Abstract
BACKGROUND AND AIMS:
Cirrhosis is associated with changes in intestinal microbiota that can lead to hepatic encephalopathy (HE) and infections, especially with antibiotic-resistant organisms. However, the impact of gut microbial antibiotic resistance gene (ARG) burden on clinical outcomes is unclear. The aims of the study were to determine the impact of ARGs in cirrhosis-related gut metagenome on outcomes and disease progression, study the effect of rifaximin on ARG burden, and compare ARGs in cirrhosis with chronic kidney disease (CKD) and diabetes.
METHODS:
In outpatients with cirrhosis who underwent metagenomics, we evaluated change in ARG abundances with progression and their multivariable impact on 90-day hospitalizations and deaths over 1 year. We also studied ARGs pre- and 8 weeks post-rifaximin in patients with compensated cirrhosis in an open-label trial. Finally, ARGs from CKD and diabetes studies were compared with cirrhosis on machine learning.
RESULTS:
A total of 163 patients with cirrhosis (43 compensated, 20 ascites-only, 30 HE-only, 70 both) and 40 controls were included. ARG abundances were higher in cirrhosis versus controls and worsened with advancing cirrhosis severity; 44 patients were hospitalized and 14 died. ARG abundances were associated with hospitalizations and mortality while controlling for cirrhosis complications, medications, and demographics. Rifaximin trial: ARG abundance patterns were minimally affected in 19 patients post-rifaximin. CKD/diabetes comparison: ARG abundance patterns in cirrhosis are distinguishable on machine learning and include more gram-positive ARGs.
CONCLUSIONS:
Cirrhosis is associated with high gut microbial ARG gene burden compared with controls, which worsens with disease progression and may be different from CKD and diabetes. ARGs are not affected by rifaximin and are associated with hospitalizations and death.
Keywords: Ascites, Hepatic Encephalopathy, Rifaximin, Infections, Machine Learning
Graphical Abstract
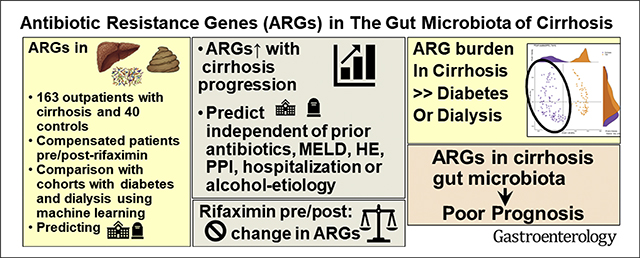
Patients with cirrhosis have a high likelihood of antibiotic resistance carriage and higher relative abundance of bacteria with pathogenic potential or “pathobionts” that is associated with poor outcomes.1–4 These poor outcomes and higher mortality could be related to hepatic encephalopathy (HE) and infections, especially with drug-resistant organisms.5,6 The summation of all antibiotic resistance genes (ARGs) and their precursors within a microbial community is termed a “resistome”; however, its clinical evaluation is currently limited to culture-based techniques.7,8 Prior studies in cirrhosis have shown altered composition and function of intestinal microbiota, which can affect clinical outcomes.9–11 With advancing cirrhosis, there is a higher relative abundance of pathobionts12 that could express ARGs.13–15 In addition, these ARGs can represent survival and quorum-sensing strategies that can predate antibiotic exposure,16 regulate the microbial ecological dynamics in the gut, and determine survival in complex multiorganismal mucosal surfaces17; however, whether these are unique to cirrhosis or are a sequelae of chronic diseases per se is unknown. The knowledge of species with ARGs that are associated with poor outcomes could be used as prognosticators and facilitate development of novel or targeted therapeutic strategies directed toward them rather than the broad-spectrum antimicrobial treatments that dominate clinical practice today.18 However, the impact of rifaximin, a nonabsorbable, gut-specific antibiotic needs to be studied in the overall metagenomic context.19
We hypothesized that ARG burden would increase with advancing cirrhosis and provide prognostic information regarding hospitalizations and death independent of disease severity and prior antibiotic exposure. We also hypothesized that unlike traditional broad-spectrum antibiotics, rifaximin therapy would not increase the ARG burden.20,21 Last, we hypothesized that the gut microbiota ARG burden is different in cirrhosis as compared with other chronic diseases associated with dysbiosis, such as chronic kidney disease on dialysis (CKD) and diabetes.
Materials and Methods
Subjects
Cross-sectional HE and hospitalization/death study.
Outpatients with cirrhosis and healthy controls between 21 and 75 years of age underwent informed consent before sample collection (Supplementary Figure 1, n = 203). Cirrhosis was defined by liver biopsy, endoscopic or radiological evidence of varices or porto-systemic shunting in chronic liver disease, frank decompensation, or through transient elastography. Patients unable to provide consent or samples, those with HIV infection, prior transplantation, those with alcohol abuse or probiotic use within the prior 8 weeks, with other organ failures (chronic kidney disease on dialysis [CKD], congestive heart failure, chronic obstructive pulmonary disease), cancer, those with other gastrointestinal diseases, or those in whom the diagnosis of cirrhosis was unclear were excluded. Patients were divided into compensated (no prior or current history of HE or ascites) and decompensated (ascites-only, HE-only, or both). Those with HE were further subdivided into those on lactulose only (Cirr-L), and those on lactulose and rifaximin (Cirr-LR). Hospitalizations and antibiotic exposures 6 months before sample collection were analyzed. Healthy controls were recruited through word of mouth and through community advertising and were all Virginia-based following a Western diet. Only individuals who were free of chronic diseases, including metabolic syndrome, autoimmunity, and intestinal disorders, and were not on prescription medications or chronic over-the-counter medications, including proton pump inhibitors (PPIs), were considered healthy controls. In addition to fecal samples, data pertaining to cirrhosis severity and concomitant medications were also recorded. All patients with cirrhosis were followed for 90 days for development of nonelective hospitalizations and for 1 year for risk of death using chart review. Details of hospitalizations were also noted. In keeping with clinical practice, all included patients were seen in clinic at least every 6 months. Based on our prior study using 16S ribosomal RNA (rRNA) and logistic regression, we were able to predict the role of microbiota on 90-day hospitalizations that occurred in 25% of subjects with 145 patients with cirrhosis.22 This was used as our minimal sample size.
Pre/post-rifaximin study.
Compensated outpatients with cirrhosis between 18 and 65 years of age without prior or current HE, with cognitive impairment or minimal HE on paper-pencil tests and without allergies to rifaximin or current/prior therapy for HE, prior episodes of HE, prior transjugular intrahepatic porto-systemic shunting, those unable to give informed consent, were recruited after informed consent in an open-label trial (Supplementary Figure 1). The clinical trial results and microbiome profile of these participants as measured by 16S rRNA gene profile are previously published.23 Cirrhosis-associated clinical details and stool for microbiota were collected at baseline. They were administered 550-mg capsules of rifaximin twice a day for 8 weeks, at which point adherence was evaluated and repeat stool sample was collected.
The protocols and biorepository were approved by the institutional review boards at the Virginia Commonwealth University and Richmond VA Medical Center, and all subjects gave written informed consent before participation for the procedures and for the biorepository. Remaining methods are in the supplement.
Analyses of ARGs
Metagenomic analyses were performed at Diversigen (www.diversigen.com) and reads after quality trimming were mapped against antimicrobial resistance (AMR) accessions available CARD v.3.1.0 (Comprehensive Antibiotic Resistance Database https://card.mcmaster.ca/).24,25 The CARD includes well-characterized, peer-reviewed resistance determinants and associated antibiotics, which is updated monthly. This database includes 88 pathogens, 9560 chromosomes, 21,362 plasmids, 102,181 whole-genome sequencing assemblies, and 222,011 alleles. The outputs are organized by the Antibiotic Resistance Ontology (ARO) and AMR gene detection models. The database also determines computer-generated resistome predictions for the sequenced genomes, plasmids, and whole-genome shotgun assemblies available at the National Center for Biotechnology Information for these 88 pathogens. These resistomes include sequence variants beyond those reported in the scientific literature, as predicted by the Resistance Gene Identifier.25
Bio-informatics Analysis
We analyzed ARG changes between compensated, HE-only, ascites-only, and patients with both on false discovery rate (FDR)-corrected Kruskal-Wallis tests. Using BiomMiner, which used DESeq2 and Kruskal-Wallis tests (FDR-corrected), we compared patients with cirrhosis with controls, then HE versus no-HE, and finally those who required hospitalizations at 90 days and death in 1 year26,27 (Supplementary Materials). We compared ARG patterns (ARO terms, resistomes, and AMR genes). Then we performed these analyses using MaAsLin2 for hospitalization and death for the ARG patterns including clinical variables such as age, gender, alcohol etiology, diabetes, PPIs, lactulose, rifaximin, decompensation status (compensated/HE-only, ascites-only, both) and model for end-stage liver disease (MELD) score.28 Separate analyses were also performed for PPI use. Finally, a similar analysis of bacterial species and ARG patterns were performed for patients pre- and post-rifaximin.29
Comparison With Papers on CKD and Diabetes
We analyzed metagenomic outputs from 2 other articles: Qin et al30 for type 2 diabetes (T2D; n = 170), and Wang et al31 for CKD on dialysis (n = 223) that had similar demographic profiles and metagenomic analyses details (Supplementary Material). Last, an Orange cross-validation modeling and prediction workflow (Supplementary Figures 2 and 3) was used to differentiate the study outputs based on these ARG patterns.32 Specifically, we developed a prediction pipeline using the Orange data mining tool to determine the predictive powers of the best performing classifiers. The ARG samples were split using a WEKA workflow into training datasets (80%) for modeling and a naïve hold-out datasets (20%) to test the predictive accuracy of the trained model. The model was trained using 5-fold cross-validation on the training dataset and then the Orange prediction function was used on each blinded sample in the naive hold-out set to classify it.
Results
Forty healthy controls and 163 patients with cirrhosis (43 compensated, 30 HE-only, 20 ascites-only, and 70 with both, Table 1) were included. When comparing vis-à-vis HE, 63 were without prior HE, and 100 had prior HE, of whom 43 were Cirr-L and 57 were Cirr-LR (Supplementary Table 1). Patients with Ascites+HE had a higher MELD and greater alcohol-related etiology, PPI use, spontaneous bacterial peritonitis (SBP) prophylaxis, lactulose and rifaximin use, and prior hospitalizations/antibiotic use compared with the rest. Demographics, dietary characteristics, and diabetes were similar. All patients were seen in clinic as standard of care at least once in 6 months before and 59 patients had required an upper endoscopy for variceal surveillance or eradication within the past year. Of the 49 people hospitalized 6 months before sample collection, most were in the ascites+HE group who were admitted a median of 1 (interquartile range 0–2) times. Most hospitalizations were due to liver-related reasons (ascites n = 13, HE n = 17, acute kidney injury n = 11, others n = 8). Exposures to antibiotics were also highest in ascites+HE, equivalent across the HE-only/ascites-only and lowest in compensated patients. Most antibiotics were administered for short courses (<14 days) within hospitalizations. The remaining were administered for outpatient urinary tract infections or suspected upper respiratory tract infections. None were diagnosed with Clostridioides difficile infection or required vancomycin; 14 patients received fluoroquinolones, 16 received cephalosporins, 3 amoxicillin-clavulanate, 3 metronidazole, 3 macrolides, and 3 trimethoprim-sulfamethoxazole. Patients with HE had a greater rate of PPI use and alcoholic etiology of cirrhosis and were more likely to be men (Supplementary Table 1). Age and diabetes prevalence were similar regardless of HE/no-HE. When Cirr-L and Cirr-LR groups were compared, we did not find significant differences on demographics, PPI use, MELD score, or alcohol-related etiology. All participants were Virginia-based and were on similar Western diet and on 7-day dietary recall had similar caloric intake (Table 1). On follow-up, 44 patients were hospitalized over 90 days and 14 died over 1 year (details later in this article). A separate group of patients with compensated cirrhosis were included in the rifaximin trial (Supplementary Figure 1).
Table 1.
Characteristics of Subject Groups
| All cirrhosis (n = 163) |
||||||
|---|---|---|---|---|---|---|
| Decompensated cirrhosis (n = 120) |
||||||
| Controls (n = 40) | Compensated (n = 43) | HE only (n = 30) | Ascites only (n = 20) | Both (n = 70) | P value | |
|
| ||||||
| Age (y) | 58.6±10.3 | 60.3±7.2 | 61.0±11.4 | 59.2±10.1 | 60.1±8.9 | .12 |
| White/Black/Other | 26/13/1 | 31/12/0 | 23/7/0 | 15/5/0 | 49/18/3 | .85 |
| Latinx/Not | 5/35 | 2/41 | 3/27 | 2/18 | 7/63 | .46 |
| Gender (male) | 24 (60) | 36 (84) | 15 (50) | 16 (80) | 59 (84) | .30 |
| Proton pump inhibitor | 0 (0) | 16 (37) | 14 (47) | 12 (60) | 49 (70) | <.0001 |
| Type 2 diabetes | 0 (0) | 22 (51) | 9 (30) | 7 (35) | 23 (33) | .35 |
| Hospitalized 6 mo prior | 0 (0) | 3 (7) | 6 (20) | 2 (10) | 38 (54) | <.0001 |
| Antibiotic exposure 6 mo prior | 0 (0) | 3 (7) | 5 (17) | 4 (20) | 32 (46) | <.0001 |
| MELD score | NA | 8.3±2.6 | 9.7±3.4 | 12.4±4.5 | 14.4±4.8 | <.0001 |
| Lactulose | NA | 0 (0) | 30 (100) | 0 (0) | 70 (100) | <.0001 |
| Rifaximin | NA | 0 (0) | 16 (53) | 0 (0) | 41 (59) | <.0001 |
| SBP prophylaxis | NA | 0 (0) | 0 (0) | 1 (5) | 6 (9) | 1.0a |
| Alcohol-related etiology | NA | 7 (16) | 8 (27) | 6 (30) | 35 (50) | <.0001 |
| Daily caloric intake | 2229±239 | 2120±421 | 2094±406 | 2010±522 | 2151±510 | .57 |
| Future hospitalizations in 90 d | 0 (0) | 2 (5) | 2 (10) | 6 (30) | 34 (48) | <.0001 |
| Death in 1 y | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 14 (20) | <.0001 |
NOTE. Data presented as raw number (%) or mean±SD. ANOVA, χ2 tests as appropriate.
MELD, model for end-stage liver disease; NA, not applicable; SBP, spontaneous bacterial peritonitis.
Comparison within both/ascites only using Fisher’s exact test.
Cirrhosis Is Associated With Greater Burden of ARGs Relative to Healthy Controls
Bacterial species that were highest in controls, which reduced over the disease spectrum, belonged to Faecali-bacterium, Alistipes, Eubacterium, and other short-chain fatty acids producers, such as Dorea, Subdoligranulum, and Roseburia (Supplementary Table 2). Patients with cirrhosis had greater abundance of resistomes associated with pathobionts belonging to Enterobacteriaceae, as well as Streptococcus, Enterococcus, and Acinetobacter spp (Supplementary Tables 3–5). These resulted in greater abundance of resistance patterns focused on beta-lactamases, macrolide, quinolone, glycopeptide, fosfomycin, and tetracycline resistance, and those focused on generic AMR pathways compared with controls (Supplementary Tables 3–5, Figure 1). There was a statistically significant difference in the species distribution based on the Bray-Curtis permutational multivariate analysis of variance (PERMANOVA) analysis (P = .001).
Figure 1.
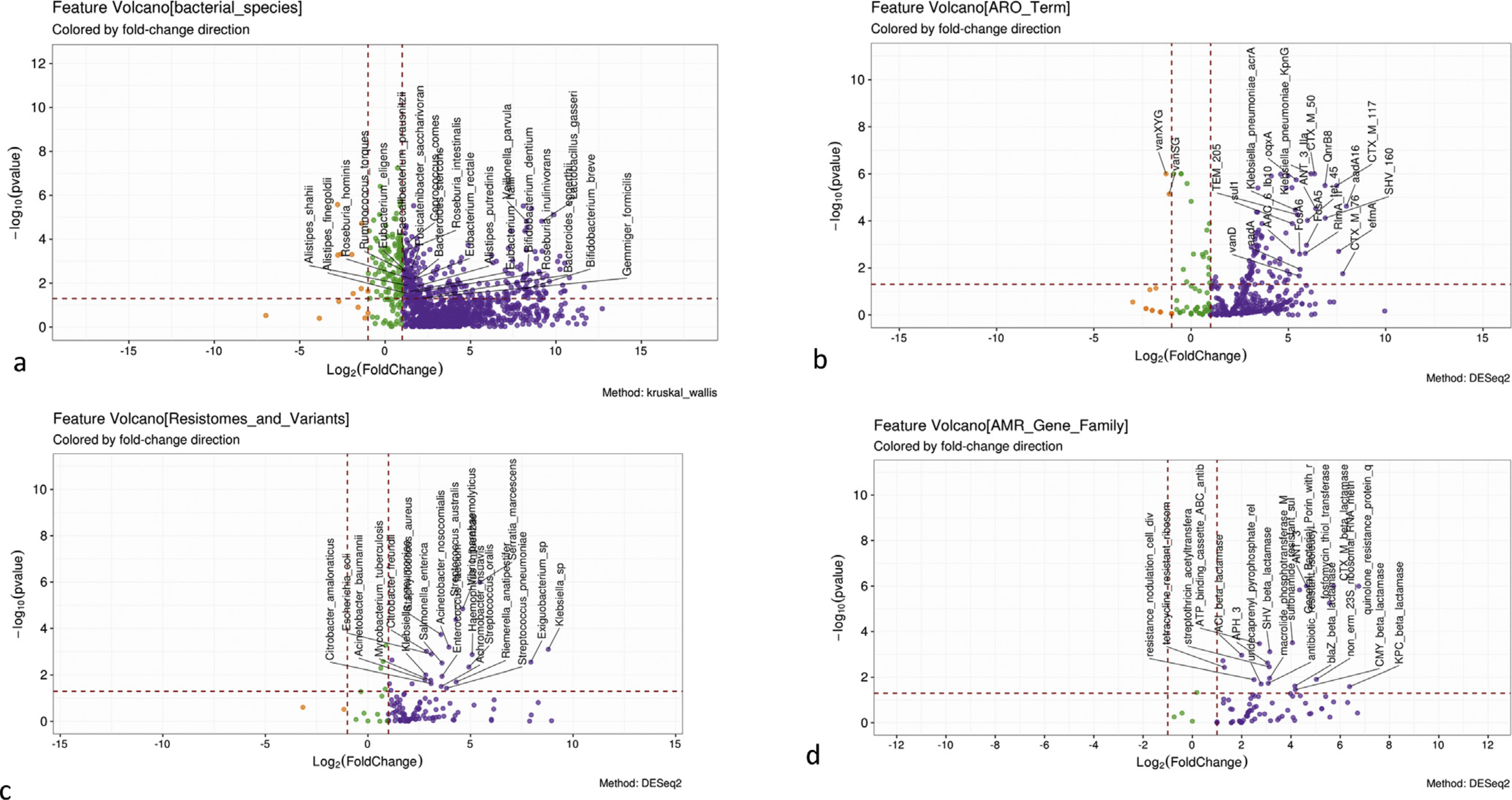
Comparison of healthy controls and cirrhosis. For all comparisons, purple is higher in cirrhosis, orange is higher in controls. (A) Volcano plot of Kruskal-Wallis comparison of bacterial species. (B) Volcano plot of DESeq2 lineage of ARO terms. (C) Volcano plot of DESeq2 lineage of resistomes and variants. (D) Volcano plot of DESeq2 lineage of AMR gene families.
ARG Patterns Across Decompensating Events
On Kruskal-Wallis (Table 2), AMR genes higher in ascites and both belonged to aminoglycoside (ANT, APH), sulfonamide resistance and selected beta-lactamases (SHV, CTX-M, SRT) and efflux pumps. Others were found only in decompensated patients regardless of complication (porins). Aminocoumarin-resistant parY, lincosamide resistance, ileS, RpoB, and PDC beta-lactamase were higher in patients with HE regardless of ascites. These patterns were reflected in ARO terms with greater membrane fusion pump efflux complex (Mex) and Klebsiella-related genes in decompensated patients. Streptomyces spp, Lactococcus, Bifidobacterium, and Staphylococcus aureus resistomes were higher were higher in patients with HE, whereas Klebsiella and Shigella spp were lower. Ascites, regardless of HE, was associated with greater abundance of Pseudomonas, Serratia, and Clostridium perfringens.
Table 2.
Kruskal-Wallis Comparisons of ARG Patterns Between Groups
| Log-2-fold changes |
|||||
| Variables | P value | Comp | HE only | Ascites only | Both |
|
| |||||
| AMR gene family | |||||
| PDC beta-lactamase | 4.55E-04 | −0.68 | 0.48 | −1.14 | 1.34 |
| Llm 23S ribosomal RNA methyltransferase | 7.59E-04 | −0.16 | 0.73 | −0.4 | −0.16 |
| Multidrug toxic compound extrusion transporter | 9.19E-04 | −0.72 | 0.18 | 0.35 | 0.19 |
| CMY beta-lactamase | .001 | −3.43 | 1.11 | 1.33 | 1 |
| sulfonamide resistant sul | .005 | −1.41 | −0.5 | 0.95 | 0.96 |
| CTX-M beta-lactamase | .006 | −1.9 | −1.93 | 3.13 | 0.7 |
| SRT beta-lactamase | .007 | −2.74 | −2.08 | 4.12 | 0.7 |
| DHA beta-lactamase | .008 | 0.23 | −1.11 | 1.99 | −1.11 |
| APH(6) | .009 | 1.5 | −1.81 | −1.24 | 1.55 |
| ANT(2”) | .011 | −1.35 | −0.87 | 1.25 | 0.96 |
| SHV beta-lactamase | .012 | −0.95 | −1.7 | 2.57 | 0.07 |
| Aminocoumarin resistant parY | .016 | −0.81 | 1.04 | −0.68 | 0.45 |
| Major facilitator superfamily antibiotic efflux pump | .020 | −0.3 | −1.02 | 0.36 | 0.96 |
| TEM beta-lactamase | .022 | −0.11 | −0.42 | 0.13 | 0.4 |
| Antibiotic resistant isoleucyl-tRNA synthetase | .036 | −0.89 | 0.33 | −0.9 | 1.47 |
| General Bacterial Porin with ↓permeability to β-lactams | .043 | −2.21 | 0.23 | 1.23 | 0.75 |
| APH(3”) | .044 | −0.96 | −0.64 | 1.03 | 0.57 |
| Rifamycin-resistant beta-subunit of RNA polymerase (rpoB) | .045 | −1.09 | 1.88 | −0.88 | 0.1 |
|
| |||||
| ARO terms | Comp | HE only | Ascites only | Both | |
|
| |||||
| arlS | .007 | −1.11 | 0.8 | −0.35 | 0.66 |
| CTX-M-141 | .013 | −1.97 | 2.81 | −1.97 | 1.13 |
| MexE | .013 | −1.21 | 0.55 | 0.12 | 0.55 |
| Klebsiella pneumoniae KpnE | .016 | −1.37 | 0.94 | 0.39 | 0.04 |
| vanSG | .016 | 1.06 | 1.06 | −1.06 | −1.06 |
| cdeA | .022 | −0.69 | 0.52 | 0.11 | 0.07 |
| CTX-M-3 | .023 | −0.33 | 1 | −0.33 | −0.33 |
| MexJ | .024 | −0.65 | 0.25 | 0.29 | 0.68 |
| SHV-126 | .027 | −0.08 | −0.26 | 0.12 | 0.22 |
| MexC | .030 | −0.58 | 0.39 | 0.91 | 1.09 |
|
| |||||
| Resistomes and variants | Comp | HE only | Ascites only | Both | |
|
| |||||
| Staphylococcus aureus | .002 | −0.78 | 0.35 | −1.17 | 1.6 |
| Klebsiella sp | .002 | −0.44 | −0.44 | 1.33 | −0.44 |
| Pseudomonas fluorescens | .003 | −0.35 | −0.1 | 0.09 | 0.36 |
| Serratia marcescens | .007 | −2.13 | −1.15 | 1.86 | 1.42 |
| Shigella flexneri | .01 | −1.58 | 0.82 | −0.21 | 0.98 |
| Lactococcus lactis | .02 | −2.32 | 1.1 | −0.21 | 1.42 |
| Streptomyces rishiriensis | .02 | −0.81 | 1.04 | −0.68 | 0.45 |
| Bifidobacterium bifidum | .03 | −0.86 | 0.32 | −0.91 | 1.45 |
| Clostridium perfringens | .04 | −0.69 | −1.74 | 2.13 | 0.3 |
| Streptomyces niveus | .04 | 0.53 | 1.21 | −1.36 | 0.38 |
NOTE. P values are FDR corrected, Comp: compensated, Log2fold changes: negative value indicates lower presence in that specific category and vice-versa.
ARG, antibiotic resistance gene; ARO, antibiotic resistance ontology.
ARG Patterns in Patients With Prior HE Compared With Those Without HE
Specific microbial changes showed lower abundance of species belonging to Lachnospiraceae and Ruminococcaceae in those with HE, whereas Streptococcus, Lactobacillus, Enterococcus, and Escherichia spp were higher in those without HE (Supplementary Table 6, Supplementary Figure 4). When ARGs were analyzed, patients with HE had greater abundance of resistomes focused on Staphylococcus, Listeria, Streptococcus, Pseudomonas, and Bifidobacterium spp relative to no-HE patients (Supplementary Tables 7–9, Supplementary Figure 4). ARG abundance of beta-lactamase, vancomycin resistance, as well as RbpA bacterial RNA polymerase binding protein were higher in HE, whereas quinolone resistance genes were higher in those without HE. These patterns were also followed when ARO terms were analyzed between patients with and without HE. Despite these changes on DESeq2, species distribution based on the Bray-Curtis PERMANOVA analysis did not show significant differences in ARG patterns (P = .11, ARO term, P = .113 AMR gene family, and P = .12 resistomes) between patients with/without HE.
ARG Patterns in Patients on PPI Compared With Those Without PPI
Because patients on PPIs had more advanced cirrhosis vs the rest (Table 1, Supplementary Table 1), we performed MaAsLin2 for ARG terms. We found that PPI use was associated with Enterococcus faecalis- and Enterococcus faecium-related genes, that is, higher Vancomycin resistance (VanYB, VanRB, VanHB, VanB) ARO terms and VanH, VanX, VanY AMR gene families. However, none of the resistomes survived FDR.
Hospitalizations
Forty-four patients needed hospitalizations at 90 days, which was most frequent in more advanced patients (Table 1). The major reasons were HE (n = 19) followed by acute kidney injury and electrolyte disturbances (n = 7), infection (n = 11), gastrointestinal bleeding (n = 3), and others (n = 9). Of the 11 infections, methicillin-resistant Staphylococcus aureus was found in 3 patients (2 bacteremia and 1 SBP), Candida spp in 3 patients, Streptococcus viridans bacteremia in 1 patient, and no organism isolated in 4 patients (cellulitis and pneumonia in 2 each). Three patients had 2 infections during the same hospitalization (SBP followed by urinary tract infections).
Bacterial species distribution based on the Bray-Curtis PERMANOVA analysis was not significantly different (P = .121) and there was no difference in the Shannon diversity between groups (2.87 ± 0.75 not hospitalized vs 2.83 ± 0.69 hospitalized, P = .178). Potentially beneficial taxa belonging to species in Ruminococcaceae and Lachnospiraceae associated with lower risk of hospitalizations (Supplementary Figure 5, Supplementary Table 10 and 11). Pathobionts belonging to Enterobacter, Pseudomonas, Yersinia, and Enterococcus spp remained associated with a greater risk of hospitalizations despite controlling for clinical factors by MAAsLin2 (Supplementary Table 11). Also, using MAAsLin2 (Table 3), aminoglycoside-2-O-nucleotidyltransferase (ANT [2]) gene, one of the most common determinants of enzyme-dependent aminoglycoside resistance prevalent in in gram-negative bacteria were associated with higher hospitalization risk, whereas generic AMR genes related to rifamycin, aminocoumarin, and lincosamide ribosomal RNA methyltransferase were associated with lower hospitalization risk. ARO term associated with hospitalizations independent of clinical factors were dfrA12, InUA, MexE, OXY beta-lactamase, and VanVB. Of these, dfrA12 is present in several pathogenic gram-negative species (Acinetobacter baumannii, Citrobacter, Enterobacter, Escherichia, Klebsiella, Proteus, Shigella, and Morganella spp), MexE is present in Pseudomonas spp, and is a multidrug efflux complex whereas OXY 1–6 beta-lactamase is found in Klebsiella spp. VanVB is a vancomycin-resistant ARG found in E faecalis, whereas InuA is a plasmid-mediated nucleotidyl-transferase found in several pathobionts belong to Enterococcus, Staphylococcus, Listeria, and Escherichia spp. These data were found in resistomes as well, where gram-negative bacteria, including Citrobacter were associated with hospitalization, whereas Streptomyces spp were protective independent of clinical factors. Regardless of whether composite of decompensation (compensated, ascites-only, HE-only, or both) or no-HE or HE were considered, the ARG analyses contribution toward hospitalizations was similar (Table 3, Supplementary Table 12).
Table 3.
Hospitalizations According to ARG Patterns Using MaAsLin2
| AMR gene family | Coefficient | SE | P value | Q value |
|
| ||||
| MELD score | 0.412904688 | 0.064851856 | 2.00E-09 | 2.18E-07 |
| Composite score of decompensation | 0.69654277 | 0.124371937 | 9.26E-08 | 5.05E-06 |
| ANT(2__) | 3.962029525 | 0.965979207 | 6.55E-05 | 0.002 |
| Lactulose use | 0.554979205 | 0.161860073 | .000773425 | 0.021 |
| Llm_23S_ribosomal_RNA_methyltransferase | −1.362069117 | 0.446233892 | .002664409 | 0.028 |
| Aminocoumarin resistant parY | −0.667160276 | 0.23270564 | .004709751 | 0.029 |
| SBP prophylaxis | 1.949305738 | 0.823698373 | .019167732 | 0.045 |
| Rifamycin resistant beta subunit of RNA polymerase (rpoB) | −0.563369541 | 0.234559872 | .01747475 | 0.048 |
|
| ||||
| ARO term | Coefficient | SE | P value | Q value |
|
| ||||
| MELD score | 0.412904688 | 0.064851856 | 2.00E-09 | 2.48E-06 |
| Composite score of decompensation | 0.69654277 | 0.124371937 | 9.26E-08 | 5.73E-05 |
| dfrA12 | 5.219724894 | 1.174708864 | 1.66E-05 | 0.0068 |
| aadA9 | 3.466531159 | 0.841747734 | 6.14E-05 | 0.0115 |
| ANT(2__) | 3.962029525 | 0.965979207 | 6.55E-05 | 0.0115 |
| lnuA | 3.758216836 | 0.892344383 | 4.25E-05 | 0.0115 |
| MexE | 5.602615552 | 1.356726994 | 5.87E-05 | 0.0115 |
| cfrC | 5.603541801 | 1.423029531 | .000123069 | 0.019 |
| CMY_48 | 5.525229197 | 1.422226328 | .000150225 | 0.0207 |
| vanVB | 3.698116695 | 0.992895908 | .000271825 | 0.034 |
| OXY_1_6 | 5.02332093 | 1.359142234 | .000301604 | 0.035 |
| OXY_1_4 | 4.706700245 | 1.288924643 | .000353835 | 0.037 |
|
| ||||
| Resistomes | Coefficient | SE | P value | Q value |
|
| ||||
| MELD score | 0.412904688 | 0.064851856 | 2.00E-09 | 3.60E-07 |
| Composite score of decompensation | 0.69654277 | 0.124371937 | 9.26E-08 | 8.34E-06 |
| Clostridium_botulinum | 5.982816577 | 1.341559342 | 1.55E-05 | 0.0009 |
| Lactulose use | 0.554979205 | 0.161860073 | .000773425 | 0.035 |
| Gram_negative_bacterium | 4.238461957 | 1.356837413 | .002124458 | 0.04 |
| Citrobacter_freundii | 1.048760055 | 0.359106171 | .004006676 | 0.042 |
| Nocardia_farcinica | −0.601249962 | 0.207076231 | .004218298 | 0.043 |
| Streptomyces_niveus | −1.483564927 | 0.518466411 | .004788398 | 0.045 |
| Streptomyces_rishiriensis | −0.66716 | 0.232706 | .00471 | 0.048 |
NOTE. Composite score of decompensation: 0 = compensated, 1 = HE only, 2 = Ascites only, 3 = both HE and ascites.
AMR, antimicrobial resistance; ARG, antibiotic resistance gene; ARO, antibiotic resistance ontology; HE, hepatic encephalopathy; MELD, model for end-stage liver disease; SBP, spontaneous bacterial peritonitis.
Deaths Over 1 Year
Fourteen patients died; all of whom had both HE and ascites. All deaths occurred due to liver-related reasons: infections in 10, variceal bleeding in 2, and the rest with cancer. As shown in Supplementary Table 13, patients who died were more likely to have pathobionts (Pseudomonas, Serratia, Klebsiella, Proteus spp) with Lactobacillus spp, and relatively lower autochthonous taxa (Lachnospira spp, Prevotella copri; Supplementary Figure 5). Bray-Curtis PERMANOVA analysis demonstrated P = .05 for bacterial species between those who died vs survived but no changes in Shannon diversity were seen (survived 2.86 ± 0.71 vs died 2.77 ± 0.89, P = .75). Microbial changes focused on Clostridium, Pseudomonas, Lactobacillus, and Neisseria spp were associated with death, whereas Streptococcus, Veillonella, Lachnospiraceae, and Bacteroides spp were associated with protection on MAAslin2 (Supplementary Table 14).
Similar to hospitalizations, ANT (2”) was the AMR gene associated with death (Table 4). ANT, MexE/M, and dfrA12 were higher in those who died, with beta-lactamase (TEM), TriB (P aeruginosa), LnUp (Lincosamide resistance), tetS (tetracycline ribosomal protection protein in E faecalis), and tet(B) present in gram-negative bacteria (Supplementary Table 15). Only CfxA6, which is a beta-lactamase from an uncultured bacterium, was associated with lower death. As expected, MELD score and greater decompensation were associated with death. When resistomes were considered, MELD score, HE, and lactulose use were associated with death, along with Legionella, gram-negative bacteria, and Enterobacter spp. Firmicutes members belonging to Desulfitobacterium spp were protective. Similar to hospitalizations, ARG patterns associated with death were similar regardless of whether composite of decompensation or no-HE or HE were considered (Table 4, Supplementary Table 15).
Table 4.
Death According to ARG Patterns Using MaAsLin2
| AMR gene family | Coefficient | SE | P value | Q value |
|
| ||||
| ANT(2) | 4.947526707 | 0.800392205 | 5.20E-09 | 5.66E-07 |
| MELD score | 0.486367013 | 0.106357407 | 9.66E-06 | 0.0005 |
| Composite score of decompensation | 0.686274301 | 0.164688078 | 5.06E-05 | 0.002 |
|
| ||||
| ARO term | Coefficient | SE | P value | Q value |
|
| ||||
| TriB | 17.01623506 | 1.918959351 | 1.51E-15 | 1.87E-12 |
| ANT(2__)_Ia | 4.947526707 | 0.800392205 | 5.20E-09 | 2.14E-06 |
| MexE | 6.358167931 | 1.01571941 | 3.48E-09 | 2.14E-06 |
| dfrA12 | 6.455393429 | 1.057135985 | 7.60E-09 | 2.35E-06 |
| cfrC | 6.839210335 | 1.269810454 | 2.57E-07 | 5.30E-05 |
| tetS | 2.932463416 | 0.541967397 | 2.29E-07 | 5.30E-05 |
| MELD score | 0.486367013 | 0.106357407 | 9.66E-06 | 0.0017 |
| AAC(6_)_Ia | 4.948180187 | 1.107617659 | 1.50E-05 | 0.0023 |
| tet(B) | 2.384584309 | 0.54127897 | 1.94E-05 | 0.0027 |
| Composite score of decompensation | 0.686274301 | 0.164688078 | 5.06E-05 | 0.006 |
| LnuP | 3.651573402 | 0.871787447 | 4.65E-05 | 0.006 |
| CfxA6 | −3.140165729 | 0.852606443 | .000316043 | 0.03 |
| TEM_48 | 4.580083552 | 1.254330205 | .000354129 | 0.03 |
| mexM | 3.608731994 | 1.005280679 | .000441217 | 0.039 |
| TEM_219 | 1.776788748 | 0.500650074 | .000509585 | 0.042 |
| ACT_38 | 3.59641316 | 1.026742741 | .000599018 | 0.046 |
| mdsB | 2.13115354 | 0.61161402 | .000638267 | 0.046 |
|
| ||||
| Resistomes | Coefficient | SE | P value | Q value |
|
| ||||
| MELD score | 0.486367013 | 0.106357407 | 9.66E-06 | 0.002 |
| Composite score of decompensation | 0.686274301 | 0.164688078 | 5.06E-05 | 0.004 |
| Legionella pneumophila | 2.128854381 | 0.574804015 | .000293338 | 0.014 |
| Gram negative bacterium | 3.925435914 | 1.152970009 | .000839916 | 0.03 |
| Desulfitobacterium hafniense | −3.209096819 | 0.988378431 | .001425081 | 0.043 |
NOTE. Composite score of decompensation: 0 = compensated, 1 = HE only, 2 = Ascites only, 3 = both HE and ascites.
AMR, antimicrobial resistance; ARG, antibiotic resistance gene; ARO, antibiotic resistance ontology; HE, hepatic encephalopathy; MELD, model for end-stage liver disease.
Pre/post-Rifaximin Trial Did Not Show Major Changes in ARG Patterns
Rifaximin was well-tolerated and safe as published.23 One sample could not be located so we analyzed samples of 19 subjects. Shannon diversity or beta-diversity in bacterial species (PERMANOVA P = .199) was not changed from baseline. On DESeq2, post-rifaximin there was a significant increase in autochthonous species such as Blautia, Butyricimonas, Lactobacillus, and Eubacterium and lower E faecium and Clostridium scindens after rifaximin therapy (Figure 2). Rifaximin did not significantly change AMR gene abundance and only reduced Klebsiella oxytoca resistome abundance. On ARO terms, there was a reduction in gram-negative resistance patterns (Escherichia coli acrA, marA, and H_NS, which are involved in antibiotic efflux) and beta-lactamases (SRT2 and TEM219). Only VanI marginally increased post-rifaximin.
Figure 2.
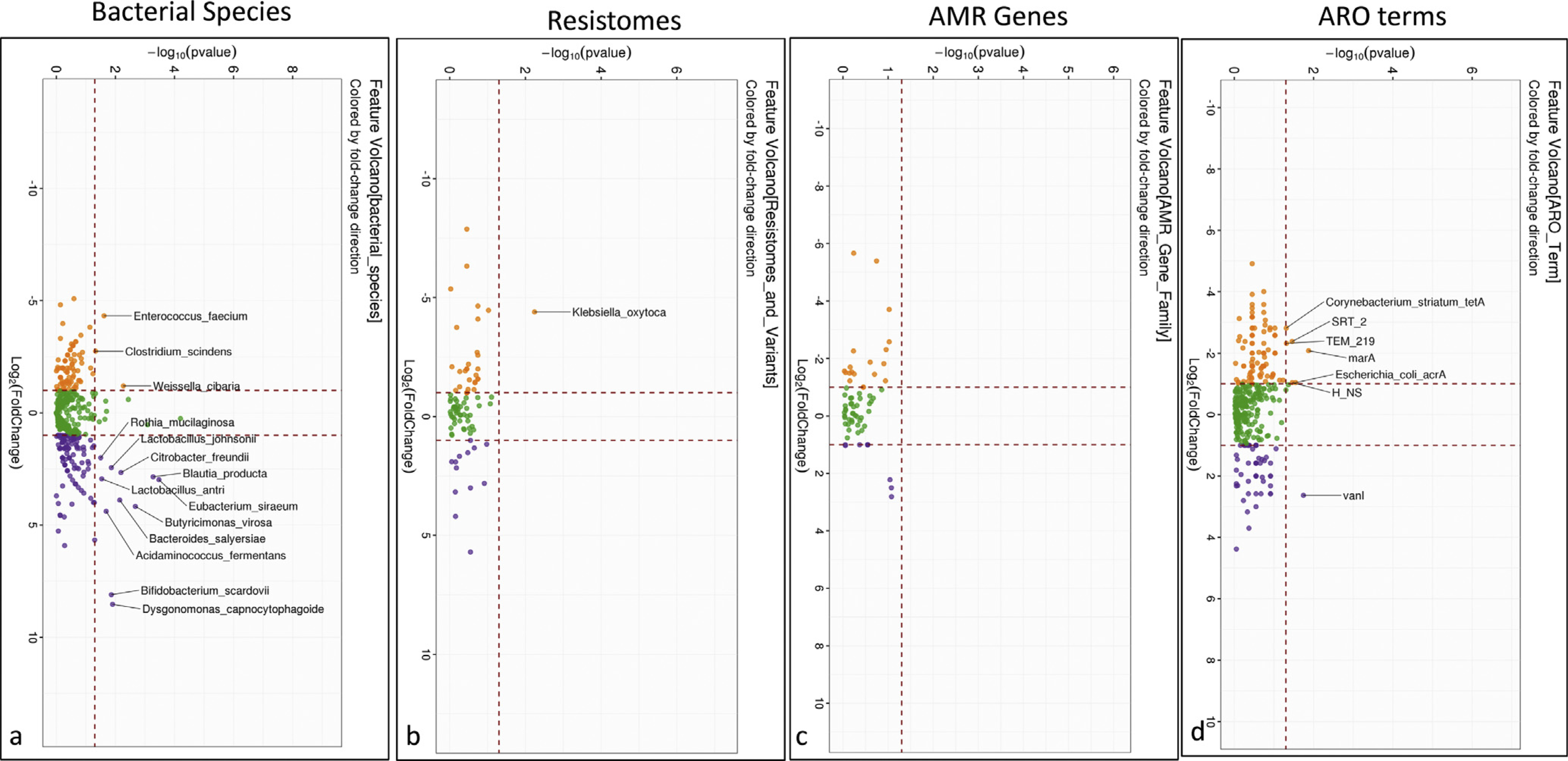
Comparison of microbial and ARG changes before and after rifaximin. (A) Volcano plot of DESeq2 lineage of bacterial species compared between pre- (orange) and post-rifaximin (purple). (B) Volcano plot of Kruskal-Wallis comparison of resistomes that changed between pre- (orange) and post-rifaximin (purple) showing higher Klebsiella oxytoca pre, which was not found post-rifaximin. (C) Volcano plot of Kruskal-Wallis comparison of ARO terms that changed between pre- (orange) and post-rifaximin (purple) showing no significant change between the time-points. (D) Volcano plot of Kruskal-Wallis comparison of AMR gene families that changed between pre- (orange) and post-rifaximin (purple) showed reduction in baseline AMR gene expressions after rifaximin.
Cirrhosis Is Associated With Higher Burden of ARGs Relative to CKD, and Diabetes, Whereas Controls Are Largely Similar
Type 2 diabetes.
Qin et al30 studied 170 Chinese patients with T2D with similar age as our patients. Figure 3 shows a significant separation on principal coordinates analysis between the groups, which were significant on PERMANOVA (all P < .001) for ARG pattern comparisons. This separation was maintained even when patients with cirrhosis with or without diabetes were compared with patients with diabetes alone (Supplementary Figures 6 and 7). Patients with cirrhosis had a higher number of ARGs compared with diabetes, with resistomes being higher in cirrhosis belonging to a wide range of gram-positive and negative microbes with pathogenic potentials, whereas patients with diabetes had a relatively narrower range of resistome representation (Supplementary Figures 8 and 9, Supplementary Tables 16–18). AMR gene families and ARO terms were relatively similarly spread between the 2 conditions spanning vancomycin, beta-lactamase, and quinolone resistance. There were only 6 ARGs different between the cirrhosis controls and the T2D controls (Supplementary Figure 10A).
Figure 3.
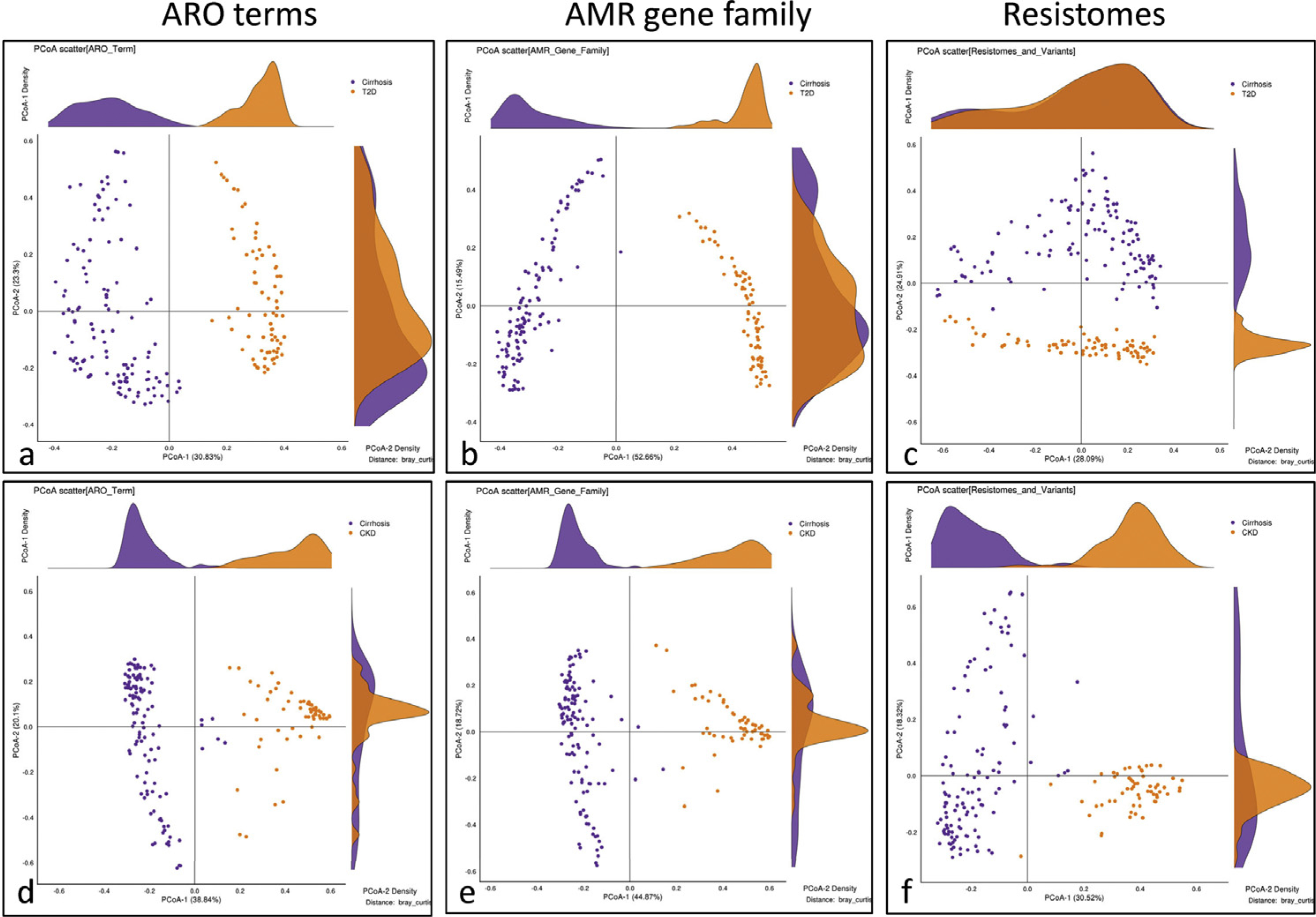
Principal coordinate analysis of cirrhosis compared to other chronic diseases. (A-C) Comparison of cirrhosis (purple) with T2D (orange, Qin et al30) showing clear separation between the groups on resistome, AMR gene family, and ARO term abundances. (D-F) Comparison of cirrhosis (purple) with CKD on dialysis (orange, Wang et al31) showing clear separation between the groups on resistome, AMR gene family, and ARO term abundances.
Chronic kidney disease.
Wang et al31 studied 223 patients with CKD on dialysis. The authors excluded recent antibiotic use and those with major nonrenal diseases including liver disease. Although patients were from China, their demographics and nonvegetarian diet intake were largely similar to our cohort. As shown in Figure 3, there was a significant separation on principal coordinates analysis between cirrhosis and CKD with PERMANOVA P < .001 for all 3 comparisons. Compared with CKD, patients with cirrhosis had a greater number of AMR, ARO, and resistome log-fold changes (Supplementary Figures 8 and 9, Supplementary Tables 19–21). Despite this, several important pathobionts were higher in CKD, such as Klebsiella pneumoniae, Acinetobacter, Enterobacter, and Legionella. Patients with cirrhosis had higher Escherichia, Staphylococcus, Enterococcus, C difficile, Klebsiella oxytoca, and Streptomyces spp. Reflecting these, there were beta-lactamase genes in both conditions but glycopeptide, vancomycin, cephalosporinase, and rifamycin resistance genes were higher in cirrhosis. Patients with CKD also had greater ARO abundances belonging to a broad spectrum of gram-negative and -positive pathobionts. There were only 7 ARGs different between the cirrhosis controls and the CKD controls, indicating minimal confounders (Supplementary Figure 10B). On Kruskal-Wallis analyses, vancomycin and efflux pumps were seen higher in cirrhosis on AMR and ARO terms (Supplementary Figures 11 and 12). Several ARO terms belonging to multidrug efflux pumps along with beta-lactamases, macrolide, aminoglycoside, quinolone, and tetracycline resistance were uniquely higher in cirrhosis. Resistomes higher in cirrhosis were gram-negative pathobionts, Streptococcus spp, and C difficile. Streptomyces spp were also increased in cirrhosis compared with diabetes and CKD (Supplementary Figure 12).
Naïve machine-learning prediction model.
The average accuracy for all the naïve samples was for each class is presented in Supplementary Table 22 and Supplementary Figure 13). For ARO terms, AMR gene families and resistomes, there was an excellent separation from samples derived from our patients compared with the other study outputs based on models created. However, random forest was the best method to separate the groups and true positivity rate in cirrhosis on the naive samples was 97.9% for AMR gene families, 99% for ARO terms, and 100% for resistomes. It should be noted that the blinded naïve samples are not used in the model training and thus represent the true accuracy of the prediction model and could be used as an accurate diagnostic of new naïve samples.
Discussion
The results of the current study demonstrate that ARG abundances are higher in cirrhosis compared with healthy controls, increase with worsening disease regardless of ascites and HE, and unlike previously described with absorbable antibiotics, are not affected by rifaximin therapy. We also found that greater abundance of ARG is related to hospitalizations and death independent of cirrhosis severity, prior antibiotic exposure, hospitalizations, or concomitant medications. Moreover, the ARG profile of cirrhosis is distinct compared with outputs from 2 articles studying CKD and diabetes.
The underlying immune deficits, exaggerated inflammatory response, liver dysfunction, and multiple hospitalizations make cirrhosis a prime candidate for suffering these negative consequences.6,33 Therefore, the carriage rate and potential impact and determinants of ARGs needs to be defined in cirrhosis. Metagenomic and 16S rRNA gene analyses have consistently demonstrated a higher proportion of pathobionts in cirrhosis compared with controls, which worsen with progression of disease.1,9,10,12,34–36 However, not all strains of potential pathobionts have ARGs associated with them, which is why we focused on those in the CARD database that have ARG genes mapped.
We confirmed prior metagenomic studies of patients with cirrhosis and determined that the relative abundances of pathobionts were higher compared with controls and worsened with advancing cirrhosis complexity. We extended prior studies by defining AMR gene families and their corresponding resistomes that were associated with this progression. In cirrhosis compared with controls, there was a higher abundance of beta-lactamase, vancomycin resistance, and quinolone resistance. This trend worsened with development of ascites, HE, and progression of cirrhosis.37 As expected, decompensated patients had relatively higher ARO and AMR gene abundance, along with resistomes belonging to pathobionts. These could be due to exposure to the health care environment, because over the past 6 months they had been hospitalized and/or exposed to antibiotics or because of the greater abundance of organisms with these genes that may be used as a survival mechanism independent of antibiotics.16
In addition, most of the decompensated patients were on rifaximin and some on SBP prophylaxis. This is important because unlike a prior study in which ciprofloxacin, amoxicillin, and metronidazole exposure over 5 days significantly increased the ARG burden,21 the use of rifaximin per se was not associated with this. This confirms and extends prior studies of rifaximin that demonstrate a low resistance footprint into the cirrhosis realm as well.38,39 We also found that rifaximin was associated with greater abundance of potentially beneficial taxa, and reduction in resistomes of Klebsiella spp as well as gram-negative ARG abundance in the small trial. This was reiterated by finding similar changes in cross-sectional subjects with HE-only or HE+ascites patients who showed lower Shigella and Klebsiella and higher Streptomyces resistomes compared with ascites-only patients. The Streptomyces spp resistome increase in cirrhosis with HE, and in cirrhosis compared with CKD and diabetes is interesting because these organisms are the source of rifamycin, from which rifaximin is derived.
The potential beneficial effect of rifaximin against hospitalizations that has been noted in HE and other gut-derived outcomes, such as SBP, could be the reason why Streptomyces resistomes,40 even though higher in cirrhosis and HE, were associated with protection from hospitalization.20,41 The favorable effect on gram-negative resistomes with rifaximin could potentially be one of the reasons behind the reduction in hospitalizations because of HE and potentially other complications of cirrhosis with rifaximin use, and association with protection against C difficile infection42 and traveler’s diarrhea.43 It also clarifies that the worsening ARG burden with cirrhosis progression reflects the underlying disease process and is not a rifaximin-related epiphenomenon.
The occurrence of ARGs could be due to exposure to health care systems and antibiotics and/or the selection of these genes as a means to enhance trans-kingdom and quorum-sensing communications.16 These are supported by studies in antibiotic-unexposed and natural systems and in organisms exposed to sub-MIC antibiotic concentrations of antibiotics show ARG expressions that are distinct from the effect seen after exposure to adequate antibiotic concentrations.40,44–46 Therefore, despite controlling for prior antibiotic exposure and hospitalizations, ARG patterns were associated with poor outcomes in cirrhosis but most of these outcomes were not antibiotic-resistant infections. However, the gut remains a major reservoir of these organisms.15,47 Unique patterns for HE and ascites-related ARG carriage and PPI use were found but on multivariable analysis, several of the genes found to be higher in decompensated patients on Kruskal-Wallis tests were also associated with hospitalizations and death. Prominently aminoglycoside resistance (ANT2) and membrane fusion pump efflux complex (MexE) found in gram-negative taxa were higher in those with negative outcomes while those potentially associated with rifaximin use (rpoB) and Streptomyces spp were protective against negative outcomes. Therefore, the presence of specific ARGs are additive prognosticators of a hostile gut milieu that can predict negative infectious and noninfectious outcomes despite controlling for clinical factors.
The focus on cirrhosis is necessitated by the comparison with several other diseases that are often comorbid or complicate the course of this disease. Diabetes is often found in cirrhosis, and cirrhosis can result in renal impairment and requirement for dialysis.48 None of our patients with cirrhosis were on dialysis. Notwithstanding differences in cohorts, the greater ARG burden in cirrhosis as well as major separation between the CKD compared with cirrhosis likely reflects the major role of liver in the regulation of the gut-liver axis and gastrointestinal immune response.49 Our finding of higher gram-positive resistomes and vancomycin resistance ARGs in cirrhosis extends prior studies of alcohol-related liver disease into the cirrhosis realm and could reflect the key role of the liver in clearing grampositive bacterial translocation.49–51 The higher ARG burden in cirrhosis vs CKD is striking because these conditions are associated with high use of antibiotics, impaired systemic immune response,52,53 and high carriage of resistant organisms.54,55 We found that ARG patterns were different between cirrhosis and diabetes but, unlike that in CKD, this was spread out. The frequent coexistence of diabetes and cirrhosis may make this differentiation less relevant patho-physiologically.48 Regardless of the comparison, we found a unique signature of ARGs in cirrhosis consisting of several gram-negative rods and C difficile and Streptococcus spp that are associated with infections and poor prognosis.56–58
These findings as well as the contribution of ARGs toward negative outcomes in cirrhosis demonstrate that this burden is clinically relevant and could be harnessed to enhance prognostication. Therapies that beneficially modulate the gut microbiota, such as fecal microbiota transplant, can reduce the ARG burden in patients with and without cirrhosis.18,21 Therefore, focusing on patients with cirrhosis that have a high ARG burden can not only improve the prognostication but also potentially select them for therapeutic options.
Our study is limited by the cross-sectional sampling, relatively small number of patients pre/post-rifaximin, and using previously published metagenomic datasets for comparison. However, our recent short-term longitudinal follow-up of patients over 15 to 30 days who were randomized to placebo or standard-of-care arms in fecal microbiota transplant trials did not show appreciable changes in ARG abundance.21 Also, several factors such as HE, prior or current antibiotics, PPIs, and other medications can affect the ARG burden. We controlled for these using individual comparisons, FDR, and multivariable analyses and found consistent changes across groups. This increases confidence in the generalization of these results into the clinical population, which often have all these factors as part of their treatment regimen. Although patients in CKD and diabetes studies were from China, the use of metagenomic libraries was similar and their demographics and other data were largely comparable to our dataset and there were few differences between their controls and ours. This was like earlier US and China data on 16S rRNA sequencing in cirrhosis vs controls59,60; however, systematic differences, including diet and socio-cultural impacts cannot be excluded. Finally, these data demonstrate association but not causation of the role of ARGs in disease progression.
We conclude that patients with cirrhosis have a high burden of ARGs compared with controls, which worsen with disease progression. Rifaximin modulates ARGs favorably, unlike absorbable antibiotics. ARGs focused on gram-negative rods are associated with 90-day hospitalizations and death over 1 year independent of clinical factors, which could refine prognostication. This ARG burden in cirrhosis is different and may be higher from that found in diabetes and CKD based on outputs from 2 previous studies. Strategies that focus on detection of ARGs for prognosis and predicting outcomes and targeting them for therapy in this era of rampant antibiotic overuse could improve the prognosis in cirrhosis.
Supplementary Material
Supplementary Figure 1. Flow chart of patients with cirrhosis in the cross-sectional study (S1A) and pre/post rifaximin open-label trial (S1B).
Supplementary Figure 2. Orange cross-validation modeling and prediction workflow
Supplementary Figure 3. Orange cross-validation modeling and prediction workflow.
Supplementary Figure 4. Comparison of cirrhosis with type 2 diabetes (T2D, purple) with patients with T2D alone (orange, Qin et al) showing clear separation between the groups on resistome, AMR gene family and ARO term abundances.
Supplementary Figure 5. Comparison of cirrhosis without type 2 diabetes (T2D, purple) with patients with T2D alone (orange, Qin et al) showing clear separation between the groups on resistome, AMR gene family and ARO term abundances.
Supplementary Figure 6. Top 20 log2Fold change differences of abundances between all patients with cirrhosis (purple) with patients with chronic kidney disease on dialysis (CKD, orange, Wang et al) with respect to resistome, AMR gene family and ARO term abundances.
Supplementary Figure 7. Heatmap of differences of resistome, AMR gene family and ARO term abundances comparing cirrhosis (purple) with chronic kidney disease on dialysis (CKD, orange)
Supplementary Figure 8. Cirrhosis vs CKD fold change.
Supplementary Figure 9. Cirrhosis vs CKD heatmap.
Supplementary Figure 10. Comparison of controls from Wang et al and Qin et al to our controls
Supplementary Figure 11. Top 20 log2Fold change differences of abundances between all patients with cirrhosis regardless of type 2 diabetes (T2D, purple) with patients with T2D alone with respect to resistome, AMR gene family and ARO term abundances.
Supplementary Figure 12. Heatmap of differences of resistome, AMR gene family and ARO term abundances comparing cirrhosis (purple) with type 2 diabetes (T2D, orange).
Supplementary Figure 13. Top 20 log2Fold change differences of abundance comparison between healthy controls in our study with those in Wang et al (S9A) and Qin et al (S9B) on ARO terms showed minimal differences.
WHAT YOU NEED TO KNOW.
BACKGROUND AND CONTEXT
Cirrhosis is associated with gut microbial dysbiosis and a growing burden of antibiotic-resistant infections. However, the impact of antibiotic resistance genes on cirrhosis-related outcomes is unclear.
NEW FINDINGS
Cirrhosis is associated with high burden of gut microbial antibiotic resistance genes abundance compared with controls, which worsens with disease progression and may be different from other diseases. Antibiotic resistance genes, which are impacted by most common antibiotics, are not affected by rifaximin therapy and are associated with hospitalizations and death independent of clinical factors.
LIMITATIONS
Cross-sectional analysis of cirrhosis and small sample size in patients pre- and post-rifaximin. Comparisons with other diseases based out of studies from geographically disparate populations.
IMPACT
Strategies that focus on detection of antibiotic resistance genes for prognosis and predicting outcomes, encouraging use of nonabsorbable antibiotics, such as rifaximin, and development of therapeutic strategies to limit antibiotic resistance gene burden this era of rampant antibiotic overuse could improve the prognosis in cirrhosis.
Acknowledgments
Microbial sequences, which are the basis for the figures, will be deposited in a publicly accessible database before publication. The SRA IDs are as follows; current study: PRJNA678582, Qin et al Type 2 diabetes: PRJNA422434, Wang et al chronic kidney disease: PRJNA449784. Due to institutional review board restrictions, metadata that are potentially identifiable according to US law are not available from our dataset.
Funding
This work was partly supported by VA merit review 2I0CX00176, NCATS R21TR002024, and R21TR003095.
Abbreviations used in this paper:
- AMR
antimicrobial resistance
- ARG
antibiotic resistance gene
- ARO
antibiotic resistance ontology
- CKD
chronic kidney disease
- FDR
false discovery rate
- HE
hepatic encephalopathy
- MELD
model for end-stage liver disease
- PERMANOVA
permutational multivariate analysis of variance
- PPI
proton pump inhibitor
- rRNA
ribosomal RNA
- T2D
type 2 diabetes
- SBP
spontaneous bacterial peritonitis
Footnotes
Conflict of interest
Jasmohan S. Bajaj’s institution has received grants from Bausch health (USA), Grifols, and Kaleido. No other conflicts exist.
CRediT Authorship Contributions
Amirhossein Shamsaddini, BS (Formal analysis: Equal; Methodology: Equal; Validation: Equal; Writing – review & editing: Equal).
Patrick Gillevet, PhD (Data curation: Lead; Formal analysis: Lead; Investigation: Equal; Supervision: Lead; Visualization: Equal; Writing – review & editing: Equal).
Chathur Acharya, MD (Formal analysis: Supporting; Investigation: Supporting).
Andrew Fagan, BS (Investigation: Equal; Project administration: Equal). Edith Gavis, RN (Investigation: Supporting; Project administration: Supporting; Supervision: Supporting).
Masoumeh Sikaroodi, PhD (Formal analysis: Equal; Methodology: Equal; Validation: Equal).
Sara McGeorge, BS (Formal analysis: Supporting; Investigation: Supporting; Resources: Supporting).
Alexander Khoruts, MD (Formal analysis: Supporting; Investigation: Supporting; Visualization: Equal; Writing – review & editing: Equal).
Somaya Albhaisi, MD (Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting).
Michael Fuchs, MD (Investigation: Supporting; Supervision: Supporting; Writing – review & editing: Supporting).
Richard Sterling, MD (Investigation: Supporting; Writing – review & editing: Supporting).
Jasmohan Singh Bajaj, MD (Conceptualization: Lead; Formal analysis: Equal; Funding acquisition: Lead; Supervision: Equal; Writing - original draft: Lead; Writing – review & editing: Supporting).
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2021.04.013.
References
- 1.Bajaj JS, Vargas HE, Reddy KR, et al. Association between intestinal microbiota collected at hospital admission and outcomes of patients with cirrhosis. Clin Gastroenterol Hepatol 2019;17:756–765.e3. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez J, Prado V, Trebicka J, et al. Multidrug-resistant bacterial infections in patients with decompensated cirrhosis and with acute-on-chronic liver failure in Europe. J Hepatol 2019;70:398–411. [DOI] [PubMed] [Google Scholar]
- 3.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008;453:620–625. [DOI] [PubMed] [Google Scholar]
- 4.Casadevall A. The pathogenic potential of a microbe. mSphere 2017;2:e00015–e00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bajaj JS, O’Leary JG, Tandon P, et al. Nosocomial infections are frequent and negatively impact outcomes in hospitalized patients with cirrhosis. Am J Gastroenterol 2019;114:1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tranah TH, Edwards LA, Schnabl B, et al. Targeting the gut-liver-immune axis to treat cirrhosis. Gut 2021; 70:982–994. [DOI] [PubMed] [Google Scholar]
- 7.D’Costa VM, McGrann KM, Hughes DW, et al. Sampling the antibiotic resistome. Science 2006;311:374–377. [DOI] [PubMed] [Google Scholar]
- 8.Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol 2007; 5:175–186. [DOI] [PubMed] [Google Scholar]
- 9.Bajaj JS, Heuman DM, Hylemon PB, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol 2014;60:940–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sung CM, Lin YF, Chen KF, et al. Predicting clinical outcomes of cirrhosis patients with hepatic encephalopathy from the fecal microbiome. Cell Mol Gastroenterol Hepatol 2019;8:301–318.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trebicka J, Bork P, Krag A, et al. Utilizing the gut microbiome in decompensated cirrhosis and acute-on-chronic liver failure. Nat Rev Gastroenterol Hepatol 2021;18:167–180. [DOI] [PubMed] [Google Scholar]
- 12.Qin N, Yang F, Li A, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014;513:59–64. [DOI] [PubMed] [Google Scholar]
- 13.Barger M, Blodget E, Pena S, et al. VRE in cirrhotic patients. BMC Infect Dis 2019;19:711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puchter L, Chaberny IF, Schwab F, et al. Economic burden of nosocomial infections caused by vancomycin-resistant enterococci. Antimicrob Resist Infect Control 2018;7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gargiullo L, Del Chierico F, D’Argenio P, et al. Gut microbiota modulation for multidrug-resistant organism decolonization: present and future perspectives. Front Microbiol 2019;10:1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pöntinen AK, Top J, Arredondo-Alonso S, et al. Apparent nosocomial adaptation of Enterococcus faecalis predates the modern hospital era. Nat Commun 2021; 12:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aminov RI. The role of antibiotics and antibiotic resistance in nature. Environ Microbiol 2009;11:2970–2988. [DOI] [PubMed] [Google Scholar]
- 18.Saha S, Tariq R, Tosh PK, et al. Faecal microbiota transplantation for eradicating carriage of multidrug-resistant organisms: a systematic review. Clin Microbiol Infect 2019;25:958–963. [DOI] [PubMed] [Google Scholar]
- 19.Caraceni P, Vargas V, Sola E, et al. The use of rifaximin in patients with cirrhosis [published online ahead of print January 9, 2021]. Hepatology 10.1002/hep.31708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bass NM, Mullen KD, Sanyal A, et al. Rifaximin treatment in hepatic encephalopathy. N Engl J Med 2010; 362:1071–1081. [DOI] [PubMed] [Google Scholar]
- 21.Bajaj JS, Shamsaddini A, Fagan A, et al. Fecal microbiota transplant in cirrhosis reduces gut microbial antibiotic resistance genes: analysis of two trials. Hepatol Commun 2020;5:258–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bajaj JS, Thacker LR, Fagan A, et al. Gut microbial RNA and DNA analysis predicts hospitalizations in cirrhosis. JCI Insight 2018;3:e98019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bajaj JS, Heuman DM, Sanyal AJ, et al. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS One 2013;8: e60042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia B, Raphenya AR, Alcock B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2017;45:D566–D573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alcock BP, Raphenya AR, Lau TTY, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 2020; 48:D517–D525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shamsaddini A, Dadkhah K, Gillevet PM. BiomMiner: an advanced exploratory microbiome analysis and visualization pipeline. PLoS One 2020;15:e0234860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Huttenhower Lab. MaAsLin2. Available at: https://huttenhower.sph.harvard.edu/maaslin/. Accessed February 24, 2021.
- 29.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 2009;5: e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012; 490:55–60. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Yang S, Li S, et al. Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents. Gut 2020;69:2131–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dadkhah E, Sikaroodi M, Korman L, et al. Gut microbiome identifies risk for colorectal polyps. BMJ Open Gastroenterol 2019;6:e000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adolph TE, Grander C, Moschen AR, et al. Liver-microbiome axis in health and disease. Trends Immunol 2018; 39:712–723. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Guo J, Qian G, et al. Gut dysbiosis in acute-on-chronic liver failure and its predictive value for mortality. J Gastroenterol Hepatol 2015;30:1429–1437. [DOI] [PubMed] [Google Scholar]
- 35.Sole C, Guilly S, Da Silva K, et al. Alterations in gut microbiome in cirrhosis as assessed by quantitative metagenomics. relationship with acute-on-chronic liver failure and prognosis. Gastroenterology 2021;160:206–218.e13. [DOI] [PubMed] [Google Scholar]
- 36.Bajaj JS, Sikaroodi M, Shamsaddini A, et al. Interaction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy [published onlie ahead of print September 30, 2020]. Gut 10.1136/gutjnl-2020-322470. [DOI] [PubMed] [Google Scholar]
- 37.Schuppan D, Afdhal NH. Liver cirrhosis. Lancet 2008; 371:838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farrell DJ. Rifaximin in the treatment of irritable bowel syndrome: is there a high risk for development of antimicrobial resistance? J Clin Gastroenterol 2013;47:205–211. [DOI] [PubMed] [Google Scholar]
- 39.Calanni F, Renzulli C, Fogli MV, et al. Comment on: Rifaximin in the treatment of irritable bowel syndrome. is there a high risk for development of antimicrobial resistance? J Clin Gastroenterol 2013;47:814. [DOI] [PubMed] [Google Scholar]
- 40.Hiltunen T, Virta M, Laine AL. Antibiotic resistance in the wild: an eco-evolutionary perspective. Philos Trans R Soc Lond B Biol Sci 2017;372:20160039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salehi S, Tranah TH, Lim S, et al. Rifaximin reduces the incidence of spontaneous bacterial peritonitis, variceal bleeding and all-cause admissions in patients on the liver transplant waiting list. Aliment Pharmacol Ther 2019; 50:435–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neff GW, Jones M, Jonas M, et al. Lack of Clostridium difficile infection in patients treated with rifaximin for hepatic encephalopathy: a retrospective analysis. J Clin Gastroenterol 2013;47:188–192. [DOI] [PubMed] [Google Scholar]
- 43.Calanni F, Renzulli C, Barbanti M, et al. Rifaximin: beyond the traditional antibiotic activity. J Antibiot (Tokyo) 2014;67:667–670. [DOI] [PubMed] [Google Scholar]
- 44.Allen HK, Donato J, Wang HH, et al. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 2010;8:251–259. [DOI] [PubMed] [Google Scholar]
- 45.Forsberg KJ, Patel S, Gibson MK, et al. Bacterial phylogeny structures soil resistomes across habitats. Nature 2014;509:612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toprak E, Veres A, Michel JB, et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet 2011;44:101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Penders J, Stobberingh EE, Savelkoul PH, et al. The human microbiome as a reservoir of antimicrobial resistance. Front Microbiol 2013;4:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elkrief L, Rautou PE, Sarin S, et al. Diabetes mellitus in patients with cirrhosis: clinical implications and management. Liver Int 2016;36:936–948. [DOI] [PubMed] [Google Scholar]
- 49.Albillos A, Lario M, Alvarez-Mon M. Cirrhosis-associated immune dysfunction: distinctive features and clinical relevance. J Hepatol 2014;61:1385–1396. [DOI] [PubMed] [Google Scholar]
- 50.Wheeler R, Chevalier G, Eberl G, et al. The biology of bacterial peptidoglycans and their impact on host immunity and physiology. Cell Microbiol 2014;16:1014–1023. [DOI] [PubMed] [Google Scholar]
- 51.Duan Y, Llorente C, Lang S, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 2019;575:505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ha CWY, Martin A, Sepich-Poore GD, et al. Translocation of viable gut microbiota to mesenteric adipose drives formation of creeping fat in humans. Cell 2020; 183:666–683.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boland BS, He Z, Tsai MS, et al. Heterogeneity and clonal relationships of adaptive immune cells in ulcerative colitis revealed by single-cell analyses. Sci Immunol 2020;5:eabb4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su G, Xu H, Riggi E, et al. Association of kidney function with infections by multidrug-resistant organisms: an electronic medical record analysis. Sci Rep 2018; 8:13372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zacharioudakis IM, Zervou FN, Ziakas PD, et al. Vancomycin-resistant enterococci colonization among dialysis patients: a meta-analysis of prevalence, risk factors, and significance. Am J Kidney Dis 2015;65:88–97. [DOI] [PubMed] [Google Scholar]
- 56.Bajaj JS, Ananthakrishnan AN, Hafeezullah M, et al. Clostridium difficile is associated with poor outcomes in patients with cirrhosis: a national and tertiary center perspective. Am J Gastroenterol 2009;105:106–113. [DOI] [PubMed] [Google Scholar]
- 57.Horvath A, Rainer F, Bashir M, et al. Biomarkers for oralization during long-term proton pump inhibitor therapy predict survival in cirrhosis. Sci Rep 2019; 9:12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bajaj JS, Acharya C, Fagan A, et al. Proton pump inhibitor initiation and withdrawal affects gut microbiota and readmission risk in cirrhosis. Am J Gastroenterol 2018;113:1177–1186. [DOI] [PubMed] [Google Scholar]
- 59.Chen Y, Yang F, Lu H, et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 2011;54:562–572. [DOI] [PubMed] [Google Scholar]
- 60.Bajaj JS, Ridlon JM, Hylemon PB, et al. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol 2012;302:G168–G175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Flow chart of patients with cirrhosis in the cross-sectional study (S1A) and pre/post rifaximin open-label trial (S1B).
Supplementary Figure 2. Orange cross-validation modeling and prediction workflow
Supplementary Figure 3. Orange cross-validation modeling and prediction workflow.
Supplementary Figure 4. Comparison of cirrhosis with type 2 diabetes (T2D, purple) with patients with T2D alone (orange, Qin et al) showing clear separation between the groups on resistome, AMR gene family and ARO term abundances.
Supplementary Figure 5. Comparison of cirrhosis without type 2 diabetes (T2D, purple) with patients with T2D alone (orange, Qin et al) showing clear separation between the groups on resistome, AMR gene family and ARO term abundances.
Supplementary Figure 6. Top 20 log2Fold change differences of abundances between all patients with cirrhosis (purple) with patients with chronic kidney disease on dialysis (CKD, orange, Wang et al) with respect to resistome, AMR gene family and ARO term abundances.
Supplementary Figure 7. Heatmap of differences of resistome, AMR gene family and ARO term abundances comparing cirrhosis (purple) with chronic kidney disease on dialysis (CKD, orange)
Supplementary Figure 8. Cirrhosis vs CKD fold change.
Supplementary Figure 9. Cirrhosis vs CKD heatmap.
Supplementary Figure 10. Comparison of controls from Wang et al and Qin et al to our controls
Supplementary Figure 11. Top 20 log2Fold change differences of abundances between all patients with cirrhosis regardless of type 2 diabetes (T2D, purple) with patients with T2D alone with respect to resistome, AMR gene family and ARO term abundances.
Supplementary Figure 12. Heatmap of differences of resistome, AMR gene family and ARO term abundances comparing cirrhosis (purple) with type 2 diabetes (T2D, orange).
Supplementary Figure 13. Top 20 log2Fold change differences of abundance comparison between healthy controls in our study with those in Wang et al (S9A) and Qin et al (S9B) on ARO terms showed minimal differences.


