Abstract
The immune checkpoint blockade (ICB) targeting on PD-1/PD-L1 has shown remarkable promise in treating cancers. However, the low response rate and frequently observed severe side effects limit its broad benefits. It is partially due to less understanding of the biological regulation of PD-L1. Here, we systematically and comprehensively summarized the regulation of PD-L1 from nuclear chromatin reorganization to extracellular presentation. In PD-L1 and PD-L2 highly expressed cancer cells, a new TAD (topologically associating domain) (chr9: 5,400,000–5,600,000) around CD274 and CD273 was discovered, which includes a reported super-enhancer to drive synchronous transcription of PD-L1 and PD-L2. The re-shaped TAD allows transcription factors such as STAT3 and IRF1 recruit to PD-L1 locus in order to guide the expression of PD-L1. After transcription, the PD-L1 is tightly regulated by miRNAs and RNA-binding proteins via the long 3′UTR. At translational level, PD-L1 protein and its membrane presentation are tightly regulated by post-translational modification such as glycosylation and ubiquitination. In addition, PD-L1 can be secreted via exosome to systematically inhibit immune response. Therefore, fully dissecting the regulation of PD-L1/PD-L2 and thoroughly detecting PD-L1/PD-L2 as well as their regulatory networks will bring more insights in ICB and ICB-based combinational therapy.
KEY WORDS: Immune checkpoint blockade, PD-L1, PD-L2, Topologically associating domain, Transcription, Post-transcriptional regulation, Post-translational regulation, Exosome
Abbreviations: 3′-UTR, 3′-untranslated region; ADAM17, a disintegrin and metalloprotease 17; APCs, antigen-presenting cells; AREs, adenylate and uridylate (AU)-rich elements; ATF3, activating transcription factor 3; CD273/274, cluster of differentiation 273/274; CDK4, cyclin-dependent kinase 4; CMTM6, CKLF like MARVEL transmembrane domain containing 6; CSN5, COP9 signalosome subunit 5; CTLs, cytotoxic T lymphocytes; dMMR, deficient DNA mismatch repair; EMT, epithelial to mesenchymal transition; EpCAM, epithelial cell adhesion molecule; FACS, fluorescence-activated cell sorting; GSDMC, Gasdermin C; GSK3β, glycogen synthase kinase 3 beta; Hi-C, high throughput chromosome conformation capture; HSF1, heat shock transcription factor 1; ICB, immune checkpoint blockade; IFN, interferon; IL-6, interleukin 6; irAEs, immune related adverse events; IRF1, interferon regulatory factor 1; JAK, Janus kinase 1; NFκB, nuclear factor kappa B; NSCLC, non-small cell lung cancer; OTUB1, OTU deubiquitinase, ubiquitin aldehyde binding 1; PARP1, poly(ADP-ribose) polymerase 1; PD-1, programmed cell death-1; PD-L1, programmed death-ligand 1; PD-L2, programmed death ligand 2; SP1, specificity protein 1; SPOP, speckle-type POZ protein; STAG2, stromal antigen 2; STAT3, signal transducer and activator of transcription 3; T2D, type 2 diabetes; TADs, topologically associating domains; TFEB, transcription factor EB; TFs, transcription factors; TNFα, tumor necrosis factor-alpha; TTP, tristetraprolin; UCHL1, ubiquitin carboxy-terminal hydrolase L1; USP22, ubiquitin specific peptidase 22
Graphical abstract
The level and characteristics of PD-L1/L2 in patients largely determine therapeutic efficacy to immune checkpoint blockade. PD-L1/L2 are precisely regulated and their characteristics are established by a complicated network.
1. Introduction
Immunotherapy augments the host immune system to generate an effective antitumor immunity, and along with chemotherapy, radiation and surgery become standard of care in the treatment of many malignancies1. Among different types of immunotherapy, neutralization of immune checkpoints (called immune checkpoint blockade, ICB) that limit antitumor response, has resulted in unprecedented responses in patients with a variety of cancers, and some patients even showed long-lasting effect2. The inhibitory molecules including ligand PD-L1 (programmed death-ligand 1) and receptor PD-1 (programmed cell death-1) attract great attention because targeting them causes more beneficial tumor response-to-toxicity3. However, the overall patient response rate is still low, and has no significant improvement in recent years4. Furthermore, in a portion of patients, the ICB treatment can cause the immune system to attack healthy tissue (immune related adverse events, irAEs) and eventually leads to discontinuing the lifesaving treatment5. Severe irAEs frequently occur at gastrointestinal, pulmonary and dermatological organs, which show considerable expression of PD-L1 and PD-L2 (programmed death ligand 2) in healthy condition6, 7, 8. However, patients who experienced irAEs show a better prognosis6, 7, 8. It suggests that PD-1/PD-L1 based immune checkpoint blockade not only evoke anti-tumor immunity but also destroy the immune homeostasis in certain organs especially organs showing high expression of PD-L1 or PD-L29,10. The low response rate, severe side effect and unpredictable outcome are probably due to the complicated biological regulation of PD-L1 and PD-L2. For example, deglycosylation of cancer tissue samples significantly improves PD-L1 quantification and prediction of clinical outcome11. Suppression of exosomal PD-L1, instead of soluble or membrane PD-L1, induces systemic anti-tumor immunity12. These recent findings pinpoint a new insight that simple examination of PD-L1 by single method or at single level will not fully reflect the nature of PD-L1 in patients. Therefore, it is important to fully understand the complicated regulatory network of PD-L1 by precisely detection of PD-L1 itself and its regulatory network. By considering this, we summarized the known regulatory networks of PD-L1/PD-L2 and emphasized more efforts to explore the regulation of PD-L1 and its twin ligand PD-L2.
2. Nuclear chromatin organization and DNA regulatory elements in the induction of PD-L1 and PD-L2
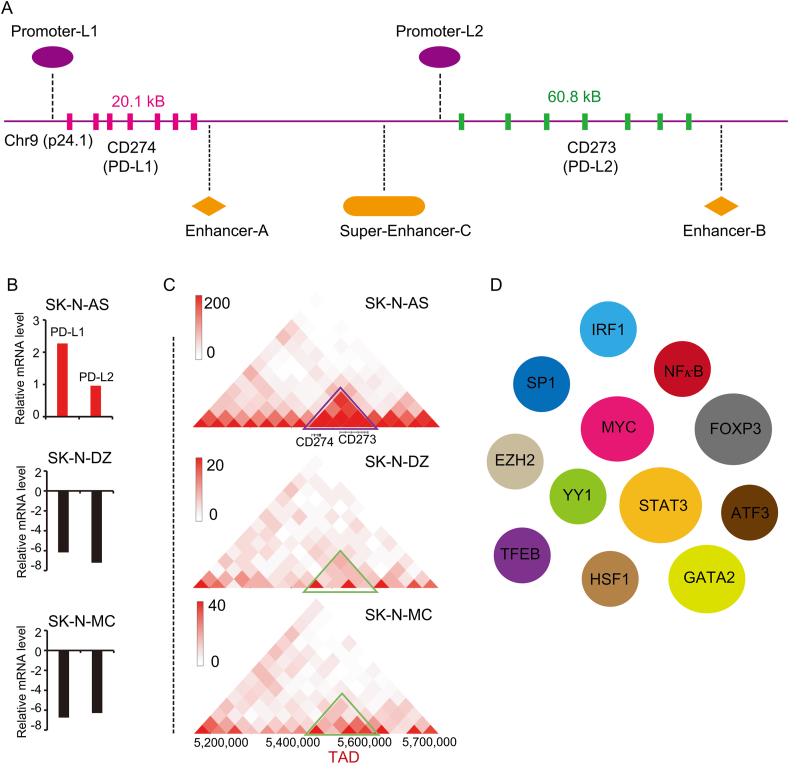
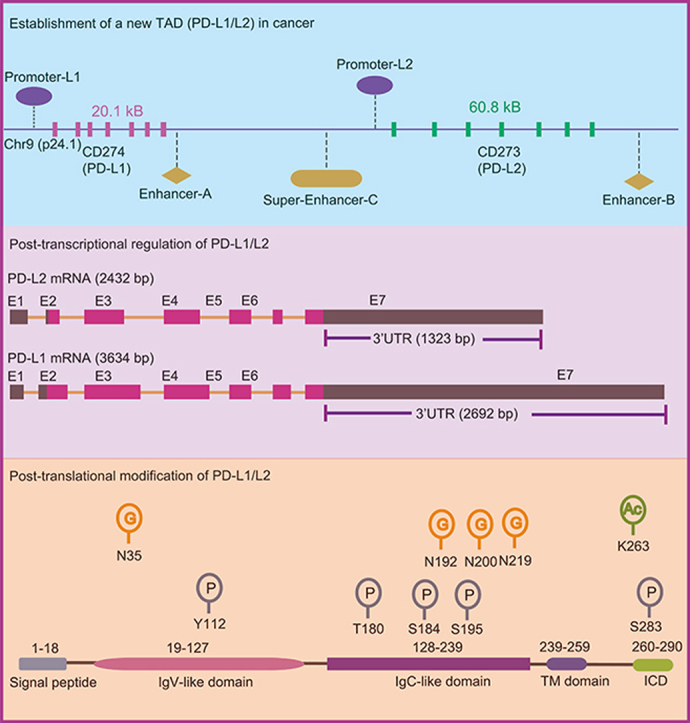
Eukaryotic genomes are extraordinarily packaged and dynamically organized within the nuclear space13,14. The nuclear 3D genome architecture is the fundamental base to achieve spatiotemporal gene expression for cell identity15. The position of a specific gene locus and its surrounding DNA regulatory elements determine the transcription of this locus. At the smaller scales, chromatin is organized into insulated spatial neighborhoods referred to as topologically associating domains (TAD), within which dynamic DNA interactions are relatively restrained by the boundaries16. In many cancers, the distribution of TADs is significantly changed17. For some cancers, such as breast cancer, the generation of new TAD boundaries results in an increased TAD number18. In the initiation and progress of cancer, cancer cells gradually hijack their favored loci and re-shape surrounding context to establish new TAD for their needs. The coding genes of PD-L1 and PD-L2 are CD274 (cluster of differentiation 274) and CD273 (cluster of differentiation 273) respectively. Both CD274 and CD273 locate on chromosome 9p24.1 (Fig. 1A). In healthy individuals, PD-L1 and PD-L2 mainly express in immune cells such as antigen presenting cells (APCs), B cells and activated T cells19,20. But a significant portion of cancer cells is able to re-organize the genetic locus of CD274 and CD273 (Fig. 1C). High-resolution chromosome conformation capture (Hi-C) is a powerful tool to explore the genome-wide interaction and TAD. By exploring the 3D genome browser21 (http://3dgenome.fsm.northwestern.edu/), the expression of PD-L1 and PD-L2 in SK-N-AS is higher than it in SK-N-DZ and SK-N-MC cells (Fig. 1B). Interestingly, comparing to SK-N-DZ and SK-N-MC cells, SK-N-AS cells re-shape a new TAD (chr9: 5,400,000–5,600,000) around CD274 and CD273 locus (Fig. 1C). It is consistent with our recent finding that there is a super-enhancer (PD-L1L2-SE, Super-enhancer-C; chr9: 5,496,361–5,504,556) between CD274 and CD273 (Fig. 1A)22. We defined this super-enhancer as PD-L1L2-SE, which is required for constitutive expression of PD-L1 and PD-L2. Genetic knockout of PD-L1L2-SE in cancer cells is sufficient to disrupt the PD-L1-mediated immune evasion22.
Figure 1.
Genomic alternation and the transcriptional regulation of CD274 and CD273. (A) CD274 and CD273 are the encoding genes of PD-L1 and PD-L2 respectively. The genomic location of CD274 and CD273 locus as well as the identified promoters, enhancer-A, enhancer-B and super-enhancer-C are depictured. (B) The expression of PD-L1 and PD-L2 in SK-N-AS, SK-N-DZ and SK-N-MC cells is indicated. The expression of PD-L1 and PD-L2 is higher in SK-N-AS cells than it in SK-N-DZ and SK-N-MC cells. The data was searched from the 3D genome browser. (C) The HiC-data (the genome-wide chromosome conformation capture) around CD274 and CD273 locus was explored. The TAD around CD274 and CD273 (topologically associating domains) in SK-N-AS, SK-N-DZ, SK-N-MC cells is shown in triangle. The data was generated from the 3D genome browser. (D) The reported transcription factors that regulate the expression of PD-L1 are demonstrated. These TFs include IRF1, GATA2, SP1, NFκB, MYC, EZH2, FOXP3, YY1, STAT3, TFEB, HSF1 and ATF3.
Besides super-enhancer, there is an enhancer (called enhancer-9 or enhancer B, chr9: 5,580,709–5,504,556) located on 140 kb downstream of CD274 (Fig. 1A)23. Deletion of this enhancer-9 results in great reduction of PD-L1 expression, but the effect on PD-L2 is not examined. In addition, another enhancer (called enhancer-A) around the 3′-UTR of CD274 increases the expression of PD-L1. The enhancer-A interacts with the promoter of CD274, which can increase the promoter activity when constructed in the same plasmid (Fig. 1A)24. Intriguingly, the activity of super-enhancer (PD-L1L2-SE) is independent on interferon γ (IFNγ). Thus, in a significant portion of cancers that show constitutive expression of PD-L1 and PD-L2, the achievement of CD274/CD273-associated TAD (CD274-TAD) might increase the interactions between enhancer/super-enhancer and promoter. The CD274-TAD might be the three-dimensional genomic foundation for the expression of PD-L1 and PD-L2 in cancers as well as immune evasion. However, the tumor-specificity and generation of CD274-TAD as well as how does CD274-TAD cooperate with other factors still need to be further explored.
3. Abnormal activity of transcriptional factors (TFs) in the up-regulation of PD-L1 and PD-L2
The generation of newly established chromatin domain and subsequently performing its function rely on DNA associated factors. A very large proportion of DNA associated proteins are TFs that bind DNA helix at specific regulatory sequences. The abnormal activity of TFs is involved in a large number of human diseases including cancer25. Lambert et al.25 reported a list of 294 oncogenic TFs by investigating the list of 1571 known oncogenic proteins from the Network of Cancer Genes NCG5.0 (http://ncg.kcl.ac.uk/statistics.php)26 and the list of 1988 human TFs as well as their regulators. The transcription of PD-L1 and PD-L2 is tightly regulated and many TFs have been reported to regulate PD-L1 in different context (Fig. 1D). MYC is the well-studied oncogenic transcription factor that had also been reported to bind directly to the promoter of PD-L1 and CD4727. Suppression of MYC in cancer cells results in reduced mRNA level of PD-L1 and enhanced anti-tumor immune response. Enforced expression of PD-L1 and CD47 in the context of MYC inactivation can suppress anti-tumor immune response and support tumor grow27. It was reported that MYC-induced transcription of PD-L1 also contributes to cisplatin-resistance in ovarian cancer28. Thus, transcription factor MYC not only transforms cells in the initiation of tumor but also drives tumor cells to escape anti-tumor immunity by directly induction of PD-L1.
Besides oncogenic signals inside cancer cells, factors in tumor microenviroment also largely contribute on the expression of PD-L1 in tumor mass. Interferon gamma (IFNγ) in tumor microenvironment is critical in primary and acquired resistance to ICB therapy by regulating PD-L1 expression4,29,30. IFNγ upregulates PD-L1 on the surface of tumor cells, then tumor cell-associated PD-L1 subsequently increases apoptosis of antigen-specific T cells30. Interferons can be divided into two types (Type I: alpha, beta and omega; Type II: gamma). Although it was firstly described in 1950s as molecules that interfere with viral replication31, IFN shows strongest effect on the induction of PD-L1 expression30,32. Upon IFN binds to interferon receptors, it transudes signal through Janus kinases (JAKs), signal transducer and activators of transcriptions (STATs)33. The interferon regulatory factor-1 is the downstream TF of STATs that is prerequisite to the IFNγ-induced upregulation of PD-L134. By systematically shRNA-based screening of IFN-associated molecules, the TFs including STAT1, STAT2, STAT3 and IRF1 are important in the regulation of PD-L1 promoter30,35. Slightly different with PD-L1, both IFNγ and IFNβ show strong effect on the induction of PD-L2. Further analysis reveals STAT3 and IRF1 are important transcriptional factors that bind to PD-L2 promoter under interferon receptor signaling35. The role of STAT3 and IRF1 in the regulation of PD-L1 has been extensively studied and consistently observed by several groups36, 37, 38. In addition, STAT3 activation can be modulated by several factors that thus regulate PD-L1 and PD-L2. For example, PARP1 poly (ADP-ribosyl)ates STAT3 and subsequently reduces STAT3 phosphorylation as well as STAT3-mediated PD-L1 expression39. Furthermore, a portion of cancer cells achieves direct functional mutations (D427H and E616G) in STAT3 and its regulators such as SOCS1 to induce PD-L1 expression in the absence of cytokine40.
In addition, several other TFs have been reported to play critical role in the regulation of PD-L1. RelA is a member of NFκB family that translocates to nucleus and binds to several NFκB binding sites at the promoter of PD-L1 to induce transcription upon cytokine stimulation41,42. In the promoter of PD-L1, there is a polymorphism (rs10815225:G/C) that is associated with the mRNA level of PD-L1 and the occurrence of gastric cancer. This polymorphism locates in the binding-site of Sp1, and transcription factor Sp1 favors G-allelic instead of C-allelic to induce PD-L1 expression43. The transcription factor EB (TFEB) positively correlates with PD-L1 expression in renal cell carcinoma (RCC). TFEB binds to PD-L1 promoter and the nuclear translocation of TFEB can be regulated by mTOR44. TFEB can also induce the expression of PD-L2 for immune evasion and drug resistance45. In brain tumor, ectopic expression of GATA2 is sufficient to induce the expression of PD-L1 and PD-L2. In such tumor, GATA2 is also essential for constitutive high expression of PD-L246. Heat shock transcription factor 1 (HSF1), the master regulator of proteotoxic stress response, can also bind to PD-L1 promoter and subsequently enhance the expression of PD-L147. Inhibition of the adenosine A1 receptor (ADORA1) suppresses tumor development in vivo in immune-deficient xenografts, but this effect is greatly reduced in an immune-competent mouse model due to upregulation of PD-L1. Deletion of ADORA1 upregulates ATF3 and then upregulates PD-L1 via the PD-L1 promoter. Combination of PD-L1 mAb treatment and ADORA1 antagonist could significantly improve the effect of each treatment alone48. FOXP3 is a master transcription factor that guides the differentiation of Treg cells. A recent report showed that FOXP3 expresses in pancreatic ductal adenocarcinoma cells and activates the transcription of PD-L149. Thus, the TFs that contribute to PD-L1 expression in different cancer types are quite different and largely dependent on the context of tumor. However, TFs that were explored previously in regulation of PD-L1 expression are focus on the promoter region of PD-L1. Given the essential role of super-enhancer (PD-L1L2-SE) in IFNγ-independent upregulation of PD-L1 and PD-L222, it is necessary to identify transcription factors that bind to PD-L1L2-SE and their functional interaction with promoter-associated transcription factors.
4. Increased mRNA stability to promote expression of PD-L1
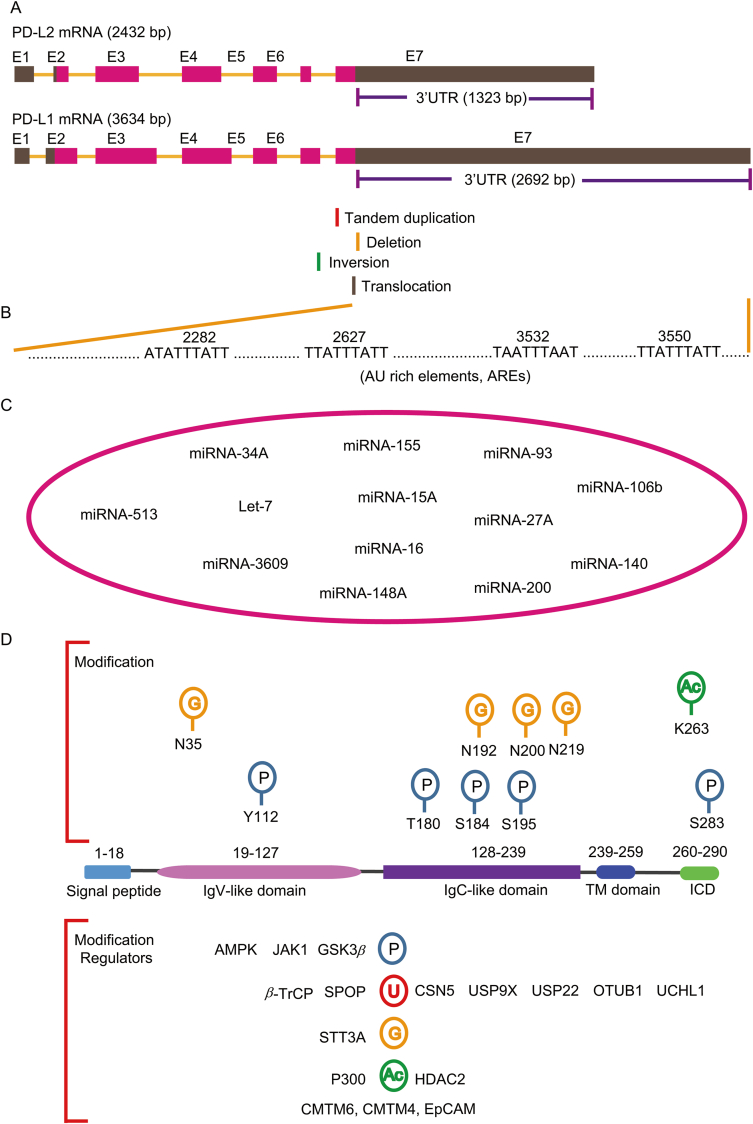
The length of PD-L1 mRNA is 3634 bp containing seven exons. However, the coding sequence of PD-L1 mRNA is relative short and majority of the sequence composes the 3′ untranslated region (3′-UTR) (2692 bp) (Fig. 2A). Comparing to PD-L1, the PD-L2 encoding mRNA contains relative short 3′-UTR (1323bp) (Fig. 2A). The abundant 3′-UTR in PD-L1 mRNA indicates the existence of a sophisticated regulatory mechanism at post-transcriptional level. Indeed, to achieve high expression of PD-L1 and immune evasion, cancer cells develop a mechanism to disrupt the 3′-UTR of PD-L1 transcripts. Genome-wide mapping reveals frequent breakpoints at a 3.1 kilobase region within the 3′ region of the PD-L1 locus (Fig. 2A)50. These breakpoints caused by diverse structural variations such as large deletions, tandem duplications, inversions and translocations are significantly associated with high expression of PD-L1 mRNA (Fig. 2A). The entire PD-L1 open reading frame (ORF) is completely preserved or slightly affected at C-terminus, thus extracellular receptor-binding and transmembrane domains of PD-L1 are not affected. These 3′-UTR-associated structural variations are frequently observed in adult T-cell leukaemia lymphoma but less common in other cancers50. In the PD-L1 3′-UTR, there are at least four AU-rich elements (AREs) with core ATTTA motif in an AU-rich context (Fig. 2B). These AU-rich elements can be recognized by AU-rich element-binding protein tristetraprolin (TTP), which negatively regulates PD-L1 mRNA level. However, in cancer cells, activated RAS signaling inhibits TTP via MK2-mediated phosphorylation and thus rescues PD-L1 mRNA from TTP51.
Figure 2.
Post-transcriptional and post-translational regulation of PD-L1. (A) The diagram showing the PD-L1 encoding gene CD274 and CD273. The exons and the 3′-untranslated regions (3′-UTR) of CD274 and CD273 are listed. The frequently observed breakpoints in CD274 resulting from tandem duplication, deletion, inversion and translocation around 3′UTR region are indicated. (B) Four potential AU rich regions in 3′-UTR of CD274 are shown. (C) The reported miRNAs that target on 3′UTR to regulate PD-L1 are summarized. These miRNAs include but not limited to miRNA-513, miRNA-34A, miRNA-3609, Let-7, miRNA-155, miRNA-148A, miRNA-15A, miRNA-16, miRNA-200, miRNA-93, miRNA-27A, miRNA-106b and miRNA-140. (D) The protein modification of PD-L1 and the associated enzymes are demonstrated. The upper panel shows the protein modification and the modified amino acids including glycosylation at N35, N192, N200, N219 and phosphorylation at Y112, T180, S184, S195 and S283 as well as acetylation at K263. The low panel shows the associated enzymes that regulate protein modification. The PD-L1 associated regulators include but not limited to AMPK, JAK1, GSK3β, β-TrCP, SPOP, CSN5, USP9X, USP22, OTUB1, UCHL1, STT3A, P300, HDAC2, CMTM6, CMTM4 and EpCAM.
Besides RNA binding proteins, there are multiple miRNAs that have been identified to target on PD-L1 mRNA (Fig. 2C). For example, miRNA-513 represses PD-L1 translation by targeting on PD-L1 3′UTR. MiRNA-513 also plays critical role in IFNγ-induced PD-L1 expression because it is regulated by IFNγ52. In acute myeloid leukemia, an inverse correlation between PD-L1 and miR-34a expression was observed. MiR-34a binds at PD-L1-3′UTR and downregulates the expression of PD-L153. Upon IFNγ and TNFα treatment, miR-155 is among the most highly up-regulated miRNAs. The PD-L1-3′UTR contains two functional miR-155 binding sites54. Endogenous miR-155 controlled the kinetics and maximal levels of PD-L1 induction after treatment with IFNγ and TNFα54. In miR-25-93-106b cluster, PD-L1 is a novel target of miR-93 and miRNA-106b. The expression of PD-L1 can be diminished upon over-expression of miR-93 and miR106b mimics55. In non-small cell lung cancer (NSCLC), the low expression of miR-140 is associated with high expression of PD-L1. Over-expression of miR-140 suppresses the expression of PD-L1 by directly binding at PD-L1 3′-UTR56. miRNA let-7 post-transcriptionally suppresses PD-L1 expression and the biogenesis of let-7 can be inhibited by RNA binding protein LIN2857. miR-3609 is down-regulated in resistant breast cancer cells and mediated the chemoresistance to adriamycin. Transfection of a miR-3609 mimic markedly suppresses PD-L1 expression in MDA-MB-231 and MDA-MB-468 cells58. MiR-148A identified in a comprehensive miRNA screening using The Cancer Genome Atlas (TCGA) dataset in colorectal cancer is a negative regulator of PD-L1 expression particularly in mismatch repair deficiency (dMMR) subset. MiR-148A directly binds to the 3′-UTR of PD-L1 and regulates immunosuppression59. miR-200 is a cellular autonomous suppressor of EMT and it also targets PD-L1. The ZEB1 and other EMT activators repress miRNA-200 expression and relieve inhibitory effect of miR-200 on PD-L160. In addition to tumor intrinsic miRNAs, exosome-containing miRNAs such as miR-27A, miRAN-15A and miRNA-16 can also target and modulate the expression of PD-L161, 62, 63. Interestingly, a Epstein–Barr virus encoding miRNA named miR-BHRF1-2-5p binds to both PD-L1 and PD-L2 3′-UTR for inhibiting their surface expression64. Although many regulators have been identified in regulation of PD-L1 via its 3′-UTR, the comprehensive regulating network in the regulation of PD-L1 3′-UTR is still poorly understood. Due to the critical role of PD-L1 in immune hemostasis, it is reasonable to set up a precise network to control PD-L1 in different setting. However, dissecting this regulatory network is challenging but is important to understand immune evasion and ICB-based therapy.
5. Post-translational modification and regulation of PD-L1 protein stability
PD-L1 protein also undergoes diverse post-translational modifications including glycosylation, phosphorylation, ubiquitination, sumoylation, and acetylation to regulate its stability (Fig. 2D). Glycosylation of membrane proteins is important for protein–protein interactions and proper activities. N-Glycosylation occurs on asparagine (N) at position 192,200 and 219 (N192, N200 and N219) of PD-L165. N-Glycosylated PD-L1 is stabilized by blocking β-TrCP-mediated protein degradation. GSK3β binds to non-N-glycosylated PD-L1 and phosphorylates PD-L1 at threonine 180 (T180) and serine 184 (S184), through which to promote β-TrCP-mediated PD-L1 degradation (Fig. 2D)65. The N-glycosylation of PD-L1 is further confirmed by other mass spectrometry analysis66. Phosphorylation at Tyr112 of PD-L1 by IL-6-activated JAK1 recruits the endoplasmic reticulum-associated N-glycosyltransferase STT3A to catalyze PD-L1 glycosylation and maintain PD-L1 stability67. Metformin is a widely used oral medication to treat type 2 diabetes (T2D) and also maintains high cytotoxic T lymphocyte (CTL) activity in tumor tissues68. It is recently found that metformin induces the activation of AMPK to directly phosphorylate S195 of PD-L1 that results in abnormal PD-L1 glycosylation and ER-associated protein degradation69. In some cases, the N-linked glycosylation mask PD-L1 to be recognized by antibodies at immunohistochemical assay, which leads to unsatisfied prediction by using PD-L1 immunohistochemical readout. Thus removal of N-linked glycosylation by a recombinant glycosidase (peptide-N-glycosidase F; PNGase F) enhances PD-L1 detection and prediction of anti-PD-L1/PD-1 therapeutic efficacy11. Due to critical role of glycosylation on PD-L1 function and antibody recognition, targeting on PD-L1 glycosylation and its associated pathways is a very promising filed in cancer therapy and clinical diagnosis70.
The protein abundance of PD-L1 fluctuates during cell cycle progression, peaking in M phase, followed by reduction in G1 and S phases71. This dynamic change of PD-L1 in cell cycle is regulated by cyclin D-CDK4 and the cullin-3-SPOP E3 ligase via proteasome mediated degradation. CDK4 mediates phosphorylation of speckle-type POZ protein (SPOP) and thereby inhibits SPOP degradation by the anaphase-promoting complex activator FZR1 (Fig. 2D). The E3 ligase SPOP interacts with PD-L1 via the last eight amino acids of PD-L1 (C-tail: 283–290) and ubiquitinates PD-L171. The ubiquitination of PD-L1 can be reversed by deubiqutinase CSN5. TNFα-induced CSN5 inhibits the degradation of PD-L1 and the CSN5 inhibitor curcumin sensitizes cancer cells to ICB therapy72. In oral squamous cell carcinoma, USP9X deubiquitinates and stabilizes PD-L173. Besides these two deubiquitinatying enzymes, USP22, OTUB1 and UCHL1 have also been identified as PD-L1 deubiquitinases to stabilize PD-L1 in different cancer74, 75, 76, 77. As such, the stability of PD-L1 is tightly regulated by E3 ligases and deubiquitinases (Fig. 2D). The abnormal function of these enzymes plays critical role to stabilize PD-L1 and immune evasion in cancer.
Furthermore, PD-L1 also undergoes reversible acetylation mediated by P300 and HDAC2, which is critical for its nuclear translocation78. Besides direct post-translational modification, other factors also play important role in regulating the stability of PD-L1. Two groups independently identify CMTM6 (CKLF like MARVEL transmembrane domain containing 6) as a critical factor in the regulation of PD-L1 protein stability by genome-wide sgRNA library screening79,80. CMTM6 is widely expressed in many tissues, but the function is not investigated. In haploid HAP1 cells, IFNγ induces high PD-L1 expression and CMTM6 is a key regulator of PD-L1 protein expression identified by FACS (fluorescence activated cell sorting) sorted sgRNAs targeting80. CMTM6 localizes at cell surface and binds with PD-L1 protein by reducing the ubiquitination of PD-L180. Using another sgRNA library screen in BxPC-3 cells, CMTM6 was also identified as a critical regulator of PD-L1 expression by preventing PD-L1 from lysosome-mediated degradation79. The regulation of PD-L1 protein level by CMTM6 and CMTM4 is widely conserved in multiple cancer types. In energy deprivation condition, aberrant energy status activates AMPK to phosphorylate PD-L1 on Ser283. The phosphorylated PD-L1 on Ser283 disrupts CMTM4-mediated degradation and subsequently triggers its degradation81 (Fig. 3). The EGF-like domain I within the extracellular domain of EpCAM (EpEX) can be released by ADAM17 and γ-secretase-mediated cleavage. EpEX binds EGFR and activates MAPK-ERK signaling to stabilize PD-L1 protein82. Thus, PD-L1 is tightly regulated at post-translational level and the abnormality involved in these pathways is deleterious in immune evasion.
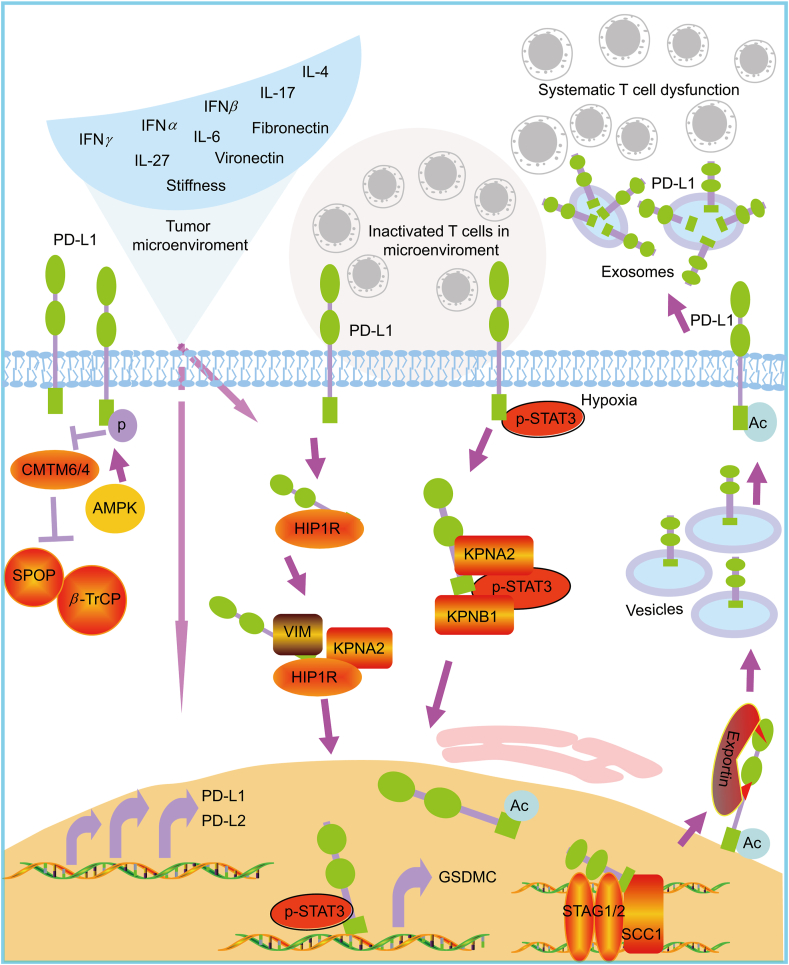
Figure 3.
The regulation of PD-L1 in tumor microenviroment and the dynamic cellular distribution. In tumor microenviroment, there are lots of reported factors, such as IFNγ, IFNα, IFNβ, IL-17, IL-4, IL-27, IL-6, Fibronectin, Vironectin and the stiffness, that can induce the expression of PD-L1 and PD-L2 via different mechanism at different levels. Expressed PD-L1 and PD-L2 resident on membrane to inactivate T cells locally. The membrane PD-L1 can translocate to nucleus through HIP1R/KPNA2/KPNB1 pathway. Nuclear PD-L1 is acetylated and translocates to membrane via exportin. Intracellular PD-L1 can secrete to tumor microenviroment or even long distance via exosome. Unlike the membrane resident PD-L1 inactivates T cells locally, the exosome-containing PD-L1 can systematically inactivate T cell function. The protein stability of PD-L1 was regulated by CMTM6 or CMTM4 by blocking SPOP- and β-TrCP-mediated degradation. However, in low energy condition, the activated AMPK can phosphorylate PD-L1 and disrupt the interaction between PD-L1 and CMTM4. Thus, the tumor microenviroment can substantially affects the distribution and function of PD-L1 not only locally but also systematically.
6. Enhanced membrane presentation and cellular transportation of PD-L1
Engagement of PD-1 by PD-L1 leads to the inhibition of T cell receptor-mediated lymphocyte proliferation and cytokine secretion. Thus, the cellular transportation and membrane presentation of PD-L1 in cancer cells is an important aspect to determine the inhibitory activity of PD-L1. Chemotherapy is a first line therapy for many cancers especially advanced cancers. However, it is recently found that chemopreventive agents such as paclitaxel, etoposide and 5-fluorouracil can greatly induce the surface expression of PD-L1 and thus might play a role in immune evasion83. Besides, chemotherapeutic agent, pemetrexed, can also increase PD-L1 levels by activating both mTOR/P70S6K and STAT3 pathways84.
In addition, nuclear staining of PD-L1 have been reported in cancer tissues, circulating tumor cells and doxorubicin-treated breast cancer cells, but the function and translocation of nuclear PD-L1 (nPD-L1) are unclear85,86. Recently, it is found that hypoxia-induced p-STAT3 physically interacts with PD-L1 and facilitates its nuclear translocation. Nuclear PD-L1 enhances the transcription of GSDMC (gasdermin C), which is specifically cleaved by caspase-8 upon TNFα treatment. The cleaved GSDMC generates a GSDMC N-terminal domain that forms pores on cell membrane and induces pyroptosis instead of apoptosis in hypoxia (Fig. 3)87. Nuclear PD-L1 also regulates sister chromatid cohesion by directly interacting with cohesion subunit STAG2 and SCC1. It allows PD-L1 to compete with WAPL binding to PDS5B and secures proper sister chromatid cohesion and segregation88. Deletion of nPD-L1 impairs the subcellular distribution of STAG1 and provokes chromosome segregation errors89. Furthermore, the nuclear import of PD-L1 is dependent on importin pathway including KPNA2 and KPNB1 (Fig. 3)90. Thus, PD-L1 not only acts as an immune checkpoint but also has non-immune checkpoint function, which is less explored. The complicated cellular function of PD-L1 makes the clinic prediction be more difficult.
7. The regulation of secretion of exosomal PD-L1 and its role in systematic anti-tumor immune suppression
In addition to contact suppression of T cell function by membrane PD-L1, tumor cells also secrete exosomal PD-L1 to suppress immune response at distance or even systematically. It is recently reported that metastatic melanoma releases a high level of exosomes that carry PD-L1 on their surface (Fig. 3)91. The containing of PD-L1 on exosomes is further increased by IFNγ. The high level of exosomal PD-L1 in patients before treatment is a marker for poor prognosis. However, the high magnitudes of increased exosomal PD-L1 after treatment is an indicator of adaptive response to T cell re-invigoration91. Systemically introduced exosomal PD-L1 can promote the growth of tumors eventhough these tumors are unable to secrete their own12. Tumor cells obtain the active defense ability to anti-tumor immunity by exosomal PD-L1. The combination of exosome secretion inhibition and ICB has the potential to improve anti-tumor response in clinic92. The exosomal PD-L1 but not soluble PD-L1 in plasma is an independent prognostic factor in gastric cancer. It seems that exosomal PD-L1 is more powerful than membrane PD-L1 to induce T-cell dysfunction due to stability of exosomal PD-L1 and MHC-I93. In follow up analysis, PD-L1 levels in circulating exosomes seem to be a more reliable marker than PD-L1 expression in tumor biopsies. It might be useful to predict the response to ICB treatment and clinical outcome94,95. In the help of technique development, exosomal PD-L1 can be examined by a dual-target-specific aptamer combined with droplet digital PCR (ddPCR)96. It is interesting to explore the nature of exosomal PD-L1 in vivo and its potential in the prediction of response as well as be a direct therapeutic target.
8. Extracellular signals orchestrate oncogenic network to generate PD-L1 and PD-L2 as well as immune evasion
In tumor mass, tumor cells and the surrounding blood vessels, immune cells, fibroblasts, signaling molecules and the extracellular matrix composed the microenvironment. In tumor microenvironment, a number of factors have been identified to induce the expression of PD-L1 and PD-L2 in tumor cells (Fig. 3). One of the most studied molecules is the inflammatory cytokine IFNγ. IFNγ is produced by activated T cells and subsequently induces PD-L1 expression on tumor cells via JAK–STAT pathway, preferentially through STAT1/IRF1. The IFNγ-induced PD-L1 feedback turns off the activated T cells and renders cancer cells survival97. Like IFNγ, type-I interferons, i.e., IFNα and IFNβ, also induce PD-L1 expression but with less effective98,99. Other inflammatory stimuli such as IL-17, TNFα, IL-4 and IL-27 can up-regulate PD-L1 expression in different cancers, too100, 101, 102 (Fig. 3).
Although IL-6 does not directly induce the transcription of PD-L1, it promotes endoplasmic reticulum-associated N-glycosylation of PD-L1 and PD-L1 stability67. The combination of blocking antibodies targeting both IL-6 and PD-L1 reduces progression of pancreatic cancer103. Besides cytokines, the matrix in tumor microenvironment such as fibronectin and vironectin can regulate constitutive and IFNγ induced PD-L1 expression via αvβ3-integrin104 (Fig. 3). Increased deposition of extracellular matrix can strengthen stiffness and affect the phenotypes and properties of cancer cells. It was recently observed that the expression of PD-L1 is enhanced as the stiffness of substrates is increased105. These features make the therapy of solid tumor become more difficulty.
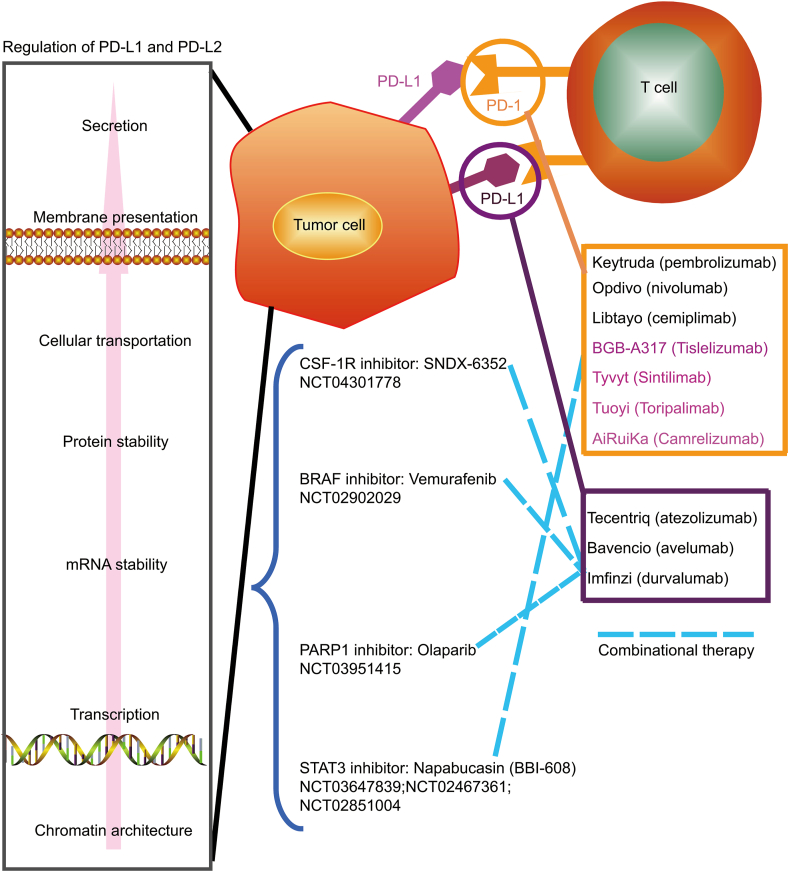
Since the first anti-CTLA4 (cytotoxic T lymphocyte antigen-4) antibody was approved by U.S. food and drug administration (FDA), there are six more ICTs targeting on PD-1 and PD-L1 have been approved106. Among them, three antibodies targeting on PD-1 (pembrolizumab, nivolumab, and cemiplimab) and others targeting on PD-L1 (atezolizumab, avelumab, and durvalumab) (Fig. 4)107. In addition, four antibodies targeting on PD-1 including tislelizumab, sintilimab, toripalimab and camrelizumab were approved by NMPA (National Medical Products Administration) at China (Fig. 4). To avoid side effects, overcome resistance and improve therapeutic efficiency, combination therapy based on ICIs is an emerging trend in tumor immunology108 (Fig. 4). However, the clinical value of combination immunotherapy remains controversial and the design of combinational trial is challenging especially the precise recruitment of patients. Simultaneously targeting the regulation pathway of PD-L1/PD-L2 and ICIs might reduce therapeutic doses to attenuate side effects. Ideally, unique molecular mechanism regarding the regulation of PD-L1/PD-L2 in cancer might provide novel ways to specifically evoke anti-tumor immune response but leave immune homeostasis intact. Currently, several combinational clinical trials targeting PD-1/PD-L1 and their regulators such as STAT3 (NCT03647839, NCT02467361, NCT02851004), PARP1 (NCT03951415), BRAF (NCT02902029), CSF-1R (NCT04301778) are ongoing (www.clinicaltrials.gov) (Fig. 4). Results from the clinical trial targeting STAT3 (napabucasin) and PD-1 (pembrolizumab) show anti-tumor activity in metastatic colorectal cancer with microsatellite stable, while the single medicine show no anti-tumor activity in such patients109. Although the results did not meet the primary end point, it is still very promising by considering no biomarkers were used in recruiting patients. Thus, comprehensive understanding the regulation of PD-L1 and PD-L2 in cancer particularly dissecting the difference between normal and cancerous cells will not only provide novel targets but also biomarkers for combinational therapy.
Figure 4.
The approved PD-1/PD-L1 antibodies and its combinational therapy with inhibitors targeting on the regulatory pathway of PD-L1 and PD-L2. Currently, there are 10 antibodies that have been approved in clinic including 7 anti-PD1 antibodies (pembrolizumab, nivolumab, cemiplimab, tislelizumab, sintilimab, toripalimab, and amrelizumab) and three anti-PD-L1 antibodies (atezolizumab, avelumab, and durvalumab). ICIs-based combinational therapy with other inhibitors is an emerging and promising trend. Several clinic trials have been already ongoing to test the inhibitors targeting the PD-L1/PD-L2 regulators such as STAT3. The number of 6 clinic trials are listed. Results from the clinical trial targeting STAT3 (napabucasin) and PD-1 (pembrolizumab) show anti-tumor activity in metastatic colorectal cancer with microsatellite stable. Although the results did not meet the primary end point, it is still very promising to consider no biomarkers were used in recruiting patients. Thus, ICI-based combinational therapy with inhibitors targeting regulatory factors of PD-L1/PD-L2, especially cancer-specific regulatory pathways, is deserved to be extensively examined by considering patients' recruitment with designed biomarkers.
9. Conclusions
The immune checkpoint blockade targeting on PD-1/PD-L1 has revolutionized cancer treatment and demonstrated the significant impact on patient outcomes, including durable response and extended survival110. However, it is still a long way to broad the benefit for more cancer patients. The current treatment design for different patients is inadequate. Thus, providing a precision therapy by design of different ICB treatment strategies along with other cancer treatment methods will be benefit to more cancer patients. However, precision of immune-checkpoint blockade is not an easy way, which is largely dependent on the fully understanding on the regulation of PD-L1 and PD-L2 in cancer as well as in normal tissue. PD-L1 broadly expressed in normal organ tissues including spleen, thymus, stomach, prostate, esophagus, liver, testis, small intestine, skin, lung, bone, pancreas and rectum111,112. However the expression of PD-L2 is restricted to several normal tissues such as lymph nodes, thymus, testis and small intestine111,112. Upon autoimmune responses as well as stimulations such as LPS (lipopolysaccharide), IFNγ, IL-4, GM-CSF or polyinosinic-polycytidylic acid, the expression of PD-L1 and PD-L2 were up-regulated in normal tissues113. Studies indicate that the expression of PD-L1 and PD-L2 largely relies on STAT1, STAT6, PU.1 and IRF4 in noncancerous tissues111, 112, 113, 114. Recent years, although researchers have made great progress in understanding PD-L1, it is still not complete or largely unknown how do other treatments affect the regulation of PD-L1 and PD-L2. Thus, it is important to take more efforts to further explore novel regulation of PD-L1 in different level and in different context as well as different characteristics between noncancerous and cancerous tissues. In this paper, we provide a relatively comprehensive summary of known PD-L1/PD-L2 regulatory networks and emphasize more efforts to add new knowledge into this PD-L1/PD-L2 regulatory panorama.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31970616; 31770935; 81873531; and 82070505), the Distinguished Professorship Program of Jiangsu Province to Yihui Fan, the Distinguished Professorship Program of Jiangsu Province to Renfang Mao, The National Undergraduate Training Programs for Innovation (202010304109Y, China). We would like to acknowledge the 3D genome browser (http://3dgenome.fsm.northwestern.edu/) for free use. We apologize to colleagues whose work was not cited in this review owing to space constraints.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Contributor Information
Yuanyuan Wu, Email: wyy84@ntu.edu.cn.
Renfang Mao, Email: maorenfang@ntu.edu.cn.
Yihui Fan, Email: fanyihui@ntu.edu.cn.
Author contributions
Zhiwei Fan, Changyue Wu, Yuanyuan Wu, Renfang Mao and Yihui Fan were responsible for the conception and design of the review. Zhiwei Fan and Changyue Wu collected the related papers and were the major contributors in writing the manuscript. Zhiwei Fan, Changyue Wu, Miaomiao Chen, Yongying Jiang, Yuanyuan Wu, Renfang Mao and Yihui Fan generated the data and wrote the manuscript. Yuanyuan Wu, Renfang Mao and Yihui Fan initiated the study and revised the manuscript. Zhiwei Fan, Changyue Wu, Renfang Mao and Yihui Fan prepared the figures. All authors read and approved the final manuscript.
Conflicts of interest
The authors claim that the researchers in this study have no conflict of interest.
References
- 1.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribas A., Wolchok J.D. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanmamed M.F., Chen L. A paradigm shift in cancer immunotherapy: from enhancement to normalization. Cell. 2018;175:313–326. doi: 10.1016/j.cell.2018.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaretsky J.M., Garcia-Diaz A., Shin D.S., Escuin-Ordinas H., Hugo W., Hu-Lieskovan S., et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Postow M.A., Sidlow R., Hellmann M.D. Immune-related adverse events associated with immune checkpoint blockade. N Engl J Med. 2018;378:158–168. doi: 10.1056/NEJMra1703481. [DOI] [PubMed] [Google Scholar]
- 6.Ouyang T., Cao Y., Kan X., Chen L., Ren Y., Sun T., et al. Treatment-related serious adverse events of immune checkpoint inhibitors in clinical trials: a systematic review. Front Oncol. 2021;11:621639. doi: 10.3389/fonc.2021.621639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonpavde G.P., Grivas P., Lin Y., Hennessy D., Hunt J.D. Immune-related adverse events with PD-1 versus PD-L1 inhibitors: a meta-analysis of 8730 patients from clinical trials. Future Oncol. 2021;17:2545–2558. doi: 10.2217/fon-2020-1222. [DOI] [PubMed] [Google Scholar]
- 8.Grimaud F., Penaranda G., Stavris C., Retornaz F., Brunel V., Cailleres S., et al. Adverse events induced by PD-1/PD-L1 inhibitors: a real-world single-center experience with a management-based approach. Ther Clin Risk Manag. 2021;17:669–677. doi: 10.2147/TCRM.S308194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dougan M., Pietropaolo M. Time to dissect the autoimmune etiology of cancer antibody immunotherapy. J Clin Invest. 2020;130:51–61. doi: 10.1172/JCI131194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei S.C., Meijers W.C., Axelrod M.L., Anang N.A.S., Screever E.M., Wescott E.C., et al. A genetic mouse model recapitulates immune checkpoint inhibitor-associated myocarditis and supports a mechanism-based therapeutic intervention. Cancer Discov. 2021;11:614–625. doi: 10.1158/2159-8290.CD-20-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H.H., Wang Y.N., Xia W., Chen C.H., Rau K.M., Ye L., et al. Removal of N-linked glycosylation enhances PD-L1 detection and predicts anti-PD-1/PD-L1 therapeutic efficacy. Cancer Cell. 2019;36:168–178. doi: 10.1016/j.ccell.2019.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poggio M., Hu T., Pai C.C., Chu B., Belair C.D., Chang A., et al. Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell. 2019;177:414–427. doi: 10.1016/j.cell.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaban H.A., Seeber A. Monitoring the spatio-temporal organization and dynamics of the genome. Nucleic Acids Res. 2020;48:3423–3434. doi: 10.1093/nar/gkaa135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinodoz S.A., Ollikainen N., Tabak B., Palla A., Schmidt J.M., Detmar E., et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell. 2018;174:744–757. doi: 10.1016/j.cell.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stadhouders R., Filion G.J., Graf T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature. 2019;569:345–354. doi: 10.1038/s41586-019-1182-7. [DOI] [PubMed] [Google Scholar]
- 16.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kantidze O.L., Gurova K.V., Studitsky V.M., Razin S.V. The 3D genome as a target for anticancer therapy. Trends Mol Med. 2020;26:141–149. doi: 10.1016/j.molmed.2019.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barutcu A.R., Lajoie B.R., McCord R.P., Tye C.E., Hong D., Messier T.L., et al. Chromatin interaction analysis reveals changes in small chromosome and telomere clustering between epithelial and breast cancer cells. Genome Biol. 2015;16:214. doi: 10.1186/s13059-015-0768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Messal N., Serriari N.E., Pastor S., Nunes J.A., Olive D. PD-L2 is expressed on activated human T cells and regulates their function. Mol Immunol. 2011;48:2214–2219. doi: 10.1016/j.molimm.2011.06.436. [DOI] [PubMed] [Google Scholar]
- 20.Dong H., Strome S.E., Matteson E.L., Moder K.G., Flies D.B., Zhu G., et al. Costimulating aberrant T cell responses by B7-H1 autoantibodies in rheumatoid arthritis. J Clin Invest. 2003;111:363–370. doi: 10.1172/JCI16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Song F., Zhang B., Zhang L., Xu J., Kuang D., et al. The 3D genome browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19:151. doi: 10.1186/s13059-018-1519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y., Wu Y., Zhang S., Ma P., Jin X., Wang Z., et al. A tumor-specific super-enhancer drives immune evasion by guiding synchronous expression of PD-L1 and PD-L2. Cell Rep. 2019;29:3435–3447. doi: 10.1016/j.celrep.2019.10.093. [DOI] [PubMed] [Google Scholar]
- 23.Chen H., Li C., Peng X., Zhou Z., Weinstein J.N., Liang H. A pan-cancer analysis of enhancer expression in nearly 9000 patient samples. Cell. 2018;173:386–399. doi: 10.1016/j.cell.2018.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong W., Deng H., Huang C., Zen C., Jian C., Ye K., et al. MLL3 enhances the transcription of PD-L1 and regulates anti-tumor immunity. Biochim Biophys Acta Mol Basis Dis. 2019;1865:454–463. doi: 10.1016/j.bbadis.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 25.Lambert M., Jambon S., Depauw S., David-Cordonnier M.H. Targeting transcription factors for cancer treatment. Molecules. 2018;23:1479. doi: 10.3390/molecules23061479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An O., Dall'Olio G.M., Mourikis T.P., Ciccarelli F.D. NCG 5.0: updates of a manually curated repository of cancer genes and associated properties from cancer mutational screenings. Nucleic Acids Res. 2016;44:D992–D999. doi: 10.1093/nar/gkv1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casey S.C., Tong L., Li Y., Do R., Walz S., Fitzgerald K.N., et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng Q., Zhang Y., Wang Z., Ding J., Song Y., Zhao W. Cisplatin-mediated down-regulation of miR-145 contributes to up-regulation of PD-L1 via the c-Myc transcription factor in cisplatin-resistant ovarian carcinoma cells. Clin Exp Immunol. 2020;200:45–52. doi: 10.1111/cei.13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J., Shi L.Z., Zhao H., Chen J., Xiong L., He Q., et al. Loss of IFN-gamma pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167:397–404. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong H., Strome S.E., Salomao D.R., Tamura H., Hirano F., Flies D.B., et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 31.Isaacs A., Lindenmann J. Pillars article: virus interference. I. The interferon. Proc R soc lond B biol sci. 1957. 147: 258-267. J Immunol. 2015;195 1911-20. [PubMed] [Google Scholar]
- 32.Matsumoto K., Inoue H., Nakano T., Tsuda M., Yoshiura Y., Fukuyama S., et al. B7-DC regulates asthmatic response by an IFN-gamma-dependent mechanism. J Immunol. 2004;172:2530–2541. doi: 10.4049/jimmunol.172.4.2530. [DOI] [PubMed] [Google Scholar]
- 33.Darnell J.E., Jr., Kerr I.M., Stark G.R. Jak–STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 34.Lee S.J., Jang B.C., Lee S.W., Yang Y.I., Suh S.I., Park Y.M., et al. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-gamma-induced upregulation of B7-H1 (CD274) FEBS Lett. 2006;580:755–762. doi: 10.1016/j.febslet.2005.12.093. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Diaz A., Shin D.S., Moreno B.H., Saco J., Escuin-Ordinas H., Rodriguez G.A., et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19:1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zerdes I., Wallerius M., Sifakis E.G., Wallmann T., Betts S., Bartish M., et al. STAT3 activity promotes programmed-death ligand 1 expression and suppresses immune responses in breast cancer. Cancers (Basel) 2019;11:1479. doi: 10.3390/cancers11101479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoyen-Ermis D., Tunali G., Tavukcuoglu E., Horzum U., Ozkazanc D., Sutlu T., et al. Myeloid maturation potentiates STAT3-mediated atypical IFN-gamma signaling and upregulation of PD-1 ligands in AML and MDS. Sci Rep. 2019;9:11697. doi: 10.1038/s41598-019-48256-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venkatraman S., Meller J., Hongeng S., Tohtong R., Chutipongtanate S. Transcriptional regulation of cancer immune checkpoints: emerging strategies for immunotherapy. Vaccines (Basel) 2020;8:735. doi: 10.3390/vaccines8040735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding L., Chen X., Xu X., Qian Y., Liang G., Yao F., et al. PARP1 suppresses the transcription of PD-L1 by poly(ADP-ribosyl)ating STAT3. Cancer Immunol Res. 2019;7:136–149. doi: 10.1158/2326-6066.CIR-18-0071. [DOI] [PubMed] [Google Scholar]
- 40.Song T.L., Nairismagi M.L., Laurensia Y., Lim J.Q., Tan J., Li Z.M., et al. Oncogenic activation of the STAT3 pathway drives PD-L1 expression in natural killer/T-cell lymphoma. Blood. 2018;132:1146–1158. doi: 10.1182/blood-2018-01-829424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang G., Wen Q., Zhao Y., Gao Q., Bai Y. NF-kappaB plays a key role in inducing CD274 expression in human monocytes after lipopolysaccharide treatment. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antonangeli F., Natalini A., Garassino M.C., Sica A., Santoni A., Di Rosa F. Regulation of PD-L1 expression by NF-kappaB in cancer. Front Immunol. 2020;11:584626. doi: 10.3389/fimmu.2020.584626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao L.H., Zhou X.R., Li F.C., Chen Q., Meng F.Y., Mao Y., et al. A polymorphism in the promoter region of PD-L1 serves as a binding-site for SP1 and is associated with PD-L1 overexpression and increased occurrence of gastric cancer. Cancer Immunol Immunother. 2017;66:309–318. doi: 10.1007/s00262-016-1936-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang C., Duan Y., Xia M., Dong Y., Chen Y., Zheng L., et al. TFEB mediates immune evasion and resistance to mTOR inhibition of renal cell carcinoma via induction of PD-L1. Clin Cancer Res. 2019;25:6827–6838. doi: 10.1158/1078-0432.CCR-19-0733. [DOI] [PubMed] [Google Scholar]
- 45.Liu W., Wang Y., Xie Y., Dai T., Fan M., Li C., et al. Cisplatin remodels the tumor immune microenvironment via the transcription factor EB in ovarian cancer. Cell Death Discov. 2021;7:136. doi: 10.1038/s41420-021-00519-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu Y., Liu C.J., Kobayashi D.K., Johanns T.M., Bowman-Kirigin J.A., Schaettler M.O., et al. GATA2 regulates constitutive PD-L1 and PD-L2 expression in brain tumors. Sci Rep. 2020;10:9027. doi: 10.1038/s41598-020-65915-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang T., Ren C., Lu C., Qiao P., Han X., Wang L., et al. Phosphorylation of HSF1 by PIM2 induces PD-L1 expression and promotes tumor growth in breast cancer. Cancer Res. 2019;79:5233–5244. doi: 10.1158/0008-5472.CAN-19-0063. [DOI] [PubMed] [Google Scholar]
- 48.Liu H., Kuang X., Zhang Y., Ye Y., Li J., Liang L., et al. ADORA1 inhibition promotes tumor immune evasion by regulating the ATF3–PD-L1 axis. Cancer Cell. 2020;37:324–339. doi: 10.1016/j.ccell.2020.02.006. [DOI] [PubMed] [Google Scholar]
- 49.Wang X., Li X., Wei X., Jiang H., Lan C., Yang S., et al. PD-L1 is a direct target of cancer-FOXP3 in pancreatic ductal adenocarcinoma (PDAC), and combined immunotherapy with antibodies against PD-L1 and CCL5 is effective in the treatment of PDAC. Signal Transduct Target Ther. 2020;5:38. doi: 10.1038/s41392-020-0144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kataoka K., Shiraishi Y., Takeda Y., Sakata S., Matsumoto M., Nagano S., et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature. 2016;534:402–406. doi: 10.1038/nature18294. [DOI] [PubMed] [Google Scholar]
- 51.Coelho M.A., de Carne Trecesson S., Rana S., Zecchin D., Moore C., Molina-Arcas M., et al. Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity. 2017;47:1083–1099. doi: 10.1016/j.immuni.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gong A.Y., Zhou R., Hu G., Li X., Splinter P.L., O’Hara S.P., et al. microRNA-513 regulates B7-H1 translation and is involved in IFN-gamma-induced B7-H1 expression in cholangiocytes. J Immunol. 2009;182:1325–1333. doi: 10.4049/jimmunol.182.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X., Li J., Dong K., Lin F., Long M., Ouyang Y., et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015;27:443–452. doi: 10.1016/j.cellsig.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 54.Yee D., Shah K.M., Coles M.C., Sharp T.V., Lagos D. microRNA-155 induction via TNF-alpha and IFN-gamma suppresses expression of programmed death ligand-1 (PD-L1) in human primary cells. J Biol Chem. 2017;292:20683–20693. doi: 10.1074/jbc.M117.809053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cioffi M., Trabulo S.M., Vallespinos M., Raj D., Kheir T.B., Lin M.L., et al. The miR-25-93-106b cluster regulates tumor metastasis and immune evasion via modulation of CXCL12 and PD-L1. Oncotarget. 2017;8:21609–21625. doi: 10.18632/oncotarget.15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie W.B., Liang L.H., Wu K.G., Wang L.X., He X., Song C., et al. miR-140 expression regulates cell proliferation and targets PD-L1 in NSCLC. Cell Physiol Biochem. 2018;46:654–663. doi: 10.1159/000488634. [DOI] [PubMed] [Google Scholar]
- 57.Chen Y., Xie C., Zheng X., Nie X., Wang Z., Liu H., et al. LIN28/let-7/PD-L1 pathway as a target for cancer immunotherapy. Cancer Immunol Res. 2019;7:487–497. doi: 10.1158/2326-6066.CIR-18-0331. [DOI] [PubMed] [Google Scholar]
- 58.Li D., Wang X., Yang M., Kan Q., Duan Z. miR3609 sensitizes breast cancer cells to adriamycin by blocking the programmed death-ligand 1 immune checkpoint. Exp Cell Res. 2019;380:20–28. doi: 10.1016/j.yexcr.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 59.Ashizawa M., Okayama H., Ishigame T., Thar Min A.K., Saito K., Ujiie D., et al. MiRNA-148a-3p regulates immunosuppression in DNA mismatch repair-deficient colorectal cancer by targeting PD-L1. Mol Cancer Res. 2019;17:1403–1413. doi: 10.1158/1541-7786.MCR-18-0831. [DOI] [PubMed] [Google Scholar]
- 60.Chen L., Gibbons D.L., Goswami S., Cortez M.A., Ahn Y.H., Byers L.A., et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao X., Tu Y., Xu Y., Guo Y., Yao F., Zhang X. Endoplasmic reticulum stress-induced exosomal miR-27a-3p promotes immune escape in breast cancer via regulating PD-L1 expression in macrophages. J Cell Mol Med. 2020;24:9560–9573. doi: 10.1111/jcmm.15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Z., Suo B., Long G., Gao Y., Song J., Zhang M., et al. Exosomal miRNA-16-5p derived from M1 macrophages enhances T cell-dependent immune response by regulating PD-L1 in gastric cancer. Front Cell Dev Biol. 2020;8:572689. doi: 10.3389/fcell.2020.572689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu L., Yu T., Jin Y., Mai W., Zhou J., Zhao C. microRNA-15a carried by mesenchymal stem cell-derived extracellular vesicles inhibits the immune evasion of colorectal cancer cells by regulating the KDM4B/HOXC4/PD-L1 axis. Front Cell Dev Biol. 2021;9:629893. doi: 10.3389/fcell.2021.629893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cristino A.S., Nourse J., West R.A., Sabdia M.B., Law S.C., Gunawardana J., et al. EBV microRNA-BHRF1-2-5p targets the 3′UTR of immune checkpoint ligands PD-L1 and PD-L2. Blood. 2019;134:2261–2270. doi: 10.1182/blood.2019000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li C.W., Lim S.O., Xia W., Lee H.H., Chan L.C., Kuo C.W., et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morales-Betanzos C.A., Lee H., Gonzalez Ericsson P.I., Balko J.M., Johnson D.B., Zimmerman L.J., et al. Quantitative mass spectrometry analysis of PD-L1 protein expression, N-glycosylation and expression stoichiometry with PD-1 and PD-L2 in human melanoma. Mol Cell Proteomics. 2017;16:1705–1717. doi: 10.1074/mcp.RA117.000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chan L.C., Li C.W., Xia W., Hsu J.M., Lee H.H., Cha J.H., et al. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J Clin Invest. 2019;129:3324–3338. doi: 10.1172/JCI126022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eikawa S., Nishida M., Mizukami S., Yamazaki C., Nakayama E., Udono H. Immune-mediated antitumor effect by type 2 diabetes drug, metformin. Proc Natl Acad Sci U S A. 2015;112:1809–1814. doi: 10.1073/pnas.1417636112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cha J.H., Yang W.H., Xia W., Wei Y., Chan L.C., Lim S.O., et al. Metformin promotes antitumor immunity via endoplasmic-reticulum-associated degradation of PD-L1. Mol Cell. 2018;71:606–620. doi: 10.1016/j.molcel.2018.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y.N., Lee H.H., Hsu J.L., Yu D., Hung M.C. The impact of PD-L1 N-linked glycosylation on cancer therapy and clinical diagnosis. J Biomed Sci. 2020;27:77. doi: 10.1186/s12929-020-00670-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rousseau H., Joffre F., Raillat C., Julian M., Pierragi M.T., Chalabi F., et al. Self-expanding endovascular stent in experimental atherosclerosis. Work in progress. Radiology. 1989;170:773–778. doi: 10.1148/radiology.170.3.2916030. [DOI] [PubMed] [Google Scholar]
- 72.Lim S.O., Li C.W., Xia W., Cha J.H., Chan L.C., Wu Y., et al. Deubiquitination and stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30:925–939. doi: 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jingjing W., Wenzheng G., Donghua W., Guangyu H., Aiping Z., Wenjuan W. Deubiquitination and stabilization of programmed cell death ligand 1 by ubiquitin-specific peptidase 9, X-linked in oral squamous cell carcinoma. Cancer Med. 2018;7:4004–4011. doi: 10.1002/cam4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y., Sun Q., Mu N., Sun X., Wang Y., Fan S., et al. The deubiquitinase USP22 regulates PD-L1 degradation in human cancer cells. Cell Commun Signal. 2020;18:112. doi: 10.1186/s12964-020-00612-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang X., Zhang Q., Lou Y., Wang J., Zhao X., Wang L., et al. USP22 deubiquitinates CD274 to suppress anticancer immunity. Cancer Immunol Res. 2019;7:1580–1590. doi: 10.1158/2326-6066.CIR-18-0910. [DOI] [PubMed] [Google Scholar]
- 76.Zhu D., Xu R., Huang X., Tang Z., Tian Y., Zhang J., et al. Deubiquitinating enzyme OTUB1 promotes cancer cell immunosuppression via preventing ER-associated degradation of immune checkpoint protein PD-L1. Cell Death Differ. 2021;28:1773–1789. doi: 10.1038/s41418-020-00700-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mao R., Tan X., Xiao Y., Wang X., Wei Z., Wang J., et al. Ubiquitin C-terminal hydrolase L1 promotes expression of programmed cell death-ligand 1 in non-small-cell lung cancer cells. Cancer Sci. 2020;111:3174–3183. doi: 10.1111/cas.14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gao Y., Nihira N.T., Bu X., Chu C., Zhang J., Kolodziejczyk A., et al. Acetylation-dependent regulation of PD-L1 nuclear translocation dictates the efficacy of anti-PD-1 immunotherapy. Nat Cell Biol. 2020;22:1064–1075. doi: 10.1038/s41556-020-0562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burr M.L., Sparbier C.E., Chan Y.C., Williamson J.C., Woods K., Beavis P.A., et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–105. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mezzadra R., Sun C., Jae L.T., Gomez-Eerland R., de Vries E., Wu W., et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:106–110. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dai X., Bu X., Gao Y., Guo J., Hu J., Jiang C., et al. Energy status dictates PD-L1 protein abundance and anti-tumor immunity to enable checkpoint blockade. Mol Cell. 2021;81:2317–2331. doi: 10.1016/j.molcel.2021.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen H.N., Liang K.H., Lai J.K., Lan C.H., Liao M.Y., Hung S.H., et al. EpCAM signaling promotes tumor progression and protein stability of PD-L1 through the EGFR pathway. Cancer Res. 2020;80:5035–5050. doi: 10.1158/0008-5472.CAN-20-1264. [DOI] [PubMed] [Google Scholar]
- 83.Zhang P., Su D.M., Liang M., Fu J. Chemopreventive agents induce programmed death-1-ligand 1 (PD-L1) surface expression in breast cancer cells and promote PD-L1-mediated T cell apoptosis. Mol Immunol. 2008;45:1470–1476. doi: 10.1016/j.molimm.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 84.Cavazzoni A., Digiacomo G., Alfieri R., La Monica S., Fumarola C., Galetti M., et al. Pemetrexed enhances membrane PD-L1 expression and potentiates T cell-mediated cytotoxicity by anti-PD-L1 antibody therapy in non-small-cell lung cancer. Cancers (Basel) 2020;12:666. doi: 10.3390/cancers12030666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Satelli A., Batth I.S., Brownlee Z., Rojas C., Meng Q.H., Kopetz S., et al. Potential role of nuclear PD-L1 expression in cell-surface vimentin positive circulating tumor cells as a prognostic marker in cancer patients. Sci Rep. 2016;6:28910. doi: 10.1038/srep28910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ghebeh H., Lehe C., Barhoush E., Al-Romaih K., Tulbah A., Al-Alwan M., et al. Doxorubicin downregulates cell surface B7-H1 expression and upregulates its nuclear expression in breast cancer cells: role of B7-H1 as an anti-apoptotic molecule. Breast Cancer Res. 2010;12:R48. doi: 10.1186/bcr2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hou J., Zhao R., Xia W., Chang C.W., You Y., Hsu J.M., et al. PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis. Nat Cell Biol. 2020;22:1264–1275. doi: 10.1038/s41556-020-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu J., Qin B., Moyer A.M., Nowsheen S., Tu X., Dong H., et al. Regulation of sister chromatid cohesion by nuclear PD-L1. Cell Res. 2020;30:590–601. doi: 10.1038/s41422-020-0315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang W., Jin J., Wang Y., Fang L., Min L., Wang X., et al. PD-L1 regulates genomic stability via interaction with cohesin-SA1 in the nucleus. Signal Transduct Target Ther. 2021;6:81. doi: 10.1038/s41392-021-00463-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Du W., Zhu J., Zeng Y., Liu T., Zhang Y., Cai T., et al. KPNB1-mediated nuclear translocation of PD-L1 promotes non-small cell lung cancer cell proliferation via the Gas6/MerTK signaling pathway. Cell Death Differ. 2021;28:1284–1300. doi: 10.1038/s41418-020-00651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen G., Huang A.C., Zhang W., Zhang G., Wu M., Xu W., et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang Y., Li C.W., Chan L.C., Wei Y., Hsu J.M., Xia W., et al. Exosomal PD-L1 harbors active defense function to suppress T cell killing of breast cancer cells and promote tumor growth. Cell Res. 2018;28:862–864. doi: 10.1038/s41422-018-0060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fan Y., Che X., Qu J., Hou K., Wen T., Li Z., et al. Exosomal PD-L1 retains immunosuppressive activity and is associated with gastric cancer prognosis. Ann Surg Oncol. 2019;26:3745–3755. doi: 10.1245/s10434-019-07431-7. [DOI] [PubMed] [Google Scholar]
- 94.Cordonnier M., Nardin C., Chanteloup G., Derangere V., Algros M.P., Arnould L., et al. Tracking the evolution of circulating exosomal-PD-L1 to monitor melanoma patients. J Extracell Vesicles. 2020;9:1710899. doi: 10.1080/20013078.2019.1710899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li J.W., Wei P., Guo Y., Shi D., Yu B.H., Su Y.F., et al. Clinical significance of circulating exosomal PD-L1 and soluble PD-L1 in extranodal NK/T-cell lymphoma, nasal-type. Am J Cancer Res. 2020;10:4498–4512. [PMC free article] [PubMed] [Google Scholar]
- 96.Lin B., Tian T., Lu Y., Liu D., Huang M., Zhu L., et al. Tracing tumor-derived exosomal PD-L1 by dual-aptamer activated proximity-induced droplet digital PCR. Angew Chem Int Ed Engl. 2021;60:7582–7586. doi: 10.1002/anie.202015628. [DOI] [PubMed] [Google Scholar]
- 97.Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 98.Eppihimer M.J., Gunn J., Freeman G.J., Greenfield E.A., Chernova T., Erickson J., et al. Expression and regulation of the PD-L1 immunoinhibitory molecule on microvascular endothelial cells. Microcirculation. 2002;9:133–145. doi: 10.1038/sj/mn/7800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garcia-Diaz A., Shin D.S., Moreno B.H., Saco J., Escuin-Ordinas H., Rodriguez G.A., et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;29:3766. doi: 10.1016/j.celrep.2019.11.113. [DOI] [PubMed] [Google Scholar]
- 100.Wang X., Yang L., Huang F., Zhang Q., Liu S., Ma L., et al. Inflammatory cytokines IL-17 and TNF-alpha up-regulate PD-L1 expression in human prostate and colon cancer cells. Immunol Lett. 2017;184:7–14. doi: 10.1016/j.imlet.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Quandt D., Jasinski-Bergner S., Muller U., Schulze B., Seliger B. Synergistic effects of IL-4 and TNFalpha on the induction of B7-H1 in renal cell carcinoma cells inhibiting allogeneic T cell proliferation. J Transl Med. 2014;12:151. doi: 10.1186/1479-5876-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carbotti G., Barisione G., Airoldi I., Mezzanzanica D., Bagnoli M., Ferrero S., et al. IL-27 induces the expression of Ido and PD-L1 in human cancer cells. Oncotarget. 2015;6:43267–43280. doi: 10.18632/oncotarget.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mace T.A., Shakya R., Pitarresi J.R., Swanson B., McQuinn C.W., Loftus S., et al. IL-6 and PD-L1 antibody blockade combination therapy reduces tumour progression in murine models of pancreatic cancer. Gut. 2018;67:320–332. doi: 10.1136/gutjnl-2016-311585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vannini A., Leoni V., Barboni C., Sanapo M., Zaghini A., Malatesta P., et al. αvβ3-integrin regulates PD-L1 expression and is involved in cancer immune evasion. Proc Natl Acad Sci U S A. 2019;116:20141–20150. doi: 10.1073/pnas.1901931116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miyazawa A., Ito S., Asano S., Tanaka I., Sato M., Kondo M., et al. Regulation of PD-L1 expression by matrix stiffness in lung cancer cells. Biochem Biophys Res Commun. 2018;495:2344–2349. doi: 10.1016/j.bbrc.2017.12.115. [DOI] [PubMed] [Google Scholar]
- 106.Twomey J.D., Zhang B. Cancer immunotherapy update: FDA-approved checkpoint inhibitors and companion diagnostics. AAPS J. 2021;23:39. doi: 10.1208/s12248-021-00574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Davis A.A., Patel V.G. The role of PD-L1 expression as a predictive biomarker: an analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J Immunother Cancer. 2019;7:278. doi: 10.1186/s40425-019-0768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu Y., Shi H., Jiang M., Qiu M., Jia K., Cao T., et al. The clinical value of combination of immune checkpoint inhibitors in cancer patients: a meta-analysis of efficacy and safety. Int J Cancer. 2017;141:2562–2570. doi: 10.1002/ijc.31012. [DOI] [PubMed] [Google Scholar]
- 109.Kawazoe A., Kuboki Y., Shinozaki E., Hara H., Nishina T., Komatsu Y., et al. Multicenter phase I/II trial of napabucasin and pembrolizumab in patients with metastatic colorectal cancer (EPOC1503/SCOOP Trial) Clin Cancer Res. 2020;26:5887–5894. doi: 10.1158/1078-0432.CCR-20-1803. [DOI] [PubMed] [Google Scholar]
- 110.Clarke J.M., George D.J., Lisi S., Salama A.K.S. Immune checkpoint blockade: the new frontier in cancer treatment. Target Oncol. 2018;13:1–20. doi: 10.1007/s11523-017-0549-7. [DOI] [PubMed] [Google Scholar]
- 111.Miao Y.R., Thakkar K.N., Qian J., Kariolis M.S., Huang W., Nandagopal S., et al. Neutralization of PD-L2 is essential for overcoming immune checkpoint blockade resistance in ovarian cancer. Clin Cancer Res. 2021;27:4435–4448. doi: 10.1158/1078-0432.CCR-20-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liang S.C., Latchman Y.E., Buhlmann J.E., Tomczak M.F., Horwitz B.H., Freeman G.J., et al. Regulation of PD-1, PD-L1, and PD-L2 expression during normal and autoimmune responses. Eur J Immunol. 2003;33:2706–2716. doi: 10.1002/eji.200324228. [DOI] [PubMed] [Google Scholar]
- 113.Loke P., Allison J.P. PD-L1 and PD-L2 are differentially regulated by Th1 and Th2 cells. Proc Natl Acad Sci U S A. 2003;100:5336–5341. doi: 10.1073/pnas.0931259100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Inaba K., Yashiro T., Hiroki I., Watanabe R., Kasakura K., Nishiyama C. Dual roles of PU.1 in the expression of PD-L2: direct transactivation with IRF4 and indirect epigenetic regulation. J Immunol. 2020;205:822–829. doi: 10.4049/jimmunol.1901008. [DOI] [PubMed] [Google Scholar]