Abstract
In an effort to discover novel, noncarbohydrate inhibitors of influenza virus neuraminidase we hypothesized that compounds which contain positively charged amino groups in an appropriate position to interact with the Asp 152 or Tyr 406 side chains might be bound tightly by the enzyme. Testing of 300 α- and β-amino acids led to the discovery of two novel neuraminidase inhibitors, a phenylglycine and a pyrrolidine, which exhibited Ki values in the 50 μM range versus influenza virus A/N2/Tokyo/3/67 neuraminidase but which exhibited weaker activity against influenza virus B/Memphis/3/89 neuraminidase. Limited optimization of the pyrrolidine series resulted in a compound which was about 24-fold more potent than 2-deoxy-2,3-dehydro-N-acetylneuraminic acid in an anti-influenza cell culture assay using A/N2/Victoria/3/75 virus. X-ray structural studies of A/N9 neuraminidase-inhibitor complexes revealed that both classes of inhibitors induced the Glu 278 side chain to undergo a small conformational change, but these compounds did not show time-dependent inhibition. Crystallography also established that the α-amino group of the phenylglycine formed hydrogen bonds to the Asp 152 carboxylate as expected. Likewise, the β-amino group of the pyrrolidine forms an interaction with the Tyr 406 hydroxyl group and represents the first compound known to make an interaction with this absolutely conserved residue. Phenylglycine and pyrrolidine analogs in which the α- or β-amino groups were replaced with hydroxyl groups were 365- and 2,600-fold weaker inhibitors, respectively. These results underscore the importance of the amino group interactions with the Asp 152 and Tyr 406 side chains and have implications for anti-influenza drug design.
The catalytic power of enzymes arises from their ability to bind the altered substrate in the transition state much more tightly than the substrate in its unaltered, ground state form (28, 29). The magnitude of this binding affinity discrimination can be estimated from a comparison of the rate constants for the enzymatic and nonenzymatic reactions (29) and appears to range between 108- and 1017-fold for most enzymes (7, 30). Any stable compound whose chemical structure resembles that of the transition state should capture some fraction of this binding affinity advantage, resulting in a potent and specific inhibitor of the target enzyme. Compounds believed to mimic the structure of the transition state or of an intermediate in the reaction pathway have been described for well over 100 different enzymes (29, 31).
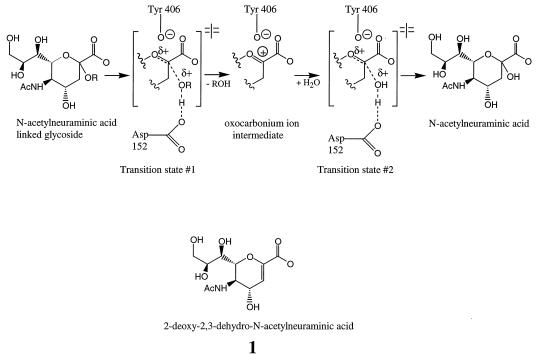
Influenza neuraminidase is one enzyme for which a putative transition state analog has been described (10). This enzyme is present on the viral surface and functions to cleave terminal α-ketosidically linked N-acetylneuraminic acid residues from glycoproteins, glycolipids, and oligosaccharides in a reaction which is essential for effective replication of the influenza virus (20). Enzyme kinetic isotope effect studies (5) have established that the neuraminidase hydrolytic reaction proceeds through a planar, oxocarbonium ion intermediate (Fig. 1). Compound 1, a stable analog of the oxocarbonium ion intermediate, exhibits a Ki value of 5 μM (15), which is four- to eightfold lower than the Km value for the substrate. However, a comparison of the neuraminidase enzymatic rate constants (10) with the nonenzymatic rate constants for glycoside hydrolysis (1, 32) suggests that the transition state for this reaction should exhibit a nominal Kd value in the range of 10−14 to 10−21 M with neuraminidase. Thus, the binding affinity of compound 1 falls far short of that expected for an ideal transition state analog, even though the chemical structure of compound 1 contains all of the functional groups and geometrical constraints present in the oxocarbonium ion intermediate.
FIG. 1.
Possible enzymatic reaction mechanism for influenza virus neuraminidase. Note the presence of partial or full positive charges on the transition state and oxocarbonium ion structures which may be stabilized by interactions with the Asp 152 and Tyr 406 residues of neuraminidase.
One of the features that is present in the chemical structure of the high energy oxocarbonium ion intermediate but is missing in the chemical structure of compound 1 is a positive charge on the ring oxygen. Burmeister and colleagues (4) have suggested that this positive charge is likely stabilized by an ionized Tyr 406 phenolate side chain from neuraminidase. Since this electrostatic interaction does not occur when substrates or products are bound, it seems likely that this interaction is a major contributor to the tight binding of the transition states and oxocarbonium ion intermediate. It also seems clear that neuraminidase must either make additional interactions with the transition states which flank the oxocarbonium ion intermediate or greatly strengthen an existing interaction, because the transition state must be the most tightly bound moiety to occur during enzyme catalysis (29). The bond breaking and bond making that occur in the transition states would be catalyzed if neuraminidase was able to donate a proton to the glycosidic oxygen in the bond-breaking step and remove a proton from the attacking water molecule in the bond-making step. This acid-base catalytic role could be fulfilled by the Asp 152 side chain (26) or perhaps mediated by Asp 152 via an intervening water molecule (5). In either case the glycosidic oxygen might develop a partial positive charge in the transition states, a feature which is not found in the substrate, product, or oxocarbonium ion intermediate. It was our hypothesis that the Tyr 406 and Asp 152 side chains were critical for recognizing the positively charged features of the transition states and high-energy intermediate from the ground state substrate and compound 1. We reasoned that compounds containing positively charged amino groups in appropriate positions to interact with the Tyr 406 and Asp 152 side chains might be good transition-state analog inhibitors. This hypothesis led us to test 300 α- and β-amino acids for inhibition of influenza virus neuraminidase. Our goal was to identify novel, noncarbohydrate neuraminidase inhibitors which could serve as lead structures for a program to develop an anti-influenza drug therapy.
MATERIALS AND METHODS
Neuraminidase.
The catalytically active head domains were purified from A/Tokyo/3/67 and B/Memphis/3/89 influenza viruses as described previously (11). Briefly, purified virus was treated with a protease for a period of time, and then the solution was ultracentrifuged to pellet the viral cores. The supernatant was then either chromatographed using a size exclusion column (for the A/ Tokyo/3/67 enzyme) or subjected to sucrose density gradient centrifugation and dialysis (for the B/Memphis/3/89 enzyme).
Test compounds.
Compound 1 was obtained from Boehringer Mannheim. Compound 2 was synthesized by reacting 2-chloro-4-t-butylphenol with α-hydroxy hippuric acid in 10% H2SO4–acetic acid overnight at room temperature. The benzoyl group was removed from the α-amino group of the adduct via treatment with 6 N HCl and heated under vacuum to generate the HCl salt of compound 2. Compound 3 was synthesized by a six-step procedure which began by reacting Boc-glycine methyl ester with sodium hydride and allyl bromide in tetrahydrofuran (THF) at 0°C. The methyl ester of the resulting bis-substituted amide was reduced to the aldehyde using diisobutylaluminum hydride in methylene chloride at −78°C. This material was reacted with a previously formed solution of N-benzylhydroxyamine hydrochloride and potassium carbonate in toluene at room temperature to generate the bicyclic product. The benzyl protecting group and the nitrogen-oxygen bond were cleaved by catalytic hydrogenation using Pd(OH) and carbon at 1 atm of H2 overnight to yield an enantiomeric mixture of (3R, 4R) and (3S, 4S) 3-amino-4-hydroxymethyl-N-butoxycarbonyl pyrrolidine. The 3-amino group was protected with carbobenzoxy (Cbz)-Cl and triethylamine in THF at 0°C. The hydroxymethyl group was then oxidized to the carboxylic acid using the Jones reagent in acetone at 0°C. The Cbz group was removed from the 3-amino group using Pd-C and 1 atm of H2 in ethanol to afford the (3R, 4R) and (3S, 4S) enantiomeric mixture of compound 3. Compound 4 was synthesized by a multistep procedure beginning with the condensation of N-Boc-glycine methyl ester and tert-butyl acrylate effected by potassium t-butoxide to give (±) 4-oxo-pyrrolidine-1,3-dicarboxylic acid di-tert-butyl ester. Hydroxylation at the 3 position was effected with m-chloroperoxybenzoic acid in dichloromethane at 0°C to room temperature. The resulting crude hydroxy ketone was reacted with hydroxylamine in ethanol to give the corresponding 4-oxime. Hydrogenation of the 4-oxime with Raney Nickel at 4 atm in ethanol gave a mixture of two 4-amino diastereomers that were isolated after protection with N-benzyloxycarbonyloxy-succinimide and separation by chromatography on silica gel. Selective deprotection of pyrrolidine nitrogen was accomplished in formic acid (96%) to give 4-benzyloxycarbonylamino-3-hydroxy-pyrrolidine-3-carboxylic acid tert-butyl ester. Reaction of the pyrrolidine nitrogen with N,N-isopropylethylcarbamoyl chloride and N,N-diisopropylethylamine in dichloromethane gave 4-benzyloxycarbonylamino-1-(ethyl-isopropyl-carbamoyl)-3-hydroxy-pyrrolidine-3-carboxylic acid tert-butyl ester. Simultaneous deprotection of the 3-carboxyl and 4-amino groups occurred with concentrated HCl at 85°C to give 4-amino-1-(ethyl-isopropyl-carbamoyl)-3-hydroxy-pyrrolidine-3-carboxylic acid hydrochloride (compound 4). Compound 6 was obtained in four steps from the hydroxy ketone described above. Reduction with sodium borohydride in ethanol gave 3,4-dihydroxy-pyrrolidine-1,3-dicarboxylic acid di-tert-butyl ester. Selective functionalization of the pyrrolidine nitrogen and deprotection (as previously described) produced compound 6. The stereochemistry of the 4-hydroxyl group was confirmed by X-ray crystallography. Compound 5 was synthesized from 5-tert-butyl-3-chloro-2-hydroxy-benzaldehyde by reaction with potassium cyanide in acetic acid to form the cyanohydrin which was subsequently hydrolyzed to compound 5 with aqueous sodium hydroxide and hydrogen peroxide.
Enzyme inhibition assays.
(i) Compounds were initially tested for inhibition of the influenza virus A/Tokyo/3/67 neuraminidase. Reaction mixtures contained a final concentration of 30 μM 4-methylumbelliferyl sialic acid substrate (Sigma), 0.5 mM test compound in 200 μl of freshly prepared 50 mM sodium citrate, 10 mM CaCl2, 0.1 mg of bovine serum albumin/ml, and 5% dimethyl sulfoxide or ethanol (pH 6.0) buffer. Reactions were started by the addition of neuraminidase to reaction mixtures which were contained in the wells of white 96-well plates. Fluorescence measurements were obtained each minute for 30 min using a Fluoroskan II fluorescence plate reader (Titertek Instruments) equipped with excitation and emission filters of 355 ± 35 nm and 460 ± 25 nm, respectively. DeltaSoft II software (Biometallics) controlled the plate reader. Reaction velocities were calculated from the linear region of the progress curves and compared to uninhibited control velocities in order to identify inhibitors. Experiments to test compounds for inhibition of B/Memphis/3/89 neuraminidase were conducted similarly, except that the substrate concentration was 20 μM. (ii) Experiments to establish the kinetic mechanism of inhibition were conducted with A/Tokyo/3/67 neuraminidase using the plate reader as outlined above, except that both substrate and inhibitor concentrations were varied. (iii) A high-performance liquid chromatography assay was established to confirm the inhibition observed in the plate reader assay. Enzyme inhibition reactions were conducted as described above but were then quenched with 1 mM compound 1 and were immediately flash frozen in dry ice for subsequent analysis. Immediately after thawing, a 100-μl aliquot of the quenched reaction mixture was injected onto a 4.6- by 50-mm Lichrosphere reverse-phase C18 column (E. Merck) which had been equilibrated with 0.1% acetic acid in water with 18% acetonitrile. Reaction components were separated with a linear gradient from 18 to 32% acetonitrile in 0.1% acetic acid from 0 to 10 min at a flow rate of 1 ml/min. The neuraminidase reaction product, methylumbelliferone, eluted at about 6 min under these conditions, as judged by UV detection at a wavelength of 352 nm. Methylumbelliferone peak areas from compound-treated reaction mixtures were compared to those of untreated reaction mixtures in order to quantitate inhibition of the neuraminidase enzymatic reaction. (iv) The Ki values for compounds 4 to 6 versus those of A/Tokyo/3/67 neuraminidase as well as all Ki values versus those of B/Memphis/3/89 neuraminidase were obtained from initial velocity measurements which were then fit to the following equation (21) by nonlinear regression using Kaleidagraph software:
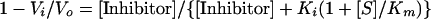 |
where Vi and Vo represent inhibited and uninhibited steady-state reaction velocities, respectively, and [S] is substrate concentration. Km values of 38 and 17 μM were used in this equation for the calculation of Ki values against A/Tokyo and B/Memphis neuraminidases, respectively. Ki values for enantiomeric mixtures were not corrected for enantiomeric purity.
X-ray crystallographic studies.
Isolation, purification, and crystallization of type A N9/tern/Australia/G70c/75 neuraminidase were performed as reported by Laver et al. (14). Crystals were soaked in a solution containing 0.93 M KH2PO4, 1.0 M K2HPO4, and 3% dimethyl sulfoxide at pH 6.7 containing millimolar concentrations of test compound. Crystals containing compound 2 were serially transferred into buffer plus compound 2 containing 0, 10, 20, and 27% glycerol for 15 min at each step. The crystals were then frozen in a stream of −140°C nitrogen. Data were collected using a MAR image plate system on a Rigaku RU-2000 rotating anode source operating at 100 mA and 50 kV. Data were processed to 1.9 Å using DENZO (18) and refined in XPLOR (3). Data from crystals soaked with compound 3 were collected on a Rigaku RTP 300 RC rotating anode source operating at 100 mA and 50 kV at 160 K using an Oxford cryosystem. The data were collected using a RAXISII detector with a MAR image plate and were reduced using the HKL package (19). The model 2BAT from the Brookhaven protein data bank was used for initial phasing, and the structures were refined with XPLOR using a combination of simulated annealing maximum likelihood refinement and individual B-factor refinement. Electron density maps were inspected on a Silicon Graphics INDIGO2 workstation using the program package QUANTA 97 from Molecular Simulations.
Anti-influenza activity in cell culture.
The anti-influenza activity assays were conducted by Southern Research Institute, Birmingham, Ala. The assay procedure tests six concentrations of each compound in triplicate against influenza virus using Madin-Darby canine kidney (MDCK) cells grown in monolayers in a 96-well microtiter plate format. The wells were initially seeded with 4 × 105 cells/well, and then the cells were grown for 3 days in order to reach confluent state. The antiviral efficacy for test compounds was assessed from wells that contained MDCK cells, 0.1 ml of the test compound solution, and 0.1 ml of an influenza virus stock solution. The titer of the influenza virus inoculum was 126 cell culture 50% infectious doses (CCID50) per ml for the A/Victoria/3/75 (H3N2) virus and 1,000 CCID50/ml for the B/Hong Kong/5/72 influenza virus. In addition, each plate contained cell controls that contained medium only, virus-infected cell controls that contained medium and virus, compound cytotoxicity controls that contained medium and each compound concentration, reagent controls that contained compound and medium but no cells, and compound colorimetric controls that contained compound and medium but no cells. The plates were incubated at 37°C in a humidified atmosphere containing 5% carbon dioxide for 3 days until maximum cytopathogenic effects were observed in the untreated virus control cultures. The cytopathogenic effects were quantitated using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) uptake procedure (17). Briefly, 20 μl of a 5-mg/ml solution of MTT in Eagle's minimal essential medium and 5% fetal bovine serum was added to each of the plate wells. The plates were then incubated at 37°C for 6 h, and then 40 μl of a solution containing 30% sodium dodecyl sulfate in 0.02 N HCl was added to each well. The plates were incubated at 37°C overnight, and then the absorbance from each well was measured spectrophotometrically at 570 nm. Absorbance values from compound-treated wells were compared to those obtained from virus and cell controls to establish the percent reduction in cytopathogenic effects due to the compound treatment. The 50% effective concentration, which represents the compound dose that reduced viral cytopathogenic effects by 50%, was calculated by using a regression analysis program for semilog curve fitting.
RESULTS
Discovery of lead compounds 2 and 3.
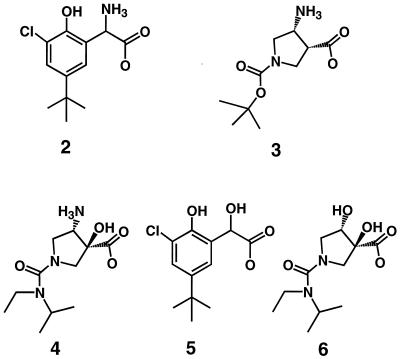
We tested approximately 300 α- and β-amino acids as potential inhibitors of the neuraminidase catalytic domain from A/N2/Tokyo3/67 influenza virus. We chose to test this group of compounds because we felt that the positively charged amino group could mimic the partial positive charge which we hypothesize occurs on the glycosidic oxygen in the transition state during hydrolysis of N-acetylneuraminic acid substrates by neuraminidase. Although several amino acid inhibitors were identified using this approach, the two most potent compounds were a phenylglycine (compound 2) and a pyrrolidine (compound 3) (Fig. 2).
FIG. 2.
Novel α- and β-amino acid inhibitors of influenza virus neuraminidases.
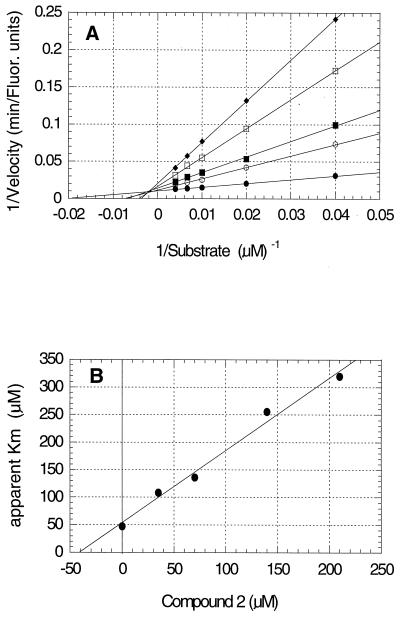
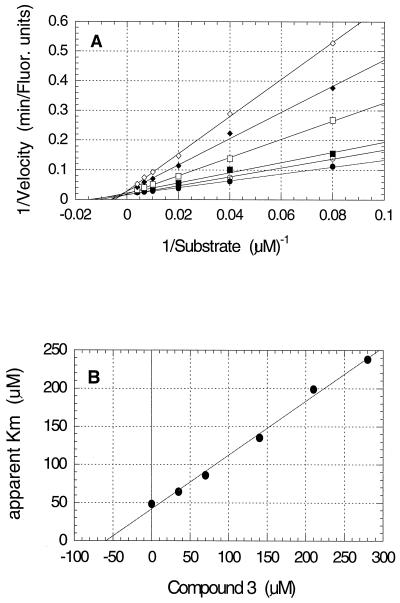
The results from studies to establish the kinetic mechanism of inhibition for compounds 2 and 3 are shown in Fig. 3A and B and 4A and B, respectively. Both compounds appear to be competitive inhibitors relative to the substrate, with Ki values of 41 μM for compound 2 and 59 μM for compound 3. The Ki′ values, derived from replots of 1/Vmax versus inhibitor concentration, were at least 10-fold larger than the Ki values (data not shown) which identified these compounds as competitive inhibitors. No evidence for time-dependent inhibition, sometimes called slow binding inhibition (16), was observed.
FIG. 3.
Kinetics of inhibition of A/Tokyo/3/67 influenza virus neuraminidase by compound 2. (A) Double reciprocal plot showing competitive inhibition of the reaction by compound 2 final concentrations of 0 (●), 35 (○), 70 (▪), 140 (□), and 210 (⧫) μM. Fluor., fluorescence. (B) Replot of the slopes from panel A against inhibitor concentration to determine the Ki value of 41 μM from the abscissa intercept.
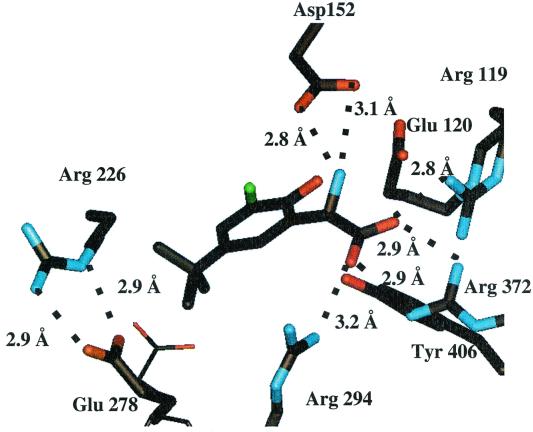
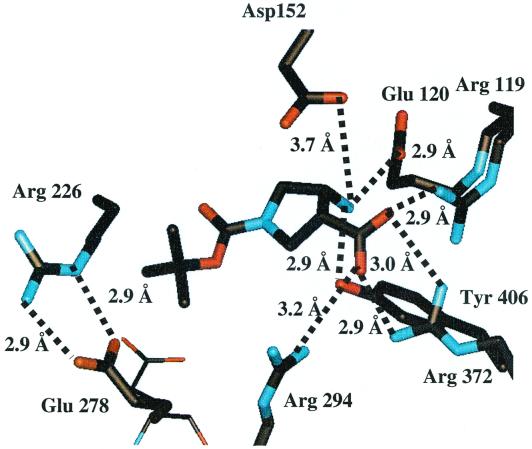
The X-ray crystal structures were solved for compounds 2 and 3 bound to the neuraminidase catalytic domain from A/N9 influenza virus. As shown in Fig. 5, the carboxylate of inhibitor 2 was well stabilized through the formation of two hydrogen bonds with the Arg 372 residue and single hydrogen bonds with residues Arg 294 and Arg 119. The α-amino group of inhibitor 2 forms two hydrogen bonds with Asp 152, which is the residue believed to interact with the glycosidic oxygen in the transition state. The hydroxyl and chloro groups on the phenyl ring of inhibitor 2 make no apparent interactions with the protein. Two of the methyl carbons present on the t-butyl group of inhibitor 2 appear to make van der Waals contacts with the beta and gamma methylene carbons of the Glu 278 side chain. Significantly, the Glu 278 residue adopts a new conformation in order to accommodate the t-butyl group of inhibitor 2. This induced conformation permits the carboxylate oxygens of the Glu 278 side chain to form two new hydrogen bonds to the side chain of Arg 226.
FIG. 5.
X-ray crystal structure of compound 2 bound to A/N9 neuraminidase. The conformation of the Glu 278 side chain of the native enzyme is shown in thin lines.
The structure of inhibitor 3 bound to N9 neuraminidase is shown in Fig. 6. The carboxylate oxygens of inhibitor 3 form hydrogen bonds with arginine residues 294, 372, and 119 in a fashion which is similar to the interactions of inhibitor 2 with the enzyme. The β-amino group is also well satisfied with hydrogen bonds to Glu 120 and Tyr 406. The carboxylate side chain of Asp 152 is 3.7 Å from the β-amino group and also is at a poor angle for a productive hydrogen bond. However, we cannot rule out a small contribution to binding affinity from an electrostatic interaction between these two oppositely charged groups. Once again, the side chain of Glu 278 appears in its induced conformation, exposing the methylene carbons to form hydrophobic interactions with the t-butyl side chain of inhibitor 3.
FIG. 6.
X-ray crystal structure of compound 3 bound to A/N9 neuraminidase. The conformation of the Glu 278 side chain of the native enzyme is shown in thin lines.
In vitro evaluation of compound 4.
The crystal structure of pyrrolidine compound 3 suggested that a compound containing a hydroxyl group in an α position in relation to the carboxylate might form good hydrogen bonds with the Asp 152 side chain. Compound 4 differs from compound 3 in that compound 4 contains the desired hydroxyl group as well as an N-ethyl-N-isopropyl urea linkage to the pyrrolidine nitrogen. Compound 4 exhibited a Ki value of 0.36 μM against A/Tokyo neuraminidase, which is a 160-fold improvement in binding affinity relative to that of compound 3. The crystal structure of compound 4 bound to N9 neuraminidase (not shown) is essentially identical to that shown in Fig. 6 for compound 3, except that the α-hydroxyl group of compound 4 is centrally positioned between the two oxygen atoms of the Asp 152 carboxyl group, with distances of 3.2 and 3.4 Å. Thus, compound 4 was found to bind to the enzyme in a manner which was predicted by computer modeling. The results for the in vitro cell culture evaluation of compound 4 are shown in Table 1. Compound 4 shows a clear antiviral effect against the A/Victoria/3/75 influenza virus and appears to be about 24-fold more active than the reference neuraminidase inhibitor 1. However, compound 4 is only about twofold more active than inhibitor 1 against the B/Hong Kong/5/72 virus. It should be noted that the cytopathogenicity assay used here provides no information about the ability of the compounds in Table 1 to reduce plaque size or plaque number. Compound 4 shows no detectable cytotoxicity at concentrations of as high as 1 mM.
TABLE 1.
Inhibition of influenza virus replication by test compoundsa
| Compound | A/Victoria/3/75 (H3N2)
|
B/Hong Kong/5/72
|
||
|---|---|---|---|---|
| EC50 | TC25 | EC50 | TC25 | |
| 4 | 2.0 ± 1.8 | >1,000 | 305 ± 15 | >1,000 |
| 1 | 48 ± 26 | >3,200 | 650 ± 320 | >3,200 |
| Ribavirin | 5.9 ± 2.7 | >130 | 6.3 ± 3.6 | >130 |
EC50, the concentration (μM) of compound that reduced virus-induced cytopathogenic effects by 50% in MDCK cells. Values are means ± standard deviations of triplicate measurements. TC25, the concentration (μM) of compound that resulted in the death of 25% of the MDCK cells.
Inhibitory activity of compounds 2 to 4 against B-strain neuraminidase.
Compounds 2 to 4 were tested for their inhibitory activity against the neuraminidase catalytic domain from B/Memphis/3/89 influenza virus, and the results are shown in Table 2. Compounds 2 to 4 were found to be 17- to 580-fold weaker inhibitors of the B-strain neuraminidase than of the A/N2 strain enzyme.
TABLE 2.
Inhibition of influenza virus neuraminidases
| Compound |
Ki (μM) for:
|
|
|---|---|---|
| A/Tokyo/3/67 | B/Memphis/3/89 | |
| 1 | 1.5 | 1.4 |
| 2 | 41 | 690 |
| 3 | 59 | 2,500 |
| 4 | 0.36 | 210 |
| 5 | 15,000 (estimated) | Not tested |
| 6 | 940 | Not tested |
Replacement of the amino groups of compounds 2 and 4 with a hydroxyl group.
Compound 5 has a chemical structure similar to that of compound 2, except that compound 5 contains a hydroxyl group instead of an amino group at the α position in relation to the carboxylate. Compound 5 was synthesized in order to understand the importance of the amino group for the binding of compound 2. Compound 5 showed 10% inhibition at 2.5 mM, which extrapolates to an estimated Ki value of 15 mM, or a nearly 365-fold weaker binding affinity than that of compound 2 against the A/N2/Tokyo neuraminidase. Likewise, compound 6 is similar to compound 4, except that the β-amino group of compound 4 has been replaced with a hydroxyl group in compound 6. A comparison of the binding affinities of compounds 4 and 6 in Table 2 shows that the binding affinity of compound 6 is 2,600-fold weaker than that of compound 4.
DISCUSSION
The hypothetical reaction scheme shown in Fig. 1 was adapted from the X-ray structural analysis of neuraminidase bound to sialic acid (26). The Asp 152 side chain has been suggested to act as a general acid during catalysis by either protonating the glycosidic oxygen directly or through an intervening water molecule (5). In either case, the glycosidic oxygen would likely develop a partial positive charge in the transition state. In principle, this partial positive charge could contribute several kilocalories/mole in selective binding affinity of the transition state relative to the substrate and planar intermediate. For example, biological specificity studies have shown that hydrogen bonds in which one partner is charged have bond strengths of 3.5 to 4.5 kcal/mol, whereas hydrogen bonds involving uncharged partners have bond strengths only in the 0.5 to 1.5 kcal/mol range (6). It seemed likely that compounds containing a positively charged amino group rather than a hydroxyl group might be stable analogs of this putative transition state structure for the neuraminidase reaction. Screening of a limited number of α- and β-amino acids led to the discovery of the phenylglycine compound 2 and pyrrolidine compound 3 as neuraminidase inhibitors. These two compounds represent the first influenza virus neuraminidase inhibitors to contain an amino group functionality in an α or β position in relation to the carboxylic acid group. For reference, the potent neuraminidase inhibitors GG167 (27), GS4071 (12), and BCX-1812 (2) contain their basic functional groups in a γ position relative to the carboxylic acid group.
X-ray crystallography has shown that the amino group of compound 2 interacts directly with the Asp 152 side chain. This α-amino–Asp 152 side chain interaction is an important contributor to the binding, as shown by the observation that the α-amino compound 2 is a 365-fold more potent inhibitor than the α-hydroxyl-substituted compound 4. This improvement is slightly better than the 100-fold difference reported for compound 1 versus its 4-amino counterpart (27). We note that the hydrogen bond lengths between the α-amino group of compound 2 and Asp 152 are 2.8 and 3.1 Å, whereas hydrogen bond lengths from Asp 152 and Glu 120 to the 4-position of compound 1 are 3.1 and 3.4 Å, respectively (26). The closer hydrogen bond distances to the α-amino group of compound 2 are a possible explanation for the larger amino group effect observed for compound 2 relative to the 4-amino counterpart of compound 1.
Pyrrolidine compound 3 is the first neuraminidase inhibitor reported to date to make a hydrogen bond to the hydroxyl group of the strictly conserved Tyr 406 residue. The Tyr 406 hydroxyl group has been proposed to ionize during enzymatic turnover to stabilize the positively charged oxocarbonium ion intermediate compound 1 (4). Site-directed mutagenesis studies provide further support for this critical role, since the Tyr406Phe mutant is devoid of enzymatic activity (8). The 2.9-Å distance between the Tyr 406 hydroxyl group and the β-amino group of compound 3 or 4 is identical to that observed between the Tyr 406 hydroxyl group and the α-carbon of compound 1 and, by extension, the presumed oxocarbonium ion intermediate shown in Fig. 1. Our studies suggest that the electrostatic interaction between Tyr 406 and the oxocarbonium ion intermediate may be significant because replacement of the positively charged amino group of compound 4 with a neutral hydroxyl group of compound 5 leads to a 2,600-fold loss of binding affinity. The interpretation of these results is complicated somewhat by the fact that the amino group of compound 4 also makes an interaction with Glu 120. Thus, the 2,600-fold replacement effect results from altered interactions with both active-site amino acids. Nevertheless, the replacement effect is quite large and underscores the importance of charged interactions between the Tyr 406 and Glu 120 side chains and bound ligands.
It has been suggested that the time-dependent inhibition of influenza virus neuraminidase by GS4071 results from the side chain of Glu 278 undergoing a slow conformational change upon binding of this inhibitor (25). However, compounds 2 to 4 also induce the Glu 278 side chain conformational change but do not show time-dependent inhibition. These results indicate that the Glu 278 conformational change does not necessarily lead to time-dependent inhibition. We have previously suggested that the time-dependent inhibition phenomenon associated with GS4071 and GG167 (9) results from the low rate of dissociation (koff values in the 10−4 s−1 range) of these very potent compounds from the enzyme (11). The koff values for compounds 2 to 4 are too fast to be measured using our routine assays, but we estimate that the values must be larger than 0.005 s−1. Thus, the koff values for compounds 2 to 4 are at least 20-fold faster than those measured for GS4071 and GG167, and so one might not expect compounds 2 to 4 to exhibit slow binding kinetics if this phenomenon resulted from low off rates.
Compounds 2 to 4 were 17- to 580-fold weaker inhibitors of B-strain neuraminidase than of A-strain enzyme. These results are consistent with previous reports that compounds which induce the Glu 278 side chain conformational change in A-strain enzymes are often 10- to 1,000-fold weaker inhibitors of B-strain neuraminidases (13, 22, 23). Although the active-site residues are strictly conserved between A and B strains of neuraminidase, the positions of the α-carbons and side chains do not overlie exactly in three dimensions. Whereas the Glu 278 conformational change can occur quite easily in the A-strain enzyme in order to accommodate a ligand, a similar conformational change in the B-strain Glu 278 results in unfavorable steric interactions and a distortion of the protein backbone (22). The net result is that the Glu 278 conformational change is less energetically favorable in B-strain neuraminidase, accounting for the binding affinity differences between A- and B-strain enzymes. Attempts to further optimize the binding affinity of compounds 2 to 4 against B-strain neuraminidases will be challenged by the reluctance of B-strain enzymes to undergo the induced Glu 278 conformational change. However, both the phenylglycine and pyrrolidine series offer the potential for significant improvements in potency, since neither series contains an N-acetyl side chain in their chemical structures. The N-acetyl side chain appears to be critical for potent neuraminidase inhibition as shown by its presence in the chemical structures of the three most potent neuraminidase inhibitors, GG167 (27), GS4071 (12), and BCX-1812 (2). Furthermore, structure-activity relationship studies have shown that the N-acetamido side chain contributes a surprising 5 orders of magnitude to the binding affinity of GG167 (24). Likewise, optimization of the hydrophobic substituent on the GS4071 core improved inhibitory activity by 6,300-fold (12). Thus, the addition of an N-acetamido side chain to the phenylglycine and pyrrolidine cores and/or further optimization of the hydrophobic substituents of compounds 2 and 3 could, in principle, lead to the discovery of novel, potent neuraminidase inhibitors.
In conclusion, we have used enzyme mechanistic information to form a hypothesis about specific interactions which might be important for the tight binding of ligands to the active site of influenza neuraminidase. We used that hypothesis to test a limited number of compounds with the appropriate chemical substructure and identified two classes of compounds with modest inhibitory potency. Using analogs, we demonstrated that interactions with Asp 152 and Tyr 406 and Glu 120 are important contributors to the binding affinities of these compounds. The Asp 152 and Tyr 406 residues are particularly attractive for targeted inhibitor design, because they are strictly conserved and lab-generated mutant enzymes at these positions exhibit poor enzymatic activity (8). Thus, drug resistance may be less likely to develop for compounds which interact with these residues. The phenylglycine and pyrrolidine core structures will serve as the foundation of our program to discover effective anti-influenza drugs.
FIG. 4.
Kinetics of inhibition of A/Tokyo/3/67 influenza virus neuraminidase by compound 3. (A) Double reciprocal plot showing competitive inhibition of the reaction by compound 3 final concentrations of 0 (●), 35 (○), 70 (▪), 140 (□), 210 (⧫), and 280 (◊) μM. (B) Replot of the slopes from panel A against inhibitor concentration to determine the Ki value of 59 μM from the abscissa intercept.
ACKNOWLEDGMENTS
We gratefully acknowledge Louise Westbrook and Lois Allen of Southern Research Institute for their anti-influenza cell culture test results.
REFERENCES
- 1.Ashwell M, Guo X, Sinnott M I. Pathways for the hydrolysis of glycosides of N-acetylneuraminic acid. J Am Chem Soc. 1992;114:10158–10166. [Google Scholar]
- 2.Babu Y S, Chand P, Bantia S, Kotian P, Dehghani A, El-Kattan Y, Lin T-H, Hutchison T L, Elliott A J, Parker C D, Ananth S L, Horn L L, Laver G W, Montgomery J A. BCX-1812 (RWJ-270201): discovery of a novel, highly potent, orally active, and selective influenza neuraminidase inhibitor through structure-based drug design. J Med Chem. 2000;43:3482–3486. doi: 10.1021/jm0002679. [DOI] [PubMed] [Google Scholar]
- 3.Brunger A T. X-PLOR Version 3.1. A system for X-ray crystallography and NMR. New Haven, Conn: Yale University Press; 1992. [Google Scholar]
- 4.Burmeister W P, Henrissat B, Bosso C, Cusack S, Ruigrok R W H. Influenza B virus neuraminidase can synthesize its own inhibitor. Structure. 1993;1:19–26. doi: 10.1016/0969-2126(93)90005-2. [DOI] [PubMed] [Google Scholar]
- 5.Chong A K J, Pegg M S, Taylor N R, von Itzstein M. Evidence for a sialosyl cation transition state complex in the reaction of sialidase from influenza virus. Eur J Biochem. 1992;207:335–343. doi: 10.1111/j.1432-1033.1992.tb17055.x. [DOI] [PubMed] [Google Scholar]
- 6.Fersht A R, Shi J-P, Knill-Jones J, Lowe D M, Wilkinson A J, Blow D M, Brick P, Carter P, Waye M M Y, Winter G. Hydrogen bonding and biological specificity analyzed by protein engineering. Nature. 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- 7.Frick L, MacNeela J P, Wolfenden R. Transition state stabilization by deaminases: rates of nonenzymatic hydrolysis of adenosine and cytidine. Bioorg Chem. 1987;15:100–108. [Google Scholar]
- 8.Ghate A A, Air G M. Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design. Eur J Biochem. 1998;258:320–331. doi: 10.1046/j.1432-1327.1998.2580320.x. [DOI] [PubMed] [Google Scholar]
- 9.Hart G J, Bethell R C. 2,3-Didehydro-2,4-dideoxy-4-guanidino-N-acetyl-D-neuraminic acid is a slow-binding inhibitor of sialidase from both influenza A virus and influenza B virus. Mol Biol Int. 1995;36:695–703. [PubMed] [Google Scholar]
- 10.Janakiraman M N, White C L, Laver W G, Air G M, Luo M. Structure of influenza virus neuraminidase B/Lee/40 complexed with sialic acid and a dehydro analog at 1.8-Ang. resolution: implications for the catalytic mechanism. Biochemistry. 1994;33:8172–8179. doi: 10.1021/bi00193a002. [DOI] [PubMed] [Google Scholar]
- 11.Kati W M, Saldivar A S, Mohamadi F, Sham H L, Laver W G, Kohlbrenner W E. GS4071 is a slow-binding inhibitor of influenza neuraminidase from both A and B strains. Biochem Biophys Res Commun. 1998;244:408–413. doi: 10.1006/bbrc.1998.8282. [DOI] [PubMed] [Google Scholar]
- 12.Kim C U, Lew W, Williams M A, Liu H, Zhang L, Swaminathan S, Bischofberger N, Chen M S, Mendel D B, Tai C Y, Laver W G, Stevens R C. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site. J Am Chem Soc. 1997;119:681–690. doi: 10.1021/ja963036t. [DOI] [PubMed] [Google Scholar]
- 13.Kim C U, Lew W, Williams M A, Wu H, Zhang L, Chen X, Escarpe P A, Mendel D B, Laver W G, Stevens R C. Structure-activity relationship studies of novel carbocyclic influenza neuraminidase inhibitors. J Med Chem. 1998;41:2451–2460. doi: 10.1021/jm980162u. [DOI] [PubMed] [Google Scholar]
- 14.Laver W G, Colman P M, Webster R G, Hinshaw V S, Air G M. Influenza virus neuraminidase with haemagglutinin activity. Virology. 1984;137:314–323. doi: 10.1016/0042-6822(84)90223-x. [DOI] [PubMed] [Google Scholar]
- 15.Meindl P, Bodo G, Palese P, Schulman J, Tuppy H. Inhibition of neuraminidase activity by derivatives of 2-deoxy-2,3-dehydro-N-acetylneuraminic acids. Virology. 1974;58:457–463. doi: 10.1016/0042-6822(74)90080-4. [DOI] [PubMed] [Google Scholar]
- 16.Morrison J F, Walsh C T. The behavior and significance of slow-binding enzyme inhibitors. Adv Enzymol Rel Areas Mol Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 17.Mossman T. Rapid colorimetric assay for cellular growth and survival: application to proliferative and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski Z. Oscillation data reduction program. In: Sawyer L, Isaacs N, Bailey S, editors. Proceedings of the CCCP study weekend: data collection and processing. Daresbury, England: SERC Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Palese P, Tobita K, Ueda M, Compans R W. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology. 1974;61:397–410. doi: 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- 21.Segel I H. Enzyme kinetics—behavior and analysis of rapid equilibrium and steady-state enzyme systems. New York, N.Y: John Wiley and Sons; 1975. pp. 100–107. [Google Scholar]
- 22.Smith P W, Sollis S L, Howes P D, Cherry P C, Cobley K N, Taylor H, Whittington A R, Scicinski J, Bethell R C, Taylor N, Skarzynski A C, Singh O, Wonacott A, Varghese J, Colman P. Novel inhibitors of influenza sialidase related to GG167: structure-activity, crystallographic and molecular dynamics studies with 4H-pyran-2-carboxylic acid 6-carboxamides. Bioorg Med Chem Lett. 1996;6:2931–2936. [Google Scholar]
- 23.Sollis S L, Smith P W, Howes P D, Cherry P C, Bethell R C. Novel inhibitors of influenza sialidase related to GG167: synthesis of 4-amino and guanidine-4H-pyran-2-carboxylic acid-6-propylamides; selective inhibitors of influenza A virus sialidase. Bioorg Med Chem Lett. 1996;6:1805–1808. [Google Scholar]
- 24.Starkey I D, Mahmoudian M, Noble D, Smith P W, Cherry P C, Howes P D, Sollis S L. Synthesis and influenza virus sialidase inhibitory activity of the 5-desacetamido analogue of 2,3-didehydro-2,4-dideoxy-4-guanidinyl-N-acetylneuraminic acid (GG167) Tetrahedron Lett. 1995;36:299–302. [Google Scholar]
- 25.Tai C Y, Escarpe P A, Sidwell R W, Williams M A, Lew W, Wu H, Kim C U, Mendel D B. Characterization of human influenza virus variants selected in vitro in the presence of the neuraminidase inhibitor GS4071. Antimicrob Agents Chemother. 1998;42:3234–3241. doi: 10.1128/aac.42.12.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varghese J N, McKimm-Breschkin J L, Caldwell J B, Kortt A A, Colman P. The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor. Proteins. 1992;14:327–332. doi: 10.1002/prot.340140302. [DOI] [PubMed] [Google Scholar]
- 27.von Itzstein M, Wu W-Y, Kok G B, Pegg M S, Dyason J C, Jin B, Phan T V, Smythe M L, White H F, Oliver S W, Colman P M, Varghese J N, Ryan D M, Woods J M, Bethell R C, Hotham V J, Cameron J M, Penn C R. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 28.Wolfenden R. Transition state analogues for enzyme catalysis. Nature. 1969;223:704–705. doi: 10.1038/223704a0. [DOI] [PubMed] [Google Scholar]
- 29.Wolfenden R. Transition state analog inhibitors and enzyme catalysis. Annu Rev Biophys Bioeng. 1976;5:271–306. doi: 10.1146/annurev.bb.05.060176.001415. [DOI] [PubMed] [Google Scholar]
- 30.Wolfenden R. Conformational aspects of inhibitor design: enzyme-substrate interactions in the transition state. Bioorg Med Chem. 1999;7:647–652. doi: 10.1016/s0968-0896(98)00247-8. [DOI] [PubMed] [Google Scholar]
- 31.Wolfenden R, Frick L. Transition state affinity and the design of enzyme inhibitors. In: Page M I, Williams A, editors. Enzyme mechanisms. London, United Kingdom: Royal Society of Chemistry; 1986. pp. 97–122. [Google Scholar]
- 32.Wolfenden R, Lu X, Young G. Spontaneous hydrolysis of glycosides. J Am Chem Soc. 1998;120:6814–6815. [Google Scholar]