Summary
Covalent tRNA modifications play multi-faceted roles in tRNA stability, folding, and recognition, as well as the rate and fidelity of translation, and other cellular processes such as growth, development, and stress responses. Mutations in genes that are known to regulate tRNA modifications lead to a wide array of phenotypes and diseases including numerous cognitive and neurodevelopmental disorders, highlighting the critical role of tRNA modification in human disease. One such gene, THUMPD1, is involved in regulating tRNA N4-acetylcytidine modification (ac4C), and recently was proposed as a candidate gene for autosomal-recessive intellectual disability. Here, we present 13 individuals from 8 families who harbor rare loss-of-function variants in THUMPD1. Common phenotypic findings included global developmental delay, speech delay, moderate to severe intellectual deficiency, behavioral abnormalities such as angry outbursts, facial dysmorphism, and ophthalmological abnormalities. We demonstrate that the bi-allelic variants identified cause loss of function of THUMPD1 and that this defect results in a loss of ac4C modification in small RNAs, and of individually purified tRNA-Ser-CGA. We further corroborate this effect by showing a loss of tRNA acetylation in two CRISPR-Cas9-generated THUMPD1 KO cell lines. In addition, we also show the resultant amino acid substitution that occurs in a missense THUMPD1 allele identified in an individual with compound heterozygous variants results in a marked decrease in THUMPD1 stability and RNA-binding capacity. Taken together, these results suggest that the lack of tRNA acetylation due to THUMPD1 loss of function results in a syndromic form of intellectual disability associated with developmental delay, behavioral abnormalities, hearing loss, and facial dysmorphism.
Keywords: THUMPD1, tRNA modifications, N4-acetylcytidine, ac4C, intellectual disability, developmental disorder, NAT10, RNA acetylation, tRNA biology
Introduction
Across all organisms, well more than 100 types of transfer RNA (tRNA) modifications exist and tRNAs contain on average 13 modifications per molecule.1, 2, 3, 4, 5, 6 Post-transcriptional modifications of tRNAs play multi-faceted roles in tRNA stability, folding, and recognition, as well as the rate and fidelity of translation, and other cellular processes such as growth, development, and stress responses.7, 8, 9, 10
An increasing number of genes involved in tRNA modifications are implicated in intellectual disability. Variants in at least ten different genes11 involved in tRNA modifications, including NOP2/Sun RNA Methyltransferase 2 (NSUN2 [MIM: 610916]),12 pseudouridine synthase 3 (PUS3 [MIM: 616283]),13,14 adenosine deaminase tRNA specific 3 (ADAT3 [MIM: 615302]),15,16 DALR anticodon binding domain containing 3 (DALRD3 [MIM: 618904]),17 tRNA Methyltransferase 1 (TRMT1 [MIM: 611669]),18 Cytosolic Thiouridylase Subunit 2 (CTU2 [MIM: 617057]),19 AlkB Homolog 8, tRNA Methyltransferase (ALKBH8 [MIM: 613306]),20 Pseudouridine Synthase 7 (PUS7 [MIM: 616261]),21,22 and WD Repeat Domain 4 (WDR4 [MIM: 605924]),23 have now been shown to be associated with neurological and neurodevelopmental disorders, emphasizing an increasingly apparent role for the importance of tRNA modifications in human neurodevelopment.
N4-acetylcytidine (ac4C) is a modification present in the wobble position of E. coli tRNA-Met,24,25 in rRNAs in a range of organisms26,27 and specifically at two conserved positions in human 18S rRNA (helix 34 and 45),28, 29, 30, 31 and at position 12 in eukaryotic serine and leucine tRNAs, with tRNA acetylation being conserved from yeast to mammals.31, 32, 33, 34, 35 ac4C modification is catalyzed by the enzyme N-acetyltransferase 10 (NAT10) or its orthologs, and in humans, acetylation of serine and leucine tRNAs requires an additional adaptor protein known as THUMP-domain containing protein 1 (THUMPD1, or Tan1p in yeast).31,35 This protein contains a single THUMP domain, named for its presence in thiouridine synthases, RNA methylases, and pseudouridine synthases across all domains of life.36 The THUMP domain is an RNA-binding domain35 and serves to recruit RNA molecules for subsequent modification by catalytic domains within the same polypeptide or by partner proteins.31,37, 38, 39 In yeast, loss of the THUMPD1 ortholog, Tan1p, results in hypoacetylation of serine and leucine tRNAs,31,35,40 and a decrease in tRNA stability that leads to a decrease in tRNA serine and leucine levels.30,40, 41, 42 This results in a temperature-sensitive growth phenotype,35,40 but it is unclear whether this effect is conserved in humans.
Recently, Maddirevula et al. proposed THUMPD1 (MIM: 616662) as a candidate gene for syndromic autosomal-recessive intellectual disability based on findings from a single individual.43 Here, we report the clinical phenotypes associated with bi-allelic variants in THUMPD1 in 13 individuals from 8 families showing syndromic intellectual disability. We demonstrate that the variants identified cause loss of function of THUMPD1 and that this defect results in a loss of ac4C modification in small RNAs and of individually purified tRNA-Ser-CGA. We further corroborate this effect by showing a loss of tRNA acetylation in two CRISPR-Cas9-generated THUMPD1 KO cell lines. Taken together, these results suggest that the lack of tRNA acetylation due to THUMPD1 loss of function results in syndromic intellectual disability.
Material and methods
Study participants
Individuals were recruited via an international, multi-center collaboration with research and diagnostic sequencing laboratories and medical genetics departments. Individuals were clinically evaluated in separate centers and contact between researchers was facilitated by use of the web-based tool GeneMatcher.44 Inclusion criteria were rare bi-allelic variants in THUMPD1 classified as pathogenic or likely pathogenic according to the American College of Medical Genetics and Genomics (ACMG) guidelines for the interpretation of sequence variants carried by individuals with compatible phenotypes.45 Photographs and clinical data reports were provided by each attending physician and/or a detailed review of medical records.
Ethics statement
Informed consent to participate in the study was obtained from all individuals, or their parents in case of minor individuals, in accordance with local institutional review boards. Samples and clinical information were obtained for each individual following local procedures and ethical standards. When needed, additional written consent to use photographs in this publication was also obtained.
Sequencing and bioinformatics analysis
Whole-exome sequencing (WES) was performed on blood-derived DNA samples using each center’s analysis platforms and next-generation sequencing (NGS) pipeline (see supplemental methods). Exome data were interpreted in agreement with local practices. cDNA and protein sequence variants are described in accordance with the recommendations of the Human Genome Variation Society using NCBI reference sequence GenBank: NM_001304550.1 and genomic coordinates from GRCh37/hg19. Priority was given to rare exonic or donor/acceptor splicing variants in accordance with the pedigree and phenotype, fitting a recessive (homozygous or compound heterozygous) model and/or variants in genes previously linked to developmental delay, intellectual disability, and other neurological disorders. Upon whole-exome sequencing analysis, THUMPD1 variants were selected and all the other candidate variants were further verified using Sanger sequencing.
Lymphoblast cell culture
Lymphoblast cells lines were generated by EBV immortalization of primary human lymphocytes isolated from whole blood of a 23-year-old male with intellectual disability, dysmorphic features, hypertension, and diabetes (from Saudi Arabia) with bi-allelic (homozygous) variant in THUMPD1 containing a GenBank: NM_001304550.1: c.706C>T (p.Gln236∗) nonsense mutation in the corresponding mRNA43 (case ID 15DG1395, individual F2:II.1 herein) as well as from whole blood of two independent control unrelated healthy Saudi individuals. Lymphoblast cells were grown in RPMI medium (Lonza) containing 25 mM glucose, 1× GlutaMAX Supplement (GIBCO), and 15% bovine calf serum (HyClone), in presence of ampicillin and streptomycin.
HeLa and HEK293T cell culture and knockout generation
HeLa cervical carcinoma cells and HEK293T human embryonic kidney cell lines were purchased from ATCC. NAT10−/− HeLa cells were obtained from Dr. Shalini Oberdoerffer (Laboratory of Receptor Biology and Gene Expression, National Cancer Institute, NIH, Bethesda, MD).46 HeLa and HEK293T THUMPD1 knockout (KO) cells were generated using a CRISPR-Cas9 Gene Knockout kit (Synthego). Briefly, purified SpCas9 and synthetic gRNA targeting exon 2 were complexed and electroporated into cells using the Neon Transfection System (Invitrogen) according to the manufacturer’s instructions. At 48 h post-transfection, cells were seeded in dilutions into 10-cm dishes. Single colonies were tested for depletion of THUMPD1 by western blot (see western blot analysis for more details). To determine the presence of insertions or deletions in THUMPD1 targeted clones, genomic DNA was isolated using a Wizard SV genomic DNA purification System (Promega) and exon 2 of THUMPD1 was PCR amplified using the primers listed in Table S1. PCR products were sequenced by Sanger sequencing and analyzed by ICE (Synthego). Clones with mutations in both THUMPD1 alleles were identified and one clone was selected for further experiments. Additional confirmation of THUMPD1 KO status was determined by sub-cloning PCR products into pCR2.1-TOPO using the TOPO TA cloning Kit (Thermo Fisher Scientific) and subsequent Sanger sequencing of individual clones. THUMPD1 protein in knockout cell lines was evaluated by western blots with corresponding cell extracts probed by anti-THUMPD1 antibodies (Bethyl Labs and/or Thermo Fisher Scientific). GAPDH and Tubulin antibodies served as a loading control for all cell protein extracts (Abcam). All other transfection experiments were performed using JetPRIME transfection reagent (PolyPlus) according to the manufacturer’s instructions.
Construction of THUMPD1 WT and p.Pro164Ser E. coli expression plasmids
Wild-type full-length human THUMPD1 in pDonr221 was obtained from the CCSB Human ORFeome collection and subcloned into pHGWA (which contains an N-terminal hexa-histidine tag followed by a TEV protease site) using standard Gateway Cloning (Thermo Fisher Scientific). Using this resultant plasmid as a template, THUMPD1 p.Pro164Ser construct was generated using standard site-directed mutagenesis procedure using the primers listed in Table S1. The resultant constructs were verified by Sanger sequencing. These plasmids have been deposited to Addgene.
Recombinant THUMPD1 E. coli overexpression and purification
To purify THUMPD1 WT GenBank: NP_001291479.1 and THUMPD1 p.Pro164Ser proteins, the corresponding 6× His-THUMPD1 constructs were transformed into E. coli Rosetta 2(DE3) strain (Novagen). Cells were grown in LB media supplemented with 0.5% w/v glucose at 37°C and protein expression was induced by addition of 0.5 mM IPTG followed by incubation at 16°C overnight. Cell lysis was conducted by sonication in lysis buffer (50 mM HEPES [pH 7.0], 1 M NaCl, 30 mM imidazole, 5% (v/v) glycerol, 1 mM DTT, 0.5 mM PMSF, EDTA-free protease inhibitor [Roche], 400 μg of RNase A [Sigma] per gram of cell pellet). The cell lysate was clarified by centrifugation and metal ion affinity purification was carried out on the soluble fraction using an AKTA Pure FPLC instrument (Cytiva) with a HisTrap 1 mL column (Cytiva). THUMPD1-containing fractions were dialyzed overnight in gel filtration buffer (20 mM HEPES [pH 7.0], 200 mM KCl, 5% glycerol, 1 mM DTT) with 30 mM imidazole and TEV protease. The next day, a HisTrap column (Cytiva) was used to capture the cleaved 6× His-tag, and the THUMPD1-containing flowthrough was concentrated. Any residual impurities in the samples were removed by size exclusion chromatography on AKTA Pure FPLC instrument (Cytiva) with a Superdex 200 10/300 GL column (Cytiva), using gel filtration buffer. Fractions containing only THUMPD1 were collected, concentrated, flash-frozen in liquid nitrogen, and stored at −80°C.
Thermo-fluorometry (thermal shift assays)
THUMPD1 WT and THUMPD1 p.Pro164Ser protein thermal shift assays were performed using a QuantStudio 5 Real-Time PCR System instrument (Applied Biosystems). 10 μL reactions were carried out in MicroAmp Optical 384-well reaction plates (Thermo) containing 2 μM protein in unfolding buffer (20 mM HEPES [pH 7.5], 100 mM KCl, 5 mM MgCl2, 1 mM DTT) and 2.5 μL of 1:50 diluted SYPRO Orange dye (Sigma-Aldrich). The assays were performed by heating from 25°C to 95°C at 1°C/min with a 2 min hold at each temperature, and a fluorescence read at each temperature increment. All protein samples were quantified in triplicates. Protein dissociation curves were plotted from the data using GraphPad Prism 9 software.
Preparation of 5′-labeled RNAs
RNA used for electrophoretic mobility shift assays (EMSA) were produced in the T7 RNA polymerase system with DNA template listed in Table S1. Recombinant T7 RNA polymerase protein was expressed and purified as described in Rio.47 In vitro transcription reactions contained 1–2 μg of linearized DNA template, transcription buffer (30 mM Tris-Cl [pH 8.1], 25 mM MgCl2, 0.01% (v/v) Triton X-100, 2 mM spermidine), 5 mM of each rNTPs, 10 mM DTT, and 20 μg of purified T7 RNA polymerase. After 3 h, 2U RQ1 RNase-Free DNase (Promega) and 10 mM MgCl2 were added to the transcription reactions for 1 h, then reactions were terminated by addition of one volume of 2× RNA Gel-Loading Buffer (95% deionized formamide, 5 mM EDTA [pH 8.0], 0.025% SDS, 0.025% bromophenol blue, 0.025% xylene cyanol). Transcription reactions were resolved by 15% denaturing PAGE and corresponding bands containing RNA were extracted from the gel and precipitated with ethanol. The 5′-ends of the RNA were radiolabeled with γ-[32P]ATP (3,000 Ci/mmol, PerkinElmer) using T4 polynucleotide kinase (NEB) and purified from unincorporated label using G-25 spin columns (Cytiva).
EMSA assays
Complexes containing THUMPD1 bound to RNA were prepared by incubating 0 to 1 μM of THUMPD1 and constant amount (0.2 to 1 nM) of 5′-labeled RNA in binding buffer (20 mM HEPES [pH 7.5], 100 mM KCl, 5% glycerol, 0.01% v/v Igepal CA-630, 5 mM MgCl2, 30 μg/mL heparin, 1 mM DTT) for 60 min at 37°C. Complexes assembled in each reaction condition were resolved on a 8% native PAGE gel in a cold room and visualized using a Typhoon 9410 Trio Phosphoimager (Cytiva) after drying and exposing the gel. Radioactive signals were quantitated with ImageQuant TL (Cytiva).
RNA preparation
Total RNA was extracted from human cell lines using TRIzol Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. For experiments requiring the separation of small (<100 nt) and large (>100 nt) RNA fractions, mirVana miRNA Isolation Kit (Thermo Scientific) was used according to the manufacturer’s instructions. RNA concentrations were determined by Nanodrop 1000 (Thermo Scientific).
qRT-PCR
Total or large RNA samples were reverse transcribed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and random hexamer priming, according to manufacturer’s instructions. To quantify levels of THUMPD1 mRNA expression, the PCR reaction was carried out using SsoAdvanced Universal SYBR Green Supermix (Biorad) with the primers listed in Table S1. The reactions were monitored using a QuantStudio 5 Real-Time PCR System (Applied Biosystems). The ΔΔCT method was used to quantify relative THUMPD1 mRNA expression changes between control cells and THUMPD1 KO / variant cells, using GAPDH as a housekeeping control. We designed the experiment with three biological and three technical replicates to validate the results (n = 3).
Northern blot analysis
A standard northern blot procedure was used to evaluate tRNA or U6 RNA levels from small RNA samples. Briefly, equivalents of 200 ng of RNA in triplicates for each sample were denatured in 1× RNA formamide loading buffer (Thermo Fisher Scientific) at 95°C for 10 min and separated on 10% PAGE gel containing 7M Urea for 45 min at constant 150V, then transferred onto Amersham Hybond N+ nylon membrane (Cytiva) in 1× TBE for 1 h at constant 400 mA. Membranes were air-dried, UV-crosslinked in Stratalinker 1800 (Stratagene/Agilent), and probed with γ-[32P]ATP oligonucleotide specific to tRNA-Ser-CGA (Table S1) overnight at 42°C in ULTRAhyb Ultrasensitive Hybridization Buffer (Ambion). After washing membranes in 2× SSC followed by 0.2 × SSC, the bound probe bands were detected using a Typhoon Prosphoimager. Before re-probing membranes with U6 RNA- or tRNA-Ala-specific γ-[32P]ATP labeled oligonucleotides, the membranes were stripped in 0.5% (w/v) SDS buffer at 60°C. tRNA-probed results were normalized to U6 RNA loading control signal to calculate the relative tRNA levels in control versus THUMPD1 or NAT10 KO RNA samples.
Western blot analysis
Protein cell extracts were prepared by lysing the cells on ice for 30 min in 1× PBS containing 1% NP-40 detergent, 1 mM DTT, and protease inhibitor cocktail (Thermo Fisher Scientific) with the addition of DNase I and RNase A. SDS-sample denaturing buffer was added to 1× concentration and samples heated at 95°C for 10 min. 30 μg of total protein was loaded onto 10% SDS-PAGE gel and separated at 200V for 45 min. Protein transfer to Amersham Protran 0.45 μm nitrocellulose membrane (Cytiva) was performed in Trans-Blot Turbo instrument (BioRad) using the turbo-option. Membranes were treated according to antibody manufacturer recommendations. We used the following antibodies: mouse monoclonal α-THUMPD1 (4A11, Thermo Fisher Scientific); rabbit polyclonal α-THUMPD1 (A304-643A, Bethyl Labs); rabbit polyclonal α-GAPDH (G9545, Sigma-Aldrich); and rat monoclonal α-tubulin (ab6160, Abcam) at suggested dilutions. Secondary HRP-conjugated goat anti-mouse and anti-rabbit antibodies were obtained BioRad and goat anti-rat from KPL. To visualize the protein bands on membranes, we used SuperSignal West Pico PLUS Chemiluminescent Substrate reagents from Thermo Fisher Scientific and Molecular Imager GelDoc XR+ (BioRad).
Detection of ac4C modifications by ImmunoNorthern blot
10 μg total RNA and large RNA or 2 μg of small RNA samples were denatured in formaldehyde loading buffer, heated to 65°C for 15 min, and separated on 1% or 1.5% (for small RNA) agarose denaturing gel for 3 h at constant 70V. Gels were stained in 0.5 μg/L ethidium bromide and documented by UV imaging before transfer. RNA was transferred onto Amersham Hybond-N+ membranes (Cytiva) by capillary transfer using 20× SSC buffer overnight. Membranes were crosslinked twice in Stratalinker, blocked with Western Blot Blocking Reagent (BioRad) in 1× PBS containing 0.05% Tween-20 (PBS-T) for 30 min at room temperature, and probed with α-ac4C antibody (ab25215, Abcam) at a 1:1,000 dilution in 5% BSA at 4°C overnight. Then membranes were washed three times with PBS-T and incubated with HRP-conjugated secondary goat anti-rabbit antibody (BioRad) at a 1:5,000 dilution in 1× PBS-T at 4°C overnight followed by four membrane washes with 1× PBS-T and treatment with SuperSignal West Pico PLUS Chemiluminescent Substrate reagents. Chemiluminescence was detected using Molecular Imager GelDoc XR+.
Purification of individual tRNA-Ser-CGA1-1 and tRNA-Ala-AGC9-1 species
Biotin-conjugated single-stranded DNA oligonucleotides containing sequences complementary to tRNA-Ser-CGA1-1 or tRNA-Ala-AGC9-1 (Table S1) were immobilized onto Pierce streptavidin magnetic beads (Thermo Fisher Scientific) as suggested by manufacturer and after a final wash were resuspended in 2.4 M tetraethylammonium chloride (TEAC). 2 mg of total RNA prepared from control or variant-containing lymphoblasts in 2.4 M TEAC was heated at 75°C for 5 min and quickly chilled on ice before addition of 50 μL aliquot of immobilized magnetic beads. The mixture was heated 55°C for 3 min and hybridization was set at 20°C for 90 min. Then magnetic beads were separated using Invitrogen DynaMag-2 Magnet, washed 3 times with 2.4 M TEAC, and bound RNA was eluted by incubation with 100 μL of 2.4M TEAC at 20°C followed by standard precipitation with ethanol. To evaluate the quality of purified tRNAs 5 ng aliquots were ran on 2100 Bioanalyzer (Agilent) at the Genomics Research Center University of Rochester or 20 ng aliquots were separated onto 10% denaturing acrylamide gel and stained with Diamond Nucleic Acid Dye (Promega).
RNA hydrolysis
RNA sample hydrolysis for liquid chromatography-mass spectrometry (LC-MS) was conducted as described in Su et al.48 Briefly, 10 μg of total RNA or 2 μg of small RNA was treated with a mixture of benzonase nuclease (Sigma-Aldrich), phosphodiesterase (US Biological), and alkaline phosphatase (Sigma-Aldrich) for 3 h at 37°C and then spin-filtered using Amicon 10K MWCO membranes (Millipore) at 16,000 × g for 15 min in cold microfuge. Resultant filtrates were analyzed by LC-MS.
LC-MS/MS analysis of RNA modifications
RNA modification analysis was carried out by adapting the method developed by Su et al.,48 using a Dionex Ultimate 3000 UHPLC coupled to a Q Exactive Plus mass spectrometer (Thermo Scientific). After purification, analytes were separated on a Hypersil Gold 2.1 × 150 mm column, protected by a 2.1 × 10 mm Hypersil Gold guard column (Thermo Scientific). The mobile phases were A: 0.1% formic acid in water, and B: 0.1% formic acid in acetonitrile. The flow rate was set to 400 μL/min, and the column oven was set to 36°C. 8 μL of each sample was injected, and the analytes were eluted using a multi-step gradient as follows: 0.00 min, 0% B; 6.00 min, 0% B; 7.65 min, 1% B; 9.35 min, 6% B; 10.00 min, 6% B; 12.00 min, 50% B; 14.00 min, 75% B; 17.00 min, 75%; 17.50 min, 0%; and 20.00 min, 0%.The Q Exactive Plus was operated in positive mode with a heated electrospray ionization (HESI) source. The spray voltage was set to 3.5 kV, the sheath gas flow rate was set to 40, and the auxiliary gas flow rate set to 7, while the capillary temperature was set to 320°C. A parallel reaction monitoring (PRM) method was used to quantify the modifications. Precursor ions were isolated using a 1.5 m/z isolation width, and then fragmented in the collision cell with an NCE of 40. The maximum injection time was set to 80 ms, while the AGC target was 2e5. Retention time scheduling was employed to ensure enough scans across the peak for each analyte. The resulting fragment ions were detected in the Orbitrap with a resolution of 17,500 at m/z 200. Peak areas from the fragment ions were extracted with a 10 ppm mass tolerance using the LC Quan node of the XCalibur software (Thermo Scientific). All detected unmodified and modified nucleosides, m/z values, and their respective retention time is presented in Table S2.
Statistical analysis
The statistical analyses (Welch’s t test, Brown-Forsythe, and Welch one-way ANOVA with Benjamini Hochberg correction, and two-way ANOVA with Benjamini Hochberg correction) were carried out using GraphPad Prism 9 (GraphPad, La Jolla, CA).
Results
Bi-allelic variants of THUMPD1 result in syndromic intellectual disability
We collected 13 individuals from 8 families originating from Europe, and the Middle East (8 males, 5 females) with rare bi-allelic variants in THUMPD1 (F1–F8). Nine individuals were born from consanguineous parents. Age at last visit ranged from 2 to 23 years old (median: 7 years old). Table 1 summarizes the core clinical features of affected individuals. All evaluated individuals had global developmental delay (1 individual could not be evaluated) and intellectual disability (1 individual could not be evaluated), which varied in severity from mild to severe. All showed a delayed language development. Motor development was less impaired, with a median age of walking of 1.5 years old. Microcephaly was observed in 4/10 of the individuals and individual F5:II.1 had a head circumference of 46 cm at 3.5 years (−1.94 SD). Mild to severe hearing loss was detected in 4 families representing 6/8 of the individuals. Ophthalmological anomalies were detected in all individuals except two, represented by a variety of anomalies (eye position anomalies, strabismus, hypermetropia, esotropia, nystagmus, myopia, bilateral zonular cataract, pale optic discs, patchy pigmented fundus, hyperopia, and/or astigmatism). Seven individuals had brain MRIs with one individual presenting agenesis of the corpus callosum. Individual F8:II.1 brain MRI was compatible with white matter injury of prematurity while her sister (F8:II.3) had a thick and foreshortened corpus callosum, with a subtle deficiency of the rostral end, bulky caudate heads, and unusually deep sella (V shaped). Facial dysmorphism included up/down-slanting palpebral fissures, epicanthus, ptosis, hypo/hypertelorism, wide sparse eyebrows, high nasal root, hypoplastic columella, broad and flat philtrum, thin upper lip, prognathism with mild limitation of jaw opening, wide mouth, low-set ears, sparse hair, scaphocephaly, frontal bossing, and discolored dentition (Figure 1). Behavioral traits associated with the phenotype included hyperactivity, aggressive behavior, and angry outbursts. Other physical features included clinodactyly of both hands, bifid and short uvula, high arched palate, cubitus and genu valgum, mild joint laxity, pes-cavus bilateral, pes-planus, and hypoplastic scrotum.
Table 1.
Summary of phenotype information for NDD-affected individuals with THUMPD1 bi-allelic pathogenic variants
| Case index | F1:II.1 | F1:II.2 | F2:II.1 | F2:II.2 | F3:II:1 | F4:II.1 | F5:II.1 | F6:II.1 | F7:II.1 | F7:II.2 | F7:II.3 | F8:II.1 | F8:II.3 | Summary |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age at last examination (y) | 14 | 7.5 | 23 | 16 | 8 | 2 | 3.5 | 4 | 10 | 16 | 19 | 7 | 14 | |
| Sex | M | M | M | M | F | M | F | M | M | F | M | F | F | |
| Consanguinity | − | − | + | + | + | N/A | + | − | + | + | + | + | + | 9/12 |
| Neurological problems | ||||||||||||||
| Developmental delay | + | + | + | + | + | + | + | + | + | + | N/A | + | + | 12/12 |
| Microcephaly | − | − | − | N/A | + | + | − | − | + | − | N/A | + | N/A | 4/10 |
| Delayed sentences | + | + | + | + | + | N/A | + | + | + | N/A | N/A | + | + | 10/10 |
| Age at first words (y) | N/A | 2 | N/A | N/A | 1 | N/A | non verbal | 2 | 2 | N/A | N/A | N/A | 2.5 | |
| Age of walking (m) | 17 | 16 | 24 | N/A | 18 | N/A | 36 | 18 | 36 | N/A | N/A | 19 | 15 | |
| Intellectual disability | + | + | + | + | + | N/A | + | + | + | + | + | + | + | 12/12 |
| Behavior abnormality | N/A | N/A | + | + | + | N/A | + | + | + | + | N/A | − | − | 7/9 |
| Febrile seizures | N/A | + | − | − | + | N/A | − | − | + | + | + | − | + | 6/11 |
| Brain MRI abnormality | N/A | N/A | N/A | N/A | − | + | − | − | − | N/A | N/A | + | + | 3/7 |
| Systemic problems | ||||||||||||||
| Short stature | − | − | − | N/A | + | N/A | − | − | + | − | N/A | + | − | 3/10 |
| Ophtalmological abnormality | + | + | + | − | + | + | + | + | + | + | + | + | − | 11/13 |
| Congenital heart defect | − | − | N/A | N/A | N/A | + | N/A | N/A | N/A | N/A | N/A | + | N/A | 2/4 |
| Hearing impairment | + | + | N/A | N/A | N/A | N/A | N/A | + | − | + | − | + | + | 6/8 |
Symbols and abbreviations: AR, autosomal recessive; CH, compound heterozygous; N/A, no data or not reported; ADHD, attention deficit hyperactivity disorder
Figure 1.
Clinical features of individuals with bi-allelic THUMPD1 variants
Photographs of five individuals from our cohort, showing facial dysmorphisms. Each individual is referenced with the corresponding identifier used throughout the manuscript: Individuals F1:II:1 (A, B) and F1:II:2 (C) are brothers and show down-slanting palpebral fissures, broad and flat philtrum, hypoplastic columella, and low-set ears. Individual F6:II:1 (F) has a wide mouth, epicanthus, ptosis, and narrow palpebral fissures. Individual F8:II.1 (G, H) shows scaphocephaly, frontal bossing, deep-set ears, hypotelorism, and intermittent bilateral esotropia.
All affected individuals harbor bi-allelic variants in THUMPD1 (GenBank: NM_001304550.1) (Figure 2). 12 individuals are homozygous for the following variants: two brothers (F1:II.1, F1:II.2) are homozygous for a frameshift variant, c.495dup (p.Ser166Leufs∗24), three (F2:II.1, F2:II.3, F3:II.1) are homozygous for variant c.706C>T (p.Gln236∗), one (F4:II.1) is homozygous for variant c.341T>G (p.Leu114∗), one (F5:II.1) is homozygous for variant c.303_306del (p.Glu102Leufs∗10), three (F7:II.1, F7:II.2, F7:II.3) are homozygous for variant c.774_776del (p.Leu258del), two sisters (F8:II.1, F8.II.3) are homozygous for variant c.634dup (p.Glu212Glyfs∗18), and lastly one individual (F6:II.1) has two variants: c.469C>T (p.Arg157∗) and c.490C>T (p.Pro164Ser) resulting in a compound heterozygous state. Individual F2:II.6 has a different disease (Rahman syndrome [MIM: 617537]), which is caused by mutations in HISTH1E, but is able to attend school with borderline performance, good behavior, and normal development for his age. It should be noted that F2:II.1 and F2:II do not share the Rahman syndrome-associated variant. These THUMPD1 variants were absent or very rare in the gnomAD Exome database (maximum allele frequency of 0.00001998). All variants lie either within the THUMP domain or the predicted α-helical N-terminal region of THUMPD1 (Figure 2B). Additionally, we identified a family with three affected brothers carrying the homozygous variant c.133G>A (p.Gly45Ser) (Table S3, family 9). Although their phenotype is in line with the other identified individuals, the missense variant is outside the known functional domains of THUMPD1, so in absence of functional data we still consider this variant of unknown significance.
Figure 2.
Family pedigrees of individuals with bi-allelic THUMPD1 variants and a schematic of THUMPD1 with positions of the corresponding mutated amino acid residues indicated
(A) Pedigrees of eight unrelated families with affected members indicated as filled circles (females) and squares (males) with denoted THUMPD1 variants and the corresponding protein alterations. Double horizontal lines indicate consanguinity. Individual F2:II.6 has a different disease (Rahman syndrome) but is able to attend school with borderline performance, good behavior, and normal development for his age.
(B) Schematic representation of the human THUMPD1 protein sequence with THUMP-domain highlighted in green. The nature and positions of the variants are shown above the primary sequence diagram (colored blue for missense mutations, orange for nonsense mutations, and red for frameshift variants).
Lymphoblasts from an individual homozygous for a c.706C>T variant exhibit a loss of THUMPD1 mRNA and protein expression, and a loss of tRNA acetylation
We obtained the lymphoblasts from one affected individual (F2:II.1; 15DG1395 from Maddirevula et al.43) who is homozygous for THUMPD1 variant c.706C>T (p.Gln236∗). Western blot analysis of the cell extracts prepared from these lymphoblasts showed that no full-length THUMPD1 was produced compared to two unrelated control individuals (Figure 3A). This is expected due to the p.Gln236∗ premature stop codon and because we probed with an anti-THUMPD1 antibody that was raised against C-terminal epitope (amino acids 303–353) of THUMPD1. Moreover, RT-qPCR analysis showed a significant decrease of THUMPD1 mRNA levels in THUMPD1 variant cells, expressing ∼20% of control #1 THUMPD1 mRNA levels (Figure 3B). Using the most abundant THUMPD1 transcript based on GTEx (GenBank: NM_001304550.1), we note that the premature terminal codon (PTC) for this variant resides in the second-last exon with the final intron within the 3′ UTR, and these kind of PTC-containing transcripts have been generally shown in most cases (but not all) to escape nonsense-mediated decay.49 Thus, to more thoroughly test whether this variant (p.Gln236∗) is an exception to this general rule, we carried out additional qPCR analysis comparing the expression levels of THUMPD1 pre-mRNA to mature mRNA in control and p.Gln236∗ variant samples and found no significant difference in pre-mRNA levels but again a reduction in mature mRNA levels in the affected individual (Figure S1). This is highly indicative that this transcript is unable to escape NMD. This also suggests that other PTC or frameshift variants that reside in this second to last exon (e.g., variant p.Glu212Glyfs∗18 found in F8:II.1 and F8.II.3) may also be sensitive to NMD.
Figure 3.
Bi-allelic c.706C>T THUMPD1 (p.Gln236∗) causes a lack of ac4C modification in tRNA
(A) Western blot analysis of the protein cell extracts prepared from two control individuals and individual F2:II.1 (15DG1395) lymphoblasts probed with anti-THUMPD1 antibody and anti-tubulin antibody (as loading control).
(B) THUMPD1 mRNA expression in two control individuals and individual F2:II.1 (15DG1395) lymphoblasts as determined by RT-qPCR of the corresponding RNA samples (relative to control #1). Samples were analyzed in triplicate, error bars represent ± SD. ∗p(adjusted) < 0.05, ∗∗∗p(adjusted) < 0.001 as determined by a Brown-Forsythe and Welch one-way ANOVA with Benjamini-Hochberg correction (FDR < 5%).
(C) LC-MS nucleoside modification analysis of small RNA samples from individual F2:II.1 (15DG1395) lymphoblasts compared (normalized) to control #1 small RNA samples. Alterations (fold change) for each nucleoside modification was calculated as a ratio of the corresponding value in variant-containing RNA to control sample RNA normalized based on the abundance of all four standard non-modified nucleosides (A, C, G, and U). Samples were run in duplicate, error bars represent ± SD.
(D) LC-MS nucleoside modification analysis of the purified tRNA-Ser-CGA1-1 samples prepared from control #1 (two top panels) and individual F2:II.1 (15DG1395) (two bottom panels) lymphoblasts. The ac4C peaks are shown in green and unmodified cytidine peaks are in blue (as controls). LC elution times are shown above each peak.
Given THUMPD1’s role in tRNA acetylation, we next examined whether the lack of full-length THUMPD1 may affect acetylation of RNA in THUMPD1 GenBank: NP_001291479.1 (p.Gln236∗) variant lymphoblasts. We extracted and digested small RNAs (predominantly containing tRNAs and small nuclear RNAs) from THUMPD1 p.Gln236∗ variant and control individual lymphoblasts and subjected the extracts to liquid chromatography coupled with mass spectrometry analysis (LC-MS) (see Material and methods). This analysis showed that compared to control individual samples, the THUMPD1 p.Gln236∗ variant individual small RNA sample contained no detectable level of ac4C (Figures 3C and S2) while most other normal and modified nucleosides remained at similar levels to control sample levels (Figure 3C). A complete list of modified nucleosides identified by LC-MS in our experiments is presented in Table S2. To further confirm that the loss of ac4C in this sample was specific to a known NAT10/THUMPD1 tRNA substrate, we specifically purified tRNA-Ser-CGA and tRNA-Ala-AGC (as a control tRNA species known not to contain ac4C) from control and THUMPD1 p.Gln236∗ variant lymphoblasts, performed LC-MS analysis of hydrolyzed RNA, and observed a >100-fold loss of ac4C signal in the tRNA-Ser-CGA sample from THUMPD1 p.Gln236∗ variant cells when compared a tRNA-Ser-CGA sample from control cells (Figure 3D). As expected, control tRNA-Ala-AGC samples had similar, background-level ac4C values to the THUMPD1 p.Gln236∗ variant cells (Figure S3). Taken together, our observations suggest that a loss of THUMPD1 protein expression results in a loss of ac4C modification of small RNAs.
Targeted THUMPD1 knockouts in human cell lines results in a loss of ac4C modification of small RNAs
To ensure that the loss of THUMPD1 in individuals with bi-allelic THUMPD1 variants specifically affects small RNA ac4C modification and was not due to any indirect secondary effects, we generated THUMPD1 knockouts (KO) in HEK293T and HeLa cell lines. THUMPD1 KO cell lines were generated using a gRNA targeting the coding sequence in exon 2 (HEK293T, Figure 4A; HeLa, Figure S4A), cloned cell colonies were subjected to sequence analysis to detect premature termination codon (PTC)-inducing indels (HEK293T, Figure 4B; HeLa, Figure S4B) and appropriate clones in each background were selected for further experiments (HEK293T, Figure 4C; HeLa, Figure S4C). THUMPD1 KO cells were tested for THUMPD1 abundance via western blot and we observed a clear absence of the full-length THUMPD1 protein in THUMPD1 KO cells compared to WT HEK293T and HeLa cells (HEK293T, Figure 4D; HeLa, Figure S4D). In addition, RT-qPCR showed that THUMPD1 mRNA levels in both KO cells were substantially reduced, exhibiting ∼20% of the expression levels observed in WT cell lines (HEK293T, Figure 4E; HeLa, Figure S4E), very similar to what was observed in THUMPD1 p.Gln236∗ variant lymphoblasts.
Figure 4.
THUMPD1 KO in HEK293T cells causes loss of ac4C modification in small RNA samples
(A) Schematic representation of the THUMPD1 KO in HEK293T cell line. sgRNA was designed to disrupt THUMPD1 nucleotide sequence in exon 2 to produce an aberrant PTC-containing transcript (see Table S1 for gRNA sequence).
(B) Sanger DNA sequencing data of the THUMPD1 exon 2 genomic region obtained from selected THUMPD1 KO clone (top) and WT HEK293T cells (bottom). Underlined DNA sequence in WT HEK293T sample is complementary to the designed sgRNA. Dotted vertical line in both sequences indicates a Cas9 target cleavage site.
(C) ICE analysis (Synthego) results of the THUMPD1 exon 2 genomic DNA region from a selected THUMPD1 KO clone. Target nucleotide sequence complementary to sgRNA is in red font and dotted vertical line indicates a Cas9 target cleavage site. The indel type and percentage of detected sequence are shown on the left.
(D) Western blot analysis of the protein cell extracts prepared from HEK293T control and two independent THUMPD1 KO cell lines probed with anti-THUMPD1 antibody and anti-GAPDH antibody (as loading control).
(E) THUMPD1 mRNA expression in HEK293T control and THUMPD1 KO cells determined by RT-qPCR of the corresponding RNA samples. Samples were analyzed in triplicate, error bars represent ± SD. ∗∗∗∗p < 0.0001, as determined by a Welch’s t test.
(F) Northern immunoblot analysis of the large and small RNA samples prepared from HEK293T control (WT) and THUMPD1 KO cells. Positions of 18S rRNA and tRNA on the gel are shown by the arrows on the left.
(G) LC-MS nucleoside modification analysis of the small RNA samples prepared from HEK293T control and THUMPD1 KO cells. Alterations (fold change) for each nucleoside modification was calculated as a ratio of the corresponding value in THUMPD1 KO RNA to HEK293T control RNA normalized based on the abundance of all standard non-modified nucleosides (A, C, G, and U). Samples were run in duplicate, error bars represent ± SD.
Importantly, much like what was observed in THUMPD1 p.Gln236∗ variant lymphoblasts, THUMPD1 disruption in HEK293T and HeLa cells led to a complete loss of ac4C modification of small RNAs. Here we used two independent procedures to confirm this observation. First, immuno-northern blots of large and small RNA samples prepared from HEK293T WT and KO cells (Figure S5) probed with an anti-ac4C antibody demonstrated a loss of ac4C modification signal of small RNAs from THUMPD1 KO cells (Figure 4F). In contrast, THUMPD1 KO did not affect the ac4C status of 18S rRNA species present in the large RNA sample, similar to what is observed in yeast.31,35 As a control, we also showed that in NAT10 KO HeLa cells there is a complete loss of ac4C modification in both rRNA and tRNA (Figure S4F). Second, LC-MS analysis of hydrolyzed small RNA samples prepared from WT and KO cells revealed that THUMPD1 KO cells exhibited an undetectable level of ac4C modification in small RNAs while most of the other nucleoside modifications remained unchanged as compared to WT cell samples (HEK293T, Figure 4G; HeLa, Figure S4G); this contrasts with the levels of ac4C in the large RNA fraction, which were much less affected (Figure S6). Next, to determine whether the loss of ac4C modification on tRNA-Ser-CGA results in a downstream reduction of tRNA-Ser-CGA levels, we carried out northern blot experiments on small RNA samples from HeLa WT, THUMPD1 KO, and NAT10 KO with specific probes that recognize tRNA-Ser-CGA, or tRNA-Ala-AGC or U6 snRNA as a control. Interestingly, we did not detect any large differences in steady-state levels of tRNA-Ser-CGA in either KO samples (Figure S7). It is possible that in HeLa cells under these growth conditions, the Rapid tRNA Decay (RTD) surveillance pathway is not measurably active against hypoacetylated tRNA-Ser-CGA.
Taken together, the specific knockout of THUMPD1 in HEK293T and HeLa cell lines recapitulates our observation of the substantial loss of ac4C modification of small RNAs and tRNA-Ser-CGA present in THUMPD1 p.Gln236∗ variant lymphoblasts obtained from individual F2:II.1 (15DG1395) in our study.
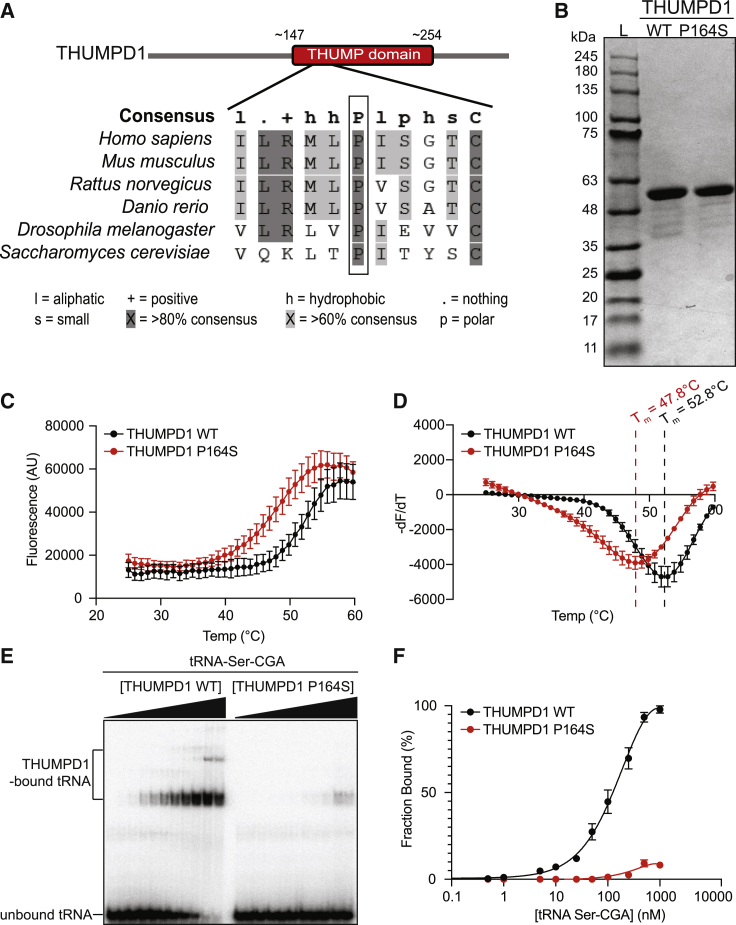
Recombinant THUMPD1 p.Pro164Ser exhibits a marked decrease in thermal stability and RNA-binding capacity
A single missense variant was identified in our cohort, p.Pro164Ser in individual F5.II.1. Pro164 is strictly conserved within THUMPD1 orthologs across eukaryotes (Figure 5A) and in architecturally similar THUMP domain-containing proteins in archaea,36 suggesting it is important for protein structure and function.
Figure 5.
THUMPD1 p.Pro164Ser affects its stability and binding to RNA
(A) Proline at position 164 (outlined with a black box) within THUMP domain (positions 147–254 [in red] is highly conserved in primary sequences of THUMPD1 orthologs), including model organisms Homo sapiens, Mus musculus, Rattus norvegicus, Danio rerio, Drosophila melanogaster, and Saccharomyces cerevisiae.
(B) Coomassie blue-stained gel showing purified WT and p.Pro164Ser (P164S) mutant proteins. Protein molecular marker sizes are shown on the left side of the gel.
(C) Thermal denaturation profile from a protein thermal shift assay of purified THUMPD1 WT (black line) and P164S mutant (red line) proteins.
(D) A plot first derivative of the fluorescence emission as a function of temperature (−dF/dT) (from the data in C). The protein melting temperature (Tm) of the P164S mutant protein (red line) is ∼5°C lower than the WT protein (black line).
(E) EMSA gel image testing the binding of purified THUMPD1 WT protein (left) and P164S mutant protein (right) to tRNA-Ser-CGA. The ascending triangles in black show an increasing concentration of THUMPD1 in the EMSA assay samples from 0 nM to 1,000 nM.
(F) Quantitation of the EMSA of THUMPD1 binding to tRNA-Ser-CGA from (E) showed that purified THUMPD1 WT protein (black line) had substantially higher affinity/binding to tRNA than P164S mutant (red line). Samples were run in duplicate, error bars represent ± SD.
To this end, we overexpressed and purified recombinant WT and mutant p.Pro164Ser THUMPD1 to homogeneity (Figure 5B) and tested thermal stability and RNA-binding capacity, using fluorescent thermal melt assays and electrophoretic mobility shift assays (EMSAs), respectively. We observed that THUMPD1 p.Pro164Ser exhibited a marked decrease in protein thermal stability with an ∼5°C decrease in melting temperature relative to WT THUMPD1 (Figures 5C and 5D), suggesting that the p.Pro164Ser substitution results in less stably folded THUMPD1. To test whether this loss of stability affects THUMPD1’s capacity to bind RNA, we carried out RNA-binding EMSA assays against an in vitro transcribed, radiolabelled tRNA-Ser-CGA (with a CCA tail added during transcription). While WT THUMPD1 bound to tRNA-Ser-CGA with an approximate nanomolar affinity, we observed a significant loss in RNA binding capacity for the p.Pro164Ser mutant (Figures 5E and 5F), indicating that this variant may not be able to efficiently bind its tRNA targets in vivo for subsequent tRNA acetylation. It should also be noted that to our knowledge this is the first demonstration of tRNA binding by WT human THUMPD1. Taken together, these data suggest that the variant p.Pro164Ser results in a loss of function of THUMPD1. Given that this variant is absent from gnomAD database and was identified in compound heterozygous state with p.Arg157∗ in individual F5.II.1, we consider it as pathogenic.
Discussion
In this study, we identified bi-allelic variants in THUMPD1 in 13 individuals presenting with a neurodevelopmental disorder. Given the genotypic and phenotypic similarities in our case series, we propose that these bi-allelic variants in THUMPD1 should be considered as pathogenic. In addition, we present functional studies that indicate THUMPD1 loss of function in these individuals. We demonstrate that variants identified cause loss of function of THUMPD1 and that this defect results in a loss of ac4C modification in small RNAs and of individually purified tRNA-Ser-CGA. We further corroborate this effect by showing a loss of tRNA acetylation in two CRISPR-Cas9-generated THUMPD1 KO cell lines. Taken together, these results suggest that the lack of tRNA acetylation due to THUMPD1 loss of function results in syndromic intellectual disability.
tRNA modifications play an important role in neurodevelopment. For example, loss of modifications in the anticodon loops of tRNA can result in perturbations at all stages of protein translation (initiation, decoding efficiency and fidelity, termination, and so on), while loss of a range of tRNA modifications in the body of tRNA appear to primarily result in changes in tRNA tertiary structure and stability, and thus also tend to result in similar, if more indirect, perturbations of protein translation (see Ramos and Fu11 for an extensive review). We speculate that phenotypes observed in this study’s individuals are a result of a similar phenomenon by which loss of tRNA modification may result in impaired protein synthesis and thus altered proteostasis at important stages of neurodevelopment. Furthermore, the individuals in our study share a number of phenotypes common to diseases where mutations in aminoacyl-tRNA synthetases have been identified (see Ognjenović and Simonović 50 for an extensive review), further highlighting that perturbation of protein translation can result in global development delay and intellectual disability. It is also possible that the lack of tRNA-Ser or tRNA-Leu acetylation affects translation of only a limited subset of proteins that are important for neurodevelopment. Indeed, it is well understood that neurons are particularly sensitive to alterations in protein synthesis, and the multi-tissue effects we observe in the individuals in this study could be a result of the complex developmental interplay and feedback between multiple tissues and the brain that occur during development (for review see Kapur et al.51).
Specifically, given ac4C’s position within the body of Ser- and Leu-tRNAs (in the D-stem of the tRNA) and its ability to increase the base pairing strength between G:C,52,53 it is possible that ac4C helps tRNAs obtain the correct cloverleaf fold and tertiary contacts, and the lack of this modification likely results in a less stable tRNA that may be more prone to degradation. Indeed, in S. cerevisiae, the deletion of TAN1 (THUMPD1 ortholog) resulted in loss of ac4C modifications on Ser- and Leu-tRNAs, causing an increased sensitivity of tRNA-Ser-CGA, tRNA-Ser-UGA, and tRNA-Leu-GAG to the rapid tRNA decay pathway that ultimately led to reduced levels of these tRNA species.35,40, 41, 42,54 This effect is mild in a single tan1Δ deletion strain and is significantly exacerbated when a tan1Δ deletion is coupled with either a mutant tRNA-Ser-CGA35 or trm44 deletion (which encodes a tRNA 2′-O-methyltransferase that methylating position U44).40 Both of these double mutants possess a temperature-sensitive growth phenotype. In the case of THUMPD1 and NAT10 KO HeLa cell lines (Figure S5) and THUMPD1 p.Gln238∗ variant lymphoblasts (data not shown) used in this study, we did not observe any dramatic changes in tRNA-Ser-CGA expression relative to WT cells. We speculate that either changes in tRNA-Ser-CGA expression due to tRNA hypoacetylation in these cell lines are too minor to be detected, but could be more pronounced at certain neurodevelopmental stages and/or cell types in humans, or when cells are undergoing stress; or unlike the slight reduction in tRNA-Ser-CGA levels Δtan1 yeast, the loss of tRNA acetylation does not result in a reduction of corresponding tRNA levels in humans. With this in mind, we cannot disregard that ac4C may have additional roles beyond stabilizing the structure of certain tRNAs and may be required for additional tRNA processing, aminoacylation, recognition, or protein translation processes. Relatedly, it has been previously shown using large-scale RNA-binding protein mass spectrometry screens that THUMPD1 may broadly interact with mRNA55 and specific pre-microRNAs,56 although further work needs to be done to confirm whether these observations are physiologically relevant.
Taken together, we showed that bi-allelic loss-of-function variants in THUMPD1 can be added to the growing list of gene variants known to affect tRNA modification and interfere with normal neurodevelopment. Additional research is required to understand the cellular response to the loss of tRNA acetylation (and indeed many other tRNA modifications) and how it results in altered neurodevelopment. Finally, our work also highlights that quantitative LC-MS is becoming an increasingly powerful tool to dissect changes in RNA modifications in both research and potentially clinical settings.57 The ability to detect a large number of modified nucleosides in parallel enables a more systems-level understanding of RNA modification dynamics in health and disease, and may potentially assist in the development of novel biomarker approaches for research and diagnosis of genetic disorders involving RNA modification, and aid in connecting genotype to phenotype.
Acknowledgments
We thank participating families for their cooperation. We thank members of the O’Connell lab for helpful discussions. We thank Dr. Shalini Oberdoerffer for providing NAT10 KO HeLa cells. We thank Kevin Welle and the University of Rochester Mass Spectrometry Resource Laboratory for invaluable assistance with nucleoside mass spectrometry experiments. This work used the Typhoon RGB scanner at the Center for RNA Biology, University of Rochester, supported by the NIH S10 OD021489-01A1 instrumentation grant. M.R.O. is supported by the National Institute of General Medical Sciences grant R35GM133462. Sequencing and analysis for subjects from family 8 were provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung and Blood Institute grant UM1 HG008900 and in part by National Human Genome Research Institute grant R01 HG009141.
Declaration of interests
The authors declare no competing interests.
Published: February 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.02.001.
Contributor Information
Benjamin Cogné, Email: benjamin.cogne@chu-nantes.fr.
Mitchell R. O’Connell, Email: mitchell_oconnell@urmc.rochester.edu.
Data and code availability
Variants were submitted to Clinvar with accession numbers Clinvar: SCV002056133, Clinvar: SCV002056134, Clinvar: SCV002056135, Clinvar: SCV002056136, Clinvar: SCV002056137, Clinvar: SCV002056138, Clinvar: SCV002056139, Clinvar: SCV002056140, and Clinvar: SCV002056141.
Web resources
OMIM, https://www.omim.org/
Supplemental information
References
- 1.El Yacoubi B., Bailly M., de Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 2.Ontiveros R.J., Stoute J., Liu K.F. The chemical diversity of RNA modifications. Biochem. J. 2019;476:1227–1245. doi: 10.1042/BCJ20180445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018;28:395–404. doi: 10.1038/s41422-018-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Crécy-Lagard V., Jaroch M. Functions of Bacterial tRNA Modifications: From Ubiquity to Diversity. Trends Microbiol. 2021;29:41–53. doi: 10.1016/j.tim.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021;22:375–392. doi: 10.1038/s41580-021-00342-0. [DOI] [PubMed] [Google Scholar]
- 6.Phizicky E.M., Hopper A.K. tRNA processing, modification, and subcellular dynamics: past, present, and future. RNA. 2015;21:483–485. doi: 10.1261/rna.049932.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klassen R., Bruch A., Schaffrath R. Induction of protein aggregation and starvation response by tRNA modification defects. Curr. Genet. 2020;66:1053–1057. doi: 10.1007/s00294-020-01103-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ranjan N., Leidel S.A. The epitranscriptome in translation regulation: mRNA and tRNA modifications as the two sides of the same coin? FEBS Lett. 2019;593:1483–1493. doi: 10.1002/1873-3468.13491. [DOI] [PubMed] [Google Scholar]
- 9.Agris P.F., Narendran A., Sarachan K., Väre V.Y.P., Eruysal E. The Enzymes. Elsevier; 2017. The Importance of Being Modified; pp. 1–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Väre V.Y., Eruysal E.R., Narendran A., Sarachan K.L., Agris P.F. Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function. Biomolecules. 2017;7:29. doi: 10.3390/biom7010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramos J., Fu D. The emerging impact of tRNA modifications in the brain and nervous system. Biochim. Biophys. Acta. Gene Regul. Mech. 2019;1862:412–428. doi: 10.1016/j.bbagrm.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Khan M.A., Rafiq M.A., Noor A., Hussain S., Flores J.V., Rupp V., Vincent A.K., Malli R., Ali G., Khan F.S., et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012;90:856–863. doi: 10.1016/j.ajhg.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdelrahman H.A., Al-Shamsi A.M., Ali B.R., Al-Gazali L. A null variant in PUS3 confirms its involvement in intellectual disability and further delineates the associated neurodevelopmental disease. Clin. Genet. 2018;94:586–587. doi: 10.1111/cge.13443. [DOI] [PubMed] [Google Scholar]
- 14.Shaheen R., Han L., Faqeih E., Ewida N., Alobeid E., Phizicky E.M., Alkuraya F.S. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum. Genet. 2016;135:707–713. doi: 10.1007/s00439-016-1665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alazami A.M., Hijazi H., Al-Dosari M.S., Shaheen R., Hashem A., Aldahmesh M.A., Mohamed J.Y., Kentab A., Salih M.A., Awaji A., et al. Mutation in ADAT3, encoding adenosine deaminase acting on transfer RNA, causes intellectual disability and strabismus. J. Med. Genet. 2013;50:425–430. doi: 10.1136/jmedgenet-2012-101378. [DOI] [PubMed] [Google Scholar]
- 16.El-Hattab A.W., Saleh M.A., Hashem A., Al-Owain M., Asmari A.A., Rabei H., Abdelraouf H., Hashem M., Alazami A.M., Patel N., et al. ADAT3-related intellectual disability: Further delineation of the phenotype. Am. J. Med. Genet. A. 2016;170A:1142–1147. doi: 10.1002/ajmg.a.37578. [DOI] [PubMed] [Google Scholar]
- 17.Lentini J.M., Alsaif H.S., Faqeih E., Alkuraya F.S., Fu D. DALRD3 encodes a protein mutated in epileptic encephalopathy that targets arginine tRNAs for 3-methylcytosine modification. Nat. Commun. 2020;11:2510. doi: 10.1038/s41467-020-16321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang K., Lentini J.M., Prevost C.T., Hashem M.O., Alkuraya F.S., Fu D. An intellectual disability-associated missense variant in TRMT1 impairs tRNA modification and reconstitution of enzymatic activity. Hum. Mutat. 2020;41:600–607. doi: 10.1002/humu.23976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaheen R., Mark P., Prevost C.T., AlKindi A., Alhag A., Estwani F., Al-Sheddi T., Alobeid E., Alenazi M.M., Ewida N., et al. Biallelic variants in CTU2 cause DREAM-PL syndrome and impair thiolation of tRNA wobble U34. Hum. Mutat. 2019;40:2108–2120. doi: 10.1002/humu.23870. [DOI] [PubMed] [Google Scholar]
- 20.Monies D., Vågbø C.B., Al-Owain M., Alhomaidi S., Alkuraya F.S. Recessive Truncating Mutations in ALKBH8 Cause Intellectual Disability and Severe Impairment of Wobble Uridine Modification. Am. J. Hum. Genet. 2019;104:1202–1209. doi: 10.1016/j.ajhg.2019.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Brouwer A.P.M., Abou Jamra R., Körtel N., Soyris C., Polla D.L., Safra M., Zisso A., Powell C.A., Rebelo-Guiomar P., Dinges N., et al. Variants in PUS7 Cause Intellectual Disability with Speech Delay, Microcephaly, Short Stature, and Aggressive Behavior. Am. J. Hum. Genet. 2018;103:1045–1052. doi: 10.1016/j.ajhg.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaheen R., Tasak M., Maddirevula S., Abdel-Salam G.M.H., Sayed I.S.M., Alazami A.M., Al-Sheddi T., Alobeid E., Phizicky E.M., Alkuraya F.S. PUS7 mutations impair pseudouridylation in humans and cause intellectual disability and microcephaly. Hum. Genet. 2019;138:231–239. doi: 10.1007/s00439-019-01980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaheen R., Abdel-Salam G.M.H., Guy M.P., Alomar R., Abdel-Hamid M.S., Afifi H.H., Ismail S.I., Emam B.A., Phizicky E.M., Alkuraya F.S. Mutation in WDR4 impairs tRNA m(7)G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol. 2015;16:210. doi: 10.1186/s13059-015-0779-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohashi Z., Murao K., Yahagi T., Von Minden D.L., McCloskey J.A., Nishimura S. Characterization of C+ located in the first position of the anticodon of Escherichia coli tRNA Met as N4 - acetylcytidine. Biochim. Biophys. Acta. 1972;262:214. [PubMed] [Google Scholar]
- 25.Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J. Biol. Chem. 1984;259:9461–9471. [PubMed] [Google Scholar]
- 26.Bruenger E., Kowalak J.A., Kuchino Y., McCloskey J.A., Mizushima H., Stetter K.O., Crain P.F. 5S rRNA modification in the hyperthermophilic archaea Sulfolobus solfataricus and Pyrodictium occultum. FASEB J. 1993;7:196–200. doi: 10.1096/fasebj.7.1.8422966. [DOI] [PubMed] [Google Scholar]
- 27.McCarroll R., Olsen G.J., Stahl Y.D., Woese C.R., Sogin M.L. Nucleotide sequence of the Dictyostelium discoideum small-subunit ribosomal ribonucleic acid inferred from the gene sequence: evolutionary implications. Biochemistry. 1983;22:5858–5868. [Google Scholar]
- 28.Thomas G., Gordon J., Rogg H. N4-Acetylcytidine. A previously unidentified labile component of the small subunit of eukaryotic ribosomes. J. Biol. Chem. 1978;253:1101–1105. [PubMed] [Google Scholar]
- 29.Ito S., Horikawa S., Suzuki T., Kawauchi H., Tanaka Y., Suzuki T., Suzuki T. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA) J. Biol. Chem. 2014;289:35724–35730. doi: 10.1074/jbc.C114.602698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito S., Akamatsu Y., Noma A., Kimura S., Miyauchi K., Ikeuchi Y., Suzuki T., Suzuki T. A single acetylation of 18 S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae. J. Biol. Chem. 2014;289:26201–26212. doi: 10.1074/jbc.M114.593996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma S., Langhendries J.-L., Watzinger P., Kötter P., Entian K.-D., Lafontaine D.L.J. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015;43:2242–2258. doi: 10.1093/nar/gkv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zachau H.G., Dütting D., Feldmann H. The structures of two serine transfer ribonucleic acids. Hoppe Seylers Z. Physiol. Chem. 1966;347:212–235. doi: 10.1515/bchm2.1966.347.1.212. [DOI] [PubMed] [Google Scholar]
- 33.Kowalski S., Yamane T., Fresco J.R. Nucleotide sequence of the “denaturable” leucine transfer RNA from yeast. Science. 1971;172:385–387. doi: 10.1126/science.172.3981.385. [DOI] [PubMed] [Google Scholar]
- 34.Kruppa J., Zachau H.G. Multiplicity of serine-specific transfer RNAs of brewer’s and baker’s yeast. Biochim. Biophys. Acta. 1972;277:499–512. doi: 10.1016/0005-2787(72)90093-7. [DOI] [PubMed] [Google Scholar]
- 35.Johansson M.J.O., Byström A.S. The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA. 2004;10:712–719. doi: 10.1261/rna.5198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aravind L., Koonin E.V. THUMP--a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends Biochem. Sci. 2001;26:215–217. doi: 10.1016/s0968-0004(01)01826-6. [DOI] [PubMed] [Google Scholar]
- 37.Kamalampeta R., Keffer-Wilkes L.C., Kothe U. tRNA binding, positioning, and modification by the pseudouridine synthase Pus10. J. Mol. Biol. 2013;425:3863–3874. doi: 10.1016/j.jmb.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 38.Waterman D.G., Ortiz-Lombardía M., Fogg M.J., Koonin E.V., Antson A.A. Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain. J. Mol. Biol. 2006;356:97–110. doi: 10.1016/j.jmb.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 39.McCleverty C.J., Hornsby M., Spraggon G., Kreusch A. Crystal structure of human Pus10, a novel pseudouridine synthase. J. Mol. Biol. 2007;373:1243–1254. doi: 10.1016/j.jmb.2007.08.053. [DOI] [PubMed] [Google Scholar]
- 40.Kotelawala L., Grayhack E.J., Phizicky E.M. Identification of yeast tRNA Um(44) 2¢-O-methyltransferase (Trm44) and demonstration of a Trm44 role in sustaining levels of specific tRNA(Ser) species. RNA. 2008;14:158–169. doi: 10.1261/rna.811008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whipple J.M., Lane E.A., Chernyakov I., D’Silva S., Phizicky E.M. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev. 2011;25:1173–1184. doi: 10.1101/gad.2050711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dewe J.M., Whipple J.M., Chernyakov I., Jaramillo L.N., Phizicky E.M. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA. 2012;18:1886–1896. doi: 10.1261/rna.033654.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maddirevula S., Alzahrani F., Al-Owain M., Al Muhaizea M.A., Kayyali H.R., AlHashem A., Rahbeeni Z., Al-Otaibi M., Alzaidan H.I., Balobaid A., et al. Autozygome and high throughput confirmation of disease genes candidacy. Genet. Med. 2019;21:736–742. doi: 10.1038/s41436-018-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.ACMG Board of Directors ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet. Med. 2015;17:68–69. doi: 10.1038/gim.2014.151. [DOI] [PubMed] [Google Scholar]
- 46.Arango D., Sturgill D., Alhusaini N., Dillman A.A., Sweet T.J., Hanson G., Hosogane M., Sinclair W.R., Nanan K.K., Mandler M.D., et al. Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell. 2018;175:1872–1886.e24. doi: 10.1016/j.cell.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rio D.C. Expression and purification of active recombinant T7 RNA polymerase from E. coli. Cold Spring Harb. Protoc. 2013;2013 doi: 10.1101/pdb.prot078527. [DOI] [PubMed] [Google Scholar]
- 48.Su D., Chan C.T.Y., Gu C., Lim K.S., Chionh Y.H., McBee M.E., Russell B.S., Babu I.R., Begley T.J., Dedon P.C. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat. Protoc. 2014;9:828–841. doi: 10.1038/nprot.2014.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindeboom R.G.H., Supek F., Lehner B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet. 2016;48:1112–1118. doi: 10.1038/ng.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ognjenović J., Simonović M. Human aminoacyl-tRNA synthetases in diseases of the nervous system. RNA Biol. 2018;15:623–634. doi: 10.1080/15476286.2017.1330245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kapur M., Monaghan C.E., Ackerman S.L. Regulation of mRNA Translation in Neurons-A Matter of Life and Death. Neuron. 2017;96:616–637. doi: 10.1016/j.neuron.2017.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumbhar B.V., Kamble A.D., Sonawane K.D. Conformational preferences of modified nucleoside N(4)-acetylcytidine, ac4C occur at “wobble” 34th position in the anticodon loop of tRNA. Cell Biochem. Biophys. 2013;66:797–816. doi: 10.1007/s12013-013-9525-8. [DOI] [PubMed] [Google Scholar]
- 53.Bartee D., Nance K.D., Meier J.L. Site-Specific Synthesis of N4-Acetylcytidine in RNA Reveals Physiological Duplex Stabilization. Preprint at bioRxiv. 2021 doi: 10.1101/2021.11.12.468326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexandrov A., Chernyakov I., Gu W., Hiley S.L., Hughes T.R., Grayhack E.J., Phizicky E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 55.Kwon S.C., Yi H., Eichelbaum K., Föhr S., Fischer B., You K.T., Castello A., Krijgsveld J., Hentze M.W., Kim V.N. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1122–1130. doi: 10.1038/nsmb.2638. [DOI] [PubMed] [Google Scholar]
- 56.Lee H.Y., Haurwitz R.E., Apffel A., Zhou K., Smart B., Wenger C.D., Laderman S., Bruhn L., Doudna J.A. RNA-protein analysis using a conditional CRISPR nuclease. Proc. Natl. Acad. Sci. USA. 2013;110:5416–5421. doi: 10.1073/pnas.1302807110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin G., Xu M., Zou M., Duan S. The Processing, Gene Regulation, Biological Functions, and Clinical Relevance of N4-Acetylcytidine on RNA: A Systematic Review. Mol. Ther. Nucleic Acids. 2020;20:13–24. doi: 10.1016/j.omtn.2020.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Variants were submitted to Clinvar with accession numbers Clinvar: SCV002056133, Clinvar: SCV002056134, Clinvar: SCV002056135, Clinvar: SCV002056136, Clinvar: SCV002056137, Clinvar: SCV002056138, Clinvar: SCV002056139, Clinvar: SCV002056140, and Clinvar: SCV002056141.