Abstract
The development of nanomedicine has recently achieved several breakthroughs in the field of cancer treatment; however, biocompatibility and targeted penetration of these nanomaterials remain as limitations, which lead to serious side effects and significantly narrow the scope of their application. The self-assembly of intermediate filaments with arginine–glycine–aspartate (RGD) peptide (RGD-IFP) was triggered by the hydrophobic cationic molecule 7-amino actinomycin D (7-AAD) to synthesize a bifunctional nanoparticle that could serve as a fluorescent imaging probe to visualize tumor treatment. The designed RGD-IFP peptide possessed the ability to encapsulate 7-AAD molecules through the formation of hydrogen bonds and hydrophobic interactions by a one-step method. This fluorescent nanoprobe with RGD peptide could be targeted for delivery into tumor cells and released in acidic environments such as endosomes/lysosomes, ultimately inducing cytotoxicity by arresting tumor cell cycling with inserted DNA. It is noteworthy that the RGD-IFP/7-AAD nanoprobe tail-vein injection approach demonstrated not only high tumor-targeted imaging potential, but also potent antitumor therapeutic effects in vivo. The proposed strategy may be used in peptide-driven bifunctional nanoparticles for precise imaging and cancer therapy.
KEY WORDS: Nanoprobe, 7-Amino actinomycin D, Intermediate filament protein, Tumor image, Antitumor therapy, Integrin αvβ3
Graphical abstract
Super-sensitive bifunctional nanoprobe that showed strong tumor-targeted imaging and antitumor therapeutic effects has been synthesized by one-step. This nanoparticle penetrated tumor cells to induce cell cycle arresting for cytotoxicity.
1. Introduction
Cancer is a disease characterized by abnormal cell proliferation. As a leading cause of human death, it has attracted significant attention from clinical researchers worldwide1,2. Furthermore, precise imaging and diagnosis of cancer are essential for effective therapeutic intervention. Currently, drug loaded nanoprobes targeted to exact sites have been extensively studied to develop more effective approaches to tumor diagnosis and treatment3,4. A plausible strategy may include the direct delivery of chemotherapeutic drugs to tumors by fluorescent nanoprobes with the subsequent enrichment of tumor tissues through prolonged drug half-life or increased solubility of hydrophobic drugs5,6. Conventional nanocarriers tend to burst or cause premature and uncontrollable drug release, and the majority have low drug loading and exhibit non-specific drug leakage, while their limited biocompatibility may stimulate immunogenic responses in humans7, 8, 9. Therefore, an ideal nanoprobe composed of biocompatible vectors should not merely have the capacity to accurately detect tumors, but also be able to improve the specificity of its antitumor action through structural modifications.
Actinomycin D (ACTD) is a cytotoxic chemotherapeutic reagent whose 2-cyclic peptide structure facilitates its embedment into the minor groove of DNA, suppresses the function of RNA polymerase, and inhibits mRNA synthesis10,11. 7-amino actinomycin D (7-AAD) acts as a homologous analogue of ACTD with a 2-cyclic peptide structure in which the seventh hydrogen atom in the connection chain is replaced with an amidogen so as to emit red fluorescence through an intramolecular charge transfer effects12. The maximum excitation/emission wavelength of 7-AAD is 546/647 nm, and its Stokes shift is approximately 100 nm, which is much higher than those of commercially available organic dyes. These parameters can help effectively decrease dye self-quenching, while reducing non-specific background fluorescence, making 7-AAD potentially suitable for tumor diagnosis and treatment13,14. However, both ACTD and 7-AAD cannot enter living cells due to their inability to penetrate cell membranes and, therefore, have a narrow antitumor spectrum.
Non-immunogenic peptides self-assembled from natural amino acids with good biocompatibility and their further modification may solve the problem of limited tissue penetration15,16. Human vimentin intermediate filaments (IFs) are the basic component of the cellular cytoskeleton, which realize cross-linking with visible components such as microfilaments and microtubules in the form of homologous dimers17. Hence, an extremely conserved α-helical motif containing 36 amino acids (EKVELQELNDRFANYIDKVRFLEQQNKILLAELEQL) of IFs was selected for the self-assembled peptides employed in this study due to its ability to form a polymeric shape via their parallel coiled-coil “rod” domains. This structure may induce self-assembly of nanoparticles (NPs) through the formation of hydrophobic, ionic, or hydrogen bonds within and between different chains under certain conditions18,19. αvβ3 is a member of the integrin family of transmembrane glycoprotein molecules and is mainly overexpressed in activated tumor angiogenic endothelial cells and certain tumor cells, while it has little or no expression in normal or mature endothelial cells20,21. In addition, αvβ3 is an important target for tumor diagnosis and therapy, and usually participates in regulating the proliferation and metastasis of tumor cells. RGD (arginine-glycine-aspartic acid)-containing polypeptides can recognize αvβ3 integrin and bind it with highly affinity and specificity. Based on these properties, a large number of studies have employed RGD sequences in various methods to enhance oncotherapeutic targeting strategies for imaging tracers or treatment22.
Herein, owing to knowledge regarding the intriguing properties of polypeptide self-assembly and 7-AAD, a sensitive bifunctional nanoprobe with peptide-driven self-assembling NPs for tumor site imaging and therapeutic applications has been designed and synthesized (Fig. 1). The nanostructure, optical properties, in vitro and in vivo efficacy, and biosafety properties of self-assembled RGD-IFP/7-AAD nanoprobes were determined using various techniques. First, the RGD-IFP peptide self-assembled with 7-AAD into a stable polymeric nano-spherical structure by electrostatic interactions and π–π stacking between the cationic drug and peptide molecules under natural conditions. 7-AAD molecules become proximal to each other through the help of the RGD-IFP peptide and generate resonant energy transfer by mutual absorption of light, which leads to fluorescence weakening. The resulting NPs were able to penetrate the cell membrane and thus to enter living cells. Second, RGD peptide moieties that could bind to overexpressed αvβ3 integrin subunits were added to the N-terminal of the IFP peptide to promote the localization of tumor cells and enhance tumor ablation in vivo. Third, after intracellular uptake, 7-AAD was released by lysosomes into the acidic environment thereby increasing antitumor efficacy and fluorescence intensity. These processes could reduce both the systemic toxicity of 7-AAD and the fluorescent background noise of normal tissues or blood circulation. The obtained results indicate that the proposed nanoprobe considerably broadens the applied spectrum of fluorescence imaging and may be used to develop new nanomedicines for precision therapy.
Figure 1.
Schematic illustration of the synthesis of RGD-IFP/7-AAD nanoprobes with antitumor properties and their antitumor mechanism. (A) Self-assembly of RGD-IFP/7-AAD nanoprobes through the cooperative coordination of small peptides and 7-AAD by one-step method. (B) After the tail intravenous injection, RGD-IFP/7-AAD nanoprobes can be used in tumor-targeted imaging and chemotherapy due to the enhancement of their cell penetration ability and accumulation via circulation. (C) RGD-IFP/7-AAD nanoprobes target tumor cells through αvβ3 integrin subunits and become internalized by endocytosis. The acidic pH values of endosomes/lysosomes stimulate the rapid drug release at the tumor sites and killing tumor cells by blocking the cell cycle.
2. Materials and methods
2.1. Materials and reagents
All chemical reagents were purchased from Sigma–Aldrich (St. Louis, MO, USA) unless otherwise mentioned. RGD-IFP and IFP peptides were synthesized by a solid-phase method with 95 % purity (China Peptides Co., Ltd., Shanghai, China). 7-Aminoactinomycin D was purchased from J&K Scientific Ltd. (Beijing, China). Trypsin, Dulbecco's Modifed Eagle's Medium (DMEM) and fetal bovine serum (FBS) were obtained from GIBCO, Invitrogen Corp. (Carlsbad, USA). Lyso-Tracker Green fluorescence probe was purchased from Invitrogen (Carlsbad, CA, USA). DAPI and EdU Cell Proliferation Kit with Alexa Fluor 488 were purchased from Beyotime Biotechnology (Shanghai, China). CCK-8 cell proliferation kit was purchased from Dojindo Molecular Technologies (USA). All buffer solutions were prepared in ultrapure water (Millipore, Billerica, MA, USA).
2.2. DNA hypochromic effect
Calf thymus DNA solution was prepared at concentration gradients of 0, 0.1, and 0.5 mg/mL, and the 7-AAD drug solution was dissolved in 45 mmol/L Tris buffers at a final concentration of 2 μmol/L for the drug buffer. After that, 0.16 mL of the DNA solution was thoroughly mixed with 1.84 mL of the drug buffer adequately and incubated for 10 min. Ultraviolet (UV)– Visible spectra were recorded in the wavelength range of 350–600 nm.
2.3. Conjugated reaction of 7-AAD with DNA
Calf thymus DNA was purchased from Sigma–Aldrich. Its aqueous solution was mixed with 7-AAD aqueous solution at an equal ratio followed by incubation for 10 min at room temperature. The equal amounts of 7-AAD, calf thymus DNA, and the mixture of 7-AAD with calf thymus DNA were mixed with the loading buffer, and the resulting samples were loaded with 1 % agarose gel. Agarose gel electrophoresis was performed at a voltage of 100 V for 45 min, and images were taken under UV irradiation.
2.4. Design and preparation of RGD-IFP peptide
In order to ensure the successful NP self-assembly, tumor cell targeting, and controlled drug release in vivo, the amino acid sequences of designed peptides must satisfy certain requirements. In this study, RGD-IFP peptides (RGDSEEKVELQELNDRFANYIDKVRFLEQQNKILLAELEQL) that could self-assemble into a stable homodimer of the human proteome were designed. Molecular weights of RGD-IFP peptides were measured by electrospray mass spectrometry (Perkin Elmer SCIEX) and 1 mg/mL peptides were added to an oxidation buffer (100 mmol/L ammonium acetate, pH = 8.5), stirred at room temperature for 3 days, purified by reversed-phase high-performance liquid chromatography, and stored at −20 °C. Their tertiary structure was obtained from the SWISS-MODEL server (https://swissmodel.expasy.org/). Net charge and hydrophilicity analyses were performed by the Novopro online tools (https://www.novopro.cn/tools/).
2.5. Self-assembly and molecular docking of RGD-IFP/7-AAD nanoprobes
Fresh stock solutions of designed RGD-IFP peptides were obtained by dissolving the lyophilized peptides in ultrapure water at a concentration of 8 mg/mL. To avoid pre-aggregation, fresh stock solutions were prepared for each experiment. RGD-IFP/7-AAD NPs were obtained through the reaction of the RGD-IFP solutions with 7-AAD mixed at a concentration ratio of 4:1 followed by shaking for 1 min. To ensure their sufficient interaction, the resulting solutions were placed into an ultrasonication bath and microwaved at a power of 40 W and temperature of 37 °C for 1 h. After that, the solutions were centrifuged at 3,000 g for 5 min, transferred through a dialysis film, and soaked in ultrapure water for 24 h to purify the prepared RGD-IFP/7-AAD NPs.
Molecular docking analysis of the RGD-IFP/7-AAD coordination environment was performed by the Molecular Operating Environment (MOE) software containing the Schrödinger Glide package with the default parameters. The RGD-IFP crystal structure and its three-dimensional (3D) model were downloaded from the website https://www.swissmodel.expasy.org/interactive (3ssu).
2.6. Characterization of RGD-IFP/7-AAD NPs
Particle size distribution and zeta potential of prepared RGD-IFP/7-AAD NPs were measured by dynamic light scattering (DLS; Zetasizer Nano ZS ZEN3600, Malvern Instruments Ltd., Worcestershire, UK). Transmission electron microscopy (TEM) images of the NPs were obtained on air-dried carbon-coated copper grids using a JEM 2100 transmission electron microscope (JEOL, Japan). Fourier-transform infrared (FTIR) spectra of RGD-IFP/7-AAD NPs were recorded by a Nicolet IS 10 FTIR spectrometer (Thermo, USA). The fluorescence resonance energy transfer (FRET) was determined by a FluoroMax-4 spectrofluorometer (HORIBA Jobin Yvon, France) with an excitation wavelength of 546 nm and emission wavelength of 647 nm.
2.7. 7-AAD release profile from nanoprobes were measured by dialysis method
An aqueous solution of the RGD-IFP/7-AAD nanoprobes with an equivalent concentration of 7-AAD was pipetted into a dialysis membrane (Mw cutoff = 5,500 Da) and then introduced into 20 mL of phosphate-buffered saline (PBS) solution with a pH value of 5.5, 6.5, or 7.4. The resulting mixture was shaken gently in a water bath at 37 °C. During this procedure, 1 mL of the medium was removed, and 1 mL of the fresh buffer was added every 30 min. The 7-AAD concentration of the buffer was determined by fluorescence spectroscopy at an excitation wavelength of 546 nm and emission wavelength of 647 nm.
2.8. Live cells for cellular uptake imaging and flow cytometry analysis
All cell lines were purchased from the National Laboratory Cell Resource (China). A549 (human lung cancer cells) and 293T (human embryonic kidney cells) were cultured in DMEM supplemented with 10 % FBS and 1 % penicillin–streptomycin solution at 37 °C under a 5 % CO2 atmosphere according to the standard cell culture protocols.
The uptakes of RGD-IFP/7-AAD nanoprobes in cells were assessed by confocal laser scanning microscopy (CLSM, Olympus FV1000, Japan) and flow cytometry (BD Accuri C6, USA). Cells were seeded at a density of 5 × 104 per well for 24 h. After replacing the old medium with a fresh one containing RGD-IFP/7-AAD, IFP/7-AAD and free 7-AAD (with a concentration of 1 μg/mL 7-AAD), the cells were further incubated for 3 h at 37 °C. After incubation, the cells were washed three times with PBS, fixed with 4 % paraformaldehyde. To determine the intracellular locations of 7-AAD species, the cell nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI). 7-AAD and DAPI were excited at wavelengths of 546 and 405 nm, respectively. For flow cytometry analysis, cells were incubated in a fresh medium containing RGD-IFP/7-AAD, IFP/7-AAD and free 7-AAD with a concentration of 1 μg/mL for 3 h. Subsequently, the cells were harvested and subjected to flow cytometry analysis at an excitation wavelength of 546 nm. The mean fluorescence intensity corresponding to 1 × 104 cells was recorded for each sample.
2.9. Endocytosis and colocalization of cell lysosomes with RGD-IFP/7-AAD nanoprobes in vitro
A549 cells were seeded in the 35 mm glass-based dish overnight, treated with RGD-IFP/7-AAD nanoprobes (the concentration of 7-AAD was 1 μg/mL) in the complete medium at 37 °C for 3 h. After washing three times with PBS, the cell lysosomes were stained with 50 nmol/L Lyso-Tracker Green fluorescence probe for 30 min. Finally, the cells were washed three more times, and their CLSM images were obtained at excitation wavelengths of 488 and 546 nm, respectively (Olympus FV1000, Japan).
2.10. Cytotoxicity assay
Cell viability was measured by Cell Counting Kit-8 (CCK-8) according to the manufacturer's instructions (Dojindo Molecular Technologies). Cells were seeded into 96-well plates at a density of 1 × 103 per well and maintained for 24 h. In the next step, they were incubated with IFP, free 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD for 24 h at different concentrations, respectively. Then, the incubation medium was replaced with a fresh one, and cells continued to be cultured for 4 h before conducting cell viability analysis using CCK-8. Cell viability was evaluated at an absorption wavelength of 450 nm by a microplate absorbance reader (Bio-rad iMark, USA). Each experiment was repeated at least three times.
2.11. EdU cell proliferation assay
Cell proliferation was detected by EdU cell proliferation kit according to the manufacturer's instructions (Beyotime Biotechnology). Cells were seeded at a density of 5 × 104 per well overnight, after which a fresh medium containing free 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD NPs replaced the old one followed by further incubation for 3 h using PBS buffer as a control. EdU (5-ethynyl-2′-deoxyuridine) was added in the amount of 10 μmol/L to every detecting well, and cells were further cultured for 2 h. After labeling S-phase cells, the culture solutions were replaced with fixation solutions followed by its removal and washing three times. Using the liquids of enhancing membrane penetrability for 10–15 min and abandoned, then washing three times, EdU colored substrate solution (containing 488 azide) was incubated for 30 min and kept in a dark place. After the EdU labeling and DAPI counterstaining steps, the cells were analyzed by CLSM imaging. All results were obtained by conducting at least three independent experiments.
2.12. Flow cytometry analysis of the cell cycle
Cells were seeded at a density of 1 × 106 per well and incubated overnight. The cellular incomplete medium for serum removal replaced the old medium after 12 h to synchronize the cell cycle. Subsequently, the cells were washed thrice with PBS and incubated in a new growth medium containing free 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD for 3 h. Subsequently, cells were washed and fixed in 70 % ethanol overnight at 4 °C under dark conditions. Ethanol-fixed cells were centrifuged at 300 ×g for 5 min, washed thrice with cold PBS, and incubated in PBS with 100 mg/mL DNase-free RNase A for 30 min on ice. Next, the cell pellet was suspended in 1 mL of DAPI staining solution for 10 min in the dark. Finally, the cells were analyzed in a flow cytometer (BD FACSAria™ III, USA). DAPI fluorescence was detected using UV laser light, and the raw data were processed by the FlowJo10.0 software.
2.13. Biodistribution and pharmacokinetic study of RGD-IFP/7-AAD nanoprobes in vivo
Briefly, 6-week-old female athymic nude mice were subcutaneously injected with 1 × 107 A549 cells into the right hips to establish a xenograft model. Tumor-bearing mice with tumor volumes of 100 mm3 were injected with free 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD nanoprobes through their caudal veins at a 7-AAD concentration of 120 μg/kg. In vivo real-time images of the anesthetized mice and their sacrificed organs were obtained by an IVIS Spectrum imaging system (Perkin Elmer). Acquisition and analysis of the raw data were performed by the Living Image 4.2 Software.
To determine pharmacokinetics study of nanoprobes, 8-week-old Sprague–Dawley rats (n = 9) were randomly divided into three groups: free 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD nanoprobes. Pharmacokinetics was determined by drawing 0.2 mL blood samples from the rats at 0.5, 1, 2, 4, 6, 8 and 24 h after injection. The concentration of 7-AAD were determined by high performance liquid chromatography (HPLC) assay and non-compartmental pharmacokinetic parameters were analyzed by DAS2.0 software.
2.14. In vivo antitumor efficacy of RGD-IFP/7-AAD nanoprobes
A549 nude mice were randomly divided into three groups (n = 5 per group) and injected intravenously with saline (control), free 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD with a 7-AAD concentration of 60 μg/kg every 6 d after the tumor size reached an approximate value of 50 mm3. Tumor sizes (tumor volume = length × width2/2) and mice weight were recorded daily. After 24 d of treatment, mice were sacrificed, and their tumors and major organs were collected into a 4 % formaldehyde solution. All procedures and experimental protocols were reviewed and approved by the Animal Experimental Ethical Inspection Form of Southeast University (Jiangsu, China).
2.15. Immunohistochemistry and histological analysis
At the end of the experiment, the sacrificed tumor tissues were cut into 10 μm slices and incubated with rabbit anti-mouse Ki67 antibodies at 4 °C overnight. After washing thrice with cold PBS buffer, horseradish peroxidase-conjugated secondary antibody (1:1000) was added to tissues, which were subsequently incubated for 2 h at room temperature. The results of terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) experiments were analyzed according to the standard protocols. The major mouse organs including hearts, livers, spleens, lungs, and kidneys were embedded in paraffin for hematoxylin–eosin (H&E) histological analysis.
2.16. Statistical analysis
All values are expressed as mean ± standard deviation (SD). All statistical analysis was performed by student's t tests or one-way ANOVA test and the nominal P value for each comparison without adjusting for multiple testing. A P value < 0.05 was considered statistically significant.
3. Results and discussion
3.1. Characteristics and molecular docking of 7-AAD and RGD-IFP
The synthetic procedures for 7-AAD are shown in Fig. 2A. The seventh hydrogen bond of ACTD is substituted by amidogen and carries a positive charge. Protons present on the amidogen enable 7-AAD to emit fluorescence through intramolecular charge transfer effects, whereas ACTD does not emit fluorescence, as shown in Fig. 2B. Whether 7-AAD also can insert into the DNA double helix to bind and affect its function remains to be determined23,24. To evaluate its effects on DNA function, 7-AAD was mixed with specified DNA concentrations, and their conjugated reaction was studied by UV–Visible spectroscopy and agarose gel electrophoresis. The degree of intercalation between small molecules and DNA can be determined from the red shift of the absorption peak obtained for the chromophore group in the visible spectrum and decreases in its absorption intensity are due to hypochromic effects25. The results presented in Fig. 2C show that after the addition of calf thymus DNA, the UV–Visible absorption intensity decreased significantly due to the hypochromic effect, and the λmax value moved towards longer wavelengths (i.e. red-shifted). The observed trends became more pronounced as the DNA concentration increased. DNA is negatively charged and 7-AAD is positively charged; their corresponding molecules migrate to opposite electrodes during electrophoresis26. A binding reaction occurred after 7-AAD was inserted into the larger molecule of calf thymus DNA, and they get together slowly migrated to the positively charged side, as shown in lane 3 of Fig. 2D.
Figure 2.
Formation and function of 7-AAD. (A) Formation of ACTD into 7-AAD through a series of steps. The red dotted circled indicates the site of hydrogen atom replacement position, and the amino substitution indicates the amino proton that provides a fluorescence signal from intramolecular charge transfer effect. Fluorescence spectra of (B) ACTD and 7-AAD. The 7-AAD conjugation with DNA was confirmed by the (C) UV–Visible spectra, fluorescence red shift, and hypochromic effect. (D) Agarose gel electrophoresis data showing the change in the charge migration pathway after the 7-AAD insertion into the DNA structure. Lanes 1–4: 7-AAD, DNA, 7-AAD+DNA, DL2000 DNA Marker.
Molecular dynamics docking procedures can identify two molecules with a stable configuration and desired energy. It determines not only their possible physical interactions, but also feasible complex configurations27. The RGD-IFP peptide is composed of three functional sequences, and its tertiary structure is a dimer consisting of α-helices (Fig. 3B). The designed main sequence included 36 amino acids with an α-helix that formed self-assembled nanospheres. The SEE sequence zone with a negative charge (Isoelectric point = 3.79) tends to attract many positively charged molecules through electrostatic interactions, while the RGD targeted sequence can bind to integrin αvβ3 subunits overexpressed in various tumor cells22,28,29. A dimer model of RGD-IFP peptide constructed via SWISS-MODEL is shown in Fig. S1, and the net charge and hydrophilicity are presented in Fig. S2. The binding free energy reflecting interactions between the ligand and receptor included torsional free energy and intermolecular energy30,31. Fig. 3C shows the interactions between the 7-AAD and RGD-IFP peptide molecules in a 2D model; meanwhile they assemble into a stable polymeric NPs by electrostatic interactions and π–π stacking. Related thermodynamic analysis of molecular docking between 7-AAD and RGD-IFP are also shown in Table S1. The Gibbs free energy was −12.16 kcal/mol, which could reflect the theoretical feasibility of self-assembly. Here, the ketone group of 7-AAD forms a hydrogen bond with the Lys 168 group of RGD-IFP peptides; the hydrophilic groups (Arg170, Glu172, Thr165, Arg170, Arg175, and Asn166) interact with 7-AAD species, and the Ala169 and Val173 groups interact hydrophobically with the opposite sides of 7-AAD molecules. In addition, the lowest torsional potential energies of 7-AAD and the RGD-IFP dimer (Fig. 3A and B) were calculated by MOE32, and the configuration with the lowest binding energy corresponded to the most stable 3D docking structure (assuming that the majority of all possible combinations of 7-AAD with IFP peptide are presented in Fig. 3C).
Figure 3.
Computer docking model of 7-AAD and RGD-IFP peptide. The 3D structure with the lowest torsional potential energy obtained for (A) 7-AAD and the dimer form of (B) RGD-IFP peptide. (C) A molecular docking model describing the coordination conformations of 7-AAD and RGD-IFP peptide in the 2D and 3D structures. (D) A diagram illustrating the interactions between all amino acids in 7-AAD molecules and RGD-IFP peptide.
3.2. Synthesis and characterization of RGD-IFP/7-AAD nanoprobes
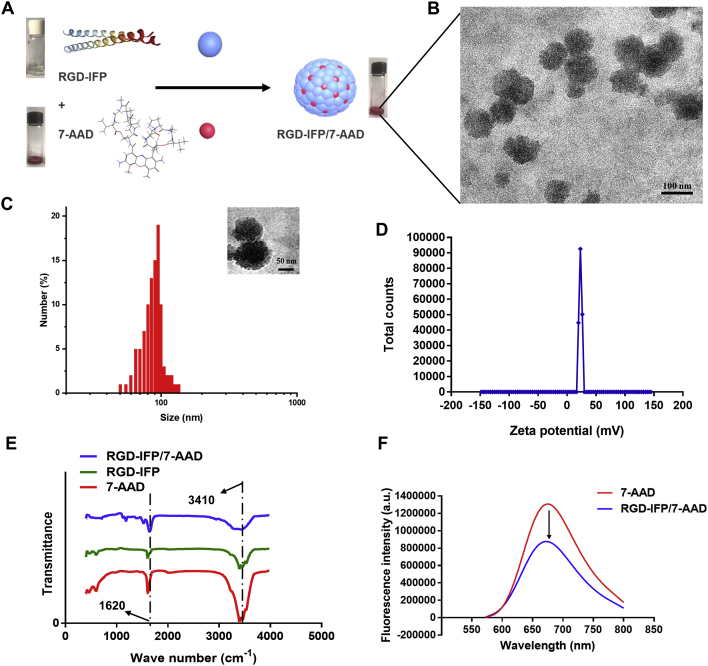
RGD-IFP/7-AAD nanoprobes were synthesized by a one-step method (details of this procedure are provided in the Materials and Methods section). Fig. 4A shows the hydrophobic 7-AAD species coated with amphiphilic RGD-IFP peptides and the turbid solution containing successfully self-assembled suspended nanospheres. Representative TEM images of the as-synthesized NPs revealed that the latter formed spherical structures with sizes of approximately 92.5 ± 26.7 nm (Fig. 4B), which was consistent with the DLS data presented in Fig. 4C. The size of NPs is less than 120 nm, indicating that these regular aggregates may be suitable for intravenous (i.v.) injection and accumulation in tumors owing to their enhanced permeability and retention properties33, 34, 35.
Figure 4.
Preparation and characterization of RGD-IFP/7-AAD nanoprobes. (A) Schematic illustration of the self-assembly of RGD-IFP/7-AAD nanoprobes performed by a one-step method. (B) TEM images of RGD-IFP/7-AAD NPs. (C) A photographic image of RGD-IFP/7-AAD NPs and their size distribution obtained from the DLS profiles. (D) Zeta potentials, (E) FTIR spectra, and (F) fluorescence spectra of 7-AAD and RGD-IFP/7-AAD NPs before and after self-assembly.
The amino groups located on NP surfaces may contribute to positive electrical potential, and such electropositivity can protect NPs from being adsorbed by positively charged proteins in the blood after intravenous injection36. Fig. 4D shows that the Zeta (ζ) potential of RGD-IFP/7-AAD NPs is 23.20 ± 2.35 mV at pH 7.4, which helps them maintain a fixed shape. The size distribution and ζ potential of RGD-IFP/7-AAD NPs remained unchanged within 15 days, confirming their stability at pH 7.4 in PBS and 10 % FBS (Fig. S3). Because FRET can be used for studying intermolecular functional groups and characteristic structures37, RGD-IFP peptide, 7-AAD, and the purified RGD-IFP/7-AAD NPs were analyzed using this method. The FTIR spectra depicted in Fig. 4E have two characteristic bands centered at 1620 and 3410 cm−1. Compared with the corresponding FTIR band of the RGD-IFP spectrum, the RGD-IFP/7-AAD band was shifted to lower wavenumbers, which indicated that RGD-IFP and 7-AAD peaks were detected for RGD-IFP/7-AAD NPs and that RGD-IFP was successfully coordinated with 7-AAD. In addition, fluorescence decrease was clearly observed for RGD-IFP/7-AAD nanoprobes by FRET, which is of great significance for high sensitivity fluorescence detection in vivo (Fig. 4F).
3.3. Living cell uptake and efficient targeting of αvβ3-expressing cells in vitro
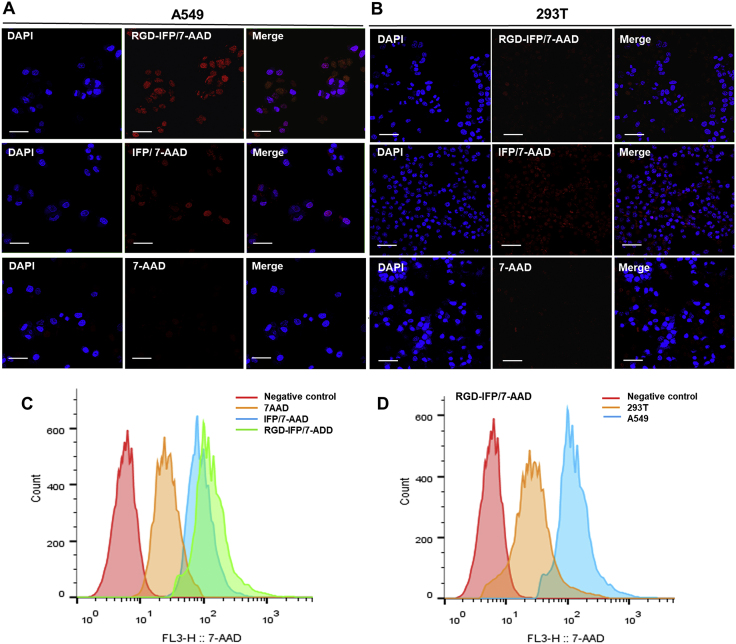
7-AAD is a fluorescent derivative of ACTD, which cannot enter living cells, and is commonly used to stain dead or apoptotic cells, so as to distinguish them from living cells by flow cytometry23,38. Once RGD-IFP/7-AAD NPs that could penetrate cell membranes were successfully fabricated, human A549 cells were used to evaluate their degree of internalization and cellular targeting efficiency. Fig. 5A shows that after incubation with IFP/7-AAD, RGD-IFP/7-AAD nanoprobes and free 7-AAD for 3 h, red fluorescence was mainly localized in A549 nuclei with RGD-IFP/7-AAD nanoprobes, while IFP/7-AAD nanoprobes were partially detected and free 7-AAD groups were not detectable. Furthermore, flow cytometric analysis was performed to compare the cellular uptake of RGD-IFP/7-AAD nanoprobes with that of IFP/7-AAD and free 7-AAD. The fluorescence signals obtained for RGD-IFP/7-AAD groups in A549 cells were clearly shifted to the furthest right of the fluorescence enhancement zone (Fig. 5C). Hence, we conclude that RGD-IFP peptides promote cellular uptake and that RGD-IFP/7-AAD NPs can be delivered directly into living tumor cells. Meanwhile, red fluorescence was detected for both groups containing IFP/7-AAD and RGD-IFP/7-AAD nanoprobes or free 7-AAD co-incubated for 12 h (Fig. S4). According to Fig. S4, cellular morphology was substantially changed due to apoptosis or necroptosis resulting from the long-term effects of 7-AAD groups; therefore free 7-AAD stained apoptotic and/or dead cells.
Figure 5.
In vitro imaging of A549 cells and enhanced internalization. In vitro imaging and targeting efficacy of RGD-IFP/7-AAD nanoprobes determined by the CLSM imaging of the (A) A549 and (B) 293T cell lines incubated with these nanoprobes and free 7-AAD for 3 h. Flow cytometry analyses of the (C) A549 cells incubated with IFP/7-AAD, RGD-IFP/7-AAD nanoprobes and free 7-AAD for 3 h to measure the cell uptake efficiency, and (D) A549/293T cell lines incubated with RGD-IFP/7-AAD nanoprobes for 3 h to detect integrin αvβ3 units. Scale bar = 20 μm.
The RGD peptide that binds to αvβ3 integrin is overexpressed in A549 cells, while it is poorly expressed in 293T cells39. As shown in Fig. 5D, the signals generated by RGD-IFP/7-AAD nanoprobes in A549 cells were significantly shifted to the right of the fluorescence enhancement zone as compared with their position for 293T cells (similar results were obtained by CLSM imaging; see Fig. 5A and B). Thus, RGD peptide enabled delivery of a greater number of RGD-IFP/7-AAD NPs into A549 cells with higher integrin αvβ3 expression levels than those of 293T cells. DAPI is a fluorescent dye that has binding affinity for the adenine- and thymine-rich DNA regions40. The high fluorescence intensity of RGD-IFP/7-AAD combined with the nuclei of A549 cells may prevent DAPI from binding to DNA molecules (Fig. 5A), owing to the released 7-AAD competing with DAPI for nuclear DNA staining sites.
3.4. Release and cytotoxicity of RGD-IFP/7-AAD nanoprobes
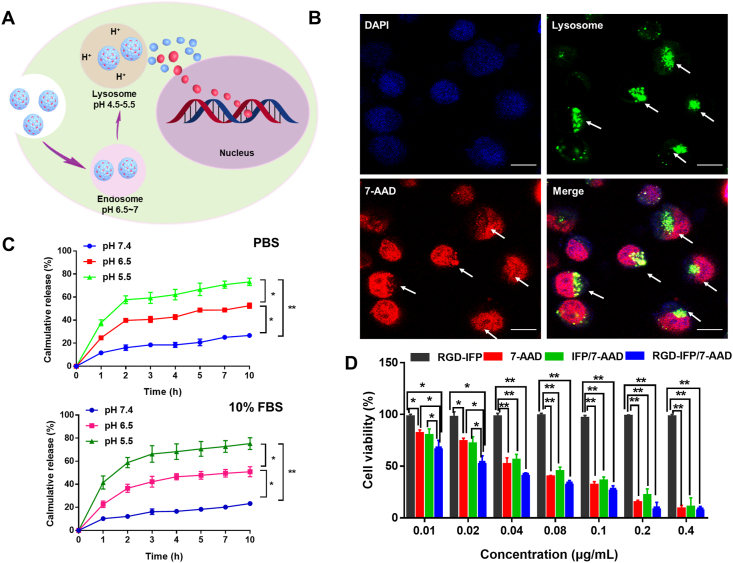
Previous research studies have shown that the NPs with sizes of 50–200 nm can be uptake by cells via endocytosis41,42. A schematic diagram of the transport and pH-controlled release of NPs is shown in Fig. 6A. NPs are ingested in the intracellular endocytosis pathway and stimulated by the acidic environments of endosomes (pH 6.5–7.0) and lysosomes (pH 4.5–5.5) to release 7-AAD from the NPs43,44. Furthermore, co-localization of RGD-IFP/7-AAD nanoprobes (red fluorescence) and the specific probe related to lysosomes (green fluorescence) is depicted in Fig. 6B. Here, arrows pointing to the yellow merged fluorescence indicate that RGD-IFP/7-AAD nanoprobes can be delivered to lysosomes through endocytosis with the simultaneous release of drugs into nuclei.
Figure 6.
Release of RGD-IFP/7-AAD nanoprobes through endosome/lysosome pathways and their in vitro cytotoxicity. (A) Schematic representation of the RGD-IFP/7-AAD nanoprobes transported and released in tumor cells. (B) Representative images of the lysosomes containing RGD-IFP/7-AAD NPs. The green fluorescence denotes the lysosomes from the Lyso-Tracker probe; the red fluorescence indicates the 7-AAD components of nanoprobes; the blue fluorescence indicates DAPI staining; and the yellow fluorescence is a merged image originated from the colocalization of nanoprobes and lysosomes. Scale bars = 10 μm. (C) 7-AAD release profiles of RGD-IFP/7-AAD nanoprobes obtained at pH values of 7.4, 6.5, and 5.5 in vitro. The bars represent the mean ± standard deviation (SD) values obtained via triplicate determination (n = 3). ∗P < 0.05, ∗∗P < 0.01. (D) Viability of the A549 cells treated with IFP, 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD nanoprobes at different concentrations. The bars represent the mean ± standard deviation (SD) values obtained via triplicate determination (n = 3). ∗P < 0.05, ∗∗P < 0.01.
As mentioned earlier, pH values of 7.4, 6.5, and 5.5 were utilized to simulate the intracellular pH conditions for drug release in vitro. According to Fig. 6C, in PBS and 10 % FBS, the synthesized RGD-IFP/7-AAD NPs were stable at pH 7.4 (<25 % drug release), while approximately 50 % or 70 % of the drugs were released when the pH decreased to 6.5 or 5.5, respectively. With the decrease of pH (from 7.4 to 5.5) of medium solution, the TEM images of nanoparticles were degraded gradually and the drug release response increasingly (Fig. S5). The viability of A549 cells treated with IFP, 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD NPs at different concentrations were compared by using the CCK-8 assays. In Fig. 6D, 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD NPs demonstrated significant dose-dependent cell killing effects on A549 cells after 24 h of treatment, while IFP alone had a negligible impact on cell viability. Owing to RGD targeting, the cell killing effect of RGD-IFP/7-AAD NPs was much stronger than that of free 7-AAD and IFP/7-AAD in A549 cells at low doses; however, after 4 h of treatment, 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD produced little effects on cell activity (Fig. S6), suggesting that the nanoprobes depicted in Fig. 4A were not caused by cell necrosis, killing the tumor cells required some time.
3.5. RGD-IFP/7-AAD nanoprobes inhibit cell proliferation via blocking cell cycle
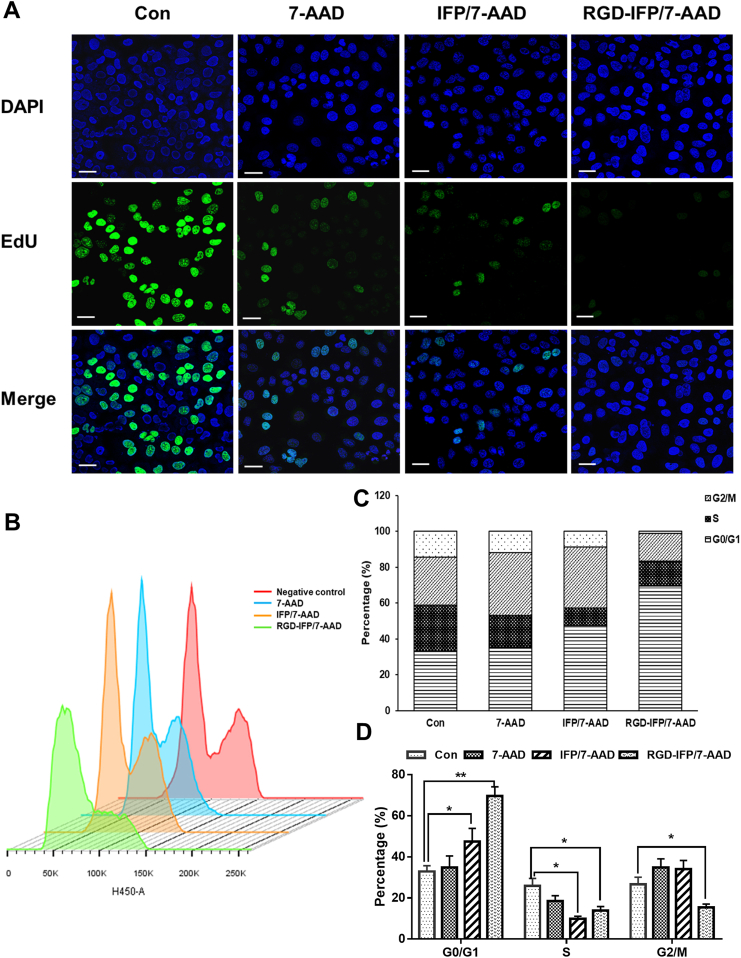
The results present in Fig. 2C and D suggested that similar to ACTD, 7-AAD may bind to DNA to thereby affecting the cell cycle, indicating its high cytotoxicity. Owing to their high targeting efficacy and small nanoscale size, RGD-IFP/7-AAD nanoprobes demonstrated a much stronger inhibition of cell proliferation after interaction with A549 cells for 3 h as compared with the effect produced by free 7-AAD and IFP/7-AAD nanoprobes (Fig. 7A). This phenomenon was observed because IFP/7-AAD and RGD-IFP/7-AAD NPs entered the tumor cells and bound to nuclear DNA, which consequently blocked the early phase of the cell replication cycle in the G0/G1 phase and effectively inhibited cell proliferation (Fig. 7B-D). Remarkably, free 7-AAD molecules mainly entered dying or late apoptotic cells and were relatively hysteretic for cell cycle blocked which was different from NPs. Cell cycle fitting details are shown in Fig. S7.
Figure 7.
Release of 7-AAD from the RGD-IFP/7-AAD nanoprobes induced a G1 phase cell cycle arrest in A549 cells for proliferative inhibition. A549 cells treated with free 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD nanoprobes were detected by the (A) EdU cell proliferation assay and (B) flow cytometry analysis of the cell cycle via DAPI staining. (C) The cell cycle division scale diagram, and (D) quantitative analysis of the percentage of the cell cycle stage based on the results of the flow cytometry studies. The bars represent the mean ± standard deviation (SD) values obtained via triplicate determination (n = 3). ∗P < 0.05, ∗∗P < 0.01. Scale bar = 20 μm.
3.6. In vivo biodistribution and antitumor efficacy of RGD-IFP/7-AAD nanoprobes
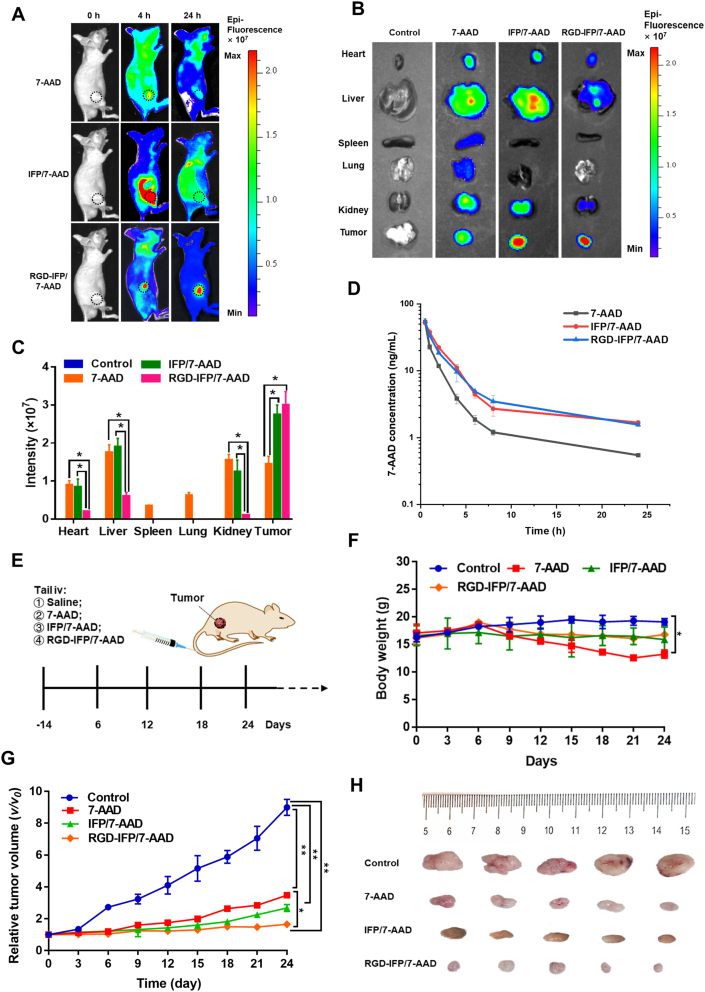
To determine the imaging and therapeutic efficiency of RGD-IFP/7-AAD nanoprobes in vivo, we intravenously injected saline, 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD NPs via mouse tail veins. Fluorescence was employed to image the delivery and accumulation of free 7-AAD and nanoprobes as well as their in vivo biodistribution using an IVIS Lumina II imaging system. In Fig. 8A and B, the RGD-IFP/7-AAD group generated a much stronger fluorescence signal in the tumor region within 4 h and 24 h as compared with that of the 7-AAD group, and showed significant tumor targeting compared to IFP/7-AAD, suggesting that RGD-IFP/7-AAD nanoprobes could be effectively delivered and released into tumor tissues. For comparison, self-assembled targeted peptide nanomaterials increased the effective loading retention at the therapeutic site while reducing their accumulation and toxicity toward other organs45,46. Four hours post-injection, the mice injected with RGD-IFP/7-AAD nanoprobes exhibited strong fluorescence at tumor sites, while fluorescence signals in liver and kidney were apparently weaker as compared with free 7-AAD and IFP/7-AAD group, indicating the super-sensitive efficiency and selective distribution of this nanoprobe. Meanwhile, αvβ3 and 7-AAD co-localized fluorescence signals were found in tumor tissues (Fig. S8). In addition, a small quantity of red fluorescence signals was found in the heart, liver, and kidney tissues. These signals were mainly distributed in abundant vascular sites, which may be related to the expression of αvβ3 or the transport and excretion of nanoprobes. Statistical analysis of the measured fluorescence intensities is presented in Fig. 8C; the obtained results confirmed the superiority of RGD-IFP/7-AAD nanoprobes for tumor imaging and localization. Pharmacokinetics results (Fig. 8D) showed that the elimination half-life time of RGD-IFP/7-AAD nanoprobes was 2.43 h, distinctly longer than the systemic circulation time of free 7-AAD. Details of pharmacokinetic parameters are shown in Table S2.
Figure 8.
Fluorescence imaging and antitumor efficacy of RGD-IFP/7-AAD nanoprobes in vivo. The xenograft model mice were divided randomly into three groups and treated with saline (control), free 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD nanoprobes via the tail intravenous injection. (A) Fluorescence images of the A549 tumor-bearing mice showing the 7-AAD and nanoprobes signals at 4/24 h post-injection. RGD-IFP/7-AAD nanoprobes promote the accumulation of a larger number of 7-AAD molecules at the tumor sites as compared with that produced by free 7-AAD. (B) Fluorescence images of the relevant major organs and tumor tissues of the sacrificed mice. (C) Quantitative analysis of the fluorescence intensities obtained for the major mouse organs and tumor tissues. (D) In vivo pharmacokinetic study after caudal vein injection. (E) Schematic diagram of the treatment timeline and profile. (F) Body weight of the different groups of mice treated for 24 d and their (G) tumor volumes (n = 5). (H) Photographic images of the isolated tumors at the end of experiment. The bars represent the mean ± SD values obtained via triplicate determination. ∗P < 0.05, ∗∗P < 0.01.
The antitumor efficacy of the proposed nanoprobe was also evaluated in vivo, and the required treatment was performed according to the diagram presented in Fig. 8E. No apparent differences in body weight were observed between the RGD-IFP/7-AAD NP-treated and the control groups, indicating that RGD-IFP/7-AAD NPs exhibited potentially high therapeutic efficacy and low internal toxicity, while treatment with free 7-AAD caused a significant decrease in body weight, with poor physical condition at the end of the experiments (Fig. 8F). The average tumor volume profiles depicted in Fig. 8G show that 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD NPs effectively inhibited tumor growth as compared with the results obtained for the control group. Solid tumors removed from nude mice also revealed that RGD-IFP/7-AAD NPs inhibited tumor growth more effectively than IFP/7-AAD or 7-AAD alone (Fig. 8H). Hence, this RGD-IFP/7-AAD nanoprobe exhibited superior antitumor efficacy and prominent fluorescence localization in vivo.
3.7. Histological analysis and biosafety evaluation
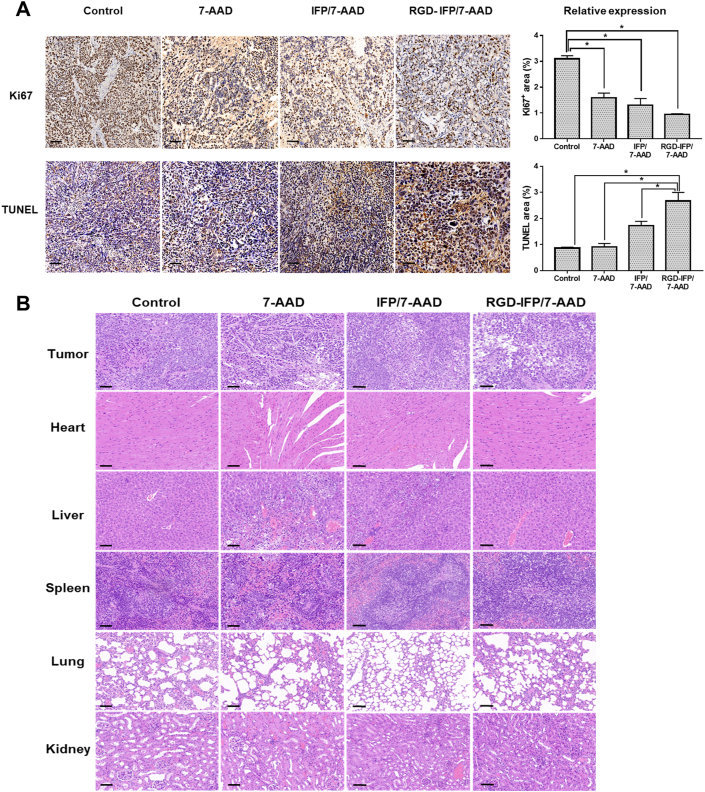
Immunohistochemical staining and H&E staining were performed to identify pathological changes to the tumors after different treatments. Tumor tissues were mostly exhibited hypercellularity and contained pleomorphic and hyperchromatic nuclei47,48, which was consistent with H&E staining results obtained from controls (Fig. 9B). It is noteworthy that 7-AAD and NPs groups had various degrees of tumor cell damage; in particular, mice treated with RGD-IFP/7-AAD NPs exhibited the highest levels. These findings were validated by performing Ki67 immunohistochemical staining and TUNEL assays to access tumor cell proliferation and apoptosis (Fig. 9A). Compared with the control group, 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD decreased the number of Ki67-positive proliferating cells. Furthermore, the Ki67-positive region of the RGD-IFP/7-AAD group was the lowest among the three studied groups, which confirmed the higher efficacy of RGD-IFP/7-AAD NPs in inhibiting tumor growth. Meanwhile, the RGD-IFP/7-AAD group demonstrated the highest TUNEL-positive apoptotic level, indicating that RGD-IFP/7-AAD NPs-treated tumors more effectively than 7-AAD and IFP/7-AAD. To evaluate the biocompatibility and potential toxicity of these nanoprobes in vivo, H&E staining of various organs was conducted. As shown in Fig. 8B, the morphologies of mouse hearts, livers, spleens, lungs, and kidneys in the control, IFP/7-AAD and RGD-IFP/7-AAD groups change very little, whereas the 7-AAD group exhibites various degrees of organic damage including hemorrhagic necrosis, fibrosis, and swelling deformations. These results suggest that RGD-IFP/7-AAD nanoprobes produced negligible side effects and possessed good biocompatibility properties.
Figure 9.
Immunohistochemical staining and histopathological analysis. After treatments with saline (control), 7-AAD, IFP/7-AAD and RGD-IFP/7-AAD nanoprobes, the tumor sections (A) were detected with immunohistochemistry to examine the proliferation (Ki67) and apoptosis (TUNEL) levels. Scale bar = 50 μm. (B) Results of the histopathological analyses of the tumor sections and major organ sections performed by their H&E staining after 24 days of treatment. The bars represent the mean ± SD values obtained via triplicate determination. ∗P < 0.05. Scale bar = 50 μm.
4. Conclusions
In summary, a super-sensitive nanoprobe composed of self-assembled RGD-IFP peptide and 7-AAD that could generate strong fluorescence signals and be potentially used in tumor-targeted chemotherapy was developed in this work. The RGD-IFP self-assembled peptide consisted of the natural amino acids present in the human body and, therefore, possessed inherent biocompatibility. The nanoprobes combined with RGD exhibited enhanced tumor targeting capability both in vitro and in vivo. 7-AAD molecules embedded into RGD-IFP peptides penetrated live cells and the released fluorescence signal intensity from the nanoprobes was significantly increased in the acidic environments of endosomes/lysosomes. Then pH-released 7-AAD molecules were quickly inserted into the double helical structure of nuclear DNA, inducing tumor cell cycle arrest and intrinsic apoptosis. Compared with free 7-AAD and IFP/7-AAD, RGD-IFP/7-AAD nanoprobes produced better tumor images and inhibited tumor growth more effectively with a significantly lower number of side effects. Therefore, intelligently designed RGD-IFP/7-AAD nanoprobes with excellent real-time tumor imaging and tumor-targeting drug delivery properties represent a promising tumor localization and treatment strategy.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81603016, 81773624, 81900453), the Natural Science Foundation of Jiangsu Province (No. BK20160706, BE2017746, China), the National Science and Technology Major Project (2018ZX09301026-005, 2020ZX09201015, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.07.020.
Contributor Information
Haiping Hao, Email: hhp_770505@hotmail.com.
Guoqiu Wu, Email: 101008404@seu.edu.cn.
Author contributions
Conception and design: Han Xiao and Guoqiu Wu; Acquisition of data: Han Xiao; Analysis and interpretation of data: Han Xiao, Rui Zhang, Xiaobo Fan, Mingyuan Zou and Xuejiao Yan; Writing, review, and/or revision of the manuscript: Han Xiao, Xinglu Jiang, Haiping Hao and Guoqiu Wu. All authors read and approved the final manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Appendix ASupplementary data
The following is the Supplementary data to this article:
References
- 1.Choi E.W., Nayak L.V., Bates P.J. Cancer-selective antiproliferative activity is a general property of some G-rich oligodeoxynucleotides. Nucleic Acids Res. 2010;38:1623–1635. doi: 10.1093/nar/gkp1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng S., Zhang Z., Qu Y., Zhang X.J., Guo H.B., Shi X.J., et al. Radiopharmaceuticals and fluorescein sodium mediated triple-modality molecular imaging allows precise image-guided tumor surgery. Adv Sci (Weinh) 2019;6:1900159. doi: 10.1002/advs.201900159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y.S., Jiang L.P., Zhang Y.W., Lu Y.M., Li J.N., Wang H., et al. Fibronectin-targeting and cathepsin B-activatable theranostic nanoprobe for MR/fluorescence imaging and enhanced photodynamic therapy for triple negative breast cancer. ACS Appl Mater Interfaces. 2020;12:33564–33574. doi: 10.1021/acsami.0c10397. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y.J., He Y., Deng Z.W., Li J.C., Huang J., Zhong S. Intelligent nanoprobe: acid-responsive drug release and in situ evaluation of its own therapeutic effect. Anal Chem. 2020;92:12371–12378. doi: 10.1021/acs.analchem.0c02099. [DOI] [PubMed] [Google Scholar]
- 5.Yoo J.W., Doshi N., Mitragotri S. Adaptive micro and nanoparticles: temporal control over carrier properties to facilitate drug delivery. Adv Drug Deliv Rev. 2011;63:1247–1256. doi: 10.1016/j.addr.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Yuan A., Qiu X.F., Tang X.L., Liu W., Wu J.H., Hu Y.Q. Self-assembled PEG-IR-780-C13 micelle as a targeting, safe and highly-effective photothermal agent for in vivo imaging and cancer therapy. Biomaterials. 2015;51:184–193. doi: 10.1016/j.biomaterials.2015.01.069. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y.Z., Jiang Y.F., Wang H.Y., Wang J.X., Shin M.C., Byun Y., et al. Curb challenges of the "Trojan Horse" approach: smart strategies in achieving effective yet safe cell-penetrating peptide-based drug delivery. Adv Drug Deliv Rev. 2013;65:1299–1315. doi: 10.1016/j.addr.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X.N., Schumann C., Albarqi H.A., Lee C.J., Alani A.W.G., Bracha S., et al. A tumor-activatable theranostic nanomedicine platform for NIR fluorescence-guided surgery and combinatorial phototherapy. Theranostics. 2018;8:767–784. doi: 10.7150/thno.21209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarkar S., Levi-Polyachenko N. Conjugated polymer nano-systems for hyperthermia, imaging and drug delivery. Adv Drug Deliv Rev. 2020;163–164:40–64. doi: 10.1016/j.addr.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Li W.J., Cai S.H., Cai L., Li X.K. Anti-apoptotic effect of hepatocyte growth factor from actinomycin D in hepatocyte-derived HL7702 cells is associated with activation of PI3K/Akt signaling. Toxicol Lett. 2006;165:142–148. doi: 10.1016/j.toxlet.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Siboni R.B., Nakamori M., Wagner S.D., Struck A.J., Coonrod L.A., Harriott S.A., et al. Actinomycin D specifically reduces Eexpanded CUG repeat RNA in myotonic dystrophy models. Cell Rep. 2015;13:2386–2394. doi: 10.1016/j.celrep.2015.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stokke T., Holte H., Simland E.B., Lie S.O., Steen H.B. Differential chromatin structure-dependent binding of 7-Aminoactinomycin D in normal and malignant bone marrow hematopoietic cells. Cancer Res. 1992;52:5007–5012. [PubMed] [Google Scholar]
- 13.Kim D., Kim J., Park Y.I., Lee N., Hyeon T. Recent development of inorganic nanoparticles for Bbiomedical imaging. ACS Cent Sci. 2018;4:324–336. doi: 10.1021/acscentsci.7b00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen S., Margel S. Engineering of near IR fluorescent albumin nanoparticles for in vivo detection of colon cancer. J Nanobiotechnol. 2012;10:36–44. doi: 10.1186/1477-3155-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negahdaripour M., Golkar N., Hajighahramani N., Kianpour S., Nezafat N., Ghasemi Y. Harnessing self-assembled peptide nanoparticles in epitope vaccine design. Biotechnol Adv. 2017;35:575–596. doi: 10.1016/j.biotechadv.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun B.F., Deng C., Meng F.H., Zhang J., Zhong Y.Z. Robust, active tumor-targeting and fast bioresponsive anticancer nanotherapeutics based on natural endogenous materials. Acta Biomater. 2016;45:223–233. doi: 10.1016/j.actbio.2016.08.048. [DOI] [PubMed] [Google Scholar]
- 17.Kaus-Drobek M., Mucke N., Szczepanowski R.H., Wedig T., Czarnocki-Cieciura M., Polakowska M., et al. Vimentin S-glutathionylation at Cys328 inhibits filament elongation and induces severing of mature filaments in vitro. FEBS J. 2020;287:5304–5322. doi: 10.1111/febs.15321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown N., Lei J.T., Zhan C.D., Shimon L.J.W., Adler-Abramovich L., Wei G.H., et al. Structural polymorphism in a self-assembled tri-aromatic peptide system. ACS Nano. 2018;12:3253–3262. doi: 10.1021/acsnano.7b07723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avitabile C., Diaferia C., Ventura B.D., Mercurio F.A., Leone M., Roviello V., et al. Self-assembling of fmoc-GC peptide nucleic acid dimers into highly fluorescent aggregates. Chem Eur J. 2018;24:4729–4735. doi: 10.1002/chem.201800279. [DOI] [PubMed] [Google Scholar]
- 20.Demircioglu F., Hodivala-Dilke K. Alphavbeta3 integrin and tumour blood vessels-learning from the past to shape the future. Curr Opin Cell Biol. 2016;42:121–127. doi: 10.1016/j.ceb.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Huang R., Rofstad E.K. Integrins as therapeutic targets in the organ-specific metastasis of human malignant melanoma. J Exp Clin Cancer Res. 2018;37:92. doi: 10.1186/s13046-018-0763-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H.J., Niu G., Wu H., Chen X.Y. Clinical application of radiolabeled RGD peptides for PET imaging of integrin alphavbeta3. Theranostics. 2016;6:78–92. doi: 10.7150/thno.13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zembruski N.C., Stache V., Haefeli W.E., Weiss J. 7-Aminoactinomycin D for apoptosis staining in flow cytometry. Anal Biochem. 2012;429:79–81. doi: 10.1016/j.ab.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Lam L.T., Pickeral O.K., Peng A.C., Rosenwald A., Hurt E.M., Giltnane J.M., et al. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biology. 2001:2. doi: 10.1186/gb-2001-2-10-research0041. research0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almaqwashi A.A., Zhou W., Naufer M.N., Riddell I.A., Yilmaz O.H., Lippard S.J., et al. DNA intercalation facilitates efficient DNA-targeted covalent binding of phenanthriplatin. J Am Chem Soc. 2019;141:1537–1545. doi: 10.1021/jacs.8b10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veselinovic J., Li Z.D., Daggumati P., Seker E. Electrically guided DNA immobilization and multiplexed DNA detection with nanoporous gold electrodes. Nanomaterials (Basel) 2018;8:351. doi: 10.3390/nano8050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C.X., Wang W.L., Xi Y.W., Wang J.W., Chen J.F., Yun J., et al. Design, preparation and characterization of cyclic RGDfK peptide modified poly(ethylene glycol)-block-poly(lactic acid) micelle for targeted delivery. Mater Sci Eng C Mater Biol Appl. 2016;64:303–309. doi: 10.1016/j.msec.2016.03.062. [DOI] [PubMed] [Google Scholar]
- 28.Camunas-Soler J., Frutos S., Bizarro C.V., Lorenzo Sd, Fuentes-Perez M.E., Ramsch R., et al. Electrostatic binding and hydrophobic collapse of peptide-nucleic acid aggregates quantified using force spectroscopy. ACS Nano. 2013;7:5102–5113. doi: 10.1021/nn4007237. [DOI] [PubMed] [Google Scholar]
- 29.Liang P.Q., Zheng J.J., Dai S.L., Wang J.Y., Zhang Z.Q., Kang T., et al. pH triggered re-assembly of nanosphere to nanofiber: the role of peptide conformational change for enhanced cancer therapy. J Control Release. 2017;260:22–31. doi: 10.1016/j.jconrel.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 30.Habtemariam B., Anisimov V.M., MacKerell A.D., Jr. Cooperative binding of DNA and CBFbeta to the Runt domain of the CBFalpha studied via MD simulations. Nucleic Acids Res. 2005;33:4212–4222. doi: 10.1093/nar/gki724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagoner J.A., Dill K.A. Opposing pressures of speed and efficiency guide the evolution of molecular machines. Mol Biol Evol. 2019;36:2813–2822. doi: 10.1093/molbev/msz190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller Y., Ma B., Nussinov R. Zinc ions promote Alzheimer Abeta aggregation via population shift of polymorphic states. Proc Natl Acad Sci U S A. 2010;107:9490–9495. doi: 10.1073/pnas.0913114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu R., Hu C., Yang Y.Y., Zhang J.Q., Gao H.L. Theranostic nanoparticles with tumor-specific enzyme-triggered size reduction and drug release to perform photothermal therapy for breast cancer treatment. Acta Pharm Sin B. 2019;9:410–420. doi: 10.1016/j.apsb.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H., Agarwal P., Zhao G., Ji G., Jewell C.M., Fisher J.P., et al. Overcoming ovarian cancer drug resistance with a cold responsive nanomaterial. ACS Cent Sci. 2018;4:567–581. doi: 10.1021/acscentsci.8b00050. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Yu G.C., Zhao X.L., Zhou J., Mao Z.W., Huang X.L., Wang Z.T., et al. Supramolecular polymer-based nanomedicine: high therapeutic performance and negligible long-term immunotoxicity. J Am Chem Soc. 2018;140:8005–8019. doi: 10.1021/jacs.8b04400. [DOI] [PubMed] [Google Scholar]
- 36.Wang S., Yu G., Wang Z.T., Jacobson O., Tian R., Lin L.S., et al. Hierarchical tumor microenvironment-responsive nanomedicine for programmed delivery of chemotherapeutics. Adv Mater. 2018 doi: 10.1002/adma.201803926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S.K., Zou Q.L., Li Y.X., Yuan C.Q., Xing R.R., Yan X.H. Smart peptide-based supramolecular photodynamic metallo-nanodrugs designed by multicomponent coordination self-assembly. J Am Chem Soc. 2018;140:10794–10802. doi: 10.1021/jacs.8b04912. [DOI] [PubMed] [Google Scholar]
- 38.Gaforio J.J., Serrano M.J., Algarra I., Ortega E., Cienfuegos GA d. Phagocytosis of apoptotic cells assessed by flow cytometry using 7-aminoactinomycin D. Cytometry. 2002;49:8–11. doi: 10.1002/cyto.10136. [DOI] [PubMed] [Google Scholar]
- 39.Asampille G., Verma B.K., Swain M., Shettar A., Rosenzweig S.A., Kondaiah P., et al. An ultra-stable redox-controlled self-assembling polypeptide nanotube for targeted imaging and therapy in cancer. J Nanobiotechnol. 2018;16:101. doi: 10.1186/s12951-018-0427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh D., Vishnoi T., Kumar A. Effect of alpha-ketoglutarate on growth and metabolism of cells cultured on three-dimensional cryogel matrix. Int J Biol Sci. 2013;9:521–530. doi: 10.7150/ijbs.4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pasparakis G., Manouras T., Vamvakaki M., Argitis P. Harnessing photochemical internalization with dual degradable nanoparticles for combinatorial photo-chemotherapy. Nat Commun. 2014;5:3623. doi: 10.1038/ncomms4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H., Agarwal P., Xiao Y.C., Peng H., Zhao S.T., Liu X.Y., et al. A nano-in-micro system for enhanced stem cell therapy of ischemic diseases. ACS Cent Sci. 2017;3:875–885. doi: 10.1021/acscentsci.7b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan L., Zhang S.L., Zhang C.Y., Yin C., Chu Z.Q., Song C.J., et al. Multidrug resistance in cancer circumvented using a cytosolic drug reservoir. Adv Sci (Weinh) 2018;5:1700289. doi: 10.1002/advs.201700289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang L.N., Sun J., Liu Q., Zhu R.R., Yang Q.N., Hua J.H., et al. Synergetic functional nanocomposites enhance immunotherapy in solid tumors by remodeling the immunoenvironment. Adv Sci (Weinh) 2019;6:1802012. doi: 10.1002/advs.201802012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chauhan K., Hernandez-Meza J.M., Rodriguez-Hernandez A.G., Juarez-Moreno K., Sengar P., Vazquez-Duhalt R. Multifunctionalized biocatalytic P22 nanoreactor for combinatory treatment of ER+ breast cancer. J Nanobiotechnol. 2018;16:17. doi: 10.1186/s12951-018-0345-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H.X., Chen J.M., Xu C., Shi L.L., Tayier M., Zhou J.H., et al. Cancer nanomedicines stabilized by pi–pi stacking between heterodimeric prodrugs enable exceptionally high drug loading capacity and safer delivery of drug combinations. Theranostics. 2017;7:3638–3652. doi: 10.7150/thno.20028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y.K., Li Y.H., Zhang X., Xu X.H., Zhang Z.J., Hu C., et al. Supramolecular PEGylated dendritic systems as pH/redox dual-responsive theranostic nanoplatforms for platinum drug delivery and NIR imaging. Theranostics. 2016;6:1293–1305. doi: 10.7150/thno.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amit M., Abdelmeguid A.S., Watcherporn T., Takahashi H., Tam S., Bell D., et al. Induction chemotherapy response as a guide for treatment optimization in sinonasal undifferentiated carcinoma. J Clin Oncol. 2019;37:504–512. doi: 10.1200/JCO.18.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.