Abstract
A combination therapy of RhizomaPolygonati (RP) with goji (Lycium chinense) has earned a long history in the prescriptions to promote male health. However, the mechanisms at both molecular and nanoscale quantum levels are unclear. Here, we found that processed RP extract induces apoptosis and cell cycle arrest in cancer cells, thereby inhibiting prostate cancer cell proliferation enhanced by processed goji extract associated with an augment of the nanoscale herbzyme of phosphatase. For network pharmacology analysis, RP-induced PI3K-AKT pathways are essential for both benign prostatic hyperplasia and prostate cancer, and the RP/goji combination induces potent pathways which include androgen and estrogen response, kinase regulation, apoptosis, and prostate cancer singling. In addition, the experimental investigation showed that the prostate cancer cells are sensitive to RP extract for inhibiting colony formation. Finally, the natural compound baicalein found in RP ingredients showed a linked activity of top-ranked signaling targets of kinases including MAPK, AKT, and EGFR by the database of cMAP and HERB. Thus, both the nanozyme and ingredients might contribute to the RP in anti-prostate cancer which can be enhanced by goji extract. The proposed nanoscale RP extract might be of significance in developing novel anti-prostate cancer agents by combining goji compositions and targeted therapy compounds.
Introduction
Rhizoma Polygonati (RP), especially the combination therapy with goji (Lycium chinense), has earned a long history in the Traditional Chinese Medicine (TCM) prescription to benefit male health. RP is a TCM that is also applied for food in the south of China but widely grown in many countries, including India, Korea, Pakistan, Iran, Japan, Russia, European countries, and America.1,2 RP has multiple functions in phytopharmaceuticals including anti-aging, anti-diabetes, anti-Parkinson disease, and anti-cancer through multiple mechanisms including kinase signaling.1,3,4
Natural RP ingredients include chemical compounds of baicalein, diosgenin, steroidal saponins, lectins, and homoisoflavanones, with a majority of polysaccharides and some proteins such as lectin.1,5,6 RP natural ingredients have been reported to play a role in anti-cancer including breast cancer, lung cancer, leukemia, gastric cancer, and prostate cancer.2 For example, polysaccharides of RP extract could augment activated autophagy signaling through Beclin-1/LC3 to prevent the growth of the prostate cancer-related fibroblasts.7 The mechanisms include the RP or family species extract-induced cell cycle arrest at G0/G1 in breast cancer cells,1 apoptosis induced by the protein component of lectin in human melanoma by MAPK or AKT,8,9 and enhanced immune response in other cancers.1
The functional protein-based natural enzyme is seldom reported in traditionally used RP, which might be due to processed RP proteins becoming denatured, thereby decreasing activity, while nanoscale particles may retain their state and form new assembly of structures. We reported the assemblies upon processing of steamed or baked herbs which can generate nanozyme activity.10 The nanozyme has been found recently and applied widely in medicine but rarely in phytomedicine because of possible artificial synthesis with metal element-enhanced enzyme activity.11 Here, we aimed to apply the natural RP with traditional processing without metal enhancement and further studied the quantum level of nanozyme–herbzyme, as defined previously,10 to explore whether and how it crosstalks in cellular response in prostate cancer signaling.
Network pharmacology analysis provided the potent expedited analysis of mechanisms of RP in phytomedicine. Recently, clinicians have applied RP in anti-COVID-19 prescriptions based on the official treatment protocols.12 We applied a network pharmacology analysis and reported the potent pathways of anti-COVID-19 including blood pressure, apoptosis, and cell signaling.6 Moreover, the anti-fatigue mechanism was analyzed by network pharmacology, and it was reported that the E2F1 and PI3K-AKT associations are essential pathways.13
Given that RP plays essential roles in male health recorded in the ancient pharmacology, we here aimed to explore the possibility, efficiency, mechanisms, and pathways of RP extract in anti-prostate cancer using benign prostatic hyperplasia (BPH) as control via experimental investigation or network pharmacology analysis with databases and experimental connectivity.
Results
The Chinese herbal medicine RP extract has been applied in anti-cancer, but how it can function in treatment is still unclear. Here, we applied RP extract to test the efficiency of targeting prostate cancer cells.
RP Extract Induces Prostate Cancer Cell Death and Inhibits Cell Cycle
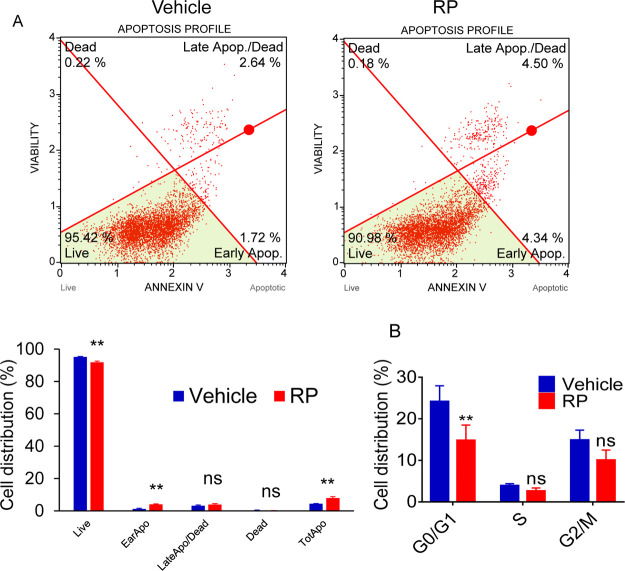
Previously, we reported that RP extract inhibits cell growth by potentially targeting kinase signaling through its phosphatase activity. To further explore the mechanisms of cell proliferation inhibition, we first tested the cell death and cell cycle regulation by RP. We found that RP induces cell growth inhibition after treatment at early cell death events indicated by annexin V-positive and Dead Cell Marker (Figure 1A). In addition, RP extract induces cell cycle arrest at G0/G1 in prostate cancer cells, as shown in Figure 1B, compared to the vehicle control.
Figure 1.
RP extract induces cell death (A) and regulates the cell cycle (B) in prostate cancer PC3 cells. ns, non-significant. *p < 0.05; **p < 0.01.
RP Extract-Induced Cell Growth Inhibition Is Enhanced by the Traditionally Prescribed Combination of Goji Associated with the Nanoscale Herbzyme of Phosphatase Activity
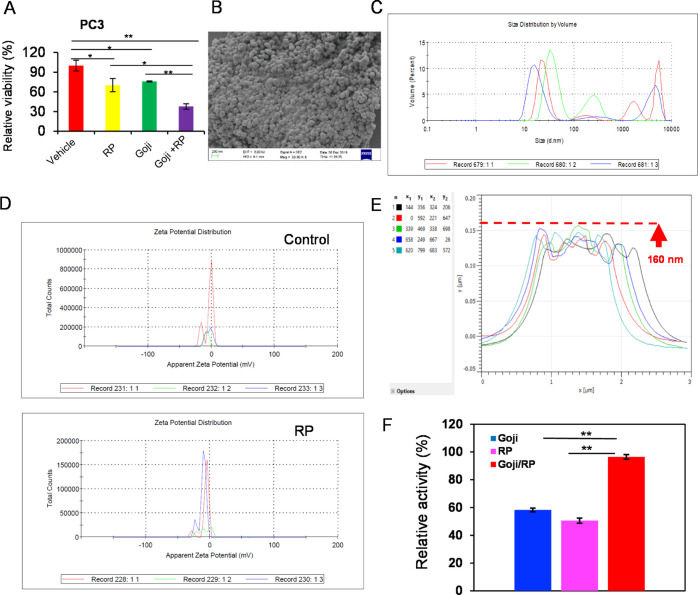
It has been shown and widely used in traditionally prescribed combinations of RP with goji in food or enhancement for male dysfunction, but with unknown effects on prostate cancer. Synergistic cooperation between RP and goji was achieved to inhibit the cancer cells growth (Figure 2A). However, the nanoscale herbzyme which is through potent phosphatase-kinase signaling and chemical components may also enhance the surface effect of the nanozyme, and chemical compounds themselves may exert the anti-cell growth function through drug targets of cell signaling. Thus, the nanoscale RP may be applied as a chemical ingredient delivery carrier in addition to the herbzyme of phosphatase activity. Both RP from Mount Tai and goji have nanoassembly of nanoflower structures as previously reported10 at a low density and reproducible at a high density (Figure 2B). We further investigated the particle size distribution and found that the Z-Average (d nm) is 238.0 and the polydispersity index (PDI) is 0.684. A majority of particles (61%) are in a size of 17.64 + 6.224 nm (Figure 2C). Next, we measured the zeta potential of RP, which is −7.95 + 2.3 mV. Goji extract showed nanoparticles of 160 nm in most sizes investigated by atomic force microscopy (AFM)10 (Figure 2E). Moreover, we tested the combined mixture effect of nanoparticles of RP and goji by measuring the synergistic cooperation on the phosphatase activity under pH 13 conditions when natural alkaline phosphatase (ALP) minimizes its function in vitro (Figure 2F). Thus, the nanoscale phosphatase may play a role in enhancing the synergistic effect.
Figure 2.
RP extract-induced cell growth inhibition in prostate cancer is enhanced by goji extract, which correlates to nanoherbzyme activity. (A) Cell viability assay. (B) SEM showing the size and cluster of RP extract at the nanoscale at a high density compared to previous reports for low-density individually separated forms.10 (C) RP nanoparticle size distribution. The Z-Average (d nm) is 238.0, and the PDI is 0.684. (D) RP nanoparticles and control zeta potential. The zeta potential of RP is −7.95 ± 2.3 mV. (E) AFM further analysis of goji extract nanoparticle size in three dimensions as previously reported.10 (F) Phosphatase assay of RP, goji, and the RP mixture with goji extract nanoparticles alone or by interactions in vitro.
Network Pharmacology Assay of RP-Induced Cell Signaling against Prostate Cancer
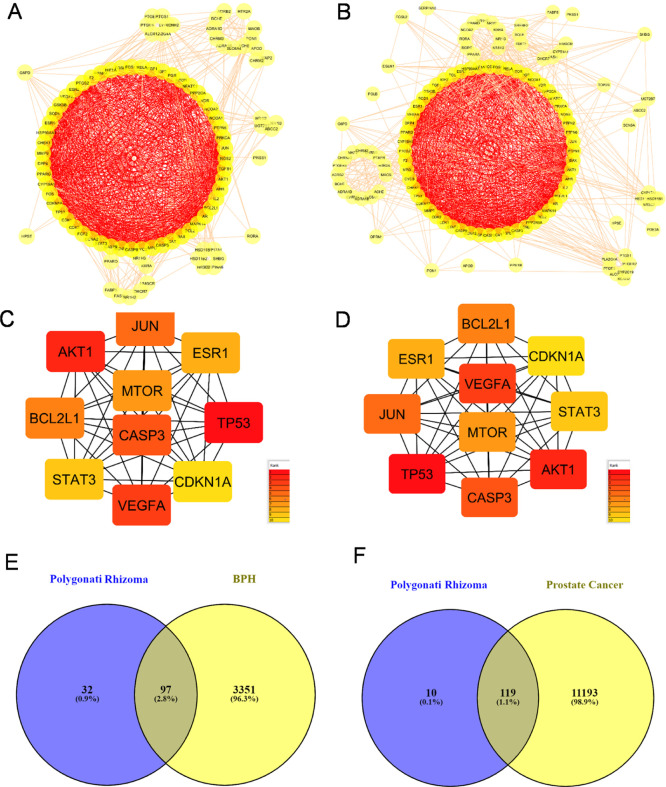
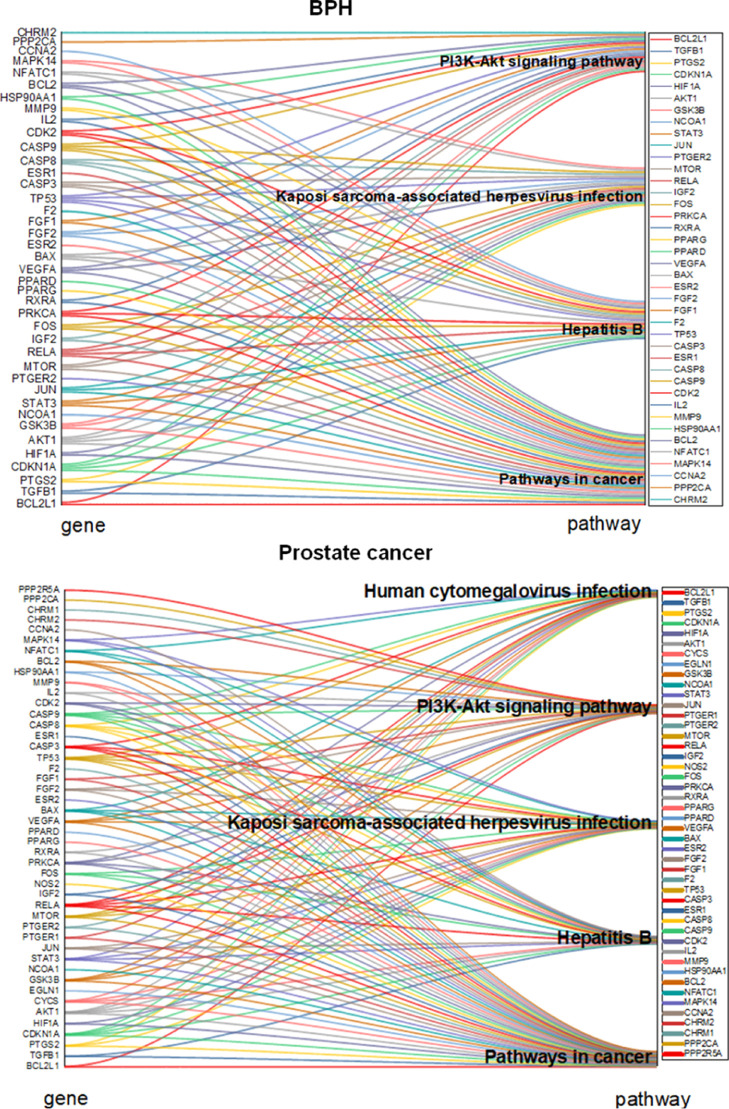
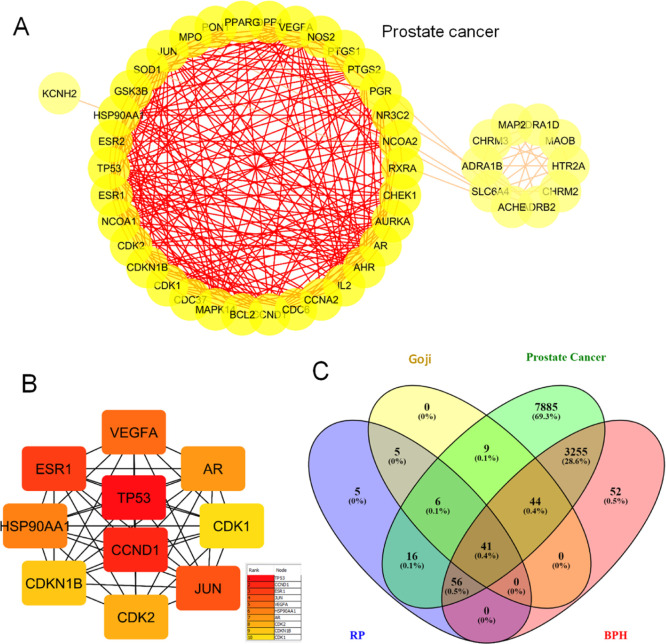
To further explore the chemical compounds assisting the complex signaling, we investigated the ingredient-mediated signaling by a network pharmacology assay (Figures 3–6). First, drug targets of potent chemical compounds were screened to search prostate cancer targets using BPH as a control. For protein–protein interaction (PPI) maps of the common targets, the top-ranked interactions involve p53, Caspase 3 apoptosis proteins, survival proteins BCL2L1, and kinase pathways AKT1 and STAT3 (Figure 3A–D). We found that the common targets of RP with prostate cancer are similar to BPH with 1.1 or 2.8%, respectively (Figure 3E,F).
Figure 3.
PPI analyses of common targets of RP with BPH (A,C,E) and prostate cancer (B,D,F). (A–D) Network data were analyzed on the ClusterONE plug-in of Cytoscape 3.8.0 to distinguish the highly interacted cluster which is given in red, and the top 10 hub targets of the PPI network are indicated. The ranking is given alongside. (E,F) Venn diagrams were constructed.
Figure 6.
Goji compound-related PPIs in a network pharmacology analysis with BPH and prostate cancer. (A,B) STRING database was used to construct the network of PPIs where the MCL clustering option was used with an MCL inflation parameter of 3. The nodes represent proteins, and edges represent their interactions, while colors represent their cluster identity. (C,D) Top hub targets of the PPI network are listed alongside.
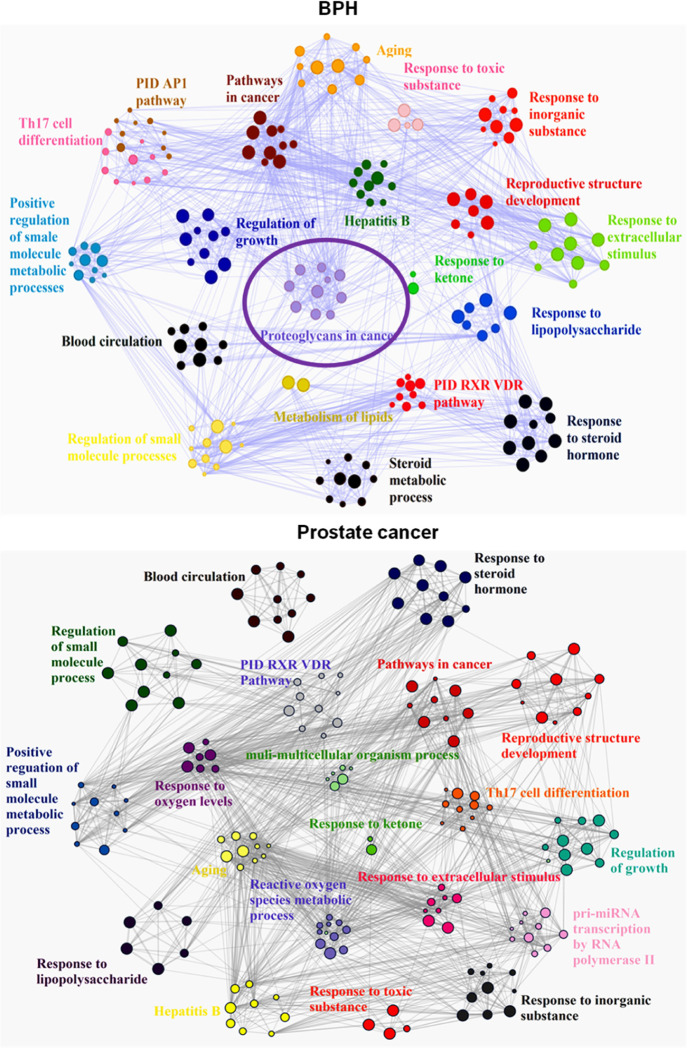
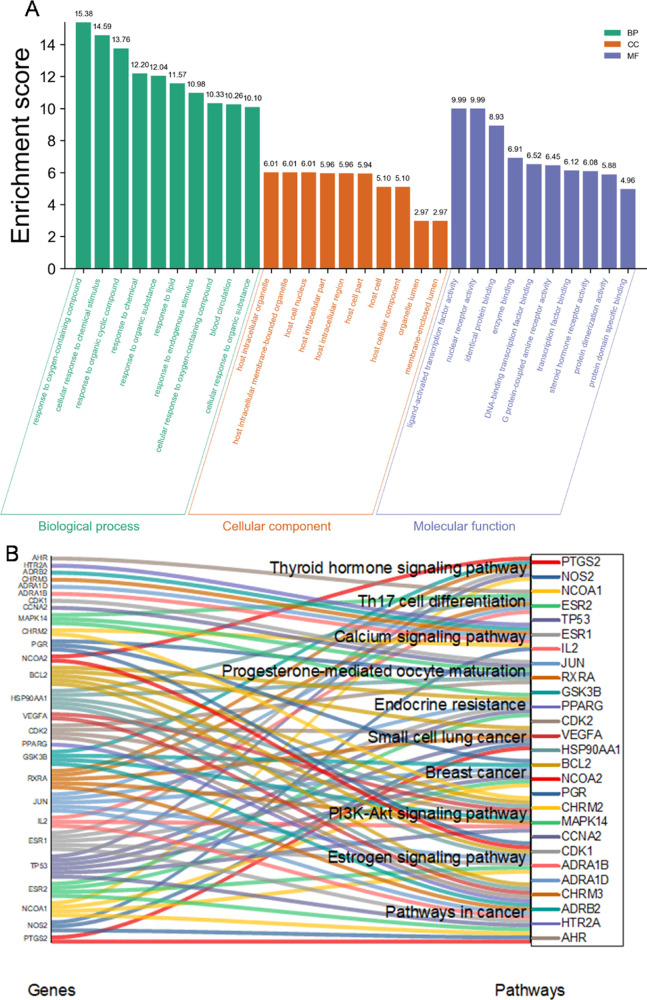
For enriched ontology clusters analysis, the set of common genes of the compound drug with prostate cancer was uploaded to the Metascape database for gene ontology (GO) enrichment analysis of biological processes. The generated network was then managed by Cytoscape 3.7.2 for further visualization. The results indicated 20 relevant biological processes including responses to hormones, lipids, immune stimulation, and toxic and extracellular stimulation, which are similar to both BPH and prostate cancer (Figure 4). For Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, we found that virus infection pathways are top in prostate cancer, and PI3K-AKT pathways are essential for both BPH and prostate cancer. The same four pathways are the PI3K-Akt signaling pathway, Kaposi sarcoma-associated herpes virus infection, and pathways in cancer and hepatitis B. In addition, RP–BPH illustrated proteoglycans in the cancer pathway, while RP-prostate cancer did not, suggesting the glycan-mediated BPH therapy potential (Figures 5 and 6).
Figure 4.
RP compound target-related PPIs with GO analysis against BPH and prostate cancer. GO cluster analysis was performed by putting the set of common genes of the compound drug with prostate cancer with the Metascape database, a gene annotation and analysis resource, for GO enrichment analysis. The generated network was then analyzed by Cytoscape 3.7.2 for further visualization. The results indicate 20 relevant biological processes. The size is related to the proportional number of input genes, and the color represents its cluster groups.
Figure 5.
RP compound target-related PPIs with KEGG pathways and functions in a network pharmacology analysis. The set of common genes was uploaded to the KOBAS database to get information about genes and their respective KEGG pathways. The results indicate that the main five pathways are enriched.
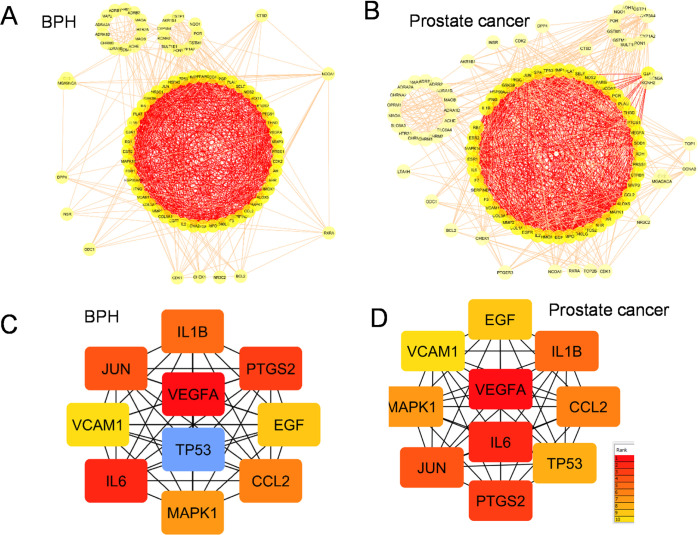
Network Pharmacology Analysis of Goji-Induced Cell Signaling against Prostate Cancer
We next performed network analyses on goji-induced PPI network signaling in prostate cancer using BPH as a control (Figures 6 and 7). We found that PPI maps showed similar ranks of nodes between BPH and prostate cancer with clusters of cytokine storm, apoptosis, kinase MAPK, growth factor EGF, and VEGF (Figure 6). Moreover, the KEGG pathway analysis showed the signaling of prostate cancer, general cancer, AGE-RAGE, and fluid shear stress and atherosclerosis pathways (Figure 7).
Figure 7.
Enrichment analyses of common targets of goji with BPH and prostate cancer. (A) BPH; (B) prostate cancer. Important biological processes were identified by implementing the ClueGO plug-in of Cytoscape software.
Network Pharmacology Analysis of RP–Goji Combinatorial Induction on Cell Signaling against Prostate Cancer
We further analyzed the RP/goji combinatorial application-induced pathways by searching the common or crosstalk signaling by the network analysis (Figures 8–10). First, we found that both RP and goji target-related PPI maps have a high rank of nodes related to hormone receptor AR and ER, heat shock protein HSP90AA1, CDK kinases, VEGF, and Jun (Figure 8A,B). In addition, for the number of common targets of RP/goji to prostate cancer versus BPH, BPH showed about 4-fold more targets than prostate cancer, suggesting the combinatorial therapy effect on BPH with multiple mechanisms in addition to cancer signaling (Figure 8).
Figure 8.
PPI analyses of common targets of goji, RP, BPH, and prostate cancer. (A) Network data were analyzed via the ClusterONE plug-in of Cytoscape 3.8.0 to distinguish the highly interacted cluster which is given in red. (B) Maximal Clique Centrality by cytoHubba plug-in of Cytoscape was used to determine the top 10 hub targets of the PPI network. The ranking is given alongside. (C) Venn diagram was constructed.
Figure 10.
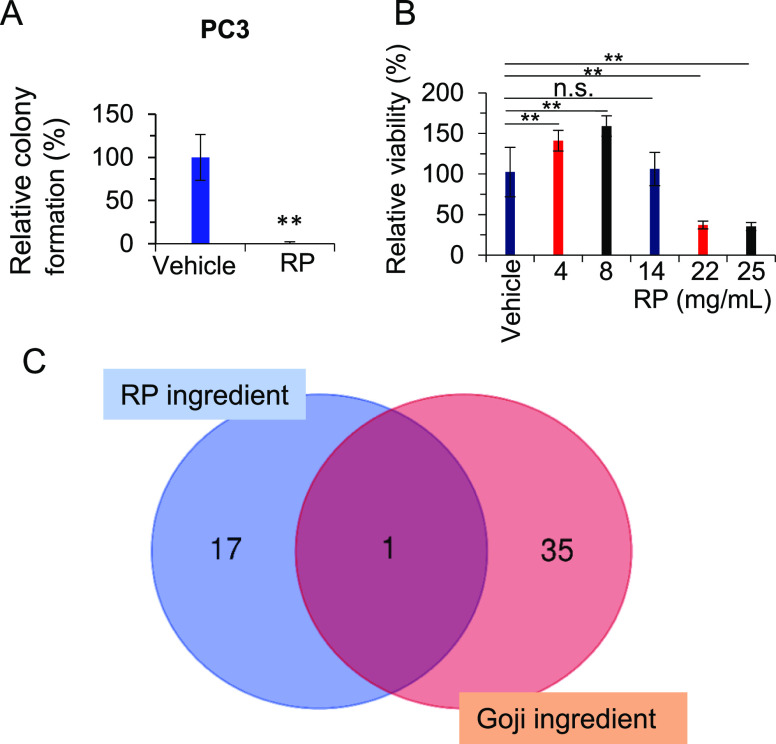
RP-induced cell growth inhibition in PC3 cells. (A) PC3 cells were sensitive to RP extract (0.5 mg/mL) for colony formation in a soft agar assay. (B) MDA-MB-231 cells are resistant to RP at a low dose. (C) Venn diagram of the ingredients of RP and goji. *p < 0.05; **p < 0.01.
For GO analysis, the top-ranked biological processes include blood circulation, response to chemical stimulation, and lipids, suggesting that the combinatorial treatment may be related to the lipid metabolism and bloodstream dynamics for prevention of prostate disease or cancer (Figure 9). For molecular functions, the top-ranked lists are transcription, enzyme binding, nuclear receptors and androgen receptor (AR) binding, DNA binding, and protein binding, suggesting the transcriptional regulation and potent kinase signaling-mediated functions.
Figure 9.
GO enrichment and KEGG pathway enrichment analyses of common targets of goji, RP, BPH, and prostate cancer. (A) Metascape database provided main GO results including biological processes. (B) Data of the KEGG pathway enrichment were selected from the Kobas database based on the corrected p-values and a κ score of 0.4 and visualized. Main 10 pathways are illustrated.
For KEGG pathway analysis, the hormone signaling, immune-related Th 17 differentiation, and PI3K-AKT pathways and stem cells are top lists, especially with calcium signaling (Figure 9). In AR tracing expression cells of PC3, cell growth are more sensitive to RP treatment at 0.5 mg/mL, which can inhibit 3D-cell growth of colony formation (Figure 10A). In contrast, based on estrogen signaling prediction in prostate cancer, we applied a breast cancer cell line MDA-MB-231 but with estrogen depleted and we found the significant resistance to the RP extract treatment as high as 22 mg/mL for inhibition of cell growth (Figure 10B). In details, at a low dose, the MDA-MB-231 cells showed resistance to the RP extract. Given that colony formation is related to stem cell signaling, our data suggest the anti-stem-like potential of RP in anti-cancer.
Experimental Database of Connectivity Analysis of RP Ingredient-Induced Signaling in Gene Profiling of Prostate Cancer Cells
Based on RP-mediated nanoscale phosphatase activity by self-assembly at pH 13, we assumed that some ingredients may play essential roles, which may create a basic surface for RP nanoparticles of the extract. Given berberine is a benzylisoquinoline alkaloid, which can induce self-assembly and anti-infection functions,14 the RP ingredient of baicalein is related to AR and more prostate cancer-related gene targets through database mining. Thus, we further analyzed the experimental related connectivity of the cMAP database15 with visualizations by the HERB16 online database tool and found that baicalein-induced differentially expressed genes are similar in number for up- and down-regulation, a KEGG enrichment list of apoptosis, protein modification, and GO enrichment with the top list of cell cycles, kinase, and kinase regulation, which is consistent with the whole extract of RP in prostate common targets (online analysis results at http://herb.ac.cn/Experiments/detail/?v=HBEXP000174). Finally, the connectivity analysis showed the PC3 and VCAP prostate cancer cells, which are androgen-non-responsive cells, have linked the activity of top-ranked signaling targets of kinases including MAPK, AKT, and EGFR, which are known for androgen-independent prostate cancer key pathways (online analysis results at http://herb.ac.cn/Experiments/detail/?v=HBEXP000174).
Discussion
As RP extract has been applied in anti-aging and enhanced male health with unclear mechanisms, we tested the function in male aging-related prostate diseases of prostate cancer while using BPH as a control to explore the potential of RP in anti-cancer. BPH has an overlapping mechanism of prostate disease compared to the transformed cancer such as AR signaling,17 but how the Chinese herbal medicine can function in treatment is still unclear. In this study, we applied RP extract to test the efficiency of targeting prostate cancer cells and demonstrated that RP inhibits prostate cancer cell growth, arrests its cell cycle, and induces apoptosis. Additionally, the network pharmacology revealed that such properties of RP may be explained by its targeting of the PI3K/AKT1 signaling cascade along with other kinase signaling and hormone-related pathways. These results were supported by cell viability and colony formation assays of MDA-MB-231 or PC3 cell lines.
RP Extract Induces Prostate Cancer Cell Cycle Arrests and Cell Death
The cell cycle arrest process involved in cellular growth may induce cancer cell apoptosis.18,19 We reported in this study that RP leads to cell cycle arrest in the G0/G1 phase. Such dysregulation of the cell cycle has been associated with cancer cell growth inhibition. While some cell cycle-specific proteins regulate cell cycle positively, there are also cyclin-dependent kinases (CDKs) at different cell cycle phases with inhibitory functions.20 Previous studies demonstrate that berberine, a main ingredient of RP, induces G0/G1 phase arrest in colorectal cancer21 and hepatocellular carcinoma cells.22 It has been shown that berberine arrests cell cycles in the G0/G1 phase by up-regulating p21, a CDK inhibitor.23 Berberine’s inhibitory effect on PI3K-AKT and MAPK signaling is also reported. These are consistent with the results of RP in this study and suggest the possible explanation of how RP may induce cell cycle arrest which causes cancer cell apoptosis and further reduces prostate cancer cell growth. However, further analyses are required to elucidate the underlying RP mechanisms.
RP-Induced Anti-cancer Signaling Analyzed by Network Pharmacology Enhanced by Goji by Phosphatase Activity
The PI3K-AKT signaling plays a crucial role in cancer cell survival in response to various stresses including osmotic stress, oxidative stress, and those caused by chemotherapeutic medicines and irradiation which usually occur during the cancer treatment procedures.24 As the signaling pathway gets activated, it leads to the phosphorylation of downstream cascades (Bad, GSK3β) which control critical survival functions of cancer cells such as the regulation of cell cycle, cell growth, and apoptosis.25 Therefore, it plays a fundamental role in prostate cancer carcinogenesis and progression.26 However, this can be disturbed via the inhibition of phosphorylation of several key players such as GSK3β. This is consistent with the findings of this study, where we determined in PPI results that RP might target this signaling pathway because its top-ranked proteins include AKT1, TP53, VEGFA, BCL2L1, MTOR, and CDKN1A, which are involved in the PI3K-AKT pathway. KEGG enrichment analyses also supported these data. Because the downstream cascade of the PI3K-AKT pathway also consists of kinase pathways, we hypothesize that RP might affect them as well. In addition, we found that RP has phosphatase activity of the nanozyme/herbzyme10 which can be enhanced by goji. Thus, goji-enhanced RP functions may crosstalk with the phosphatase/kinase and regulate kinase pathways through the above signaling we found.
The recurrence of prostate cancer after the androgen deprivation therapy which is commonly used along with radio- and chemotherapies is linked to cells that do not express AR or changes in function or structure.27 Recent studies found that androgen-independent prostate cancer may be targeted via ER pathways.28 According to Chaurasiya et al.,29 ERβ1 regulates the PI3K/AKT pathway in PC3 cells and its agonists may be used to inhibit AKT signaling. As RP at 0.5 mg/mL inhibits colony formation of PC3 cells that have no much function of AR, while a 44-fold higher dose of RP (22 mg/mL) was required to inhibit cell growth of the MDA-MB-231 estrogen-depleted cell line, our data suggest the possible mechanism of RP, which may be the activation of ER to block AKT activity in PC3 cells. Additionally, our experimental assays in consistency with network pharmacology displayed that RP’s anti-cancer effect is enhanced by goji, which is commonly used in combination with RP. Goji has been reported to target prostate cancer tumors by inducing cell apoptosis.30 These may explain the synergistic effect of goji with RP.
Comparing to Other Similar Works
Our work reported an RP-induced anti-cancer function in prostate cancer. RP-induced anti-cancer has been studied in different types of cancer. Cell cycle arrest at G0/G1 by RP polysaccharides has been reported in H22 hepatocellular carcinoma transplanted tumor growth and induced caspase-mediated cell death.31 Cancer cells that are reported to be inhibited by RP include breast cancer cells (MCF-27, MDA-MB-435), esophageal cancer cells (ECA-109), gastric cancer cells (HGC-27), colorectal cancer cells (HCT-8), Hela cells, leukemia cells (HL-60), prostate cancer-associated fibroblasts, and lung cancer cells.1,5−9,31 The mechanisms are also related to the immune system response.1 In recent findings, polysaccharides extracted from Polygonatum cyrtonema Hua could arrest the Hela cells at the G2/M cell cycle phase via increased expression of CyclinD1,32 which is different from our finding of G0/G1 arrest in prostate cancer. This might be due to the special prostate cancer signaling crosstalk of distinct cancer signaling. The evidence is that in the same cervical cancer cells of Hela, another compound, methyl protodioscin, which is a steroid saponin, induced the same cycle arrest as G2/M.33
In triple-negative breast cancer, the polysaccharide extracted from Polygonatum sibiricum decreased the population of myeloid cells and hematopoietic cell expansion in the spleen, suggesting the anti-breast cancer function.34 In the P. sibiricum, there are 64 chemical compounds with main parts as rhamnetin, wogonin, chrysosplenetin B, dauriporphine, and 5-hydroxyl-7,8-panicolin, whose targets include Akt, MAPK14, PIK3CG, and GSK3 of kinases and cell death-related p53, NOS2, and SCN5A, as almost half drug-targeted compounds are related to anti-cancer by network pharmacology analysis,35 which is consistent with our current results. The screened top-ranked targets of Akt35 are similar to our results based on whole chemical component database analysis. Importantly, the top targets are related to kinases, which is interesting to our finding that RP has phosphatase activity and potentially can target kinases as a nanoscale drug in delivery. Another study on RP-mediated anti-cancer effects revealed that some fractions of extracted steroidal glycosides could inhibit growth of HepG2, A549, and Caco2 cancer cells.36 In addition, polysaccharides extracted from P. sibiricum could affect the MAPK signaling cascade in anti-cancer through cytokine-related immune enhancement.37
The most interesting finding from breast cancer cells is that the extract of homoisoflavanones promotes the breast cancer cell MCF7 growth and binds to the estrogen receptor α ligand binding domain, suggesting the estrogen-like function.38 We may propose the combination of the special extract using nano RP as a carrier to inhibit hormone-sensitive prostate cancer instead of anti-androgen therapy which may result in decreased quality of life.39 Moreover, estrogen-based therapy needs more accurately estimate of the side effects in the pandemic era.40
All the above others’ studies focused on extracting RP on cancer signaling and apoptosis effects, but the whole extract of processed RP-mediated effects, especially by nanoparticles of the extract, is unclear. Nanoparticles of the herb have anti-cancer effects by the drug carrier, encapsulation, and drug delivery, but the nanozyme effect also is unknown. Here, we reported the nanozyme function by RP extract in vitro as a potent phosphatase to inhibit cancer cell signaling of growth. Phosphatase is a stem cell marker. Therefore, the nanoscale RP’s herbzyme activity may be functional in anti-cancer besides the chemical compound-mediated anti-cancer pathways. Here, we systematically analyzed both chemical compounds and the herbzyme of the nanoscale extract effect, which is a complete understanding and prediction of the RP in prostate cancer, in addition to that reported for the sugar or steroid-mediated function of RP extract.
Overall, our study suggested an underlying mechanism of RP’s anti-prostate cancer characteristics. However, in the future, there should be an in vivo experiment to validate the results of our network pharmacology analyses as the outcome is based on the data mining and analysis of huge amounts of information via bioinformatics tools which may differ from the actual in vivo experimental results.
Conclusions
RP extract inhibits prostate cancer cell growth, which is enhanced by the traditionally prescribed combination of goji associated with nanoscale herbzymatic phosphatase activity in vitro. Network pharmacology analysis of RP/goji combinatorial induction on cell signaling against prostate cancer includes hormone receptor AR and ER, kinases, and hormone signaling, which were validated by experimental investigation on colony formation, and cancer cell growth inhibition.
Materials and Methods
Nanoparticle Processing, Measurement of Size Distribution, and Zeta Potential
RP nanoparticles were obtained by processing RP slices of commercial products (Mount Tai-RP, Taian Xianlu Food Co., Ltd., Taian, China) as described previously,10 followed by boiling in a microwave and filtering by 200 nm. RP nanoparticle size distributions were measured using a Zetasizer Nano ZS analyzer (Malvern Instrument) with disposable microcuvettes under settings of a material absorbtion of 0.010 and a temperature of 25 °C. Zeta potential was measured by Malvern Instruments at a temperature of 25 °C with a count rate (kcps) of 17.4.
Apoptosis and Cell Cycle Analysis using a Muse Analyzer
For cell apoptosis and cell cycle analysis, cells were treated by RP for 24 h under detached conditions in 6-well plates at 8.5 mg/mL. For the cell apoptosis assay, cells were collected, suspended, and stained by the protocol provided in a Muse Annexin V Dead Cell Assay Kit (Merck Millipore). In details, cells were detached by treatment with trypsin and washed by phosphate-buffered saline (PBS) buffer, followed by suspending in PBS with 1% fetal bovine serum at 4 × 105 cells/mL concentration. Finally, cells were stained with the Muse Annexin V kit reagent according to the instructions of the manufacturer of the Muse cell analyzer (Merck Millipore). Cells were counted as the four populations of non-apoptotic cells with annexin V-/7-AAD–, early apoptotic cells with annexin V+/7-AAD–, late stage apoptotic/dead cells with annexin V+/7-AAD+, and mostly nuclear debris with annexin V–/7-AAD+ (Merck Millipore protocol). The gating was performed following the manufacturer’s guidelines of Muse software. During the data analysis, it requires visually identifying the above-mentioned four different cell populations on the plot for primary data. Further, the gates were adjusted using the manufacturer’s Muse software. The gates were in the same positions for all samples to remove the debris with the threshold markers. For cell cycle analysis, cells were treated by trypsin, collected, and washed by PBS and subjected to fix by ice-cold ethanol (70%) for at least 3 h. Then, cells were washed by PBS gently and finally stained with reagent kits of the Muse Cell Cycle Assay Kit (Merck Millipore). Data analysis was performed by the protocol of the Muse cell analyzer.
Cell Survival and Soft Agar Assays
The cell survival assay was performed by treating cells for 3 days with the concentration indicated in the figure legend, followed by fixation and staining with crystal violet as described previously.41 For the soft agar assay, cells were plated in 6-well plates with 6% bottom agarose and 3% top agarose (Sigma-Aldrich) and treated with the vehicle or RP extract at 0.5 mg/mL during plating when cooling down using the standard protocol of the soft agar assay. About 4 weeks post-treatment, the colonies were counted and inhibition efficiencies were calculated by percentage.
Network Pharmacology Analysis
RP extract ingredients were shown in our previously published paper.5,6 Based on the chemical compound, we obtained the chemical-gene targets through the database of “HERB” (http://herb.ac.cn/Detail/?v=HERB005429&label=Herb).16 The PPI maps of RP or goji drug target proteins were constructed using the STRING online tool and Cytoscape software.42,43 GO and KEGG pathway enrichment were analyzed by g:Profiler (http://biit.cs.ut.ee/gprofiler/page/citing) with visualization by an online tool for data analysis and graphing (http://www.bioinformatics.com.cn) or by the HERB (http://herb.ac.cn/Detail/?v=HERB005429&label=Herb) database16 and Origin software with the following details. The Metascape database provided main biological processes which then were visualized by the GO chord tool of bioinformatics software, and the data of KEGG pathway enrichment were selected from the Kobas database based on the corrected p-values and a κ score of 0.4 and visualized by OriginLab 2020b software.44−48 The connectivity map of the herb compound to known database gene expression profiling was obtained by HERB online request16 linking to the data set of cMAP perturbagens (https://clue.io/).15
Phosphatase Herbzyme Activity Assay
The phosphatase ALP (purchased from Life Technologies) and substrate of NBT/BCIP (nitro blue tetrazolium-5-Bromo-4-chloro-3-indolyl phosphate) (Thermo Fisher) were used to measure phosphatase activity of the herbzyme as described previously.10 The ALP was also applied as a positive control.
Scanning Electron and Atomic Force Microscopies
Atomic force microscopy (AFM) analysis was carried out with a Smart SPM 1000. AFM and scanning electron microscopy (SEM) sample preparations were described previously.41 In detail, for SEM analysis, herbal RP extracts were put on foil papers and subjected to quick drying in air.10 Finally, the samples were investigated using a Zeiss SEM microscope as described previously.41
Statistical Analysis
Student’s T test was the statistical analysis method to check the significance, and p < 0.05 or 0.01 was applied for analysis.
Acknowledgments
The authors would like to thank Prof. Xugang Li, who analyzed data with additional comments; Andrey Tsoy for kind support with the use of the Muse cell analyzer; and students including Aigerim Kabulova, Saltanat Kaldar, and Aigerim Nugmanova for kind help. The authors thank the Nazarbayev University FDCRG Program with grant number 16797152 (15798117, i.e., ID 110119FD4531) and grant number 16796808 (15874919, i.e., ID 110119FD4542).
Glossary
Abbreviations
- ALP
alkaline phosphatase
- AFM
atomic force microscopy
- SEM
scanning electron microscopy
- AR
androgen receptor
- ER
estrogen receptor
- RP
Huangjing, Rhizoma Polygonati, Polygonati Rhizoma
Author Contributions
Bexultan Kazybay and Qinglei Sun are co-first authors. Bexultan Kazybay performed database analysis and wrote parts of the paper including but not limited to the methods, figure legends, results and discussion. Qinglei Sun and Na Xu performed SEM, chemical analysis of RP, and wrote a part of the paper. Yingqiu Xie created the concept, designed with preforming some experiments, analyzed data, and wrote the paper. Qian Wang processed the RP. Kanat Dukenbayev performed AFM and wrote a part of methods. Madina Razbekova, Ayan A. Nurkesh, Qing Yang, Aidana Kutzhanova, and Anar Kabylda performed experiments and analyzed data. Cuiping Ma examined the chemistry of RP and methods, revised the paper, and provided consulting.
This work was supported from the Nazarbayev University Faculty-Development Competitive Research Grants Program [grant number 16797152 (15798117), that is, ID 110119FD4531, and grant number 16796808 (15874919), that is, ID 110119FD4542 to Y.X. as PI or Co-PI] and Shandong Taishanghuangjing Biotechnology Co. Ltd (Previously named as Taian Xianlu Food Co. Ltd).
The authors declare the following competing financial interest(s): Y.X. received financial support for research and collaboration with consulting; Q.W. received a salary from Shandong Taishanghuangjing Biotechnology Co. Ltd, Taian.
References
- Cui X.; Wang S.; Cao H.; Guo H.; Li Y.; Xu F.; Zheng M.; Xi X.; Han C. Review: The Bioactivities and Pharmacological Applications of Polygonatum sibiricum polysaccharides. Molecules 2018, 23, 1170. 10.3390/molecules23051170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P.; Zhao C.; Li X.; Gao Q.; Huang L.; Xiao P.; Gao W. The genus Polygonatum: A review of ethnopharmacology, phytochemistry and pharmacology. J. Ethnopharmacol. 2018, 214, 274–291. 10.1016/j.jep.2017.12.006. [DOI] [PubMed] [Google Scholar]
- Huang S.; Yuan H.; Li W.; Liu X.; Zhang X.; Xiang D.; Luo S. Polygonatum sibiricum Polysaccharides Protect against MPP-Induced Neurotoxicity via the Akt/mTOR and Nrf2 Pathways. Oxid. Med. Cell. Longevity 2021, 2021, 8843899. 10.1155/2021/8843899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G.-f.; Zhang Z.-y.; Lu L.; Xiao D.-q.; Xiong C.-x.; Zhao Y.-x.; Zong S.-h. Protective effects of Polygonatum sibiricum polysaccharide on ovariectomy-induced bone loss in rats. J. Ethnopharmacol. 2011, 136, 224–229. 10.1016/j.jep.2011.04.049. [DOI] [PubMed] [Google Scholar]
- Mu C.; Sheng Y.; Wang Q.; Amin A.; Li X.; Xie Y. Dataset of potential Rhizoma Polygonati compound-druggable targets and partial pharmacokinetics for treatment of COVID-19. Data in Brief 2020, 33, 106475. 10.1016/j.dib.2020.106475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu C.; Sheng Y.; Wang Q.; Amin A.; Li X.; Xie Y. Potential compound from herbal food of Rhizoma Polygonati for treatment of COVID-19 analyzed by network pharmacology: Viral and cancer signaling mechanisms. J. Funct. Foods 2021, 77, 104149. 10.1016/j.jff.2020.104149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. Y.; Hu M. H.; Qi G. Y.; Ma C. X.; Wang Y. Y.; Ma F. L.; Tao N.; Qin Z. H. Polysaccharides from Polygonatum Inhibit the Proliferation of Prostate Cancer-Associated Fibroblasts. Asian Pac. J. Cancer Prev. 2016, 17, 3829. [PubMed] [Google Scholar]
- Li C.; Chen J.; Lu B.; Shi Z.; Wang H.; Zhang B.; Zhao K.; Qi W.; Bao J.; Wang Y. Molecular switch role of Akt in Polygonatum odoratum lectin-induced apoptosis and autophagy in human non-small cell lung cancer A549 cells. PLoS One 2014, 9, e101526 10.1371/journal.pone.0101526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.; Wu L.; Wang D.; Wang H.; Chen J.; Yang C.; Bao J.; Wu C. Role of reactive oxygen species-mediated MAPK and NF-κB activation in polygonatum cyrtonema lectin-induced apoptosis and autophagy in human lung adenocarcinoma A549 cells. J. Biochem. 2016, 160, 315–324. 10.1093/jb/mvw040. [DOI] [PubMed] [Google Scholar]
- Benassi E.; Fan H.; Sun Q.; Dukenbayev K.; Wang Q.; Shaimoldina A.; Tassanbiyeva A.; Nurtay L.; Nurkesh A.; Kutzhanova A.; Mu C.; Dautov A.; Razbekova M.; Kabylda A.; Yang Q.; Li Z.; Amin A.; Li X.; Xie Y. Generation of particle assemblies mimicking enzymatic activity by processing of herbal food: the case of rhizoma polygonati and other natural ingredients in traditional Chinese medicine. Nanoscale Adv. 2021, 3, 2222–2235. 10.1039/D0NA00958J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.; Li D.; Chen L.; He H.; Wang Q.; Hong C.; He J.; Gao X.; Yang Y.; Jiang B.; Nie G.; Yan X.; Gao L.; Fan K. High-Performance Self-Cascade Pyrite Nanozymes for Apoptosis-Ferroptosis Synergistic Tumor Therapy. ACS Nano 2021, 15, 5735–5751. 10.1021/acsnano.1c01248. [DOI] [PubMed] [Google Scholar]
- Luo H.; Tang Q. L.; Shang Y. X.; Liang S. B.; Yang M.; Robinson N.; Liu J. P. Can Chinese Medicine Be Used for Prevention of Corona Virus Disease 2019 (COVID-19)? A Review of Historical Classics, Research Evidence and Current Prevention Programs. Chin. J. Integr. Med. 2020, 26 (4), 243–250. 10.1007/s11655-020-3192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-F.; Hu Y. Q.; Wu Q. G.; Zhang R. Virtual Screening of Potential Anti-fatigue Mechanism of Polygonati Rhizoma Based on Network Pharmacology. Comb. Chem. High Throughput Screening 2019, 22, 612–624. 10.2174/1386207322666191106110615. [DOI] [PubMed] [Google Scholar]
- Li T.; Wang P.; Guo W.; Huang X.; Tian X.; Wu G.; Xu B.; Li F.; Yan C.; Liang X.-J.; Lei H. Natural Berberine-Based Chinese Herb Medicine Assembled Nanostructures with Modified Antibacterial Application. ACS Nano 2019, 13, 6770–6781. 10.1021/acsnano.9b01346. [DOI] [PubMed] [Google Scholar]
- Lamb J.; Crawford E. D.; Peck D.; Modell J. W.; Blat I. C.; Wrobel M. J.; Lerner J.; Brunet J.-P.; Subramanian A.; Ross K. N.; Reich M.; Hieronymus H.; Wei G.; Armstrong S. A.; Haggarty S. J.; Clemons P. A.; Wei R.; Carr S. A.; Lander E. S.; Golub T. R. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Fang S.; Dong L.; Liu L.; Guo J.; Zhao L.; Zhang J.; Bu D.; Liu X.; Huo P.; Cao W.; Dong Q.; Wu J.; Zeng X.; Wu Y.; Zhao Y. HERB: a high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Res. 2021, 49, D1197–D1206. 10.1093/nar/gkaa1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A.; Shah A. A.; K N.; Lobo R. Mechanistic targets for BPH and prostate cancer-a review. Rev. Environ. Health 2021, 36, 261. 10.1515/reveh-2020-0051. [DOI] [PubMed] [Google Scholar]
- Chen J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harbor Perspect. Med. 2016, 6, a026104. 10.1101/cshperspect.a026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucci B.; Kasten M.; Giordano A. Cell cycle and apoptosis. Neoplasia 2000, 2, 291–299. 10.1038/sj.neo.7900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R. Jr. Cyclin-dependent protein serine/threonine kinase inhibitors as anticancer drugs. Pharmacol. Res. 2019, 139, 471–488. 10.1016/j.phrs.2018.11.035. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Liu X.; Yu M.; Xu M.; Xiao Y.; Ma W.; Huang L.; Li X.; Ye X. Berberine inhibits proliferation and induces G0/G1 phase arrest in colorectal cancer cells by downregulating IGF2BP3. Life Sci. 2020, 260, 118413. 10.1016/j.lfs.2020.118413. [DOI] [PubMed] [Google Scholar]
- Li F.; Dong X.; Lin P.; Jiang J. Regulation of Akt/FoxO3a/Skp2 Axis Is Critically Involved in Berberine-Induced Cell Cycle Arrest in Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2018, 19, 327. 10.3390/ijms19020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Wang X.; Sharvan R.; Gao J.; Qu S. Berberine could inhibit thyroid carcinoma cells by inducing mitochondrial apoptosis, G0/G1 cell cycle arrest and suppressing migration via PI3K-AKT and MAPK signaling pathways. Biomed. Pharmacother. 2017, 95, 1225–1231. 10.1016/j.biopha.2017.09.010. [DOI] [PubMed] [Google Scholar]
- Sabbah D. A.; Hajjo R.; Bardaweel S. K.; Zhong H. A. Phosphatidylinositol 3-kinase (PI3K) inhibitors: a recent update on inhibitor design and clinical trials (2016-2020). Expert Opin. Ther. Pat. 2021, 31, 877. 10.1080/13543776.2021.1924150. [DOI] [PubMed] [Google Scholar]
- Nunnery S. E.; Mayer I. A. Targeting the PI3K/AKT/mTOR Pathway in Hormone-Positive Breast Cancer. Drugs 2020, 80, 1685–1697. 10.1007/s40265-020-01394-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorning B. Y.; Dass M. S.; Smalley M. J.; Pearson H. B. The PI3K-AKT-mTOR Pathway and Prostate Cancer: At the Crossroads of AR, MAPK, and WNT Signaling. Int. J. Mol. Sci. 2020, 21, 4507. 10.3390/ijms21124507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkhoff H.; Berges R. The evolving role of oestrogens and their receptors in the development and progression of prostate cancer. Eur. Urol. 2009, 55, 533–542. 10.1016/j.eururo.2008.10.035. [DOI] [PubMed] [Google Scholar]
- Bonkhoff H. Estrogen receptor signaling in prostate cancer: Implications for carcinogenesis and tumor progression. Prostate 2018, 78, 2–10. 10.1002/pros.23446. [DOI] [PubMed] [Google Scholar]
- Chaurasiya S.; Wu W.; Strom A. M.; Warner M.; Gustafsson J.-Å. Estrogen receptor β regulates AKT activity through up-regulation of INPP4B and inhibits migration of prostate cancer cell line PC-3. Proc. Natl. Acad. Sci. U.S.A. 2020, 117, 26347–26355. 10.1073/pnas.2007160117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q.; Li Z.; Yan J.; Zhu F.; Xu R.-J.; Cai Y.-Z. Lycium barbarum polysaccharides induce apoptosis in human prostate cancer cells and inhibits prostate cancer growth in a xenograft mouse model of human prostate cancer. J. Med. Food 2009, 12, 695–703. 10.1089/jmf.2008.1232. [DOI] [PubMed] [Google Scholar]
- Jiang H. Study on anti-tumor activity of Polygonatum sibirium polysaccharide. J. Nanjing Univ. Chin. Med. 2010, 26, 479–480. [Google Scholar]
- Li L.; Thakur K.; Cao Y.-Y.; Liao B.-Y.; Zhang J.-G.; Wei Z.-J. Anticancerous potential of polysaccharides sequentially extracted from Polygonatum cyrtonema Hua in Human cervical cancer Hela cells. Int. J. Biol. Macromol. 2020, 148, 843–850. 10.1016/j.ijbiomac.2020.01.223. [DOI] [PubMed] [Google Scholar]
- Ma Y.-L.; Zhang Y.-S.; Zhang F.; Zhang Y.-Y.; Thakur K.; Zhang J.-G.; Wei Z.-J. Methyl protodioscin from Polygonatum sibiricum inhibits cervical cancer through cell cycle arrest and apoptosis induction. Food Chem. Toxicol. 2019, 132, 110655. 10.1016/j.fct.2019.110655. [DOI] [PubMed] [Google Scholar]
- Xie Y.; Jiang Z.; Yang R.; Ye Y.; Pei L.; Xiong S.; Wang S.; Wang L.; Liu S. Polysaccharide-rich extract from Polygonatum sibiricum protects hematopoiesis in bone marrow suppressed by triple negative breast cancer. Biomed. Pharmacother. 2021, 137, 111338. 10.1016/j.biopha.2021.111338. [DOI] [PubMed] [Google Scholar]
- Huang Z.-z.; Du X.; Ma C.-d.; Zhang R.-r.; Gong W.-l.; Liu F. Identification of Antitumor Active Constituents in Polygonatum sibiricum Flower by UPLC-Q-TOF-MSE and Network Pharmacology. ACS Omega 2020, 5, 29755–29764. 10.1021/acsomega.0c03582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D.; Li X.; Chang W.; Han Y.; Liu B.; Chen G.; Li N. Antiproliferative steroidal glycosides from rhizomes of Polygonatum sibiricum. Phytochemistry 2019, 164, 172–183. 10.1016/j.phytochem.2019.05.013. [DOI] [PubMed] [Google Scholar]
- Long T.; Liu Z.; Shang J.; Zhou X.; Yu S.; Tian H.; Bao Y. Polygonatum sibiricum polysaccharides play anti-cancer effect through TLR4-MAPK/NF-κB signaling pathways. Int. J. Biol. Macromol. 2018, 111, 813–821. 10.1016/j.ijbiomac.2018.01.070. [DOI] [PubMed] [Google Scholar]
- Chen H.; Li Y.-J.; Li X.-F.; Sun Y.-J.; Li H.-W.; Su F.-Y.; Cao Y.-G.; Zhang Y.-L.; Zheng X.-K.; Feng W.-S. Homoisoflavanones with estrogenic activity from the rhizomes of Polygonatum sibiricum. J. Asian Nat. Prod. Res. 2018, 20, 92–100. 10.1080/10286020.2017.1343821. [DOI] [PubMed] [Google Scholar]
- Ibragimova N.; Zhanzak Z.; Meyerbekova A.; Xie Y. Re: Long-term Psychological and Quality-of-life Effects of Active Surveillance and Watchful Waiting After Diagnosis of Low-risk Localised Prostate Cancer. Eur. Urol. 2018, 73, 143–144. 10.1016/j.eururo.2017.10.019. [DOI] [PubMed] [Google Scholar]
- Kazybay B.; Xie Y. Re: Herjan J.T. Coelingh Bennink, Jean-Michel Foidart, Frans M.J. Debruyne. Treatment of Serious COVID-19 with Testosterone Suppression and High-dose Estrogen Therapy. Eur Urol. In press.. Eur Urol. 2021, 80 (5), e115–e116. 10.1016/j.eururo.2021.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurkesh A. A.; Sun Q.; Fan H.; Dukenbayev K.; Tsoy A.; Altaikyzy A.; Wang K.; Xie Y. Date Pit Carbon Dots Induce Acidic Inhibition of Peroxidase and Disrupt DNA Repair in Antibacteria Resistance. Glob. Chall. 2019, 3, 1900042. 10.1002/gch2.201900042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D.; Gable A. L.; Lyon D.; Junge A.; Wyder S.; Huerta-Cepas J.; Simonovic M.; Doncheva N. T.; Morris J. H.; Bork P.; Jensen L. J.; Mering C. V. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N. S.; Wang J. T.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros J. C.Venny. An interactive tool for comparing lists with Venn’s diagrams. 2007–2015, https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed 2021).
- Origin(Pro), Version Number (e.g. “Version 2021”); OriginLab Corporation, Northampton, MA, USA, 2021; (accessed 2021).
- Raudvere U.; Kolberg L.; Kuzmin I.; Arak T.; Adler P.; Peterson H.; Vilo J. Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heatmap was plotted by http://www.bioinformatics.com.cn, an online platform for data analysis and visualization (accessed 2021).
- Xie C.; Mao X.; Huang J.; Ding Y.; Wu J.; Dong S.; Kong L.; Gao G.; Li C.-Y.; Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]