Abstract
Malarial parasites rely on aspartic proteases called plasmepsins to digest hemoglobin during the intraerythrocytic stage. Plasmepsins from Plasmodium falciparum and Plasmodium vivax have been cloned and expressed for a variety of structural and enzymatic studies. Recombinant plasmepsins possess kinetic similarity to the native enzymes, indicating their suitability for target-based antimalarial drug development. We developed an automated assay of P. falciparum plasmepsin II and P. vivax plasmepsin to quickly screen compounds in the Walter Reed chemical database. A low-molecular-mass (346 Da) diphenylurea derivative (WR268961) was found to inhibit plasmepsins with a Ki of 1 to 6 μM. This compound appears to be selective for plasmepsin, since it is a poor inhibitor of the human aspartic protease cathepsin D (Ki greater than 280 μM). WR268961 inhibited the growth of P. falciparum strains W2 and D6, with 50% inhibitory concentrations ranging from 0.03 to 0.16 μg/ml, but was much less toxic to mammalian cells. The Walter Reed chemical database contains over 1,500 compounds with a diphenylurea core structure, 9 of which inhibit the plasmepsins, with Ki values ranging from 0.05 to 0.68 μM. These nine compounds show specificity for the plasmepsins over human cathepsin D, but they are poor inhibitors of P. falciparum growth in vitro. Computational docking experiments indicate how diphenylurea compounds bind to the plasmepsin active site and inhibit the enzyme.
Malaria, the most severe parasitic disease, infects nearly 300 million people and kills more than a million each year (28). Plasmodium falciparum and Plasmodium vivax are the two malaria species responsible for the most infections and deaths. Although several very effective antimalarial drugs have been used to control this disease, P. falciparum has developed resistance to nearly all available antimalarial drugs (27). Recently, P. vivax from Southeast Asia has developed resistance to the most widely used antimalarial drug, chloroquine. The search for novel antimalarial drugs against specific parasitic targets is thus an urgent task to pursue. In the last decade, many potential targets for new antimalarial drugs have been discovered, such as dihydropteroate synthase, hemoglobin degradation enzymes, and shikimate pathway enzymes (17). Our work focuses on the discovery of new inhibitors of hemoglobin degradation enzymes called plasmepsins.
Malarial parasites invade human erythrocytes in the asexual stage of infection. While residing in erythrocytes, the parasites rely on human hemoglobin as a food source, digesting it with a series of proteases. The aspartic proteases, called plasmepsins, are critical for hemoglobin degradation and are thus logical targets for antimalarial drug development (14, 19, 25). At least four plasmepsins have been identified and cloned from P. falciparum (26; R. Banerjee and D. E. Goldberg, Mol. Parasite Meet., MBL, Woods Hole, Mass., 1999). Active recombinant plasmepsin II has been successfully obtained in large enough quantities (3, 10) to facilitate detailed kinetic studies (12) and structural studies of this enzyme (20, 21). Recombinant plasmepsin II has kinetic behavior similar to native plasmepsin II and has been used for inhibitor screening with combinatorial libraries and structure-based drug design (1a, 2, 9). Aspartic protease-specific inhibitors, such as pepstatin, SC-50083 (5), and Ro 40–4388 (15), arrest parasite growth by interrupting the metabolism of hemoglobin. These results indicate that recombinant plasmepsins are suitable targets for antimalarial drug design and enzyme-based inhibitor screening.
We conducted a plasmepsin-based antimalarial screen with recombinant plasmepsins from P. falciparum and P. vivax. Here we report the discovery of small nonpeptidyl compounds from the Walter Reed chemical database that block P. falciparum development in vitro by inhibiting plasmepsins. Computational docking experiments indicate how these compounds bind to the plasmepsin active site and inhibit the enzyme.
MATERIALS AND METHODS
Parasite culture.
A chloroquine-sensitive D6 strain, chloroquine-resistant W2 strain, and wild-type strain (WR87) of Plasmodium falciparum were cultivated in RPMI 1640 medium with 6% human erythrocytes supplemented with 10% human serum (24). The parasites were cultured in an atmosphere of 5% CO2, 5% O2, and 90% N2 at 37°C.
Plasmepsin assay.
The substrate used for the plasmepsin assay (Bachem) is a synthetic peptide (DABCYL-Glu-Arg-Nle-Phe-Leu-Ser-Phe-Pro-EDANS) designed to mimic the cleavage site present in hemoglobin. The kinetic constants for the substrate are kcat = 0.78 s−1 and Km = 0.10 μM for P. falciparum plasmepsin, and kcat = 0.69 s−1 and Km = 0.16 μM for P. vivax plasmepsin. The substrate is conjugated with the fluorescent donor EDANS and the quencher DABCYL (13). Fluorescence is only detectable when the EDANS group is separated from the DABCYL group by cleavage of the substrate (12). We developed an automated plasmepsin assay protocol that allowed us to screen a large number of compounds within a short period of time. Compounds were manually added to 96-well plates followed by the addition of assay buffer (15 mM NaCl, 100 mM formate [pH 4.4]) by using an automated dilutor (BioMec 2000; Beckman). After thorough mixing and dilution, the contents of the plates were transferred to test plates, and plasmepsin enzyme solution was added with the dilutor. After a 10-min incubation at 37°C, background fluorescence was measured with a fluorescence plate reader (Wallac Victor2). Finally, the substrate was added (final concentration of 10 μM), and the reaction mixture was incubated for 30 min at 37°C followed by fluorescence detection. We tested each compound in this prescreen in triplicate at a concentration of 10 μg/ml. Compounds that reduced the activity of plasmepsin by 50% or more at this concentration were selected for a second screen to determine 50% inhibitory concentrations (IC50s). The best inhibitors were assayed by using a range of inhibitor and substrate concentrations to determine Ki values (ENZYME KINETICS from Trinity Software).
In vitro drug susceptibility assay.
All compounds with IC50 values below 5 μg/ml were tested in a cell-based in vitro drug susceptibility assay to determine if they were capable of interrupting Plasmodium metabolism and growth. The semiautomated microdilution technique of Desjardins et al. (4) was used to assess the sensitivity of the parasites to the selected compounds. The incorporation of [3H]hypoxanthine into the parasites was measured as a function of compound concentration to determine IC50 values.
Hemoglobin degradation assay.
Selected compounds were added to synchronized parasite cultures during early ring stage. After a 12-h incubation with the compounds, smears of the parasite culture were Giemsa stained and studied under a microscope for any morphological changes. Malaria parasites were separated from erythrocytes with 0.1% saponin. The isolated parasites were lysed, and their cellular contents were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to evaluate the amount of intact hemoglobin in the parasites as described by Rosenthal (18).
Aspartic protease selectivity assay.
Human aspartic protease cathepsin D (Calbiochem) was used to test whether our compounds were specific inhibitors of plasmepsin. The kinetic constants for the fluorogenic substrate are kcat = 0.56 s−1 and Km = 0.12 μM for cathepsin D. The protocol of Takahashi and coworkers (23) was adapted to our automated 96-well-plate screen. All assay conditions remained the same as those used in the plasmepsin assay.
Toxicity assay.
Selected compounds were tested for toxicity in vitro against two mammalian cell lines. A subclone (G8) of the murine monocyte-like macrophage line J774 was obtained from Jose Alunda, Departmento de Sanidad Animal, Facultad de Veterinaria, Universidad Complutense, Madrid, Spain. Murine cells were maintained in 75-cm2 tissue culture flasks in Dulbecco's modified Eagle medium (GIBCO) supplemented with 10% fetal calf serum, 2 mM l-glutamine, and 50 μg of gentamicin per ml under humidified 5% CO2–95% air at 37°C. A subclone (NG-108-15) of a hybrid rat neuroblastoma-mouse glioma cell line was the gift of Marshall Nirenberg, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, Md. Neuronal cells were maintained in 75-cm2 tissue culture flasks in Dulbecco's modified essential medium including hypoxanthine-aminopterin-thymidine (HAT) supplement, 10% fetal calf serum, and 50 μg of gentamicin per ml under humidified 5% CO2–95% air at 37°C.
Toxicity tests were performed in 96-well tissue culture plates with the protein-binding dye sulforhodamine B (SRB) as described previously (22). Test compounds were serially diluted and added to empty wells of the 96-well plate. The wells were immediately seeded with the appropriate cells in their respective culture media. Appropriate solvent blanks (no compound) were run in each test. After 72 h under culture conditions, cells were fixed to the plate by layering 50% trichloroacetic acid (TCA) (4°C) over the growth medium in each well to produce a final TCA concentration of 10%. After incubation for 1 h at 4°C, cultures were washed five times with tap water and air dried. Wells were stained for 30 min with 0.4% (wt/vol) SRB in 1% acetic acid and washed four times with 1% acetic acid. Cultures were air dried, and bound dye was solubilized with 10 mM Tris base (pH 10.5) for 15 min on a gyratory shaker at room temperature. A Spectra MAX Plus microtiter plate reader (Molecular Devices) was used to measure the optical density at wavelengths of 490 to 530 nm.
Molecular modeling.
The crystal structures of P. vivax (Protein Data Bank ID 1QS8) and P. falciparum plasmepsin II (20) were used as targets for a series of docking studies. Water molecules and the inhibitor pepstatin were removed from the crystal structures prior to the docking experiments. The P. falciparum enzyme was altered by shortening residue valine 78 to glycine to make the active site more accessible to ligands during the docking experiments. (P. vivax plasmepsin has a glycine at this position.) The structures of small molecule ligands were generated in InsightII (MSI) and docked to plasmepsin by using the program Autodock3 (8, 8a). Autodock3 randomly places the ligand near the active site in a random conformation. The program uses a simulated annealing algorithm coupled with the Metropolis energy test to evaluate successive random changes in position, orientation, and conformation of the ligand. We typically ran 100 independent docking runs for each compound and analyzed the 10 conformations with the lowest calculated energies.
In all docking runs, the charge state of ionizable groups was chosen to be consistent with the acidic conditions (pH 4.4) used for the plasmepsin assay. Carboxyl groups were assigned a negative charge, except for the active site aspartic acid residues (D34 and D214), which were adjusted to have a combined charge of −1 electron. Bonds in the ligand were allowed rotational freedom during docking runs, except for the N-C bonds of the urea moiety, which were frozen in a planar conformation. Although the benzamidine moiety found in WR268961 has a preference for near-planar conformations (10 to 35°), the bond between the benzene ring and the amidine was allowed to rotate during docking.
RESULTS
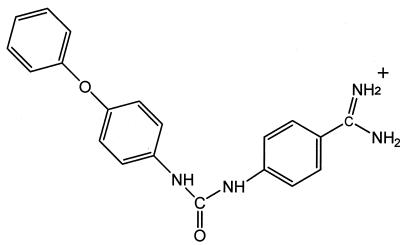
We selected 168 compounds from the Walter Reed chemical inventory that were designed as protease inhibitors, but were structurally dissimilar, and tested them in our P. vivax plasmepsin prescreen assay. One compound, WR268961, significantly inhibited plasmepsin activity at a concentration of 10 μg/ml. The Ki values of this compound against P. vivax plasmepsin and against P. falciparum plasmepsin II are similar: 1.2 and 6.1 μM, respectively. WR268961 is a small (346 Da), nonpeptidyl compound with relatively high solubility in aqueous solution. Structurally, it belongs to a class of compounds containing a diphenylurea moiety [1-(4-amidinophenyl)-3-(4-phenoxyphenyl) urea] (Fig. 1).
FIG. 1.
Molecular structure of Walter Reed compound WR268961.
We tested WR268961 to see if it could inhibit the growth of P. falciparum. Our in vitro drug susceptibility assay showed that WR268961 significantly abolished parasite proliferation, with IC50 values ranging from 0.03 (chloroquine-resistant W2 strain) to 0.16 (chloroquine-sensitive D6-strain) μg/ml (Table 1). We also tested WR268961 for general toxicity to mammalian cells. Cultured neuronal cells and macrophages were 25 to 100 times less sensitive to WR268961 than the parasites (Table 1). One concern about protease inhibitors is that they often inhibit closely related mammalian proteases, such as cathepsin D, as well as the intended target protease. We tested human liver cathepsin D and found that WR268961 displayed almost no inhibition (Table 1).
TABLE 1.
Plasmepsin inhibition, antiparasitic activity, mammalian cell toxicity, and protease selectivity of Walter Reed compound WR268961
| Assay and target enzyme or cell type | IC50 (μg/ml) | Ki (μM) |
|---|---|---|
| Plasmepsin inhibition | ||
| P. falciparum | 5.8 | 6.1 |
| P. vivax | 2.5 | 1.2 |
| P. falciparum drug susceptibility | ||
| Strain W2 | 0.03 | NAa |
| Strain D6 | 0.16 | NA |
| Mammalian cell toxicity | ||
| Neuronal cells (NG-108-15) | 4 | NA |
| Macrophages (J774) | 4 | NA |
| Protease selectivity | ||
| Human cathepsin D | >450 | >280 |
NA, not applicable.
Malaria parasites display distinct morphological changes when hemoglobin digestion is blocked. Rosenthal (18) showed that treatment of P. falciparum parasites with the falcipain inhibitor E-64 (E-64 inhibits the hemoglobin-digesting enzyme falcipain, but not plasmepsins) resulted in enlargement of parasite food vacuoles and the accumulation of undigested hemoglobin in them. In a similar experiment, we incubated ring-stage P. falciparum parasites overnight with WR268961 (plasmepsin inhibitor), with E-64 (falcipain inhibitor), or with no drug. Figure 2 shows representative micrographs of Giemsa-stained schizont-stage parasite smears taken 12 h later. Parasites incubated with WR268961 or with E-64 displayed abnormally enlarged food vacuoles relative to the control with no drug (Fig. 2, top). Morphological abnormalities were even more pronounced during erythrocytic rupture (Fig. 2, bottom). The inhibitor-treated parasites were isolated from infected erythrocytes and resolved by SDS-PAGE to evaluate the accumulation of intact hemoglobin. As shown in Fig. 3, the E-64-treated parasites contain more undigested hemoglobin than the WR268961-treated parasites. The low level of intact hemoglobin in the WR268961-treated parasites may indicate that WR268961 does not inhibit the initial step of hemoglobin degradation, but instead blocks the further processing of partially digested hemoglobin.
FIG. 2.
Micrographs of Giemsa-stained schizont-stage parasites. The top panel shows abnormally enlarged food vacuoles in inhibitor-treated parasites. The lower panel shows even more pronounced morphological abnormalities during erythrocytic rupture.
FIG. 3.
Parasite food vacuole contents visualized by Coomassie-stained SDS-PAGE. Intact hemoglobin accumulates in parasites treated with the falcipain inhibitor E-64, but not in parasites treated with WR268961.
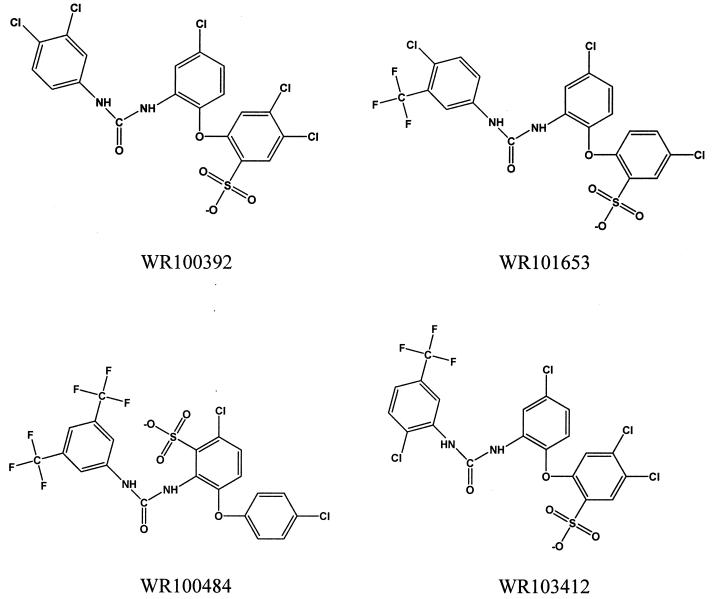
The Walter Reed chemical database contains hundreds of thousands of compounds. We searched the database for compounds that contain the core diphenylurea found in WR268961 and identified 1,508 compounds. We assayed 346 of these diphenylurea-containing compounds in our automated P. vivax plasmepsin assay. Nine of these compounds inhibit plasmepsin with an IC50 lower than 5 μg/ml (Table 2). Interestingly, all nine compounds contain a phenoxyl group similar to the one found in WR268961; four of the nine compounds have an ortho-phenoxyl group (Fig. 4), while the others have a para-phenoxyl group (Fig. 5). This finding suggests that diphenylurea with a phenoxyl side chain may be the functional structure that is responsible for plasmepsin inhibition. Unlike WR268961, all nine plasmepsin inhibitors contain an acidic sulfonic acid group instead of the basic amidine group found in WR268961.
TABLE 2.
IC50 values and Ki values for diphenylurea derivatives containing a sulfonic acid group
| Walter Reed compound | Plasmepsin inhibition (Ki [μM])
|
IC50 (μg/ml)
|
Protease selectivity (Ki [μM]) | ||||
|---|---|---|---|---|---|---|---|
|
P. falciparum drug susceptibility
|
Mammalian cell toxicity
|
||||||
| P. falciparum | P. vivax | Strain W2 | Strain D6 | Neuronal cells | Macrophage cells | Human cathepsin D | |
| WR100392 | 0.09 | 0.08 | 16 | 30 | 15 | 10 | 3.5 |
| WR101653 | 0.12 | 0.43 | 20 | 21 | 78 | 65 | 35 |
| WR100484 | 0.20 | 0.05 | 6.6 | 7.1 | 43 | 39 | 3.3 |
| WR103412 | 0.05 | 0.25 | 15 | 20 | 36 | 22 | 83 |
| WR100081 | 0.18 | 0.08 | 6.5 | 13 | 27 | 72 | 26 |
| WR099444 | 0.68 | 0.15 | 16 | 25 | 30 | >100 | 16 |
| WR099445 | 0.19 | 0.27 | 20 | 26 | 73 | >100 | 191 |
| WR100080 | 0.06 | 0.06 | 16 | 23 | 6.2 | 4.0 | 2.7 |
| WR100400 | 0.11 | 0.59 | 21 | 20 | 27 | 43 | 157 |
FIG. 4.
Structures of active ortho-phenoxyl diphenylurea compounds.
FIG. 5.
Structures of active para-phenoxyl diphenylurea compounds.
We tested the nine new inhibitors in our in vitro drug susceptibility assay to determine whether they were capable of interrupting parasite growth. Despite their ability to inhibit plasmepsin, all of these compounds displayed weak potency in blocking P. falciparum growth, with IC50 values greater than 6 μg/ml (Table 2). The nine compounds have similar IC50 values when tested on neuronal cells and macrophages, demonstrating a lack of specificity between parasite and mammalian cells (Table 2). Interestingly, these nine WR268961 analogues inhibit the plasmepsins 17 to 1,000 times better than human cathepsin D, demonstrating specificity between parasite and mammalian proteases (Table 2).
We conducted a series of docking experiments to understand how our compounds inhibit plasmepsin. We docked the diphenylurea core found in our inhibitors into the crystal structures of P. falciparum plasmepsin II (20) and P. vivax plasmepsin (Protein Data Bank ID 1QS8) by using the program Autodock3 (8, 8a). Only one low-energy conformation of diphenylurea was found despite whether P. falciparum or P. vivax plasmepsin was used as the docking target. Diphenylurea mimics the core region (the statine residue) of the peptidyl inhibitor pepstatin observed in the crystal structures of both plasmepsins. Figure 6 compares the crystal structure of bound pepstatin (Fig. 6A) to that in the model of the docked diphenylurea (Fig. 6B).
FIG. 6.
Crystal structure of pepstatin bound to plasmepsin (A) compared with the model of diphenylurea bound to plasmepsin (B). The compounds are colored by atom type (carbon is gray, oxygen is red, and nitrogen is blue), and the plasmepsin enzyme is represented by a molecular surface that is colored based on the surface potential (red is negative and blue is positive). Diphenylurea mimics the core region (the statine residue) of pepstatin.
We also docked WR268961into the plasmepsin active site and found that the diphenylurea core of WR268961 binds in the same manner as we observed with diphenylurea alone. Figure 7 compares the bound conformation of pepstatin observed in the crystal structure (Fig. 7A) with the conformation of WR268961 determined by docking (Fig. 7B). WR268961 mimics the hydrogen bonds formed by pepstatin with G78 and G36 (Fig. 7), but WR268961 only forms one hydrogen bond to the active site aspartic acid residues (D34 and D214) instead of the two possible hydrogen bonds formed by pepstatin with these residues. This binding mode potentially leaves space for the catalytic water molecule, displaced by the statine hydroxyl upon pepstatin binding, which is often found between the active site aspartic acid residues in aspartic proteases.
FIG. 7.
Crystal structure of pepstatin bound to plasmepsin (A) compared with the model of WR268961 bound to plasmepsin (B). The compounds are colored by atom type, and the main chain of the plasmepsin enzyme is represented by a semitransparent yellow worm. The side chains of four plasmepsin residues involved in hydrogen bonds (purple dashed lines) are colored by atom type.
The WR268961 phenoxyl side chain occupies an active site pocket that is not involved in pepstatin binding. Interestingly, the basic amidine group of WR268961 was not found to interact with the acidic active site aspartic acid residues (D34 and D214) and may be more important for solubility. In the bound conformation of WR268961, the amidine group does not contact any plasmepsin atoms (distance less than 3.5 Å) when rotated through the likely range of benzamidine dihedral angles (10 to 35°).
DISCUSSION
The hemoglobin degradation enzymes of malaria parasites have been validated as targets for antimalarial drug development and discovery. Of these enzymes, only the aspartic proteases, called plasmepsins, have been produced recombinantly and structurally characterized. Several peptide-like compounds, such as pepstatin, SC-50083, and Ro40–4388, display potent inhibition of plasmepsins, but they are much less effective at inhibiting parasite growth, perhaps due to the difficulty these large compounds have gaining access to the parasitic food vacuoles in which hemoglobin degradation takes place. We discovered a small nonpeptidyl compound, WR268961, that inhibits P. vivax and P. falciparum plasmepsins as well as inhibiting P. falciparum growth in vitro at concentrations between 0.03 and 0.16 μg/ml. WR268961 is a small compound with relatively high aqueous solubility that appears to be an excellent lead for the design of more potent antiparasitic compounds. Preliminary data indicate that WR268961 is specific for plasmepsins versus mammalian aspartic proteases and is specific for malaria parasites versus mammalian cells.
Malaria parasites cultured in vitro display distinct morphological changes after incubation with WR268961. The parasite food vacuoles become abnormally enlarged in schizont-stage parasites, and even more pronounced abnormalities are evident during erythrocyte rupture (Fig. 2). Similar morphological changes are seen in parasites incubated with E-64, an inhibitor of the hemoglobin-degrading cysteine protease falcipain (Fig. 2). However, when the protein contents of parasites incubated with WR268961 are compared by SDS-PAGE with the contents of parasites incubated with E-64, a distinct difference can be seen. Parasites treated with WR268961 do not seem to accumulate intact hemoglobin as do parasites treated with E-64 (Fig. 3). Instead, WR268961-treated parasites may contain partially digested hemoglobin fragments that are not further processed due to plasmepsin inhibition. This result suggests that E-64 inhibits the initial step of hemoglobin digestion, while WR268961 inhibits subsequent steps. Currently there is a lack of consensus on which hemoglobinase initiates hemoglobin degradation, with some evidence pointing to plasmepsins (5–7) and some evidence pointing to falcipain (1, 15, 18).
We found nine compounds from the Walter Reed chemical inventory that are similar in structure to WR268961 and that also inhibit P. falciparum in our in vitro drug susceptibility assay. These compounds all contain a diphenylurea core with a phenoxyl side chain (Fig. 4 and 5), and they all inhibit P. falciparum plasmepsin II and P. vivax plasmepsin, with Ki values between 0.05 and 0.68 μM (Table 2). These compounds seem to be specific for plasmepsins versus mammalian aspartic proteases, but they do not seem to be specific for malaria parasites versus mammalian cells (Table 2). More importantly, these compounds are not potent inhibitors of malaria parasite growth and have IC50 values about 100 times greater than that of WR268961. Although these compounds contain the phenoxyl diphenylurea structure found in WR268961, they differ from WR268961 in net charge. All nine compounds contain an acidic sulfonic acid group, whereas WR268961 contains a basic amidine group. Docking experiments suggest that the diphenylurea core of WR268961 interacts specifically with the plasmepsin active site, whereas the amidine group does not. Instead, the amidine group of WR268961 and the sulfonic acid groups of the other nine inhibitors may be more important for solubility. The pKa values of the plasmepsin inhibitors may have an impact on the in vitro inhibition of parasite growth. Although all of our compounds inhibit plasmepsin with similar values, only the basic WR268961 is a potent inhibitor of parasite growth. This result may make sense in light of the fact that several successful antimalarials contain basic chemical groups.
REFERENCES
- 1.Bailly E, Jambou R, Savel J, Jaureguiberry G. Plasmodium falciparum: differential sensitivity in vitro to E-64 (cysteine protease inhibitor) and pepstatin A (aspartyl protease inhibitor) J Protozool. 1992;39:593–599. doi: 10.1111/j.1550-7408.1992.tb04856.x. [DOI] [PubMed] [Google Scholar]
- 1a.Carroll C D, Orlowski M. Screening aspartyl proteases with combinatorial libraries. Adv Exp Med Biol. 1998;436:375–380. doi: 10.1007/978-1-4615-5373-1_52. [DOI] [PubMed] [Google Scholar]
- 2.Carroll C D, Johnson T O, Tao S, Lauri G, Orlowski M, Gluzman I Y, Goldberg D E, Dolle R E. Evaluation of a structure-based statine cyclic diamino amide encoded combinatorial library against plasmepsin II and cathepsin D. Bioorg Med Chem Lett. 1998;8:3203–3206. doi: 10.1016/s0960-894x(98)00554-x. [DOI] [PubMed] [Google Scholar]
- 3.Dame J B, Reddy G R, Yowell C A, Dunn B M, Kay J, Berry C. Sequence, expression and modeled structure of an aspartic proteinase from the human malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 1994;64:177–190. doi: 10.1016/0166-6851(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 4.Desjardins R E, Canfield C J, Haynes J D, Chulay J D. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob Agents Chemother. 1979;16:710–718. doi: 10.1128/aac.16.6.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francis S E, Gluzman I Y, Oksman A, Knickerbocker A, Mueller R, Bryant M L, Sherman D R, Russell D G, Goldberg D E. Molecular characterization and inhibition of Plasmodium falciparum aspartic hemoglobinase. EMBO J. 1994;13:306–317. doi: 10.1002/j.1460-2075.1994.tb06263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gluzman I Y, Francis S E, Oksman A, Smith C E, Duffin K L, Goldberg D E. Order and specificity of the Plasmodium falciparum hemoglobin degradation pathway. J Clin Investig. 1994;93:1602–1608. doi: 10.1172/JCI117140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldberg D E, Andrew A F G, Cerami A, Henderson G B. Hemoglobin degradation in the malaria parasite Plasmodium falciparum: an ordered process in a unique organelle. Proc Natl Acad Sci USA. 1990;87:2931–2935. doi: 10.1073/pnas.87.8.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodsell D S, Olson A J. Automated docking of substrates to proteins by simulated annealing. Proteins. 1990;8:195–202. doi: 10.1002/prot.340080302. [DOI] [PubMed] [Google Scholar]
- 8a.Goodsell D S, Morris G M, Olson A J. Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 9.Haque T S, Skillman A G, Lee C E, Habashita H, Gluzman I Y, Ewing T J, Goldberg D E, Kuntz I D, Ellman J A. Potent, low-molecular-weight non-peptide inhibitors of malarial aspartyl protease plasmepsin II. J Med Chem. 1999;42:1428–1440. doi: 10.1021/jm980641t. [DOI] [PubMed] [Google Scholar]
- 10.Hill J, Tyas L, Phylip L H, Kay J, Dunn B M, Berry C. High level expression and characterization of plasmepsin II, an aspartic proteinase from Plasmodium falciparum. FEBS Lett. 1994;32:155–158. doi: 10.1016/0014-5793(94)00940-6. [DOI] [PubMed] [Google Scholar]
- 11.Kamchonwongpaisan S, Samoff E, Meshnick S R. Identification of hemoglobin degradation products in Plasmodium falciparum. Mol Biochem Parasitol. 1997;86:179–186. doi: 10.1016/s0166-6851(97)02855-7. [DOI] [PubMed] [Google Scholar]
- 12.Luker K E, Francis S E, Gluzman I Y, Goldberg D E. Kinetic analysis of plasmepsin I and II aspartic proteases of the Plasmodium falciparum digestive vacuole. Mol Biochem Parasitol. 1996;79:71–78. doi: 10.1016/0166-6851(96)02651-5. [DOI] [PubMed] [Google Scholar]
- 13.Matayoshi E D, Wang G T, Krafft G A, Erickson J. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science. 1990;247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- 14.Moon R P, Bur D, Loetscher H, D'Arcy A, Tyas L, Oefner C, Gruenninger-Leitch F, Mona D, Rupp K, Dorn A, Matile H, Certa U, Berry C, Kay J, Ridley R G. Studies on plasmepsin I and II from the malarial parasite Plasmodium falciparum and their exploitation as drug targets. Adv Exp Med Biol. 1998;436:397–406. doi: 10.1007/978-1-4615-5373-1_56. [DOI] [PubMed] [Google Scholar]
- 15.Moon R P, Tyas L, Certa U, Rupp K, Bur D, Jacquet C, Matile H, Loetscher H, Grueninger-Leitch F, Kay J, Dunn B M, Berry C, Ridley R G. Expression and characterization of plasmepsin I from Plasmodium falciparum. Eur J Biochem. 1997;244:552–560. doi: 10.1111/j.1432-1033.1997.00552.x. [DOI] [PubMed] [Google Scholar]
- 16.Morris G M, Goodsell D S, Huey R, Olson A J. Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J Comput-Aided Mol Des. 1996;10:293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 17.Roberts F, Roberts C W, Johnson J J, Kyle D E, Krell T, Coggins J R, Coombs G H, Milhous W K, Tzipori S, Ferguson D J, Chakrabarti D, McLeod R. Evidence for the shikimate pathway in apicomplexan parasites. Nature. 1998;393:801–805. doi: 10.1038/31723. [DOI] [PubMed] [Google Scholar]
- 18.Rosenthal P J. Plasmodium falciparum: effects of proteinase inhibitors on globin hydrolysis by cultured malaria parasites. Exp Parasitol. 1995;80:272–281. doi: 10.1006/expr.1995.1033. [DOI] [PubMed] [Google Scholar]
- 19.Rosenthal P J. Proteases of malaria parasites: new targets for chemotherapy. Emerg Infect Dis. 1998;4:49–57. doi: 10.3201/eid0401.980107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva A M, Lee A Y, Gulnik S V, Maier P, Collins J, Bhat T N, Collins P J, Cachau R E, Luker K E, Gluzman I Y, Francis S E, Oksman A, Goldberg D E, Erickson J W. Structure and inhibition of plasmepsin II, a hemoglobin-degrading enzyme from Plasmodium falciparum. Proc Natl Acad Sci USA. 1996;93:10034–10039. doi: 10.1073/pnas.93.19.10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silva A M, Lee A Y, Erickson J W, Goldberg D E. Structural analysis of plasmepsin II. A comparison with human aspartic proteases. Adv Exp Med Biol. 1998;436:363–373. [PubMed] [Google Scholar]
- 22.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren J T, Bokesch H, Kenney S, Boyd M R. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi T, Dehdarani A H, Onezawa S Y, Tang J. Porcine spleen cathepsin B is an exopeptidase. J Biol Chem. 1986;261:9375–9381. [PubMed] [Google Scholar]
- 24.Trager W, Jensen J B. Human malaria parasites in continuous culture. Science. 1976;193:674–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 25.Tyas L R, Moon P, Loetscher H, Dunn B M, Kay J, Ridley R G, Berry C. Plasmepsin I and II from the malarial parasite Plasmodium falciparum. Adv Exp Med Biol. 1998;436:407–411. doi: 10.1007/978-1-4615-5373-1_57. [DOI] [PubMed] [Google Scholar]
- 26.Westling J, Yowell C A, Majer P, Erickson J W, Dame J B, Dunn B M. Plasmodium falciparum, P. vivax, and P. malariae: a comparison of the active site properties of plasmepsins cloned and expressed from three different species of the malaria parasite. Exp Parasitol. 1997;87:185–193. doi: 10.1006/expr.1997.4225. [DOI] [PubMed] [Google Scholar]
- 27.White N J. Drug resistance in malaria. Br Med Bull. 1998;54:703–715. doi: 10.1093/oxfordjournals.bmb.a011721. [DOI] [PubMed] [Google Scholar]
- 28.World Health Organization. Control of tropical diseases (CTD): malaria control. Geneva, Switzerland: World Health Organization Office of Information; 1995. [Google Scholar]