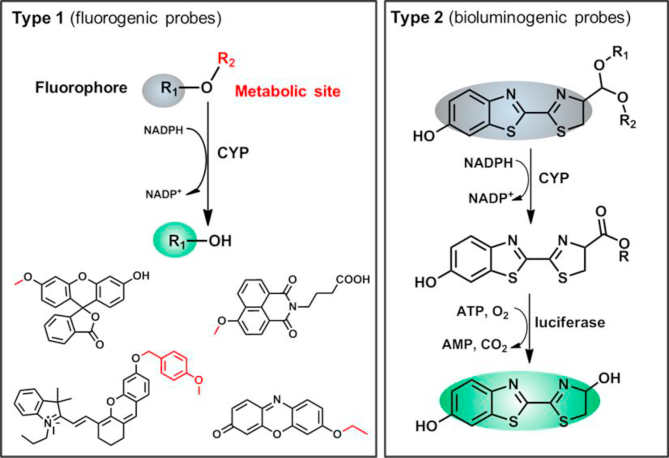
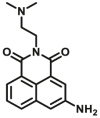
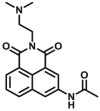
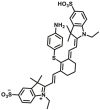
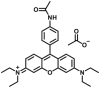
Abstract
Drug-metabolizing enzymes (DMEs), a diverse group of enzymes responsible for the metabolic elimination of drugs and other xenobiotics, have been recognized as the critical determinants to drug safety and efficacy. Deciphering and understanding the key roles of individual DMEs in drug metabolism and toxicity, as well as characterizing the interactions of central DMEs with xenobiotics require reliable, practical and highly specific tools for sensing the activities of these enzymes in biological systems. In the last few decades, the scientists have developed a variety of optical substrates for sensing human DMEs, parts of them have been successfully used for studying target enzyme(s) in tissue preparations and living systems. Herein, molecular design principals and recent advances in the development and applications of optical substrates for human DMEs have been reviewed systematically. Furthermore, the challenges and future perspectives in this field are also highlighted. The presented information offers a group of practical approaches and imaging tools for sensing DMEs activities in complex biological systems, which strongly facilitates high-throughput screening the modulators of target DMEs and studies on drug/herb‒drug interactions, as well as promotes the fundamental researches for exploring the relevance of DMEs to human diseases and drug treatment outcomes.
KEY WORDS: Optical substrates, Drug-metabolizing enzymes (DMEs), Fluorescence-based assay, High-throughput screening
Graphical abstract
The design principals and recent adavances in the development and applications of optical substrates for human DMEs, have been comprehensively summarized and discussed.
1. Introduction
Drug-metabolizing enzymes (DMEs) are a class of enzymes that catalyze the biotransformation of a great deal of drugs. Besides, DMEs have also been reported responsible for the metabolism of endogenous compounds (such as bile acids, steroids, and prostaglandins) and other xenobiotic chemicals (such as pollutants, carcinogens, food toxicants, as well as pesticides)1. The enzymatic reactions catalyzed by human DMEs mainly include oxidation, reduction, hydrolysis, and conjugation, which also divided to functionalization (phase I) and phase II reactions2. Phase I reaction including oxidation, reduction, and hydrolysis reaction, which often involve enhancing the hydrophilicity of the substrate compound by either introducing to or removal from it of a functional group, such as–OH, –COOH, –NH2, or –SH1. Generally, phase I reactions often occur through the two following ways. One is by introduction of the functional group, such as aliphatic and aromatic hydroxylation. The other one is through modifying the existing functionalities, for example oxidative N-, O-, or S-dealkylation, oxidation of alcohols to acids, reduction of azo and nitro compounds, reduction of ketones and aldehydes to alcohols, and hydrolysis of esters or amides1, 2, 3. In humans, the main phase I drug-metabolizing enzymes including carboxylesterase (CES), cytochrome P450s (CYPs), monoamine oxidases (MAOs), flavincontaining monooxygenases (FMOs), and xanthine oxidase/aldehyde oxidase (XO/AO)2,3. The phase II reactions are conjugative reactions, including glucuronidation, methylation, sulfonation, acetylation, glutathione and amino acid conjugation. In mammals, uridine 5′-diphospho (UDP)-glucuronosyltransferases (UGTs), methyl (N-methyl-, thiomethyl-, and thiopurinemethyl-) transferases, N-acetyltransferases (NATs), sulfotransferases (SULTs), and glutathione S-transferases (GSTs) are several important phase II enzyme classes4, 5, 6.
The abundance and activities of DMEs can directly affect the fate of drugs in humans, thereby have been recognized as the critical determinants of drug safety and efficacy7,8. Notably, the expression and activity levels of DMEs can vary significantly among individuals, as well as be affected by age, genetic, environmental, physiological, and pathophysiological factors9, 10, 11. Over the past few decades, a group of highly specific substrates (also termed probe substrates) for individual DMEs have been reported, which offer reliable tools to both academic and industry scientists for deciphering and understanding the key roles of individual DMEs in drug metabolism and toxicity, as well as screening and characterizing the interactions of human DMEs with xenobiotics. Unfortunately, most of the known probe substrates for DMEs are endogenous substances (such as testosterone as the probe substrate for CYP3A) or drugs (such as paclitaxel as the probe substrate for CYP2C8). These endogenous or drug substrates as well as their corresponding metabolites are generally quantified by using mass spectrometry-based techniques, which always require complicated sample pretreatment, expensive instruments and professional operators12, 13, 14. To overcome these shortcomings, both academic and industry scientists strongly call for the development of reliable, practical, easy to operate, time- and cost-effective assays for sensing target DME(s) in biological samples15.
In contrast to mass spectrometry-based assays, fluorescence-based assays have been widely used for sensing and real-time imaging of enzymes in a broad range of biological systems, due to their great superiority including rapid response, easy to conduct, ultrahigh sensitivity, in situ imaging, and applicable for high-throughput detection5,16, 17, 18, 19, 20. These fluorescent substrates can be used in a range of modern analytical instruments, such as liquid chromatography equipped with fluorescence detector, fluorescence microplate reader, fluorescence microscope and laser confocal microscope system. Through about ten years quickly development, a great deal of fluorogenic and bioluminescent substrates have been developed for detection the activity levels of target DME(s) in various biological systems, which offer a variety of practical and powerful tools for both basic researches and translational studies. No doubt, these tools have greatly facilitated the in vitro and in vivo DME-related studies, including the visual dynamic monitoring of enzyme activities in disease states, and deciphering the relevance of target DME(s) to various human diseases21. Moreover, parts of the reported optical tools for sensing DME(s) have been successfully applied in high-throughput discovery of leading compounds and in drug/herb‒drug iterations (DDIs/HDIs) studies, which have efficiently incorporated with information from other studies to facilitate drug research and development21. This review covers the design principals and recent Research progress in the development of highly specific optical substrates for a range of target human DMEs, as well as the biomedical applications of these optical compounds. Additionally, the challenges, perspective, and future reserch important in this field are also emphasized.
2. Optical substrates for phase I drug-metabolizing enzymes
2.1. Substrates for carboxylesterases
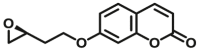
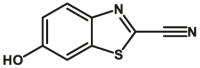
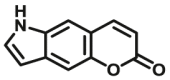
Carboxylesterase enzymes (CES) belong to the serine hydrolase superfamily, which responsible for the cleavage of various esters. Mammalian CES have been reported hydrolysis of a large number of xenobiotics or endogenous compounds bearing ester-, amide-, carbamate- or thioester-bonds. As important DMEs, CES play a crucial role of activating a panel of prodrugs bearing amide-, ester-, or amide-bond(s) to their active products5. In humans, human carboxylesterase 1A (hCES1A) and human carboxylesterase 2A (hCES2A) are two predominant CES members, but these two enzymes display notable differences in tissue distribution and substrate preferences. Generally, hCES1A is mainly expressed in the liver while hCES2A is primarily exists in the intestine. Since the important role in drug metabolism, the dysfunction or strong inhibition of CES might affect the in vivo biotransformation of their substrate-drugs. Constructing the highly selective and ultra-sensitive optical substrates for sensing CES1A or CES2A would greatly promote the discovery of CES inhibitors. Based on the understanding of the substrate preference of human CES, numerous of probes for CES, as well as the highly specific fluorescent probe substrates for hCES subtypes have been designed over the past decades (Table 1). As depicted in Table 1, probe CES-F1 to CES-F7 are non-specific substrates for hCES1A or hCES2A, due to these compounds could be hydrolyzed by both hCES1A and hCES2A. With the help of the studies on the substrate preference of both hCES1A and hCES2A, series of highly specific optical substrates for either hCES1A or hCES2A have been designed and developed in the past decade. As shown in Fig. 1 and Table 1, all of the specific drug- or optical substrates for CES1A are esters consisted of a large acyl and a small alcohol group (CES1A-B1, B2 and CES1A-F1 to CES1A-F3). By contrast, the specific substrates for CES2A are esters that are consisted of a large alcohol and small acyl group. Of note, CES1A-B1 was an ultra-sensitive and highly specific substrate for CES1A, which could be used to detect the trace amount of CES1A in plasma. CES1A-F2 was a relative long-wavelength fluorescent substrate for highly selective sensing of CES1A in living cells and animals, such as zebrafish22. Meanwhile, a number of fluorogenic substrates for CES2A including two-photon (CES2A-F5) and NIR fluorescent substrates (CES2A-F6) have been developed for monitoring CES2A functions in complex biological systems23,24. With these subtype-specific probes in hand, it's feasible to sense endogenous CES in living systems, to quantify CES activity levels in complex matrix, as well as to use these tools for high-throughput screening CES modulators.
Table 1.
Optical substrates for human CES.
| Enzyme | No. | Substrate | Metabolite | Km (μmol/L) | Vmax (μmol/min/mg) | λex/λem (nm) | Ref. |
|---|---|---|---|---|---|---|---|
| CES | CES-F1 |  |
 |
N.A. | N.A. | 571/585 | 163 |
| CES | CES-F2 | 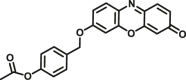 |
 |
N.A. | N.A. | 550/585 | 164 |
| CES | CES-F3 |  |
 |
N.A. | N.A. | 670/705 | 165 |
| CES | CES-F4 |  |
 |
4.4 | N.A. | 820/521 | 166 |
| CES | CES-F5 |  |
 |
N.A. | N.A. | 670/706 | 167 |
| CES | CES-F6 |  |
 |
18 | N.A. | 520/575 | 168 |
| CES | CES-F7 | 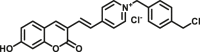 |
N.A. | N.A. | 810/620 530/620 |
169 | |
| CES1A | CES1A-B1 |  |
 |
71.3 | 7882 | Biolumincence | 155 |
| CES1A | CES1A-B2 | 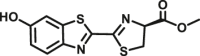 |
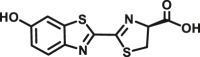 |
4.51 | 1208 | Biolumincence | 154 |
| CES1A | CES1A-F1 | 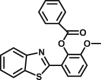 |
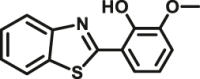 |
12.18 | 93,070 | 304/488 | 170 |
| CES1A | CES1A-F2 | 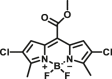 |
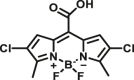 |
2.1 | 59.7 | 530/595 | 22 |
| CES1A | CES1A-F3 | 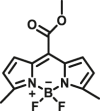 |
 |
2.9 | 0.117 | 505/560 | 156 |
| CES2A | CES2A-F1 | 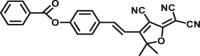 |
 |
4.82 | 14600 | 483/525 | 171 |
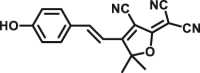
| CES2A | CES2A-F2 |  |
 |
3.06 | 5084 | 560/612 | 172 |
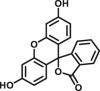
| CES2A | CES2A-F3 |  |
 |
N.A. | N.A. | 452/564 | 173 |
| CES2A | CES2A-F4 | 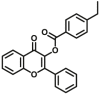 |
 |
N.A. | N.A. | 346/528 | 174 |
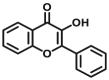
| CES2A | CES2A-F5 | 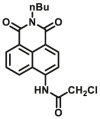 |
 |
8.58 | 34.87 | 430/542 | 24 |
| CES2A | CES2A-F6 |  |
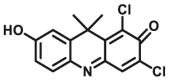 |
1.92 | 20.4 | 600/662 | 23 |
| CES2A | CES2A-F7 |  |
 |
6.6 | N.A. | 548/605 | 175 |
| CES2A | CES2A-F8 | 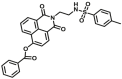 |
 |
3.48 | 0.027 | 414/560 | 162 |
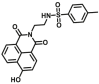
| CES2A | CES2A-F9 |  |
 |
4.33 | 280.2 | 373/540 | 176 |
Figure 1.
Design strategy for constructing CES optical substrates and the representative optical substrates.
2.2. Optical substrates for CYPs
The CYP enzymes are most important classes of DMEs that could catalyze oxygen insertion into a vast array of substrates, including lipids, steroids, drugs and environmental toxins, and play a pivotal role in the detoxification of xenobiotics, cellular metabolism and homeostasis25. Up to now, 57 CYP genes have been identified in humans, which are classified into 18 families and 44 subfamilies based on sequence similarity25,26. Among all identified CYP families in humans, CYP1, CYP2, and CYP3 are primarily responsible for the metabolism of xenobiotics, and are the so-called ‘xenobiotic-metabolizing’ CYPs, which are highly expressed in the liver and intestine. By contrast, the other CYP families are usually called as ‘endobiotic-metabolizing’ CYPs and are characterized with tissue-specific distributions. For example, CYP4 family is mainly involved in the metabolism of fatty acids and eicosanoids, which is highly expressed in the liver. CYP24 and CYP27 families are involved in the metabolism of vitamins, which are mainly distributed in the kidney. CYP7, CYP17, CYP19, CYP21, CYP39 and CYP46 families are primarily involved in the transformations of sterols. CYP7 and CYP39 families are rich in the liver, CYP21 is mainly distributed in steroidogenic tissues, and CYP46 family is a central neuro-specific metabolic enzyme26. In particular, CYP2W1 is specifically distributed in tumor tissue, while its physiologic function remains unknown27. The ‘endobiotic-metabolizing’ CYPs are always associated with the etiology of some diseases.
Currently, a variety of optical substrates (Table 2) have been developed for sensing CYP activities and for high-throughput screening of CYP modulators. Generally, the optical substrates or sensors for CYP(s) can be classified as substrate-type sensors, electrochemical biosensors and so on28,29. Amongst these, substrate-type sensors are widely used in characterization of CYP modulators, and other drug metabolism studies29. Substrate-type CYP sensors are substrate of target CYP(s), which can readily record the changes in CYPs activities in the presence of any tested compound or herbal extract, which strongly facilitate the discovery of CYP inhibitors or inducers, which in turn, decrease the rates of drug development failure30.
Table 2.
Reported optical substrates for human CYPs.
| Target enzyme | No. | Substrate | Metabolite | Km or S50 (μmol/L) | Vmax or Kcat | λex (nm)/λem (nm) | Ref. |
|---|---|---|---|---|---|---|---|
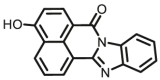
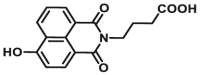
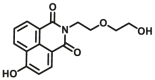
| CYP1A1 | CYP1A1-F1 | 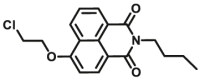 |
 |
0.84 | 18.99a | 450/562 | 32 |
| CYP1A1-F2 | 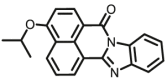 |
 |
0.29 | 5.54a | 470/525 | 35 | |
| CYP1A1-F3 |  |
 |
11.99 | 17.44a | 450/564 | 153 | |
| CYP1A1-F4 |  |
 |
N.A. | N.A. | 458/552 | 177 | |
| CYP1A1-F5 | 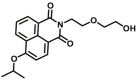 |
 |
N.A. | N.A. | 446/550 | 178 | |
| CYP1A1-F6 | 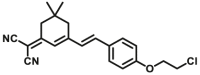 |
 |
N.A. | N.A. | 555/673 | 179 | |
| CYP1A1-F7 | 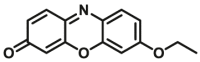 |
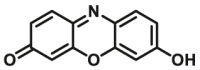 |
2.80 | 18.2a | 530/590 | 180 | |
| CYP1A1-F8 |  |
 |
0.278 | 9.17a | 540/590 | 33 | |
| CYP1A1-F9 | 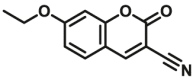 |
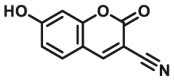 |
1.2 | 24.2a | 409/460 | 34 | |
| CYP1A2 | CYP1A2-F1 |  |
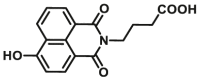 |
9.8 | 32.06a | 450/564 | 153 |
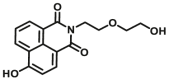
| CYP1A2-F2 | 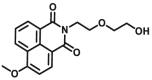 |
 |
N.A. | N.A. | 458/552 | 177 | |
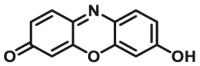
| CYP1A2-F3 | 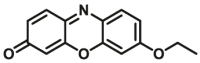 |
 |
3.0 | 0.23a | 530/590 | 180 | |
| CYP1A2-F4 |  |
 |
1.14 | 7.13a | 409/460 | 34 | |
| CYP1A2-F5 |  |
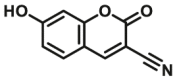 |
11 | 21.0c | 415/460 | 181 | |
| CYP1A2-F6 | 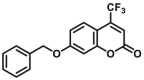 |
 |
4.1 | 11.1a | 410/510 | 182 | |
| CYP1B1 | CYP1B1-F1 |  |
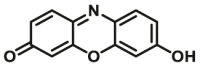 |
1.0 | 1.33a | 530/590 | 180 |
| CYP2A6 | CYP2A6-F1 | 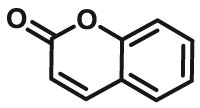 |
 |
2.1 | 0.79b | 390/460 | 34 |
| CYP2B6 | CYP2B6-F1 |  |
 |
8.5 | 14.0a | 409/530 | 38 |
| CYP2B6-F2 | 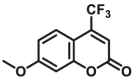 |
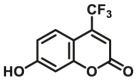 |
6.79 | 6.48a | 409/530 | 34 | |
| CYP2B6-F3 |  |
 |
52 | 66.0c | 415/460 | 181 | |
| CYP2B6-F4 | 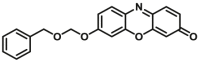 |
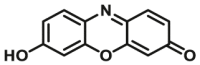 |
0.73 | 0.80c | 550/590 | 181 | |
| CYP2C8 | CYP2C8-F1 | 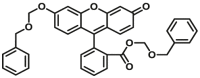 |
 |
3.6 | 2.10 c | 490/520 | 181 |
| CYP2C9 | CYP2C9-F1 |  |
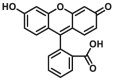
 |
43 | 2.10c | 425/460 | 181 |
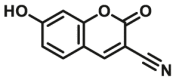
| CYP2C9-F2 |  |
 |
4.5 | 4.50c | 490/520 | 181 | |
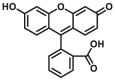
| CYP2C9-F3 | 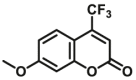 |
 |
54.7 | 0.56a | 409/530 | 34 | |
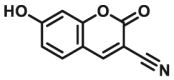
| CYP2C19 | CYP2C19-F1 |  |
 |
9.11 | 0.84a | 409/460 | 34 |
| CYP2C19-F2 |  |
 |
16.6 | 5.70c | 415/460 | 181 | |
| CYP2C19-F3 | 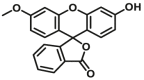 |
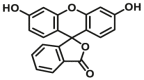 |
1.14 | 0.01b | 485/530 | 31 | |
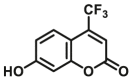
| CYP2D6 | CYP2D6-F1 | 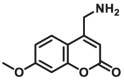 |
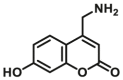 |
11.5 | 2.81c | 390/460 | 183 |
| CYP2D6-F2 | 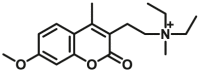 |
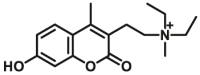 |
4.81 | 6.19a | 390/460 | 34 | |
| CYP2D6-F3 |  |
 |
5.0 | 0.60c | 405/490 | 181 | |
| CYP2E1 | CYP2E1-F1 |  |
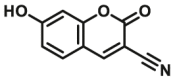 |
22 | 2.10a | 415/460 | 184 |
| CYP2J2 | CYP2J2-F1 |  |
 |
4.2 | 0.44d | 656/718 | 185 |
| CYP2J2-F2 | 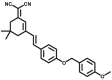 |
 |
N.A. | N.A. | 555/673 | 186 | |
| CYP3A | CYP3A-F1 |  |
 |
89 for 3A4 N.A. for 3A5 |
10.0c for 3A4 N.A. for 3A5 |
415/460 | 187 |
| CYP3A-F2 | 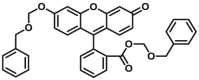 |
 |
7.8b for 3A4 N.A. for 3A5 |
16.0c for 3A4 N.A. for 3A5 |
490/520 | 187 | |
| CYP3A-F3 | 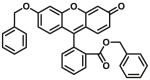 |
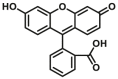 |
1.03 for 3A4 2.17 for 3A5 |
0.55 for 3A4 1.28 for 3A5 |
490/520 | 34 | |
| CYP3A4 | CYP3A4-F1 | 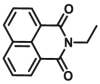 |
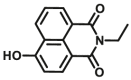 |
59.8 | 2.18a | 450/558 | 36 |
| CYP3A4-F2 |  |
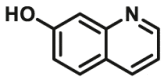 |
70 | 3.39b | 420/530 | 37 | |
| CYP3A4-F3 |  |
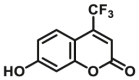 |
26 | 61.2c | 410/510 | 182 | |
| CYP3A4-F4 |  |
 |
7.8 | 1.6 | 490/520 | 187 | |
| CYP3A4-F5 |  |
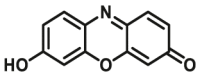 |
1.4 | 1.1 | 550/590 | 181 | |
| CYP3A4-F6 |  |
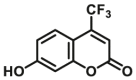 |
4.6 | 0.02b | 410/510 | 37 | |
| CYP1A1 | Luciferin-1A1 | 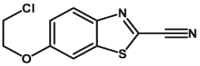 |
 |
3.0 | N.A. | 560 | 188 |
| CYP1A2 | Luciferin-1A2 | 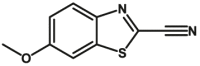 |
 |
6.0 | N.A. | 560 | 188 |
| CYP2B6 | Luciferin-2B6 |  |
 |
3.0 | N.A. | 560 | 189 |
| CYP2C9 | Luciferin-H |  |
 |
100 | N.A. | 560 | 188 |
| CYP2C19 | Luciferin-H EGE |  |
 |
10 | N.A. | 560 | 189 |
| CYP2D6 | Luciferin-ME EGE | 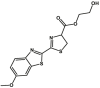 |
 |
30 | N.A. | 560 | 189 |
| CYP2J2 | Luciferin-2J2/4F12 |  |
 |
1.0 | N.A. | 560 | 188 |
| CYP3A4 | Luciferin-IPA |  |
 |
3.0 | N.A. | 560 | 188 |
| CYP3A7 | Luciferin-3A7 |  |
 |
33b | N.A. | 560 | 188 |
| CYP4A11 | Luciferin-4A |  |
 |
80 | N.A. | 560 | 188 |
| CYP4F3B | Luciferin-4F2/3 |  |
 |
∼10b | N.A. | 560 | 188 |
| CYP4F12 | Luciferin-4F12 | 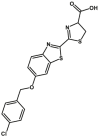 |
 |
10 | N.A. | 560 | 188 |
NA.: not available or not assayed.
Vmax values were in nmol/min/nmol P450 for CYP enzymes.
Vmax values were in nmol/min/mg human liver microsome.
kcat or Vmax values were in min‒1.
Vmax values were in nmol/min/mg CYP2J2; λex: excitation wavelength; λem: emission wavelength.
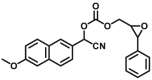
The reported optical substrates for human CYPs have been summarized in Table 2. It is notable that most of the reported optical CYP substrates showed poor selectivity with the exception of CYP1A1-F1 (CYP1A1), CYP1A1-F2 (CYP1A1), CYP1A1-F5 (CYP1A1), CYP1A1-F6 (CYP1A1), CYP2A6-F1 (CYP2A6), CYP3A4-F1 (CYP3A4), CYP3A4-F2 (CYP3A4), and CYP2J2-F1 (CYP2J2). The non-specific optical substrates of CYPs are limited to be used for screening CYP inhibitors in vitro by using individual recombinant human CYP enzyme. As shown in Table 2, CYP2C19-F3, a so-called fluorogenic substrate of CYP2C19, can also be metabolized by CYP1A1 and CYP2C931. Coincidentally, CYP3A4-F6, a fluorogenic substrate of CYP3A4, can also be metabolized by both CYP1A231.
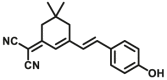
In recent years, the highly-selective optical probes for several subtypes of CYPs have been rapidly developed and widely applied. The milestone is the development of probe fluorescent probe CYP1A1-F1, a first isoform-specific fluorescent probe of hCYP1A1 which is first reported by Dai et al.32. hCYP1A1 and hCYP1A2 are two CYP1A isoforms in humans and have high sequence homology. The volume of the catalytic cavity of hCYP1A1 (524 Å3) is larger than that of hCYP1A2 (375 Å3). Interestingly, hCYP1A2 prefers to catalyze O-demethylation of the substrates with O-methyl group(s), while hCYP1A1 prefers to catalyze O-dealkylation of relatively large ethers, such as O-ethyl or O-chloroethyl33. These subtle characteristics inspire the researchers to design and develop a panel of other specific optical substrates for CYP1A1 via adjusting the volume of the leaving ether group(s), such as CYP1A1-F2 and CYP1A1-F834. Inspired by the development of CYP1A1-F1, Ning et al.35 have reported a new two-photon fluorescent substrate (CYP1A1-F2) for CYP1A1, which was designed based on the substrate preferences of CYP1A1 and the principle of intramolecular charge transfer (ICT). Recently, a resorufin-based substrate (CYP1A1-F8) for highly selective and sensitive sensing CYP1A1 activities has been developed by Jin et al.33. CYP3A4-F1 was developed by a novel two-dimensional strategy. In the first dimension, docking-based virtual screening was used to select a suitable two-proton fluorophore as the substrate candidate for CYP3A4. In the second dimensional design, chemical modifications on the selected fluorophore (N-substituted 1,8-naphthalimide) were performed to optimize the catalytic activity and isoform-selectivity. Another case is the selective optical probe for CYP2J2, such as CYP2J2-F1 and CYP2J2-F2, which are designed based on the characteristics of narrow substrate channel of CYP2J236,37. Besides of the highly selective fluorescent substrates for CYP1A1 and CYP2J2, another major breakthrough is the development of an CYP3A4 isoform-specific two-photon fluorescent substrate CYP3A4-F138. CYP3A4-F1 shows excellent specificity for sensing the activity of CYP3A4 in biological systems, which can distinguish the very similar subtype CYP3A5 from CYP3A4.
In addition to the fluorescence-based CYP activity assays, a panel of bioluminescent-based CYP activity assays has also been developed in the past few decades. In contrast to fluorescence-based assays, bioluminescent-based assays do not need the external light excitation, owing to the luminescent signals are initiated by ATP and luciferase. In the field of biological imaging, bioluminescent probes are very popular with the researchers, owing to the high signal to background imaging and almost no tissue autofluorescence. As shown in Table 2, the relatively selective luminogenic substrates for various human CYP isoenzymes have been reported. Similar to fluorescent probes, most of the bioluminogenic substrates showed poor selectivity towards target enzyme except for Luciferin-H and Luciferin-IPA.
The CYP-metabolic reaction types include hydrocarbon hydroxylation, O-/S-/N-dealkylation; N-/S-oxidation, epoxidation, and dehydrogenation, etc. The electron donor ability of hydroxyl (‒OH) is stronger than that of the ether bond (‒O‒), which enhance the ICT process and result in red-shifted ratiometric or off-on fluorescence response39. Hence, most of optical probes for CYPs are designed based on the dealkylation reaction (Fig. 2). In general, O-dealkylation is a preferential reaction for CYP1A and other CYPs rather than CYP3A4, while CYP3A4 has the extraordinary ability to mediate the aromatic hydroxylation36. Notably, the exception is the development of hydroxylation-based fluorescent probe of CYP3A4 (CYP3A4-F1) recently reported by Ning et al.36. This probe was designed by introducing the alkyl substituent groups to the non-reaction site of 1,8-naphthalimide, resulting the aromatic hydroxylation reaction of CYP3A4-F1.
Figure 2.
Design strategies for constructing CYP substrates and the representative optical substrates.
Overall, the highly selective probes are crucial for activity sensing and screening of selective modulators of the target human CYPs. Besides of ‘xenobiotic-metabolizing’ enzymes in the CYP1-3 families, ‘endobiotic-metabolizing’ CYPs are also getting more and more attentions, since these enzymes are closely related to the pathological conditions.
2.3. Optical substrates for monoamine oxidases (MAOs)
Monoamine oxidases (MAOs) are a family of mitochondrial-bound enzymes with high expression levels in neuronal and gastro-intestinal tissues. Two major isoforms: MAO-A and MAO-B have attracted wide attention. These isoforms share over 70% sequence similarity but differ in enzyme activity, substrate preference and distribution. They catalyze the oxidative deamination of a range of monoamines and have major roles in metabolizing released neurotransmitters. Increasing evidences have shown that dysregulation of MAO-A is related to depressive disorders and neuropsychiatric illnesses, while irregular amounts of MAO-B contribute mainly to neurodegenerative diseases. To better monitor the activity of MAOs, many optical substrates, especially small-molecule fluorescent ones (Table 3), are developed, which serve as powerful tools to visualize MAOs in living cells and even organisms.
Table 3.
Reported optical substrates for human MAOs.
| Enzyme | No. | Substrate | Metabolite | Km (μmol/L) | Vmax or Kcat | λex/λem (nm) | Ref. |
|---|---|---|---|---|---|---|---|
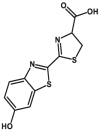
| MAOs | MAOs-F1 | 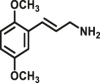 |
 |
218 | Kcat = 7.25 S‒1 | 343/393 | 40 |
| MAOs | MAOs-F2 |  |
 |
510 for MAO-B; 31 for MAO-A |
Kcat=0.35 S‒1 Kcat=0.0083 S‒1 |
335/524 | 41 |
| MAOs | MAOs-F3 |  |
 |
MR1: 7.6 mmol/L for MAO A, 1.8 mmol/L for MAO B MR2: 6.3 mmol/L for MAO-A, 3.4 mmol/L for MAO-B | N.A. | 550/583 | 42 |
| MAOs | MAOs-F4 |  |
 |
146.1±7.21 for MAO-A; 106.8 ±5.06 for MAO-B | Kcat = 0.16 S‒1 for MAO-A; 0.14 S‒1 for MAO-B | 360/530 | 43 |
| MAOs | MAOs-F5 | 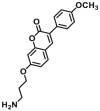 |
 |
N.A. | N.A. | 345/465 | 44 |
| MAOs | MAOs-F6 |  |
 |
Substrate1: 11.4 for MAO-A, 7.7 for MAO-B; Substrate2: 8.9 for MAO-A, 6.8 for MAO-B; Substrate3: 20.6 for MAO-A, 11.4 for MAO-B; Substrate4: 11.4 for MAO-A, 10.5 for MAO-B | N.A. | 470/535 | 45 |
| MAOs | MAOs-F7 | 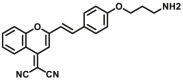 |
 |
N.A. | N.A. | 420/664 | 46 |
| MAOs | MAOs-F8 | 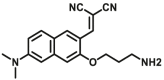 |
 |
68±8 for MAO-A; 74 6 for MAO-B | N.A. | 448/585 | 47 |
| MAOs | MAOs-F9 | 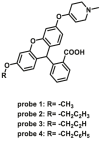 |
 |
Substrate1: 257.9 for MAOA, 47.1 for MAOB; Substrate2: 236.8 for MAOA, 43.9 for MAOB; Substrate3: 217.0 for MAOA, 42.9 for MAOB; Substrate4: 246.2 for MAOA, 44.6 for MAOB | N.A. | 470/515 | 190 |
| MAOs | MAOs-F10 | 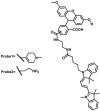 |
 |
Substrate1: 647.2 for MAO-A, 157.1 for MAO-B; Substrate2: 82.1 for MAO-A, 13.8 for MAO-B | N.A. | 475/570 | 52 |
| MAO-A | MAO-A-F1 | 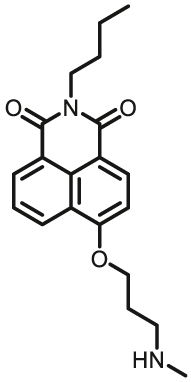 |
 |
N.A. | N.A. | 425/550 | 53 |
| MAO-A | MAO-A-F2 |  |
 |
N.A. | N.A. | 425/550 | 54 |
| MAO-A | MAO-A-F3 |  |
 |
2.9 | N.A. | 550/586 | 55 |
| MAO-A | MAO-A-F4 |  |
 |
4.5 | N.A. | 530/675 | 56 |
| MAO-A | MAO-A-F5 | 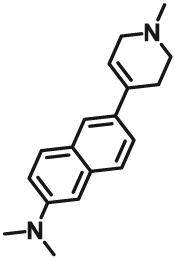 |
 |
11.96 | Vmax=0.42 nmol/min/mg | 430/618 | 58 |
| MAO-B | MAO-B-F1 |  |
 |
N.A. | N.A. | 375/456 | 48 |
| MAO-B | MAO-B-F2 |  |
 |
8.33 | Kcat = 0.0051 S‒1 | 304/449 | 49 |
| MAO-B | MAO-B-F3 | 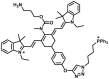 |
 |
10.13 | Vmax=3.55 nmol/min/mg | 650/750 730/803 |
50 |
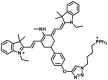
| MAO-B | MAO-B-F4 |  |
 |
59.63 | Vmax=1.42 nmol/min/mg | 360/460 | 51 |
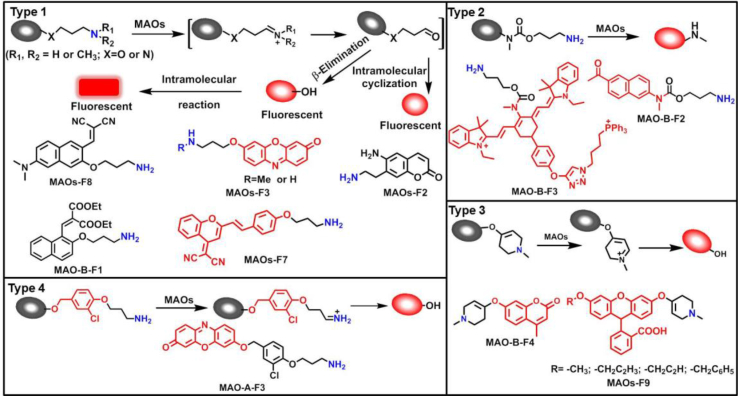
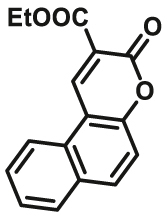
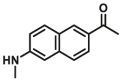
Currently, most of reported MAO substrates were designed on the basis of tandem amine oxidation/β-elimination mechanism (Fig. 3, type 1). The first reaction-based fluorescent substrate (MAOs-F1) for MAOs, reported by the Silverman group40 in 1996, was based on amine oxidation followed by iminium intermediate hydrolysis. After that, they modified the sensing scheme and developed a turn-on fluorescent substrate (MAOs-F2) that underwent a sequence of oxidation and intramolecular cyclization by MAOs41. On this basis, Chang and co-workers42 developed a substrate-tethered activity substrate (MAOs-F3) by utilizing a tandem amine oxidation/β-elimination mechanism to detect MAOs activity in vitro and in living cells directly and specifically without the need of additional enzymes or other activating reagents. Subsequently, a large number of fluorescent dyes were applied to improve the selectivity and sensitivity of detection. Aw et al.43 chose 2-(2′-hydroxy-5′-chlorophenyl)-6-chloro-4-(3H)-quinazolinone (HPQ) as fluorescent reporters, and HPQ fluorophores are alkylated with aminopropyl groups (MAOs-F4) for MAO enzymatic reactions. Upon addition of MAO, the amine oxidation and subsequent β-elimination lead to the release of acrolein and a green fluorescent product, HPQ, thus allowing a direct and effective detection of MAOs activity in vitro and living cells. Other fluorophores including coumarin (MAOs-F5)44 and fluorescein (MAOs-F6)45 were also applied for sensing MAOs activity in complex system, these activity-based substrates displayed great advantage in cell imaging of monoamine oxidases. Fluorescent substrates with emission wavelength in NIR region (>650 nm) have a great advantage in avoiding interference from biological matrix autofluorescence, minimizing photo-damage to biological samples, and penetrating relatively deeper tissues. Inspired by these advantages, Li et al.46, developed the first red emission fluorescent substrates (MAOs-F7) for the detection of MAOs with high sensitivity (LOD = 1.2 μg/mL), large Stokes shift (227 nm), solid-state fluorescence and excellent imaging ability in sensing MAOs in living cells. For the activation strategy described previously, fluorescence is produced by directly cleavage of the masking group to generate a fluorescent signal. But it limits this reaction only to the fluorophores containing –OH groups. Hence, many other types of MAOs fluorescent substrates were further developed by chemists based on cascade reactions. Chen et al.41 designed and synthesized a novel fluorogenic substrate (MAOs-F2) for MAOs on the basis of the oxidation–intramolecular cyclization cascade sequence. Later, Kim et al.47 designed a selective MAO substrate according to another cascade reaction (MAOs-F8). In brief, after hydrolysis of aliphatic amine, intramolecular nucleophilic attack of the phenolic oxygen on the electron withdrawing group results in the cyclized and highly fluorescent compound IminoPOS. Qin et al.48 reported a similar reaction (MAO-B-F1) for MAO detection by using coumarin and carboxylic acid as the electron withdrawing group. Upon MAO-B treatment, the re-exposed oxygen anion will attack the nearest carbonyl carbon atom, leading to the formation of the fluorescent product and the emission of fluorescent signals. Some chemists used carbamate group as a linker between fluorophore and sensing group (propylamine) (MAO-B-F2, F3)49,50, after metabolized by MAO, the carbamate group would automatically hydrolyze to produce the turn-on fluorescent dye (Fig. 3, type 2).
Figure 3.
Design strategies for constructing the fluorescent substrates for MAOs and the representative probes.
Unlike most reported MAO probes based on oxidation/β-elimination, Long et al.54 designed and developed a new class of fluorogenic substrates based on monoamine oxidase-triggered oxidative C–O bond cleavage. Fluorophores were conjugated with N-alkylated tetrahydropyridine moiety as recognition group, upon α-carbon oxidation on the group to form the corresponding dihydropyridinium intermediate, which was rapidly hydrolyzed to yield an aminoenone byproduct, liberating the free fluorophore (Fig. 3, type 3). According to the above mechanism, their team used some other simple fluorophores and successfully developed several MAOs specific fluorescent probes (MAOs-F9, F10, MAO-B-F4)51,52.
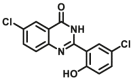
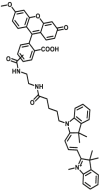
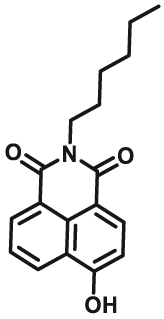
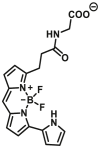
Notably, the development of highly selective substrates for sensing MAO-A is extremely challenging. Previously reported fluorescent substrates for MAO are only suitable for sensing either MAO-B or the total activity levels of the two major MAO isoforms, and fluorescent substrates for highly-selective sensing MAO-A are still lacking. Ma and his team53 reported two new ratiometric fluorescence substrates for highly selective sensing MAO-A rather than MAO-B with high sensitivity. The substrates were designed by combining a recognition group of propylamine with the fluorescent skeleton of 1,8-naphthalimide, and the sensing mechanism is based on amine oxidation and β-elimination to liberate the fluorophore (4-hydroxy-N-butyl-1,8-naphthalimide). The two substrates showed an outstanding ratiometric fluorescence response for MAO-A but not MAO-B, indicating their superior ability to distinguish MAO-A from MAO-B. Moreover, substrate 1 (MAO-A-F1), displaying higher sensitivity than substrate 2, has been applied to imaging the activity of endogenous MAO-A in different cell lines successfully. Meng et al.54 further modified the fluorophore and constructed a new two photon fluorescent substrate for MAO-A (MAO-A-F2) with high reaction speed and good temperature stability, and this probe was successfully used to image this enzyme in living cell and zebrafish. Additionally, they proposed a new design strategy for a specific fluorescent substrate for MAO-A by applying the characteristic structure of the enzymatic inhibitor with propylamine as a recognition group (Fig. 3, type 4)55. The high specificity of the representative substrate (MAO-A-F3) is demonstrated by sensing MAO-A in different living cells such as SH-SY5Y (high level of MAO-A) and HepG2 (high level of MAO-B), as further validated by both control substrate and Western blot analyses. The use of the characteristic structure of an inhibitor may serve as a general strategy to design a specific recognition moiety of fluorescent substrates for an enzyme. Yang et al.56 chose DCM as near-infrared fluorophores and designed a near-infrared fluorescent substrate (MAO-A-F4) based on inhibitor (clorgyline) structure-guided for the specific detection of MAO-A in cells and in vivo.
Some chemists have also begun to explore the functions of enzymes in subcellular organelles and their role in the occurrence and development of diseases. Kim et al.57 devised a two-photon substrate for highly selective targeting and sensing the MAO-A in prostate cancer, and this probe showed efficiency to visualize MAO-A in living cells and inhibit their growth and metastasis potential. Fang et al.58 reported the first MAO-A-specific two-photon fluorogenic substrate (MAO-A-F5), capable of selective and sensitive imaging of endogenous MAO-A activities in MAO-A expressing neuronal SY-SY5Y cells, the brain of tumor-bearing mice and human glioma tissues by using two-photon fluorescence microscopy (TPFM) with high resolution.
2.4. Optical probes for aldehyde dehydrogenases (ALDHs)
Aldehyde dehydrogenases (ALDHs) are a series of evolutionarily conserved enzymes with pyridine nucleotide-dependent oxidoreductase activity, which performs a variety of critical cellular processes. There are currently 19 known members of the ALDH superfamily, among which ALDH1A1 and ALDH2 are the more commonly studied ALDH isoenzymes in mammals. ALDH1A1 is a cytosolic enzyme responsible for the oxidation of retinal to retinoic acid, which is essential for normal development and physiological homeostasis. ALDH2 is mainly mitochondrial acetaldehyde dehydrogenase and plays a major role in an organism's survival when exposed to various hazards. For the past few decades, ALDH has been studied as a potential universal marker for normal and cancer stem cells, many methods have been made to identify and isolate viable, functionally active ALDH pos cells to effectively study the function of ALDH.
The previous commonly used method to study ALDH activity is to monitor NADH production through changes in absorption or fluorescence. However, this assay is lack of sensitive or specific for ALDH, because NADH fluorescence assays can be used for any enzyme that consumes or creates NADH. In order to provide an alternative approach for selective detecting ALDH activity, more and more researchers tend to design specific optical probes based on the catalytic properties of ALDH itself (Fig. 4, Table 4). Jacek Wierzchowski et al.59 identified 7-methoxy-I-naphthaldehyde as a fluorogenic synthetic substrate (ALDHs-F1) for human “low Km” aldehyde dehydrogenase (ALDH) isozymes. Oxidation of the naphthaldehyde by ALDH and NAD+ leads to a highly fluorescent naphthoate, which can be detected and quantified selectively on the basis of its unique spectral characteristics. The sensitivity of the proposed ALDH assay is sufficient to measure the catalytic activity of the enzyme in diluted blood lysates without any pre-purification by a continuous method. Later, they found similar structure 6-methoxy-2-naphthaldehyde was selective oxidized by class III ALDH present in human saliva60.
Figure 4.

Design strategy for constructing the fluorescent substrates for ALDHs and the representative probes.
Table 4.
Reported optical probes for human ALDHs.
| Enzyme | No. | Substrate | Metabolite | Km (μmol/L) | Vmax | λex/λem (nm) | Ref. |
|---|---|---|---|---|---|---|---|
| ALDHs | ALDHs-F1 | 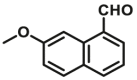 |
 |
0.55 for ALDH1 | N.A. | 330/390 | 60 |
| ALDHs | ALDHs-F2 | 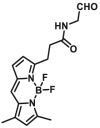 |
 |
N.A. | N.A. | 488/512 | 61 |
| ALDHs | ALDHs-F3 | 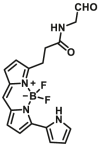 |
 |
N.A. | N.A. | 493/560 | 62 |
| ALDH1 | ALDH1-F1 | 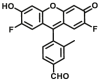 |
 |
N.A. | N.A. | 496/516 | 63 |
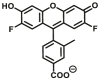
| ALDH1 | ALDH1-F2 | 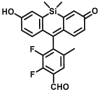 |
 |
N.A. | N.A. | 594/614 | 64 |
| ALDH1/ALDH2 | ALDH1-B1 |  |
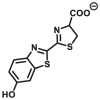 |
38 for ALDH1 | N.A. | Biolumincence | 65 |
| ALDH3A1 | ALDH3A1-F1 | 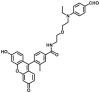 |
 |
3.49 | N.A. | 494/516 | 61 |
The currently available commercial assay for identifying ALDH bright cells (cells with high ALDH activity) is based on enzymatic hydrolysis of a fluorescent substrate ALDEFLUOR (ALDHs-F2) to produce green fluorescence61. This assay is highly sensitive, reproducible, nontoxic, and easy to conduct, and has been used for sensing ALDH activity levels in multiple biological systems. Minn et al.62 described a new red-shifted fluorescent ALDH substrate (AldeRed-588-A) (ALDHs-F3) to overcome the limitations of green fluorescence emission of ALDEFLUORTM reagent. To improve the probe selectivity and minimize leakage of the metabolized probe molecule, Atsushi Yagishita et al.61 selected a more hydrophilic fluorophore, TokyoGreen (TG) to develop ALDH3A1-specific fluorescent probes (ALDH3A1-F1) through a series of experimental screening. This probe enabled the visualization of ALDH3A1-positive cells by fluorescence microscopy as well as isolation of ALDH3A1-positive viable cells from a human Caucasian esophageal squamous cell line (OE21) that heterogeneously expresses ALDH3A1 by flow cytometry. Thanks to the development of these probes, the study of ALDH function and related diseases has been greatly progressed. However, these fluorogenic substrates still suffer from major drawbacks. For example, BODIPY-aminoacetaldehyde (BAAA) is equally fluorescent compared to its turned-over carboxylate product, cancer stem cells are identified based on their ability to retain the BAAA product relative to the un-activated probe using efflux pump inhibitors. In addition, BAAA exhibits cross reactivity with several ALDH isoforms, which makes the interpretation of experimental results challenging. To overcome these problems, Anorma et al.63 have designed an activity-based sensing (ABS) probe for ALDH1A1, termed as AlDeSense (ALDH1-F1), which is based on the photostable Pennsylvania Green dye platform and is equipped with a pendant benzaldehyde moiety. AlDeSense exhibits a 20-fold fluorescent enhancement when incubated with ALDH1A1, and display excellent selectivity for ALDH1A1 over other ALDH isoforms and common biological relevant species63. These excellent characteristics make it successful for monitoring cancer stem cells plasticity in a tumor model of live mice. Using the same design strategy, they subsequently developed a red-shifted analog (red-AlDeSense) (ALDH1-F2), a cell-permeable, red-shifted activity-based sensor for ALDH1A1 based on the TokyoMagenta dye platform64.
Fluorescent probes are sometimes susceptible to spontaneous fluorescence interference from biological matrices, in order to provide an alternative approach for sensing ALDH activity, Sarah J. Duellman65 developed a bioluminescent assay (ALDH1-B1) that provides many advantages over fluorogenic substrates, including greater sensitivity and a lower false hit rate. The proluciferin derivative that is converted from an acetal form to an aldehyde through acid hydrolysis, and then the aldehyde moiety on this molecule was converted by ALDH to a carboxylic acid to form luciferin. This method is more sensitive and the signal stability provided allows convenient assay set-up in batch mode-based high throughput screening.
2.5. Optical probes for NAD(P)H quinone oxidoreductases (NQOs)
NAD(P)H quinone oxidoreductase 1 (NQO1) is a kind of cytoflavin protease widely distributed in human body. It can catalyze a two-electron reduction of multiple quinones to hydroquinone in a manner dependent on NAD(P)H. Human NAD(P)H quinone oxidoretase-1 (hNQO1) as an important metabolic enzyme plays an important role in detoxification of endogenous quinone derivatives, stabilization of metabolic function and regulation of tumor suppressor (P53, P33) and plays a protective role in oxidative stress. Thus, NQO1 has become a specific biomarker and valuable target for early diagnosis.
Generally, NQO1 activated fluorescent substrates were designed basing on its property of quinone bio-reduction, where the fluorescent probe combined with a quinone-based NQO1 substrate as a trigger group to makes the fluorophore in a quenched state (Fig. 5 and Table 5). The strong fluorescence of these probes occurs when the quinone part is specifically activated by NQO1. The “trimethyl lock” containing quinone propionic acid (QPA) has been extensively used as a universal NQO1 activated trigger group because of its rapid and selective reduction by NQO1 to form hydroquinone analog, which underwent lactonization to release fluorescent probe (Fig. 5, type 1)66. Consequently, McCarley group and his team67, 68, 69 have developed several QPA-based fluorescent substrates for highly selective imaging of NQO1 in cancer cells (NQO1-F1-F3). Other fluorophores include naphthalimide (NQO1-F4,F11)70,71, acedan (NQO1-F5,F6)72,73, coumarin (NQO1-F7)74, hemicyanine (NQO1-F8,F10)75,76, cyanine 7 amine (NQO1-F9)75, 2-hydroxyphenyl substituted benzimidazole (HBI)/benzoxazole (HBO)/benzothiazole (HBT) (NQO1-F12)77, 2-dicyanomethylene-3-cyano-4,5,5-trimethyl-2,5-dihydrofuran (TCF) (NQO1-F13)78, dicyanoisophorone (NQO1-F14)79, and 7-nitro-2,1,3-benzoxadiazole (NBD) (NQO1-F15)80 were also used in designing NQOs triggered sensors. Some chemists add some self-leaving groups to the above strategy, upon NQOs treatment, the linking group leaves and releases the fluorescent group (Fig. 5, types 2 and 3, NQO1 F16–F19)81, 82, 83, 84. For example, Fei et al.84 constructed a small fluorescent probe NQO1-F16 by attaching an NQOs trigger (a trimethyl-locked quinone propionic acid) to a 6-hydroxylphenol-BODIPY platform through a p-aminobenzyl alcohol linker. After treatment with NQOs, reductant-mediated conversion of the trimethyl-locked quinone to hydroquinone triggered lactonization to release p-aminobenzyloxycarbonate, which then underwent self-immolation to deliver the red fluorescent 6-hydroxylphenol-BODIPY dye.
Figure 5.
Design strategies for constructing the fluorescent substrates for NQOs and the representative probes.
Table 5.
Reported optical probes for human NQOs.
| Enzyme | No. | Substrate | Metabolite | Km (μmol/L) | Vmax (μmol/min/mg) | λex/λem (nm) | Ref. |
|---|---|---|---|---|---|---|---|
| NQO1 | NQO1-F1 |  |
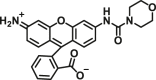 |
23.7 | 2.14 | 485/520 | 67 |
| NQO1 | NQO1-F2 |  |
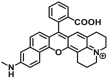 |
13.9 | 3.14 | 585/624 | 68 |
| NQO1 | NQO1-F3 | 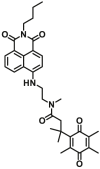 |
 |
3.77 | 0.24 | 374/490 | 69 |
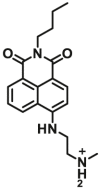
| NQO1 | NQO1-F4 |  |
 |
2.17±0.46 | 4.01 | 410/540 | 191 |
| NQO1 | NQO1-F5 | 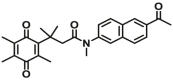 |
 |
14.6 | Vmax=0.9 μmol/min/L | 360/502 | 72 |
| NQO1 | NQO1-F6 | 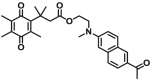 |
 |
10.68 | 0.962 | 400/520 | 73 |
| NQO1 | NQO1-F7 |  |
 |
5.06 ± 1.31 | 101.5 | 360/450 | 74 |
| NQO1 | NQO1-F8 | 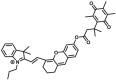 |
 |
N.A. | N.A. | 670/710 | 75 |
| NQO1 | NQO1-F9 | 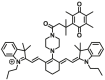 |
 |
1.42 ± 0.18 | 1.89 | 730/800 | 75 |
| NQO1 | NQO1-F10 |  |
 |
3.73 | 1.22 | 680/704 | 76 |
| NQO1 | NQO1-F11 | 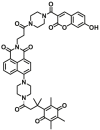 |
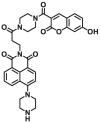 |
1.80 ± 0.37 | 0.267 | 345/540 | 70 |
| NQO1 | NQO1-F12 | 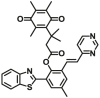 |
 |
3.79 | Vmax=0.126 μmol/min/L | 397/600 | 77 |
| NQO1 | NQO1-F13 | 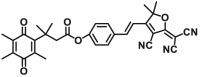 |
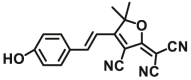 |
N.A. | N.A. | 444/561 | 78 |
| NQO1 | NQO1-F14 | 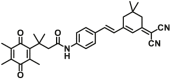 |
 |
33.82 ± 0.45 | 0.00419 | 460/646 | 79 |
| NQO1 | NQO1-F15 |  |
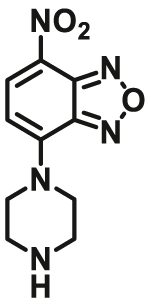 |
6.82 | 197.76 | 470/550 | 80 |
| NQO1 | NQO-F16 | 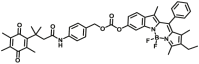 |
 |
6.77 | Vmax=0.043 μmoL/min/L | 510/585 | 84 |
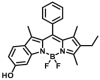
| NQO1 | NQO1-F17 |  |
 |
10.4 | 0.00225 | 440/525 | 83 |
| NQO1 | NQO1-F18 |  |
 |
1.1 ± 0.5 | 0.31 | 606/755 | 82 |
| NQO1 | NQO1-F19 |  |
 |
N.A. | N.A. | 435/490 | 192 |
2.6. Optical probes for epoxide hydrolases (EHs)
Epoxide hydrolases (EHs) have traditionally been regarded as detoxifying enzymes for metabolizing highly active epoxides with mutagenic and carcinogenic properties into less active corresponding diols. Two distinct mammalian xenobiotic epoxide hydrolases are well known, a membrane-bound microsomal enzyme (mEH) and the soluble one (sEH). To determine epoxide hydrolase activity, various methods such as liquid chromatography (LC), gas chromatography (GC), or UV/Vis spectroscopic and fluorophotometric are used.
Previously used assays determine the hydrolysis of epoxides by monitoring only the decrease or increase of UV absorption, by using substances such as 4-nitrophenyl (2S,3S)-2,3-epoxy-3-phenylpropyl carbonate85, 1-naphtyl-trans-2,3-epoxyphenyl-propyl-carbonate), 1,2-epoxy-1-(p-nitrophenyl)pentane, 1,2-epoxy-1-(2-quinolyl)pentane86, 2-(4-chlorophenyl)oxirane (EHs-U1)87, etc. Considering that this method is usually low in sensitivity, susceptible to interference from biological mechanisms, and poor selectivity, it is currently of less use.
Generally, fluorophotometric based assays of EHs are based on the changes in fluorescence spectra before and after the reaction. In the past few decades, many teams have developed several specific fluorescent substrates (summarized in Table 6) for epoxide hydrolase activity detection and inhibitor screening based on the ability of enzymes to hydrolyze epoxides. Jocelyne et al.88 developed a new method to access the enantioselectivity (E) of epoxide hydrolases by applying chiral fluorogenic probes (EHs-F1). Hammock and his team devised a fluorogenic substrate for mammalian sEH activity and inhibition studies, which is based on the enzymatic hydrolysis of a readily synthesized β-epoxycarbonate (sEH-F1) that results in the production of a fluorescent aldehyde89. Shen et al.90 designed and synthesized a resorufin-based fluorogenic substrate (mEH-F1), 7-(2-(oxiran-2-yl)ethoxy) resorufin, which was hydrolyzed by microsomal epoxide hydrolase to form the corresponding diol, which upon further treatment with sodium periodate and liberated the strongly fluorescent resorufin. The probe displays good biological compatibility and favorable photophysical properties, such as high quantum yield and a wide working pH range. Christophe Morisseau91 developed a series of absorbent and fluorescent substrates, and found that cyano (6-methoxy-naphthalen-2-yl)methyl glycidyl carbonate (mEH-F2,F3) displayed the highest activity towards eEH. The new fluorescence-based assay was also successfully applied for the discovery of structure–activity relationships among mEH inhibitors. At present, the research and development of EHs optical probes is still in its infancy. Most probe reactions require two or more reaction steps, which is cumbersome to operate. In the future, it is necessary to develop a one-step reaction with high sensitivity for the study of EHs functions.
Table 6.
Reported optical probes for human EHs.
3. Optical substrates for phase II drug metabolizing enzymes
3.1. Optical substrates for uridine-diphosphate glucuronosyltransferases (UGTs)
Uridine-diphosphate (UDP) glucuronosyltransferases (UGTs; EC 2.4.1.17) superfamily is one of the most important phase II drug metabolizing enzymes which catalyze the covalent addition of glucuronic acid from the sugar donor UDP-glucuronic acid (UDPGA) to a wide range of lipophilic substances that contain a suitable acceptor functional group, such as hydroxyl, carboxyl, carbonyl, sulfuryl, and amine groups. In human, UGT superfamily is classified into four major families: UGT1, 2, 3 and 8, of which the UGT1 and 2 families being most important for glucuronidation reactions. The substrate spectra of human UGT enzymes (particularly UGT1A members) are highly overlapped to each other5,92, 93, 94, 95. Accumulating evidence indicates that UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A9, UGT2B7, UGT2B10, and UGT2B15 play crucial roles in hepatic glucuronidation of xenobiotics, whereas intestinal UGT1A10 plays a pivotal role in pre-systemic clearance.
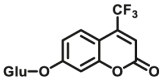
Over the past decade, considerable progress has been made in the design and development of optical substrates for several UGT isoforms including UGT1A1 (Table 7). The fluorescent substrates originally used for human UGTs are typical “on-off” fluorescent probes, which usually use a phenolic fluorophore as a substrate and the fluorescence signals will be turned off after glucuronidation. Many reported fluorescent substrates are lack of specificity, which can be metabolized by multiple UGT isoforms, for example 7-hydroxy-4-trifluoromethylcoumarin (HFC) (UGTs-F1)96 and 1-naphthol (UGTs-F2)97,98. Terai et al.99 have reported a series of “on-off” fluorescent substrates (UGT1A1-F1) for UGT1A1 based on the mechanism of donor-excited photoinduced electron transfer (d-PET) using TokyoGreen as the fluorophores. More recently, Juvonen et al.100 designed and developed several highly specific and sensitive fluorescent probes (UGT1A10-F1-F4) for UGT1A10 by applying molecular docking simulations and phenotypic screening. Though these “on-off” fluorescent probes are applicable for high throughput screening of potential inhibitors using recombinant UGTs, they are not conducive to the sensitive and accurate determination of the corresponding glucuronides in complex biological samples.
Table 7.
Reported optical probes for human UGTs.
| Enzyme | No. | Substrate | Metabolite | Km (μmol/L) | Vmax (μmol/min/mg) or Kcat | λex/λem (nm) | Ref. |
|---|---|---|---|---|---|---|---|
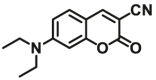
| UGTs | UGTs-F1 | 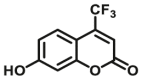 |
 |
260 for UGT1A6; 8.0 for UGT1A7; 28 for UGT1A10; 4.5 for UGT2A1 | 0.052 for UGT1A6; 0.0027 for UGT1A7; 0.0108 for UGT1A10; 0.0062 for UGT2A1 | 364/498 | 96 |
| UGTs | UGTs-F2 |  |
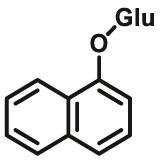 |
3.1 for UGT1A6; 87 for UGT1A8; 2.0 for UGT1A10 | 0.0196 for UGT1A6; 0.001 for UGT1A8; 0.0013 | 290/330 | 97 |
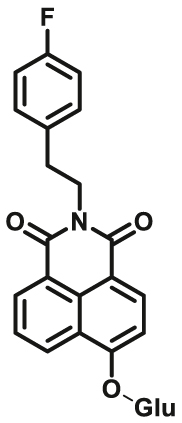
| UGTs | UGTs-F3 | 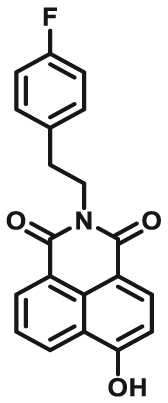 |
 |
N.A. | N.A. | 360/450 | 101 |
| UGT1A10 | UGT1A10-F1 |  |
 |
3.4 | 16.4 | 405/460 | 100 |
| UGT1A10 | UGT1A10-F2 |  |
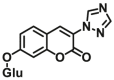 |
7.0 | 5.3 | 405/460 | 100 |
| UGT1A10 | UGT1A10-F3 | 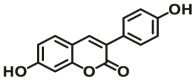 |
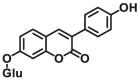 |
2.2 | 5.9 | 405/460 | 100 |
| UGT1A10 | UGT1A10-F4 | 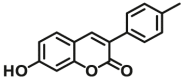 |
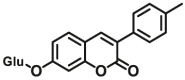 |
3.9 | 22.4 | 405/460 | 100 |
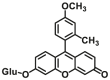
| UGT1A1 | UGT1A1-F1 | 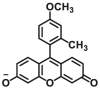 |
 |
2.7 for UGT1A1 | Vmax = 0.966 μmol/min/L | N.A. | 99 |
| UGT1A1 | UGT1A1-F2 | 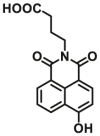 |
 |
126.7 | 1303 | 450/564 362/450 |
102 |
| UGT1A1 | UGT1A1-F3 |  |
 |
0.7 | 561 | 370/520 | 103 |
| UGT1A1 | UGT1A1-F4 | 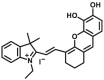 |
 |
N.A. | N.A. | 670/720 | 106 |
| UGT1A7c | UGT1A7c-F1 |  |
 |
N.A. | N.A. | 544/612 | 104 |
| UGT1A8 | UGT1A8-F1 |  |
 |
1.35 | N.A. | 500/580 | 193 |
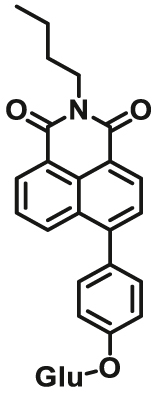
To overcome the limitation of “on-off” fluorescent probes, several groups are dedicated to the development of “turn on” or ratiometric fluorescent substrates for monitoring human UGTs. Recently, our group also designed a universal fluorescent probe (UGTs-F3) of UGTs and developed a fluorescence-based microplate assay to simultaneously determine the inhibitory effects of the tested compound(s) on ten human UGT isoforms101. To obtain an UGT1A1-specific probe, we designed a panel of UGTs potential fluorescent substrates based on enzymatic characteristics and its substrate preference. By phenotypic screening, we found N-3-carboxy propyl-4-hydroxy-1,8-naphthalimide (NCHN, UGT1A1-F2) displayed the best combination of selectivity, sensitivity and ratiometric fluorescence response following UGT1A1-catalyzed glucuronidation102. However, the affinity of NCHN for UGT1A1 is very low, and the binding site of NCHN on UGT1A1 is distinct from that of bilirubin. We further modified the fluorophore 4-hydroxy-1,8-naphthalimide (HN), and subsequently developed a practical and high-affinity fluorescent substrate (N-butyl-4-(4-hydroxyphenyl)-1,8-naphthalimide, NHPN, UGT1A1-F3) for UGT1A1. NHPN displayed good reactivity and very high affinity for human UGT1A1 (Km = 0.7 μmol/L), and could bind on UGT1A1 at the same ligand-binding site as bilirubin, which could serve as a good surrogate for bilirubin103. Guided by the structure–activity relationships, Kim et al.104 reported a fluorescent probe (UGT1A7c-F1) to detect mouse UGT1A7c activity which was the only member enriched in microglia. This substrate was used as a practical tool for labeling microglia in both cell cultures and live rodent brains, and be used to identify microglia in neural disorders. Recently, Feng et al.105 developed a red emission fluorescent substrate (UGT1A8-F1) for highly selective sensing UGT1A8 in living cells and cancer tissue. Later, they propose a novel “molecular-splicing strategy” to construct enzyme-activated fluorescent probes and designed a highly selective NIR fluorescent probe (UGT1A1-F4) for UGT1A1106. This fluorescent substrate has been successfully used for sensing UGT1A1 in complex systems, and has been applied for monitoring the bile excretion function in living cells and animal models.
3.2. Optical substrates for catechol-O-methyltransferase (COMT)
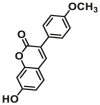
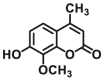
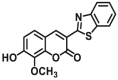
Catechol-O-methyltransferases (COMT, EC 2.1.1.6) are a class of Mg2+ dependent conjugative enzymes that catalyze the transfer of a methyl group from common methyl donor S-adenosyl-l-methionine (AdoMet or SAM) to one of hydroxyl groups of catechol substrates. COMT plays an important role in the biotransformation of a wide range of endogenous compounds (such as dopamine, noradrenaline, and adrenaline) and exogenous compounds (such as protocatechuic acid, caffeic acid, 5,6-dihydroxyindoles, and flavonoids). Two forms of COMT are found in tissues, a soluble form (S-COMT) and a membrane-bound form (MB-COMT). Previously, several colorimetric substrates such as epinephrine (E, COMT-F1)107, nitrocatechol (NC, COMT-F2)108, and 3,4-dihydroxyacetophenone (DHAP, COMT-F3)109 have been used to study COMT activity and inhibitors. The development of COMT fluorescent probes is very challenging, mainly because the addition of a methyl group to the hydroxyl group of the fluorophore usually does not change the fluorescence spectrum. In the past, many fluorescent substrates of COMT, for example, norepinephrine (NE, COMT-F4)110, dopamine (DA, COMT-F5)111, 3,4-dihydroxybenzaldehyde (DHBAd, COMT-F6)112, 2-(3,4-dihydroxyphenyl) naptho-[1,2-d]-thiazole (DNT, COMT-F7)113, 5,6-dihydroxyindole-2-carboxylic acid (5,6-DHI2C, COMT-F8)114, need to be separated from the products by liquid chromatography to achieve enzyme activity detection110,111. Up to now, only two specific fluorescent probes (COMT-F9 and COMT-F10) (Table 8) have been reported for highly selective sensing catechol-O-methyltransferase activities in living cells and tissues115,116. Notably, 3-(benzo[d]thiazol-2-yl)-7,8-dihydroxy-2H-chromen-2-one (3-BTD, COMT-F10) was a practical two-photon fluorescent substrate for mammalian COMT, and this agent was successfully applied for two-photon imaging of COMT in living cells and tissue slices with high resolution. This fluorescent probe was rationally designed on the basis of the region-selective 8-O-methylation of 7,8-dihydroxycoumarin by human COMT and the beneficial effect of the introduction of electron-withdrawing groups at the C-3 site of courmarin on its fluorescence properties.
Table 8.
Reported optical probes for mammalian COMT.
| Enzyme | No. | Substrate | Metabolite | Km (μmol/L) | Vmax (μmol/min/mg) | λex/λem (nm) | Ref. |
|---|---|---|---|---|---|---|---|
| COMT | COMT-F1 |  |
 |
N.A. | 0.0924×10‒3 | N.A. | 107 |
| COMT | COMT-F2 |  |
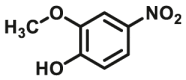 |
142 | N.A. | N.A. | 108 |
| COMT | COMT-F3 | 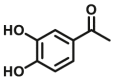 |
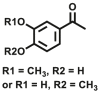 |
8.04 | N.A. | N.A. | 109 |
| COMT | COMT-F4 | 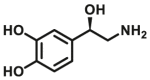 |
 |
366 for S-COMT; 12 for MB-COMT | 22.9 for S-COMT; 4.62 for MB-COMT | 430/505 | 110 |
| COMT | COMT-F5 |  |
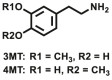 |
510 | 0.0062×10‒3 | 430/505 | 111 |
| COMT | COMT-F6 | 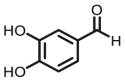 |
 |
N.A. | N.A. | 349/393;347/389 | 112 |
| COMT | COMT-F7 | 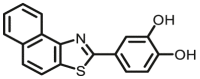 |
 |
2.1 for m-MNT 3.2 for p-MNT |
N.A. | 348/390 | 113 |
| COMT | COMT-F8 | 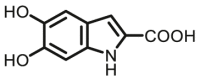 |
 |
44 | 0.295×10‒3 | 295/340 | 114 |
| COMT | COMT-F9 |  |
 |
5.17 | 0.123 | 320/520 | 115 |
| COMT | COMT-F10 | 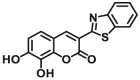 |
 |
0.79 | 0.0445 | 390/510 | 116 |
3.3. Optical substrates for sulfotransferases (SULTs)
Sulfotransferases are an important class of enzymes that are responsible for catalyzing the transfer of sulfo groups from a donor (for example 3′-phosphoadenosine 5′-phosphosulfate (PAPS)) to an acceptor (such as the amino or hydroxyl groups of a small molecule, xenobiotic, carbohydrate, or peptide). These enzymes are important targets in the design of novel therapeutics for treatment of a variety of diseases. Similar to COMT, the development of highly selective fluorescent probes for SULTs also has great challenges in both molecular design and imaging applications. So far, very few of fluorescent substrates for SULTs (Table 9) have been reported117, 118, 119. Unfortunately, these fluorescent substrates display poor selectivity and emit short emission wavelength, so these agents cannot be used for sensing target enzyme in living systems or in vivo. Recently, a specific fluorescent probe for SULT1 was designed by modifying 3-hydroxy-1,8-naphthalimide and be used for sensing SULT1 activity in living cells120. In future, solid works are needed to develop more practical fluorescent probes for these conjugative enzymes.
Table 9.
Reported optical substrates for human SULTs.
3.4. Optical substrates for N-acetyltransferases (NATs)
Aromatic amine N-acetyltransferases (NATs, EC 2.3.1.5) are phase II enzymes, which physiological function is to transfer the acetyl group on acetyl-CoA to the N atom or O atom of aromatic amines/hydrazine/aromatic hydroxylamine/and aromatic hydrazine. They play an important role in the detoxification and biotransformation reactions of carcinogens, as well as the effects of some arylamine and hydrazine-containing drugs, which should be considered carefully in clinical practices. Two NATs, NAT1 and NAT2, are commonly expressed in human liver and intestine. Dysfunction of NATs is not only closely related to diseases but also affect the disposal of arylamine drugs such as dapsone, isoniazid, caffeine, etc. Therefore, the accurate detection of NATs activity is significant for disease diagnosis and personalized therapy in clinical practices.
Laurieri et al.121, developed an inhibitor based noncovalent sensor (NAT1-C1) (Table 10) for specific detection of human arylamine N-acetyltransferase 1. The solution of sensor was observed to undergo a distinctive color change from red to blue in the presence of either NAT1 or mNat2, but not in other isozymes. Later, they described the use of in silico modeling alongside structure–activity relationship studies to advance the hit compound towards a potential probe to quantify hNAT1 levels in complex systems122. These studies pave the way for the development of inhibitors with improved intrinsic sensitivity which could enable detection of NAT1 in complex biological samples and potentially serve as tools for clarifying the unknown function NAT1 plays in estrogen receptor overexpressed breast cancer. Qian et al.123 took advantage of the catalytic properties of NAT2 and developed a new ratiometric fluorescence probe (NAT2-F1) for specific detection of NAT2 in complex biological samples. On this basis, their team also designed a near-infrared fluorescence probe (NAT2-F2) for NAT2 by using cyanines as reporting part124. The probe could be used for selectively and sensitively monitoring the activity of NAT2 in enzymatic system, living mice and tissue homogenate samples. Jin et al.125 designed and synthesized a fluorescent substrate (NAT2-F3) for real-time and specific sensing bacterial NAT2. This substrate had also been successfully used for monitoring NAT2 activity in various bacteria and applied for the high-throughput screening of natural inhibitors from herbal medicines against NAT2. Considering fluorescent probes are susceptible to interference from biological components, Tetsuo Nagano developed a long-lived luminescent probe (NATs-L1) to sensitively and rapidly detect arylamine N-acetyltransferase (NATs) activity of cell homogenates126. The probe exhibited approximately 100-fold increase of luminescence after N-acetylation catalyzed by NATs, with relatively high specificity for NAT2 over NAT1.
Table 10.
Reported optical probes for NATs.
3.5. Glutathione S-transferases (GSTs)
Glutathione S-transferases (GSTs, EC 2.5.1.18) are a superfamily of phase II-drug metabolizing enzymes which catalyse the nucleophilic attack of reduced glutathione (GSH) on electrophiles, for example halonitroaromatics and α,β-unsaturated carbonyls, thereby detoxifying the endogenously generated or exogenous reactive electrophiles to protect cells from the formation of undesirable protein or nucleotide adducts. Three subclasses of cytosolic glutathione S-transferases, GSTA, GSTM and GSTP are regularly overexpressed in tumor cells, especially in those with drug-resistance. The detection and visualization of altered GSTs activity in cancer cells would therefore be useful for cancer diagnosis and assessment of the efficacy of drug treatment.
Optical substrates for GST detection in the early years are 2,4-chlorodinitrobenzene (CDNB) and 5-(pentafluorobenzoylamino) fluorescein (PFB-F), which was based on the GSH conjugation activity of GST (Fig. 6, type 1, Table 11). These substrates can be used to detect and track GSTs in cell lysates or in vitro, however, these development methods are very rough and impractical. Fujikawa et al.127 made a great improvement on the basis of previous strategy and designed the first off-on fluorogenic substrates for GSTs, isomer dinitrobenzamide fluoresceins (DNAFs) (GSTs-F1) and N-(4-(6-hydroxy-3-oxo-3H-xanthen-9-yl)-3-methylphenyl)-3,4-dinitrobenzamide (DNAT-Me), and DNAT-Me was successfully applied for sensing GSTs activity in living cells. Through phenotypic screening, they found 3,4-dinitrobenzanilide (NNBA) substituent display high selectivity towards GSTs and hence was rationally integrated into fluorophore. The newly developed fluorogenic substrates are obviously better than known substrates, and were able to detect nuclear GSH/GST activity in HuCCT1 cell lines. At present, most of the fluorescent probes of GST are based on this strategy, which use 3,4-dinitrobenzene or similar group as recognition group, for example, He et al.128, developed a new NIR fluorogenic substrate Cy-GST (GSTs-F2) by incorporating 3,4-dinitrobenzoic acid into a NIR fluorophore for the highly selective detection of GSTs in living cells and in vivo. Cui et al.129 devised a two-photon fluorescent probe (GSTP1A1-F1) based on BODIPY for the real-time detection of GSTs activities and fluorescence imaging in both cancer cells and liver tissues. Meanwhile, some off-on probes (GSTs-F4, GSTP1-F1, GSTP1-F2, GSTM1-F1) were also developed for trapping GSTs activity on the basis of the catalytic action of GSTs by introducing a GSH group into probes to trigger fluorescence emission129, 130, 131, 132.
Figure 6.
Design strategies of GSTs fluorescent substrate and representative probes.
Table 11.
Reported optical probes for GSTs.
| Enzyme | No. | Substrate | Metabolite | Km (μmol/L) | Vmax or Kcat | λex/λem (nm) | Ref. |
|---|---|---|---|---|---|---|---|
| GSTs | GSTs-F1 | 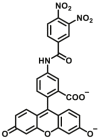 |
 |
1107 × 104 L/mol·s for hGSTA1 | N.A. | 492/516 | 127 |
| GSTs | GSTs-F2 |  |
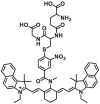 |
21.34 | Kcat = 389 S‒1 | 730/810 nm | 128 |
| GSTs | GSTs-F3 | 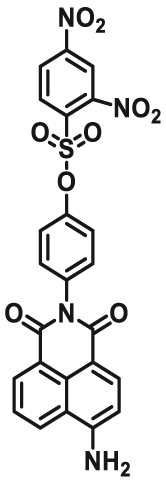 |
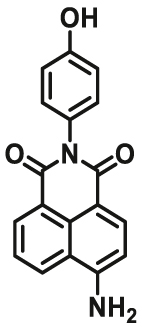 |
84 | Vmax = 3.84 μmol/min/L | 420/550 | 135 |
| GSTs | GSTs-F4 |  |
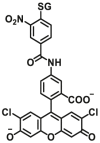 |
0.31 for hGSTM1-1 | N.A. | 505/525 | 132 |
| GSTs | GSTs-F5 |  |
 |
N.A. | N.A. | 540/620 | 133 |
| GSTs | GSTs-F6 | 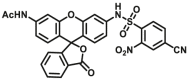 |
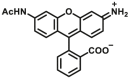 |
3.2 ± 0.3 for GSTA1-1; 4.9 ± 0.8 for GSTM2-2; 4.1 ± 0.4 for GSTP1-1 | Kcat = 11.36 ± 0.8 S‒1 for GSTA1-1; 2.09 ± 0.19 for GSTM2-2; 0.46 ± 0.03 for GSTP1-1 | 490/522 | 134 |
| GSTs | GSTs-B1 | 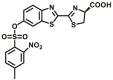 |
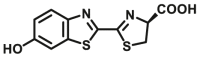 |
39.3 ± 3.1 | N.A. | Biolumincence | 136 |
| GSTs | GSTs-B2 | 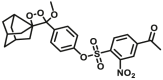 |
 |
2.8 ± 0.53 for GSTM1-1; 3.7 ± 0.59 for GSTP1-1 | Kcat = 0.0006 S‒1 for GSTM1-1; Kcat = 0.0009 S‒1 for GSTP1-1 | Biolumincence | 137 |
| MGST1 | MGST1-F1 | 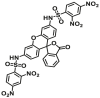 |
 |
1.7 ± 0.5 | NA | 499/522 | 194 |
| GSTP1 | GSTP1-F1 |  |
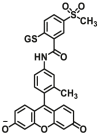 |
N.A. | N.A. | 493/510 | 130 |
| GSTP1 | GSTP1-F2 | 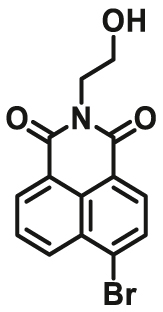 |
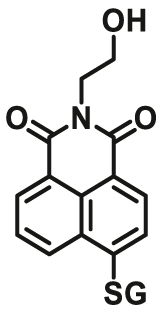 |
13.3 ± 2.8 | Kcat = 0.01±0.0017 S-1 | 389/503 | 131 |
| GSTP1 | GSTP1-F3 |  |
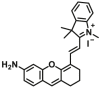 |
0.39 | Vmax=0.0357 μmol/min/L | 670/695 | 195 |
| GSTP1 | GSTP1-F4 | 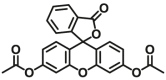 |
 |
6.4 | Kcat= 0.6 S‒1 | 490/510 | 196 |
| GSTA1-1 | GSTP1A1-F1 |  |
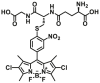 |
N.A. | N.A. | 495/553 | 197 |
| GSTM1 | GSTM1-F1 | 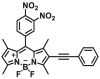 |
 |
1.55 for HLS9 | N.A. | 530/588 790/588 |
129 |
| GST | GST-F1 | 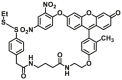 |
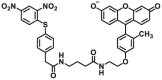 |
489.35 | Vmax=7.08 μmol/min/L | 490/510 | 198 |
In addition to using the strategies used above, some researchers tried to develop other strategies for the development of enzymatic probes (Fig. 6, type 2). Zhang et al.133 synthesized a panel of potential substrates for GSTs by incorporating 2,4-dinitrobenzenesulfonate (DNs) into a fluorophore containing an amino group (including coumarin, cresyl violet, and rhodamine) that turned out to be good substrates for various GST isoforms, and these substrates were used to monitor GST activity in cell extracts. They found GSTs-5 displayed high stability and was successfully used for fluorescence imaging of microsomal MGST1 activity in living cells. Shibata et al.134 synthesized four 4-substituted-2-nitrobenzene-sulfonyl substrates as caged fluorescent probes to investigate the influence of substituents on human GST metabolic reaction, and found that different GST subtypes responded significantly different to substrates with varied electron-withdrawing groups. Through comprehensive assessment, they revealed that the 4-cyano-2-nitro-benzenesulfonyl (CNs) caged probe (GSTs-F6) was the best tool for the detection of GST activity. Moreover, Zhang et al.135, designed a TP fluorescent probe (GSTs-F3) by introducing a 2,4-dinitrobenzenesulfonyl group to a naphthalimide derivative for sensing GST activity in cells and drug induced liver injury samples.
John et al.136 synthesized a panel of luciferin derivatives for sensing mammalian GST and found that GSTs-B1 exhibited the greatest reactivity, with substantially high affinity for GST from S. japonicum. While Ito et al.137 used 4-acetyl-2-nitrobenzenesulfonyl as the recognition sites of GST, and designed 19F NMR and bioluminescent probes (GSTs-B2), which could be used to detect GST activity in live cells, and serve as sensitive signal-amplification system by using GST as a tag protein in the pGEX vector.
4. Others
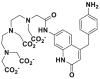
Recent years, some dipeptidyl peptidases (DPP 2, DPP 4, DPP 8, and DPP 9) have also been found pose the function of drug metabolization. The nitrile (CN) group is introduced as an efficacious pharmacophore into the increasing number of therapeutic drugs, which not only can enhance the selectivity and binding affinity of these drugs to target proteins but also can block metabolically labile sites because nitrile biotransformation is a minor metabolic pathway. Recent work proved that DPPs, especially DPP-4 partly involved in the conversion of nitrile group of some cyanopyrrolidine DPP-4 inhibitors (vildagliptin, anagliptin, and besigliptin) into carboxylic acid is their major metabolic pathway in vitro and in vivo138,139. The study of the above DPPs in drug metabolism is in their early phase. However, the study of DPP-4 as a drug target has been attracted wide attention. DPP-4 displays unique Gly–Pro hydrolytic activity which makes it is feasible to develop convenient biochemical assay(s) for highly selective sensing DPP-4 in complex biological systems140, 141, 142, 143, 144. Currently, several fluorescent substrates for DPP-4 have been reported (Table 12)145, 146, 147, 148, 149, 150, 151. The emergence of these substrates making it possible to imaging DPP-4 in live cells, subcellular structure, or accurate and sensitive detection of DPP-4 activity in biological systems including cell preparations and plasma152.
Table 12.
Reported optical probes for human DPP 4.
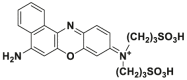
| Enzyme | No. | Substrate | Metabolite | Km (μmol/L) | Vmax or Kcat | λex/λem (nm) | Ref. |
|---|---|---|---|---|---|---|---|
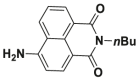
| DPP 4 | DPP 4-F1 | 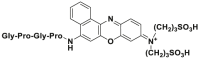 |
 |
56 | 0.63 μmol/min/L | 630/670 | 145 |
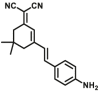
| DPP 4-F2 | 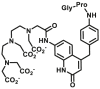 |
 |
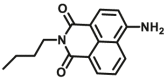
33.6 | 5.9 μmol/min/L | 330/546 | 199 | |
| DPP 4-F3 | 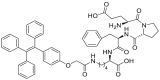 |
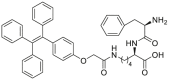 |
N.A. | N.A. | 320/450 | 146 | |
| DPP 4-F4 |  |
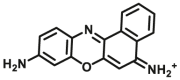 |
112 | 7.8 μmol/min/L | 585/625 | 147 | |
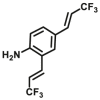
| DPP 4-F5 | 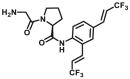 |
 |
N.A. | N.A. | 300/460 | 149 | |
| DPP 4-F6 | 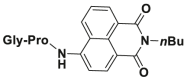 |
 |
42.93 | 15.49 μmol/min/mg | 430/535 810/535 |
148 | |
| DPP 4-F7 |  |
 |
150.1 | 15.03 μmol/min/mg | 458/658 | 149 | |
| DPP 4-F8 |  |
 |
N.A. | N.A. | 450/540 | 150 | |
| DPP 4-F9 |  |
 |
N.A. | N.A. | 595/612 | 151 | |
| DPP4-F10 | 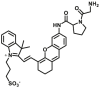 |
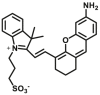 |
N.A. | N.A. | 640/690 | 200 |
5. Biomedical applications of optical substrates of DMEs
5.1. Sensing or imaging of target enzymes in biological systems
Through about ten years quickly develop, a variety of optical substrates for drug-metabolizing enzymes have been widely used to real-time monitor or in situ imaging the target enzymes in biological systems, owing to their highly sensitive, non-destructiveness, easy conduct. Quantification of targets in S9 or microsomes was the most commonly used applications with these molecular fluorescence substrates. NCMN, the specific ratiometric fluorescent substrate for CYP1A, was capable of detection of CYP1A activity of individual HLMs. And 15-fold catalytic activity variations were found in these biological samples, which was confirmed with a traditional non-florescent substrate (phenacetin)153. Besides, a bioluminescent sensor CES1A-B2 for CES1A was developed to measure the CES1A function of different human tumor cell homogenates, including HepG2, SKOV3, MCF-7, A549, and Caco-2154. The CES1A catalytic activities in different tumor cells detected by CES1A-2 were highly reliable, due to the catalytic activities was consistent with the relative abundance of CES1A in both protein expression and mRNA154. Also, CES1A-B2 was capable of real-time sensing of endogenous CES1A of living cells154. Notably, Wang et al.155 reported a more specific bioluminescent sensor for monitoring of CES1A in human plasma.
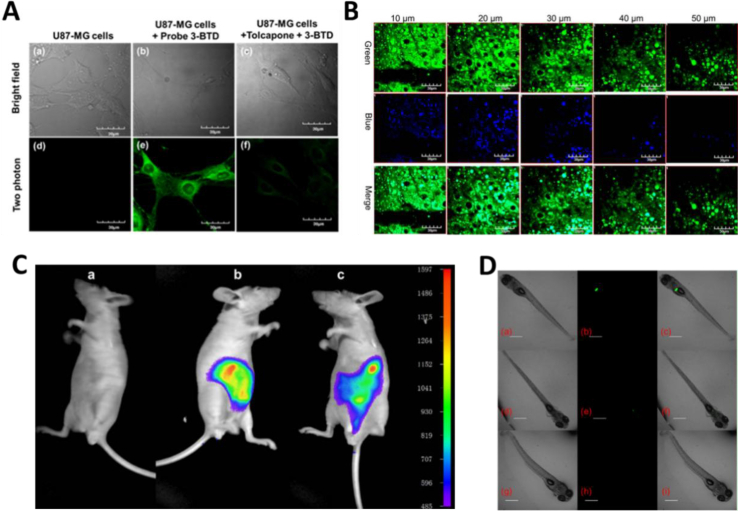
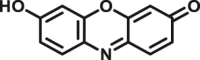
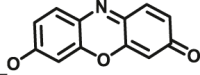
In addition to directly sensing enzyme activities in biological systems, the optical substrates are also capable of real-time imaging DMEs in living cells, tissues or even animal. Wang et al.116 reported an optimized two-photon fluorescent substrate 3-BTD (COMT-F10), which exhibited relatively low cytotoxicity to living human glioblastoma cells. Besides, by using substrate COMT-F10, fluorescence images can be obtained not only in both one-photon but also in two-photon modes (Fig. 7A). By taking advantages of the two-photons excited fluorescent substrates in the wide penetration depth, Jing et al.24 developed a two-photons fluorescent substrate CES2A-F5 for CES2A. The sensing ability of substrate CES2A-F5 for CES2 in fresh mouse liver tissue section was investigated. After incubation of NCEN with mouse liver slice for 60 min, strong fluorescence responses in green channel (Ex 800 nm/Em 495–540 nm) was observed in the section, and the depth of confocal image was 50 μm (Fig. 7B). Jin et al.23 devised a near-infrared fluorescent substrate CES2A-F6 for CES2A in another work, which has been successfully used to visualization of CES2A levels in organs and living mice and for the first time. By utilization of DDAB, the tissue distribution of CES2A in animal can be easily obtained (Fig. 7C and D). Moreover, Ding et al.156 presented a fluorescence substrate for imagining CES1A in zebrafish. In summary, the optical substrates gain a deeper insight into the activities and tissue distribution of target enzymes in multiple biological systems and facilitated associated drug discovery.
Figure 7.
In situ imaging of target DME in living systems. (A) Confocal fluorescence images of U87-MG cells with COMT-F10. Reproduced with permission from Ref. 116, copyright © 2017, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (B) Two-photon confocal fluorescent images of endogenous CES2A in a mouse liver slice stained by CES2A-F5. Reproduced with permission from Ref. 24, copyright © 2015, American Chemical Society. (C) Representative fluorescence images of visualizing CES2A levels in nude mice using CES2A-F6. Reproduced with permission from Ref. 23, copyright © 2016, Elsevier B.V. (D) Biological imaging of CES1A activity in zebrafish by using CES1A-F3. Reproduced with permission from Ref. 156, copyright © 2018, Elsevier B.V.
5.2. Screening and characterizing of the modulators of DMEs
Recently, evaluation of potent inhibitors toward DMEs has attracted increasing attentions. The developed of these highly sensitive optical substrates mentioned above promoted the discovery of inhibitors and inducers. Recently, Zhou et al.101 reported a practical method for high-throughput screening and evaluation of human UGT inhibitors by utilizing a fluorescence-based microplate assay with fluorogenic substrate 4-HN-335. Zou et al.157 evaluate of a variety of triterpenoids using two subtype specific substrate (CES1A-B2 for CES1 and CES2A-F6 for CES2, respectively). By further 3D-QSAR assay, the relationships linking between structure of these triterpenoids and inhibitory effects on CES1A was obtained. Lei et al.158 evaluated the inhibition of pyrethroids by utilization optical substrate substrates for CES1A (CES1A-B2, CES1A-F1, and CES1A-F3). The results indicated that CES1A may have two ligand-binding sites, and deltamethrin was proved can reversible inhibited CES1A. By molecular docking, the binding site of deltamethrin on CES1A was high overlap with CES1A-3. Moreover, much more focus on study potent inhibitors towards DMEs from edible herbals. Liu et al.159 proposed a “fingerprint-efficacy” method to the identification of CES2A inhibitors from white Mulberry root-bark with optical substrate for CES2A as well as LC‒DAD-ESI-MS. Notably, Xue et al.160 constructed a cell imaging based multiparametric assay strategy (also called high-content analysis) for study the CES2A inhibitors in living system. This method successfully integrated CES2A fluorescent substrate CES2A-F5 with nuclear substrates, Hoechst 33,342 and PI, which could serve as an efficient tool for the accuracy measurement inhibitory effect and cytotoxicity of compounds against CES2A in living cell system.
Besides inhibitors screening, these sensitive substrates were also used for inducer discovery, due to their excellent abilities in real-time monitoring intracellular activities. More recently, Zhu et al.161 reported an ultra-sensitive method to evaluate human UGT1A1 activities in living cells. This newly developed method was easy-to-use and can be applied for evaluating UGT1A1 inducers in living cells. Using this approach, Zhu et al.161 screened a series of flavonoids and revealed neobavaisoflavone was a potent UGT1A1 inducer. Besides, Wang et al.154 use CES1A-B1 (a newly developed bioluminescent substrate of CES1A to evaluate the induction effects of glucose to CES1A in HepG2 cells co-expressed luciferase. And the induction could be detection either by the hydrolysis activities of cell lysis or through the luminescent imaging in the intact cells154.
5.3. Indication for organ injury or disease diagnosis
In addition to participate in the metabolic clearance of a variety of drugs, DMEs are also responsible for the biotransformation of a variety of endogenous substances in the human body. Thus, some key DMEs can also serve as diagnostic markers or therapeutic targets of human diseases. Of note, numbers of DMEs have been recognized as biomarkers for indicated diseases or organ injuries due to their inherit tissue or subcellular distribution characteristics and relationship with diseases. Several DMEs have been reported related with tumors. Fang58 reported a two-photon fluorogenic probe (MAO-A-F5) that can be used for real-time sensing MAO-A activities in tissue samples. With the help of this probe, it was found that MAO-A activities were relatively high in the glioma tissues from humans, but its activity levels are relatively low in negative controls (paracancerous tissue). In future, more clinical samples from patients with various tumors should be used to confirm the ability of MAO-A as a promising biomarker of human glioma. Previous work has suggested that NAT1 may work as a candidate biomarker in breast tumor. Laurieri et al.121 developed a small molecule colorimetric reagent which can selectively bind to human NAT1, which may be subsequently applied for the diagnosis of breast tumor. Since DMEs are mainly distributed in the liver, several hepatic DMEs have been suggested as diagnostic biomarkers for liver injuries. Recently, Wang et al.154 revealed that CES1A may serve as a novel serological biomarker for hepatocyte injury by using a practical bioluminescent substrate for CES1A (Fig. 8). In this work, the results showed that serum activity levels of CES1A were elevated dramatically in liver injury mice or the patients with liver diseases. Besides, Tian et al.162 reported a ratiometric fluorescent probe which can also target to endoplasmic reticulum CES2. With the help of this probe, the activity levels of CES2 in the liver was found down-regulated in the acute liver injury mice induced by acetaminophen. These studies offered solid evidence to support that two mammalian CES can serve as novel serological indicators for hepatocyte injury. In near future, more regulated clinical trials are needed to validate the applicability of CES activity as novel indicators or biomarkers for the detection of liver injury in humans.
Figure 8.
A bioluminescent substrate reveals that CES1A is a novel serologic indicator for hepatocyte injury.
GSTs are key enzymes responsible for urinary excretion and detoxification of xenobiotics via catalyzing the GSH-conjugation reactions on the xenobiotics or their reactive metabolites that are generated in the liver. Zhang et al.135, have developed a two-photon probe GSTs-F3 for sensing GSTP activity. This substrate can be used for imaging intracellular GSTP and monitoring GSTP activities in drug-induced liver injury samples, and the results have suggested that GSTs-F3 can be served as a potential sensor for the diagnosis of DILI135. Besides, GSTs have been reported closely related with the progressing of pulmonary fibrosis. He et al.128 reported a fluorescent substrate (GSTs-F2) which can be capable of the detection intracellular GSTs concentration changes. With the help of this sensor, it was found that intracellular GSTs were significantly increased in the cells and mice models with pulmonary fibrosis. Moreover, the GSTs levels in the idiopathic pulmonary fibrosis patients were abnormal high. This work provides solid evidence to support that the GSTs levels are potential biomarkers for the diagnosis of idiopathic pulmonary fibrosis. Collectively, several optical substrates for DMEs have indicated the potential applications as promising indicators or biomarkers for the diagnosis of organ injury or other diseases. Notably, most of these key findings are generated from cell assays or animal tests, the applicability and practicability of the DME optical substrates for disease diagnosis should be carefully validated by performing a panel of well-designed randomized clinical trials.
6. Summary and perspectives
In this review article, the molecular design strategies and principles for constructing highly specific optical substrates for various human DMEs, as well as the recent advances in the development and applications of optical substrates for sensing target human DMEs, are well-summarized. More than ten informative lists of optical substrates for various classes of human DMEs including both phase I and phase II DMEs, alongside with the chemical structures of both substrate and metabolite(s), kinetic parameters and the detection conditions of the corresponding metabolite(s), are comprehensively summarized and discussed in-depth. Furthermore, the challenges for constructing practical and highly specific optical substrates for target DMEs and future applications of DME optical substrates are also highlighted by us. The information and knowledge presented here offer a group of practical approaches and imaging tools for sensing DMEs activities in complex biological systems, which will strongly facilitate high-throughput screening the modulators of DMEs and studies on drug/herb‒drug interactions, as well as the fundamental researches for exploring the relevance of DMEs to human diseases and drug treatment outcomes.
Although a variety of optical substrates for human DMEs have been developed over the past few decades, the specificity and practicability of most reported optical substrates for human DMEs are not good enough for precise sensing target enzyme in complex biological samples. Of note, the majority of reported optical substrates for human phase I DMEs are only suitable for sensing target enzyme in vitro or in several types of living cells, owing to the corresponding phase I metabolite of these optical substrates could be further metabolized by phase II DMEs (such as UGTs and SULTs). Thus, the in vivo optical substrates for human phase I DMEs with high specificity, good safety profiles and excellent optical properties (emits strong and long-wavelength lights) are always highly desirable. By contrast, the metabolites of most optical substrates for human phase II DMEs (such as UGTs and SULTs) are always the ultimate metabolites in living systems, suggesting that these substrates can be used for sensing target phase II DME(s) in vivo. Unfortunately, the practical and highly specific optical substrates for human phase II DMEs are rarely reported, owing to that in most cases, the fluorescence signals of the phase II metabolites of the fluorophores are “turned-off” or “blue-shifted”, which is undesirable for sensing the activity levels of target enzyme in complex biological systems. Compared with the traditional assays (such as drug-sbustrates based enzyme activity assays), optical substrates/probes possess multiple insurmountable advantages, such as rapid response, easy to perform, ultrahigh sensitivity, in situ imaging, and applicable for high-throughput detection. It should be noted that some optical substrates/probes cannot replace drug-substrates of target enzymes in the drug–drug interaction (DDI) study, owing to they have distinct ligand-binding sites or they have a huge difference in binding affinity. Thus, prior to DDI study, it is better to carefully investigate the ligand-binding sites of both optical substrates and drug-substrates for a target enzyme, as well as to explore the difference in binding mode or binding affinity of optical substrates and drug-substrates.
In future, extensive efforts should be made in the design and development of more practical optical substrates for highly specific and sensitive sensing target DME(s) in real samples. More design and optimization strategies should be proposed and used for improving the isoform-specificity, sensitivity, practicability and in-situ imaging ability of optical substrates for sensing a target human DMEs in living systems. To improve the spatial resolution of optical substrates for human DMEs, the targeted drug delivery systems could be used for delivering the highly specific optical substrates to tumor or a specific organ (such as the liver) or target cells (such as macrophages), which may further enhance the sensing or imaging ability to quantify the real activities of target DMEs in a specific tissue or at cellular levels. Furthermore, the specificity and response of the known optical substrates for human DMEs in different animal species are rarely reported. Therefore, most researchers do not know how to use the reported optical probes to decipher the relevance of DMEs to various diseases at animal level. In future, more in-depth investigations on species difference in the specificity and response of the optical substrates for DMEs among various animal species should be conducted, which will very helpful for the researchers to select the most suitable animals as surrogate models for a specific purpose.
Acknowledgments
This work was supported by the NSF of China (81922070, 81973286, 82073813, 81803489), the National Key Research and Development Program of China (2017YFC1700200, 2017YFC1702000), the Three-year Action Plan of Shanghai TCM Development [ZY-(2018–2020)-CCCX-5001, China], Program of Shanghai Academic/Technology Research Leader (18XD1403600, China), Shuguang Program (18SG40, China) supported by Shanghai Education Development Foundation and Shanghai Municipal Education Commission, and Drug Innovation Major Project (2018ZX09731016, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences
Contributor Information
Dandan Wang, Email: wangdandan801@126.com.
Guangbo Ge, Email: geguangbo@dicp.ac.cn.
Author contributions
Guangbo Ge and Dandan Wang designed and prepared the manuscript with contributions from all authors. Qiang Jin prepared the optical substrates for phase II drug-metabolizing enzymes. Jingjing Wu prepared the optical substrates for phase I drug-metabolizing enzymes. Yue Wu and Hongxin Li prepared the tables in this review. Moshe Finel proofread this review.
Conflicts of interest
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
References
- 1.Iyanagi T. Molecular mechanism of phase I and phase II drug-metabolizing enzymes: implications for detoxification. Int Rev Cytol. 2007;260:35–112. doi: 10.1016/S0074-7696(06)60002-8. [DOI] [PubMed] [Google Scholar]
- 2.Manikandan P., Nagini S. Cytochrome P450 structure, function and clinical significance: a review. Curr Drug Targets. 2018;19:38–54. doi: 10.2174/1389450118666170125144557. [DOI] [PubMed] [Google Scholar]
- 3.Zanger U.M., Schwab M. Cytochrome p450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Munro A.W., Girvan H.M., Mason A.E., Dunford A.J., McLean K.J. What makes a P450 tick?. Trends Biochem Sci. 2013;38:140–150. doi: 10.1016/j.tibs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Wang D., Zou L., Jin Q., Hou J., Ge G., Yang L. Human carboxylesterases: a comprehensive review. Acta Pharm Sin B. 2018;8:699–712. doi: 10.1016/j.apsb.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yiannakopoulou E.C. Pharmacogenomics of phase II metabolizing enzymes and drug transporters: clinical implications. Pharmacogenomics J. 2013;13:105–109. doi: 10.1038/tpj.2012.42. [DOI] [PubMed] [Google Scholar]
- 7.Court M.H. Interindividual variability in hepatic drug glucuronidation: studies into the role of age, sex, enzyme inducers, and genetic polymorphism using the human liver bank as a model system. Drug Metab Rev. 2010;42:209–224. doi: 10.3109/03602530903209288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Achour B., Russell M.R., Barber J., Rostami-Hodjegan A. Simultaneous quantification of the abundance of several cytochrome P450 and uridine 5′-diphospho-glucuronosyltransferase enzymes in human liver microsomes using multiplexed targeted proteomics. Drug Metab Dispos. 2014;42:500–510. doi: 10.1124/dmd.113.055632. [DOI] [PubMed] [Google Scholar]
- 9.Lin J.H., Lu A.Y. Interindividual variability in inhibition and induction of cytochrome P450 enzymes. Annu Rev Pharmacol Toxicol. 2001;41:535–567. doi: 10.1146/annurev.pharmtox.41.1.535. [DOI] [PubMed] [Google Scholar]
- 10.Sweeney B.P., Bromilow J. Liver enzyme induction and inhibition: implications for anaesthesia. Anaesthesia. 2006;61:159–177. doi: 10.1111/j.1365-2044.2005.04462.x. [DOI] [PubMed] [Google Scholar]
- 11.Papasotiriou D.G., Markoutsa S., Meyer B., Papadioti A., Karas M., Tsiotis G. Comparison of the membrane subproteomes during growth of a new pseudomonas strain on lysogeny broth medium, glucose, and phenol. J Proteome Res. 2008;7:4278–4288. doi: 10.1021/pr800192n. [DOI] [PubMed] [Google Scholar]
- 12.Hop C.E.C.A., Cole M.J., Davidson R.E., Duignan D.B., Federico J., Janiszewski J.S., et al. High throughput adme screening: practical considerations, impact on the portfolio and enabler of in silico adme models. Curr Drug Metab. 2008;9:847–853. doi: 10.2174/138920008786485092. [DOI] [PubMed] [Google Scholar]
- 13.Fallon J.K., Harbourt D.E., Maleki S.H., Kessler F.K., Ritter J.K., Smith P.C. Absolute quantification of human uridine-diphosphate glucuronosyl transferase (UGT) enzyme isoforms 1A1 and 1A6 by tandem LC‒MS. Drug metab lett. 2008;2:210–222. doi: 10.2174/187231208785425764. [DOI] [PubMed] [Google Scholar]
- 14.Coward J.K., Wu F.Y. A continuous spectrophotometric assay for catechol-O-methyltransferase. Anal Biochem. 1973;55:406–410. doi: 10.1016/0003-2697(73)90130-9. [DOI] [PubMed] [Google Scholar]
- 15.White R.E. High-throughput screening in drug metabolism and pharmacokinetic support of drug discovery. Annu Rev Pharmacol Toxicol. 2000;40:133–157. doi: 10.1146/annurev.pharmtox.40.1.133. [DOI] [PubMed] [Google Scholar]
- 16.Ung Y.T., Ong C.E., Pan Y. Current high-throughput approaches of screening modulatory effects of xenobiotics on cytochrome P450 (CYP) enzymes. High Throughput. 2018;7:29. doi: 10.3390/ht7040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lv X., Xia Y., Finel M., Wu J., Ge G., Yang L. Recent progress and challenges in screening and characterization of UGT1A1 inhibitors. Acta Pharm Sin B. 2019;9:258–278. doi: 10.1016/j.apsb.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J., Guan X., Dai Z., He R., Ding X., Yang L., et al. Molecular probes for human cytochrome P450 enzymes: recent progress and future perspectives. Coordin Chem Rev. 2021;427:213600. [Google Scholar]
- 19.Feng L., Ning J., Tian X., Wang C., Zhang L., Ma X., et al. Fluorescent probes for bioactive detection and imaging of phase II metabolic enzymes. Coordin Chem Rev. 2019;399:213026. [Google Scholar]
- 20.Tian Z., Yan F., Tian X., Feng L., Cui J., Deng S., et al. A nir fluorescent probe for vanin-1 and its applications in imaging, kidney injury diagnosis, and the development of inhibitor. Acta Pharm Sin B. 2022;12:316–325. doi: 10.1016/j.apsb.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuhr U., Hsin C.H., Li X., Jabrane W., Sorgel F. Assessment of pharmacokinetic drug‒drug interactions in humans: in vivo probe substrates for drug metabolism and drug transport revisited. Annu Rev Pharmacol Toxicol. 2019;59:507–536. doi: 10.1146/annurev-pharmtox-010818-021909. [DOI] [PubMed] [Google Scholar]
- 22.Tian Z., Ding L., Li K., Song Y., Dou T., Hou J., et al. Rational design of a long-wavelength fluorescent probe for highly selective sensing of carboxylesterase 1 in living systems. Anal Chem. 2019;91:5638–5645. doi: 10.1021/acs.analchem.8b05417. [DOI] [PubMed] [Google Scholar]
- 23.Jin Q., Feng L., Wang D.D., Wu J.J., Hou J., Dai Z.R., et al. A highly selective near-infrared fluorescent probe for carboxylesterase 2 and its bioimaging applications in living cells and animals. Biosens Bioelectron. 2016;83:193–199. doi: 10.1016/j.bios.2016.04.075. [DOI] [PubMed] [Google Scholar]
- 24.Jin Q., Feng L., Wang D.D., Dai Z.R., Wang P., Zou L.W., et al. A two-photon ratiometric fluorescent probe for imaging carboxylesterase 2 in living cells and tissues. Acs Appl Mater Inter. 2015;7:28474–28481. doi: 10.1021/acsami.5b09573. [DOI] [PubMed] [Google Scholar]
- 25.Singh D., Kashyap A., Pandey R.V., Saini K.S. Novel advances in cytochrome P450 research. Drug Discov Today. 2011;16:793–799. doi: 10.1016/j.drudis.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Guengerich F.P. In: Cytochrome p450: structure, mechanism, and biochemistry. Ortiz de Montellano P.R., editor. Springer; Boston: 2015. Human cytochrome P450 enzymes; pp. 523–785. [Google Scholar]
- 27.Li Y., Kang X., Yang G., Dai P., Chen C., Wang H. Identification of genetic polymorphisms of CYP2W1 in the three main Chinese ethnicities: Han, Tibetan, and uighur. Drug Metab Dispos. 2016;44:1510–1515. doi: 10.1124/dmd.115.069153. [DOI] [PubMed] [Google Scholar]
- 28.Schneider E., Clark D.S. Cytochrome P450 (CYP) enzymes and the development of CYP biosensors. Biosens Bioelectron. 2013;39:1–13. doi: 10.1016/j.bios.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 29.Wright A.T., Song J.D., Cravatt B.F. A suite of activity-based probes for human cytochrome P450 enzymes. J Am Chem Soc. 2009;131:10692–10700. doi: 10.1021/ja9037609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright A.T., Cravatt B.F. Chemical proteomic probes for profiling cytochrome P450 activities and drug interactions in vivo. Chem Biol. 2007;14:1043–1051. doi: 10.1016/j.chembiol.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudsakorn S., Skell J., Williams D.A., O'Shea T.J., Liu H.L. Evaluation of 3-O-methylfluorescein as a selective fluorometric substrate for CYP2C19 in human liver microsomes. Drug Metab Dispos. 2007;35:841–847. doi: 10.1124/dmd.106.014472. [DOI] [PubMed] [Google Scholar]
- 32.Dai Z.R., Feng L., Jin Q., Cheng H., Li Y., Ning J., et al. A practical strategy to design and develop an isoform-specific fluorescent probe for a target enzyme: CYP1A1 as a case study. Chem Sci. 2017;8:2795–2803. doi: 10.1039/c6sc03970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Q., Ma H.Y., Feng L., Wang P., He R.J., Ning J., et al. Sensing cytochrome P450 1A1 activity by a resorufin-based isoform-specific fluorescent probe. Chin Chem Lett. 2020;31:2945–2949. [Google Scholar]
- 34.Ghosal A., Hapangama N., Yuan Y., Lu X.W., Horne D., Patrick J.E., et al. Rapid determination of enzyme activities of recombinant human cytochromes P450, human liver microsomes and hepatocytes. Biopharm Drug Dispos. 2003;24:375–384. doi: 10.1002/bdd.374. [DOI] [PubMed] [Google Scholar]
- 35.Ning J., Tian Z., Wang B., Ge G., An Y., Hou J., et al. A highly sensitive and selective two-photon fluorescent probe for real-time sensing of cytochrome P450 1A1 in living systems. Mater Chem Front. 2018;2:2013–2020. [Google Scholar]
- 36.Ning J., Wang W., Ge G.B., Chu P., Long F.D., Yang Y.L., et al. Target enzyme-activated two-photon fluorescent probes: a case study of CYP3A4 using a two-dimensional design strategy. Angew Chem Int Edit. 2019;58:9959–9963. doi: 10.1002/anie.201903683. [DOI] [PubMed] [Google Scholar]
- 37.Renwick A.B., Lewis D.F.V., Fulford S., Surry D., Williams B., Worboys P.D., et al. Metabolism of 2,5-bis(trifluoromethyl)-7-benzyloxy-4-trifluoromethylcoumarin by human hepatic CYP isoforms: evidence for selectivity towards CYP3A4. Xenobiotica. 2001;31:187–204. doi: 10.1080/00498250110043526. [DOI] [PubMed] [Google Scholar]
- 38.Spatzenegger M., Liu H., Wang Q.M., Debarber A., Koop D.R., Halpert J.R. Analysis of differential substrate selectivities of CYP2B6 and CYP2E1 by site-directed mutagenesis and molecular modeling. J Pharmacol Exp Ther. 2003;304:477–487. doi: 10.1124/jpet.102.043323. [DOI] [PubMed] [Google Scholar]
- 39.Feng L., Ning J., Tian X., Wang C., Yu Z., Huo X., et al. Fluorescent probes for the detection and imaging of cytochrome P450. Coordin Chem Rev. 2021;437:213740. [Google Scholar]
- 40.Zhou J.J., Zhong B., Silverman R.B. Direct continuous fluorometric assay for monoamine oxidase B. Anal Biochem. 1996;234:9–12. doi: 10.1006/abio.1996.0041. [DOI] [PubMed] [Google Scholar]
- 41.Chen G., Yee D.J., Gubernator N.G., Sames D. Design of optical switches as metabolic indicators: new fluorogenic probes for monoamine oxidases (MAO A and B) J Am Chem Soc. 2005;127:4544–4545. doi: 10.1021/ja0428457. [DOI] [PubMed] [Google Scholar]
- 42.Albers A.E., Rawls K.A., Chang C.J. Activity-based fluorescent reporters for monoamine oxidases in living cells. Chem Commun (Camb) 2007;28:4647–4649. doi: 10.1039/b713190a. [DOI] [PubMed] [Google Scholar]
- 43.Aw J., Shao Q., Yang Y., Jiang T., Ang C., Xing B. Synthesis and characterization of 2-(2′-hydroxy-5′-chlorophenyl)-6-chloro-4(3H)-quinazolinone-based fluorogenic probes for cellular imaging of monoamine oxidases. Chem Asian J. 2010;5:1317–1321. doi: 10.1002/asia.201000025. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y.X., Xu Y.F., Tan S.Y., Xu L., Qian X.H. Rapid and sensitive fluorescent probes for monoamine oxidases B to a at low concentrations. Tetrahedron Lett. 2012;53:6881–6884. [Google Scholar]
- 45.Li X., Zhang H., Xie Y., Hu Y., Sun H., Zhu Q. Fluorescent probes for detecting monoamine oxidase activity and cell imaging. Org Biomol Chem. 2014;12:2033–2036. doi: 10.1039/c3ob42326c. [DOI] [PubMed] [Google Scholar]
- 46.Li L.L., Li K., Liu Y.H., Xu H.R., Yu X.Q. Red emission fluorescent probes for visualization of monoamine oxidase in living cells. Sci Rep. 2016;6:31217. doi: 10.1038/srep31217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim D., Sambasivan S., Nam H., Kim K.H., Kim J.Y., Joo T., et al. Reaction-based two-photon probes for in vitro analysis and cellular imaging of monoamine oxidase activity. Chem Commun (Camb) 2012;48:6833–6835. doi: 10.1039/c2cc32424e. [DOI] [PubMed] [Google Scholar]
- 48.Qin H.H., Li L.L., Li K., Yu X.Q. Novel strategy of constructing fluorescent probe for MAO-b via cascade reaction and its application in imaging MAO-B in human astrocyte. Chin Chem Lett. 2019;30:71–74. [Google Scholar]
- 49.Li L., Zhang C.W., Chen G.Y., Zhu B., Chai C., Xu Q.H., et al. A sensitive two-photon probe to selectively detect monoamine oxidase B activity in Parkinson's disease models. Nat Commun. 2014;5:3276. doi: 10.1038/ncomms4276. [DOI] [PubMed] [Google Scholar]
- 50.Wang R., Han X., You J., Yu F., Chen L. Ratiometric near-infrared fluorescent probe for synergistic detection of monoamine oxidase b and its contribution to oxidative stress in cell and mice aging models. Anal Chem. 2018;90:4054–4061. doi: 10.1021/acs.analchem.7b05297. [DOI] [PubMed] [Google Scholar]
- 51.Long S., Chen L., Xiang Y., Song M., Zheng Y., Zhu Q. An activity-based fluorogenic probe for sensitive and selective monoamine oxidase-B detection. Chem Commun. 2012;48:7164–7166. doi: 10.1039/c2cc33089j. [DOI] [PubMed] [Google Scholar]
- 52.Li X., Yu J., Zhu Q., Qian L., Li L., Zheng Y., et al. Visualization of monoamine oxidases in living cells using “turn-on” fluorescence resonance energy transfer probes. Analyst. 2014;139:6092–6095. doi: 10.1039/c4an01195c. [DOI] [PubMed] [Google Scholar]
- 53.Wu X., Li L., Shi W., Gong Q., Li X., Ma H. Sensitive and selective ratiometric fluorescence probes for detection of intracellular endogenous monoamine oxidase a. Anal Chem. 2016;88:1440–1446. doi: 10.1021/acs.analchem.5b04303. [DOI] [PubMed] [Google Scholar]
- 54.Meng Z.J., Yang L., Yao C.X., Li H., Fu Y., Wang K.X., et al. Development of a naphthlimide-based fluorescent probe for imaging monoamine oxidase a in living cells and zebrafish. Dyes Pigments. 2020;176:108208. [Google Scholar]
- 55.Wu X., Shi W., Li X., Ma H. A strategy for specific fluorescence imaging of monoamine oxidase a in living cells. Angew Chem Int Ed Engl. 2017;56:15319–15323. doi: 10.1002/anie.201708428. [DOI] [PubMed] [Google Scholar]
- 56.Yang Z., Li W., Chen H., Mo Q., Li J., Zhao S., et al. Inhibitor structure-guided design and synthesis of near-infrared fluorescent probes for monoamine oxidase a (MAO-A) and its application in living cells and in vivo. Chem Commun. 2019;55:2477–2480. doi: 10.1039/c8cc10084e. [DOI] [PubMed] [Google Scholar]
- 57.Kim W.Y., Won M., Salimi A., Sharma A., Lim J.H., Kwon S.H., et al. Monoamine oxidase-a targeting probe for prostate cancer imaging and inhibition of metastasis. Chem Commun. 2019;55:13267–13270. doi: 10.1039/c9cc07009e. [DOI] [PubMed] [Google Scholar]
- 58.Fang H., Zhang H., Li L., Ni Y., Shi R., Li Z., et al. Rational design of a two-photon fluorogenic probe for visualizing monoamine oxidase a activity in human glioma tissues. Angew Chem Int Edit. 2020;59:7536–7541. doi: 10.1002/anie.202000059. [DOI] [PubMed] [Google Scholar]
- 59.Wierzchowski J., Wroczynski P., Interewicz E. Selective assay of the cytosolic forms of the aldehyde dehydrogenase in rat, with possible significance for the investigations of cyclophosphamide cytotoxicity. Acta Pol Pharm. 1996;53:203–208. [PubMed] [Google Scholar]
- 60.Wierzchowski J., Wroczynski P., Laszuk K., Interewicz E. Fluorimetric detection of aldehyde dehydrogenase activity in human blood, saliva, and organ biopsies and kinetic differentiation between class i and class iii isozymes. Anal Biochem. 1997;245:69–78. doi: 10.1006/abio.1996.9921. [DOI] [PubMed] [Google Scholar]
- 61.Yagishita A., Ueno T., Esumi H., Saya H., Kaneko K., Tsuchihara K., et al. Development of highly selective fluorescent probe enabling flow-cytometric isolation of ALDH3A1-positive viable cells. Bioconjug Chem. 2017;28:302–306. doi: 10.1021/acs.bioconjchem.6b00618. [DOI] [PubMed] [Google Scholar]
- 62.Minn I., Wang H., Mease R.C., Byun Y., Yang X., Wang J., et al. A red-shifted fluorescent substrate for aldehyde dehydrogenase. Nat Commun. 2014;5:3662. doi: 10.1038/ncomms4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anorma C., Hedhli J., Bearrood T.E., Pino N.W., Gardner S.H., Inaba H., et al. Surveillance of cancer stem cell plasticity using an isoform-selective fluorescent probe for aldehyde dehydrogenase 1A1. Acs Central Sci. 2018;4:1045–1055. doi: 10.1021/acscentsci.8b00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bearrood T.E., Aguirre-Figueroa G., Chan J. Rational design of a red fluorescent sensor for aldh1a1 displaying enhanced cellular uptake and reactivity. Bioconjug Chem. 2020;31:224–228. doi: 10.1021/acs.bioconjchem.9b00723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duellman S.J., Valley M.P., Kotraiah V., Vidugiriene J., Zhou W., Bernad L., et al. A bioluminescence assay for aldehyde dehydrogenase activity. Anal Biochem. 2013;434:226–232. doi: 10.1016/j.ab.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 66.Mendoza M.F., Hollabaugh N.M., Hettiarachchi S.U., McCarley R.L. Human NAD(P)H:quinone oxidoreductase type I (HNQO1) activation of quinone propionic acid trigger groups. Biochemistry. 2012;51:8014–8026. doi: 10.1021/bi300760u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silvers W.C., Payne A.S., McCarley R.L. Shedding light by cancer redox-human NAD(P)H:quinone oxidoreductase 1 activation of a cloaked fluorescent dye. Chem Commun (Camb) 2011;47:11264–11266. doi: 10.1039/c1cc14578a. [DOI] [PubMed] [Google Scholar]
- 68.Best Q.A., Johnson A.E., Prasai B., Rouillere A., McCarley R.L. Environmentally robust rhodamine reporters for probe-based cellular detection of the cancer-linked oxidoreductase HNQO1. ACS Chem Biol. 2016;11:231–240. doi: 10.1021/acschembio.5b00792. [DOI] [PubMed] [Google Scholar]
- 69.Prasai B., Silvers W.C., McCarley R.L. Oxidoreductase-facilitated visualization and detection of human cancer cells. Anal Chem. 2015;87:6411–6418. doi: 10.1021/acs.analchem.5b01615. [DOI] [PubMed] [Google Scholar]
- 70.Park S.Y., Won M., Kang C., Kim J.S., Lee M.H. A coumarin-naphthalimide hybrid as a dual emissive fluorescent probe for HNQO1. Dyes Pigments. 2019;164:341–345. [Google Scholar]
- 71.Park S.Y., Jung E., Kim J.S., Chi S.G., Lee M.H. Cancer-specific HNQO1-responsive biocompatible naphthalimides providing a rapid fluorescent turn-on with an enhanced enzyme affinity. Sensors (Basel) 2019;20:53. doi: 10.3390/s20010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pan D., Luo F., Liu X., Liu W., Chen W., Liu F., et al. A novel two-photon fluorescent probe with a long Stokes shift and a high signal-to-background ratio for human NAD(P)H:quinone oxidoreductase 1 (HNQO1) detection and imaging in living cells and tissues. Analyst. 2017;142:2624–2630. doi: 10.1039/c7an00575j. [DOI] [PubMed] [Google Scholar]
- 73.Kwon N., Cho M.K., Park S.J., Kim D., Nam S.J., Cui L., et al. An efficient two-photon fluorescent probe for human NAD(P)H:quinone oxidoreductase (HNQO1) detection and imaging in tumor cells. Chem Commun (Camb) 2017;53:525–528. doi: 10.1039/c6cc08971b. [DOI] [PubMed] [Google Scholar]
- 74.Cuff S., Lewis R.D., Chinje E., Jaffar M., Knox R., Weeks I. An improved cell-permeable fluorogenic substrate as the basis for a highly sensitive test for NAD(P)H quinone oxidoreductase 1 (NQO1) in living cells. Free Radic Biol Med. 2018;116:141–148. doi: 10.1016/j.freeradbiomed.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 75.Zhang C., Zhai B.B., Peng T., Zhong Z., Xu L., Zhang Q.Z., et al. Design and synthesis of near-infrared fluorescence-enhancement probes for the cancer-specific enzyme HNQO1. Dyes Pigments. 2017;143:245–251. [Google Scholar]
- 76.Zheng Y., Pan D., Zhang Y., Zhang Y., Shen Y. Hemicyanine-based near-infrared fluorescent probe for the ultrasensitive detection of HNQO1 activity and discrimination of human cancer cells. Anal Chim Acta. 2019;1090:125–132. doi: 10.1016/j.aca.2019.09.012. [DOI] [PubMed] [Google Scholar]
- 77.Yang Q., Wen Y., Xu J., Shao S. An HBT-based fluorescent dye with enhanced quantum yield in water system and its application for constructing NQO1 fluorescent probe. Talanta. 2020;216:120982. doi: 10.1016/j.talanta.2020.120982. [DOI] [PubMed] [Google Scholar]
- 78.Zhu Y., Han J.L., Zhang Q., Zhao Z., Wang J., Xu X.W., et al. A highly selective fluorescent probe for human NAD(P)H:quinone oxidoreductase 1 (HNQO1) detection and imaging in living tumor cells. Rsc Adv. 2019;9:26729–26733. doi: 10.1039/c9ra05650e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Punganuru S.R., Madala H.R., Arutla V., Zhang R., Srivenugopal K.S. Characterization of a highly specific NQO1-activated near-infrared fluorescent probe and its application for in vivo tumor imaging. Sci Rep. 2019;9:8577. doi: 10.1038/s41598-019-44111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuan Z.W., Xu M.J., Wu T.Z., Zhang X.Y., Shen Y.Z., Ernest U., et al. Design and synthesis of nqo1 responsive fluorescence probe and its application in bio-imaging for cancer diagnosis. Talanta. 2019;198:323–329. doi: 10.1016/j.talanta.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 81.Qiao Z., Zhang H., Zhang Y., Wang K. Detection of lipase activity in cells by a fluorescent probe based on formation of self-assembled micelles. iScience. 2020;23:101294. doi: 10.1016/j.isci.2020.101294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shen Z., Prasai B., Nakamura Y., Kobayashi H., Jackson M.S., McCarley R.L. A near-infrared, wavelength-shiftable, turn-on fluorescent probe for the detection and imaging of cancer tumor cells. ACS Chem Biol. 2017;12:1121–1132. doi: 10.1021/acschembio.6b01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hettiarachchi S.U., Prasai B., McCarley R.L. Detection and cellular imaging of human cancer enzyme using a turn-on, wavelength-shiftable, self-immolative profluorophore. J Am Chem Soc. 2014;136:7575–7578. doi: 10.1021/ja5030707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fei Q., Zhou L., Wang F., Shi B., Li C., Wang R., et al. Rational construction of probes rendering ratiometric response to the cancer-specific enzyme NQO1. Dyes Pigments. 2017;136:846–851. [Google Scholar]
- 85.Dietze E.C., Kuwano E., Hammock B.D. Spectrophotometric substrates for cytosolic epoxide hydrolase. Anal Biochem. 1994;216:176–187. doi: 10.1006/abio.1994.1023. [DOI] [PubMed] [Google Scholar]
- 86.Wixtrom R.N., Hammock B.D. Continuous spectrophotometric assays for cytosolic epoxide hydrolase. Anal Biochem. 1988;174:291–299. doi: 10.1016/0003-2697(88)90548-9. [DOI] [PubMed] [Google Scholar]
- 87.Mateo C., Archelas A., Furstoss R. A spectrophotometric assay for measuring and detecting an epoxide hydrolase activity. Anal Biochem. 2003;314:135–141. doi: 10.1016/s0003-2697(02)00646-2. [DOI] [PubMed] [Google Scholar]
- 88.Mantovani S.M., de Oliveira L.G., Marsaioli A.J. Whole cell quick e for epoxide hydrolase screening using fluorescent probes. J Mol Catal B Enzym. 2008;52–53:173–177. [Google Scholar]
- 89.Jones P.D., Wolf N.M., Morisseau C., Whetstone P., Hock B., Hammock B.D. Fluorescent substrates for soluble epoxide hydrolase and application to inhibition studies. Anal Biochem. 2005;343:66–75. doi: 10.1016/j.ab.2005.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shen W., Zhang J., Mao G., Jiang K., Zhu Q. A long-wavelength, fluorogenic probe for epoxide hydrolase: 7-(2-(oxiran-2-yl)ethoxy) resorufin. Biol Pharm Bull. 2009;32:1496–1499. doi: 10.1248/bpb.32.1496. [DOI] [PubMed] [Google Scholar]
- 91.Morisseau C., Bernay M., Escaich A., Sanborn J.R., Lango J., Hammock B.D. Development of fluorescent substrates for microsomal epoxide hydrolase and application to inhibition studies. Anal Biochem. 2011;414:154–162. doi: 10.1016/j.ab.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou Q.H., Qin W.W., Finel M., He Q.Q., Tu D.Z., Wang C.R., et al. A broad-spectrum substrate for the human UDP-glucuronosyltransferases and its use for investigating glucuronidation inhibitors. Int J Biol Macromol. 2021;180:252–261. doi: 10.1016/j.ijbiomac.2021.03.073. [DOI] [PubMed] [Google Scholar]
- 93.Liang S.C., Ge G.B., Liu H.X., Zhang Y.Y., Wang L.M., Zhang J.W., et al. Identification and characterization of human UDP-glucuronosyltransferases responsible for the in vitro glucuronidation of daphnetin. Drug Metab Dispos. 2010;38:973–980. doi: 10.1124/dmd.109.030734. [DOI] [PubMed] [Google Scholar]
- 94.Xia Y.L., Wang J.J., Li S.Y., Liu Y., Gonzalez F.J., Wang P., et al. Synthesis and structure‒activity relationship of coumarins as potent MCL-1 inhibitors for cancer treatment. Bioorgan Med Chem. 2021;29:115851. doi: 10.1016/j.bmc.2020.115851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xia Y., Lv X., He G., Dou T., Ge G.B., Yang L. In vitro characterization of the glucuronidation pathways of d-luciferin mediated by huamn UDP-glucuronosyl-transferases. Drug Metab Pharmacok. 2017;32:S77. [Google Scholar]
- 96.Rahikainen T., Hakkinen M.R., Finel M., Pasanen M., Juvonen R.O. A high throughput assay for the glucuronidation of 7-hydroxy-4-trifluoromethylcoumarin by recombinant human UDP-glucuronosyltransferases and liver microsomes. Xenobiotica. 2013;43:853–861. doi: 10.3109/00498254.2013.783724. [DOI] [PubMed] [Google Scholar]
- 97.Uchaipichat V., Mackenzie P.I., Guo X.H., Gardnerstephen D., Galetin A., Houston J.B., et al. Human UDP-glucuronosyltransferases: isoform selectivity and kinetics of 4-methylumbelliferone and 1-naphthol glucuronidation, effects of organic solvents, and inhibition by diclofenac and probenecid. Drug Metab Dispos. 2004;32:413–423. doi: 10.1124/dmd.32.4.413. [DOI] [PubMed] [Google Scholar]
- 98.Soikkeli A., Kurkela M., Hirvonen J., Yliperttula M., Finel M. Fluorescence-based high-throughput screening assay for drug interactions with UGT1A6. Assay Drug Dev Technol. 2011;9:496–502. doi: 10.1089/adt.2010.0349. [DOI] [PubMed] [Google Scholar]
- 99.Terai T., Tomiyasu R., Ota T., Ueno T., Komatsu T., Hanaoka K., et al. Tokyogreen derivatives as specific and practical fluorescent probes for UDP-glucuronosyltransferase (UGT) 1A1. Chem Commun. 2013;49:3101–3103. doi: 10.1039/c3cc38810g. [DOI] [PubMed] [Google Scholar]
- 100.Juvonen R.O., Rauhamaki S., Kortet S., Niinivehmas S., Troberg J., Petsalo A., et al. Molecular docking-based design and development of a highly selective probe substrate for UDP-glucuronosyltransferase 1A10. Mol Pharmaceut. 2018;15:923–933. doi: 10.1021/acs.molpharmaceut.7b00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou Q.H., Lv X., Tian Z.H., Finel M., Feng L., Huo P.C., et al. A fluorescence-based microplate assay for high-throughput screening and evaluation of human UGT inhibitors. Anal Chim Acta. 2021;1153:338305. doi: 10.1016/j.aca.2021.338305. [DOI] [PubMed] [Google Scholar]
- 102.Lv X., Ge G.B., Feng L., Troberg J., Hu L.H., Hou J., et al. An optimized ratiometric fluorescent probe for sensing human UDP-glucuronosyltransferase 1A1 and its biological applications. Biosens Bioelectron. 2015;72:261–267. doi: 10.1016/j.bios.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 103.Lv X., Feng L., Ai C.Z., Hou J., Wang P., Zou L.W., et al. A practical and high-affinity fluorescent probe for uridine diphosphate glucuronosyltransferase 1A1: a good surrogate for bilirubin. J Med Chem. 2017;60:9664–9675. doi: 10.1021/acs.jmedchem.7b01097. [DOI] [PubMed] [Google Scholar]
- 104.Kim B., Fukuda M., Lee J.Y., Su D., Sanu S., Silvin A., et al. Visualizing microglia with a fluorescence turn-on UGT1A7C substrate. Angew Chem Int Edit. 2019;58:7972–7976. doi: 10.1002/anie.201903058. [DOI] [PubMed] [Google Scholar]
- 105.Zhu M.Y., Tian Z.H., Jin L.L., Huo X.K., Wang C., Cui J.N., et al. A highly selective fluorescent probe for real-time imaging of UDP-glucuronosyltransferase 1A8 in living cells and tissues. Front Chem Sci Eng. 2019;3:145–150. [Google Scholar]
- 106.Ma X., Tian X., Liu T., Ma Y., Gao J., Feng L., et al. Molecular-splicing strategy to construct a near-infrared fluorescent probe for UDP-glucuronosyltransferase 1A1. Angew Chem Int Edit. 2021;60:24566–24572. doi: 10.1002/anie.202109479. [DOI] [PubMed] [Google Scholar]
- 107.Abdel-Latif A.A. Reaction of catecholamines with hydroxylamine and its application to the assay of catechol O-methyl transferase. Anal Biochem. 1969;29:468–475. doi: 10.1016/0003-2697(69)90331-5. [DOI] [PubMed] [Google Scholar]
- 108.Herblin W.F. A simple colorimetric assay for catechol-O-methyl transferase. Anal Biochem. 1973;51:19–22. doi: 10.1016/0003-2697(73)90448-x. [DOI] [PubMed] [Google Scholar]
- 109.Borchardt R.T. A rapid spectrophotometric assay for catechol-O-methyltransferase. Anal Biochem. 1974;58:382–389. doi: 10.1016/0003-2697(74)90206-1. [DOI] [PubMed] [Google Scholar]
- 110.Masuda M., Tsunoda M., Imai K. High-performance liquid chromatography‒fluorescent assay of catechol-O-methyltransferase activity in rat brain. Anal Bioanal Chem. 2003;376:1069–1073. doi: 10.1007/s00216-003-2025-8. [DOI] [PubMed] [Google Scholar]
- 111.Shoup R.E., Davis G.C., Kissinger P.T. Determination of catechol-O-methyltransferase activity in various tissues by liquid chromatography. Anal Chem. 1980;52:483–487. doi: 10.1021/ac50053a024. [DOI] [PubMed] [Google Scholar]
- 112.Zaitsu K., Okada Y., Nohta H., Kohashi K., Ohkura Y. Assay for catechol-O-methyltransferase by high-performance liquid chromatography with fluorescence detection. J Chromat A. 1981;211:129–136. [Google Scholar]
- 113.Nohta H., Noma S., Ohkura Y., Yoo B.T. Assay for catechol-O-methyltransferase in erythrocytes using a new fluorogenic substrate, 2-(3,4-dihydroxyphenyl)naphtho[1,2-d]thiazole. J Chromatogr. 1984;308:93–100. doi: 10.1016/s0021-9673(01)87536-1. [DOI] [PubMed] [Google Scholar]
- 114.Smit N.P., Pavel S., Kammeyer A., Westerhof W. Determination of catechol O-methyltransferase activity in relation to melanin metabolism using high-performance liquid chromatography with fluorimetric detection. Anal Biochem. 1990;190:286–291. doi: 10.1016/0003-2697(90)90195-f. [DOI] [PubMed] [Google Scholar]
- 115.Qian X.K., Wang P., Xia Y.L., Dou T.Y., Jin Q., Wang D.D., et al. A highly selective fluorescent probe for sensing activities of catechol-O-methyltransferase in complex biological samples. Sensor Actuat B-Chem. 2016;231:615–623. [Google Scholar]
- 116.Wang P., Xia Y.L., Zou L.W., Qian X.K., Dou T.Y., Jin Q., et al. An optimized two-photon fluorescent probe for biological sensing and imaging of catechol-O-methyltransferase. Chemistry. 2017;23:10800–10807. doi: 10.1002/chem.201701384. [DOI] [PubMed] [Google Scholar]
- 117.Lu L.Y., Hsu Y.C., Yang Y.S. Spectrofluorometric assay for monoamine-preferring phenol sulfotransferase (SULT1A3) Anal Biochem. 2010;404:241–243. doi: 10.1016/j.ab.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 118.Chen W.T., Liu M.C., Yang Y.S. Fluorometric assay for alcohol sulfotransferase. Anal Biochem. 2005;339:54–60. doi: 10.1016/j.ab.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 119.Reinen J., Vriese E., Glatt H., Vermeulen N.P. Development and validation of a fluorescence HPLC-based screening assay for inhibition of human estrogen sulfotransferase. Anal Biochem. 2006;357:85–92. doi: 10.1016/j.ab.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 120.Baglia R.A., Mills K.R., Mitra K., Tutol J.N., Ball D., Page K.M., et al. An activity-based fluorescent sensor for the detection of the phenol sulfotransferase SULT1A1 in living cells. RSC Chem Biol. 2021;2:830–834. doi: 10.1039/d0cb00231c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Laurieri N., Crawford M.H., Kawamura A., Westwood I.M., Robinson J., Fletcher A.M., et al. Small molecule colorimetric probes for specific detection of human arylamine N-acetyltransferase 1, a potential breast cancer biomarker. J Am Chem Soc. 2010;132:3238–3239. doi: 10.1021/ja909165u. [DOI] [PubMed] [Google Scholar]
- 122.Egleton J.E., Thinnes C.C., Seden P.T., Laurieri N., Lee S.P., Hadavizadeh K.S., et al. Structure‒activity relationships and colorimetric properties of specific probes for the putative cancer biomarker human arylamine N-acetyltransferase 1. Bioorgan Med Chem. 2014;22:3030–3054. doi: 10.1016/j.bmc.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 123.Cui L., Zhong Y., Zhu W., Xu Y., Qian X. Selective and sensitive detection and quantification of arylamine N-acetyltransferase 2 by a ratiometric fluorescence probe. Chem Commun. 2010;46:7121–7123. doi: 10.1039/c0cc01000f. [DOI] [PubMed] [Google Scholar]
- 124.Wang X., Cui L., Zhou N.N., Zhu W.P., Wang R., Qian X.H., et al. A highly selective and sensitive near-infrared fluorescence probe for arylamine N-acetyltransferase 2 in vitro and in vivo. Chem Sci. 2013;4:2936–2940. [Google Scholar]
- 125.Jin Y.Z., Tian Z.H., Tian X.G., Feng L., Cui J.N., Ma X.C. A highly selective fluorescent probe for real-time imaging of bacterial NAT2 and high-throughput screening of natural inhibitors for tuberculosis therapy. Mater Chem Front. 2019;3:145–150. [Google Scholar]
- 126.Terai T., Kikuchi K., Urano Y., Kojima H., Nagano T. A long-lived luminescent probe to sensitively detect arylamine N-acetyltransferase (NAT) activity of cells. Chem Commun (Camb) 2012;48:2234–2246. doi: 10.1039/c2cc17622j. [DOI] [PubMed] [Google Scholar]
- 127.Fujikawa Y., Urano Y., Komatsu T., Hanaoka K., Kojima H., Terai T., et al. Design and synthesis of highly sensitive fluorogenic substrates for glutathione S-transferase and application for activity imaging in living cells. J Am Chem Soc. 2008;130:14533–14543. doi: 10.1021/ja802423n. [DOI] [PubMed] [Google Scholar]
- 128.He N., Bai S., Huang Y., Xing Y., Chen L., Yu F., et al. Evaluation of glutathione S-transferase inhibition effects on idiopathic pulmonary fibrosis therapy with a near-infrared fluorescent probe in cell and mice models. Anal Chem. 2019;91:5424–5432. doi: 10.1021/acs.analchem.9b00713. [DOI] [PubMed] [Google Scholar]
- 129.Tian Z., Tian X., Feng L., Tian Y., Huo X., Zhang B., et al. A highly sensitive and selective two-photon fluorescent probe for glutathione S-transferase detection and imaging in living cells and tissues. J Mater Chem B. 2019;7:4983–4989. doi: 10.1039/c9tb00834a. [DOI] [PubMed] [Google Scholar]
- 130.Mori M., Fujikawa Y., Kikkawa M., Shino M., Sawane M., Sato S., et al. A highly selective fluorogenic substrate for imaging glutathione S-transferase P1: development and cellular applicability in epigenetic studies. Chem Commun (Camb) 2019;55:8122–8125. doi: 10.1039/c9cc03064f. [DOI] [PubMed] [Google Scholar]
- 131.Fujikawa Y., Terakado K., Nampo T., Mori M., Inoue H. 4-Bromo-1,8-naphthalimide derivatives as fluorogenic substrates for live cell imaging of glutathione S-transferase (GST) activity. Talanta. 2019;204:633–640. doi: 10.1016/j.talanta.2019.06.059. [DOI] [PubMed] [Google Scholar]
- 132.Fujikawa Y., Morisaki F., Ogura A., Morohashi K., Enya S., Niwa R., et al. A practical fluorogenic substrate for high-throughput screening of glutathione S-transferase inhibitors. Chem Commun (Camb) 2015;51:11459–11462. doi: 10.1039/c5cc02067k. [DOI] [PubMed] [Google Scholar]
- 133.Zhang J., Shibata A., Ito M., Shuto S., Ito Y., Mannervik B., et al. Synthesis and characterization of a series of highly fluorogenic substrates for glutathione transferases, a general strategy. J Am Chem Soc. 2011;133:14109–14119. doi: 10.1021/ja205500y. [DOI] [PubMed] [Google Scholar]
- 134.Shibata A., Nakano Y., Ito M., Araki M., Zhang J., Yoshida Y., et al. Fluorogenic probes using 4-substituted-2-nitrobenzenesulfonyl derivatives as caging groups for the analysis of human glutathione transferase catalyzed reactions. Analyst. 2013;138:7326–7330. doi: 10.1039/c3an01339a. [DOI] [PubMed] [Google Scholar]
- 135.Zhang J., Jin Z., Hu X.X., Meng H.M., Li J., Zhang X.B., et al. Efficient two-photon fluorescent probe for glutathione S-transferase detection and imaging in drug-induced liver injury sample. Anal Chem. 2017;89:8097–8103. doi: 10.1021/acs.analchem.7b01659. [DOI] [PubMed] [Google Scholar]
- 136.Zhou W., Shultz J.W., Murphy N., Hawkins E.M., Bernad L., Good T., et al. Electrophilic aromatic substituted luciferins as bioluminescent probes for glutathione S-transferase assays. Chem Commun (Camb) 2006:4620–4622. doi: 10.1039/b610682j. [DOI] [PubMed] [Google Scholar]
- 137.Ito M., Shibata A., Zhang J., Hiroshima M., Sako Y., Nakano Y., et al. Universal caging group for the in-cell detection of glutathione transferase applied to 19F NMR and bioluminogenic probes. Chembiochem. 2012;13:1428–1432. doi: 10.1002/cbic.201200242. [DOI] [PubMed] [Google Scholar]
- 138.Kong F., Pang X., Zhao J., Deng P., Zheng M., Zhong D., et al. Hydrolytic metabolism of cyanopyrrolidine DPP-4 inhibitors mediated by dipeptidyl peptidases. Drug Metab Dispos. 2019;47:238–248. doi: 10.1124/dmd.118.084640. [DOI] [PubMed] [Google Scholar]
- 139.Asakura M., Fujii H., Atsuda K., Itoh T., Fujiwara R. Dipeptidyl peptidase-4 greatly contributes to the hydrolysis of vildagliptin in human liver. Drug Metab Dispos. 2015;43:477–484. doi: 10.1124/dmd.114.062331. [DOI] [PubMed] [Google Scholar]
- 140.Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Marguet D., Baggio L., Kobayashi T., Bernard A.M., Pierres M., Nielsen P.F., et al. Enhanced insulin secretion and improved glucose tolerance in mice lacking CD26. Proc Natl Acad Sci U S A. 2000;97:6874–6879. doi: 10.1073/pnas.120069197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Morimoto C., Schlossman S.F. The structure and function of CD26 in the T-cell immune response. Immunol Rev. 1998;161:55–70. doi: 10.1111/j.1600-065x.1998.tb01571.x. [DOI] [PubMed] [Google Scholar]
- 143.Takeda Y., Kume H., Takafuji K., Hirata H., Kijima T., Tomonaga T., et al. Proteomic profiling of peripheral exosomes leads to the identification of novel biomarker candidates for emphysema. Am J Resp Crit Care. 2016;193 [Google Scholar]
- 144.Ogawa Y., Kanai-Azuma M., Akimoto Y., Kawakami H., Yanoshita R. Exosome-like vesicles with dipeptidyl peptidase IV in human saliva. Biol Pharm Bull. 2008;31:1059–1062. doi: 10.1248/bpb.31.1059. [DOI] [PubMed] [Google Scholar]
- 145.Lai K.S., Ho N.H., Cheng J.D., Tung C.H. Selective fluorescence probes for dipeptidyl peptidase activity―fibroblast activation protein and dipeptidyl peptidase IV. Bioconjugate Chem. 2007;18:1246–1250. doi: 10.1021/bc0603586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang Y., Wu X., Cheng Y., Zhao X. A fluorescent switchable AIE probe for selective imaging of dipeptidyl peptidase-4 in vitro and in vivo and its application in screening DPP-4 inhibitors. Chem Commun. 2016;52:3478–3481. doi: 10.1039/c5cc08921b. [DOI] [PubMed] [Google Scholar]
- 147.Gong Q., Shi W., Li L., Wu X., Ma H. Ultrasensitive fluorescent probes reveal an adverse action of dipeptide peptidase IV and fibroblast activation protein during proliferation of cancer cells. Anal Chem. 2016;88:8309–8314. doi: 10.1021/acs.analchem.6b02231. [DOI] [PubMed] [Google Scholar]
- 148.Zou L.W., Wang P., Qian X.K., Feng L., Yu Y., Wang D.D., et al. A highly specific ratiometric two-photon fluorescent probe to detect dipeptidyl peptidase IV in plasma and living systems. Biosens Bioelectron. 2017;90:283–289. doi: 10.1016/j.bios.2016.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liu T., Ning J., Wang B., Dong B., Li S., Tian X., et al. Activatable near-infrared fluorescent probe for dipeptidyl peptidase IV and its bioimaging applications in living cells and animals. Anal Chem. 2018;90:3965–3973. doi: 10.1021/acs.analchem.7b04957. [DOI] [PubMed] [Google Scholar]
- 150.Xing J., Gong Q., Zhang R., Sun S., Zou R., Wu A. A novel non-enzymatic hydrolytic probe for dipeptidyl peptidase IV specific recognition and imaging. Chem Commun. 2018;54:8773–8776. doi: 10.1039/c8cc05048a. [DOI] [PubMed] [Google Scholar]
- 151.Ogasawara A., Kamiya M., Sakamoto K., Kuriki Y., Fujita K., Komatsu T., et al. Red fluorescence probe targeted to dipeptidylpeptidase-IV for highly sensitive detection of esophageal cancer. Bioconjugate Chem. 2019;30:1055–1060. doi: 10.1021/acs.bioconjchem.9b00198. [DOI] [PubMed] [Google Scholar]
- 152.Ma H., Qian X.K., Zhang J., Jin Q., Zou L.W., Liu S.Q., et al. Accurate and sensitive detection of dipeptidyl peptidase-IV activity by liquid chromatography with fluorescence detection. Anal Methods. 2020;12:848–854. [Google Scholar]
- 153.Dai Z.R., Ge G.B., Feng L., Ning J., Hu L.H., Jin Q., et al. A highly selective ratiometric two-photon fluorescent probe for human cytochrome P450 1A. J Am Chem Soc. 2015;137:14488–14495. doi: 10.1021/jacs.5b09854. [DOI] [PubMed] [Google Scholar]
- 154.Wang D.D., Jin Q., Zou L.W., Hou J., Lv X., Lei W., et al. A bioluminescent sensor for highly selective and sensitive detection of human carboxylesterase 1 in complex biological samples. Chem Commun. 2016;52:3183–3186. doi: 10.1039/c5cc09874b. [DOI] [PubMed] [Google Scholar]
- 155.Wang D.D., Zou L.W., Jin Q., Guan X.Q., Yu Y., Zhu Y.D., et al. Bioluminescent sensor reveals that carboxylesterase 1A is a novel endoplasmic reticulum-derived serologic indicator for hepatocyte injury. ACS Sens. 2020;5:1987–1995. doi: 10.1021/acssensors.0c00384. [DOI] [PubMed] [Google Scholar]
- 156.Ding L., Tian Z., Hou J., Dou T., Jin Q., Wang D., et al. Sensing carboxylesterase 1 in living systems by a practical and isoform-specific fluorescent probe. Chin Chem Lett. 2019;30:558–562. [Google Scholar]
- 157.Zou L.W., Dou T.Y., Wang P., Lei W., Weng Z.M., Hou J., et al. Structure‒activity relationships of pentacyclic triterpenoids as potent and selective inhibitors against human carboxylesterase 1. Front Pharmacol. 2017;8:435. doi: 10.3389/fphar.2017.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lei W., Wang D.D., Dou T.Y., Hou J., Feng L., Yin H., et al. Assessment of the inhibitory effects of pyrethroids against human carboxylesterases. Toxicol Appl Pharm. 2017;321:48–56. doi: 10.1016/j.taap.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 159.Liu Y.J., Li S.Y., Hou J., Liu Y.F., Wang D.D., Jiang Y.S., et al. Identification and characterization of naturally occurring inhibitors against human carboxylesterase 2 in white mulberry root-bark. Fitoterapia. 2016;115:57–63. doi: 10.1016/j.fitote.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 160.Xue L.J., Qian X.K., Jin Q., Zhu Y.D., Wang X.Y., Wang D.D., et al. Construction and application of a high-content analysis for identifying human carboxylesterase 2 inhibitors in living cell system. Anal Bioanal Chem. 2020;412:2645–2654. doi: 10.1007/s00216-020-02494-y. [DOI] [PubMed] [Google Scholar]
- 161.Zhu Y.D., Guan X.Q., Chen J., Peng S., Finel M., Zhao Y.Y., et al. Neobavaisoflavone induces bilirubin metabolizing enzyme UGT1A1 via PPARα and PPARSγ. Front Pharmacol. 2020;11:628314. doi: 10.3389/fphar.2020.628314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tian X., Yan F., Zheng J., Cui X., Feng L., Li S., et al. Endoplasmic reticulum targeting ratiometric fluorescent probe for carboxylesterase 2 detection in drug-induced acute liver injury. Anal Chem. 2019;91:15840–15845. doi: 10.1021/acs.analchem.9b04189. [DOI] [PubMed] [Google Scholar]
- 163.Hakamata W., Machida A., Oku T., Nishio T. Design and synthesis of an ER-specific fluorescent probe based on carboxylesterase activity with quinone methide cleavage process. Bioorg Med Chem Lett. 2011;21:3206–3209. doi: 10.1016/j.bmcl.2011.04.066. [DOI] [PubMed] [Google Scholar]
- 164.Zhang Y., Chen W., Feng D., Shi W., Li X., Ma H. A spectroscopic off-on probe for simple and sensitive detection of carboxylesterase activity and its application to cell imaging. Analyst. 2012;137:716–721. doi: 10.1039/c2an15952j. [DOI] [PubMed] [Google Scholar]
- 165.Li D., Li Z., Chen W., Yang X. Imaging and detection of carboxylesterase in living cells and zebrafish pretreated with pesticides by a new near-infrared fluorescence off-on probe. J Agric Food Chem. 2017;65:4209–4215. doi: 10.1021/acs.jafc.7b00959. [DOI] [PubMed] [Google Scholar]
- 166.Wang J., Chen Q., Tian N., Zhu W., Zou H., Wang X., et al. A fast responsive, highly selective and light-up fluorescent probe for the two-photon imaging of carboxylesterase in living cells. J Mater Chem B. 2018;6:1595–1599. doi: 10.1039/c8tb00147b. [DOI] [PubMed] [Google Scholar]
- 167.Li M., Zhai C., Wang S., Huang W., Liu Y., Li Z. Detection of carboxylesterase by a novel hydrosoluble near-infrared fluorescence probe. Rsc Adv. 2019;9:40689–40693. doi: 10.1039/c9ra08150j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhou H., Tang J., Zhang J., Chen B., Kan J., Zhang W., et al. A red lysosome-targeted fluorescent probe for carboxylesterase detection and bioimaging. J Mater Chem B. 2019;7:2989–2996. [Google Scholar]
- 169.Jiang A., Chen G., Xu J., Liu Y.X., Zhao G.H., Liu Z.J., et al. Ratiometric two-photon fluorescent probe for in situ imaging of carboxylesterase (CE)-mediated mitochondrial acidification during medication. Chem Commun. 2019;55:11358–11361. doi: 10.1039/c9cc05759e. [DOI] [PubMed] [Google Scholar]
- 170.Liu Z.M., Feng L., Ge G.B., Lv X., Hou J., Cao Y.F., et al. A highly selective ratiometric fluorescent probe for in vitro monitoring and cellular imaging of human carboxylesterase 1. Biosens Bioelectron. 2014;57:30–35. doi: 10.1016/j.bios.2014.01.049. [DOI] [PubMed] [Google Scholar]
- 171.Feng L., Liu Z.M., Xu L., Lv X., Ning J., Hou J., et al. A highly selective long-wavelength fluorescent probe for the detection of human carboxylesterase 2 and its biomedical applications. Chem Commu (Camb) 2014;50:14519–14522. doi: 10.1039/c4cc06642a. [DOI] [PubMed] [Google Scholar]
- 172.Wang J., Williams E.T., Bourgea J., Wong Y.N., Patten C.J. Characterization of recombinant human carboxylesterases: fluorescein diacetate as a probe substrate for human carboxylesterase 2. Drug Metab Dispos. 2011;39:1329–1333. doi: 10.1124/dmd.111.039628. [DOI] [PubMed] [Google Scholar]
- 173.Liu Z., Feng L., Hou J., Lv X., Ning J., Ge G., et al. A ratiometric fluorescent sensor for highly selective detection of human carboxylesterase 2 and its application in living cells. Sensors Actuat B-Chem. 2014;205:151–157. [Google Scholar]
- 174.Feng L., Liu Z.M., Hou J., Lv X., Ning J., Ge G.B., et al. A highly selective fluorescent esipt probe for the detection of human carboxylesterase 2 and its biological applications. Biosens Bioelectron. 2015;65:9–15. doi: 10.1016/j.bios.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 175.Kailass K., Sadovski O., Capello M., Kang Y., Fleming J.B., Hanash S.M., et al. Measuring human carboxylesterase 2 activity in pancreatic cancer patient-derived xenografts using a ratiometric fluorescent chemosensor. Chem Sci. 2019;10:8428–8437. doi: 10.1039/c9sc00283a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Park S.J., Kim Y.J., Kang J.S., Kim I.Y., Choi K.S., Kim H.M. Carboxylesterase-2-selective two-photon ratiometric probe reveals decreased carboxylesterase-2 activity in breast cancer cells. Anal Chem. 2018;90:9465–9471. doi: 10.1021/acs.analchem.8b02101. [DOI] [PubMed] [Google Scholar]
- 177.Zhang X., Zhou Y., Gu X., Cheng Y., Hong M., Yan L., et al. Synthesis of a selective ratiometric fluorescent probe based on naphthalimide and its application in human cytochrome P450 1A. Talanta. 2018;186:413–420. doi: 10.1016/j.talanta.2018.04.079. [DOI] [PubMed] [Google Scholar]
- 178.Ji H., Zhang X., Dai Y., Xue T., Misal S., Qi Z. A highly selective ratiometric fluorescent probe based on naphthalimide for detection and imaging of CYP1A1 in living cells and zebrafish. Analyst. 2019;144:7390–7397. doi: 10.1039/c9an01767d. [DOI] [PubMed] [Google Scholar]
- 179.Xue T., Dai Y., Zhang X., Cheng Y., Gu X., Ji H., et al. Ultrasensitive near-infrared fluorescent probe with large Stokes shift for real-time tracing of CYP1A1 in living cells and zebrafish model. Sensor Actuat B-chem. 2019;293:265–272. [Google Scholar]
- 180.Shimada T., Gillam E.M., Sutter T.R., Strickland P.T., Guengerich F.P., Yamazaki H. Oxidation of xenobiotics by recombinant human cytochrome P450 1B1. Drug Metab Dispos. 1997;25:617–622. [PubMed] [Google Scholar]
- 181.Makings L., Zlokarnik G., Life Technologies Corp, assignee . 4 Feb 2003. Optical molecular sensors for cytochrome P450 activity.https://patents.google.com/patent/US6514687B1/en US006514687B1. Available from: [Google Scholar]
- 182.Renwick A.B., Surry D., Price R.J., Lake B.G., Evans D.C. Metabolism of 7-benzyloxy-4-trifluoromethyl-coumarin by human hepatic cytochrome P450 isoforms. Xenobiotica. 2000;30:955–969. doi: 10.1080/00498250050200113. [DOI] [PubMed] [Google Scholar]
- 183.Venhorst J., ter Laak A.M., Commandeur J.N.M., Funae Y., Hiroi T., Vermeulen N.P.E. Homology modeling of rat and human cytochrome P450 2D (CYP2D) isoforms and computational rationalization of experimental ligand-binding specificities. J Med Chem. 2003;46:74–86. doi: 10.1021/jm0209578. [DOI] [PubMed] [Google Scholar]
- 184.Marks B.D., Smith R.W., Braun H.A., Goossens T.A., Christenson M., Ozers M.S., et al. A high throughput screening assay to screen for CYP2E1 metabolism and inhibition using a fluorogenic vivid P450 substrate. Assay Drug Dev Technol. 2002;1:73–81. doi: 10.1089/154065802761001329. [DOI] [PubMed] [Google Scholar]
- 185.Ning J., Liu T., Dong P., Wang W., Ge G., Wang B., et al. Molecular design strategy to construct the near-infrared fluorescent probe for selectively sensing human cytochrome P450 2J2. J Am Chem Soc. 2019;141:1126–1134. doi: 10.1021/jacs.8b12136. [DOI] [PubMed] [Google Scholar]
- 186.Dai Y., Xue T., Ji H., Zhang P., Zhang D., Qi Z. In situ target enzyme-activated near-infrared fluorescent probe: a case study of CYP2J2 using three-fragmentary molecular assembly engineering. Sensor Actuat B-Chem. 2021;328:129034. [Google Scholar]
- 187.Foti R., Wienkers L., Wahlstrom J. Application of cytochrome P450 drug interaction screening in drug discovery. High T Scr. 2010;13:145–158. doi: 10.2174/138620710790596718. [DOI] [PubMed] [Google Scholar]
- 188.Cali J.J., Ma D., Wood M.G., Meisenheimer P.L., Klaubert D.H. Bioluminescent assays for adme evaluation: dialing in CYP selectivity with luminogenic substrates. Expert Opin Drug Met. 2012;8:1115–1130. doi: 10.1517/17425255.2012.695345. [DOI] [PubMed] [Google Scholar]
- 189.Promega Corporation. P450-GloTM assay technical bulletin. Available from: https://www.promega.com.cn/resources/protocols/technical-bulletins/101/p450-glo-assays-protocol/.
- 190.Xiang Y.M., He B.Y., Li X.F., Zhu Q. The design and synthesis of novel "turn-on" fluorescent probes to visualize monoamine oxidase-B in living cells. Rsc Adv. 2013;3:4876–4879. [Google Scholar]
- 191.Sun Y.P., Jung E., Kim J.S., Chi S.G., Min H.L.J.S. Cancer-specific HNQO1-responsive biocompatible naphthalimides providing a rapid fluorescent turn-on with an enhanced enzyme affinity. Sensors. 2019;20:53. doi: 10.3390/s20010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Han J., Cheng L., Zhu Y., Xu X., Ge C. Covalent-assembly based fluorescent probes for detection of HNQO1 and imaging in living cells. Front Chem. 2020;8:756. doi: 10.3389/fchem.2020.00756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhu M., Tian Z., Jin L., Huo X., Wang C., Cui J., et al. A highly selective fluorescent probe for real-time imaging of UDP-glucuronosyltransferase 1A8 in living cells and tissues. Front Chem Sci Eng. 2022;16:103–111. [Google Scholar]
- 194.Alander J., Johansson K., Heuser V.D., Farebo H., Jarvliden J., Abe H., et al. Characterization of a new fluorogenic substrate for microsomal glutathione transferase 1. Anal Biochem. 2009;390:52–56. doi: 10.1016/j.ab.2009.03.046. [DOI] [PubMed] [Google Scholar]
- 195.Song A., Shen X., Feng T., Gai S., Wei H., Li X., et al. Optimized fluorescent probe for specific imaging of glutathione S-transferases in living cells and mice. Chem Asian J. 2020;15:1464–1468. doi: 10.1002/asia.202000152. [DOI] [PubMed] [Google Scholar]
- 196.Fujikawa Y., Nampo T., Mori M., Kikkawa M., Inoue H. Fluorescein diacetate (FDA) and its analogue as substrates for pi-class glutathione S-transferase (GSTP1) and their biological application. Talanta. 2018;179:845–852. doi: 10.1016/j.talanta.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 197.Tian X., Tian Z., Wang Y., Hou J., Feng L., Song L., et al. A highly selective fluorescent probe for detecting glutathione transferases to reveal anticancer-activity sensitivity of cisplatin in cancer cells and tumor tissues. Sensor Actuat B-Chem. 2018;277:423–430. [Google Scholar]
- 198.Song A., Feng T., Shen X., Gai S., Zhai Y., Chen H. Fluorescence detection of glutathione S-transferases in a low gsh level environment. Chem Commun (Camb) 2019;55:7219–7222. doi: 10.1039/c9cc02702e. [DOI] [PubMed] [Google Scholar]
- 199.Kawaguchi M., Okabe T., Terai T., Hanaoka K., Kojima H., Minegishi I., et al. A time-resolved fluorescence probe for dipeptidyl peptidase 4 and its application in inhibitor screening. Chemistry. 2010;16:13479–13486. doi: 10.1002/chem.201001077. [DOI] [PubMed] [Google Scholar]
- 200.Guo X., Mu S., Li J., Zhang Y., Liu X., Zhang H., et al. Fabrication of a water-soluble near-infrared fluorescent probe for selective detection and imaging of dipeptidyl peptidase IV in biological systems. J Mater Chem B. 2020;8:767–775. doi: 10.1039/c9tb02301a. [DOI] [PubMed] [Google Scholar]