-TASSER website.46 The complex between FBXW7 and CYCLIN E1 was then modeled with Schrodinger (2020-3). The highest-resolution experimental X-ray structures of FBXW7 (aa 263–706, PDB: 2OVR)47 and the modeled CYCLIN E1 from i-TASSER were used for building the complex. A restraint docking approach was applied in Schrodinger. There were four restraints (between 4 and 6 Å) that were applied to the residues between FBWX7 and CYCLIN E1, namely Ser384(CYCLIN E1)-Arg479(FBXW7), Thr380(CYCLIN E1)-Arg505(FBXW7), Thr380(CYCLIN E1)-Arg465(FBXW7), and Thr380(CYCLIN E1)-Arg479(FBXW7).47 We then screened the top solutions to evaluate them by their ability to satisfy the experimental data.
FBXW7 missense variants were first annotated for predicted consequences via the Variant Effect Predictor (release 101) including dbNSFP (4.1a) output.48,49 MTR scores were included from the MTR-Viewer. We selected a number of these scores to capture conservation, physicochemical properties, and genic intolerance. We examined structural properties by using the mCSM suite to manually map the missense variants to the homology-modeled complex of FBXW7 with CYCLIN E1 bound. We used mCSM to predict changes to thermodynamic stability (ΔΔG) and mCSM-PPI2 to predict changes to binding affinity.50,51 Additionally, changes to charge, volume, and residue nature were reported for each substitution.
Functional analysis of FBXW7 variants
The open reading frame of FBXW7 variants (GenBank: NM_001349798.2; c.1267G>A [p.Gly423Arg]; c.1439A>G [p.Asp480Gly]; c.1631T>G [p.Val544Gly]; c.1920C>A [p.Ser640Arg]; c.2020C>T [p.Arg674Trp]; c.2021G>C [p.Arg674Pro]; and c.2066G>A [p.Arg689Gln]); and known substrates E1 CYCLIN (GenBank: NM_001238.4) and E2 CYCLIN (GenBank: NM_057749.3) were synthesized, their sequences were verified, and they were cloned inframe into C-terminal- epitope-tagged vectors pcDNA3.1/Myc-His (ThermoFisher, V80020) and pcDNA3.1/V5-His (ThermoFisher, V81020), respectively (Integrated DNA Technologies).
HEK293T cells (American Type Culture Collection CRL-3216) were transiently transfected with an FBXW7 variant alone or in combination with either a known substrate or empty vector through the use of Fugene HD (Promega, E2311) and harvested at 60–72 h after transfection. Where indicated, cells were treated with 5 μM MG-132 (Merck, 1474790) or DMSO (Sigma, D2650) at 48 h after transfection and harvested after 16 h. Protein lysates were obtained by resuspension and sonication (Digital Sonifier Cell Disruptor 250, Branson) in 2% SDS, 10 mM TRIS (pH 7.5) with 1× Complete Protease Inhibitor Cocktail (Roche, 11697498001) followed by protein estimation (ThermoFisher, 23225).
Immunoblots were performed on 50 μg of total protein via the Criterion TGX system (BioRad) and probed sequentially with antibodies to anti-c-Myc (9E10, Abcam, AB32, 1:5000); anti-V5 (ThermoFisher, R960, 1:5000), and GAPDH (1D4, Novus Biologicals, NB300-221, 1:5000). Primary antibodies were detected with goat anti-mouse IgG (H+L, Jackson ImmunoResearch, 115-005-003, 1:10000), and bands were visualized with the Clarity Western ECL Substrate (BioRad, 1705061) and the Amersham Imager 680 (GE Health, 29270772).
Densitometry of detected bands was recorded for semiquantitative analysis between samples. Lanes and bands were identified automatically and then manually modified where appropriate; the rolling-ball method was used for background correction. Individual sample values were first determined by normalization of the intensity of the protein of interest to the housekeeping control protein for each individual sample. To control for individual blot variation, we then normalized each sample to the intensity of the signal of the FBXW7 WT sample before combining samples for statistical significance testing by a two-sample, two-tailed Student’s t test; p < 0.05 was considered significant.
Drosophila ago knockdown models
Two Drosophila UAS-RNAi lines (RNAi-1, BL34802; and RNAi-2, BL31501), both previously validated52,53 and carrying inducible RNAi constructs against the FBXW7 homolog archipelago (ago; CG15010; FBgn0041171), and the matching genetic background control (BL36303) were obtained from the Bloomington Drosophila Stock Center. Drosophila stocks were maintained at room temperature on standard Drosophila diet (sugar, cornmeal, agar, and yeast).
The efficiency and relative strength of ago RNAi-1 and ago RNAi-2 constructs were determined by quantitative real-time-PCR (qPCR) analysis. The ago RNAi-1 and ago RNAi-2 lines and their genetic background controls were crossed to the ubiquitous Act-Gal4/TM3 Sb Tb driver, and mRNA was extracted from wandering L3 larva of the appropriate genotype with QIAGEN’s Rneasy Lipid Tissue Mini Kit. DNase treatment was performed with QIAGEN’s RNase-Free DNase Set, and cDNA was synthesized with the Bio-Rad iScript cDNA synthesis kit according to the manufacturer’s protocols. PCRs were performed with primers targeting ago (5′-GGCCACGACGATCATGTG-3′ and 5′-GACTTTGAGCGTGCGATCC-3′) and β′COP (5′-AACTACAACACCCTGGAGAAGG-3′ and 5′-ACATCTTCTCCCAATTCCAAAG-3′) with the GoTaq qPCR Master Mix (Promega) on an Applied Biosystem Fast 7500 Real-Time machine. The initial denaturation was performed for 10 min at 95°C, followed by 15 s at 95°C and 30 s at 60°C for 40 cycles (qPCR data collection). The products were then denatured at 95°C for 1 min and cooled to 65°C for 1 min (melt curve data collection). For each condition, three biological and three technical replicates were analyzed. Differential gene expression was calculated via the 2ΔΔCt method.54 The average Ct value for each sample was calculated and subtracted from the Ct value of the reference gene so that the ΔCt value could be calculated.55 A two-sample t test (equal variance) comparing the 2ΔΔCt values of the RNAi line and genetic background control was performed in Microsoft Excel for calculation of p values).
For inducing neuronal knockdown, the UAS-RNAi lines were crossed to either of two panneuronal promotor lines: (1) elav(III)-Gal4 with genotype “w1118; 2xGMR-wIR; elav-Gal4, UAS-Dicer-2" and to (2) elav(I) – Gal4 with genotype “c155-Gal4, GMR-wIR; +; +". The latter is a strong Gal4 insertion into the endogenous elav locus. Crosses were maintained at 25°C, 70% humidity in a 12 h:12 h light:dark cycle. Habituation learning and basal motor function were tested in the light-off jump-reflex habituation and fatigue assays, as previously described.56 In brief, three- to four-day-old males were individually placed in semi-transparent vials enclosed by two microphones. The filled vials were inserted into two independent 16-unit light-off jump-habituation systems (Aktogen) and left to acclimatize for 5 min before the start of the habituation paradigm assay, in which 32 flies were simultaneously exposed to 100 light-off pulses of 15 ms with a 1 s inter-trial-interval. The noise amplitude produced by wing vibrations was recorded for 500 ms after each light-off pulse. The measured sound amplitudes were filtered with a threshold to remove background noise, leading to the annotation of a jump at amplitude above 0.8. The jumps were collected and analyzed by a custom-made Labview Software (National Instruments). A high initial jump response to the light-off pulse decreased with the increase of the number of repeated pulses. A fly was considered to have habituated when it failed to jump for five consecutive light-off pulses (no jump criterion). The last jump was then stated as the number of trials needed to reach the no-jump criterion (trials to criterion, TTC). If the fraction of flies jumping to at least one of the first five light-off pulses (initial jump response) was <50%, genotypes were classified as non-performers on the basis of reduced motor performance of the tested population. Habituation per genotype was quantified as the mean trials to criterion (mTTC) of all flies of the same genotype.
The fatigue assay was performed after the habituation assay, which was equivalent to the habituation assay but involved two adaptations; (1) increased inter-trial-interval from 1 to 5 s and (2) shortened trial length from 100 to 50 light-off pulses. The increased inter-trial-interval prevented the flies from habituating and thereby elicited a jump response at each light-off trial. As for the habituation assay, the no-jump criterion was five consecutive pulses without a jump. Failing to jump for five consecutive light-off pulses in this assay was identified as a basal failure to execute jumping and was deemed to be due to increased fatigue. The last jump was scored as the number of trials it took to reach the no-jump criterion (TTC). The TTCs of the simultaneously measured flies of the same genotype were averaged (mean TTC (mTTC)).
Statistics
Protein density and qPCR: statistical significance was assessed by a two-sample, two-tailed Student’s t test, and p < 0.05 was considered significant.
Drosophila behavior: the effect of the genotype on habituation and fatigue was scored by comparison of log-transformed TTC values of the mutant versus the control flies after correction for the experimental day and system via a linear-model regression analysis with R statistical software (v.3.0.0).56
Results
Monoallelic FBXW7 variants are associated with neurodevelopmental disability, brain anomalies, hypotonia, and gastrointestinal issues
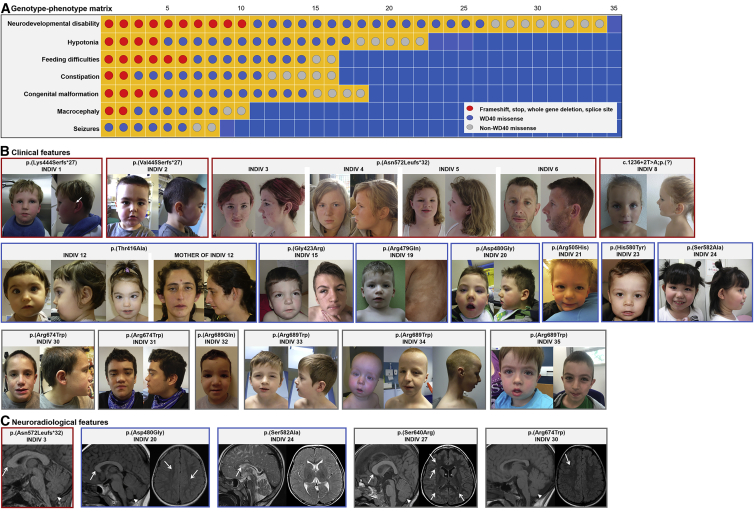
Using clinical or research-based chromosomal microarrays, genomic sequencing (trio genome or exome), and the global matchmaking platforms Matchmaker Exchange39 and GeneMatcher,40 we have identified 35 individuals (26 male, 74.3%) from 32 families with 28 distinct variants in FBXW7. The variants arose de novo in 30 individuals, including two individuals displaying mosaicism and two showing familial transmission from an affected parent (Table S1). The clinical phenotype is characterized by neurodevelopmental disability (34/35; 97.1%), including global developmental delay and intellectual disability ranging from borderline to severe, language disorder, and hypotonia (22/35; 62.9%); individual 21 was severely affected and had episodes of developmental regression and progressive spasticity. Seizures of varying types were reported in 8/35 (22.9%) individuals. Feeding difficulties and constipation were each reported in 16/35 (45.7%) individuals. Growth was generally within normal limits, but macrocephaly was noted in 10/35 (28.6%) and microcephaly in 2/35 (5.7%) individuals. Congenital anomalies were diverse and included palatal, uvular, or laryngeal anomalies (11/35, 31.4%); cardiac anomalies (11/35, 31.4%); and cryptorchidism (5/26 males, 19.2%) (Figure 1, Table 1 and Table S2).
Figure 1.
Characteristics of FBXW7 neurodevelopmental syndrome
(A) Genotype-phenotype matrix of clinical features of key phenotypes associated with FBXW7 neurodevelopmental variants. Each square represents an individual overlaid with variant class, and each row represents a clinical feature (affected—yellow; unaffected—blue). Variant types are depicted by dots: red (frameshift, stop, whole-gene deletion), blue (missense affecting WD40 domain), and gray (missense not in a WD40 domain).
(B) Clinical features of affected individuals depicting phenotype by variant type: individual 1, aged 3 years, frontal and lateral, with arrow marking preauricular pit; individual 2, aged 3 years 2 months, frontal and lateral; individual 3, aged 14 years 9 months, from family 1, frontal and lateral; individual 4, aged 11 years 9 months, from family 1, frontal and lateral; individual 5, aged 6 years 3 months, from family 1, frontal and lateral; individual 6, aged 44 years, father of individuals 3–5 from family 1, frontal and lateral (note midface retrusion with class III malocclusion); individual 8, aged 5 years, frontal; individual 12 at 12 months, frontal, lateral, and at 26 months, frontal; mother of individual 12, aged 34 years, frontal and lateral; individual 15 at 3 years and 15 years; individual 19 at age 6 years, frontal, and with cutaneous Blaschkoid dyspigmentation suggestive of somatic mosaicism; individual 20 at 5 years, frontal and lateral; individual 21 at 3 years; individual 23 at 3 years; individual 24 at 5 years, frontal and lateral; individual 30 aged 15 years, frontal and lateral; individual 31 aged 15 years, frontal and lateral; individual 32 aged 2 years; individual 33 aged 10 years; individual 34 aged 1 year, frontal, and 12 years, frontal and lateral; individual 35 aged 3 years and 7 years, frontal. Deeply set eyes with upper eyelid fullness are evident in individuals 1, 2, 3, 5, 15, 21, 24, 32 (also in individual 21, not pictured).
(C) Neuroradiological features of selected individuals; sagittal images of T1-weighted brain MRI scans of individuals 3, 20, 27, and 30 and T2-weighted brain scan of individual 24, displaying large cerebellar vermis with tonsillar ectopia (white arrowheads) and thick callosal genu (arrows)—note the generally thinned corpus callosum in individuals 24 and 30; axial T1-weighted brain MRI scans of individuals 20, 27, and 30 and T2-weighted brain scan of individual 24 displaying scattered subcortical white-matter hyperintensities and severely delayed myelination, equivalent to 7–10 months.
Table 1.
Demographic and clinical features of affected individuals
| Demographic features | |
| Sex | 26 male/9 female |
| Age range | 23 months–44 years, 6 months |
| Medical history | |
| Prenatal history | Normal; only one premature birth |
| Neurologic or CNS features | |
| Hypotonia (HP: 0001252) | 22/35 (62.9%) |
| Seizures (HP: 0001250) | 8/35 (22.9%) |
| Ataxia (HP: 0001251) | 2/35 (5.7%) |
| Developmental regression (HP: 0002376) | 1/35 (2.9%) |
| Abnormality of brain morphology (HP: 0012443) | 13/17 (76.5%) |
| Macrocephaly (HP: 0000256) | 10/35 (28.6%) |
| Microcephaly (HP: 0000252) | 2/35 (5.7%) |
| Development, cognition, and psychiatric features | |
| Neurodevelopmental abnormality (HP: 0012759) | 34/35 (97.1%) |
| Mild-moderate developmental delay or intellectual disability (HP: 0011342, HP: 0011343, HP: 0001256, and HP: 0002342) | 27/35 (77.1%) |
| Severe global developmental delay or intellectual disability (HP: 0011344, HP: 0010864) | 3/35 (8.6%) |
| Delayed speech and language development only (HP: 0000750) | 1/35 (2.9%) |
| Specific learning disability (HP: 0001328) | 2/35 (5.7%) |
| No neurodevelopmental abnormality | 1/35 (2.9%) |
| Ophthalmologic features | |
| Strabismus (HP: 0000486) | 5/35 (14.3%) |
| Abnormality of refraction (HP: 0000539) | 6/35 (17.1%) |
| Astigmatism (HP: 0000483) | 1/35 (2.9%) |
| Cerebral visual impairment (HP: 0100704) | 1/35 (2.9%) |
| Audiology and hearing | |
| Mixed hearing impairment (HP: 0000410) | 2/35 (5.7%) |
| Oral, dentition, and other ENT features | |
| Abnormal palate or uvula morphology (HP: 0000174), (HP: 0000172) | 10/35 (28.6%) |
| Laryngeal cleft (HP: 0008751) | 1/35 (2.9%) |
| Cardiac features | |
| Abnormal heart morphology (HP: 0001627) | 11/35 (31.4%) |
| Respiratory features | |
| Recurrent pneumonia (HP: 0006532) | 3/35 (8.6%) |
| Gastrointestinal and feeding features | |
| Feeding difficulties, including difficulties with nasogastric tube feeding (HP: 0011968, HP: 0040288) | 16/35 (45.7%); 5/16 (31.3%) |
| Constipation (HP: 0002019) | 16/35 (45.7%) |
| Gastresophageal reflux (HP: 0002020) | 7/35 (20.0%) |
| Renal and genitourinary features | |
| Cryptorchidism (HP: 0000028) | 5/26 (19.2%) |
| Hematologic features | |
| Neutropenia (HP: 0001875) | 2/35 (5.7%) |
n = 35; the frequency of clinical features is expressed as a fraction (and percentage) of the number assessed for that feature.
There was no recognizable facial gestalt; however, we noted deeply set eyes with upper eyelid fullness in 9/35 (25.7%) individuals. Other craniofacial features in some individuals included cleft (overt and submucous) or high palate (10/35, 28.6%), midface retrusion with class III malocclusion (1/35, 2.9%), and a tall or broad forehead (4/35, 11.4%). In individual 19 with somatic mosaicism of the FBXW7 variant, we observed cutaneous Blaschkoid dyspigmentation.
Neuroimaging was undertaken in 17 individuals (15 by MRI, one by CT, and one by both modalities); brain anomalies were identified in 13/17 (76.5%) individuals and included an absent, hypoplastic, or dysplastic corpus callosum (7/17; 41.2%); an abnormal cerebellum (5/17; 29.4%); delayed myelination (2/17; 11.7%); a thick brainstem (2/17; 11.7%); and polymicrogyria (2/17; 11.7%) (Table 1 and Table S2). Scattered small subcortical calcifications were noted on a computed tomography brain scan of individual 22. Ten brain MRI scans of seven individuals (3, 18, 19, 21, 25, 28, and 31) were available for systematic review by a pediatric neuroradiologist (S.M.). The most common anomalies were related to the posterior fossa, where the cerebellum was enlarged or at the upper limit of the normal range, except in individual 19, who had severe cerebellar atrophy with large folia and a thick dysmorphic corpus callosum and brainstem. Notably, this individual, previously reported as patient IV.1,57 also has a familial CACNA1A pathogenic variant of variable expressivity, c.835C>T (p.Arg279Cys) (GenBank: NM_023035.2). Although FBXW7 is a known tumor suppressor, none of the individuals in our cohort has so far developed cancer; the oldest individual is 44 years old. Notably, 13 of the 28 variants observed in this cohort are also reported in somatic form in the COSMIC database, which has collated 1,481 (440 unique) known somatic variants that span the entire coding region of FBXW7 in various cancer types (Figure S1 and Table S1).
Germline FBXW7 missense variants identified in this cohort cluster within the substrate-binding surface of the WD40 domain
We identified 28 germline FBXW7 variants in 35 individuals (Figure 2 and Table S1). Two individuals had large chromosomal deletions encompassing FBXW7. One individual had a canonical splice-site variant, c.1236+2T>A, which is predicted to result in donor-site loss. Seven individuals (three de novo and four familial) had frameshift variants affecting the longest transcript (GenBank: NM_001349798.2). Two variants, c.1331_1332del (p.Lys444Serfs∗27) and c.1332dup (p.Val445Serfs∗27), are predicted to undergo nonsense-mediated decay (NMD) with presumed loss of function (LoF). In contrast, c.1331_1332del (p.Asn572Leufs∗32) and c.1939A>T (p.Lys647∗) are within the 54 bp upstream of the final intron/exon junction and are predicted to escape NMD.58 These truncated proteins might be targeted for degradation via the UPS. The remaining 25 individuals had 21 unique missense variants clustering at the carboxy-terminal half of the protein, and 16/21 (76.2%) of these variants occurred within the WD40 domain. Three variants, c.1267G>A (p.Gly423Arg); c.2020C>T (p.Arg674Trp); and c.2065C>T (p.Arg689Trp), were recurrent in unrelated individuals.
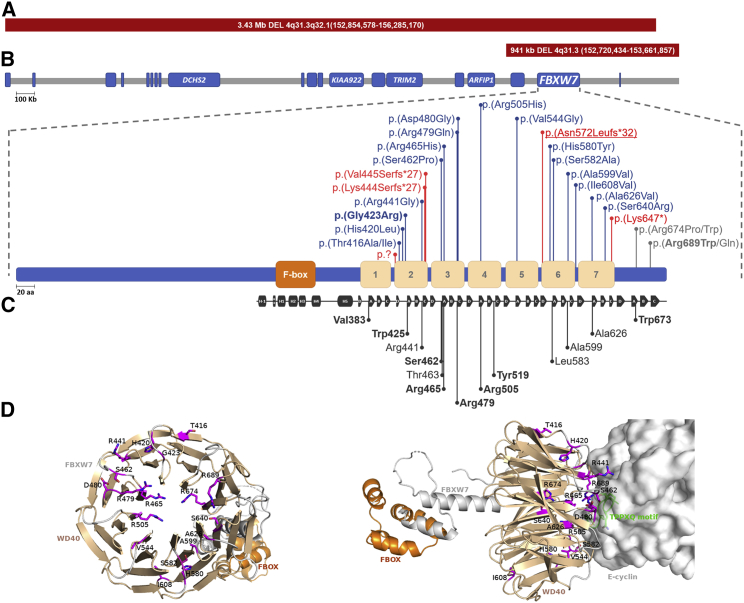
Figure 2.
FBXW7 variants detected in this cohort cluster within the substrate-binding surface of the WD40 domain
(A) The gene structure surrounding FBXW7 on chromosome 4 (GRCh37: 4q31–3q32.1) demonstrates the genomic position of two large genomic deletions identified in individuals 8 and 9 (thick red bars above chromosome).
(B) Missense FBXW7 variants identified in this study cluster within the WD40-repeat domain. Frameshift, stop-gain, or splice-site (red) and missense (blue—within the WD40 domain; and gray—outside the WD40 domain) variants are shown above the protein. Recurrent non-familial (bold); recurrent familial (underlined); F-box domain (orange); and a WD40-repeat domain (beige) derived from DECIPHER.
(C) Representation of the resolved structure of FBXW7 when in complex with a CYCLIN E1 degron (residues 360–390) demonstrating that the residues of FBXW7 that directly interact with CYCLIN E1 span similar residues as disease variants identified in this cohort. The positions of FBXW7 residues that directly interact with CYCLIN E1 are shown below the schematic depiction. F-box helices (rectangles H-1, H0, and H1–H3), linker α-helical domain (rectangles H4–H5), and the canonical eight-bladed β-propeller structure of the WD40 domain with each blade consisting of four antiparallel β strands (arrows [A–D] are shown,47 Amino acids in bold have also been shown to directly interact with DISC1.59
(D) FBXW7 variants associated with neurodevelopmental disorder are predominantly located at the substrate-binding surface of the WD40 repeat domain. The location of residues (in sticks with carbon atoms in purple, nitrogen atoms in blue, and oxygen atoms in red) impacted by mutations is shown on the tertiary structure of FBXW7 (cartoon) in configuration with CYCLIN E1 (surface in gray). The docking location of the conserved FBXW7 substrate-binding TPPXQ motif (cartoon in green) of CYCLIN E1 is demonstrated in close proximity to many of the impacted residues. Figure S1 provides an overlay of the variant residue with the wild-type residue for each individual variant, allowing identification of the change predicted in interaction for each missense variant.
The crystal structure of the FBXW7 and SKP1 complex has been determined with the substrates CYCLIN E1 and DISC1.47,59 The F-box domain located in the N-terminal half of FBXW7 mediates interaction with SKP1, whereas the WD40 domain forms a canonical eight-bladed β-propeller structure. Thirteen residues positioned at the top surface of the propeller directly interact with CYCLIN E1 (seven of these also interact with DISC1). The position of the variants identified in this study aligns with the residues required for this interaction: Arg441, Ser462, Arg465, Arg479, Arg505, and Ala599. A further four variants, c.1267G>A ((p.Gly423Arg)); c.1744T>G (p.Ser582Ala); c.2021G>C (p.Arg674Pro); and c.2020C>T (p.Arg674Trp), impact residues adjacent to critical residues Trp425, Leu583, Trp673, respectively.
To investigate the potential functional impact of the variants observed in this cohort, we mapped the amino acid position to the tertiary structure previously resolved for FBXW7 by crystallography (amino acids 263–706).47 This demonstrated fthat the amino acids implicated in disease cluster at the surface of the substrate-binding interface (Figure 2 and Figure S2). Using the mutation Cutoff Scanning Matrix (mCSM) suite, we tested the predicted impact of each missense variant on the stability of FBXW7. Our tests demonstrated that 16/21 (76.2%) variants are predicted to decrease FBXW7 stability (average −0.735 ± 1.05 ΔΔG; Table S3). Next, we assessed the distance to the interface and the binding affinity to determine the potential of the variants to impact the interaction with CYCLIN E1. This demonstrated that FBXW7 missense variants identified in this cohort are positioned very close to the interaction interface (average 7.80 ± 5.24 Å) and that 13 (65%) are predicted to decrease the binding affinity of FBXW7 to CYCLIN E1 (average −0.39 ± 0.74 ΔΔG).
None of the variants detected in this cohort were observed in the population database, gnomAD v2.1 (140k exomes and genomes); however, for each of three variants—c.1394G>A (p.Arg465His), c.1436G>A (p.Arg479Gln), and c.1796C>T (p.Ala599Val) —an ultra-rare (allele frequency < 0.000005) alternative amino acid substitution, c.1393C>T (p.Arg465Cys), c.1435C>G (p.Arg479Gly), and c.1796C>G (p.Ala599Gly), respectively, has been observed. Nevertheless, these gnomAD substitutions were detected with allele balance rates of ≤45% where mosaicism could not be excluded (Table S1). To further assess how the variants observed in our cohort differed from variants reported in gnomAD, we investigated the impact of the 78 gnomAD missense variants on the resolved FBXW7 crystal structure. Although the majority of gnomAD variants (69; 89%) are predicted to have a destabilizing effect on the protein (average −0.723 ± 0.616 ΔΔG), these variants are dispersed throughout the structure and positioned much farther from the interface with CYCLIN E1 (average 29.24 ± 17.24 Å) than the variants identified in this study (Figure S1 and Table S4). Furthermore, the gnomAD variants are predicted to have a very small effect on the binding affinity to CYCLIN E1 (average −0.064 ± 0.25 ΔΔG).
Disease-associated variants impair the ability of FBXW7 to degrade substrates CYCLIN E1 and CYCLIN E2
To experimentally determine the functional consequences of FBXW7 variants observed in this cohort, we cloned a subset into a mammalian expression vector with a C-terminal Myc tag and exogenously expressed in HEK293T cells. We selected variants within the WD40 domain (p.Gly423Arg, p.Asp480Gly, p.Val544Gly, and p.Ser640Arg) and outside the WD40 domain (p.Arg674Trp, p.Arg674Pro, and p.Arg689Gln) for cloning. The steady-state amount of FBXW7 protein was assessed by immunoblot, and all mutant proteins were detected (Figure 3A). Relative to FBXW7wild type, FBXW7Arg674Trp and FBXW7Arg689Gln demonstrated a decrease in steady-state protein amount, 0.42-fold and 0.16-fold, respectively, but only FBXW7Arg689Gln reached statistical significance (p = 0.007) (Figure 3B). After treatment with UPS inhibitor MG-132, only FBXW7Arg689Gln was found to have a steady-state protein amount that was increased relative to those of FBXW7wild type (3.4-fold, p = 0.003; Figure 3C). This suggests that most missense variants tested (six of seven) are unlikely to cause protein instability or consequent degradation by the UPS in vivo.
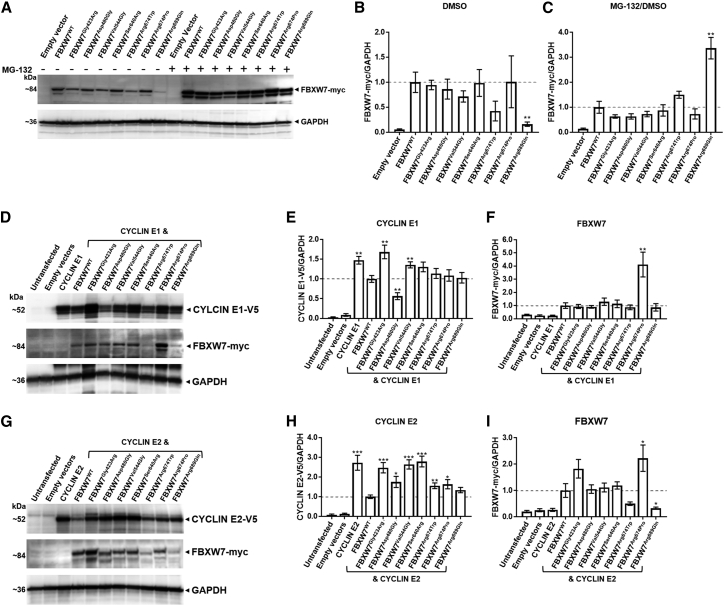
Figure 3.
Disease-associated variants impair the ability of FBXW7 to degrade substrates CYCLIN E1 and CYCLIN E2
(A) The majority of disease-associated FBXW7 variants do not impact steady-state protein amounts. Representative immunoblots of wild-type or mutant FBXW7 with and without inhibition of the ubiquitin proteasome system are shown. HEK293T cells exogenously expressing wild-type FBXW7 or mutant FBXW7 with a C-terminal Myc tag for 32 h were treated with 5 μM MG-132 for 16 h (four independent replicates).
(B) Quantification of FBXW7:GAPDH from DMSO-treated samples of mutant FBXW7 protein in (A) relative to FBXW7wild type; statistical support for altered steady-state amounts of the mutant FBXW7 protein was only evident for FBXW7Arg689Gln (p = 0.007).
(C) Quantification of FBXW7:GAPDH in MG-132-treated cells and versus their DMSO-treated counterpart in (A), demonstrating the change in steady-state mutant FBXW7 protein relative to FBXW7wild type protein after UPS inhibition; statistical support for altered steady-state protein amount was only evident for FBXW7Arg689Gln (p = 0.003).
(D) Certain FBXW7 mutant proteins demonstrate impaired CYCLIN E1 substrate degradation. Representative immunoblots of wild-type or mutant FBXW7 co-expressed with the substrate CYCLIN E1 are shown. Whole-cell lysates extracted from HEK293T cells that exogenously expressed wild-type FBXW7 or mutant FBXW7 with a C-terminal Myc tag and CYCLIN E1 with a C-terminal V5 tag for 48 h are shown (nine independent replicates).
(E) Quantification of CYCLIN E1:GAPDH in (D) for samples expressing mutant FBXW7 versus FBXW7wild type protein; statistical support for altered steady-state protein amount of CYCLIN E1 was evident for FBXW7Gly423Arg (p = 0.002), FBXW7Asp480Gly (p = 0.002), and FBXW7Val544Gly (p = 0.007).
(F) Quantification of FBXW7:GAPDH in (D) for FBXW7 mutant proteins versus FBXW7wild type protein when cells were co-transfected with CYCLIN E1. Statistical support for altered steady-state protein amount was evident only for FBXW7Arg674Pro (p = 0.005).
(G) The majority of FBXW7 mutant proteins demonstrate impaired CYCLIN E2 substrate degradation. Representative immunoblots of wild-type or mutant FBXW7 co-expressed with the substrate CYCLIN E2 are shown. Whole-cell lysates extracted from HEK293T cells that exogenously expressed wild-type FBXW7 or mutant FBXW7 with a C-terminal Myc tag and CYCLIN E2 with a C-terminal V5 tag for 48 h are shown (ten independent replicates).
(H) Quantification of CYCLIN E2:GAPDH in (G) for samples expressing FBXW7 mutant protein versus FBXW7wild type protein; statistical support for altered steady-state protein amount of CYCLIN E2 was evident for FBXW7Gly423Arg (p = 0.00005), FBXW7Asp480Gly (p = 0.02), FBXW7Val544Gly (p = 0.000005), FBXW7Ser640Arg (p = 0.000007), FBXW7Arg674Trp (p = 0.005), and FBXW7Arg674Pro (p = 0.02).
(I) Quantification of FBXW7:GAPDH in (G) for FBXW7 mutant proteins versus FBXW7wild type protein when cells were co-transfected with CYCLIN E2; statistical support for altered steady-state protein amount was evident for FBXW7Arg674Pro (p = 0.04) and FBXW7Arg674Pro (p = 0.02). All graphs present mean ± SEM. Student’s t test: ∗p < 0.05; ∗∗p < 0.01; and ∗∗∗p < 0.001.
Next, we assessed the functional impact of these missense variants by co-expressing them with C-terminal V5-tagged substrates CYCLIN E1 and CYCLIN E2 in HEK293T cells. As expected, steady-state protein amounts of CYCLIN E1 and CYCLIN E2 were reduced when co-expressed with FBXW7wild type (p = 0.002 and p = 0.0003, respectively). This confirmed that exogenously expressed wild-type FBXW7 retains its ability to ubiquitinate and degrade CYCLIN E1 and CYCLIN E2 in vitro and is provides a suitable way to assess variant effects. Collectively, variants within the WD40 domain appear to have a greater impact on the ability of FBXW7 to degrade CYCLIN E1 and CYCLIN E2 than the variants outside the WD40 domain (Figures 3D and 3E and Figures 3G/and 3H, respectively). FBXW7Gly423Arg, FBXW7Val544Gly, and FBXW7Ser640Arg were less efficient at degrading substrate; CYCLIN E1 steady-state protein amounts were elevated by 1.7-fold (p = 0.002), 1.4-fold (p = 0.007), and 1.3-fold (p = 0.06), respectively, in comparison to amounts seen with FBXW7wildtype (Figure 3E). Similarly, CYCLIN E2 steady-state protein amounts were elevated by 2.5-fold (p = 5 × 10−5), 2.6-fold (p = 4.6 × 10−6), and 2.8-fold (p = 7.5 × 10−6), respectively. By contrast, FBXW7Asp480Gly did not have a consistent effect on steady-state protein amounts of the two substrates. Although it was more efficient at degrading CYCLIN E1 than FBXW7wildtype (0.6-fold, p = 0.002) it was less efficient at degrading CYCLIN E2 (1.8-fold, p = 0.02).
The variants outside the WD domain have a more subtle impact on CYCLIN E1 and CYCLIN E2 steady-state protein amounts. Although CYCLIN E1 steady-state protein amounts were slightly increased when co-expressed with FBXW7Arg674Trp and FBXW7Arg674Pro, this did not achieve statistical significance. However, the steady-state protein amounts of CYCLIN E2 was found to be elevated 1.6-fold (p = 0.005) and 1.6-fold (p = 0.02), respectively compared to FBXW7wildtype. Notably, FBXW7Arg689Gln, which was found to be turned over by the UPS, had CYCLIN E1 and CYCLIN E2 steady-state protein amounts comparable to FBXW7wildtype, suggesting that the variant protein is able to efficiently degrade CYCLIN E1 and CYCLIN E2. FBXW7 mutant protein steady state protein amounts when co-expression with CYCLIN E1 or CYCLIN E2 were also assessed and it was found that FBXW7Arg674Pro steady-state protein amounts were increased by 4.1-fold (p = 0.2) and 2.2-fold (p = 0.02) relative to FBXW7wildtype, respectively (Figure 3F and 3I). These studies provide evidence in an in vivo cell culture model that FBXW7 missense variants identified in this cohort may destabilize the mutant protein and impact the ability of FBXW7 to degrade target substrates.
Neuronal knockdown of the FBXW7 Drosophila ortholog archipelago causes cognitive and severe neurological deficits
To address the consequences of partial loss of FBXW7 function (as seen for most of the investigated variants) in vivo, we turned to Drosophila melanogaster as a model. The Drosophila genome encodes a one-to-one FBXW7 ortholog termed archipelago (ago). The two proteins share 61% amino acid similarity, and the F-box domain and the seven WD40 repeats are highly conserved (Figure S3).60 The E3 ubiquitin ligase function of FBXW7, its role in cell-cycle progression and growth, and its substrate cyclin e (ortholog of CYCLIN E1/2) have been confirmed in flies.60,61 Partial loss of ago function was attempted with the UAS-Gal4 system,62 and two previously validated lines carrying ago-specific UAS-RNA interference constructs (RNAi-1 and RNAi-2).52,53 We first determined efficiency and relative strength of ago RNAi-1 and ago RNAi-2 constructs by quantitative RT-PCR upon ubiquitous knockdown by using the Act-GAL4 driver. The driver crossed to the genetic background of both RNAi lines served as a control in all experiments. Both lines led to lower levels of ago, albeit to different degrees. The expression level of ago relative to control levels was 19% in ago RNAi-1 (p = 0.005) and 67% (p = 0.11) in ago RNAi-2 (Figure S5). Because the latter was also previously shown to be effective in downregulating ago,52 we crossed both lines and the control to the pan-neuronal promotor line elav-Gal4(III) to generate neuron-specific ago knockdown and control animals. Progeny of the appropriate genotypes were selected and subjected to characterization of basal motor function and habituation, a simple form of learning frequently defective in Drosophila models of intellectual disability,56,63 in the light-off jump reflex habituation paradigm (Figure 4A). In this assay, individual flies are exposed to 100 light-off pulses (trials) with a 1 s inter-trial interval. Wild-type flies will initially startle in response the light-off stimulus, but they learn to suppress their escape response upon repeated, non-harmful stimuli.
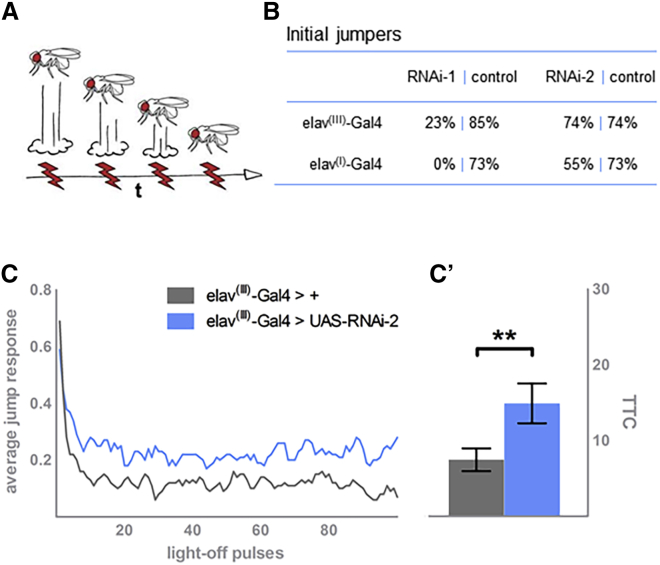
Figure 4.
Knockdown of the FBXW7 Drosophila ortholog ago, specifically in neurons, can lead to deficits in habituation learning deficits and more severe neuronal dysfunction
(A) Simplified scheme of the habituation assay, used for assessing the stimulus-induced escape response of individual flies upon repeated exposure. In controls, as depicted, the initial high jump response gradually wanes. Of note, in reality, the amplitude of jumps does not wane, but the frequency decreases in the tested population.
B) Knockdown of ago with RNAi-1 and either elav(I)-Gal4 or elav(III)-Gal4 severely impairs jumping. Knockdown of ago with RNAi-2 driven by either driver is less detrimental, allowing assessment of habituation learning.
(C) Neuronal knockdown of ago by elav(III)-Gal4 and RNAi-2 reduces the ability of flies to habituate to the stimulus (in blue); in ccontrast to their genetic-background controls (in gray), they keep jumping with increased frequency throughout the course of the experiment.
(D) Quantification of habituation according to mean trials to no-jump criterion (mTTC). Precise genotypes tested in (C) and (D): w/Y; 2xGMR-wIR/+; elav-Gal4(III), UAS-Dicer-2/ UAS-RNAi-2 (in blue; n = 71, mTTC = 14.91, p = 0.0015). Genetic background control w/Y; 2xGMR-wIR/+; elav-Gal4(III), UAS-Dicer-2/+ (in gray; n = 71, mTTC = 7.46). Statistical significance was assessed by a linear-model regression analysis on the log-transformed mTTC values; ∗p = 0.05, ∗∗p = 0.01, and ∗∗∗p < 0.001.
Elav-Gal4(III)-mediated neuronal knockdown of ago with the strong RNAi-1 line severely affected the flies’ ability to jump and participate in the assay (23% of initial jumpers, Figure 4B), revealing moderate neurological defects, which precluded an assessment of habituation learning. Knockdown of ago with the mild RNAi-2 line was less detrimental and did not impair the jump response (74% of initial jumpers, Figure 4B), yet caused a deficit in habituation learning (Figure 4C): ago knockdown flies (in blue) adapted incompletely and more slowly to the light-off stimuli in comparison to their genetic background controls (gray). Quantification of habituation via the mean trials to no-jump criterion (mTTC, see “subjects and methods”) demonstrated this defect to be significant; ago knockdown flies need on average twice as many trials as their controls to succeed in suppressing their jump response (n = 71, p = 0.002, Figure 4D). These results are in agreement with the qPCR results. Using an independent, stronger pan-neuronal promotor line (elav c155(I)-GAL4)64 further confirmed this finding. Elav c155(I)-GAL4-induced RNAi-1 knockdown completely abolished jumping (0%, Figure 4B), whereas the combination with RNAi-2 affected initial jumping (55%, Figure 4B) but still resulted in sufficient performance for habituation testing. This combination resulted in a faster decline of the jump response with decreased mTTC in comparison to the control (p = 1.6 × 10−3; Figure S4). Further experiments using a fatigue regime (see “subjects and methods”) revealed that this premature decay in the jump response was due to impaired neuronal function, not to faster adaptation (p = 1.4 × 10−5; Figure S4). Together, our results showed that loss of the FBXW7 fly ortholog ago affected learning or compromised neuronal function more severely the higher its level of knockdown.
Discussion
FBXW7 variants are associated with a variable neurodevelopmental syndrome
Here we provide detailed clinical and functional characterization of the neurodevelopmental syndrome associated with germline monoallelic variants in FBXW7. In support of our finding, FBXW7 was recently identified as one of 28 developmental-disorder-associated genes in a large multi-center cohort by bioinformatic analysis for gene-specific enrichment of de novo mutations, but without deep phenotyping.65 The neurodevelopmental phenotype involves mild to severe global developmental delay and intellectual disability. At the mildest end of the spectrum, isolated speech delay (n = 1) and learning difficulties or borderline intellect (n = 2) were observed; only one individual was reported to have no neurodevelopmental issues (but had hypotonia). In contrast, the majority of the cohort had mild to moderate intellectual disability (n = 27), and severe neurodevelopmental disability was observed in three individuals, including one with an additional diagnosis of familial CACNA1A-related disorder57 and another with episodic developmental regression. However, in the latter proband, no other candidate genomic variants were identified as an alternative cause, and the reason for the regressive episodes remains unclear in this individual. After neurodevelopmental disability, the next most frequently observed neurologic feature was hypotonia, common also to the other F-box-related neurodevelopmental syndromes associated with germline pathogenic variants in FBXL4. FBXO11, FBXW11, and FBXO28,9, 10, 11,13,16,17, but not in FBXL3.18
The neurodevelopmental phenotype associated with FBXW7 has substantial clinical overlap with that of FBXW11; areas of overlap include mild to severe neurodevelopmental disability, speech and language delay, micro- or macrocephaly, and brain anomalies, including corpus callosum hypoplasia, dilated ventricles, and white matter atrophy. However, in contrast to individuals with FBXW11 variants,9 those in the FBXW7 cohort did not commonly display autistic and/or stereotypical behaviors, psychiatric features, or ocular abnormalities.
It is notable that the FBXO11-related neurodevelopmental phenotype is just as variable; its severity ranges from normal cognition to profound disability.10,11,66 Individuals with FBXO11 variants were also found to have variability in head size, a similar observation made of the FBXW7 cohort, although we found that macrocephaly was more common than microcephaly. Macrocephaly has been observed in an individual with focal segmental glomerulosclerosis, Wilms tumor, invasive ductal breast carcinoma, and a 157 kb partial chromosomal deletion of FBXW7, but her neurodevelopmental phenotype was not reported.67 The DECIPHER database lists five individuals with copy-number losses that are various sizes and involve FBXW7. Three of these individuals have neurodevelopmental disability, and one experiences constipation. Another individual was reported with a 120.84 kb deletion including FBXW7, as well as two deletions in homozygous form on chromosomes 9 and 14, but the only listed phenotype was T cell acute lymphoblastic leukemia.
FBXW7 is a critical tumor suppressor and one of the most commonly mutated genes in human cancer (it is identified in 3.5% of all cancers).68 More recently, truncating variants in FBXW7 have been suggested to predispose the carrier to Wilms tumor in four individuals, and a missense variant was identified in an individual with a rhabdoid tumor; however, the individual’s neurodevelopmental phenotype was not well described.69 The oldest individual in this cohort is 44 years old, and although no cancer has so far been observed, longer-term follow-up will be necessary if we are to determine whether there is any cancer predisposition risk.
When we compared the frequency of key clinical features between variant types, no genotype-phenotype correlation was apparent, similar to findings for the comparison undertaken in a FBXO11 cohort.10 We observed three variants (p.Gly423Arg, p.Arg674Trp, and p.Arg689Trp) recurring in unrelated individuals. The degree of neurodevelopmental disability and hypotonia appeared to be consistent between individuals with the same variant. However, there was some variability in head size, and macrocephaly was inconsistently observed in each genotype. The familial cases demonstrated intra-familial variability. For instance, the family carrying the p.Asn572Leufs∗32 variant (individuals 3–6) were ascertained from a speech-and-language-disorders cohort. The proband (individual 3) had cleft palate and neurodevelopmental disability, but her sisters were less severely affected. Their father (individual 6) only had borderline-low verbal IQ. Neuroimaging was only undertaken in the proband, but it would be interesting to investigate whether her sisters and father also had similar brain anomalies. Furthermore, the p.Asn572Leufs∗32 variant is likely to escape NMD, as is the p.Lys647∗ variant identified in individual 7, whose phenotype is relatively mild compared to those of the individuals with missense variants. This observation suggests that a truncated FBXW7 might lead to a milder phenotype. We did not identify any individuals with variants affecting FBXW7’s N-terminal region, including the F-box. It is possible that the phenotypic consequences of variants in this region are either lethal or sub-clinical, although the latter appears to be more likely given the lack of regional missense constraint relative to the WD40 domain in gnomAD. Addressing this possibility, along with the possibility that milder phenotypes might emerge over time, will require the study of additional affected individuals.
Another explanation for variable expressivity among individuals carrying pathogenic variants in the same gene is the possibility of multiple diagnoses.70 This is well illustrated in individuals 19 and 31. Individual 19, in addition to carrying the mosaic FBXW7 variant, is heterozygous for a maternally inherited CACNA1A pathogenic variant.57 This combination is responsible for his more severe phenotype compared to that of his relatives carrying the CACNA1A variant alone and that of the other individuals in the FBXW7 cohort, and is likely to also explain the difference in his cerebellar abnormalities. Individual 31 also has a de novo likely pathogenic variant in KMT2D, and this is reflected in his facial features, including long palpebral features characteristic of Kabuki syndrome and the deeply set eyes and upper eyelid fullness observed in other individuals in the FBXW7 cohort. We also considered whether mosaicism for the FBXW7 variant might account for phenotypic attenuation or variable expressivity. We found that individuals 19 and 25 had clinical and genomic features suggestive of mosaicism, yet their phenotype was typical and no less severe than the rest of the cohort, which probably reflects the variable consequences of mosaicism. The emerging phenotype associated with variants in FBX genes appears to be characterized by neurodevelopmental disability with variable involvement of other systems. We speculate that these FBX proteins might function in convergent molecular and/or developmental pathways and that other FBX-related genes might subsequently be identified as playing a role in neurodevelopmental disorders.
FBXW7 missense variants identified in this cohort impair substrate turnover
Individuals harboring germline FBXW7 variants in this study demonstrate considerable phenotypic heterogeneity. FBXW7 encodes three isoforms; FBXW7α, FBXW7β, and FBXW7γ. All three contain the F-box and WD40 domain, but they differ at the N-terminal sequences that dictate their subcellular location; nucleus, cytoplasm, and nucleolus, respectively.71 Studies in mice indicate that the isoforms also demonstrate different tissue specificity.72 All variants identified in this study, whole-gene deletions as well as LoF, truncating, and missense variants, are predicted to affect the function of all three FBXW7 isoforms. In addition, FBXW7 has been shown to undergo multiple post-translational modifications, including auto-ubiquitination, de-ubiquitination, and dimerization (reviewed in8). There are numerous reported FBXW7 substrates, including CYCLIN E1/E2,61,73,74 PSEN1,75 NOTCH1/2/4,76,77 MYC,78 JUN,79,80 REV-ERBα,81,82 KLF5,79 DISC1,59 MCL-1,81 CCDC6,83 and mTOR.84 FBXW7 recognizes substrates upon phosphorylation at a conserved Cdc4 phosphodegron, a short linear motif that is inert until phosphorylated.8 Substrate binding occurs when the degron phosphorylations interact with two FBXW7 β-propeller pockets and upstream phosphodegron residues fit into a hydrophobic groove.85 Our in silico protein modeling suggests the amino acids implicated in this neurodevelopmental syndrome mainly cluster at the surface of the substrate-binding interface and are likely to impair substrate binding (Figure 2). We have demonstrated that some FBXW7 missense variants can affect steady-state FBXW7 protein amounts, suggesting that in some cases protein instability might lead to degradation of the mutant protein via the UPS. However, the majority of mutant proteins investigated were not turned over by the UPS more than the wild-type protein but demonstrated reduced capacity to turn over known substrates CYCLIN E1 and CYCLIN E2 (Figure 3). Fascinatingly, one mutant, FBXW7Asp480Gly, demonstrated divergent effects: it was less efficient at degrading CYCLIN E2 but more efficient at degrading CYCLIN E1 in comparison to FBXW7wild type and thus acted in a substrate-dependent manner. Collectively, our data suggest that the neurodevelopmental-disability-associated variants observed in this cohort are likely to alter binding affinity to substrates, and we hypothesize that other known substrates are likely to also be impacted.
Within a cell of an individual harboring a neurodevelopmental-disease-associated FBXW7 missense variant, FBXW7 can exist as a monomer (wild type or mutant) or as dimers (either as wild type only and mutant only or as both wild type and mutant) that can function at the cytosol, nucleus, or nucleolus. The impact on protein stability, binding affinity, and ubiquitin-ligase activity is likely to be variant specific, and although we propose that the phenotype is largely reflective of haploinsufficiency or loss of function (evident in those individuals with whole-gene deletions and NMD-predicted variants), we cannot rule out the possibility that some variants might have alternative mechanisms. Interrogation of gnomAD demonstrates that FBXW7 is intolerant of loss-of-function variation (pLI = 1.00), further supporting the notion that haploinsufficiency or loss of function is the predominant mechanism of disease. Some of the phenotypic variability of FBXW7 neurodevelopmental disability might, at least in part, be reflective of the functional consequence of the genotype or other, yet-unidentified modifiers. Identification of additional individuals with a broader spectrum of FBXW7 variants (including predicted LoF variants) associated with neurodevelopmental disability and further molecular characterization will most likely lead to a deeper understanding of genotype-phenotype correlation.
Animal models support a role for FBXW7 in development broadly and specifically in the nervous system
FBXW7 is a critical tumor suppressor and one of the most commonly deregulated ubiquitin-proteasome-system proteins in human cancer. However, this clinical cohort clearly demonstrates that FBXW7 also functions in human neurological development. Animal Fbxw7 models—knockout, haploinsufficient, and knock-in—also support a fundamental role for FBXW7 in development broadly, and in the brain specifically. Fbxw7-knockout mice die in utero at embryonic day 10.5, and they manifest hematopoietic abnormalities as well as abnormalities of vascular development and heart-chamber maturation.86,87 Heterozygous knock-in FBXW7 human-cancer mutations p.Arg465Cys and p.Arg482Gln (in mice, affecting residues Arg468 and Arg482) lead to perinatal lethality as a result of abnormal lung development, open eyes at birth (43%), and/or cleft palate (30%).87 Notably, heterozygous null animals show no lung abnormality, demonstrating that these missense variants are distinct from null alleles.88 Furthermore, heterozygous conditional gut-specific deletion of Fbxw7 (villin-Cre) result in an impaired differentiation of intestinal goblet cells,88 providing support for a role for FBXW7 in the development and function of the gut, which is significant because nearly half of individuals in our cohort manifested constipation (16/35, 45.7%).
Fbxw7nestin-Cre-knockout mice that lack expression exclusively in the central and peripheral nervous system (including in precursors of neuronal and glial cells) die in the perinatal period as a result of defective suckling and present with several morphological brain abnormalities, including third-ventricle dilation and distortion, hypoplastic pons, an abnormal cerebellum, and markedly reduced cellularity of the cortex. Notch, a key regulator of glial and neuronal cell fate in the brain, accumulates in cells and skews radial glia differentiation toward the astrocytic lineage by increasing apoptosis of neuronal precursors.33,36Fbxw7 haploinsufficiency in the mouse nervous system has also been investigated with a nestin-Cre system and has been shown to be associated with impaired differentiation of neural stem cells; such an impairment has also been shown to occur via a Notch-dependent mechanism. During development, it is proposed that Fbxw7 haploinsufficiency leads to alterations of Notch-mediated lateral inhibition, an interaction between adjacent cells that drives them toward different final states.88 Collectively, these studies provide evidence that Fbxw7 is a key regulator of neural-stem-cell differentiation and maintenance in the brain, and we speculate that dysregulation of Notch lateral inhibition during brain development might underpin the broad spectrum of brain abnormalities identified in FBXW7 neurodevelopmental syndrome.
Several studies have also investigated the role of FBXW7 orthologs in myelination. Fbxw7Cre-dhh-knockout mice that lack expression in Schwann cells of the peripheral nervous system demonstrated enhanced myelination —these mice made thicker myelin sheaths and in some cases, unexpectedly, myelinated multiple axons in a manner similar to the way in which oligodendrocytes of the central nervous system myelinate axons. In addition, Fbxw7Cre-dhh knockout led to an early increase in Schwann cells, smaller Remak bundles, and hypermyelination. These effects could be ameliorated by knockout of the substrate mTOR, but had no effect on the myelination of multiple axons.32 In the zebrafish central nervous system, fbxw7vu56 homozygous-mutant larvae and morpholino knockdown of fbxw7 demonstrate excessive oligodendrocyte cells and hypermyelination as a result of elevated Notch and mTOR signaling.31,34
In this study, in light of the fact that neurodevelopmental disability was the major hallmark of this cohort, we specifically aimed to support the notion of the role of FBXW7 in an intellectual-disability- and cognition-relevant assay. Specifically, we asked whether FBXW7, in addition to its functions in neural stem cells and glia, could also be required more directly, in postmitotic neurons, for basal neuronal and cognitive function. To address this, and also because, as in mice, ago null animals are embryonic lethal,60,89 we targeted the gene in a tissue-specific manner. We used two pan-neuronal promotor lines and two independent RNAi lines to generate an allelic series. Our results revealed that the FBXW7 ortholog ago is indispensable in postmitotic neurons; animals with the stronger promotor and UAS-RNAi line were severely impaired, whereas less-stringent conditions kept locomotor function intact and revealed deficits in habituation learning. The gene therefore joins a steeply increasing number of intellectual-disability- and autism-spectrum-disorder-associated genes56,63,90 that are implicated in this fundamental form of learning that is crucial for information processing, sensory filtering, and cognition. These also include mTOR-pathway genes such as PTEN & TSC1,56 to which FBXW7 has already been connected.31 Further studies should aim to dissect FBXW7 targets that are mediating the defects in cognitive functioning and study their reversibility; the established Drosophila model is suitable for this purpose. Collectively, the multiple lines of evidence presented herein converge to support the identification of FBXW7 variants as causal for a human neurodevelopmental disorder.
Acknowledgments
The authors thank the affected individuals and all family members for participating in this research. Please see the supplemental information for a complete list of Acknowledgments and funding.
Declaration of interests
I.E.S. has served on scientific advisory boards for UCB, Eisai, GlaxoSmithKline, BioMarin, Nutricia, Rogcon, Chiesi, Encoded Therapeutics, Xenon Pharmaceuticals, and Knopp Biosciences; has received speaker honoraria from GlaxoSmithKline, UCB, BioMarin, Biocodex, and Eisai; has received funding for travel from UCB, Biocodex, GlaxoSmithKline, Biomarin and Eisai; has served as an investigator for Zogenix, Zynerba, Ultragenyx, GW Pharma, UCB, Eisai, Anavex Life Sciences, Ovid Therapeutics, Epygenyx, Encoded Therapeutics and Marinus; and has consulted for Zynerba Pharmaceuticals, Atheneum Partners, Ovid Therapeutics, Care Beyond Diagnosis, Epilepsy Consortium and UCB. She may accrue future revenue on pending patent WO2009/086591; her patent for SCN1A testing is held by Bionomics and is licensed to various diagnostic companies; and she has a patent for a molecular diagnostic/therapeutic target for benign familial infantile epilepsy (BFIE) (PRRT2), WO/2013/059884. She receives and/or has received research support from the National Health and Medical Research Council of Australia, Medical Research Future Fund, Health Research Council of New Zealand, CURE, Australian Epilepsy Research Fund, and the National Institute of Neurological Disorders and Stroke of the National Institutes of Health. J.P. is co-chief scientific officer for Global Gene Corp. All other authors declare no competing interests.
Published: April 7, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.03.002.
Data and code availability
The datasets supporting the current study have not been deposited in a public repository because of restrictions related to patient consent, but they are available from the corresponding author on request.
Web resources
DECIPHER, https://www.deciphergenomics.org/
GeneMatcher, https://genematcher.org/
Github, https://github.com
Human Phenotype Ontology, https://hpo.jax.org/app/
Online Mendelian Inheritance in Man, http://www.omim.org
UCSC Genome Browser, https://genome.ucsc.edu
1000 Genomes, http://www.internationalgenome.org
Supplemental information
References
- 1.Deciphering Developmental Disorders S., Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemke J.R. Predicting incidences of neurodevelopmental disorders. Brain. 2020;143:1046–1048. doi: 10.1093/brain/awaa079. [DOI] [PubMed] [Google Scholar]
- 3.Wilfert A.B., Sulovari A., Turner T.N., Coe B.P., Eichler E.E. Recurrent de novo mutations in neurodevelopmental disorders: properties and clinical implications. Genome Med. 2017;9:101. doi: 10.1186/s13073-017-0498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruel A.L., Vitobello A., Tran Mau-Them F., Nambot S., Sorlin A., Denommé-Pichon A.S., Delanne J., Moutton S., Callier P., Duffourd Y., et al. Next-generation sequencing approaches and challenges in the diagnosis of developmental anomalies and intellectual disability. Clin. Genet. 2020;98:433–444. doi: 10.1111/cge.13764. [DOI] [PubMed] [Google Scholar]
- 5.Colas P. Cyclin-dependent kinases and rare developmental disorders. Orphanet J. Rare Dis. 2020;15:203. doi: 10.1186/s13023-020-01472-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LiCausi F., Hartman N.W. Role of mTOR Complexes in Neurogenesis. Int. J. Mol. Sci. 2018;19:E1544. doi: 10.3390/ijms19051544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen K.M., Busino L. The Biology of F-box Proteins: The SCF Family of E3 Ubiquitin Ligases. Adv. Exp. Med. Biol. 2020;1217:111–122. doi: 10.1007/978-981-15-1025-0_8. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z., Hu Q., Xu W., Liu W., Liu M., Sun Q., Ye Z., Fan G., Qin Y., Xu X., et al. Function and regulation of F-box/WD repeat-containing protein 7. Oncol. Lett. 2020;20:1526–1534. doi: 10.3892/ol.2020.11728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holt R.J., Young R.M., Crespo B., Ceroni F., Curry C.J., Bellacchio E., Bax D.A., Ciolfi A., Simon M., Fagerberg C.R., et al. De Novo Missense Variants in FBXW11 Cause Diverse Developmental Phenotypes Including Brain, Eye, and Digit Anomalies. Am. J. Hum. Genet. 2019;105:640–657. doi: 10.1016/j.ajhg.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregor A., Sadleir L.G., Asadollahi R., Azzarello-Burri S., Battaglia A., Ousager L.B., Boonsawat P., Bruel A.L., Buchert R., Calpena E., et al. University of Washington Center for Mendelian Genomics. DDD Study De Novo Variants in the F-Box Protein FBXO11 in 20 Individuals with a Variable Neurodevelopmental Disorder. Am. J. Hum. Genet. 2018;103:305–316. doi: 10.1016/j.ajhg.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jansen S., van der Werf I.M., Innes A.M., Afenjar A., Agrawal P.B., Anderson I.J., Atwal P.S., van Binsbergen E., van den Boogaard M.J., Castiglia L., et al. De novo variants in FBXO11 cause a syndromic form of intellectual disability with behavioral problems and dysmorphisms. Eur. J. Hum. Genet. 2019;27:738–746. doi: 10.1038/s41431-018-0292-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balak C., Belnap N., Ramsey K., Joss S., Devriendt K., Naymik M., Jepsen W., Siniard A.L., Szelinger S., Parker M.E., et al. A novel FBXO28 frameshift mutation in a child with developmental delay, dysmorphic features, and intractable epilepsy: A second gene that may contribute to the 1q41-q42 deletion phenotype. Am. J. Med. Genet. A. 2018;176:1549–1558. doi: 10.1002/ajmg.a.38712. [DOI] [PubMed] [Google Scholar]
- 13.Schneider A.L., Myers C.T., Muir A.M., Calvert S., Basinger A., Perry M.S., Rodan L., Helbig K.L., Chambers C., Gorman K.M., et al. FBXO28 causes developmental and epileptic encephalopathy with profound intellectual disability. Epilepsia. 2021;62:e13–e21. doi: 10.1111/epi.16784. [DOI] [PubMed] [Google Scholar]
- 14.Au P.Y., Argiropoulos B., Parboosingh J.S., Micheil Innes A. Refinement of the critical region of 1q41q42 microdeletion syndrome identifies FBXO28 as a candidate causative gene for intellectual disability and seizures. Am. J. Med. Genet. A. 2014;164A:441–448. doi: 10.1002/ajmg.a.36320. [DOI] [PubMed] [Google Scholar]
- 15.Cassina M., Rigon C., Casarin A., Vicenzi V., Salviati L., Clementi M. FBXO28 is a critical gene of the 1q41q42 microdeletion syndrome. Am. J. Med. Genet. A. 2015;167:1418–1420. doi: 10.1002/ajmg.a.37033. [DOI] [PubMed] [Google Scholar]
- 16.Bonnen P.E., Yarham J.W., Besse A., Wu P., Faqeih E.A., Al-Asmari A.M., Saleh M.A., Eyaid W., Hadeel A., He L., et al. Mutations in FBXL4 cause mitochondrial encephalopathy and a disorder of mitochondrial DNA maintenance. Am. J. Hum. Genet. 2013;93:471–481. doi: 10.1016/j.ajhg.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gai X., Ghezzi D., Johnson M.A., Biagosch C.A., Shamseldin H.E., Haack T.B., Reyes A., Tsukikawa M., Sheldon C.A., Srinivasan S., et al. Mutations in FBXL4, encoding a mitochondrial protein, cause early-onset mitochondrial encephalomyopathy. Am. J. Hum. Genet. 2013;93:482–495. doi: 10.1016/j.ajhg.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ansar M., Paracha S.A., Serretti A., Sarwar M.T., Khan J., Ranza E., Falconnet E., Iwaszkiewicz J., Shah S.F., Qaisar A.A., et al. Biallelic variants in FBXL3 cause intellectual disability, delayed motor development and short stature. Hum. Mol. Genet. 2019;28:972–979. doi: 10.1093/hmg/ddy406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charng W.L., Karaca E., Coban Akdemir Z., Gambin T., Atik M.M., Gu S., Posey J.E., Jhangiani S.N., Muzny D.M., Doddapaneni H., et al. Exome sequencing in mostly consanguineous Arab families with neurologic disease provides a high potential molecular diagnosis rate. BMC Med. Genomics. 2016;9:42. doi: 10.1186/s12920-016-0208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labonne J.D., Lee K.H., Iwase S., Kong I.K., Diamond M.P., Layman L.C., Kim C.H., Kim H.G. An atypical 12q24.31 microdeletion implicates six genes including a histone demethylase KDM2B and a histone methyltransferase SETD1B in syndromic intellectual disability. Hum. Genet. 2016;135:757–771. doi: 10.1007/s00439-016-1668-4. [DOI] [PubMed] [Google Scholar]
- 21.Yokotsuka-Ishida S., Nakamura M., Tomiyasu Y., Nagai M., Kato Y., Tomiyasu A., Umehara H., Hayashi T., Sasaki N., Ueno S.I., Sano A. Positional cloning and comprehensive mutation analysis identified a novel KDM2B mutation in a Japanese family with minor malformations, intellectual disability, and schizophrenia. J. Hum. Genet. 2021;66:597–606. doi: 10.1038/s10038-020-00889-4. [DOI] [PubMed] [Google Scholar]
- 22.Yeh C.H., Bellon M., Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol. Cancer. 2018;17:115. doi: 10.1186/s12943-018-0857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sailo B.L., Banik K., Girisa S., Bordoloi D., Fan L., Halim C.E., Wang H., Kumar A.P., Zheng D., Mao X., et al. FBXW7 in Cancer: What Has Been Unraveled Thus Far? Cancers (Basel) 2019;11:E246. doi: 10.3390/cancers11020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song Y., Lai L., Chong Z., He J., Zhang Y., Xue Y., Xie Y., Chen S., Dong P., Chen L., et al. E3 ligase FBXW7 is critical for RIG-I stabilization during antiviral responses. Nat. Commun. 2017;8:14654. doi: 10.1038/ncomms14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang C., Chen F., Feng L., Shan Q., Zheng G.H., Wang Y.J., Lu J., Fan S.H., Sun C.H., Wu D.M., et al. FBXW7 suppresses HMGB1-mediated innate immune signaling to attenuate hepatic inflammation and insulin resistance in a mouse model of nonalcoholic fatty liver disease. Mol. Med. 2019;25:29. doi: 10.1186/s10020-019-0099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onoyama I., Suzuki A., Matsumoto A., Tomita K., Katagiri H., Oike Y., Nakayama K., Nakayama K.I. Fbxw7 regulates lipid metabolism and cell fate decisions in the mouse liver. J. Clin. Invest. 2011;121:342–354. doi: 10.1172/JCI40725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izumi N., Helker C., Ehling M., Behrens A., Herzog W., Adams R.H. Fbxw7 controls angiogenesis by regulating endothelial Notch activity. PLoS ONE. 2012;7:e41116. doi: 10.1371/journal.pone.0041116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pronk M.C.A., Majolée J., Loregger A., van Bezu J.S.M., Zelcer N., Hordijk P.L., Kovačević I. FBXW7 regulates endothelial barrier function by suppression of the cholesterol synthesis pathway and prenylation of RhoB. Mol. Biol. Cell. 2019;30:607–621. doi: 10.1091/mbc.E18-04-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao W., Guo N., Zhao S., Chen Z., Zhang W., Yan F., Liao H., Chi K. FBXW7 promotes pathological cardiac hypertrophy by targeting EZH2-SIX1 signaling. Exp. Cell Res. 2020;393:112059. doi: 10.1016/j.yexcr.2020.112059. [DOI] [PubMed] [Google Scholar]
- 30.Thompson B.J., Jankovic V., Gao J., Buonamici S., Vest A., Lee J.M., Zavadil J., Nimer S.D., Aifantis I. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J. Exp. Med. 2008;205:1395–1408. doi: 10.1084/jem.20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kearns C.A., Ravanelli A.M., Cooper K., Appel B. Fbxw7 Limits Myelination by Inhibiting mTOR Signaling. J. Neurosci. 2015;35:14861–14871. doi: 10.1523/JNEUROSCI.4968-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harty B.L., Coelho F., Pease-Raissi S.E., Mogha A., Ackerman S.D., Herbert A.L., Gereau R.W., 4th, Golden J.P., Lyons D.A., Chan J.R., Monk K.R. Myelinating Schwann cells ensheath multiple axons in the absence of E3 ligase component Fbxw7. Nat. Commun. 2019;10:2976. doi: 10.1038/s41467-019-10881-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto A., Onoyama I., Sunabori T., Kageyama R., Okano H., Nakayama K.I. Fbxw7-dependent degradation of Notch is required for control of “stemness” and neuronal-glial differentiation in neural stem cells. J. Biol. Chem. 2011;286:13754–13764. doi: 10.1074/jbc.M110.194936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snyder J.L., Kearns C.A., Appel B. Fbxw7 regulates Notch to control specification of neural precursors for oligodendrocyte fate. Neural Dev. 2012;7:15. doi: 10.1186/1749-8104-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jandke A., Da Costa C., Sancho R., Nye E., Spencer-Dene B., Behrens A. The F-box protein Fbw7 is required for cerebellar development. Dev. Biol. 2011;358:201–212. doi: 10.1016/j.ydbio.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 36.Hoeck J.D., Jandke A., Blake S.M., Nye E., Spencer-Dene B., Brandner S., Behrens A. Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nat. Neurosci. 2010;13:1365–1372. doi: 10.1038/nn.2644. [DOI] [PubMed] [Google Scholar]
- 37.Ko Y.U., Kim C., Lee J., Kim D., Kim Y., Yun N., Oh Y.J. Site-specific phosphorylation of Fbxw7 by Cdk5/p25 and its resulting decreased stability are linked to glutamate-induced excitotoxicity. Cell Death Dis. 2019;10:579. doi: 10.1038/s41419-019-1818-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ko Y.U., Song H.Y., Kim W.K., Yune T.Y., Yun N., Oh Y.J. Calpain-mediated cleavage of Fbxw7 during excitotoxicity. Neurosci. Lett. 2020;736:135265. doi: 10.1016/j.neulet.2020.135265. [DOI] [PubMed] [Google Scholar]
- 39.Philippakis A.A., Azzariti D.R., Beltran S., Brookes A.J., Brownstein C.A., Brudno M., Brunner H.G., Buske O.J., Carey K., Doll C., et al. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reese M.G., Eeckman F.H., Kulp D., Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 43.Hebsgaard S.M., Korning P.G., Tolstrup N., Engelbrecht J., Rouzé P., Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brunak S., Engelbrecht J., Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- 45.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 46.Yang J., Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43(W1):W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hao B., Oehlmann S., Sowa M.E., Harper J.W., Pavletich N.P. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 48.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X., Jian X., Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pires D.E., Ascher D.B., Blundell T.L. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics. 2014;30:335–342. doi: 10.1093/bioinformatics/btt691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodrigues C.H.M., Myung Y., Pires D.E.V., Ascher D.B. mCSM-PPI2: predicting the effects of mutations on protein-protein interactions. Nucleic Acids Res. 2019;47(W1):W338–W344. doi: 10.1093/nar/gkz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li L., Anderson S., Secombe J., Eisenman R.N. The Drosophila ubiquitin-specific protease Puffyeye regulates dMyc-mediated growth. Development. 2013;140:4776–4787. doi: 10.1242/dev.096941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nam S., Cho K.O. Wingless and Archipelago, a fly E3 ubiquitin ligase and a homolog of human tumor suppressor FBW7, show an antagonistic relationship in wing development. BMC Dev. Biol. 2020;20:14. doi: 10.1186/s12861-020-00217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 55.Qureshi R., Sacan A. A novel method for the normalization of microRNA RT-PCR data. BMC Med. Genomics. 2013;6(Suppl 1):S14. doi: 10.1186/1755-8794-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fenckova M., Blok L.E.R., Asztalos L., Goodman D.P., Cizek P., Singgih E.L., Glennon J.C., IntHout J., Zweier C., Eichler E.E., et al. Habituation Learning Is a Widely Affected Mechanism in Drosophila Models of Intellectual Disability and Autism Spectrum Disorders. Biol. Psychiatry. 2019;86:294–305. doi: 10.1016/j.biopsych.2019.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Angelini C., Van Gils J., Bigourdan A., Jouk P.S., Lacombe D., Menegon P., Moutton S., Riant F., Sole G., Tournier-Lasserve E., et al. Major intra-familial phenotypic heterogeneity and incomplete penetrance due to a CACNA1A pathogenic variant. Eur. J. Med. Genet. 2019;62:103530. doi: 10.1016/j.ejmg.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 58.Pawlicka K., Kalathiya U., Alfaro J. Nonsense-Mediated mRNA Decay: Pathologies and the Potential for Novel Therapeutics. Cancers (Basel) 2020;12:E765. doi: 10.3390/cancers12030765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yalla K., Elliott C., Day J.P., Findlay J., Barratt S., Hughes Z.A., Wilson L., Whiteley E., Popiolek M., Li Y., et al. FBXW7 regulates DISC1 stability via the ubiquitin-proteosome system. Mol. Psychiatry. 2018;23:1278–1286. doi: 10.1038/mp.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moberg K.H., Bell D.W., Wahrer D.C., Haber D.A., Hariharan I.K. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature. 2001;413:311–316. doi: 10.1038/35095068. [DOI] [PubMed] [Google Scholar]
- 61.Koepp D.M., Schaefer L.K., Ye X., Keyomarsi K., Chu C., Harper J.W., Elledge S.J. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 62.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 63.Stessman H.A., Xiong B., Coe B.P., Wang T., Hoekzema K., Fenckova M., Kvarnung M., Gerdts J., Trinh S., Cosemans N., et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 2017;49:515–526. doi: 10.1038/ng.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gonzales E.D., Tanenhaus A.K., Zhang J., Chaffee R.P., Yin J.C. Early-onset sleep defects in Drosophila models of Huntington’s disease reflect alterations of PKA/CREB signaling. Hum. Mol. Genet. 2016;25:837–852. doi: 10.1093/hmg/ddv482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaplanis J., Samocha K.E., Wiel L., Zhang Z., Arvai K.J., Eberhardt R.Y., Gallone G., Lelieveld S.H., Martin H.C., McRae J.F., et al. Deciphering Developmental Disorders Study Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature. 2020;586:757–762. doi: 10.1038/s41586-020-2832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fritzen D., Kuechler A., Grimmel M., Becker J., Peters S., Sturm M., Hundertmark H., Schmidt A., Kreiß M., Strom T.M., et al. De novo FBXO11 mutations are associated with intellectual disability and behavioural anomalies. Hum. Genet. 2018;137:401–411. doi: 10.1007/s00439-018-1892-1. [DOI] [PubMed] [Google Scholar]
- 67.Roversi G., Picinelli C., Bestetti I., Crippa M., Perotti D., Ciceri S., Saccheri F., Collini P., Poliani P.L., Catania S., et al. Constitutional de novo deletion of the FBXW7 gene in a patient with focal segmental glomerulosclerosis and multiple primitive tumors. Sci. Rep. 2015;5:15454. doi: 10.1038/srep15454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Consortium A.P.G., AACR Project GENIE Consortium AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7:818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mahamdallie S., Yost S., Poyastro-Pearson E., Holt E., Zachariou A., Seal S., Elliott A., Clarke M., Warren-Perry M., Hanks S., et al. Identification of new Wilms tumour predisposition genes: an exome sequencing study. Lancet Child Adolesc. Health. 2019;3:322–331. doi: 10.1016/S2352-4642(19)30018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Posey J.E., Harel T., Liu P., Rosenfeld J.A., James R.A., Coban Akdemir Z.H., Walkiewicz M., Bi W., Xiao R., Ding Y., et al. Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation. N. Engl. J. Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Welcker M., Orian A., Grim J.E., Eisenman R.N., Clurman B.E. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr. Biol. 2004;14:1852–1857. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 72.Matsumoto A., Onoyama I., Nakayama K.I. Expression of mouse Fbxw7 isoforms is regulated in a cell cycle- or p53-dependent manner. Biochem. Biophys. Res. Commun. 2006;350:114–119. doi: 10.1016/j.bbrc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 73.Strohmaier H., Spruck C.H., Kaiser P., Won K.A., Sangfelt O., Reed S.I. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 74.Klotz K., Cepeda D., Tan Y., Sun D., Sangfelt O., Spruck C. SCF(Fbxw7/hCdc4) targets cyclin E2 for ubiquitin-dependent proteolysis. Exp. Cell Res. 2009;315:1832–1839. doi: 10.1016/j.yexcr.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 75.Li J., Pauley A.M., Myers R.L., Shuang R., Brashler J.R., Yan R., Buhl A.E., Ruble C., Gurney M.E. SEL-10 interacts with presenilin 1, facilitates its ubiquitination, and alters A-beta peptide production. J. Neurochem. 2002;82:1540–1548. doi: 10.1046/j.1471-4159.2002.01105.x. [DOI] [PubMed] [Google Scholar]
- 76.Wu G., Lyapina S., Das I., Li J., Gurney M., Pauley A., Chui I., Deshaies R.J., Kitajewski J. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 2001;21:7403–7415. doi: 10.1128/mcb.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fukushima H., Shimizu K., Watahiki A., Hoshikawa S., Kosho T., Oba D., Sakano S., Arakaki M., Yamada A., Nagashima K., et al. NOTCH2 Hajdu-Cheney Mutations Escape SCFFBW7-Dependent Proteolysis to Promote Osteoporosis. Mol. Cell. 2017;68:645–658.e5. doi: 10.1016/j.molcel.2017.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Popov N., Herold S., Llamazares M., Schülein C., Eilers M. Fbw7 and Usp28 regulate myc protein stability in response to DNA damage. Cell Cycle. 2007;6:2327–2331. doi: 10.4161/cc.6.19.4804. [DOI] [PubMed] [Google Scholar]
- 79.Zhao D., Zheng H.Q., Zhou Z., Chen C. The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 2010;70:4728–4738. doi: 10.1158/0008-5472.Can-10-0040. [DOI] [PubMed] [Google Scholar]
- 80.Nateri A.S., Riera-Sans L., Da Costa C., Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 81.Tong J., Tan S., Nikolovska-Coleska Z., Yu J., Zou F., Zhang L. FBW7-Dependent Mcl-1 Degradation Mediates the Anticancer Effect of Hsp90 Inhibitors. Mol. Cancer Ther. 2017;16:1979–1988. doi: 10.1158/1535-7163.Mct-17-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao X., Hirota T., Han X., Cho H., Chong L.W., Lamia K., Liu S., Atkins A.R., Banayo E., Liddle C., et al. Circadian Amplitude Regulation via FBXW7-Targeted REV-ERBα Degradation. Cell. 2016;165:1644–1657. doi: 10.1016/j.cell.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morra F., Luise C., Merolla F., Poser I., Visconti R., Ilardi G., Paladino S., Inuzuka H., Guggino G., Monaco R., et al. FBXW7 and USP7 regulate CCDC6 turnover during the cell cycle and affect cancer drugs susceptibility in NSCLC. Oncotarget. 2015;6:12697–12709. doi: 10.18632/oncotarget.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Okazaki H., Matsunaga N., Fujioka T., Okazaki F., Akagawa Y., Tsurudome Y., Ono M., Kuwano M., Koyanagi S., Ohdo S. Circadian regulation of mTOR by the ubiquitin pathway in renal cell carcinoma. Cancer Res. 2014;74:543–551. doi: 10.1158/0008-5472.CAN-12-3241. [DOI] [PubMed] [Google Scholar]
- 85.Welcker M., Larimore E.A., Swanger J., Bengoechea-Alonso M.T., Grim J.E., Ericsson J., Zheng N., Clurman B.E. Fbw7 dimerization determines the specificity and robustness of substrate degradation. Genes Dev. 2013;27:2531–2536. doi: 10.1101/gad.229195.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ikenoue T., Terakado Y., Zhu C., Liu X., Ohsugi T., Matsubara D., Fujii T., Kakuta S., Kubo S., Shibata T., et al. Establishment and analysis of a novel mouse line carrying a conditional knockin allele of a cancer-specific FBXW7 mutation. Sci. Rep. 2018;8:2021. doi: 10.1038/s41598-018-19769-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Davis H., Lewis A., Spencer-Dene B., Tateossian H., Stamp G., Behrens A., Tomlinson I. FBXW7 mutations typically found in human cancers are distinct from null alleles and disrupt lung development. J. Pathol. 2011;224:180–189. doi: 10.1002/path.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sancho R., Blake S.M., Tendeng C., Clurman B.E., Lewis J., Behrens A. Fbw7 repression by hes5 creates a feedback loop that modulates Notch-mediated intestinal and neural stem cell fate decisions. PLoS Biol. 2013;11:e1001586. doi: 10.1371/journal.pbio.1001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mortimer N.T., Moberg K.H. The archipelago ubiquitin ligase subunit acts in target tissue to restrict tracheal terminal cell branching and hypoxic-induced gene expression. PLoS Genet. 2013;9:e1003314. doi: 10.1371/journal.pgen.1003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McDiarmid T.A., Belmadani M., Liang J., Meili F., Mathews E.A., Mullen G.P., Hendi A., Wong W.R., Rand J.B., Mizumoto K., et al. Systematic phenomics analysis of autism-associated genes reveals parallel networks underlying reversible impairments in habituation. Proc. Natl. Acad. Sci. USA. 2020;117:656–667. doi: 10.1073/pnas.1912049116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the current study have not been deposited in a public repository because of restrictions related to patient consent, but they are available from the corresponding author on request.