Abstract
Cyclin-dependent kinase 9 (CDK9) activity is correlated with worse outcomes of triple-negative breast cancer (TNBC) patients. The heterodimer between CDK9 with cyclin T1 is essential for maintaining the active state of the kinase and targeting this protein–protein interaction (PPI) may offer promising avenues for selective CDK9 inhibition. Herein, we designed and generated a library of metal complexes bearing the 7-chloro-2-phenylquinoline CˆN ligand and tested their activity against the CDK9–cyclin T1 PPI. Complex 1 bound to CDK9 via an enthalpically-driven binding mode, leading to disruption of the CDK9–cyclin T1 interaction in vitro and in cellulo. Importantly, complex 1 showed promising anti-metastatic activity against TNBC allografts in mice and was comparably active compared to cisplatin. To our knowledge, 1 is the first CDK9–cyclin T1 PPI inhibitor with anti-metastatic activity against TNBC. Complex 1 could serve as a new platform for the future design of more efficacious kinase inhibitors against cancer, including TNBC.
KEY WORDS: Metal complex, Kinase inhibitor, Protein–protein interaction, Epigenetics, Triple-negative breast cancer, Metastasis
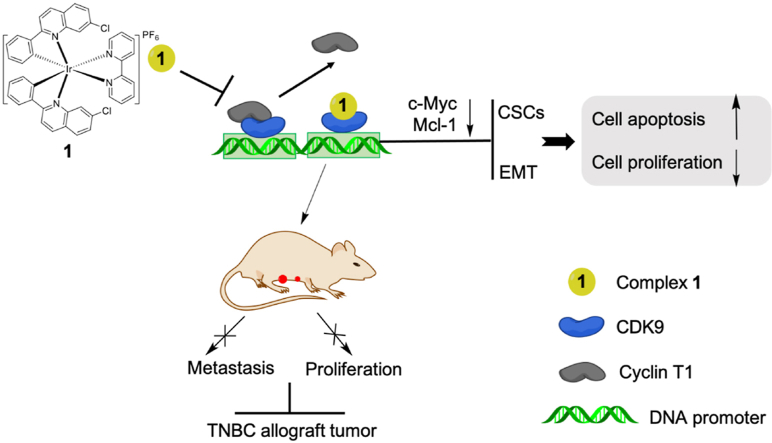
Graphical abstract
Inhibition of the CDK9–cyclin T1 protein–protein interaction as a new approach against triple-negative breast cancer. Complex 1 inhibited the CDK9–cyclin T1 interaction and suppressed metastasis of TNBC tumors in mice.
1. Introduction
Triple-negative breast cancer (TNBC) is one of the most aggressive subtypes of breast cancer and exhibits high relapse risk, high metastasis, and low overall survival1, 2, 3. Due to a dearth of clinical anti-TNBC drugs, the treatment of metastatic TNBC is extremely challenging4. Cyclin-dependent kinase 9 (CDK9) is a regulator of transcription and is often dysregulated in breast cancer5,6. High CDK9 expression is associated with worse overall survival among patients with TNBC7. The CDK9–cyclin T1 complex regulates the transcriptional elongation of RNA polymerase II and increases the transcription activity of cellular myelocytomatosis oncogene (c-Myc) and myeloid cell leukemia 1 (Mcl-1), which are oncogenes that increase metastasis by maintaining cancer stem cells (CSCs) and epithelial-mesenchymal transition (EMT)8,9. Most pre-clinical/clinical molecularly-targeted drugs cannot effectively reduce c-Myc and Mcl-1 level10,11. Additionally, specifically and directly targeting c-Myc and Mcl-1 has proven challenging11,12.
Flavopiridol is a CDK kinase inhibitor that targets the ATP-binding site and has entered phase II trials for cancer13, but it exhibited dose-limiting toxicities due to its off-target effects on other CDK kinases14. Due to the conserved sequence similarity of the ATP-binding region of CDK family members, it is difficult to selectively target CDKs15,16. The strategy of directly disrupting protein–protein interactions (PPI) has emerged as an alternative avenue to selectively inhibit kinases via targeting the less highly-conserved regions of the protein17,18. Previous research has identified small molecule or peptide inhibitors targeting CDK9–cyclin T1 interaction by virtual screening2,19,20. Recently, our laboratory has identified a tetrahydroisoquinoline-based ATP-competitive CDK9–cyclin T1 PPI inhibitor using in silico docking, and this molecule could inhibit proliferation and migration in TNBC cells.
Quinoline scaffolds have been reported as an important construction motif in heterocyclic compounds21. Quinolines appear frequently in anticancer drugs22. Importantly, quinoline scaffolds have previously been reported as CDK kinase, type II tyrosine kinase, and PPI inhibitors23, 24, 25, 26. For instance, the 2-phenylquinoline motif is found in the clinical candidate OSI-906, which is a selective kinase inhibitor of IGF-1R27. In addition, previous research has demonstrated that quinoline compounds have potential inhibitory activity against CDK9–cyclin T1 PPI20. Meanwhile, organometallic complexes have attracted recent attention as scaffolds for developing highly potent and selective kinase inhibitors28. Small variations in the structure of auxiliary ligands can lead to significant changes in selectivity for different targets. For example, Pim1 half-sandwich complex that functioned as a GSK3 inhibitor was transformed into an inhibitor of PAK1 kinase by replacing the cyclopentadienyl moiety with a chloride and sterically demanding diamine ligands29,30. In addition, we have previously reported a different Rh(III) complex contain the 7-chloro-2-phenylquinoline CˆN ligand as possessing Wee1 kinase inhibitory activity in TP53-mutated TNBC cells18.
In this study, we have identified an Ir(III)-based complex 1 containing the 7-chloro-2-phenylquinoline CˆN and 2,2′-bipyridine NˆN ligands as anti-cancer agent against TNBC. Complex 1 engages CDK9 to disrupt the CDK9–cyclin T1 PPI and inhibit CDK9–cyclin T1 enzymatic activity as demonstrated using multiple biochemical and physical assays. In cells, complex 1 transcriptionally reduces levels of c-Myc and Mcl-1, activates the pro-apoptotic regulators, and reverses EMT and the ratio of CSCs. Finally, complex 1 suppresses TNBC proliferation and metastasis both in vitro and in vivo. To date, 1 is the first reported metal-based CDK9–cyclin T1 PPI inhibitor with anti-metastatic activity against TNBC. Our work also provides insight into the role of the metal-based PPI inhibitors against TNBC and highlights the feasibility and significance of CDK9–cyclin T1 PPI in TNBC target therapy for metastasis.
2. Results
2.1. Design and synthesis of iridium(III) and rhodium(III) complexes 1–19 as CDK9–cyclin T1 PPI inhibitors
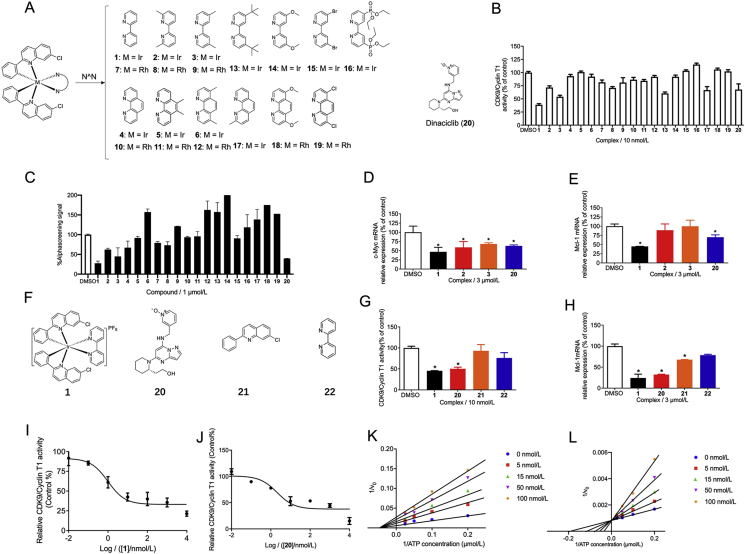
As described earlier, the 2-phenylquinoline motif has appeared in various kinase inhibitors in the literature. Therefore, we were interested to explore whether the incorporation of this structural motif into kinetically-inert Ir(III) and Rh(III) complexes could generate candidate complexes that were active against the CDK9–cyclin T1 interaction. Complexes 1–19 all contain the 7-chloro-2-phenylquinoline CˆN ligand, which was also a key structural component of a different Rh(III) complex that we previously reported to display Wee1 kinase inhibitory activity in TP53-mutated TNBC cells18. Meanwhile, 2,2′-bipyridine and 1,10-phenanthroline-based NˆN ligands are widely-used ligand in metal complexes36,37. Ir(III) complexes 1 and 4, Rh(III) complexes 7 and 10 are the parent complexes which contain 2,2′-bipyridine and 1,10-phenanthroline-based NˆN ligands respectively. To generate structure–activity relationship (SAR) data, we explored the effect of changing the number and position of methyl groups (2, 3, 5, 6, 8, 9, 11–13, 17), variations in the substituents on 2,2′-bipyridine ligand (14–16) or 1,10-phenanthroline (18–19) ligands (Fig. 1A). The known CDK9 inhibitor dinaciclib (20) was used as a positive control38.
Figure 1.
The in cellulo and in vitro activity of complex 1 and selected complexes and ligands. (A) The chemical structures of the Ir(III) and Rh(III) complexes 1–19 and positive control 20 (dinaciclib) evaluated in this study. (B) Effects of 1–20 on CDK9/cyclin T1 activity were determined by a chemiluminescence assay. (C) Inhibition of the CDK9–cyclin T1 PPI by 1–20 as measured using the AlphaScreen assay (C). (D) c-Myc mRNA and (E) Mcl-1 mRNA levels by compounds (3 μmol/L) in MDA-MB-231 cells as measured by qPCR assay. (F) Structure of 1, 20, and ligands 21, 22. (G) Effect of compounds and ligands (10 nmol/L) on CDK9–cyclin T1 activity. (H) Effect of compounds and ligands (3 μmol/L) on Mcl-1 mRNA levels. (I) 1 inhibits CDK9–cyclin T1 activity in a dose-dependent manner. (J) 20 inhibits CDK9–cyclin T1 activity in a dose-dependent manner. (K) Double-reciprocal plot of initial rate against ATP concentration in the presence of complex 1 at 0, 5, 15, 50, 100 nmol/L. (L) Double-reciprocal plot of initial rate against ATP concentration in the presence of compound 20 at 0, 5, 15, 50, 100 nmol/L. ∗P < 0.05 compared to DMSO group by the t-test.
Complexes 1–19 were screened in a preliminary in vitro chemiluminescence assay to measure their ability to inhibit CDK9–cyclin T1 enzymatic activity. At 10 nmol/L, the Ir(III) complex 1 had the highest potency against CDK9–cyclin T1 activity (55.9% inhibition), and was comparably potent to the positive control, dinaciclib (20) (31.0% inhibition) (Fig. 1B). In addition, the AlphaScreen assay was also performed to assess the ability of the complexes to disrupt the CDK9–cyclin T1 PPI. The results showed that at 1 μmol/L, complex 1 showed the highest ability to disrupt the CDK9–cyclin T1 PPI in vitro (72.4% inhibition) and was similarly potent to dinaciclib (20) (60.4% inhibition) (Fig. 1C).
To generate more robust data for SAR analysis, EC50 values for were obtained for complexes 1–19 and positive control 20 from the CDK9–cyclin T1 enzymatic activity assay (Supporting Information Table S1). Dose-response curves for complex 1 (EC50 = 3.3 ± 1.2 nmol/L) and the positive control 20 (EC50 = 10 ± 2.1 nmol/L) are displayed in Fig. 1I and J respectively. These complexes all carried the same 7-chloro-2-phenylquinoline CˆN ligand, which has been previously demonstrated to be beneficial for anti-PPI activity but possessed different NˆN ligands. The Ir(III) complexes 1–6 (EC50 = 3.3–180.6 nmol/L) are generally more potent than their corresponding Rh(III) congeners 7–12 (EC50 = 234.4–>1000 nmol/L) even though they bear the same NˆN ligands and charge. Additionally, the Ir(III) complexes 1 (3.3 nmol/L) and 3 (13.6 nmol/L) were more potent than complexes 4 (234.4 nmol/L) and 6 (>1000 nmol/L), respectively, indicating that the 2,2′-bipyridine NˆN scaffold was superior to the 1,10-phenanthroline NˆN scaffold. A similar trend was observed when comparing the Rh(III) 2,2′-bipyridine complexes 7 (162.2 nmol/L) and 9 (180.6 nmol/L) with the Rh(III) 1,10-phenanthroline NˆN scaffold complexes 10 (>1000 nmol/L) and 12 (1000 nmol/L). As metal complex 1 (with the unsubstituted 2,2′-bipyridine NˆN ligand) showed the highest potency (3.3 nmol/L), this suggested that substitution on the NˆN ligand is undesirable for CDK9–cyclin T1 inhibitory activity. After complex 1, the second-most active compound was the Ir(III) complex 3 (13.6 nmol/L) which carried the 4,4′-dimethyl-2,2′-bipyridine NˆN ligand. The position and number of the methyl groups were important for potency, as Ir(III) metal complex 2 (with 6,6′-dimethyl-2,2′-bipyridine) (50.1 nmol/L) showed lower potency than 3 despite the fact that they have the same number of the methyl groups. Interestingly, extending the methyl group of 3 into the t-butyl substituent of 13 led to only a slight decrease of potency. However, polar groups (14, 16, 18) (>1000 nmol/L) or chlorine atoms (19) (>1000 nmol/L) on the NˆN ligand detracted from potency, although interestingly bromine atoms (15) (63.1 nmol/L) were somewhat tolerated. Finally, having a single methyl group on the 1,10-phenanthroline ligand also appeared to be tolerated in the Ir(III) complex 17 (316.4 nmol/L). Taken together, the results indicated that the 2,2′-bipyridine NˆN ligand and the character of the metal center are both important determinants of CDK9–cyclin T1 inhibitory activity.
To study the in cellulo activity of complex 1, its ability to inhibit Mcl-1 and c-Myc transcription, which are under the control of CDK9–cyclin T1, were assessed in MDA-MB-231 TNBC cells. Selected complexes (1–3) and positive control (20) were also tested to extend the SAR to in cellulo results. Complex 1 (at 3 μmol/L) significantly inhibited c-Myc (Fig. 1D) and Mcl-1 (Fig. 1E) mRNA levels and was more potent than the positive control. Moreover, the individual ligands of complex 1, 7-chloro-2-phenylquinoline (21), and 2,2′-bipyridine (22) (Fig. 1F), were also examined for their effects on CDK9–cyclin T1 activity and Mcl-1 transcription. The isolated ligands showed much lower activity against CDK9–cyclin T1 activity in vitro (Fig. 1G) and against Mcl-1 transcription in cellulo (Fig. 1H). This suggests that the Ir(III) center plays a key role in arranging the ligands into an active configuration.
Finally, the kinetics of enzyme inhibition was studied to evaluate the inhibitory mechanism of 1 and 20. As shown in the double-reciprocal plots, 1 functioned as a non-ATP-competitive antagonist of CDK9–cyclin T1, with a Ki value of 3.3 nmol/L (Fig. 1I and K). This suggests that 1 engages CDK9 at different site to the ATP-binding site. Meanwhile, compound 20 functioned an ATP-competitive inhibitor of CDK9 with a Ki value of 10.0 nmol/L (Fig. 1J and L), which is consistent with previous reports in the literature39,40.
2.2. Iridium(III) complex 1 selectively binds with CDK9 to inhibit CDK9–cyclin T1 PPI
The mode of action of complex 1 was further studied using a GST pull-down assay. The results showed complex 1 could inhibit the interaction between CDK9 and cyclin T1 in vitro (Fig. 2A). Next, a co-IP experiment was performed to verify the ability of 1 to disrupt the interaction between CDK9 and cyclin T1 in cellulo (Fig. 2B). In MDA-MB-231 cells, 1 reduced the amount of cyclin T1 co-precipitating with CDK9 (Fig. 2C). Hence, these results demonstrate that complex 1 could disrupt the CDK9–cyclin T1 PPI both in vitro and in cellulo.
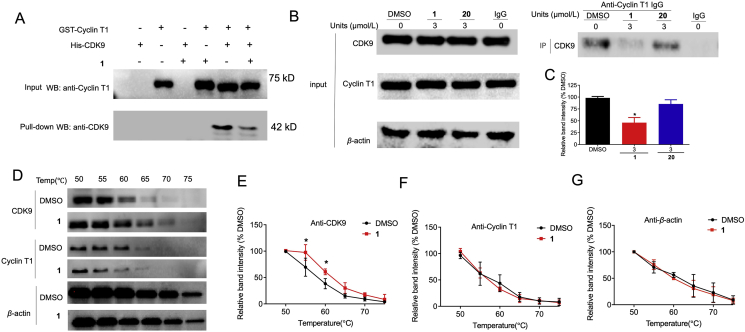
Figure 2.
Complex 1 disrupts the interaction between CDK9 and cyclin T1 by targeting CDK9. (A) GST pull-down assay of cyclin T1 and CDK9. GST-tag cyclin T1 protein was immobilized in the gravity flow column and then incubated with 300 μL CDK9 and/or complex 1. Eluted samples were analyzed by Western blotting. (B) The CDK9–cyclin T1 PPI was examined by co-IP in MDA-MB-231 cells. MDA-MB-231 cells were treated with DMSO and 3 μmol/L 1 or 20 for 4 h, and cell lysates were subject to immunoprecipitation with the indicated antibody. (C) Relative densitometry analysis of CDK9. (D) MDA-MB-231 lysates were treated with 3 μmol/L compound 1 or DMSO for 30 min, followed by CETSA analysis to evaluate CDK9, cyclin T1, and β-actin stabilization. (E–G) Densitometry analysis of CDK9, cyclin T1, and β-actin content in the soluble fraction. ∗P < 0.05 compared to DMSO group by the t-test.
Cyclin T2 can also associate with CDK9 and the CDK9–cyclin T2 possess transcription elongation activity41. Meanwhile, CDK9 and CDK7 have overlapping functions on transcriptionally active Pol II42, with CDK7 recognizing cyclin H43. To investigate the selectivity of 1, further co-IP experiments were conducted (Supporting Information Fig. S1). 1 did not significantly decrease cyclin T2 co-precipitating with CDK9 or cyclin H co-precipitating with CDK7 was observed in MDA-MB-231 cells, suggesting that it had no effect on the CDK9–cyclin T2 and CDK7–cyclin H PPIs.
To investigate whether complex 1 targets CDK9 or cyclin T1 to achieve PPI inhibition, the cellular thermal shift assay (CETSA) was performed (Fig. 2D). In MDA-MB-231 cell lysates treated with 1, the melting temperature of CDK9 was significantly increased from 55 to 60 °C (Fig. 2E). However, it had no significant effect on the stability of cyclin T1 or β-actin (Fig. 2F and G). This suggests that 1 could bind with CDK9 to disrupt the CDK9–cyclin T1 PPI even in the complex cellular environment. CDK9 is homologous with CDK1, CDK2, CDK5, CDK1244. Thus, we measured the binding strength of 1 for CDK1, CDK2, CDK5, and CDK12 with CETSA (Supporting Information Fig. S2a). The results showed that these other CDK members were not significantly stabilized by complex 1. Moreover, the kinase inhibitory activity of 1 against CDK1, CDK2, CDK4/6, CDK5, CDK12 was also evaluated (Fig. S2b). These results showed that the enzymatic activities of these other CDK members were not significantly inhibited by complex 1. Taken together, these results indicate that 1 can selectively engage and inhibit CDK9 over other CDK family members.
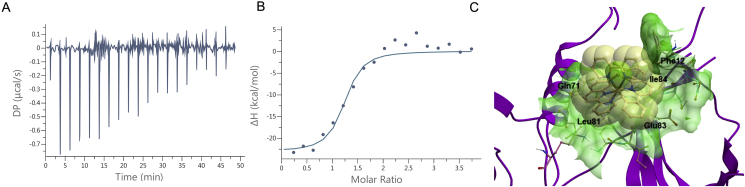
To study the thermodynamics of the interaction between 1 and CDK9, we conducted isothermal titration calorimetric (ITC) analysis (Fig. 3A and B). The stoichiometry of the complex was determined to be N = 1.17 ± 0.05, suggesting that CDK9 possesses one binding site for complex 1. The binding interaction between 1 and CDK9 was calculated to be strongly exothermic (ΔH = −23.2 ± 1.34 kcal/mol), which compensated for an unfavorable entropic contribution (−TΔS = 13.8 kcal/mol). This indicates that the interaction is enthalpically-driven, which is advantageous for pharmaceutical drug development. The binding affinity (Kd) value was calculated as 134 ± 68.8 nmol/L, indicating a low single-digit micromolar affinity of interaction. For the blank control group, no significant interaction was detectable upon injection of 100 μmol/L of 1 into PBS buffer (Supporting Information Fig. S3). To further study the mode of action of complex 1, we used molecular docking to explore the binding mode of complex 1 to CDK9 (Fig. 3C). Docking was performed using the Molsoft ICM method (ICM version 3.9-1b molecular docking software) using a model of CDK9 derived from the crystal structure of the CDK9/cyclin T1 complex (PDB code: 3BLH)45. Complex 1 is predicted to bind to CDK9 in its interface region with cyclin T1. The NˆN ligand forms a partial p–π interaction with Phe12 and van der Waals interaction with the sidechain of Gln71. Meanwhile, one of the CˆN ligands forms extensive van der Waals interactions with Glu83 and Ile84, while another CˆN ligand extends into a small pocket to interact with Leu81.
Figure 3.
The binding model of the complex 1 against the CDK9–cyclin T1 PPI. (A–B) ITC analysis of the 1–CDK9 interaction. (A) Raw data for 19 sequential injections (2 μL per injection) of a solution of 1 (100 μmol/L) into a solution of CDK9 (5 μmol/L). (B) “Net” heat effects obtained by subtracting the dilution heat from the reaction heat, which was fitted by using the “one set of sites” binding model. N (sites) = 1.17 ± 0.05; Kd = 134 ± 68.2 nmol/L; ΔG = −9.38 kcal/mol; ΔH = −23.2 ± 1.4 kcal/mol; −TΔS = 13.4 kcal/mol. (C) Binding model of complex 1 with CDK9 (PDB code: 3BLH) which is depicted as a space-filling representation showing carbon (yellow), nitrogen (blue), chlorine (green) atoms. The binding pocket of the CDK9 is represented as a translucent green surface.
2.3. Iridium(III) complex 1 exhibits cytotoxicity, anti-proliferation, and anti-metastasis activity in TNBC cells
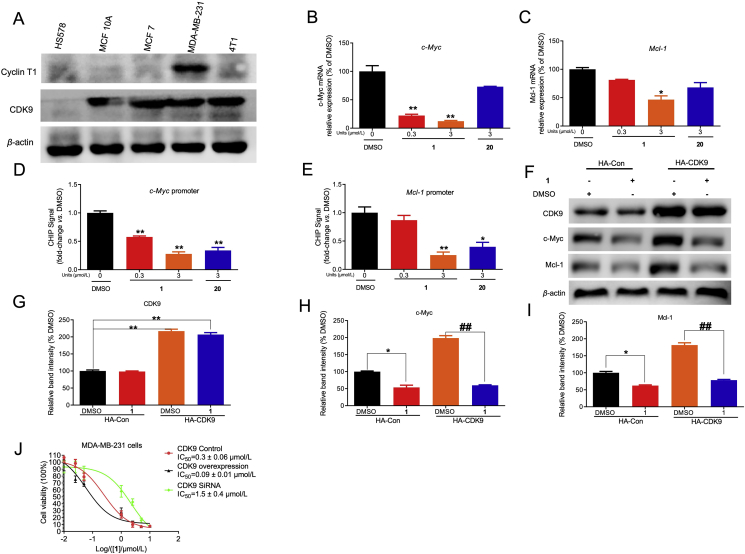
Many cancer cell lines, including TNBC, overexpress CDK930,46,47. The anticancer activity of complex 1 was assessed in a number of breast cancer (MCF-10A, MCF-7, MDA-MB-231, and 4T1) and normal (HEK293T, LO2, and HUVEC) cell lines using the MTT assay. The results showed that the antiproliferative activity of complex 1 was higher in the breast cancer cell lines MDA-MB-231 (IC50: 0.21 ± 0.01 μmol/L), 4T1 (IC50: 0.40 ± 0.03 μmol/L), MCF-10A (IC50: 0.72 ± 0.02 μmol/L), and MCF-7 (IC50: 0.68 ± 0.13 μmol/L) than in the normal human cell lines HUVEC (IC50: 2.20 ± 0.19 μmol/L), HEK293T (IC50: 3.60 ± 0.50 μmol/L) and LO2 (IC50: 1.07 ± 0.21 μmol/L) (Supporting Information Fig. S4). Additionally, immunoblotting experiments showed that CDK9 is overexpressed in breast cancer cell lines (4T1, MDA-MB-231, MCF7) compared to human normal breast cell lines (MCF-10A and HS578) (Fig. 4A). These results suggest that the cytotoxicity exhibited by 1 may be associated with CDK9 expression status. Oncogenic c-Myc and Mcl-1 level are under the control of CDK9–cyclin T148,49. Inhibition of CDK9 prevents productive transcription of c-Myc which regulates the proliferation and survival of cancer cells50. In addition, Mcl-1 is another gene that is directly regulated by CDK9 which enables tumor metastasis and apoptosis in breast cancer51. We hypothesized that 1 downregulated c-Myc and Mcl-1 via blocking the interaction between CDK9 and the c-Myc and Mcl-1 promoters. Chromatin immunoprecipitation (ChIP) and quantitative polymerase chain reaction (qPCR) analysis indicated that 1 (at 3.0 μmol/L) blocked the binding of CDK9 to the c-Myc and Mcl-1 gene promoters, leading to decreased mRNA production (Fig. 4B–E).
Figure 4.
The cytotoxicity of complex 1 in CDK9 overexpressed breast cancer cells. (A) The CDK9 protein level was evaluated in different breast cancer cell lines (MCF7, 4T1, MDA-MB-231) and human normal breast cell lines (MCF 10A, HS578). CDK9, cyclin T1, and β-actin were blotted to control for total protein levels. (B–C) Inhibition effects of 1 or 20 on the c-Myc and Mcl-1 mRNA level in MDA-MB-231 cells were determined by qPCR. (D–E) Inhibition effect of 1 or 20 on the binding ability of CDK9 to c-Myc and Mcl-1 promoter in MDA-MB-231 cells by ChIP-qPCR. Indicated concentrations of 1 or 20 treated with cells for 4 h. Then cells were collected to perform ChIP-qPCR. (F) CDK9 plasmid treatment led to efficient overprotein level in MDA-MB-231 cells. CDK9, c-Myc, Mcl-1, and β-actin were blotted to control for total protein levels. (G–I) Relative densitometry analysis of CDK9 (G), c-Myc (H), and Mcl-1 (I). (J) The cytotoxicity effect of 1 on control, CDK9 knockdown, and CDK9 overexpressing MDA-MB-231 cells. 1 inhibited the growth of on control, CDK9 knockdown, overexpression-treatment MDA-MB-231 cells with an IC50 value of 0.3, 1.5 and 0.09 μmol/L, respectively. Data are presented as the mean ± SD. ∗P < 0.05, ∗∗P < 0.01 vs DMSO group, ##P < 0.01 compared to DMSO group by the t-test.
To study the role of CDK9 in complex 1-induced suppression of c-Myc and Mcl-1, CDK9 was knocked down by siRNA. Both CDK9 knockdown or complex 1 treatment reduced c-Myc and Mcl-1 protein level in MDA-MB-231 cells (Supporting Information Fig. S5). Moreover, treating CDK9 knockdown cells with 1 did not cause a further significant decrease of c-Myc and Mcl-1 level. Next, CDK9 was overexpressed by transfection with CDK9 plasmid. Overexpression of CDK9 increased the sensitivity of MDA-MB-231 cells to c-Myc and Mcl-1 inhibition by 1, suggesting that 1 acted through CDK9 to inhibit oncogenic expression (Fig. 4F–I). Moreover, the cytotoxicity of 1 in CDK9-overexpressing MDA-MB-231 cells was higher relative to control cells (IC50 = 0.09 vs 0.3 μmol/L) (Fig. 4J). In contrast, knockdown of CDK9 using siRNA reduced the cytotoxicity of the cells towards 1 (IC50 = 1.5 μmol/L). Taken together, these results indicate that the cytotoxicity and anti-proliferation activity of 1 is, at least in part, CDK9-dependent.
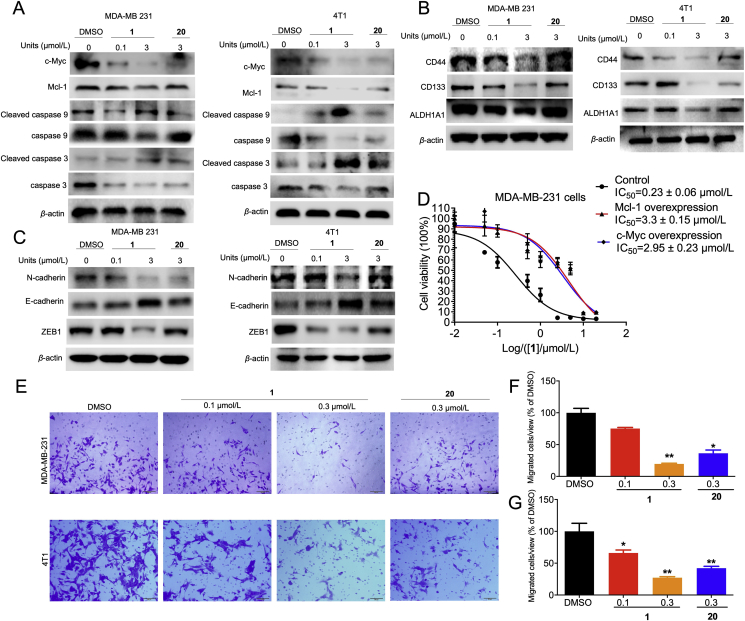
Next, we further studied the downstream effects of c-Myc and Mcl-1 in the CDK9–cyclin T1 pathway. Mcl-1 prevents the activation of the pro-apoptotic regulators, caspases 3 and 952,53. As expected, treatment of MDA-MB-231 and 4T1 cells with 1 led to an increase in cleaved caspase 3/9 protein levels, suggesting that 1 could promote apoptosis presumably through inhibiting Mcl-1 (Fig. 5A). The effect of 1 on CSC and EMT biomarkers regulated by c-Myc were also investigated. Complex 1 inhibited the protein level of both CSC biomarkers CD44, CD133, and ALDH1A (Fig. 5B) and EMT biomarkers N-cadherin and ZEB1 (Fig. 5C). Moreover, 1 augmented the protein level of E-cadherin, which is a repressor of EMT. Finally, the role of c-Myc and Mcl-1 in mediating the effects of 1 was confirmed by the observation that c-Myc and Mcl-1 transfection partially rescued MDA-MB-231 cells from 1-induced antiproliferative activity (Fig. 5D). Cell migration is required for the metastatic ability of cancer cells and is directly regulated by c-Myc and Mcl-154, 55, 56. The ability of complex 1 to inhibit TNBC migration was studied using a transwell migration assay (Fig. 5E). Complex 1 (0.3 μmol/L) inhibited MDA-MB-231 and 4T1 migration by 80% and 73%, respectively, and was more potent than the positive control (20) which showed 64% and 58% inhibition at the same concentration (Fig. 5F and G). Taken together, these results suggest that complex 1 exerts pro-apoptotic, anti-proliferative, and anti-migration effects in cellulo by mediating downstream signaling pathways of c-Myc and Mcl-1.
Figure 5.
Complex 1 induced cell apoptosis and inhibited cell migration through inhibition of c-Myc and Mcl-1 protein level. (A) MDA-MB-231 and 4T1 cells were treated with the indicated concentrations of 1 or 20 treated for 4 h. Cell lysates were collected and immunoblotted with the anti-apoptosis corresponding antibodies. (B–C) The effect of 1 on CSC (B) and EMT (C) biomarkers levels in MDA-MB-231 and 4T1 cells. DMSO or indicated concentrations of 1 or 20 were treated with MDA-MB-231 and 4T1 cells for 4 h. Then cells were collected and incubated with appropriate antibodies. (D) The anti-proliferative effect of 1 in MDA-MB-231 cells after transfection with c-Myc and Mcl-1. Cells were transfected with c-Myc, Mcl-1, or control plasmid for 48 h, then treated with DMSO or 1 for 48 h. Cell viability was detected by the MTT assay. (E) 1 inhibited the migration of TNBC cell lines. Indicated concentrations of 1 or 20 treated with MDA-MB-231 and 4T1 cells for 24 h and evaluated by transwell. (F–G) Quantitative analysis of migration cells. Data are presented as the mean ± SD. ∗P < 0.05, ∗∗P < 0.01 compared to DMSO group by the t-test.
2.4. Iridium(III) complex 1 suppresses tumor growth and inhibit lung metastasis in TNBC allograft model
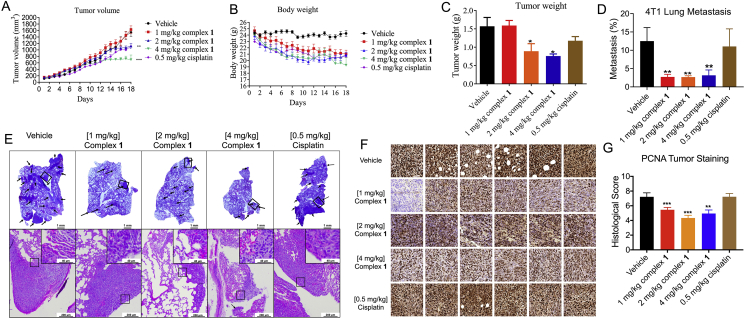
Given the promising anti-apoptosis and anti-metastasis effect exhibited by 1 in vitro, the in vivo efficacy of 1 was studied using a murine model of TNBC. The anti-TNBC chemotherapeutic drug, cisplatin, was used as a positive control. Female BALB/c mice were injected subcutaneously with 4T1 (murine TNBC) cells for tumor establishment, followed by intraperitoneal daily injection with 1, 2, or 4 mg/kg of complex 1 or 0.5 mg/kg of cisplatin. Encouragingly, treatment with 4 mg/kg of 1 led to a significant decrease of tumor volume relative to the negative control starting from day 13 (Fig. 6A). The effects of complex 1 and cisplatin on body weight were similar (Fig. 6B). At sacrifice, complex 1 reduced average tumor weight by 43.3% and 51.7% in the 2 and 4 mg/kg treatment group, respectively, compared to 25.2% for the cisplatin group (Fig. 6C). To assess the effect of 1 on the metastatic activity of tumors, H&E stained lung sections were examined to visualize metastatic lesions. At all dosages of 1 tested, the incidence of lung metastases was significantly reduced compared to the untreated and 0.5 mg/kg cisplatin-treated cohorts (Fig. 6D). By observation of visible tumor nodules in the lungs, the percentage of metastatic burden area was significantly reduced by 1 even in the 1 mg/kg group (Fig. 6E). Proliferating cell nuclear antigen (PCNA) is a classic biomarker of cell proliferation and is correlated with the degree of distant metastasis and tumor survival57. To further study mechanism of 1 in vivo, PCNA levels were analyzed by immunohistochemistry. The results showed that 1 decreased PCNA levels in orthotopic tumors (Fig. 6F and G).
Figure 6.
Complex 1 suppresses tumor growth, lung metastasis, and proliferation in a TNBC mouse model. Female BALB/c mice were subcutaneously inoculated with 1.0 × 106 4T1 cells, and after the tumor volume reached 60 mm3, injected intraperitoneally daily with 1 or 2 or 4 mg/kg of complex 1 or 0.5 mg/kg of cisplatin for 18 days. (A) Tumor volume of 4T1 tumor-bearing mice after control (vehicle), 1 (1, 2 or 4 mg/kg) and cisplatin (0.5 mg/kg) treatment (n = 6–8). (B) Body weight of tumor-bearing mice over the course of treatment. (C) Comparison of tumor weight after sacrifice. (D) The percentage of metastatic burden area. (E) Representative photograph images of lung metastatic tissues and H&E stained lung sections in vehicle control, complex 1, and cisplatin groups after 18 days. Scale bar = 200 μm (×40). (F) Six random PCNA-stained areas of a single tumor from each group are shown at ×40 magnification. (G) PCNA histological score is displayed. Six random areas of each tumor section are enrolled, and each dot represents the score of one area. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 compared to vehicle by the t-test.
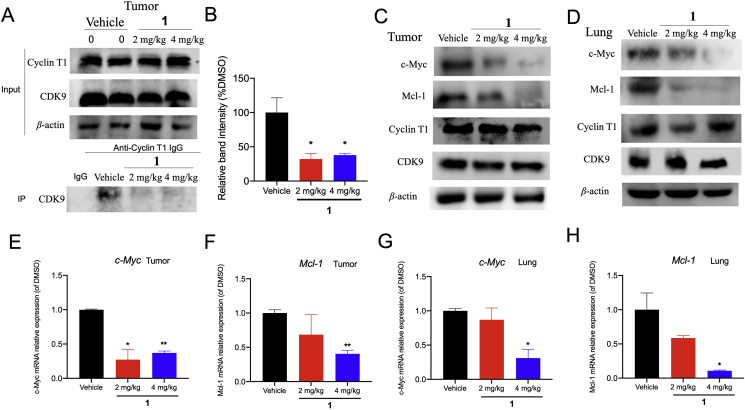
To validate the mechanism of action of 1 in the animal models, tumor and lung tissues were examined after sacrifice. Consistent with the cellular results, the co-IP assay showed that 2 mg/kg and 4 mg/kg of 1 treatment impaired the interaction between CDK9 and cyclin T1 in vivo, as indicated by a reduction of cyclin T1 co-precipitated with CDK9 in tumor tissues (Fig. 7A and B). Moreover, the protein expression of c-Myc, Mcl-1, cyclin T1, and CDK9 were elevated in tumor and lung tissues after treatment, suggesting that 1 could inhibit c-Myc and Mcl-1 expression without influencing CDK9–cyclin T1 levels in vivo (Fig. 7C and D). Moreover, tumors and lung in complex 1-treated mice showed the decrease of c-Myc and Mcl-1 mRNA level in a dose-dependent fashion (Fig. 7E‒H). Taken together, the data suggest that 1 could disrupt CDK9–cyclin T1 PPI in vivo, which could account for its anti-tumor effects in the animal model.
Figure 7.
1 suppressed the CDK9–cyclin T1 PPI and decreased c-Myc and Mcl-1 levels in vivo. (A–B) The CDK9–cyclin T1 PPI was examined by co-IP in 2 mg/kg and 4 mg/kg of complex 1-treated mice. (C–D) Protein levels of c-Myc, Mcl-1, cyclin T1, CDK9, and β-actin in tumor and lung tissues treated with vehicle or 2 and 4 mg/kg of 1 were measured by Western blotting. (E–H) 1 impaired c-Myc and Mcl-1 transcription in tumor and lung tissues. Data are represented as mean ± SD. ∗P < 0.05, ∗∗P < 0.01 vs Vehicle group, using Student's t-test.
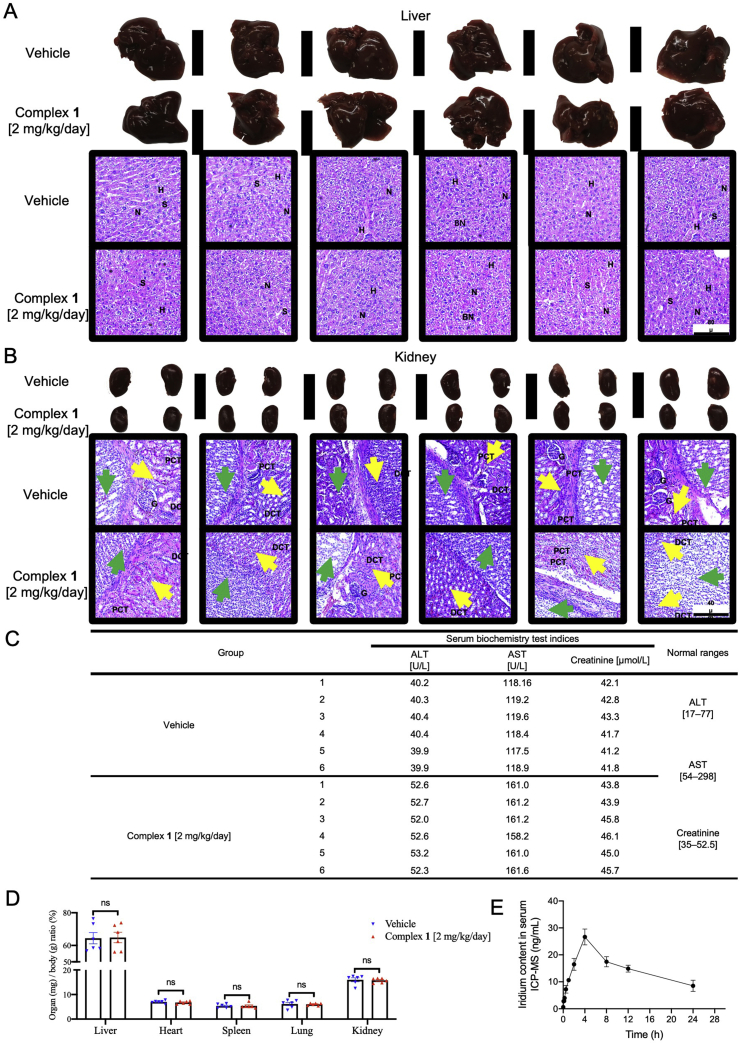
We also evaluated the toxicity of complex 1 in 4T1 mice model. In Fig. 8A and B, liver and kidney tissues from complex 1 (2 mg/kg/day) treated animal showed homogeneous histology characteristics, compared with that from vehicle control animal. None of the most common poisoning signs such as swelling, hyperemia, nor tissue adhesion were found in liver and kidney tissues. Additionally, no significant toxic or pathological changes were identified in H&E histological examination. Serum alanine transaminase (ALT) and aspartate transaminase (AST) are indices for liver toxicity, and creatinine is the index of renal injury. The normal ranges of ALT, AST, and creatinine are 17–77 U/L, 54–298 U/L, and 35–52.5 μmol/L, respectively58,59. As shown in Fig. 8C, mice treated with 14 days of 1 (2 mg/kg/day) only showed slight increases of ALT, AST, and creatinine and were still within the normal range. Values of organ weight normalized to body weight indicated no significant changes between vehicle and 1 (2 mg/kg/day) groups (Fig. 8D). Collectively, these data that 1 is non-toxic to the animals at the applied dosage. The pharmacokinetic characteristics of intraperitoneally injected 1 at 2.0 mg/kg in mice was determined by assaying iridium content over time (0, 0.08, 0.17, 0.25, 0.5, 1, 2, 4, 8, 12, and 24 h) using ICP-MS (Fig. 8E). The Cmax value occurred between 2 and 8 h, and the maximum mean peak area (27 ng/mL) of iridium content of 1 in serum was obtained at 4 h.
Figure 8.
The toxicity and pharmacokinetic profiles of complex 1. Twelve female mice were randomly divided into two groups and were treated with 1 (2 mg/kg/day) for 18 days. Histological examination of liver and kidney injury. (A) Liver specimens and corresponding H&E staining images indicated the hepatic architecture with nucleus (N), hepatocytes (H), and sinusoids (S) in both vehicle and 1 (2 mg/kg/day) treated group. None of significant hepatic centrilobular mononuclear cell infiltration, fatty changes, nor hepatic cell necrosis were observed, except for sporadic binucleated hepatocytes (BN) in both groups. Scale bar = 80 μm (×10). (B) Kidney specimens and corresponding H&E staining images showed the histological structure with glomerulus (G), proximal convoluted tubules (PCT), and distal convoluted tubules (DCT) in both vehicle and 1 (2 mg/kg/day) treated group. Yellow arrows indicate the renal medulla; green arrows indicate renal pelvis. None pyknotic cells, distal tubular necrosis, sloughing of tubular epithelial cells, nor proximal hemorrhage in interstitial tissue were identified. Scale bar = 40 μm (×20). (C) Serum alanine transaminase (ALT), aspartate transaminase (AST), creatinine levels, and corresponding normal ranges of vehicle and 1 (2 mg/kg/day) treated mice. (D) Normalized organ weights as percentages of body weight. Mice given vehicle and 1 (2 mg/kg/day) were sacrificed at Day 18, the normalized ratio were calculated: organs weight divided by body weight as mg/g, not significant (ns). (E) Pharmacokinetic profile of 1 at different time points. BALB/c mice were intraperitoneally injected with 1 (2 mg/kg/day). The concentrations of 1 in the serum were measured at 0, 0.08, 0.17, 0.25, 0.5, 1, 2, 4, 8, 12, and 24 h. All data were mean ± SEM from 6 mice per treatment group. Significance was determined using one-way ANOVA analysis.
3. Discussion
Until now, chemotherapy is still the mainstay of treatment in TNBC, as there is a critical lack of targeted treatments that have been approved for treating TNBC60. Hence, new targeted therapeutic approaches for TNBC are urgently required. Protein kinases are often implicated in cancer development61, and represent the largest set of pharmacological targets after G-protein-coupled receptors15. CDK9 is a protein kinase that is dysregulated in breast cancer, including TNBC7. CDK9 acts as a transcriptional activator in cancer to promote cancer cell growth, metastasis, and proliferation5. Hence, inhibition of CDK9 is emerging as a promising strategy for targeted therapy in TNBC. However, most reported CDK9 inhibitors are limited by their side effects due to lack of selectivity16. This is exacerbated by the high sequence and structural similarity among CDK family members, which leads to toxicity and off-target effects15. In particular, CDK1, CDK2, CDK5, CDK12 share a high degree of homology with CDK944. To overcome the selectivity issue of kinases, targeting less-conserved regulatory subunits may be a viable strategy. In particular, cyclin T1 forms a complex with CDK9 to activate the kinase activity of CDK945.
Metal complex inhibitors are emerging as attractive scaffolds for developing protein kinases due to their diverse structures. In addition, previous research has demonstrated that the quinoline motif may have CDK9–cyclin T1 PPI inhibitory activity20. In this study, we have synthesized Ir(III) and Rh(III) complexes containing the 7-chloro-2-phenylquinoline NˆN ligand as anti-tumor agents. In in vitro chemiluminescence and AlphaScreen assays, the Ir(III) complex 1 potently inhibited CDK9–cyclin T1 kinase activity and disrupted the CDK9–cyclin T1 PPI. CETSA results demonstrated the target selectivity of 1 for CDK9 over the related CDK1, CDK2, CDK5, and CDK12 kinases, while in enzymatic assays, 1 exhibited no obvious inhibition of CDK1, CDK2, CDK4/CDK6, CDK5, CDK12 activity. ITC experiments confirmed that complex 1 engaged CKD9 via an enthalpically-driven binding mode with CDK9. The GST-pull down and co-IP assay revealed that 1 could disrupt CDK9 and cyclin T1 PPI in cellulo. Complex 1 showed potent cytotoxicity of 1 against different breast cancer cell lines (include TNBC cell lines which show high levels of CDK9 expression). Significantly, complex 1 showed enhanced potency against CDK9-overexpressing TNBC cells but reduced potency against CDK9-knockdown TNBC cells, suggesting that the cytotoxicity exhibited by 1 could be associated with CDK9 expression status. While the effect of complex 1 on other targets cannot be ruled out, the above data provide strong evidence that the CDK9–cyclin T1 PPI plays a key role in mediating the biological effects of 1.
c-Myc and Mcl-1 are two important oncogenes regulated by CDK9 that regulate for cell proliferation, apoptosis, and metastasis9,62. Complex 1 blocked the binding of CDK9 to the c-Myc and Mcl-1 gene promoters in order to generate pro-apoptotic, anti-proliferative, and anti-migration effects in cellulo. The effect of 1 on CDK9–cyclin T1 PPI and anti-tumor activity were further investigated in a TNBC mice model. 1 significantly inhibited tumor growth, tumor proliferation, and metastasis compared to cisplatin-treated cohorts, without causing observable toxicity to the mice. We presume that the anti-tumor activity of complex 1 is due to its inhibition of the CDK9–cyclin T1 PPI, resulting in the downregulation of c-Myc and Mcl-1 in vivo. The potency of 1 against TNBC allografts was comparable to the clinically used anticancer drug, cisplatin.
4. Conclusions
In conclusion, we have synthesized the novel Ir(III) complex 1 as a potent CDK9–cyclin T1 PPI inhibitor. Complex 1 engages CDK9 via an enthalpically-driven binding mode to disrupt the CDK9–cyclin T1 interaction as demonstrated by multiple biochemical experiments, causing the reduction of oncogenic c-Myc and Mcl-1 expression. Moreover, complex 1 exhibited antiproliferative activity and inhibited EMT and CSC biomarkers in both MDA-MB-231 and 4T1 TNBC cell lines, as well as inducing apoptosis and decreasing migration. In a TNBC allograft model, complex 1 showed more potent inhibition of tumor growth and metastasis compared to the clinically used anticancer drug, cisplatin. This study also provides understanding into how the CDK9–cyclin T1 interaction mediates in TNBC and showcases the potential of targeting this PPI for TNBC therapy.
5. Experimental
5.1. Chemicals and reagents
Compounds 21 and 22 (commercially available, purity >95%), and cisplatin were purchased from J&K Scientific Ltd., Hong Kong, China. Compound 20 was purchased from Medchemexpress (Shanghai, China), and complexes 1–19 were synthesized. The purity of all complexes were validated by combustion using automatic analyzers (Atlantic Microlab Inc., Norcross, GA, USA) to confirm >95% purity. All the complexes were dissolved in dimethyl sulfoxide (DMSO). Lipo3000, MTT kit were purchased from Sigma–Aldrich (Santa Clara, CA, USA). Plasmids c-Myc, Mcl-1, HA-CDK9 were purchased from Addgene. TurboFect™ Transfection Reagent was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Proteins bands were detected using enhanced chemiluminescent Plus reagents (GE Healthcare) and analyzed by Image Lab. Dulbecco's modified Eagle's medium (DMEM), Fetal bovine serum (FBS) were obtained from Gibco BRL (Gaithersburg, MD, USA). All other reagents and chemicals were obtained from commercial sources.
5.2. Chemical synthesis
5.2.1. General experimental
Mass spectrometry was performed at the Mass Spectroscopy Unit at the Department of Chemistry, Hong Kong Baptist University, Hong Kong, China. Deuterated solvents for NMR purposes were obtained from Armar and used as received. 1H and 13C NMR were recorded on a Bruker Advance 400 spectrometer operating at 400 MHz (1H) and 100 MHz (13C). 1H and 13C chemical shifts were referenced internally to solvent shift (acetonitrile-d3 (δ): 1H 1.94, 13C 118.7; acetone-d6 (δ): 1H 2.05, 13C 29.7). Chemical shifts (d) are quoted in ppm, the downfield direction being defined as positive. Uncertainties in chemical shifts are typically ±0.01 ppm for 1H and ±0.05 for 13C. Coupling constants are typically ±0.1 Hz for 1H–1H and ±0.5 Hz for 1H–13C couplings. The following abbreviations are used for convenience in reporting the multiplicity of NMR resonances: s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet. All NMR data were acquired and processed using standard Bruker software (Topspin).
5.2.2. Synthesis of iridium(III) and rhodium(III) complexes
General synthetic procedure. Ir/Rh complexes were synthesized into two steps based on our previous report31. Firstly, the cyclometalated dichloro-bridged dimer of the general formula M2(CˆN)4Cl2 (where M = Ir or Rh) was first synthesized. In brief, MCl3·3H2O (1 eq) and cyclometallated CˆN ligands (2.1 eq) were mixed and heated to 130 °C in a mixture of 2-methoxyethanol and deionized water (3:1) under nitrogen protection overnight. The mixture was allowed to cool to room temperature and filtered. The precipitate was filtered and washed with several portions of water followed by diethyl ether and was dried to yield the dimers for next step. The final complexes were obtained from the reaction between the dimers and the corresponding NˆN ligands. In brief, a suspension of the dimers (1 eq) and the corresponding NˆN ligands (1.1 eq) were mixed and stirred in dichloromethane/methanol (v/v, 1/1) overnight under ambient temperature. To the resulting solution was added by excess ammonium hexafluorophosphate, and then the solution was stirred another 0.5 h. The solution was filtered to remove undissolved impurities, while the solvent was removed by rotary evaporation. The solid was redissolved into acetone, and then the product was precipitated by adding diethyl ether, and filtered to yield the desired solid. The complexes that were not pure by 1H NMR were further purified by preparative silica thin-layer chromatography (20 cm × 20 cm), which were fully characterized by 1H NMR, 13C NMR, HRMS and elemental analysis.
Complex 1: Yield: 76%. 1H NMR (400 MHz, acetone-d6) δ 8.60 (dt, J = 8.2, 1.1 Hz, 2H), 8.57 (s, 4H), 8.35 (ddd, J = 5.5, 1.6, 0.8 Hz, 2H), 8.28 (dd, J = 8.0, 1.3 Hz, 2H), 8.21 (td, J = 7.9, 1.6 Hz, 2H), 7.99 (dd, J = 8.4, 0.7 Hz, 2H), 7.78 (ddd, J = 7.7, 5.5, 1.2 Hz, 2H), 7.52–7.44 (m, 4H), 7.20 (ddd, J = 8.1, 7.3, 1.2 Hz, 2H), 6.88 (td, J = 7.5, 1.3 Hz, 2H), 6.57 (dd, J = 7.7, 1.1 Hz, 2H). 13C NMR (101 MHz, acetone-d6) δ 172.24, 156.46, 152.04, 149.02, 148.50, 146.42, 141.23, 140.89, 137.85, 134.94, 132.08, 132.02, 129.61, 128.85, 128.11, 127.38, 125.36, 125.02, 124.01, 119.41. MALDI-TOF-HRMS: Calcd. for C40H26Cl2IrN4 [M–PF6]+: 825.1146, Found: 825.1186. (C40H26Cl2IrN4PF6) C, H, N: Calcd. 49.49, 2.70, 5.77; Found 49.57, 2.61, 5.72.
Complex 2: Yield: 68%. 1H NMR (400 MHz, acetone-d6) δ 8.60 (dd, J = 8.9, 0.8 Hz, 2H), 8.40 (d, J = 8.8 Hz, 2H), 8.14–7.99 (m, 8H), 7.82 (d, J = 2.0 Hz, 2H), 7.62–7.49 (m, 4H), 7.08 (t, J = 7.2 Hz, 2H), 6.83 (t, J = 7.5 Hz, 2H), 6.44 (dd, J = 7.8, 1.1 Hz, 2H), 1.87 (s, 6H). 13C NMR (101 MHz, acetone-d6) δ 171.66, 163.20, 158.13, 147.90, 147.75, 145.40, 140.33, 140.09, 136.70, 132.75, 131.16, 130.86, 128.60, 127.64, 127.30, 126.56, 124.37, 122.87, 121.75, 117.88, 23.48. MALDI-TOF-HRMS: Calcd. for C42H30Cl2IrN4 [M–PF6]+: 853.1477, Found: 853.1410. (C42H30Cl2IrN4PF6) C, H, N: Calcd. 50.51, 3.03, 5.61; Found 50.75, 3.06, 5.64.
Complex 3: Yield: 77%. 1H NMR (400 MHz, acetone-d6) δ 8.56 (d, J = 1.0 Hz, 4H), 8.50–8.40 (m, 2H), 8.26 (dd, J = 7.9, 1.3 Hz, 2H), 8.14 (d, J = 5.7 Hz, 2H), 8.00 (d, J = 8.6 Hz, 2H), 7.59 (ddd, J = 5.7, 1.8, 0.8 Hz, 2H), 7.48 (dd, J = 8.6, 2.0 Hz, 2H), 7.43 (d, J = 2.0 Hz, 2H), 7.18 (ddd, J = 8.1, 7.2, 1.2 Hz, 2H), 6.86 (td, J = 7.5, 1.3 Hz, 2H), 6.55 (dd, J = 7.7, 1.1 Hz, 2H), 2.51 (s, 6H). 13C NMR (101 MHz, acetone-d6) δ 206.53, 206.33, 206.13, 172.38, 156.22, 153.39, 152.59, 148.65, 148.35, 146.51, 141.17, 137.87, 134.97, 132.02, 132.00, 130.22, 128.82, 128.10, 127.43, 125.87, 125.12, 123.88, 119.35, 21.17. MALDI-TOF-HRMS: Calcd. for C42H30Cl2IrN4 [M–PF6]+: 853.1459, Found: 853.1447. (C42H30Cl2IrN4PF6) C, H, N: Calcd. 50.56, 2.93, 5.62; Found 50.29, 2.98, 5.78.
Complex 4: Yield: 58%. 1H NMR (400 MHz, acetone-d6) δ 8.84 (d, J = 8.4 Hz, 2H), 8.74 (d, J = 5.0 Hz, 2H), 8.66–8.48 (m, 4H), 8.34 (d, J = 8.0 Hz, 2H), 8.23 (s, 2H), 8.16 (t, J = 5.1 Hz, 2H), 7.88 (d, J = 8.4 Hz, 2H), 7.37 (s, 2H), 7.34–7.22 (m, 3H), 6.94 (t, J = 7.7 Hz, 2H), 6.69 (d, J = 7.7 Hz, 2H). 13C NMR (101 MHz, acetone-d6) δ 172.39, 151.71, 149.85, 148.70, 147.53, 146.72, 141.19, 140.07, 137.56, 135.31, 132.13, 132.08, 131.86, 129.20, 128.89, 128.13, 127.99, 127.24, 124.57, 124.16, 119.48. MALDI-TOF-HRMS: Calcd. for C42H26Cl2IrN4 [M–PF6]+: 849.1146, Found: 849.1187; Anal.: (C42H26Cl2F6IrN4P) C, H, N: Calcd. 50.71, 2.63, 5.63; Found 50.88, 2.76, 5.78.
Complex 5: Yield: 61%. 1H NMR (400 MHz, acetone-d6) δ 8.94 (d, J = 8.6 Hz, 2H), 8.67 (d, J = 5.1, Hz, 2H), 8.63–8.50 (m, 4H), 8.33 (d, J = 7.9 Hz, 2H), 8.12 (dd, J = 8.6, 5.1 Hz, 2H), 7.88 (d, J = 8.6 Hz, 2H), 7.35–7.33 (m, 2H), 7.30 (d, J = 8.6 Hz, 2H), 7.24 (t, J = 7.5 Hz, 2H), 6.91 (t, J = 7.5 Hz, 2H), 6.65 (d, J = 7.7 Hz, 2H), 2.77 (s, 6H). 13C NMR (101 MHz, acetone-d6) δ 172.45, 152.35, 148.71, 148.48, 146.74, 146.70, 141.16, 137.58, 136.76, 135.26, 133.56, 132.40, 132.07, 131.86, 128.91, 128.01, 127.84, 127.24, 124.62, 124.10, 119.48, 15.44. MALDI-TOF-HRMS: Calcd. for C44H30Cl2IrN4 [M–PF6]+: 877.1477, Found: 877.1432; Anal.: (C44H30Cl2IrN4PF6 + 1.5 H2O) C, H, N: Calcd. 50.34, 3.17, 5.34; Found 50.12, 2.97, 5.67.
Complex 6: Yield: 69%. 1H NMR (400 MHz, acetone-d6) δ 8.62–8.48 (m, 6H), 8.34 (s, 2H), 8.31 (dd, J = 8.0, 1.3 Hz, 2H), 7.97 (dd, J = 5.3, 1.0 Hz, 2H), 7.88 (d, J = 8.6 Hz, 2H), 7.36 (d, J = 2.0 Hz, 2H), 7.31 (dd, J = 8.6, 2.0 Hz, 2H), 7.23 (t, J = 8.1 Hz, 2H), 6.91 (t, J = 7.5 Hz, 2H), 6.65 (d, J = 7.7 Hz, 2H), 2.94 (d, J = 0.8 Hz, 6H). 13C NMR (101 MHz, acetone-d6) δ 172.49, 152.53, 150.51, 149.20, 148.77, 147.35, 146.71, 141.11, 137.57, 135.25, 132.02, 131.83, 131.43, 128.88, 128.60, 127.96, 127.25, 125.42, 124.61, 123.99, 119.44, 18.96. MALDI-TOF-HRMS: Calcd. for C44H30Cl2IrN4 [M–PF6]+: 877.1477, Found: 877.1170. Anal.: (C44H30Cl2F6IrN4P) C, H, N: Calcd. 51.67, 2.96, 5.48; Found 51.38, 3.08, 5.64.
Complex 7: Yield: 76%. 1H NMR (400 MHz, acetone-d6) δ 8.66 (d, J = 8.8 Hz, 2H), 8.59 (d, J = 8.8 Hz, 2H), 8.53 (dt, J = 8.2, 1.0 Hz, 2H), 8.48–8.41 (m, 2H), 8.35 (dd, J = 7.9, 1.4 Hz, 2H), 8.19 (td, J = 7.9, 1.6 Hz, 2H), 8.00 (d, J = 8.7 Hz, 2H), 7.72 (ddd, J = 7.7, 5.4, 1.2 Hz, 2H), 7.58 (d, J = 1.9 Hz, 2H), 7.47 (dd, J = 8.7, 2.0 Hz, 2H), 7.28 (ddd, J = 8.0, 7.3, 1.1 Hz, 2H), 6.97 (td, J = 7.6, 1.4 Hz, 2H), 6.61 (dt, J = 7.7, 1.0 Hz, 2H). 13C NMR (101 MHz, acetone-d6) δ 169.03, 155.21, 149.14, 147.91, 146.14, 141.64, 141.08, 140.80, 137.34, 135.75, 131.77, 131.67, 128.97, 128.59, 128.25, 127.68, 125.05, 124.89, 119.67. MALDI-TOF-HRMS: Calcd. for C40H26Cl2F6N4PRh [M–PF6]+: 735.0584, Found: 735.0620. (C40H26Cl2F6N4PRh + 1.5H2O) C, H, N: Calcd. 52.88, 3.22, 6.17; Found 52.77, 2.98, 6.25.
Complex 8: Yield: 69%. 1H NMR (400 MHz, acetone-d6) δ 8.69 (dd, J = 8.8, 0.8 Hz, 2H), 8.42 (d, J = 8.8 Hz, 2H), 8.18–8.08 (m, 6H), 7.96 (t, J = 7.8 Hz, 2H), 7.89–7.80 (m, 2H), 7.57 (dd, J = 8.7, 2.0 Hz, 2H), 7.48 (dd, J = 7.7, 1.1 Hz, 2H), 7.16 (ddd, J = 8.0, 7.2, 1.1 Hz, 2H), 6.91 (ddd, J = 7.9, 7.2, 1.4 Hz, 2H), 6.47 (dt, J = 7.8, 1.1 Hz, 2H), 1.95 (s, 6H). 13C NMR (101 MHz, acetone-d6) δ 168.89, 163.38, 157.76, 154.27, 148.14, 146.32, 141.03, 140.98, 137.27, 134.88, 131.79, 128.96, 128.44, 127.74, 127.73, 125.46, 124.88, 122.33, 119.17, 114.18, 23.87. MALDI-TOF-HRMS: Calcd. for C42H30Cl2N4Rh [M–PF6]+: 763.0903, Found: 762.0282. Anal.: (C42H30Cl2N4RhPF6 + H2O) C, H, N: Calcd. 54.39, 3.48, 6.04; Found 54.46, 3.45, 6.13.
Complex 9: Yield: 51%. 1H NMR (400 MHz, CDCl3) δ 8.47 (m, 4H), 8.35 (d, J = 8.0 Hz, 1H), 8.15 (d, J = 4.0 Hz, 2H), 8.01 (d, J = 8.0 Hz, 1H), 7.93 (d, J = 8.0 Hz, 1H), 7.70 (d, J = 8.0 Hz, 1H), 7.22 (d, J = 12.0 Hz, 2H), 7.14 (m, 2H), 7.07 (d, J = 4.0 Hz, 3H), 6.84 (t, J = 8.0 Hz, 1H), 2.37 (s, 8H). 13C NMR (100 MHz, CDCl3) 167.69, 167.39, 165.87, 154.88, 153.01, 152.47, 147.89, 147.25, 146.17, 145.86, 143.74, 138.60, 136.16, 134.10, 129.83, 129.23, 127.31, 126.73, 126.05, 125.32, 124.45, 123.70, 123.35, 123.06, 121.03, 116.91, 20.19. MALDI-TOF-HRMS: Calcd. for C42H30Cl2IrN4Rh [M–PF6]+: 763.0897, Found: 763.0485.
Complexes 10–12: Reported18.
Complex 13: Yield: 74%. 1H NMR (400 MHz, acetone-d6) δ 8.62 (d, J = 2.0 Hz, 2H), 8.57 (s, 4H), 8.28 (dd, J = 7.9, 1.3 Hz, 2H), 8.21 (d, J = 5.9 Hz, 2H), 8.00 (d, J = 8.6 Hz, 2H), 7.80 (dd, J = 6.0, 2.0 Hz, 2H), 7.48 (dd, J = 8.7, 2.0 Hz, 2H), 7.38 (d, J = 2.0 Hz, 2H), 7.19 (ddd, J = 8.0, 7.2, 1.2 Hz, 2H), 6.86 (td, J = 7.4, 1.3 Hz, 2H), 6.55 (dd, J = 7.8, 1.1 Hz, 2H), 1.32 (s, 18H). 13C NMR (101 MHz, acetone-d6) δ 165.37, 155.53, 154.19, 152.96, 148.67, 148.58, 147.95, 146.48, 141.14, 137.74, 135.41, 134.93, 131.97, 128.81, 128.08, 126.81, 125.02, 123.86, 122.41, 119.36, 36.35, 30.36. MALDI-TOF-HRMS: Calcd. for C48H42Cl2IrN4 [M–PF6]+: 937.2399, Found: 937.2361. (C48H42Cl2IrN4PF6) C, H, N: Calcd. 53.23, 3.91, 5.17; Found 52.95, 3.76, 5.12.
Complex 14: Yield: 67%. 1H NMR (400 MHz, acetone-d6) δ 8.62–8.51 (m, 4H), 8.25 (dd, J = 7.9, 1.3 Hz, 2H), 8.14–7.97 (m, 6H), 7.54–7.46 (m, 4H), 7.31 (dd, J = 6.5, 2.6 Hz, 2H), 7.17 (ddd, J = 8.1, 7.3, 1.2 Hz, 2H), 6.84 (td, J = 7.5, 1.3 Hz, 2H), 6.56 (dd, J = 7.7, 1.1 Hz, 2H), 4.00 (s, 6H). 13C NMR (101 MHz, DMSO-d6) δ 170.89, 167.74, 156.50, 151.89, 148.23, 147.15, 145.40, 140.37, 136.15, 133.56, 131.35, 131.07, 128.17, 127.11, 126.18, 123.54, 122.71, 118.59, 114.90, 111.03, 56.93. MALDI-TOF-HRMS: Calcd. for C42H30Cl2IrN4O2 [M–PF6]+: 885.1357, Found: 885.1381. (C42H30Cl2IrN4O2PF6) C, H, N: Calcd. 48.94, 2.93, 5.44; Found 48.87, 2.77, 5.39.
Complex 15: Yield: 65%. 1H NMR (400 MHz, acetone-d6) δ 8.95 (d, J = 2.0 Hz, 2H), 8.65–8.54 (m, 4H), 8.27 (dd, J = 7.9, 1.3 Hz, 2H), 8.23 (d, J = 6.0 Hz, 2H), 8.06 (dd, J = 6.0, 2.0 Hz, 2H), 8.02 (d, J = 8.7 Hz, 2H), 7.50 (dd, J = 8.7, 2.0 Hz, 2H), 7.36 (d, J = 2.0 Hz, 2H), 7.21 (ddd, J = 8.1, 7.2, 1.2 Hz, 2H), 6.88 (td, J = 7.5, 1.3 Hz, 2H), 6.57 (dd, J = 7.8, 1.1 Hz, 2H). 13C NMR (101 MHz, MeOD) δ 172.6, 167.0, 159.5, 156.9, 154.2, 149.9, 145.1, 141.6, 138.8, 138.2, 135.3, 133.6, 132.3, 129.7, 129.2, 128.6, 127.9, 125.8, 125.2, 124.7, 119.6, 66.9. MALDI-TOF-HRMS: Calcd. for C40H24Br2Cl2IrN4 [M–PF6]+: 982.9334, Found: 982.9384. (C40H24Br2Cl2IrN4PF6) C, H, N: Calcd 50.83, 2.64, 5.65; Found 50.86, 2.65, 5.75.
Complex 16: Yield: 71%. 1H NMR (400 MHz, acetone-d6) δ 8.94 (dt, J = 13.6, 1.2 Hz, 2H), 8.63–8.54 (m, 6H), 8.30 (dd, J = 7.9, 1.2 Hz, 2H), 8.06 (ddd, J = 12.2, 5.6, 1.4 Hz, 2H), 8.00 (d, J = 8.6 Hz, 2H), 7.47 (dd, J = 8.7, 2.0 Hz, 2H), 7.36 (d, J = 2.0 Hz, 2H), 7.23 (ddd, J = 8.0, 7.2, 1.1 Hz, 2H), 6.90 (td, J = 7.5, 1.3 Hz, 2H), 6.58 (dd, J = 7.8, 1.1 Hz, 2H), 4.19–4.04 (m, 8H), 1.28–1.19 (m, 12H). 13C NMR (100 MHz, CDCl3) 170.82, 154.79, 149.62, 148.77, 148.65, 147.36, 144.74, 142.62, 140.76, 140.38, 137.60, 134.18, 131.82, 130.90, 130.30, 130.23, 128.09, 127.57, 126.24, 125.78, 123.92, 118.66, 64.21, 16.41. MALDI-TOF-HRMS: Calcd. for C48H44Cl2IrN4O6P2 [M–PF6]+: 1097.1725, Found: 1097.1759. Anal.: (C48H44Cl2IrN4O6P2PF6 + 2.5H2O) C, H, N: Calcd. 46.38, 3.57, 4.51; Found 46.27, 3.74, 4.56.
Complex 17: Yield: 78%. 1H NMR (400 MHz, acetone-d6) δ 8.83 (dd, J = 8.3, 1.5 Hz, 1H), 8.74 (d, J = 8.4 Hz, 1H), 8.66–8.57 (m, 2H), 8.56–8.46 (m, 2H), 8.32 (dd, J = 5.2, 1.5 Hz, 1H), 8.27–8.16 (m, 4H), 7.99 (dd, J = 8.3, 5.3 Hz, 2H), 7.94 (dd, J = 8.6, 4.5 Hz, 2H), 7.86 (d, J = 2.0 Hz, 1H), 7.40 (dd, J = 8.7, 2.0 Hz, 1H), 7.31 (dd, J = 8.6, 2.0 Hz, 1H), 7.19 (ddd, J = 8.0, 7.3, 1.1 Hz, 1H), 7.12 (ddd, J = 8.1, 7.2, 1.2 Hz, 1H), 6.88 (dtd, J = 17.7, 7.5, 1.3 Hz, 2H), 6.81 (d, J = 2.0 Hz, 1H), 6.55 (dd, J = 7.7, 1.1 Hz, 1H), 6.32 (dd, J = 7.8, 1.0 Hz, 1H), 2.55 (s, 3H). 13C NMR (101 MHz, acetone-d6) δ 173.83, 171.73, 166.04, 152.96, 150.09, 149.28, 148.74, 148.49, 148.42, 147.68, 147.25, 146.04, 141.52, 141.31, 140.39, 140.27, 137.70, 137.18, 133.87, 132.94, 132.74, 132.50, 132.37, 131.94, 131.91, 130.64, 129.53, 129.44, 129.04, 128.77, 128.27, 128.15, 127.70, 127.54, 127.38, 127.28, 124.89, 124.44, 124.41, 123.55, 119.35, 119.26, 26.98. MALDI-TOF-HRMS: Calcd. for C43H28Cl2IrN4 [M–PF6]+: 863.1303, Found: 863.1329. (C43H28Cl2IrN4PF6) C, H, N: Calcd. 51.20, 2.80, 5.55; Found 51.25, 2.92, 5.58.
Complex 18: Yield: 77%. 1H NMR (400 MHz, acetone-d6) δ 8.61 (t, J = 2.8 Hz, 4H), 8.54 (dd, J = 6.1, 2.5 Hz, 2H), 8.43–8.33 (m, 2H), 8.21 (d, J = 2.4 Hz, 2H), 7.90 (dd, J = 8.6, 2.5 Hz, 2H), 7.55 (dt, J = 4.9, 2.4 Hz, 4H), 7.30 (ddd, J = 10.4, 6.9, 2.4 Hz, 4H), 7.06–6.93 (m, 2H), 6.68 (dd, J = 7.7, 2.3 Hz, 2H), 4.20 (d, J = 2.4 Hz, 8H). 13C NMR (101 MHz, acetone-d6) δ 169.51, 168.39, 165.11, 151.21, 148.16, 146.58, 146.27, 140.58, 136.95, 136.00, 131.51, 131.41, 128.46, 128.03, 127.53, 124.88, 124.70, 123.45, 121.32, 119.56, 107.89, 57.89. MALDI-TOF-HRMS: Calcd. for C44H30Cl2N4O2Rh [M–PF6]+: 819.0795, Found: 819.0872. (C44H30Cl2N4O2RhPF6 + H2O) C, H, N: Calcd. 53.73, 3.28, 5.70; Found 53.55, 3.17, 6.05.
Complex 19: Yield: 66%. 1H NMR (400 MHz, acetone-d6) δ 8.83 (d, J = 5.5 Hz, 2H), 8.62 (d, J = 2.3 Hz, 4H), 8.51 (d, J = 1.5 Hz, 2H), 8.40 (dd, J = 7.9, 1.5 Hz, 2H), 8.30 (d, J = 5.5 Hz, 2H), 7.90 (d, J = 8.7 Hz, 2H), 7.37 (d, J = 2.0 Hz, 2H), 7.36–7.28 (m, 4H), 7.01 (td, J = 7.5, 1.5 Hz, 2H), 6.70 (dt, J = 7.8, 1.0 Hz, 2H). 13C NMR (101 MHz, acetone-d6) δ 168.39, 168.29, 168.08, 150.70, 148.06, 146.76, 146.59, 146.20, 140.93, 137.14, 136.08, 131.75, 129.87, 128.81, 128.53, 128.17, 127.70, 125.84, 125.33, 124.42, 119.79. MALDI-TOF-HRMS: Calcd. for C42H24Cl4N4Rh [M–PF6]+: 795.0174, Found: 828.9445. (C42H24Cl4N4RhPF6 + H2O) C, H, N: Calcd. 50.83, 2.64, 5.65; Found 50.86, 2.65, 5.75.
5.3. Cell lines and culture
Breast cancer cell lines MCF-10A, MCF-7, MDA-MB-231, 4T1, and human normal cell lines LO2, HUEVC, HEK-293T were purchased from American Type Culture Collection (Manassas, VA, USA). Cells were cultured in DMEM, with 10% heat-inactivated FBS (Gibco, Grand Island, NY, USA), 100 units/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL amphotericin B.
5.4. CDK9/cyclin T1 activity inhibition assay
The ability of compounds 1–22 to inhibit CDK9 and cyclin T1 activity was evaluated using the CDK9 Assay Kit (BPS Bioscience, San Diego, CA, USA) according to the manufacturer's instruction. The concentration of compounds was 10 nmol/L.
5.5. AlphaScreen assay
10 μL of 300 nmol/L His-CDK9 protein and 2 μL of compounds 1–20 were mixed and incubated in a OptiPlate-384 assay plate (PerkinElmer) at room temperature for 30 min. Then, 10 μL of 300 nmol/L GST-cyclin T1 protein was added and the mixture was incubated for a further 30 min. The acceptor beads and donor beads were diluted with 1 × assay buffer (0.1% BSA, pH = 7.4). 10 μL of the acceptor beads were added to each well and the mixture was incubated for 60 min at room temperature. Next, 10 μL of donor beads was added to each well and the mixture incubated at room temperature for 60 min. Finally, the signal was read with the SpectraMax Paradigm Platform with AlphaScreen 384 Detection Cartridge.
5.6. Co-immunoprecipitation (co-IP) assay
The method was performed as previously described32. Briefly, MDA-MB-231 cells were treated with the 3 μmol/L complex 1 and 20 for 4 h. Cells were lysed and collected protein samples. Then calculate the concentration of protein samples through bicinchoninic acid method. 20 μg of each protein sample were incubated overnight with 10 μL pre-incubated anti-cyclin T1 magnetic beads. The complex was washed 3 times to elute non-specific and non-cross-linked antibodies. Then, the precipitated proteins were subjected to 10% SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting with the indicated antibodies. The antibodies against CDK9 (1:1000, cat# ab76320), cyclin T1 (1:1000, cat# ab184703), and β-actin (1:1000, cat# ab8227) purchased from Abcam (Cambridge, MA, USA). CDK7 (1:1000, cat# abs113221) was purchased from Absin. Cyclin H (1:1000, cat#AF2419) was purchased from Beyotime. Cyclin T2 (1:1000, cat#sc812443) was purchased from Santa Cruz Biotechnology. Proteins bands were detected using enhanced chemiluminescent Plus reagents (GE Healthcare) and analyzed by Image Lab.
5.7. GST pull-down assay
The method was performed as previously described33. The pull-down assay was performed by using a gravity flow column with GST-agarose beads. Cyclin T1 expressed as a GST-cyclin T1 fusion protein and CDK9 expressed as a His-CDK9 fusion protein for the pull-down experiment. GST-cyclin T1 fusion protein was immobilized on a GST-gravity column, then His-tagged purified CDK9 and complex 1 were added into the column. After washing the column three times with wash buffer, 10 mmol/L reduced glutathione was then used to elute the proteins which were analyzed using Western blotting.
5.8. Cellular thermal shift assay
MDA-MB-231 cells were seeded in a 100 cm2 dish at a density of 1 × 105/mL. Cell lysates were collected and incubated with 3 μmol/L complex 1 or DMSO for 30 min at room temperature. Then, the treated lysates were divided into 50 μL in PCR tubes and heated individually at different temperatures (Veriti thermal cycler, Applied Biosystems/Life Technologies). The heated lysates were centrifuged and the supernatants were analyzed by 10% SDS-polyacrylamide gel electrophoresis followed by immunoblotting analysis by probing with the indicated antibody.
5.9. High-sensitivity isothermal titration calorimetry (ITC)
ITC was used to characterize the binding affinity of metal complex 1 with CDK9 protein. The experiments were performed on MicroCal PEAQ-ITC microcalorimeter (Malvern Panalytical GmbH, Germany). Both interaction partners were dissolved in PBS. Experiments were carried out at 25 °C. In routine experiments, CDK9 (5 μmol/L) was titrated in 2 μL-steps into 1 solution (100 μmol/L) in the calorimeter cell. Time intervals between the injections were adjusted to 200 s, which was sufficient for the heat signal to return to baseline. Reaction mixtures were continuously stirred at 750 rpm. Dilution heats associated with the 1 addition into the buffer (determined in separate control experiments) had small constant values that were negligible to measured binding heats. The instrument software (MicroCal PEAQ-ITC Analysis) was used for baseline adjustment, peak integration, and normalization of the reaction heats with respect to the molar amount of injected ligand as well as for data fitting and binding parameter evaluation.
5.10. Plasmid transfection assay
MDA-MB-231 cells were seeded in a 6-well plate with a density of 2 × 105 cell/mL. DMEM mixed with empty vector control or plasmids combined with TurboFect for 15 min at 37 °C, before adding to cells. Then analyze transgene expression after 24–48 h.
5.11. Knockdown assay
The knockdown assay was performed as previously described34. Briefly, MDA-MB-231 cells were seeded in a 6-well plate at 70% confluences in DMEM medium for 24 h. CDK9 siRNA (5′-GGAGAAUUUUACUGUGUUUtt-3) and Lipo3000 regent were gently mixed and incubated for 20 min at room temperature. Remove growth medium from cells and replace with 0.5 mL of fresh medium. Then the mixture of 500 μL of Lipo3000 and CDK9 siRNA were added to each well. Cells were incubated at 37 °C in a CO2 incubator for 48 h post-transfection before the further research.
5.12. Animal experiment
Female BALB/c mice were purchased from The Chinese University of Hong Kong. All the experiments were carried out according to the “Institutional Animal Care and User Committee guidelines” of the Macau University of Science and Technology, China. 4T1 cells were orthotopically injected under the second pair of breasts. Mice were randomly divided into five groups, and monitored for body weight and tumor volumes as previously described18. Mice were subcutaneously inoculated with 1.0 × 106 4T1 cells, and after the tumor volume reached 60 mm3, injected intraperitoneally daily with 1 or 2 or 4 mg/kg of complex 1 or 0.5 mg/kg of cisplatin for 18 days. Tumor and lung samples were collected, fixed with 4% paraformaldehyde for 48 h, processed with paraffin, and cut into sections at 5 μmol/L thickness. H&E-stained lung sections were examined by microscope for metastatic lesions. Lung tissues were imaged by Leica DFC310 FX camera and lung areas were captured & analyzed by Leica Application Suit V4.4 software. The percentage of metastatic lung area was calculated as metastatic burden area/lung area. All tumor sections for the immunohistochemistry detection of tumor marker, PCNA, were performed on the Dako Link 48 System. Antigen retrieval was completed by using Dako Target Retrieval Solution in low pH (6.1) at 97 °C for 20 min. PCNA antibody (sc-25280, Santa Cruz) was diluted in 1:4000 and the PCNA signal was detected by Dako Flex + (Polymer system) with DAB as chromogen. The assessment of PCNA based on a mean score considering both the intensity of staining and the proportion of tumor cells with an unequivocal positive reaction35. Positive reactions were defined as those showing brown signals in the cell nucleus. For PCNA, a staining index (values, 0–12) was determined by multiplying the score for staining intensity with the score for positive area. The intensity was scored as follows: 0, negative; 1, weak; 2, moderate; and 3, strong. The frequency of positive cells was defined as follows: 0, less than 5%; 1, 5%–25%; 2, 26%–50%; 3, 51%–75%; and 4, greater than 75%. For example, a specimen containing 75% tumor cells with moderate intensity (3 × 2 = 6) and another 25% tumor cells with weak intensity (1 × 1 = 1) received a final score of 6 + 1 = 7. For statistical analysis, scores of 0–7 were considered low expression and scores of 8–12 considered high expression. Six areas of each PCNA stained tumor section were randomly captured by Leica Application Suit V4.4 software. Mann–Whitney t-test was used to statistically analyze the results of tumor metastasis and PCNA assignment.
5.13. Statistical analysis
All statistical tests were performed using GraphPad Prism version 8.0 (Graph Pad, San Diego, CA, USA). Statistical significance was determined using the Student's t-test for experiments comparing two groups. Comparisons among groups were analyzed using analysis of variance (ANOVA). Unless stated otherwise, P values were 2-tailed and considered significant if P < 0.05. Error bars represent SEM of three experiments unless stated otherwise.
Acknowledgments
This research is supported by Hong Kong Baptist University, the Health and Medical Research Fund (HMRF/14150561), the National Natural Science Foundation of China (22077109 and 21775131), the Science and Technology Development Fund, Macau SAR (File no. 0007/2020/A), SKL-QRCM(UM)-2020-2022, the University of Macau (MYRG2019-00002-ICMS, China), Foshan Medicine Dengfeng Project of China (2019–2021), 2020 Guangdong Provincial Science and Technology Innovation Strategy Special Fund (Guangdong-Hong Kong-Macau Joint Lab, No: 2020B1212030006, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2021.10.024.
Contributor Information
Vincent Kam Wai Wong, Email: bowaiwong@gmail.com.
Dik-Lung Ma, Email: edmondma@hkbu.edu.hk.
Chung-Hang Leung, Email: duncanleung@um.edu.mo.
Author contributions
Sha-Sha Cheng, Yuan-Qing Qu, Jia Wu, Guan-Jun Yang: experimental design, data acquisition and analysis, manuscript preparation. Hao Liu, Wanhe Wang, Chun-Yuen Wong, Guodong Li: data acquisition and analysis. Qi Huang: data acquisition. Feng Chen: preparation of materials. Vincent Kam Wai Wong, Dik-Lung Ma, Chung-Hang Leung: supervision and design of the project, data analysis and manuscript preparation.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lee K.L., Kuo Y.C., Ho Y.S., Huang Y.H. Triple-negative breast cancer: current understanding and future therapeutic breakthrough targeting cancer stemness. Cancers. 2019;11:1344–1380. doi: 10.3390/cancers11091334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng S.S., Yang G.J., Wang W.H., Ma D.L., Leung C.H. Discovery of a tetrahydroisoquinoline-based CDK9‒cyclin T1 protein–protein interaction inhibitor as an anti-proliferative and anti-migration agent against triple-negative breast cancer cells. Genes Dis. 2021 doi: 10.1016/j.gendis.2021.06.005. https://www.sciencedirect.com/science/article/pii/S2352304221000866 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang G.J., Wang W., Mok S.W.F., Wu C., Law B.Y.K., Miao X.M., et al. Selective inhibition of lysine-specific demethylase 5A (KDM5A) using a rhodium (III) complex for triple-negative breast cancer therapy. Angew Chem Int Ed. 2018;57:13091–13095. doi: 10.1002/anie.201807305. [DOI] [PubMed] [Google Scholar]
- 4.Al-Mahmood S., Sapiezynski J., Garbuzenko O.B., Minko T. Metastatic and triple-negative breast cancer: challenges and treatment options. Drug Deliv Transl Res. 2018;8:1483–1507. doi: 10.1007/s13346-018-0551-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLaughlin R.P., He J., van der Noord V.E., Redel J., Foekens J.A., Martens J.W.M., et al. A kinase inhibitor screen identifies a dual cdc7/CDK9 inhibitor to sensitise triple-negative breast cancer to EGFR-targeted therapy. Breast Cancer Res. 2019;21:77–92. doi: 10.1186/s13058-019-1161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cidado J., Boiko S., Proia T., Ferguson D., Criscione S.W., San Martin M., et al. AZD4573 is a highly selective CDK9 inhibitor that suppresses mcl-1 and induces apoptosis in hematologic cancer cells. Clin Cancer Res. 2020;26:922–934. doi: 10.1158/1078-0432.CCR-19-1853. [DOI] [PubMed] [Google Scholar]
- 7.Brisard D., Eckerdt F., Marsh L.A., Blyth G.T., Jain S., Cristofanilli M., et al. Antineoplastic effects of selective CDK9 inhibition with atuveciclib on cancer stem-like cells in triple-negative breast cancer. Oncotarget. 2018;9:37305–37318. doi: 10.18632/oncotarget.26468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee K.M., Giltnane J.M., Balko J.M., Schwarz L.J., Guerrero-Zotano A.L., Hutchinson K.E., et al. MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab. 2017;26:633–647. doi: 10.1016/j.cmet.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong Z., Mejia A., Veeranki O., Verma A., Correa A.M., Dokey R., et al. Targeting CDK9 and mcl-1 by a new CDK9/p-TEFb inhibitor with and without 5-fluorouracil in esophageal adenocarcinoma. Ther Adv Med Oncol. 2019;11:1–17. doi: 10.1177/1758835919864850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katsuta E., Yan L., Takeshita T., McDonald K.A., Dasgupta S., Opyrchal M., et al. High myc mRNA expression is more clinically relevant than myc DNA amplification in triple-negative breast cancer. Int J Mol Sci. 2019;21:217–229. doi: 10.3390/ijms21010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolomsky A., Vogler M., Köse M.C., Heckman C.A., Ehx G., Ludwig H., et al. Mcl-1 inhibitors, fast-lane development of a new class of anti-cancer agents. J Hematol Oncol. 2020;13:1–19. doi: 10.1186/s13045-020-01007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H., Liu H., Qing G. Targeting oncogenic myc as a strategy for cancer treatment, signal transduction and targeted therapy. Signal Transduct Target Ther. 2018;3:1–7. doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussain A., Verma C.K., Chouhan U. Identification of novel inhibitors against cyclin dependent kinase 9/cyclin T1 complex as: anti cancer agent. Saudi J Biol Sci. 2017;24:1229–1242. doi: 10.1016/j.sjbs.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deep A., Marwaha R.K., Marwaha M.G., Nandal R., Sharma A.K. Flavopiridol as cyclin dependent kinase (CDK) inhibitor: a review. New J Chem. 2018;42:18500–18507. [Google Scholar]
- 15.Bhullar K.S., Lagarón N.O., McGowan E.M., Parmar I., Jha A., Hubbard B.P., et al. Kinase-targeted cancer therapies: progress, challenges and future directions. Mol Cancer. 2018;17:48. doi: 10.1186/s12943-018-0804-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krystof V., Baumli S., Fürst R. Perspective of cyclin-dependent kinase 9 (CDK9) as a drug target. Curr Pharm Des. 2012;18:2883–2890. doi: 10.2174/138161212800672750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong H.J., Lu L., Leung K.H., Wong C.C.L., Peng C., Yan S.C., et al. An iridium (III)-based irreversible protein–protein interaction inhibitor of BRD4 as a potent anticancer agent. Chem Sci. 2015;6:5400–5408. doi: 10.1039/c5sc02321a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang G.J., Zhong H.J., Ko C.N., Wong S.Y., Vellaisamy K., Ye M., et al. Identification of a rhodium(III) complex as a wee1 inhibitor against TP53-mutated triple-negative breast cancer cells. Chem Commun (Camb) 2018;54:2463–2466. doi: 10.1039/c7cc09384e. [DOI] [PubMed] [Google Scholar]
- 19.Randjelović J., Erić S., Savić V. Computational study and peptide inhibitors design for the CDK9–cyclin T1 complex. J Mol Model. 2013;19:1711–1725. doi: 10.1007/s00894-012-1735-2. [DOI] [PubMed] [Google Scholar]
- 20.Randjelovic J., Eric S., Savic V. In silico design of small molecule inhibitors of CDK9/cyclin T1 interaction. J Mol Graph Model. 2014;50:100–112. doi: 10.1016/j.jmgm.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Jain S., Chandra V., Jain P.K., Pathak K., Pathak D., Vaidya A. Comprehensive review on current developments of quinoline-based anticancer agents. Arab J Chem. 2019;12:4920–4946. [Google Scholar]
- 22.Fischer P.M. Approved and experimental small-molecule oncology kinase inhibitor drugs: a mid-2016 overview. Med Res Rev. 2017;37:314–367. doi: 10.1002/med.21409. [DOI] [PubMed] [Google Scholar]
- 23.Dietrich J., Hulme C., Hurley L.H. The design, synthesis, and evaluation of 8 hybrid DFG-out allosteric kinase inhibitors: a structural analysis of the binding interactions of Gleevec®, Nexavar®, and BIRB-796. Bioorg Med Chem. 2010;18:5738–5748. doi: 10.1016/j.bmc.2010.05.063. [DOI] [PubMed] [Google Scholar]
- 24.Honma T., Hayashi K., Aoyama T., Hashimoto N., Machida T., Fukasawa K., et al. Structure-based generation of a new class of potent CDK4 inhibitors: new de novo design strategy and library design. J Med Chem. 2001;44:4615–4627. doi: 10.1021/jm0103256. [DOI] [PubMed] [Google Scholar]
- 25.El Newahie A.M.S., Ismail N.S.M., Abou El Ella D.A., Abouzid K.A.M. Quinoxaline-based scaffolds targeting tyrosine kinases and their potential anticancer activity. Arch Pharm. 2016;349:309–326. doi: 10.1002/ardp.201500468. [DOI] [PubMed] [Google Scholar]
- 26.Solbak S.M.Ø., Zang J., Narayanan D., Høj L.J., Bucciarelli S., Softley C., et al. Developing inhibitors of the p47phox–p22phox protein–protein interaction by fragment-based drug discovery. J Med Chem. 2020;63:1156–1177. doi: 10.1021/acs.jmedchem.9b01492. [DOI] [PubMed] [Google Scholar]
- 27.Carden C.P., Kim E.S., Jones R.L., Alam S.M., Johnson F.M., Stephens S.A.W., et al. Phase I study of intermittent dosing of OSI-906, a dual tyrosine kinase inhibitor of insulin-like growth factor-1 receptor (IGF-1R) and insulin receptor (IR) in patients with advanced solid tumors. J Clin Oncol. 2010;2007:2530. [Google Scholar]
- 28.Meggers E., Atilla-Gokcumen G.E., Bregman H., Maksimoska J., Mulcahy S.P., Pagano N., et al. Exploring chemical space with organometallics: ruthenium complexes as protein kinase inhibitors. Synlett. 2007;8:1177–1189. [Google Scholar]
- 29.Maksimoska J., Feng L., Harms K., Yi C.L., Kissil J., Marmorstein R., et al. Targeting large kinase active site with rigid, bulky octahedral ruthenium complexes. J Am Chem Soc. 2008;130:15764–15765. doi: 10.1021/ja805555a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Del Re M., Bertolini I., Crucitta S., Fontanelli L., Rofi E., De Angelis C., et al. Overexpression of TK1 and CDK9 in plasma-derived exosomes is associated with clinical resistance to CDK4/6 inhibitors in metastatic breast cancer patients. Breast Cancer Res Treat. 2019;178:57–62. doi: 10.1007/s10549-019-05365-y. [DOI] [PubMed] [Google Scholar]
- 31.Wang W.H., Yang C., Lin S., Vellaisamy K., Li G.D., Tan W., et al. First synthesis of an oridonin-conjugated iridium (III) complex for the intracellular tracking of NF-κB in living cells. Chem Eur J. 2017;23:4929–4935. doi: 10.1002/chem.201700770. [DOI] [PubMed] [Google Scholar]
- 32.Yang C., Wang W., Li G.D., Zhong H.J., Dong Z., Wong C.K., et al. Anticancer osmium complex inhibitors of the HIF-1α and p300 protein‒protein interaction. Sci Rep. 2017;7:42860–42869. doi: 10.1038/srep42860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li G.D., Ko C.N., Li D., Yang C., Wang W.H., Yang G.J., et al. A small molecule HIF-1α stabilizer that accelerates diabetic wound healing. Nat Commun. 2021;12:1–11. doi: 10.1038/s41467-021-23448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang T.S., Wang W., Zhong H.J., Dong Z.Z., Huang Q., Mok S.W., et al. An anti-prostate cancer benzofuran-conjugated iridium(III) complex as a dual inhibitor of STAT3 and NF-κB. Cancer Lett. 2017;396:76–84. doi: 10.1016/j.canlet.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 35.Brown H.M., Komorowski R.A., Wilson S.D., Demeure M.J., Zhu Y.R. Predicting metastasis of pheochromocytomas using DNA flow cytometry and immunohistochemical markers of cell proliferation: a positive correlation between MIB-1 staining and malignant tumor behavior. Cancer. 1999;86:1583–1589. [PubMed] [Google Scholar]
- 36.Kaes C., Katz A., Hosseini M.W. Bipyridine: the most widely used ligand. a review of molecules comprising at least two 2,2′-bipyridine units. Chem Rev. 2000;100:3553–3590. doi: 10.1021/cr990376z. [DOI] [PubMed] [Google Scholar]
- 37.Accorsi G., Listorti A., Yoosaf K., Armaroli N. 1,10-Phenanthrolines: versatile building blocks for luminescent molecules, materials and metal complexes. Chem Soc Rev. 2009;38:1690–1700. doi: 10.1039/b806408n. [DOI] [PubMed] [Google Scholar]
- 38.Baker A., Gregory G.P., Verbrugge I., Kats L., Hilton J.J., Vidacs E., et al. The CDK9 inhibitor dinaciclib exerts potent apoptotic and antitumor effects in preclinical models of MLL-rearranged acute myeloid leukemia. Cancer Res. 2016;76:1158–1169. doi: 10.1158/0008-5472.CAN-15-1070. [DOI] [PubMed] [Google Scholar]
- 39.Law M.E., Corsino P.E., Narayan S., Law B.K. Cyclin-dependent kinase inhibitors as anticancer therapeutics. Mol Pharmacol. 2015;88:846–852. doi: 10.1124/mol.115.099325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anshabo A.T., Milne R., Wang S., Albrecht H. CDK9: a comprehensive review of its biology, and its role as a potential target for anti-cancer agents. Front Oncol. 2021;11:1573. doi: 10.3389/fonc.2021.678559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonawane Y.A., Taylor M.A., Napoleon J.V., Rana S., Contreras J.I., Natarajan A. Cyclin dependent kinase 9 inhibitors for cancer therapy: miniperspective. J Med Chem. 2016;59:8667–8684. doi: 10.1021/acs.jmedchem.6b00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glover-Cutter K., Larochelle S., Erickson B., Zhang C., Shokat K., Fisher R.P., et al. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol. 2009;29:5455–5464. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matt Kim J., McGaughy J.T., Kent Bogle R., Ravnik S.E. Meiotic expression of the cyclin H/Cdk7 complex in male germ cells of the mouse. Biol Reprod. 2001;64:1400–1408. doi: 10.1095/biolreprod64.5.1400. [DOI] [PubMed] [Google Scholar]
- 44.De Falco G., Giordano A. CDK9: from basal transcription to cancer and AIDS. Cancer Biol Ther. 2002;1:341–346. [PubMed] [Google Scholar]
- 45.Baumli S., Lolli G., Lowe E.D., Troiani S., Rusconi L., Bullock A.N., et al. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27:1907–1918. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlafstein A.J., Withers A.E., Rudra S., Danelia D., Switchenko J.M., Mister D., et al. CDK9 expression shows role as a potential prognostic biomarker in breast cancer patients who fail to achieve pathologic complete response after neoadjuvant chemotherapy. Int J Breast Cancer. 2018;2018:1–10. doi: 10.1155/2018/6945129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu T., Qin Z., Tian Y., Wang J., Xu C., Li Z., et al. Recent developments in the biology and medicinal chemistry of CDK9 inhibitors: an update. J Med Chem. 2020;63:13228–13257. doi: 10.1021/acs.jmedchem.0c00744. [DOI] [PubMed] [Google Scholar]
- 48.Bacon C.W., D'Orso I. CDK9: a signaling hub for transcriptional control. Transcription. 2019;10:57–75. doi: 10.1080/21541264.2018.1523668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H., Pandey S., Travers M., Sun H., Morton G., Madzo J., et al. Targeting CDK9 reactivates epigenetically silenced genes in cancer. Cell. 2018;175:1244–1258. doi: 10.1016/j.cell.2018.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boffo S., Damato A., Alfano L., Giordano A. CDK9 inhibitors in acute myeloid leukemia. Cancer Res. 2018;37:36–46. doi: 10.1186/s13046-018-0704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips D.C., Jin S., Gregory G.P., Zhang Q., Xue J., Zhao X., et al. A novel CDK9 inhibitor increases the efficacy of venetoclax (ABT-199) in multiple models of hematologic malignancies. Leukemia. 2020;34:1646–1657. doi: 10.1038/s41375-019-0652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu J.Y., Li B.X., He J., Luo Q.Y., Dai L.T., Zhu X.Y., et al. A mitophagy-mediated autophagy pathway that triggers apoptotic cell death in non-small cell lung cancer (NSCLC) for anticancer chemotherapy by norcantharidin. Authorea. 2020;2020:1–12. Preprints. [Google Scholar]
- 53.Losuwannarak N., Sritularak B., Chanvorachote P. Cycloartobiloxanthone induces human lung cancer cell apoptosis via mitochondria-dependent apoptotic pathway. In Vivo. 2018;32:71–78. doi: 10.21873/invivo.11206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Entschladen F., Drell T.L., 4th, Lang K., Joseph J., Zaenker K.S. Tumour-cell migration, invasion, and metastasis: navigation by neurotransmitters. Lancet Oncol. 2004;5:254–258. doi: 10.1016/S1470-2045(04)01431-7. [DOI] [PubMed] [Google Scholar]
- 55.Chanvorachote P., Sriratanasak N., Nonpanya N. C-myc contributes to malignancy of lung cancer: a potential anticancer drug target. Anticancer Res. 2020;40:609–618. doi: 10.21873/anticanres.13990. [DOI] [PubMed] [Google Scholar]
- 56.Huang H., Shah K., Bradbury N.A., Li C., White C. Mcl-1 promotes lung cancer cell migration by directly interacting with VDAC to increase mitochondrial Ca2+ uptake and reactive oxygen species generation. Cell Death Dis. 2014;5:e1482. doi: 10.1038/cddis.2014.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guzińska-Ustymowicz K., Pryczynicz A., Kemona A., Czyzewska J. Correlation between proliferation markers: PCNA, Ki-67, MCM-2 and antiapoptotic protein Bcl-2 in colorectal cancer. Anticancer Res. 2009;29:3049–3052. [PubMed] [Google Scholar]
- 58.Joshua P.E., Nwauzor C.O., Odimegwu D.C., Ukachukwu U.G., Asomadu R.O., Ezeorba T.P.C. Experimental and molecular predictions of the adjuvanticity of snail mucin on hepatitis B vaccine in albino mice. PLoS One. 2021;16 doi: 10.1371/journal.pone.0246915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brosius F.C., Alpers C.E., Bottinger E.P., Breyer M.D., Coffman T.M., Gurley S.B., et al. Mouse models of diabetic nephropathy. J Am Soc Nephrol. 2009;20:2503–2512. doi: 10.1681/ASN.2009070721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Damaskos C., Garmpi A., Nikolettos K., Vavourakis M., Diamantis E., Patsouras A., et al. Triple-negative breast cancer: the progress of targeted therapies and future tendencies. Anticancer Res. 2019;39:5285–5296. doi: 10.21873/anticanres.13722. [DOI] [PubMed] [Google Scholar]
- 61.Gross S., Rahal R., Stransky N., Lengauer C., Hoeflich K.P. Targeting cancer with kinase inhibitors. J Clin Investig. 2015;125:1780–1789. doi: 10.1172/JCI76094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolfer A., Wittner B.S., Irimia D., Flavin R.J., Lupien M., Gunawardane R.N., et al. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc Natl Acad Sci U S A. 2010;107:3698–3703. doi: 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.