Six piperazinium salts are reported, five of them are hydrated and two crystallized as 2:2 salts. They exhibit asymmetric units of a common 4-nitrophenylpiperazine cation and different p-substituent benzoate anions. Their crystal structures mainly pack as chains stabilized by strong N—H⋯O and O—H⋯O hydrogen bonds and other weak interactions such as C—H⋯O and C—H⋯π.
Keywords: crystal structure, piperazine, benzoate anion, biological activity
Abstract
Six piperazinium salts, namely 4-(4-nitrophenyl)piperazin-1-ium 4-bromobenzoate dihydrate, C10H14N3O2
+·C7H4BrO2
−·2H2O, (I), 4-(4-nitrophenyl)piperazin-1-ium 4-iodobenzoate dihydrate, C10H14N3O2
+·C7H4IO2
−·2H2O, (II), 4-(4-nitrophenyl)piperazin-1-ium 4-hydroxybenzoate monohydrate, C10H14N3O2
+·C7H5O3
−·H2O, (III), 4-(4-nitrophenyl)piperazin-1-ium 4-methylbenzoate monohydrate, C10H14N3O2
+·C8H7O2
−·H2O, (IV), 4-(4-nitrophenyl)piperazin-1-ium 4-methoxybenzoate hemihydrate, 2C10H14N3O2
+·2C8H7O3
−·H2O, (V), and 4-(4-nitrophenyl)piperazin-1-ium 4-ethoxybenzoate, 2C10H14N3O2
+·2C9H9O3
−, (VI), have been synthesized and their crystal structures solved by single-crystal X-ray diffraction, revealing that all of them crystallize in the triclinic space group P
 except for (V), which crystallizes in the monoclinic space group P21/c and has a disordered nitro group. Compounds (I) and (II) are isostructural. The crystal packing of (I)–(V) is constructed from organic chains formed by a combination of hydrogen bonds of type N—H⋯O and/or O—H⋯O and other weak interactions of type C—H⋯O and/or C—H⋯π, forming sheets, whereas (VI) shows a cationic and anionic-based layer structure.
except for (V), which crystallizes in the monoclinic space group P21/c and has a disordered nitro group. Compounds (I) and (II) are isostructural. The crystal packing of (I)–(V) is constructed from organic chains formed by a combination of hydrogen bonds of type N—H⋯O and/or O—H⋯O and other weak interactions of type C—H⋯O and/or C—H⋯π, forming sheets, whereas (VI) shows a cationic and anionic-based layer structure.
1. Chemical context
Piperazines and substituted piperazines are important pharmacophores that can be found in many biologically active compounds used to treat a number of different diseases (Berkheij, 2005 ▸) as antifungal (Upadhayaya et al., 2004 ▸), anti-bacterial, anti-malarial and anti-psychotic agents (Choudhary et al., 2006 ▸). A valuable insight into advances on the antimicrobial activity of piperazine derivatives was given by Kharb et al. (2012 ▸). Piperazines are among the most important building blocks in current drug discovery and are found in biologically active compounds across a number of different therapeutic areas (Brockunier et al., 2004 ▸; Bogatcheva et al., 2006 ▸). Pharmacological and toxicological information for piperazine derivatives is reviewed by Elliott (2011 ▸).
4-Nitrophenylpiperazinium chloride monohydrate has been used as an intermediate in the synthesis of anticancer drugs, transcriptase inhibitors and antifungal reagents and is also an important reagent for potassium channel openers, which show considerable biomolecular current-voltage rectification characteristics (Lu, 2007 ▸). The inclusion behaviours of 4-sulfonatocalix[n]arenes (SCXn) (n = 4, 6, 8) with 1-(4-nitrophenyl)piperazine (NPP) were investigated by UV spectroscopy and fluorescence spectroscopy at different pH values (Zhang et al., 2014 ▸). The design, synthesis and biological profiling of arylpiperazine-based scaffolds for the management of androgen-sensitive prostatic disorders was reported by Gupta et al. (2016 ▸). 4-Nitrophenylpiperazine was the starting material in the synthesis and biological evaluation of novel piperazine-containing hydrazone derivatives (Kaya et al., 2016 ▸). The crystal structure of 4-nitrophenyl piperazinium chloride monohydrate was reported by Lu (2007 ▸) and that of 4,6-dimethoxypyrimidin-2-amine-1-(4-nitrophenyl)piperazine (1:1) by Wang et al. (2014 ▸) while Ayeni et al. (2019 ▸) described the synthesis and crystal structure of a Schiff base, 5-methyl-2-{[4-(4-nitrophenyl)piperazin-1-yl]methyl}phenol is published. NMR-based investigations of acyl-functionalized piperazines concerning their conformational behaviour in solution has been studied and the crystal structures of 1-(4-fluorobenzoyl)-4-(4-nitrophenyl)piperazine, 1-(4-bromobenzoyl)-4-(4-nitrophenyl)piperazine and 1-(3-bromobenzoyl)-4-(4-nitrophenyl)piperazine have been reported (Wodtke et al., 2018 ▸). We have recently reported the crystal structures of some salts of 4-methoxyphenylpiperazine (Kiran Kumar et al., 2019 ▸) and also 2-methoxyphenylpiperazine (Harish Chinthal et al., 2020 ▸), as well as some salts of piperazine derivatives (Archana et al., 2021 ▸).
In view of the importance of piperazines in general and the use of 4-nitrophenylpiperazine in particular, the present paper reports the crystal structures of some salts of 4-nitrophenylpiperazine with organic acids. The crystal structures of 4-(4-nitrophenyl)piperazin-1-ium 4-bromobenzoate dihydrate (I), 4-(4-nitrophenyl)piperazin-1-ium 4-iodobenzoate dihydrate (II), 4-(4-nitrophenyl)piperazin-1-ium 4-hydroxybenzoate monohydrate (III), 4-(4-nitrophenyl)piperazin-1-ium 4-methylbenzoate monohydrate (IV), 4-(4-nitrophenyl)piperazin-1-ium 4-methoxybenzoate hemihydrate (V) and 4-(4-nitrophenyl)piperazin-1-ium 4-ethoxybenzoate (VI) are reported herein.
2. Structural commentary
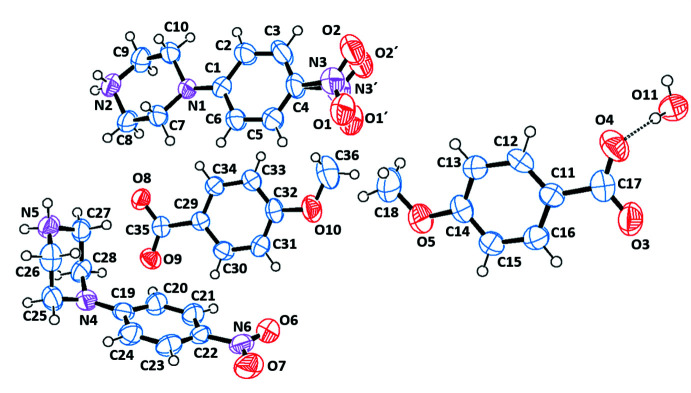
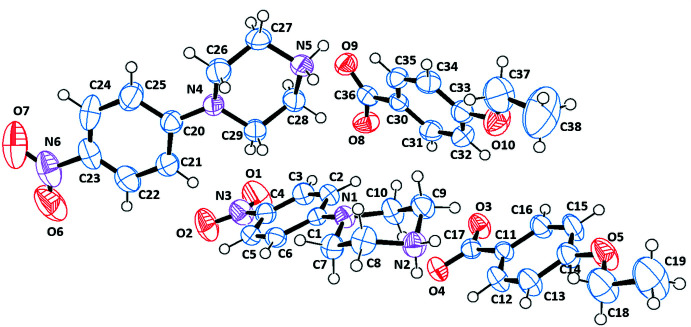
The asymmetric units of the title salts are shown in Figs. 1 ▸–6 ▸ ▸ ▸ ▸ ▸. They include 1:1 dihydrated salts [(I), (II)], 1:1 monohydrated salts [(III), (IV)], 2:2 monohydrated salt (V) and solvent-free 2:2 salt (VI). Compounds (I) and (II) are isostructural. In all salts, the cation is common and consists of a protonated chair-shaped piperazine ring (N1/N2/C7–C10), which makes dihedral angles of 10.91 (1), 12.13 (1), 14.82 (6), 3.11 (8), 5.73 (1) and 13.08 (9)°, respectively, for compounds (I)–(VI) with the nitrobenzene moiety (N3/O1/O2/C1–C6) and exhibits a maximum deviation from its mean plane at atom N2 of −0.253 (2), 0.254 (2), 0.288 (2), 0.278 (2), 0.241 (3) and 0.303 (3) Å in (I)–(VI), respectively. The piperazine rings of the additional cations (N4/N5/C25–C28) in compounds (V) and (VI) have the same conformation, making dihedral angles of 64.53 (1) and 21.70 (1)°, respectively, with the nitrobenzene moieties (N6/O6/O7/C19–C25). Within the cations, the benzene rings are almost planar, with maximum deviations from mean plane ranging from −0.016 (3) Å at atom C20 for (VI) to 0.003 (2) Å at atom C4 for (III). The p-nitro substituent groups deviate significantly from planes of the benzene rings in all compounds except the (C1–C6) ring of (VI). The anions of the title salts are formed from a benzoate anion with different p-substituents for each compound that deviate significantly from planarity, with maximum deviations of 0.045 (1) Å at Br1 for (I), 0.063 (1) Å at I1 for (II), −0.021 (2) Å at hydroxyl atom O3 for (III), −0.010 (1) Å at methyl atom C18 for (IV), −0.033 (1) and 0.034 (1) Å at methoxy atoms O5 and O10 for (V) and −0.025 (2) and −0.013 (2) Å at ethoxy atoms O5 and O10 for (VI).
Figure 1.
The independent components of compound (I) showing the atom-labelling scheme and the hydrogen bonds, drawn as dashed lines. Displacement ellipsoids are drawn at the 50% probability level.
Figure 2.
The independent components of compound (II) showing the atom-labelling scheme and the hydrogen bonds, drawn as dashed lines. Displacement ellipsoids are drawn at the 50% probability level.
Figure 3.
The independent components of compound (III) showing the atom-labelling scheme and the hydrogen bonds, drawn as dashed lines. Displacement ellipsoids are drawn at the 50% probability level.
Figure 4.
The independent components of compound (IV) showing the atom-labelling scheme and the hydrogen bonds, drawn as dashed lines. Displacement ellipsoids are drawn at the 50% probability level.
Figure 5.
The independent components of compound (V) showing the atom-labelling scheme and the hydrogen bonds, drawn as dashed lines. Displacement ellipsoids are drawn at the 50% probability level.
Figure 6.
The independent components of compound (VI) showing the atom-labelling scheme Displacement ellipsoids are drawn at the 50% probability level.
3. Supramolecular features
In the crystal structures of the two isomorphous salts (I) and (II), the ions are arranged in chains perpendicular to the a-axis direction. The water molecules play an essential role in holding the chains together, forming complex sheets in the bc plane (Figs. 7 ▸ and 8 ▸, Tables 1 ▸ and 2 ▸). The cations and anions in (III) are linked through strong O—H⋯O and N—H⋯O hydrogen bonds, forming chains along the [011] direction (Fig. 9 ▸ a, Table 3 ▸). These chains are further linked via the water molecules and C9—H9A⋯O3 interactions, generating a three-dimensional supramolecular architecture along the a axis (Fig. 9 ▸ b). The structure of (IV) is constructed from double chains running along the [101] direction. Each chain is formed by linking the molecules through a combination of N—H⋯O, O—H⋯O and C—H⋯O interactions (Fig. 10 ▸ a, Table 4 ▸); the resulting double chains are symmetrically related by an inversion center and are connected via N2—H21⋯O4 and C7—H7A⋯O4 interactions. These hydrated double chains are weakly linked into sheets lying in the bc plane by C—H⋯π (arene) interactions (Fig. 10 ▸ b). The supramolecular assembly of compound (V), which has a disordered nitro group, is built up of N2—H22N⋯O11, O11—H11O⋯O4 and N5—H51⋯O9 hydrogen bonds linking the ions into organic chains running parallel to the [010] direction (Fig. 11 ▸ a, Table 5 ▸). The chains are further connected cooperatively through other interactions of type N—H⋯O, generating a multilayer network along the b-axis direction (Fig. 11 ▸ b). In compound (VI), a set of N—H⋯O, C—H⋯O and C—H⋯π interactions (Fig. 12 ▸ a, Table 6 ▸) link the molecules into cationic and anionic layers running parallel to the b-axis direction and join these layer motifs, generating the complete molecular structure along the a axis (Fig. 12 ▸ b).
Figure 7.
(a) A general view of the main intermolecular interactions (N—H⋯O and O—H⋯O) and (b) the molecular packing of (I) with hydrogen bonds shown as dashed lines.
Figure 8.
(a) A general view of the main intermolecular interactions (N—H⋯O and O—H⋯O) in (II) and (b) the molecular packing of (II) with hydrogen bonds shown as dashed lines.
Table 1. Hydrogen-bond geometry (Å, °) for (I) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H21⋯O5i | 0.85 (2) | 1.99 (2) | 2.810 (3) | 162 (3) |
| N2—H22⋯O6ii | 0.83 (2) | 1.91 (2) | 2.707 (3) | 160 (3) |
| C3—H3⋯O1iii | 0.93 | 2.59 | 3.260 (4) | 130 |
| C13—H13⋯O4iv | 0.93 | 2.57 | 3.483 (3) | 166 |
| C15—H15⋯O2v | 0.93 | 2.47 | 3.269 (4) | 144 |
| O5—H1W⋯O3vi | 0.80 (2) | 1.97 (2) | 2.759 (2) | 169 (3) |
| O5—H2W⋯O3i | 0.80 (2) | 2.00 (2) | 2.772 (2) | 161 (3) |
| O6—H4W⋯O4 | 0.82 (2) | 2.03 (2) | 2.832 (3) | 166 (3) |
| O6—H3W⋯O4vii | 0.78 (2) | 1.99 (2) | 2.760 (3) | 169 (3) |
Symmetry codes: (i)
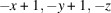 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
 ; (v)
; (v)
 ; (vi)
; (vi)
 ; (vii)
; (vii)
 .
.
Table 2. Hydrogen-bond geometry (Å, °) for (II) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H21⋯O5i | 0.86 (2) | 1.99 (2) | 2.825 (4) | 164 (4) |
| N2—H22⋯O6ii | 0.85 (2) | 1.88 (2) | 2.702 (3) | 163 (4) |
| C3—H3⋯O1iii | 0.93 | 2.59 | 3.275 (4) | 131 |
| C13—H13⋯O4iv | 0.93 | 2.62 | 3.526 (4) | 166 |
| C15—H15⋯O2v | 0.93 | 2.49 | 3.311 (4) | 147 |
| O5—H1W⋯O3vi | 0.81 (2) | 1.96 (2) | 2.756 (3) | 170 (4) |
| O5—H2W⋯O3i | 0.81 (2) | 1.96 (2) | 2.753 (3) | 166 (4) |
| O6—H4W⋯O4 | 0.80 (2) | 2.08 (2) | 2.836 (3) | 160 (4) |
| O6—H3W⋯O4vii | 0.80 (2) | 1.95 (2) | 2.728 (3) | 165 (4) |
Symmetry codes: (i)
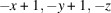 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
 ; (v)
; (v)
 ; (vi)
; (vi)
 ; (vii)
; (vii)
 .
.
Figure 9.
(a) A general view of the main intermolecular interactions (N—H⋯O and O—H⋯O) in (III) and (b) the molecular packing of (III) with hydrogen bonds shown as dashed lines.
Table 3. Hydrogen-bond geometry (Å, °) for (III) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H21⋯O5 | 0.89 (2) | 1.93 (2) | 2.819 (2) | 177 (3) |
| N2—H22⋯O4i | 0.94 (2) | 1.65 (2) | 2.583 (2) | 177 (3) |
| O3—H17⋯O6 | 0.85 (2) | 1.82 (2) | 2.669 (2) | 177 (3) |
| O6—H1W⋯O5ii | 0.83 (2) | 1.95 (2) | 2.768 (2) | 169 (3) |
| O6—H2W⋯O1iii | 0.83 (2) | 2.11 (2) | 2.944 (2) | 178 (3) |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 .
.
Figure 10.
(a) A general view of the main intermolecular interactions (N—H⋯O and O—H⋯O) in (IV) and (b) the molecular packing of (IV) with hydrogen bonds shown as dashed lines.
Table 4. Hydrogen-bond geometry (Å, °) for (IV) .
Cg3 is the centroids of the C11–C16 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H21⋯O4i | 0.89 (2) | 1.93 (2) | 2.811 (3) | 167 (4) |
| N2—H22⋯O3ii | 0.91 (2) | 1.81 (2) | 2.717 (3) | 177 (4) |
| C3—H3⋯O1iii | 0.93 | 2.54 | 3.427 (4) | 161 |
| C9—H9A⋯O5iv | 0.97 | 2.31 | 3.113 (3) | 140 |
| O5—H1W⋯O4i | 0.84 (2) | 1.92 (2) | 2.756 (3) | 171 (4) |
| O5—H2W⋯O3 | 0.85 (2) | 1.94 (2) | 2.772 (3) | 164 (4) |
| C6—H6⋯Cg3v | 0.93 | 2.93 | 3.590 (3) | 129 |
Symmetry codes: (i)
 ; (ii)
; (ii)
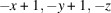 ; (iii)
; (iii)
 ; (iv)
; (iv)
 ; (v)
; (v)
 .
.
Figure 11.
(a) A general view of the main intermolecular interactions (N—H⋯O and O—H⋯O) in (V) and (b) the molecular packing of (V) with hydrogen bonds shown as dashed lines.
Table 5. Hydrogen-bond geometry (Å, °) for (V) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C7—H7B⋯O7i | 0.97 | 2.54 | 3.451 (5) | 157 |
| C9—H9B⋯O4ii | 0.97 | 2.31 | 3.270 (5) | 169 |
| C20—H20⋯O9 | 0.93 | 2.53 | 3.461 (5) | 174 |
| C25—H25A⋯O2aiii | 0.97 | 2.5 | 3.206 (10) | 130 |
| C25—H25A⋯O2′biii | 0.97 | 2.49 | 3.212 (11) | 131 |
| C27—H27A⋯O7i | 0.97 | 2.58 | 3.548 (5) | 175 |
| C28—H28B⋯O9 | 0.97 | 2.55 | 3.489 (5) | 164 |
| C36—H36C⋯O1′bii | 0.96 | 2.49 | 3.395 (14) | 158 |
| N2—H21N⋯O8iv | 0.88 (2) | 1.83 (2) | 2.697 (4) | 166 (4) |
| N2—H21N⋯O9iv | 0.88 (2) | 2.57 (3) | 3.196 (4) | 129 (3) |
| N2—H22N⋯O11v | 0.88 (2) | 1.89 (2) | 2.758 (5) | 169 (4) |
| N5—H51N⋯O9vi | 0.87 (2) | 1.93 (2) | 2.778 (5) | 164 (4) |
| N5—H52N⋯O3vii | 0.91 (2) | 1.82 (2) | 2.724 (5) | 171 (4) |
| O11—H11O⋯O4 | 0.84 (2) | 1.83 (2) | 2.663 (4) | 176 (4) |
| O11—H12O⋯O8v | 0.84 (2) | 1.92 (2) | 2.754 (4) | 173 (4) |
Symmetry codes: (i)
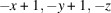 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
 ; (v)
; (v)
 ; (vi)
; (vi)
 ; (vii)
; (vii)
 .
.
Figure 12.
(a) A general view of the main intermolecular interactions (N—H⋯O, O—H⋯O and C—H⋯O) in (VI) and (b) the molecular packing of (VI) with hydrogen bonds shown as dashed lines.
Table 6. Hydrogen-bond geometry (Å, °) for (VI) .
Cg2 and Cg6 are the centroids of the C1–C6 and C30–C35 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H31N⋯O3i | 0.94 (2) | 1.68 (2) | 2.613 (3) | 172 (5) |
| N2—H31N⋯O4i | 0.94 (2) | 2.51 (4) | 3.157 (3) | 127 (4) |
| N2—H32N⋯O9ii | 0.90 (2) | 1.96 (2) | 2.843 (3) | 171 (5) |
| N5—H61N⋯O8i | 0.91 (2) | 1.78 (2) | 2.686 (3) | 175 (5) |
| N5—H61N⋯O9i | 0.91 (2) | 2.59 (4) | 3.174 (3) | 122 (4) |
| N5—H62N⋯O4iii | 0.90 (2) | 1.83 (2) | 2.708 (3) | 165 (5) |
| C22—H22⋯O2iv | 0.93 | 2.6 | 3.502 (5) | 165 |
| C27—H27B⋯O9i | 0.97 | 2.59 | 3.215 (3) | 123 |
| C28—H28B⋯O7v | 0.97 | 2.65 | 3.410 (4) | 135 |
| C29—H29B⋯O1i | 0.97 | 2.53 | 3.249 (4) | 131 |
| C35—H35⋯O4iii | 0.93 | 2.52 | 3.263 (3) | 137 |
| C10—H10A⋯Cg6 | 0.97 | 2.82 | 3.746 (3) | 159 |
| C29—H29A⋯Cg2 | 0.97 | 2.76 | 3.556 (3) | 139 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
 ; (v)
; (v)
 .
.
4. Database survey
A search of the Cambridge Structural Database (Version 2020.3, last update February 2022; Groom et al., 2016 ▸) for the phenyl piperazinium cation and para substituent benzoate anion involved in the reported six salts gave the following hits, 4-(4-methoxyphenyl)piperazin-1-ium 4-fluorobenzoate monohydrate, 4-(4-methoxyphenyl)piperazin-1-ium 4-chlorobenzoate monohydrate and 4-(4-methoxyphenyl)piperazin-1-ium 4-bromobenzoate monohydrate (FOVPOY, FOVPUE and FOVQAL; Kiran Kumar et al., 2019 ▸) and 4-(4-methoxyphenyl)piperazin-1-ium 4-iodobenzoate monohydrate (KUJPUD; Kiran Kumar et al., 2020 ▸). They exhibit a methoxy group as a substituent in the phenyl piperazinium cation rather than a nitro group as in the title compounds (I)–(VI) and they also crystallize as monohydrates similar to compounds (III)–(V). Although the title compounds (I) and (II) have halogen-based anions and chain-based structures, they are not isostructural with the above compounds, the crystal structures of which are based on differently sized chains of rings formed via a combination of hydrogen bonds of type N—H⋯O and O—H⋯O and other weak interactions of type C—H⋯O and C—H⋯π to form sheets. In 4-(4-methoxyphenyl)piperazin-1-ium 4-aminobenzoate monohydrate (IHIMEU; Kiran Kumar et al., 2020 ▸) the presence of an amino substituent on the anion, which acts as both a donor and an acceptor of hydrogen bonds, makes the supramolecular assembly of this compound more complex than for the compounds reported herein.
5. Synthesis and crystallization
Synthesis:
For the synthesis of salts (I)–(VI), a solution of commercially available (from Sigma–Aldrich) 4-nitrophenylpiperazine (100 mg, 0.483 mol) in methanol (10 ml) was mixed with equimolar solutions of the appropriate acids in methanol (10 ml) and ethyl acetate (10 ml), viz.4-bromobenzoic acid (97 mg, 0.483 mol) for (I), 4-iodobenzoic acid (120 mg, 0.483 mol) for (II), 4-hydroxybenzoic acid (67 mg, 0.483 mol) for (III), 4-methylbenzoic acid (66 mg, 0.483 mol) for (IV), 4-methoxybenzoic acid (73 mg, 0.483 mol) for (V) and 4-ethoxybenzoicacid (80 mg, 0.483 mol) for (VI). The corresponding solutions were stirred for 15 minutes at room temperature and allowed to stand at the same temperature. The products obtained were subjected to crystallization.
Crystallization: Crystallization was carried out using the slow evaporation technique. X-ray quality crystals were formed on slow evaporation in a week for all compounds, where ethanol:ethylacetate (1:1) was used for crystallization. The corresponding melting points were 430–432 K (I), 453–455 K (II), 446–448 K (III), 398–400 K (IV), 413–415 K (V) and 408–410 K (VI).
6. Refinement
Crystal data, data collection and refinement details are summarized in Table 7 ▸. C-bound H atoms were positioned with idealized geometry and refined using a riding model with C—H = 0.93 Å (aromatic), 0.96 Å (methyl) or 0.97 Å (methylene). The H atoms on the N atom were located in a difference map and later restrained to N—H = 0.86 (2) Å. All H atoms were refined with isotropic displacement parameters set at 1.2 U eq (C-aromatic, C-methylene, N) or 1.5 U eq (C-methyl) times those of the parent atom. For the disordered nitro group in (V), the component atoms were restrained to have the same Uij components and the occupancy ratio is 0.519 (6):0.481 (6).
Table 7. Experimental details.
| (I) | (II) | (III) | |
|---|---|---|---|
| Crystal data | |||
| Chemical formula | C10H14N3O2 +·C7H4BrO2 −·2H2O | C10H14N3O2 +·C7H4IO2 −·2H2O | C10H14N3O2 +·C7H5O3 −·H2O |
| M r | 444.28 | 491.28 | 363.37 |
| Crystal system, space group | Triclinic, P

|
Triclinic, P

|
Triclinic, P

|
| Temperature (K) | 293 | 293 | 293 |
| a, b, c (Å) | 7.738 (1), 9.320 (1), 13.949 (2) | 7.7652 (4), 9.2852 (5), 13.930 (1) | 9.636 (1), 10.301 (1), 10.867 (1) |
| α, β, γ (°) | 94.46 (1), 95.04 (1), 104.71 (2) | 94.985 (5), 95.331 (5), 104.875 (6) | 103.90 (1), 108.32 (1), 112.96 (1) |
| V (Å3) | 964.0 (2) | 960.09 (10) | 857.80 (17) |
| Z | 2 | 2 | 2 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα |
| μ (mm−1) | 2.17 | 1.71 | 0.11 |
| Crystal size (mm) | 0.48 × 0.44 × 0.24 | 0.48 × 0.48 × 0.2 | 0.50 × 0.32 × 0.24 |
| Data collection | |||
| Diffractometer | Oxford Diffraction Xcalibur | Oxford Diffraction Xcalibur | Oxford Diffraction Xcalibur |
| Absorption correction | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) |
| T min, T max | 0.367, 0.422 | 0.458, 0.711 | 0.959, 0.974 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 6123, 3536, 2520 | 6331, 3518, 2952 | 5342, 3140, 2342 |
| R int | 0.019 | 0.017 | 0.013 |
| (sin θ/λ)max (Å−1) | 0.602 | 0.602 | 0.602 |
| Refinement | |||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.037, 0.104, 1.04 | 0.029, 0.069, 1.03 | 0.043, 0.106, 1.05 |
| No. of reflections | 3528 | 3513 | 3135 |
| No. of parameters | 262 | 262 | 251 |
| No. of restraints | 6 | 6 | 5 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.50, −0.51 | 0.54, −0.66 | 0.19, −0.19 |
| (IV) | (V) | (VI) | |
|---|---|---|---|
| Crystal data | |||
| Chemical formula | C10H14N3O2 +·C8H7O2 −·H2O | 2C10H14N3O2 +·2C8H7O3 −·H2O | C10H14N3O2 +·C9H9O3 − |
| M r | 361.39 | 736.77 | 373.4 |
| Crystal system, space group | Triclinic, P

|
Monoclinic, P21/c | Triclinic, P

|
| Temperature (K) | 293 | 293 | 293 |
| a, b, c (Å) | 6.1136 (5), 7.6965 (7), 19.708 (2) | 15.808 (1), 7.5198 (7), 31.020 (2) | 7.874 (1), 9.263 (1), 27.996 (3) |
| α, β, γ (°) | 79.577 (8), 87.162 (8), 86.699 (8) | 90, 92.561 (7), 90 | 81.030 (6), 85.675 (6), 68.229 (5) |
| V (Å3) | 909.79 (15) | 3683.8 (5) | 1872.8 (4) |
| Z | 2 | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα |
| μ (mm−1) | 0.10 | 0.1 | 0.10 |
| Crystal size (mm) | 0.48 × 0.26 × 0.02 | 0.5 × 0.36 × 0.36 | 0.44 × 0.32 × 0.08 |
| Data collection | |||
| Diffractometer | Oxford Diffraction Xcalibur | Oxford Diffraction Xcalibur | Oxford Diffraction Xcalibur |
| Absorption correction | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) |
| T min, T max | 0.970, 0.998 | 0.958, 0.965 | 0.963, 0.992 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 5980, 3347, 1911 | 15326, 6718, 2602 | 13344, 6868, 3803 |
| R int | 0.019 | 0.066 | 0.027 |
| (sin θ/λ)max (Å−1) | 0.602 | 0.602 | 0.602 |
| Refinement | |||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.053, 0.138, 1.01 | 0.074, 0.169, 1.00 | 0.061, 0.137, 1.05 |
| No. of reflections | 3343 | 6715 | 6858 |
| No. of parameters | 248 | 507 | 501 |
| No. of restraints | 4 | 45 | 16 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.20, −0.16 | 0.27, −0.18 | 0.23, −0.22 |
Supplementary Material
Crystal structure: contains datablock(s) global, I, II, III, IV, V, VI. DOI: 10.1107/S2056989022004157/ex2056sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022004157/ex2056Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989022004157/ex2056IIsup3.hkl
Structure factors: contains datablock(s) III. DOI: 10.1107/S2056989022004157/ex2056IIIsup4.hkl
Structure factors: contains datablock(s) IV. DOI: 10.1107/S2056989022004157/ex2056IVsup5.hkl
Structure factors: contains datablock(s) V. DOI: 10.1107/S2056989022004157/ex2056Vsup6.hkl
Structure factors: contains datablock(s) VI. DOI: 10.1107/S2056989022004157/ex2056VIsup7.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
4-(4-Nitrophenyl)piperazin-1-ium 4-bromobenzoate dihydrate (I). Crystal data
| C10H14N3O2+·C7H4BrO2−·2H2O | Z = 2 |
| Mr = 444.28 | F(000) = 456 |
| Triclinic, P1 | Dx = 1.531 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.738 (1) Å | Cell parameters from 6123 reflections |
| b = 9.320 (1) Å | θ = 3.0–25.3° |
| c = 13.949 (2) Å | µ = 2.17 mm−1 |
| α = 94.46 (1)° | T = 293 K |
| β = 95.04 (1)° | Prism, yellow |
| γ = 104.71 (2)° | 0.48 × 0.44 × 0.24 mm |
| V = 964.0 (2) Å3 |
4-(4-Nitrophenyl)piperazin-1-ium 4-bromobenzoate dihydrate (I). Data collection
| Oxford Diffraction Xcalibur diffractometer | 2520 reflections with I > 2σ(I) |
| ω scans | Rint = 0.019 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | θmax = 25.3°, θmin = 3.0° |
| Tmin = 0.367, Tmax = 0.422 | h = −9→8 |
| 6123 measured reflections | k = −6→11 |
| 3536 independent reflections | l = −16→16 |
4-(4-Nitrophenyl)piperazin-1-ium 4-bromobenzoate dihydrate (I). Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: structure-invariant direct methods |
| R[F2 > 2σ(F2)] = 0.037 | Hydrogen site location: mixed |
| wR(F2) = 0.104 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0623P)2] where P = (Fo2 + 2Fc2)/3 |
| 3528 reflections | (Δ/σ)max < 0.001 |
| 262 parameters | Δρmax = 0.50 e Å−3 |
| 6 restraints | Δρmin = −0.51 e Å−3 |
| 0 constraints |
4-(4-Nitrophenyl)piperazin-1-ium 4-bromobenzoate dihydrate (I). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Nitrophenyl)piperazin-1-ium 4-bromobenzoate dihydrate (I). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.9273 (3) | 0.7891 (3) | 0.53814 (19) | 0.0873 (8) | |
| O2 | 0.6916 (3) | 0.6357 (3) | 0.57409 (17) | 0.0767 (6) | |
| N1 | 0.3360 (2) | 1.0060 (2) | 0.29246 (14) | 0.0392 (5) | |
| N2 | 0.1306 (3) | 1.1025 (2) | 0.13987 (16) | 0.0433 (5) | |
| N3 | 0.7641 (3) | 0.7395 (3) | 0.53118 (17) | 0.0537 (6) | |
| C1 | 0.4412 (3) | 0.9374 (2) | 0.34950 (16) | 0.0353 (5) | |
| C2 | 0.6297 (3) | 0.9791 (3) | 0.35426 (19) | 0.0462 (6) | |
| H2 | 0.685335 | 1.050743 | 0.316027 | 0.055* | |
| C3 | 0.7335 (3) | 0.9160 (3) | 0.4144 (2) | 0.0479 (6) | |
| H3 | 0.858174 | 0.947226 | 0.418128 | 0.057* | |
| C4 | 0.6524 (3) | 0.8067 (3) | 0.46921 (17) | 0.0401 (6) | |
| C5 | 0.4678 (3) | 0.7621 (3) | 0.46618 (18) | 0.0460 (6) | |
| H5 | 0.414057 | 0.688671 | 0.503722 | 0.055* | |
| C6 | 0.3636 (3) | 0.8264 (3) | 0.40761 (18) | 0.0430 (6) | |
| H6 | 0.239197 | 0.796118 | 0.406198 | 0.052* | |
| C7 | 0.4258 (3) | 1.1047 (3) | 0.2239 (2) | 0.0479 (6) | |
| H7A | 0.454966 | 1.044787 | 0.17075 | 0.058* | |
| H7B | 0.537417 | 1.169773 | 0.256358 | 0.058* | |
| C8 | 0.3088 (3) | 1.1978 (3) | 0.1846 (2) | 0.0493 (7) | |
| H8A | 0.29158 | 1.266385 | 0.236558 | 0.059* | |
| H8B | 0.368224 | 1.256083 | 0.136469 | 0.059* | |
| C9 | 0.0406 (3) | 1.0114 (3) | 0.2121 (2) | 0.0495 (6) | |
| H9A | −0.074749 | 0.948618 | 0.182599 | 0.059* | |
| H9B | 0.01919 | 1.076165 | 0.265103 | 0.059* | |
| C10 | 0.1561 (3) | 0.9148 (3) | 0.2506 (2) | 0.0462 (6) | |
| H10A | 0.097528 | 0.858526 | 0.29974 | 0.055* | |
| H10B | 0.168666 | 0.844357 | 0.1984 | 0.055* | |
| Br1 | 0.91659 (5) | 0.45794 (4) | 0.33416 (3) | 0.08360 (18) | |
| O3 | 0.4483 (2) | 0.78822 (19) | 0.01696 (15) | 0.0543 (5) | |
| O4 | 0.2197 (2) | 0.63974 (18) | 0.07787 (14) | 0.0505 (5) | |
| C11 | 0.5153 (3) | 0.6356 (2) | 0.13620 (18) | 0.0350 (5) | |
| C12 | 0.6851 (3) | 0.6376 (3) | 0.10986 (19) | 0.0431 (6) | |
| H12 | 0.71881 | 0.674975 | 0.052451 | 0.052* | |
| C13 | 0.8046 (3) | 0.5846 (3) | 0.1680 (2) | 0.0499 (7) | |
| H13 | 0.916747 | 0.583844 | 0.149142 | 0.06* | |
| C14 | 0.7551 (3) | 0.5332 (3) | 0.2538 (2) | 0.0466 (6) | |
| C15 | 0.5891 (3) | 0.5325 (3) | 0.28271 (19) | 0.0459 (6) | |
| H15 | 0.55828 | 0.499101 | 0.341588 | 0.055* | |
| C16 | 0.4690 (3) | 0.5821 (2) | 0.22315 (19) | 0.0424 (6) | |
| H16 | 0.35572 | 0.579557 | 0.241532 | 0.051* | |
| C17 | 0.3848 (3) | 0.6919 (2) | 0.07250 (19) | 0.0391 (6) | |
| O5 | 0.7579 (2) | 0.01738 (19) | 0.02770 (14) | 0.0468 (4) | |
| O6 | 0.0305 (3) | 0.3352 (2) | 0.06917 (16) | 0.0581 (5) | |
| H21 | 0.143 (4) | 1.051 (3) | 0.0892 (17) | 0.07* | |
| H22 | 0.075 (4) | 1.160 (3) | 0.118 (2) | 0.07* | |
| H1W | 0.674 (3) | −0.053 (3) | 0.031 (2) | 0.07* | |
| H2W | 0.720 (4) | 0.088 (3) | 0.018 (2) | 0.07* | |
| H4W | 0.095 (4) | 0.421 (2) | 0.080 (2) | 0.07* | |
| H3W | −0.041 (4) | 0.331 (3) | 0.0257 (18) | 0.07* |
4-(4-Nitrophenyl)piperazin-1-ium 4-bromobenzoate dihydrate (I). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0490 (13) | 0.0892 (16) | 0.122 (2) | 0.0142 (11) | −0.0189 (13) | 0.0497 (15) |
| O2 | 0.0701 (14) | 0.0858 (15) | 0.0809 (16) | 0.0235 (12) | −0.0004 (12) | 0.0500 (13) |
| N1 | 0.0347 (10) | 0.0406 (11) | 0.0417 (11) | 0.0091 (8) | −0.0042 (9) | 0.0124 (9) |
| N2 | 0.0459 (12) | 0.0431 (12) | 0.0435 (13) | 0.0189 (10) | −0.0065 (10) | 0.0088 (10) |
| N3 | 0.0542 (15) | 0.0524 (13) | 0.0551 (14) | 0.0173 (11) | −0.0090 (12) | 0.0161 (11) |
| C1 | 0.0396 (13) | 0.0344 (12) | 0.0327 (13) | 0.0132 (10) | −0.0010 (10) | 0.0026 (10) |
| C2 | 0.0378 (14) | 0.0518 (15) | 0.0515 (16) | 0.0111 (11) | 0.0047 (12) | 0.0229 (12) |
| C3 | 0.0361 (13) | 0.0503 (15) | 0.0583 (17) | 0.0125 (11) | −0.0001 (12) | 0.0145 (13) |
| C4 | 0.0441 (14) | 0.0411 (13) | 0.0363 (13) | 0.0156 (11) | −0.0037 (11) | 0.0060 (11) |
| C5 | 0.0489 (15) | 0.0463 (14) | 0.0429 (15) | 0.0100 (11) | 0.0016 (12) | 0.0177 (11) |
| C6 | 0.0339 (12) | 0.0498 (14) | 0.0455 (15) | 0.0093 (11) | 0.0025 (11) | 0.0145 (12) |
| C7 | 0.0416 (14) | 0.0450 (14) | 0.0524 (16) | 0.0036 (11) | −0.0082 (12) | 0.0166 (12) |
| C8 | 0.0558 (16) | 0.0385 (13) | 0.0499 (16) | 0.0096 (12) | −0.0116 (13) | 0.0117 (11) |
| C9 | 0.0392 (14) | 0.0603 (16) | 0.0517 (16) | 0.0187 (12) | −0.0040 (12) | 0.0138 (13) |
| C10 | 0.0354 (13) | 0.0487 (14) | 0.0530 (16) | 0.0077 (11) | −0.0030 (12) | 0.0159 (12) |
| Br1 | 0.0734 (3) | 0.0942 (3) | 0.0856 (3) | 0.0319 (2) | −0.02375 (19) | 0.0272 (2) |
| O3 | 0.0418 (10) | 0.0434 (10) | 0.0808 (13) | 0.0119 (8) | 0.0005 (9) | 0.0298 (9) |
| O4 | 0.0291 (9) | 0.0497 (10) | 0.0727 (13) | 0.0090 (7) | 0.0003 (8) | 0.0169 (9) |
| C11 | 0.0330 (12) | 0.0242 (11) | 0.0468 (14) | 0.0078 (9) | −0.0007 (11) | 0.0021 (10) |
| C12 | 0.0403 (14) | 0.0432 (13) | 0.0504 (16) | 0.0160 (11) | 0.0087 (12) | 0.0125 (11) |
| C13 | 0.0345 (13) | 0.0548 (15) | 0.0659 (19) | 0.0194 (11) | 0.0057 (13) | 0.0145 (14) |
| C14 | 0.0443 (15) | 0.0381 (13) | 0.0548 (17) | 0.0096 (11) | −0.0109 (13) | 0.0102 (12) |
| C15 | 0.0500 (16) | 0.0433 (14) | 0.0403 (15) | 0.0053 (11) | −0.0007 (12) | 0.0082 (11) |
| C16 | 0.0376 (13) | 0.0378 (13) | 0.0499 (16) | 0.0068 (10) | 0.0051 (12) | 0.0034 (11) |
| C17 | 0.0375 (13) | 0.0266 (11) | 0.0533 (16) | 0.0108 (10) | −0.0010 (11) | 0.0032 (11) |
| O5 | 0.0370 (10) | 0.0413 (10) | 0.0619 (12) | 0.0103 (7) | −0.0023 (9) | 0.0133 (9) |
| O6 | 0.0491 (12) | 0.0429 (10) | 0.0816 (15) | 0.0141 (8) | −0.0142 (10) | 0.0195 (10) |
4-(4-Nitrophenyl)piperazin-1-ium 4-bromobenzoate dihydrate (I). Geometric parameters (Å, º)
| O1—N3 | 1.222 (3) | C8—H8B | 0.97 |
| O2—N3 | 1.217 (3) | C9—C10 | 1.516 (3) |
| N1—C1 | 1.391 (3) | C9—H9A | 0.97 |
| N1—C7 | 1.474 (3) | C9—H9B | 0.97 |
| N1—C10 | 1.477 (3) | C10—H10A | 0.97 |
| N2—C9 | 1.476 (4) | C10—H10B | 0.97 |
| N2—C8 | 1.490 (3) | Br1—C14 | 1.906 (2) |
| N2—H21 | 0.850 (17) | O3—C17 | 1.264 (3) |
| N2—H22 | 0.833 (18) | O4—C17 | 1.257 (3) |
| N3—C4 | 1.454 (3) | C11—C16 | 1.388 (3) |
| C1—C2 | 1.405 (3) | C11—C12 | 1.391 (3) |
| C1—C6 | 1.410 (3) | C11—C17 | 1.506 (3) |
| C2—C3 | 1.376 (3) | C12—C13 | 1.385 (3) |
| C2—H2 | 0.93 | C12—H12 | 0.93 |
| C3—C4 | 1.377 (4) | C13—C14 | 1.374 (4) |
| C3—H3 | 0.93 | C13—H13 | 0.93 |
| C4—C5 | 1.378 (4) | C14—C15 | 1.378 (4) |
| C5—C6 | 1.372 (3) | C15—C16 | 1.382 (3) |
| C5—H5 | 0.93 | C15—H15 | 0.93 |
| C6—H6 | 0.93 | C16—H16 | 0.93 |
| C7—C8 | 1.502 (3) | O5—H1W | 0.802 (17) |
| C7—H7A | 0.97 | O5—H2W | 0.802 (17) |
| C7—H7B | 0.97 | O6—H4W | 0.823 (17) |
| C8—H8A | 0.97 | O6—H3W | 0.778 (18) |
| C1—N1—C7 | 117.46 (19) | N2—C8—H8B | 109.4 |
| C1—N1—C10 | 117.30 (18) | C7—C8—H8B | 109.4 |
| C7—N1—C10 | 112.08 (19) | H8A—C8—H8B | 108 |
| C9—N2—C8 | 109.8 (2) | N2—C9—C10 | 110.3 (2) |
| C9—N2—H21 | 113 (2) | N2—C9—H9A | 109.6 |
| C8—N2—H21 | 110 (2) | C10—C9—H9A | 109.6 |
| C9—N2—H22 | 115 (2) | N2—C9—H9B | 109.6 |
| C8—N2—H22 | 106 (2) | C10—C9—H9B | 109.6 |
| H21—N2—H22 | 102 (3) | H9A—C9—H9B | 108.1 |
| O2—N3—O1 | 122.4 (2) | N1—C10—C9 | 111.3 (2) |
| O2—N3—C4 | 118.8 (2) | N1—C10—H10A | 109.4 |
| O1—N3—C4 | 118.8 (2) | C9—C10—H10A | 109.4 |
| N1—C1—C2 | 121.5 (2) | N1—C10—H10B | 109.4 |
| N1—C1—C6 | 121.4 (2) | C9—C10—H10B | 109.4 |
| C2—C1—C6 | 117.1 (2) | H10A—C10—H10B | 108 |
| C3—C2—C1 | 121.2 (2) | C16—C11—C12 | 118.6 (2) |
| C3—C2—H2 | 119.4 | C16—C11—C17 | 120.5 (2) |
| C1—C2—H2 | 119.4 | C12—C11—C17 | 120.9 (2) |
| C2—C3—C4 | 119.9 (2) | C13—C12—C11 | 120.9 (2) |
| C2—C3—H3 | 120 | C13—C12—H12 | 119.5 |
| C4—C3—H3 | 120 | C11—C12—H12 | 119.5 |
| C3—C4—C5 | 120.6 (2) | C14—C13—C12 | 119.0 (2) |
| C3—C4—N3 | 119.2 (2) | C14—C13—H13 | 120.5 |
| C5—C4—N3 | 120.2 (2) | C12—C13—H13 | 120.5 |
| C6—C5—C4 | 119.8 (2) | C13—C14—C15 | 121.4 (2) |
| C6—C5—H5 | 120.1 | C13—C14—Br1 | 119.7 (2) |
| C4—C5—H5 | 120.1 | C15—C14—Br1 | 118.9 (2) |
| C5—C6—C1 | 121.4 (2) | C14—C15—C16 | 119.2 (2) |
| C5—C6—H6 | 119.3 | C14—C15—H15 | 120.4 |
| C1—C6—H6 | 119.3 | C16—C15—H15 | 120.4 |
| N1—C7—C8 | 111.5 (2) | C15—C16—C11 | 120.9 (2) |
| N1—C7—H7A | 109.3 | C15—C16—H16 | 119.6 |
| C8—C7—H7A | 109.3 | C11—C16—H16 | 119.6 |
| N1—C7—H7B | 109.3 | O4—C17—O3 | 124.3 (2) |
| C8—C7—H7B | 109.3 | O4—C17—C11 | 117.8 (2) |
| H7A—C7—H7B | 108 | O3—C17—C11 | 117.9 (2) |
| N2—C8—C7 | 111.17 (19) | H1W—O5—H2W | 108 (3) |
| N2—C8—H8A | 109.4 | H4W—O6—H3W | 109 (3) |
| C7—C8—H8A | 109.4 | ||
| C7—N1—C1—C2 | 10.7 (3) | C9—N2—C8—C7 | 58.1 (3) |
| C10—N1—C1—C2 | 148.9 (2) | N1—C7—C8—N2 | −55.1 (3) |
| C7—N1—C1—C6 | −171.5 (2) | C8—N2—C9—C10 | −58.5 (3) |
| C10—N1—C1—C6 | −33.3 (3) | C1—N1—C10—C9 | 165.8 (2) |
| N1—C1—C2—C3 | 176.8 (2) | C7—N1—C10—C9 | −53.9 (3) |
| C6—C1—C2—C3 | −1.1 (4) | N2—C9—C10—N1 | 56.8 (3) |
| C1—C2—C3—C4 | 2.0 (4) | C16—C11—C12—C13 | 1.3 (3) |
| C2—C3—C4—C5 | −1.6 (4) | C17—C11—C12—C13 | −179.4 (2) |
| C2—C3—C4—N3 | 179.0 (2) | C11—C12—C13—C14 | −1.7 (4) |
| O2—N3—C4—C3 | −174.9 (3) | C12—C13—C14—C15 | 0.4 (4) |
| O1—N3—C4—C3 | 5.0 (4) | C12—C13—C14—Br1 | 179.36 (19) |
| O2—N3—C4—C5 | 5.7 (4) | C13—C14—C15—C16 | 1.2 (4) |
| O1—N3—C4—C5 | −174.4 (3) | Br1—C14—C15—C16 | −177.75 (18) |
| C3—C4—C5—C6 | 0.5 (4) | C14—C15—C16—C11 | −1.6 (3) |
| N3—C4—C5—C6 | 179.9 (2) | C12—C11—C16—C15 | 0.4 (3) |
| C4—C5—C6—C1 | 0.3 (4) | C17—C11—C16—C15 | −178.9 (2) |
| N1—C1—C6—C5 | −177.9 (2) | C16—C11—C17—O4 | −26.9 (3) |
| C2—C1—C6—C5 | 0.0 (4) | C12—C11—C17—O4 | 153.8 (2) |
| C1—N1—C7—C8 | −166.7 (2) | C16—C11—C17—O3 | 153.3 (2) |
| C10—N1—C7—C8 | 53.0 (3) | C12—C11—C17—O3 | −26.0 (3) |
4-(4-Nitrophenyl)piperazin-1-ium 4-bromobenzoate dihydrate (I). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H21···O5i | 0.85 (2) | 1.99 (2) | 2.810 (3) | 162 (3) |
| N2—H22···O6ii | 0.83 (2) | 1.91 (2) | 2.707 (3) | 160 (3) |
| C3—H3···O1iii | 0.93 | 2.59 | 3.260 (4) | 130 |
| C13—H13···O4iv | 0.93 | 2.57 | 3.483 (3) | 166 |
| C15—H15···O2v | 0.93 | 2.47 | 3.269 (4) | 144 |
| O5—H1W···O3vi | 0.80 (2) | 1.97 (2) | 2.759 (2) | 169 (3) |
| O5—H2W···O3i | 0.80 (2) | 2.00 (2) | 2.772 (2) | 161 (3) |
| O6—H4W···O4 | 0.82 (2) | 2.03 (2) | 2.832 (3) | 166 (3) |
| O6—H3W···O4vii | 0.78 (2) | 1.99 (2) | 2.760 (3) | 169 (3) |
Symmetry codes: (i) −x+1, −y+1, −z; (ii) x, y+1, z; (iii) −x+2, −y+2, −z+1; (iv) x+1, y, z; (v) −x+1, −y+1, −z+1; (vi) x, y−1, z; (vii) −x, −y+1, −z.
4-(4-Nitrophenyl)piperazin-1-ium 4-iodobenzoate dihydrate (II). Crystal data
| C10H14N3O2+·C7H4IO2−·2H2O | Z = 2 |
| Mr = 491.28 | F(000) = 492 |
| Triclinic, P1 | Dx = 1.699 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.7652 (4) Å | Cell parameters from 6331 reflections |
| b = 9.2852 (5) Å | θ = 2.6–25.4° |
| c = 13.930 (1) Å | µ = 1.71 mm−1 |
| α = 94.985 (5)° | T = 293 K |
| β = 95.331 (5)° | Prism, brown |
| γ = 104.875 (6)° | 0.48 × 0.48 × 0.2 mm |
| V = 960.09 (10) Å3 |
4-(4-Nitrophenyl)piperazin-1-ium 4-iodobenzoate dihydrate (II). Data collection
| Oxford Diffraction Xcalibur diffractometer | 2952 reflections with I > 2σ(I) |
| ω scans | Rint = 0.017 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | θmax = 25.4°, θmin = 2.6° |
| Tmin = 0.458, Tmax = 0.711 | h = −8→9 |
| 6331 measured reflections | k = −11→9 |
| 3518 independent reflections | l = −14→16 |
4-(4-Nitrophenyl)piperazin-1-ium 4-iodobenzoate dihydrate (II). Refinement
| Refinement on F2 | 0 constraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.029 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.069 | w = 1/[σ2(Fo2) + (0.0319P)2 + 0.5892P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max < 0.001 |
| 3513 reflections | Δρmax = 0.54 e Å−3 |
| 262 parameters | Δρmin = −0.66 e Å−3 |
| 6 restraints |
4-(4-Nitrophenyl)piperazin-1-ium 4-iodobenzoate dihydrate (II). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Nitrophenyl)piperazin-1-ium 4-iodobenzoate dihydrate (II). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.9211 (3) | 0.7945 (3) | 0.5327 (2) | 0.0765 (8) | |
| O2 | 0.6869 (3) | 0.6415 (3) | 0.5706 (2) | 0.0702 (7) | |
| N1 | 0.3324 (3) | 1.0055 (2) | 0.29012 (17) | 0.0344 (5) | |
| N2 | 0.1277 (3) | 1.1016 (3) | 0.13972 (19) | 0.0404 (6) | |
| N3 | 0.7583 (4) | 0.7448 (3) | 0.52680 (19) | 0.0471 (6) | |
| C1 | 0.4369 (3) | 0.9384 (3) | 0.34660 (19) | 0.0311 (6) | |
| C2 | 0.6239 (4) | 0.9794 (3) | 0.3495 (2) | 0.0421 (7) | |
| H2 | 0.678241 | 1.049839 | 0.310458 | 0.051* | |
| C3 | 0.7273 (4) | 0.9175 (3) | 0.4087 (2) | 0.0434 (7) | |
| H3 | 0.851637 | 0.947138 | 0.411003 | 0.052* | |
| C4 | 0.6475 (4) | 0.8107 (3) | 0.4653 (2) | 0.0348 (6) | |
| C5 | 0.4638 (4) | 0.7665 (3) | 0.4644 (2) | 0.0407 (7) | |
| H5 | 0.411197 | 0.694461 | 0.502907 | 0.049* | |
| C6 | 0.3609 (4) | 0.8297 (3) | 0.4063 (2) | 0.0390 (7) | |
| H6 | 0.236893 | 0.800552 | 0.405831 | 0.047* | |
| C7 | 0.4203 (4) | 1.1048 (3) | 0.2222 (2) | 0.0412 (7) | |
| H7A | 0.448427 | 1.044706 | 0.168453 | 0.049* | |
| H7B | 0.532139 | 1.170403 | 0.255077 | 0.049* | |
| C8 | 0.3038 (4) | 1.1984 (3) | 0.1836 (2) | 0.0439 (7) | |
| H8A | 0.287569 | 1.267775 | 0.236 | 0.053* | |
| H8B | 0.361796 | 1.256375 | 0.135285 | 0.053* | |
| C9 | 0.0381 (4) | 1.0095 (4) | 0.2115 (2) | 0.0456 (7) | |
| H9A | −0.07676 | 0.945606 | 0.181239 | 0.055* | |
| H9B | 0.016429 | 1.074515 | 0.264773 | 0.055* | |
| C10 | 0.1531 (4) | 0.9144 (3) | 0.2497 (2) | 0.0413 (7) | |
| H10A | 0.096206 | 0.859686 | 0.299686 | 0.05* | |
| H10B | 0.163241 | 0.841846 | 0.19758 | 0.05* | |
| I1 | 0.92211 (3) | 0.44728 (3) | 0.33815 (2) | 0.05798 (10) | |
| O3 | 0.4459 (3) | 0.7894 (2) | 0.01595 (18) | 0.0503 (6) | |
| O4 | 0.2183 (3) | 0.6411 (2) | 0.07685 (16) | 0.0463 (5) | |
| C11 | 0.5106 (3) | 0.6368 (3) | 0.1340 (2) | 0.0321 (6) | |
| C12 | 0.6798 (4) | 0.6390 (3) | 0.1072 (2) | 0.0387 (7) | |
| H12 | 0.71341 | 0.676998 | 0.050132 | 0.046* | |
| C13 | 0.7975 (4) | 0.5851 (3) | 0.1648 (2) | 0.0411 (7) | |
| H13 | 0.909115 | 0.584052 | 0.146196 | 0.049* | |
| C14 | 0.7470 (4) | 0.5326 (3) | 0.2509 (2) | 0.0376 (7) | |
| C15 | 0.5813 (4) | 0.5331 (3) | 0.2798 (2) | 0.0398 (7) | |
| H15 | 0.550434 | 0.499714 | 0.338523 | 0.048* | |
| C16 | 0.4631 (4) | 0.5834 (3) | 0.2208 (2) | 0.0378 (7) | |
| H16 | 0.350345 | 0.581695 | 0.238875 | 0.045* | |
| C17 | 0.3825 (4) | 0.6931 (3) | 0.0705 (2) | 0.0356 (6) | |
| O5 | 0.7577 (3) | 0.0174 (2) | 0.02906 (17) | 0.0446 (5) | |
| O6 | 0.0304 (3) | 0.3341 (3) | 0.06807 (19) | 0.0529 (6) | |
| H21 | 0.144 (5) | 1.051 (4) | 0.0882 (19) | 0.063* | |
| H22 | 0.076 (4) | 1.162 (3) | 0.116 (3) | 0.063* | |
| H1W | 0.671 (4) | −0.051 (3) | 0.032 (3) | 0.063* | |
| H2W | 0.711 (5) | 0.084 (3) | 0.022 (3) | 0.063* | |
| H4W | 0.100 (4) | 0.414 (3) | 0.080 (3) | 0.063* | |
| H3W | −0.052 (4) | 0.326 (4) | 0.027 (2) | 0.063* |
4-(4-Nitrophenyl)piperazin-1-ium 4-iodobenzoate dihydrate (II). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0437 (15) | 0.0752 (18) | 0.108 (2) | 0.0122 (13) | −0.0192 (14) | 0.0400 (16) |
| O2 | 0.0633 (16) | 0.0754 (18) | 0.0786 (18) | 0.0200 (13) | 0.0033 (13) | 0.0469 (15) |
| N1 | 0.0300 (12) | 0.0321 (13) | 0.0391 (13) | 0.0054 (10) | −0.0037 (10) | 0.0090 (10) |
| N2 | 0.0420 (14) | 0.0388 (15) | 0.0413 (15) | 0.0159 (11) | −0.0066 (12) | 0.0072 (11) |
| N3 | 0.0496 (17) | 0.0450 (16) | 0.0461 (15) | 0.0151 (13) | −0.0076 (13) | 0.0084 (13) |
| C1 | 0.0328 (14) | 0.0280 (14) | 0.0316 (14) | 0.0093 (11) | −0.0008 (11) | 0.0009 (11) |
| C2 | 0.0340 (15) | 0.0433 (17) | 0.0496 (18) | 0.0067 (13) | 0.0047 (13) | 0.0196 (14) |
| C3 | 0.0279 (15) | 0.0464 (18) | 0.0548 (19) | 0.0086 (13) | −0.0010 (13) | 0.0115 (15) |
| C4 | 0.0369 (15) | 0.0344 (15) | 0.0332 (15) | 0.0118 (12) | −0.0029 (12) | 0.0049 (12) |
| C5 | 0.0422 (17) | 0.0417 (17) | 0.0383 (16) | 0.0087 (13) | 0.0047 (13) | 0.0139 (13) |
| C6 | 0.0292 (14) | 0.0443 (17) | 0.0430 (17) | 0.0073 (12) | 0.0016 (12) | 0.0118 (13) |
| C7 | 0.0353 (15) | 0.0389 (17) | 0.0459 (17) | 0.0034 (13) | −0.0040 (13) | 0.0148 (14) |
| C8 | 0.0483 (18) | 0.0350 (16) | 0.0447 (17) | 0.0078 (14) | −0.0075 (14) | 0.0095 (13) |
| C9 | 0.0354 (16) | 0.055 (2) | 0.0479 (18) | 0.0160 (14) | −0.0023 (14) | 0.0103 (15) |
| C10 | 0.0297 (15) | 0.0401 (17) | 0.0524 (18) | 0.0071 (13) | −0.0035 (13) | 0.0110 (14) |
| I1 | 0.05063 (15) | 0.06217 (17) | 0.06139 (16) | 0.01852 (11) | −0.01269 (10) | 0.01870 (11) |
| O3 | 0.0370 (11) | 0.0400 (12) | 0.0758 (16) | 0.0088 (9) | 0.0016 (11) | 0.0276 (11) |
| O4 | 0.0283 (11) | 0.0460 (12) | 0.0629 (14) | 0.0073 (9) | −0.0016 (10) | 0.0134 (10) |
| C11 | 0.0290 (14) | 0.0241 (14) | 0.0415 (16) | 0.0059 (11) | −0.0004 (12) | 0.0023 (12) |
| C12 | 0.0376 (16) | 0.0394 (16) | 0.0431 (17) | 0.0138 (13) | 0.0088 (13) | 0.0119 (13) |
| C13 | 0.0315 (15) | 0.0436 (17) | 0.0516 (19) | 0.0157 (13) | 0.0044 (13) | 0.0082 (14) |
| C14 | 0.0336 (15) | 0.0306 (15) | 0.0458 (17) | 0.0078 (12) | −0.0064 (13) | 0.0047 (13) |
| C15 | 0.0421 (17) | 0.0365 (16) | 0.0373 (16) | 0.0043 (13) | 0.0025 (13) | 0.0068 (13) |
| C16 | 0.0288 (14) | 0.0356 (16) | 0.0471 (17) | 0.0062 (12) | 0.0048 (13) | 0.0018 (13) |
| C17 | 0.0328 (15) | 0.0241 (14) | 0.0475 (17) | 0.0073 (12) | −0.0011 (13) | −0.0012 (12) |
| O5 | 0.0345 (11) | 0.0374 (13) | 0.0610 (14) | 0.0094 (9) | −0.0027 (10) | 0.0111 (11) |
| O6 | 0.0442 (13) | 0.0383 (12) | 0.0735 (17) | 0.0097 (10) | −0.0123 (11) | 0.0164 (12) |
4-(4-Nitrophenyl)piperazin-1-ium 4-iodobenzoate dihydrate (II). Geometric parameters (Å, º)
| O1—N3 | 1.222 (3) | C8—H8B | 0.97 |
| O2—N3 | 1.221 (3) | C9—C10 | 1.503 (4) |
| N1—C1 | 1.376 (3) | C9—H9A | 0.97 |
| N1—C10 | 1.462 (3) | C9—H9B | 0.97 |
| N1—C7 | 1.469 (4) | C10—H10A | 0.97 |
| N2—C8 | 1.473 (4) | C10—H10B | 0.97 |
| N2—C9 | 1.479 (4) | I1—C14 | 2.090 (3) |
| N2—H21 | 0.859 (18) | O3—C17 | 1.257 (3) |
| N2—H22 | 0.850 (18) | O4—C17 | 1.257 (3) |
| N3—C4 | 1.440 (4) | C11—C16 | 1.391 (4) |
| C1—C2 | 1.400 (4) | C11—C12 | 1.395 (4) |
| C1—C6 | 1.410 (4) | C11—C17 | 1.493 (4) |
| C2—C3 | 1.361 (4) | C12—C13 | 1.377 (4) |
| C2—H2 | 0.93 | C12—H12 | 0.93 |
| C3—C4 | 1.378 (4) | C13—C14 | 1.386 (4) |
| C3—H3 | 0.93 | C13—H13 | 0.93 |
| C4—C5 | 1.378 (4) | C14—C15 | 1.385 (4) |
| C5—C6 | 1.357 (4) | C15—C16 | 1.372 (4) |
| C5—H5 | 0.93 | C15—H15 | 0.93 |
| C6—H6 | 0.93 | C16—H16 | 0.93 |
| C7—C8 | 1.502 (4) | O5—H1W | 0.805 (18) |
| C7—H7A | 0.97 | O5—H2W | 0.805 (18) |
| C7—H7B | 0.97 | O6—H4W | 0.795 (18) |
| C8—H8A | 0.97 | O6—H3W | 0.800 (18) |
| C1—N1—C10 | 117.2 (2) | N2—C8—H8B | 109.6 |
| C1—N1—C7 | 117.8 (2) | C7—C8—H8B | 109.6 |
| C10—N1—C7 | 112.8 (2) | H8A—C8—H8B | 108.1 |
| C8—N2—C9 | 110.6 (2) | N2—C9—C10 | 110.3 (2) |
| C8—N2—H21 | 108 (3) | N2—C9—H9A | 109.6 |
| C9—N2—H21 | 115 (3) | C10—C9—H9A | 109.6 |
| C8—N2—H22 | 104 (3) | N2—C9—H9B | 109.6 |
| C9—N2—H22 | 118 (3) | C10—C9—H9B | 109.6 |
| H21—N2—H22 | 101 (3) | H9A—C9—H9B | 108.1 |
| O2—N3—O1 | 122.5 (3) | N1—C10—C9 | 111.5 (2) |
| O2—N3—C4 | 119.1 (3) | N1—C10—H10A | 109.3 |
| O1—N3—C4 | 118.4 (3) | C9—C10—H10A | 109.3 |
| N1—C1—C2 | 121.1 (2) | N1—C10—H10B | 109.3 |
| N1—C1—C6 | 121.5 (2) | C9—C10—H10B | 109.3 |
| C2—C1—C6 | 117.4 (2) | H10A—C10—H10B | 108 |
| C3—C2—C1 | 120.9 (3) | C16—C11—C12 | 119.5 (3) |
| C3—C2—H2 | 119.6 | C16—C11—C17 | 120.4 (2) |
| C1—C2—H2 | 119.6 | C12—C11—C17 | 120.1 (2) |
| C2—C3—C4 | 119.9 (3) | C13—C12—C11 | 120.4 (3) |
| C2—C3—H3 | 120.1 | C13—C12—H12 | 119.8 |
| C4—C3—H3 | 120.1 | C11—C12—H12 | 119.8 |
| C5—C4—C3 | 121.2 (3) | C12—C13—C14 | 119.0 (3) |
| C5—C4—N3 | 119.5 (3) | C12—C13—H13 | 120.5 |
| C3—C4—N3 | 119.3 (3) | C14—C13—H13 | 120.5 |
| C6—C5—C4 | 118.9 (3) | C15—C14—C13 | 121.4 (3) |
| C6—C5—H5 | 120.5 | C15—C14—I1 | 119.1 (2) |
| C4—C5—H5 | 120.5 | C13—C14—I1 | 119.6 (2) |
| C5—C6—C1 | 121.8 (3) | C16—C15—C14 | 119.2 (3) |
| C5—C6—H6 | 119.1 | C16—C15—H15 | 120.4 |
| C1—C6—H6 | 119.1 | C14—C15—H15 | 120.4 |
| N1—C7—C8 | 111.9 (2) | C15—C16—C11 | 120.5 (3) |
| N1—C7—H7A | 109.2 | C15—C16—H16 | 119.8 |
| C8—C7—H7A | 109.2 | C11—C16—H16 | 119.8 |
| N1—C7—H7B | 109.2 | O4—C17—O3 | 125.0 (3) |
| C8—C7—H7B | 109.2 | O4—C17—C11 | 116.9 (2) |
| H7A—C7—H7B | 107.9 | O3—C17—C11 | 118.1 (2) |
| N2—C8—C7 | 110.2 (2) | H1W—O5—H2W | 101 (4) |
| N2—C8—H8A | 109.6 | H4W—O6—H3W | 117 (4) |
| C7—C8—H8A | 109.6 | ||
| C10—N1—C1—C2 | 148.6 (3) | C9—N2—C8—C7 | 58.1 (3) |
| C7—N1—C1—C2 | 8.9 (4) | N1—C7—C8—N2 | −54.7 (3) |
| C10—N1—C1—C6 | −33.7 (4) | C8—N2—C9—C10 | −58.5 (3) |
| C7—N1—C1—C6 | −173.4 (3) | C1—N1—C10—C9 | 165.8 (3) |
| N1—C1—C2—C3 | 177.1 (3) | C7—N1—C10—C9 | −52.6 (3) |
| C6—C1—C2—C3 | −0.8 (4) | N2—C9—C10—N1 | 55.3 (4) |
| C1—C2—C3—C4 | 1.3 (5) | C16—C11—C12—C13 | 1.6 (4) |
| C2—C3—C4—C5 | −1.0 (5) | C17—C11—C12—C13 | −179.0 (3) |
| C2—C3—C4—N3 | 179.3 (3) | C11—C12—C13—C14 | −1.7 (4) |
| O2—N3—C4—C5 | 6.3 (4) | C12—C13—C14—C15 | 0.1 (4) |
| O1—N3—C4—C5 | −173.9 (3) | C12—C13—C14—I1 | 179.0 (2) |
| O2—N3—C4—C3 | −173.9 (3) | C13—C14—C15—C16 | 1.6 (4) |
| O1—N3—C4—C3 | 5.8 (4) | I1—C14—C15—C16 | −177.4 (2) |
| C3—C4—C5—C6 | 0.1 (5) | C14—C15—C16—C11 | −1.6 (4) |
| N3—C4—C5—C6 | 179.8 (3) | C12—C11—C16—C15 | 0.0 (4) |
| C4—C5—C6—C1 | 0.4 (5) | C17—C11—C16—C15 | −179.3 (3) |
| N1—C1—C6—C5 | −178.0 (3) | C16—C11—C17—O4 | −26.0 (4) |
| C2—C1—C6—C5 | −0.1 (4) | C12—C11—C17—O4 | 154.6 (3) |
| C1—N1—C7—C8 | −166.2 (2) | C16—C11—C17—O3 | 153.1 (3) |
| C10—N1—C7—C8 | 52.5 (3) | C12—C11—C17—O3 | −26.2 (4) |
4-(4-Nitrophenyl)piperazin-1-ium 4-iodobenzoate dihydrate (II). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H21···O5i | 0.86 (2) | 1.99 (2) | 2.825 (4) | 164 (4) |
| N2—H22···O6ii | 0.85 (2) | 1.88 (2) | 2.702 (3) | 163 (4) |
| C3—H3···O1iii | 0.93 | 2.59 | 3.275 (4) | 131 |
| C13—H13···O4iv | 0.93 | 2.62 | 3.526 (4) | 166 |
| C15—H15···O2v | 0.93 | 2.49 | 3.311 (4) | 147 |
| O5—H1W···O3vi | 0.81 (2) | 1.96 (2) | 2.756 (3) | 170 (4) |
| O5—H2W···O3i | 0.81 (2) | 1.96 (2) | 2.753 (3) | 166 (4) |
| O6—H4W···O4 | 0.80 (2) | 2.08 (2) | 2.836 (3) | 160 (4) |
| O6—H3W···O4vii | 0.80 (2) | 1.95 (2) | 2.728 (3) | 165 (4) |
Symmetry codes: (i) −x+1, −y+1, −z; (ii) x, y+1, z; (iii) −x+2, −y+2, −z+1; (iv) x+1, y, z; (v) −x+1, −y+1, −z+1; (vi) x, y−1, z; (vii) −x, −y+1, −z.
4-(4-Nitrophenyl)piperazin-1-ium 4-hydroxybenzoate monohydrate (III). Crystal data
| C10H14N3O2+·C7H5O3−·H2O | Z = 2 |
| Mr = 363.37 | F(000) = 384 |
| Triclinic, P1 | Dx = 1.407 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.636 (1) Å | Cell parameters from 5342 reflections |
| b = 10.301 (1) Å | θ = 2.6–25.3° |
| c = 10.867 (1) Å | µ = 0.11 mm−1 |
| α = 103.90 (1)° | T = 293 K |
| β = 108.32 (1)° | Rod, yellow |
| γ = 112.96 (1)° | 0.50 × 0.32 × 0.24 mm |
| V = 857.80 (17) Å3 |
4-(4-Nitrophenyl)piperazin-1-ium 4-hydroxybenzoate monohydrate (III). Data collection
| Oxford Diffraction Xcalibur diffractometer | 2342 reflections with I > 2σ(I) |
| ω scans | Rint = 0.013 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | θmax = 25.3°, θmin = 2.6° |
| Tmin = 0.959, Tmax = 0.974 | h = −11→11 |
| 5342 measured reflections | k = −12→11 |
| 3140 independent reflections | l = −13→12 |
4-(4-Nitrophenyl)piperazin-1-ium 4-hydroxybenzoate monohydrate (III). Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.043 | w = 1/[σ2(Fo2) + (0.0386P)2 + 0.3231P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.106 | (Δ/σ)max < 0.001 |
| S = 1.05 | Δρmax = 0.19 e Å−3 |
| 3135 reflections | Δρmin = −0.19 e Å−3 |
| 251 parameters | Extinction correction: SHELXL2018/3 (Sheldrick 2015b), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 5 restraints | Extinction coefficient: 0.032 (3) |
| 0 constraints |
4-(4-Nitrophenyl)piperazin-1-ium 4-hydroxybenzoate monohydrate (III). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Nitrophenyl)piperazin-1-ium 4-hydroxybenzoate monohydrate (III). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.3138 (2) | 0.37000 (19) | 0.65529 (18) | 0.0346 (4) | |
| C2 | 0.1431 (2) | 0.2527 (2) | 0.57638 (19) | 0.0418 (4) | |
| H2 | 0.068235 | 0.250847 | 0.613904 | 0.05* | |
| C3 | 0.0843 (2) | 0.1407 (2) | 0.4450 (2) | 0.0434 (5) | |
| H3 | −0.029409 | 0.063818 | 0.39416 | 0.052* | |
| C4 | 0.1942 (2) | 0.1428 (2) | 0.38907 (19) | 0.0411 (4) | |
| C5 | 0.3624 (2) | 0.2553 (2) | 0.4636 (2) | 0.0483 (5) | |
| H5 | 0.436252 | 0.255393 | 0.42532 | 0.058* | |
| C6 | 0.4208 (2) | 0.3670 (2) | 0.5942 (2) | 0.0472 (5) | |
| H6 | 0.53484 | 0.443048 | 0.64379 | 0.057* | |
| C7 | 0.5230 (2) | 0.6338 (2) | 0.8284 (2) | 0.0453 (5) | |
| H7A | 0.489341 | 0.67826 | 0.763471 | 0.054* | |
| H7B | 0.611237 | 0.616548 | 0.817407 | 0.054* | |
| C8 | 0.5933 (2) | 0.7472 (2) | 0.9784 (2) | 0.0493 (5) | |
| H8A | 0.643572 | 0.71093 | 1.044494 | 0.059* | |
| H8B | 0.681545 | 0.846719 | 0.99504 | 0.059* | |
| C9 | 0.3302 (3) | 0.6143 (2) | 0.9836 (2) | 0.0457 (5) | |
| H9A | 0.242868 | 0.625614 | 1.002896 | 0.055* | |
| H9B | 0.381743 | 0.579404 | 1.050389 | 0.055* | |
| C10 | 0.2514 (2) | 0.4953 (2) | 0.83370 (19) | 0.0403 (4) | |
| H10A | 0.176601 | 0.394834 | 0.82667 | 0.048* | |
| H10B | 0.183115 | 0.521491 | 0.76885 | 0.048* | |
| C11 | 0.1749 (2) | 0.5843 (2) | 0.29147 (19) | 0.0418 (5) | |
| C12 | 0.2469 (3) | 0.7413 (2) | 0.3258 (2) | 0.0495 (5) | |
| H12 | 0.262285 | 0.778209 | 0.258209 | 0.059* | |
| C13 | 0.2959 (2) | 0.8431 (2) | 0.4597 (2) | 0.0438 (5) | |
| H13 | 0.344454 | 0.94863 | 0.481898 | 0.053* | |
| C14 | 0.2738 (2) | 0.7906 (2) | 0.56226 (18) | 0.0364 (4) | |
| C15 | 0.2004 (2) | 0.6329 (2) | 0.52544 (19) | 0.0395 (4) | |
| H15 | 0.18337 | 0.595519 | 0.592378 | 0.047* | |
| C16 | 0.1520 (2) | 0.5299 (2) | 0.39188 (19) | 0.0410 (4) | |
| H16 | 0.104259 | 0.424435 | 0.369591 | 0.049* | |
| C17 | 0.3293 (2) | 0.9024 (2) | 0.7080 (2) | 0.0440 (5) | |
| N1 | 0.37669 (18) | 0.48418 (16) | 0.78933 (15) | 0.0372 (4) | |
| N2 | 0.4599 (2) | 0.76559 (19) | 1.00415 (18) | 0.0479 (4) | |
| N3 | 0.1322 (2) | 0.02537 (19) | 0.25010 (18) | 0.0518 (4) | |
| O1 | 0.2282 (2) | 0.04014 (19) | 0.19537 (17) | 0.0759 (5) | |
| O2 | −0.0112 (2) | −0.08579 (18) | 0.19233 (16) | 0.0715 (5) | |
| O3 | 0.1311 (2) | 0.48813 (18) | 0.15852 (15) | 0.0637 (4) | |
| O4 | 0.3985 (2) | 1.04290 (17) | 0.73256 (16) | 0.0744 (5) | |
| O5 | 0.30323 (18) | 0.85085 (16) | 0.79750 (14) | 0.0547 (4) | |
| O6 | −0.0679 (3) | 0.1917 (2) | 0.09903 (19) | 0.0809 (6) | |
| H21 | 0.412 (3) | 0.796 (3) | 0.941 (2) | 0.097* | |
| H22 | 0.513 (3) | 0.838 (3) | 1.099 (2) | 0.097* | |
| H17 | 0.070 (3) | 0.394 (2) | 0.143 (3) | 0.097* | |
| H1W | −0.140 (3) | 0.168 (3) | 0.129 (3) | 0.097* | |
| H2W | −0.112 (3) | 0.128 (3) | 0.015 (2) | 0.097* |
4-(4-Nitrophenyl)piperazin-1-ium 4-hydroxybenzoate monohydrate (III). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0348 (10) | 0.0343 (9) | 0.0377 (10) | 0.0187 (8) | 0.0167 (8) | 0.0173 (8) |
| C2 | 0.0357 (10) | 0.0427 (10) | 0.0444 (11) | 0.0159 (8) | 0.0211 (8) | 0.0168 (9) |
| C3 | 0.0356 (10) | 0.0380 (10) | 0.0439 (11) | 0.0113 (8) | 0.0148 (8) | 0.0145 (9) |
| C4 | 0.0463 (11) | 0.0347 (9) | 0.0373 (10) | 0.0191 (8) | 0.0175 (8) | 0.0119 (8) |
| C5 | 0.0429 (11) | 0.0481 (11) | 0.0505 (12) | 0.0215 (9) | 0.0255 (9) | 0.0120 (10) |
| C6 | 0.0315 (10) | 0.0440 (11) | 0.0514 (12) | 0.0139 (8) | 0.0177 (9) | 0.0081 (9) |
| C7 | 0.0379 (10) | 0.0401 (10) | 0.0487 (11) | 0.0133 (8) | 0.0209 (9) | 0.0137 (9) |
| C8 | 0.0459 (11) | 0.0395 (11) | 0.0468 (11) | 0.0154 (9) | 0.0160 (9) | 0.0117 (9) |
| C9 | 0.0590 (12) | 0.0451 (11) | 0.0434 (11) | 0.0277 (10) | 0.0295 (10) | 0.0231 (9) |
| C10 | 0.0429 (10) | 0.0421 (10) | 0.0420 (10) | 0.0213 (9) | 0.0239 (9) | 0.0211 (9) |
| C11 | 0.0398 (10) | 0.0443 (11) | 0.0361 (10) | 0.0170 (9) | 0.0194 (8) | 0.0133 (9) |
| C12 | 0.0595 (13) | 0.0525 (12) | 0.0441 (11) | 0.0248 (10) | 0.0315 (10) | 0.0269 (10) |
| C13 | 0.0494 (11) | 0.0392 (10) | 0.0482 (11) | 0.0207 (9) | 0.0274 (9) | 0.0224 (9) |
| C14 | 0.0357 (9) | 0.0405 (10) | 0.0381 (10) | 0.0210 (8) | 0.0190 (8) | 0.0183 (8) |
| C15 | 0.0410 (10) | 0.0440 (10) | 0.0378 (10) | 0.0201 (8) | 0.0208 (8) | 0.0223 (9) |
| C16 | 0.0409 (10) | 0.0356 (10) | 0.0423 (11) | 0.0151 (8) | 0.0194 (8) | 0.0167 (8) |
| C17 | 0.0482 (11) | 0.0462 (12) | 0.0409 (11) | 0.0255 (9) | 0.0218 (9) | 0.0180 (9) |
| N1 | 0.0349 (8) | 0.0348 (8) | 0.0385 (8) | 0.0157 (7) | 0.0170 (7) | 0.0130 (7) |
| N2 | 0.0616 (11) | 0.0410 (9) | 0.0425 (10) | 0.0259 (8) | 0.0257 (9) | 0.0168 (8) |
| N3 | 0.0575 (11) | 0.0430 (10) | 0.0443 (10) | 0.0214 (9) | 0.0209 (9) | 0.0124 (8) |
| O1 | 0.0806 (12) | 0.0662 (11) | 0.0599 (10) | 0.0220 (9) | 0.0434 (9) | 0.0052 (8) |
| O2 | 0.0600 (10) | 0.0523 (9) | 0.0564 (10) | 0.0087 (8) | 0.0163 (8) | 0.0010 (8) |
| O3 | 0.0759 (11) | 0.0545 (9) | 0.0430 (8) | 0.0171 (8) | 0.0337 (8) | 0.0121 (7) |
| O4 | 0.1147 (14) | 0.0416 (9) | 0.0554 (10) | 0.0278 (9) | 0.0450 (10) | 0.0145 (7) |
| O5 | 0.0721 (10) | 0.0620 (9) | 0.0425 (8) | 0.0375 (8) | 0.0324 (7) | 0.0259 (7) |
| O6 | 0.0936 (14) | 0.0547 (10) | 0.0652 (11) | 0.0142 (10) | 0.0484 (10) | 0.0041 (8) |
4-(4-Nitrophenyl)piperazin-1-ium 4-hydroxybenzoate monohydrate (III). Geometric parameters (Å, º)
| C1—N1 | 1.392 (2) | C10—H10A | 0.97 |
| C1—C6 | 1.397 (2) | C10—H10B | 0.97 |
| C1—C2 | 1.402 (2) | C11—O3 | 1.360 (2) |
| C2—C3 | 1.371 (3) | C11—C16 | 1.379 (3) |
| C2—H2 | 0.93 | C11—C12 | 1.382 (3) |
| C3—C4 | 1.373 (3) | C12—C13 | 1.374 (3) |
| C3—H3 | 0.93 | C12—H12 | 0.93 |
| C4—C5 | 1.373 (3) | C13—C14 | 1.389 (2) |
| C4—N3 | 1.446 (2) | C13—H13 | 0.93 |
| C5—C6 | 1.365 (3) | C14—C15 | 1.383 (2) |
| C5—H5 | 0.93 | C14—C17 | 1.494 (3) |
| C6—H6 | 0.93 | C15—C16 | 1.378 (2) |
| C7—N1 | 1.468 (2) | C15—H15 | 0.93 |
| C7—C8 | 1.501 (3) | C16—H16 | 0.93 |
| C7—H7A | 0.97 | C17—O4 | 1.250 (2) |
| C7—H7B | 0.97 | C17—O5 | 1.260 (2) |
| C8—N2 | 1.470 (3) | N2—H21 | 0.893 (17) |
| C8—H8A | 0.97 | N2—H22 | 0.935 (17) |
| C8—H8B | 0.97 | N3—O2 | 1.221 (2) |
| C9—N2 | 1.476 (2) | N3—O1 | 1.230 (2) |
| C9—C10 | 1.507 (3) | O3—H17 | 0.850 (17) |
| C9—H9A | 0.97 | O6—H1W | 0.832 (17) |
| C9—H9B | 0.97 | O6—H2W | 0.834 (17) |
| C10—N1 | 1.467 (2) | ||
| N1—C1—C6 | 120.65 (15) | N1—C10—H10B | 108.9 |
| N1—C1—C2 | 122.40 (15) | C9—C10—H10B | 108.9 |
| C6—C1—C2 | 116.95 (16) | H10A—C10—H10B | 107.8 |
| C3—C2—C1 | 121.37 (17) | O3—C11—C16 | 122.11 (17) |
| C3—C2—H2 | 119.3 | O3—C11—C12 | 118.10 (17) |
| C1—C2—H2 | 119.3 | C16—C11—C12 | 119.78 (17) |
| C2—C3—C4 | 119.60 (17) | C13—C12—C11 | 120.14 (18) |
| C2—C3—H3 | 120.2 | C13—C12—H12 | 119.9 |
| C4—C3—H3 | 120.2 | C11—C12—H12 | 119.9 |
| C3—C4—C5 | 120.68 (17) | C12—C13—C14 | 120.94 (17) |
| C3—C4—N3 | 119.73 (17) | C12—C13—H13 | 119.5 |
| C5—C4—N3 | 119.59 (17) | C14—C13—H13 | 119.5 |
| C6—C5—C4 | 119.67 (17) | C15—C14—C13 | 118.02 (16) |
| C6—C5—H5 | 120.2 | C15—C14—C17 | 121.42 (16) |
| C4—C5—H5 | 120.2 | C13—C14—C17 | 120.55 (16) |
| C5—C6—C1 | 121.72 (17) | C16—C15—C14 | 121.51 (17) |
| C5—C6—H6 | 119.1 | C16—C15—H15 | 119.2 |
| C1—C6—H6 | 119.1 | C14—C15—H15 | 119.2 |
| N1—C7—C8 | 113.07 (16) | C15—C16—C11 | 119.60 (17) |
| N1—C7—H7A | 109 | C15—C16—H16 | 120.2 |
| C8—C7—H7A | 109 | C11—C16—H16 | 120.2 |
| N1—C7—H7B | 109 | O4—C17—O5 | 124.14 (18) |
| C8—C7—H7B | 109 | O4—C17—C14 | 116.82 (17) |
| H7A—C7—H7B | 107.8 | O5—C17—C14 | 119.04 (17) |
| N2—C8—C7 | 110.93 (16) | C1—N1—C10 | 116.66 (14) |
| N2—C8—H8A | 109.5 | C1—N1—C7 | 115.79 (14) |
| C7—C8—H8A | 109.5 | C10—N1—C7 | 114.73 (14) |
| N2—C8—H8B | 109.5 | C8—N2—C9 | 108.67 (15) |
| C7—C8—H8B | 109.5 | C8—N2—H21 | 109.1 (18) |
| H8A—C8—H8B | 108 | C9—N2—H21 | 108.9 (18) |
| N2—C9—C10 | 110.96 (15) | C8—N2—H22 | 106.2 (17) |
| N2—C9—H9A | 109.4 | C9—N2—H22 | 110.8 (17) |
| C10—C9—H9A | 109.4 | H21—N2—H22 | 113 (2) |
| N2—C9—H9B | 109.4 | O2—N3—O1 | 122.24 (17) |
| C10—C9—H9B | 109.4 | O2—N3—C4 | 119.43 (17) |
| H9A—C9—H9B | 108 | O1—N3—C4 | 118.32 (17) |
| N1—C10—C9 | 113.15 (15) | C11—O3—H17 | 110.2 (19) |
| N1—C10—H10A | 108.9 | H1W—O6—H2W | 108 (3) |
| C9—C10—H10A | 108.9 | ||
| N1—C1—C2—C3 | −179.46 (17) | O3—C11—C16—C15 | 179.34 (17) |
| C6—C1—C2—C3 | −0.3 (3) | C12—C11—C16—C15 | 0.4 (3) |
| C1—C2—C3—C4 | 0.0 (3) | C15—C14—C17—O4 | 178.09 (18) |
| C2—C3—C4—C5 | 0.4 (3) | C13—C14—C17—O4 | −1.4 (3) |
| C2—C3—C4—N3 | −179.58 (17) | C15—C14—C17—O5 | −2.4 (3) |
| C3—C4—C5—C6 | −0.5 (3) | C13—C14—C17—O5 | 178.03 (18) |
| N3—C4—C5—C6 | 179.41 (18) | C6—C1—N1—C10 | 169.31 (17) |
| C4—C5—C6—C1 | 0.3 (3) | C2—C1—N1—C10 | −11.5 (2) |
| N1—C1—C6—C5 | 179.30 (18) | C6—C1—N1—C7 | 29.6 (2) |
| C2—C1—C6—C5 | 0.1 (3) | C2—C1—N1—C7 | −151.19 (18) |
| N1—C7—C8—N2 | −53.2 (2) | C9—C10—N1—C1 | 176.12 (15) |
| N2—C9—C10—N1 | 52.1 (2) | C9—C10—N1—C7 | −43.8 (2) |
| O3—C11—C12—C13 | −178.90 (18) | C8—C7—N1—C1 | −175.25 (16) |
| C16—C11—C12—C13 | 0.1 (3) | C8—C7—N1—C10 | 44.3 (2) |
| C11—C12—C13—C14 | −0.2 (3) | C7—C8—N2—C9 | 61.2 (2) |
| C12—C13—C14—C15 | −0.3 (3) | C10—C9—N2—C8 | −60.7 (2) |
| C12—C13—C14—C17 | 179.26 (18) | C3—C4—N3—O2 | −9.7 (3) |
| C13—C14—C15—C16 | 0.8 (3) | C5—C4—N3—O2 | 170.3 (2) |
| C17—C14—C15—C16 | −178.76 (17) | C3—C4—N3—O1 | 171.61 (19) |
| C14—C15—C16—C11 | −0.8 (3) | C5—C4—N3—O1 | −8.3 (3) |
4-(4-Nitrophenyl)piperazin-1-ium 4-hydroxybenzoate monohydrate (III). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H21···O5 | 0.89 (2) | 1.93 (2) | 2.819 (2) | 177 (3) |
| N2—H22···O4i | 0.94 (2) | 1.65 (2) | 2.583 (2) | 177 (3) |
| O3—H17···O6 | 0.85 (2) | 1.82 (2) | 2.669 (2) | 177 (3) |
| O6—H1W···O5ii | 0.83 (2) | 1.95 (2) | 2.768 (2) | 169 (3) |
| O6—H2W···O1iii | 0.83 (2) | 2.11 (2) | 2.944 (2) | 178 (3) |
Symmetry codes: (i) −x+1, −y+2, −z+2; (ii) −x, −y+1, −z+1; (iii) −x, −y, −z.
4-(4-Nitrophenyl)piperazin-1-ium 4-methylbenzoate monohydrate (IV). Crystal data
| C10H14N3O2+·C8H7O2−·H2O | Z = 2 |
| Mr = 361.39 | F(000) = 384 |
| Triclinic, P1 | Dx = 1.319 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 6.1136 (5) Å | Cell parameters from 5980 reflections |
| b = 7.6965 (7) Å | θ = 3.1–25.4° |
| c = 19.708 (2) Å | µ = 0.10 mm−1 |
| α = 79.577 (8)° | T = 293 K |
| β = 87.162 (8)° | Plate, yellow |
| γ = 86.699 (8)° | 0.48 × 0.26 × 0.02 mm |
| V = 909.79 (15) Å3 |
4-(4-Nitrophenyl)piperazin-1-ium 4-methylbenzoate monohydrate (IV). Data collection
| Oxford Diffraction Xcalibur diffractometer | 1911 reflections with I > 2σ(I) |
| ω scans | Rint = 0.019 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | θmax = 25.4°, θmin = 3.1° |
| Tmin = 0.970, Tmax = 0.998 | h = −5→7 |
| 5980 measured reflections | k = −8→9 |
| 3347 independent reflections | l = −23→21 |
4-(4-Nitrophenyl)piperazin-1-ium 4-methylbenzoate monohydrate (IV). Refinement
| Refinement on F2 | 0 constraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.053 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.138 | w = 1/[σ2(Fo2) + (0.0554P)2 + 0.2441P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.01 | (Δ/σ)max < 0.001 |
| 3343 reflections | Δρmax = 0.20 e Å−3 |
| 248 parameters | Δρmin = −0.16 e Å−3 |
| 4 restraints |
4-(4-Nitrophenyl)piperazin-1-ium 4-methylbenzoate monohydrate (IV). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Nitrophenyl)piperazin-1-ium 4-methylbenzoate monohydrate (IV). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.7597 (5) | −0.1408 (4) | 0.52918 (12) | 0.1114 (9) | |
| O2 | 1.0599 (5) | −0.2808 (4) | 0.50875 (13) | 0.1312 (11) | |
| N1 | 0.8270 (3) | 0.0591 (2) | 0.20408 (9) | 0.0426 (5) | |
| N2 | 0.8376 (3) | 0.2441 (3) | 0.06365 (10) | 0.0492 (5) | |
| N3 | 0.9029 (6) | −0.1858 (4) | 0.48992 (13) | 0.0795 (8) | |
| C1 | 0.8503 (4) | 0.0072 (3) | 0.27523 (12) | 0.0421 (6) | |
| C2 | 0.6848 (5) | 0.0416 (4) | 0.32264 (13) | 0.0620 (8) | |
| H2 | 0.559385 | 0.107832 | 0.306884 | 0.074* | |
| C3 | 0.7020 (5) | −0.0200 (4) | 0.39241 (13) | 0.0660 (8) | |
| H3 | 0.58958 | 0.005008 | 0.423273 | 0.079* | |
| C4 | 0.8844 (5) | −0.1175 (4) | 0.41582 (13) | 0.0589 (7) | |
| C5 | 1.0511 (5) | −0.1516 (4) | 0.37142 (15) | 0.0698 (8) | |
| H5 | 1.175889 | −0.217226 | 0.388096 | 0.084* | |
| C6 | 1.0361 (4) | −0.0894 (4) | 0.30170 (13) | 0.0606 (8) | |
| H6 | 1.152207 | −0.112351 | 0.271761 | 0.073* | |
| C7 | 0.6682 (4) | 0.2049 (3) | 0.18122 (12) | 0.0490 (6) | |
| H7A | 0.721093 | 0.313511 | 0.191295 | 0.059* | |
| H7B | 0.530333 | 0.182601 | 0.207033 | 0.059* | |
| C8 | 0.6287 (4) | 0.2287 (4) | 0.10502 (12) | 0.0546 (7) | |
| H8A | 0.553149 | 0.128385 | 0.095886 | 0.066* | |
| H8B | 0.535474 | 0.334375 | 0.091454 | 0.066* | |
| C9 | 0.9774 (4) | 0.0810 (3) | 0.08466 (12) | 0.0525 (7) | |
| H9A | 1.113597 | 0.087916 | 0.057129 | 0.063* | |
| H9B | 0.902753 | −0.020822 | 0.076595 | 0.063* | |
| C10 | 1.0256 (4) | 0.0595 (3) | 0.15971 (12) | 0.0499 (6) | |
| H10A | 1.110342 | −0.050791 | 0.173238 | 0.06* | |
| H10B | 1.11409 | 0.155146 | 0.166426 | 0.06* | |
| O3 | 0.2656 (3) | 0.7140 (3) | 0.07107 (9) | 0.0598 (5) | |
| O4 | 0.0053 (3) | 0.5296 (3) | 0.11199 (9) | 0.0620 (5) | |
| C11 | 0.2728 (4) | 0.5764 (3) | 0.18855 (12) | 0.0399 (6) | |
| C12 | 0.4646 (4) | 0.6537 (3) | 0.19936 (13) | 0.0484 (6) | |
| H12 | 0.533917 | 0.726876 | 0.162881 | 0.058* | |
| C13 | 0.5535 (4) | 0.6230 (3) | 0.26365 (14) | 0.0575 (7) | |
| H13 | 0.68258 | 0.675862 | 0.269499 | 0.069* | |
| C14 | 0.4567 (5) | 0.5162 (3) | 0.31955 (14) | 0.0562 (7) | |
| C15 | 0.2645 (4) | 0.4395 (4) | 0.30874 (14) | 0.0584 (7) | |
| H15 | 0.195058 | 0.366932 | 0.345395 | 0.07* | |
| C16 | 0.1748 (4) | 0.4689 (3) | 0.24468 (13) | 0.0496 (6) | |
| H16 | 0.045866 | 0.41565 | 0.238897 | 0.06* | |
| C17 | 0.1749 (4) | 0.6080 (3) | 0.11893 (13) | 0.0437 (6) | |
| C18 | 0.5550 (6) | 0.4855 (4) | 0.38973 (16) | 0.0863 (10) | |
| H18A | 0.442848 | 0.502929 | 0.424033 | 0.129* | |
| H18B | 0.667715 | 0.567332 | 0.389742 | 0.129* | |
| H18C | 0.617002 | 0.366684 | 0.400046 | 0.129* | |
| O5 | 0.7006 (3) | 0.7136 (5) | 0.02422 (13) | 0.1211 (12) | |
| H21 | 0.903 (6) | 0.338 (4) | 0.073 (2) | 0.145* | |
| H22 | 0.807 (6) | 0.256 (5) | 0.0183 (11) | 0.145* | |
| H1W | 0.792 (5) | 0.667 (5) | 0.0534 (17) | 0.145* | |
| H2W | 0.576 (4) | 0.720 (6) | 0.045 (2) | 0.145* |
4-(4-Nitrophenyl)piperazin-1-ium 4-methylbenzoate monohydrate (IV). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.145 (2) | 0.132 (2) | 0.0477 (14) | 0.0055 (18) | 0.0100 (15) | −0.0001 (14) |
| O2 | 0.153 (2) | 0.155 (3) | 0.0702 (17) | 0.044 (2) | −0.0390 (17) | 0.0148 (17) |
| N1 | 0.0427 (11) | 0.0478 (12) | 0.0358 (11) | 0.0059 (9) | −0.0018 (9) | −0.0064 (9) |
| N2 | 0.0540 (13) | 0.0562 (14) | 0.0357 (11) | 0.0002 (11) | −0.0042 (10) | −0.0041 (10) |
| N3 | 0.111 (2) | 0.078 (2) | 0.0469 (17) | −0.0053 (17) | −0.0113 (16) | −0.0005 (14) |
| C1 | 0.0489 (14) | 0.0408 (14) | 0.0377 (14) | 0.0000 (11) | −0.0047 (11) | −0.0100 (11) |
| C2 | 0.0675 (17) | 0.0685 (19) | 0.0442 (16) | 0.0202 (15) | 0.0004 (13) | −0.0028 (14) |
| C3 | 0.083 (2) | 0.072 (2) | 0.0394 (16) | 0.0130 (17) | 0.0050 (14) | −0.0057 (14) |
| C4 | 0.085 (2) | 0.0556 (17) | 0.0352 (15) | 0.0002 (16) | −0.0113 (14) | −0.0044 (13) |
| C5 | 0.0719 (19) | 0.083 (2) | 0.0515 (19) | 0.0176 (17) | −0.0203 (15) | −0.0060 (16) |
| C6 | 0.0564 (16) | 0.078 (2) | 0.0448 (16) | 0.0167 (15) | −0.0066 (13) | −0.0092 (14) |
| C7 | 0.0428 (14) | 0.0616 (17) | 0.0401 (14) | 0.0080 (12) | −0.0034 (11) | −0.0050 (12) |
| C8 | 0.0441 (14) | 0.0706 (18) | 0.0468 (16) | 0.0043 (13) | −0.0071 (12) | −0.0052 (13) |
| C9 | 0.0597 (16) | 0.0579 (17) | 0.0385 (15) | 0.0061 (13) | 0.0028 (12) | −0.0086 (12) |
| C10 | 0.0499 (15) | 0.0558 (16) | 0.0417 (15) | 0.0138 (12) | −0.0008 (12) | −0.0080 (12) |
| O3 | 0.0574 (11) | 0.0779 (13) | 0.0404 (10) | −0.0028 (10) | −0.0074 (8) | 0.0006 (9) |
| O4 | 0.0577 (11) | 0.0758 (13) | 0.0557 (12) | −0.0102 (10) | −0.0167 (9) | −0.0143 (10) |
| C11 | 0.0404 (13) | 0.0387 (13) | 0.0413 (14) | 0.0068 (11) | −0.0074 (11) | −0.0103 (11) |
| C12 | 0.0513 (15) | 0.0449 (15) | 0.0490 (16) | −0.0006 (12) | −0.0065 (12) | −0.0072 (12) |
| C13 | 0.0547 (16) | 0.0533 (17) | 0.0679 (19) | 0.0004 (13) | −0.0224 (14) | −0.0158 (15) |
| C14 | 0.0707 (18) | 0.0497 (16) | 0.0498 (17) | 0.0066 (14) | −0.0233 (14) | −0.0103 (13) |
| C15 | 0.0706 (18) | 0.0570 (17) | 0.0453 (16) | −0.0042 (14) | −0.0099 (13) | −0.0007 (13) |
| C16 | 0.0488 (15) | 0.0518 (16) | 0.0485 (16) | −0.0039 (13) | −0.0089 (12) | −0.0074 (13) |
| C17 | 0.0427 (14) | 0.0473 (15) | 0.0420 (15) | 0.0071 (12) | −0.0042 (12) | −0.0121 (12) |
| C18 | 0.114 (3) | 0.079 (2) | 0.068 (2) | −0.003 (2) | −0.0467 (19) | −0.0066 (17) |
| O5 | 0.0602 (14) | 0.210 (3) | 0.0697 (16) | −0.0101 (18) | −0.0093 (12) | 0.0407 (18) |
4-(4-Nitrophenyl)piperazin-1-ium 4-methylbenzoate monohydrate (IV). Geometric parameters (Å, º)
| O1—N3 | 1.216 (3) | C9—C10 | 1.500 (3) |
| O2—N3 | 1.204 (3) | C9—H9A | 0.97 |
| N1—C1 | 1.399 (3) | C9—H9B | 0.97 |
| N1—C10 | 1.460 (3) | C10—H10A | 0.97 |
| N1—C7 | 1.463 (3) | C10—H10B | 0.97 |
| N2—C8 | 1.480 (3) | O3—O3 | 0.000 (5) |
| N2—C9 | 1.483 (3) | O3—C17 | 1.259 (3) |
| N2—H21 | 0.892 (19) | O4—C17 | 1.254 (3) |
| N2—H22 | 0.908 (19) | C11—C12 | 1.387 (3) |
| N3—C4 | 1.468 (3) | C11—C16 | 1.389 (3) |
| C1—C2 | 1.390 (3) | C11—C17 | 1.498 (3) |
| C1—C6 | 1.391 (3) | C12—C13 | 1.379 (3) |
| C2—C3 | 1.378 (4) | C12—H12 | 0.93 |
| C2—H2 | 0.93 | C13—C14 | 1.380 (4) |
| C3—C4 | 1.360 (4) | C13—H13 | 0.93 |
| C3—H3 | 0.93 | C14—C15 | 1.387 (4) |
| C4—C5 | 1.356 (4) | C14—C18 | 1.509 (4) |
| C5—C6 | 1.377 (4) | C15—C16 | 1.377 (3) |
| C5—H5 | 0.93 | C15—H15 | 0.93 |
| C6—H6 | 0.93 | C16—H16 | 0.93 |
| C7—C8 | 1.509 (3) | C18—H18A | 0.96 |
| C7—H7A | 0.97 | C18—H18B | 0.96 |
| C7—H7B | 0.97 | C18—H18C | 0.96 |
| C8—H8A | 0.97 | O5—H1W | 0.843 (19) |
| C8—H8B | 0.97 | O5—H2W | 0.854 (19) |
| C1—N1—C10 | 117.39 (18) | N2—C9—H9A | 109.6 |
| C1—N1—C7 | 117.37 (18) | C10—C9—H9A | 109.6 |
| C10—N1—C7 | 113.94 (18) | N2—C9—H9B | 109.6 |
| C8—N2—C9 | 108.7 (2) | C10—C9—H9B | 109.6 |
| C8—N2—H21 | 107 (3) | H9A—C9—H9B | 108.2 |
| C9—N2—H21 | 110 (3) | N1—C10—C9 | 112.7 (2) |
| C8—N2—H22 | 109 (3) | N1—C10—H10A | 109.1 |
| C9—N2—H22 | 110 (3) | C9—C10—H10A | 109.1 |
| H21—N2—H22 | 112 (4) | N1—C10—H10B | 109.1 |
| O2—N3—O1 | 123.5 (3) | C9—C10—H10B | 109.1 |
| O2—N3—C4 | 118.5 (3) | H10A—C10—H10B | 107.8 |
| O1—N3—C4 | 117.9 (3) | O3—O3—C17 | 0 (10) |
| C2—C1—C6 | 116.8 (2) | C12—C11—C16 | 117.5 (2) |
| C2—C1—N1 | 121.7 (2) | C12—C11—C17 | 121.2 (2) |
| C6—C1—N1 | 121.4 (2) | C16—C11—C17 | 121.2 (2) |
| C3—C2—C1 | 121.6 (3) | C13—C12—C11 | 120.7 (2) |
| C3—C2—H2 | 119.2 | C13—C12—H12 | 119.7 |
| C1—C2—H2 | 119.2 | C11—C12—H12 | 119.7 |
| C4—C3—C2 | 119.6 (3) | C12—C13—C14 | 122.0 (2) |
| C4—C3—H3 | 120.2 | C12—C13—H13 | 119 |
| C2—C3—H3 | 120.2 | C14—C13—H13 | 119 |
| C5—C4—C3 | 120.7 (2) | C13—C14—C15 | 117.3 (2) |
| C5—C4—N3 | 119.3 (3) | C13—C14—C18 | 121.3 (3) |
| C3—C4—N3 | 120.0 (3) | C15—C14—C18 | 121.4 (3) |
| C4—C5—C6 | 120.1 (3) | C16—C15—C14 | 121.2 (3) |
| C4—C5—H5 | 119.9 | C16—C15—H15 | 119.4 |
| C6—C5—H5 | 119.9 | C14—C15—H15 | 119.4 |
| C5—C6—C1 | 121.2 (2) | C15—C16—C11 | 121.3 (2) |
| C5—C6—H6 | 119.4 | C15—C16—H16 | 119.3 |
| C1—C6—H6 | 119.4 | C11—C16—H16 | 119.3 |
| N1—C7—C8 | 112.6 (2) | O4—C17—O3 | 123.8 (2) |
| N1—C7—H7A | 109.1 | O4—C17—O3 | 123.8 (2) |
| C8—C7—H7A | 109.1 | O3—C17—O3 | 0.0 (2) |
| N1—C7—H7B | 109.1 | O4—C17—C11 | 118.1 (2) |
| C8—C7—H7B | 109.1 | O3—C17—C11 | 118.0 (2) |
| H7A—C7—H7B | 107.8 | O3—C17—C11 | 118.0 (2) |
| N2—C8—C7 | 111.1 (2) | C14—C18—H18A | 109.5 |
| N2—C8—H8A | 109.4 | C14—C18—H18B | 109.5 |
| C7—C8—H8A | 109.4 | H18A—C18—H18B | 109.5 |
| N2—C8—H8B | 109.4 | C14—C18—H18C | 109.5 |
| C7—C8—H8B | 109.4 | H18A—C18—H18C | 109.5 |
| H8A—C8—H8B | 108 | H18B—C18—H18C | 109.5 |
| N2—C9—C10 | 110.1 (2) | H1W—O5—H2W | 108 (4) |
| C10—N1—C1—C2 | 164.4 (2) | C8—N2—C9—C10 | −60.7 (3) |
| C7—N1—C1—C2 | 22.9 (3) | C1—N1—C10—C9 | 168.3 (2) |
| C10—N1—C1—C6 | −18.9 (3) | C7—N1—C10—C9 | −48.9 (3) |
| C7—N1—C1—C6 | −160.4 (2) | N2—C9—C10—N1 | 55.7 (3) |
| C6—C1—C2—C3 | −1.2 (4) | C16—C11—C12—C13 | 0.3 (3) |
| N1—C1—C2—C3 | 175.6 (3) | C17—C11—C12—C13 | −179.7 (2) |
| C1—C2—C3—C4 | −0.3 (5) | C11—C12—C13—C14 | −0.3 (4) |
| C2—C3—C4—C5 | 1.3 (5) | C12—C13—C14—C15 | 0.1 (4) |
| C2—C3—C4—N3 | −179.1 (3) | C12—C13—C14—C18 | −179.4 (3) |
| O2—N3—C4—C5 | −5.0 (4) | C13—C14—C15—C16 | 0.1 (4) |
| O1—N3—C4—C5 | 173.8 (3) | C18—C14—C15—C16 | 179.6 (3) |
| O2—N3—C4—C3 | 175.5 (3) | C14—C15—C16—C11 | −0.1 (4) |
| O1—N3—C4—C3 | −5.7 (4) | C12—C11—C16—C15 | −0.1 (4) |
| C3—C4—C5—C6 | −0.8 (5) | C17—C11—C16—C15 | 179.9 (2) |
| N3—C4—C5—C6 | 179.6 (3) | O3—O3—C17—O4 | 0.00 (14) |
| C4—C5—C6—C1 | −0.8 (5) | O3—O3—C17—C11 | 0.0 (2) |
| C2—C1—C6—C5 | 1.8 (4) | C12—C11—C17—O4 | 177.7 (2) |
| N1—C1—C6—C5 | −175.0 (3) | C16—C11—C17—O4 | −2.4 (3) |
| C1—N1—C7—C8 | −170.1 (2) | C12—C11—C17—O3 | −3.5 (3) |
| C10—N1—C7—C8 | 47.0 (3) | C16—C11—C17—O3 | 176.4 (2) |
| C9—N2—C8—C7 | 59.5 (3) | C12—C11—C17—O3 | −3.5 (3) |
| N1—C7—C8—N2 | −52.7 (3) | C16—C11—C17—O3 | 176.4 (2) |
4-(4-Nitrophenyl)piperazin-1-ium 4-methylbenzoate monohydrate (IV). Hydrogen-bond geometry (Å, º)
Cg3 is the centroids of the C11–C16 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H21···O4i | 0.89 (2) | 1.93 (2) | 2.811 (3) | 167 (4) |
| N2—H22···O3ii | 0.91 (2) | 1.81 (2) | 2.717 (3) | 177 (4) |
| C3—H3···O1iii | 0.93 | 2.54 | 3.427 (4) | 161 |
| C9—H9A···O5iv | 0.97 | 2.31 | 3.113 (3) | 140 |
| O5—H1W···O4i | 0.84 (2) | 1.92 (2) | 2.756 (3) | 171 (4) |
| O5—H2W···O3 | 0.85 (2) | 1.94 (2) | 2.772 (3) | 164 (4) |
| C6—H6···Cg3v | 0.93 | 2.93 | 3.590 (3) | 129 |
Symmetry codes: (i) x+1, y, z; (ii) −x+1, −y+1, −z; (iii) −x+1, −y, −z+1; (iv) −x+2, −y+1, −z; (v) x+1, y−1, z.
4-(4-Nitrophenyl)piperazin-1-ium 4-methoxybenzoate hemihydrate (V). Crystal data
| 2C10H14N3O2+·2C8H7O3−·H2O | F(000) = 1560 |
| Mr = 736.77 | Dx = 1.328 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 2899 reflections |
| a = 15.808 (1) Å | θ = 2.6–25.3° |
| b = 7.5198 (7) Å | µ = 0.1 mm−1 |
| c = 31.020 (2) Å | T = 293 K |
| β = 92.561 (7)° | Prism, orange |
| V = 3683.8 (5) Å3 | 0.5 × 0.36 × 0.36 mm |
| Z = 4 |
4-(4-Nitrophenyl)piperazin-1-ium 4-methoxybenzoate hemihydrate (V). Data collection
| Oxford Diffraction Xcalibur diffractometer | 2602 reflections with I > 2σ(I) |
| ω scans | Rint = 0.066 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | θmax = 25.3°, θmin = 2.6° |
| Tmin = 0.958, Tmax = 0.965 | h = −19→18 |
| 15326 measured reflections | k = −9→8 |
| 6718 independent reflections | l = −37→33 |
4-(4-Nitrophenyl)piperazin-1-ium 4-methoxybenzoate hemihydrate (V). Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.074 | w = 1/[σ2(Fo2) + (0.0554P)2 + 0.9198P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.169 | (Δ/σ)max < 0.001 |
| S = 1.00 | Δρmax = 0.27 e Å−3 |
| 6715 reflections | Δρmin = −0.18 e Å−3 |
| 507 parameters | Extinction correction: SHELXL2018/3 (Sheldrick 2015b), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 45 restraints | Extinction coefficient: 0.0029 (4) |
| 0 constraints |
4-(4-Nitrophenyl)piperazin-1-ium 4-methoxybenzoate hemihydrate (V). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Nitrophenyl)piperazin-1-ium 4-methoxybenzoate hemihydrate (V). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| C1 | 0.2115 (2) | 0.2769 (5) | 0.15386 (13) | 0.0456 (11) | |
| C2 | 0.2042 (3) | 0.2439 (6) | 0.19789 (14) | 0.0687 (14) | |
| H2 | 0.151236 | 0.21805 | 0.208209 | 0.082* | |
| C3 | 0.2734 (3) | 0.2487 (7) | 0.22634 (14) | 0.0860 (16) | |
| H3 | 0.26712 | 0.22698 | 0.255532 | 0.103* | |
| C4 | 0.3512 (3) | 0.2855 (7) | 0.21153 (15) | 0.0702 (14) | |
| C5 | 0.3613 (3) | 0.3157 (6) | 0.16906 (15) | 0.0722 (14) | |
| H5 | 0.414916 | 0.339042 | 0.159279 | 0.087* | |
| C6 | 0.2925 (3) | 0.3119 (6) | 0.14031 (13) | 0.0598 (12) | |
| H6 | 0.300335 | 0.333225 | 0.111231 | 0.072* | |
| C7 | 0.1555 (2) | 0.2450 (6) | 0.07998 (12) | 0.0599 (12) | |
| H7A | 0.166523 | 0.119002 | 0.076636 | 0.072* | |
| H7B | 0.205192 | 0.309359 | 0.07138 | 0.072* | |
| C8 | 0.0813 (3) | 0.2965 (6) | 0.05086 (13) | 0.0690 (13) | |
| H8A | 0.075136 | 0.42483 | 0.050937 | 0.083* | |
| H8B | 0.09152 | 0.259489 | 0.021595 | 0.083* | |
| C9 | −0.0117 (3) | 0.2610 (6) | 0.10976 (14) | 0.0669 (13) | |
| H9A | −0.062504 | 0.202151 | 0.118906 | 0.08* | |
| H9B | −0.020149 | 0.38837 | 0.112112 | 0.08* | |
| C10 | 0.0624 (2) | 0.2059 (6) | 0.13852 (12) | 0.0631 (13) | |
| H10A | 0.052793 | 0.242915 | 0.16785 | 0.076* | |
| H10B | 0.066876 | 0.077208 | 0.138331 | 0.076* | |
| C11 | 0.9721 (3) | 0.2971 (6) | 0.30361 (13) | 0.0510 (11) | |
| C12 | 0.8980 (3) | 0.2023 (6) | 0.30443 (12) | 0.0563 (12) | |
| H12 | 0.888658 | 0.13067 | 0.328178 | 0.068* | |
| C13 | 0.8365 (3) | 0.2095 (6) | 0.27110 (14) | 0.0632 (13) | |
| H13 | 0.78756 | 0.14142 | 0.272281 | 0.076* | |
| C14 | 0.8487 (3) | 0.3182 (6) | 0.23643 (14) | 0.0628 (12) | |
| C15 | 0.9229 (3) | 0.4129 (6) | 0.23467 (13) | 0.0649 (13) | |
| H15 | 0.93208 | 0.484494 | 0.210883 | 0.078* | |
| C16 | 0.9839 (3) | 0.4028 (6) | 0.26775 (14) | 0.0626 (13) | |
| H16 | 1.033592 | 0.46798 | 0.26601 | 0.075* | |
| C17 | 1.0355 (3) | 0.2887 (7) | 0.34129 (16) | 0.0608 (13) | |
| C18 | 0.7146 (3) | 0.2457 (8) | 0.20218 (17) | 0.118 (2) | |
| H18C | 0.681472 | 0.273815 | 0.176387 | 0.178* | |
| H18B | 0.725759 | 0.120242 | 0.20308 | 0.178* | |
| H18A | 0.684002 | 0.279268 | 0.226948 | 0.178* | |
| C19 | 0.3816 (3) | 0.5044 (6) | −0.09170 (13) | 0.0503 (11) | |
| C20 | 0.3778 (3) | 0.6432 (6) | −0.06180 (13) | 0.0582 (12) | |
| H20 | 0.326082 | 0.697741 | −0.057457 | 0.07* | |
| C21 | 0.4478 (3) | 0.6998 (6) | −0.03904 (13) | 0.0603 (12) | |
| H21 | 0.443054 | 0.790435 | −0.018877 | 0.072* | |
| C22 | 0.5251 (2) | 0.6260 (6) | −0.04523 (13) | 0.0499 (11) | |
| C23 | 0.5330 (3) | 0.4942 (6) | −0.07542 (14) | 0.0610 (12) | |
| H23 | 0.58581 | 0.445412 | −0.080183 | 0.073* | |
| C24 | 0.4629 (3) | 0.4354 (6) | −0.09836 (13) | 0.0623 (13) | |
| H24 | 0.468943 | 0.347143 | −0.119 | 0.075* | |
| C25 | 0.3099 (3) | 0.2720 (7) | −0.13618 (14) | 0.0722 (14) | |
| H25A | 0.36547 | 0.248859 | −0.147048 | 0.087* | |
| H25B | 0.269401 | 0.27516 | −0.160582 | 0.087* | |
| C26 | 0.2867 (3) | 0.1245 (6) | −0.10589 (14) | 0.0694 (13) | |
| H26A | 0.284623 | 0.011999 | −0.121195 | 0.083* | |
| H26B | 0.329107 | 0.115475 | −0.082393 | 0.083* | |
| C27 | 0.1998 (2) | 0.3414 (6) | −0.06853 (12) | 0.0579 (12) | |
| H27A | 0.237409 | 0.345068 | −0.042968 | 0.07* | |
| H27B | 0.142682 | 0.365292 | −0.059877 | 0.07* | |
| C28 | 0.2258 (2) | 0.4799 (6) | −0.10009 (13) | 0.0583 (12) | |
| H28A | 0.186099 | 0.480675 | −0.124849 | 0.07* | |
| H28B | 0.224623 | 0.596261 | −0.086644 | 0.07* | |
| C29 | 0.2315 (2) | 0.8165 (5) | 0.03520 (12) | 0.0442 (10) | |
| C30 | 0.3063 (2) | 0.9094 (5) | 0.03221 (13) | 0.0517 (11) | |
| H30 | 0.319956 | 0.957242 | 0.005761 | 0.062* | |
| C31 | 0.3609 (2) | 0.9330 (6) | 0.06743 (15) | 0.0597 (12) | |
| H31 | 0.411473 | 0.994162 | 0.064463 | 0.072* | |
| C32 | 0.3412 (3) | 0.8662 (6) | 0.10717 (15) | 0.0570 (12) | |
| C33 | 0.2661 (3) | 0.7752 (6) | 0.11108 (13) | 0.0590 (12) | |
| H33 | 0.251489 | 0.731519 | 0.137782 | 0.071* | |
| C34 | 0.2130 (2) | 0.7495 (5) | 0.07521 (13) | 0.0524 (11) | |
| H34 | 0.163238 | 0.685277 | 0.077981 | 0.063* | |
| C35 | 0.1734 (3) | 0.7879 (6) | −0.00372 (14) | 0.0459 (11) | |
| C36 | 0.3801 (3) | 0.8360 (7) | 0.18157 (17) | 0.1098 (19) | |
| H36C | 0.42611 | 0.863024 | 0.201777 | 0.165* | |
| H36B | 0.329805 | 0.895356 | 0.190161 | 0.165* | |
| H36A | 0.370656 | 0.709953 | 0.181018 | 0.165* | |
| N1 | 0.14171 (19) | 0.2823 (4) | 0.12508 (10) | 0.0475 (9) | |
| N2 | 0.0026 (2) | 0.2144 (5) | 0.06465 (13) | 0.0581 (10) | |
| N3 | 0.4265 (6) | 0.2464 (13) | 0.2410 (3) | 0.075 (2) | 0.519 (6) |
| N3' | 0.4213 (6) | 0.3345 (15) | 0.2420 (3) | 0.075 (2) | 0.481 (6) |
| N4 | 0.3106 (2) | 0.4437 (5) | −0.11425 (10) | 0.0583 (10) | |
| N5 | 0.2034 (2) | 0.1639 (5) | −0.08869 (12) | 0.0612 (10) | |
| N6 | 0.5983 (2) | 0.6796 (6) | −0.01867 (13) | 0.0650 (11) | |
| O1 | 0.4960 (7) | 0.2571 (14) | 0.2266 (4) | 0.099 (2) | 0.519 (6) |
| O1' | 0.4904 (8) | 0.3633 (14) | 0.2283 (4) | 0.099 (2) | 0.481 (6) |
| O2 | 0.4177 (6) | 0.2116 (13) | 0.2788 (3) | 0.097 (2) | 0.519 (6) |
| O2' | 0.4086 (6) | 0.3408 (15) | 0.2797 (3) | 0.097 (2) | 0.481 (6) |
| O3 | 1.1003 (2) | 0.3818 (4) | 0.33942 (9) | 0.0826 (10) | |
| O4 | 1.0196 (2) | 0.1889 (4) | 0.37194 (10) | 0.0789 (10) | |
| O5 | 0.7922 (2) | 0.3398 (5) | 0.20234 (9) | 0.0895 (11) | |
| O6 | 0.58811 (19) | 0.7838 (5) | 0.01114 (11) | 0.0827 (11) | |
| O7 | 0.66853 (19) | 0.6203 (5) | −0.02668 (10) | 0.0881 (11) | |
| O8 | 0.10807 (17) | 0.6951 (4) | 0.00037 (8) | 0.0590 (8) | |
| O9 | 0.19207 (16) | 0.8596 (4) | −0.03853 (9) | 0.0593 (8) | |
| O10 | 0.40056 (18) | 0.8950 (4) | 0.13979 (10) | 0.0790 (10) | |
| O11 | 1.00751 (19) | 0.3517 (4) | 0.44766 (10) | 0.0649 (9) | |
| H21N | −0.0388 (19) | 0.252 (5) | 0.0468 (11) | 0.078* | |
| H22N | 0.007 (3) | 0.099 (3) | 0.0609 (12) | 0.078* | |
| H51N | 0.190 (2) | 0.077 (4) | −0.0717 (11) | 0.078* | |
| H52N | 0.164 (2) | 0.153 (6) | −0.1110 (9) | 0.078* | |
| H11O | 1.010 (3) | 0.296 (5) | 0.4244 (9) | 0.078* | |
| H12O | 0.969 (2) | 0.305 (5) | 0.4618 (12) | 0.078* |
4-(4-Nitrophenyl)piperazin-1-ium 4-methoxybenzoate hemihydrate (V). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.043 (3) | 0.040 (3) | 0.054 (3) | 0.002 (2) | 0.000 (2) | −0.002 (2) |
| C2 | 0.056 (3) | 0.100 (4) | 0.051 (3) | −0.008 (3) | 0.007 (3) | 0.001 (3) |
| C3 | 0.073 (3) | 0.145 (5) | 0.039 (3) | −0.011 (4) | −0.005 (3) | 0.004 (3) |
| C4 | 0.052 (3) | 0.108 (4) | 0.050 (3) | −0.009 (3) | −0.013 (2) | 0.001 (3) |
| C5 | 0.052 (3) | 0.107 (4) | 0.057 (3) | −0.017 (3) | −0.001 (3) | 0.005 (3) |
| C6 | 0.049 (3) | 0.082 (4) | 0.048 (3) | −0.005 (3) | −0.006 (2) | 0.009 (2) |
| C7 | 0.050 (3) | 0.082 (3) | 0.047 (3) | −0.009 (3) | −0.002 (2) | 0.002 (2) |
| C8 | 0.064 (3) | 0.077 (3) | 0.064 (3) | −0.023 (3) | −0.018 (2) | 0.017 (3) |
| C9 | 0.052 (3) | 0.076 (4) | 0.073 (4) | −0.003 (3) | −0.003 (2) | −0.015 (3) |
| C10 | 0.052 (3) | 0.089 (4) | 0.048 (3) | −0.013 (3) | 0.000 (2) | −0.003 (2) |
| C11 | 0.055 (3) | 0.049 (3) | 0.049 (3) | 0.004 (3) | 0.004 (2) | 0.002 (2) |
| C12 | 0.071 (3) | 0.059 (3) | 0.039 (3) | 0.007 (3) | 0.006 (2) | 0.008 (2) |
| C13 | 0.058 (3) | 0.074 (4) | 0.058 (3) | −0.010 (3) | 0.000 (2) | 0.010 (3) |
| C14 | 0.064 (3) | 0.071 (4) | 0.051 (3) | 0.007 (3) | −0.012 (3) | 0.004 (3) |
| C15 | 0.068 (3) | 0.070 (4) | 0.056 (3) | −0.009 (3) | −0.004 (3) | 0.018 (2) |
| C16 | 0.058 (3) | 0.064 (3) | 0.065 (3) | −0.009 (3) | 0.000 (3) | 0.009 (3) |
| C17 | 0.063 (3) | 0.057 (4) | 0.062 (3) | 0.014 (3) | −0.010 (3) | −0.011 (3) |
| C18 | 0.103 (5) | 0.148 (6) | 0.100 (4) | −0.040 (4) | −0.047 (3) | 0.023 (4) |
| C19 | 0.044 (3) | 0.065 (3) | 0.042 (3) | −0.002 (3) | 0.007 (2) | 0.009 (2) |
| C20 | 0.038 (3) | 0.071 (3) | 0.066 (3) | 0.003 (3) | 0.002 (2) | −0.004 (3) |
| C21 | 0.046 (3) | 0.071 (3) | 0.064 (3) | −0.002 (3) | 0.008 (2) | −0.009 (2) |
| C22 | 0.035 (3) | 0.064 (3) | 0.051 (3) | −0.007 (2) | 0.002 (2) | 0.009 (2) |
| C23 | 0.038 (3) | 0.068 (3) | 0.077 (3) | 0.004 (3) | 0.012 (2) | 0.005 (3) |
| C24 | 0.050 (3) | 0.067 (3) | 0.071 (3) | −0.003 (3) | 0.015 (3) | −0.011 (2) |
| C25 | 0.063 (3) | 0.100 (4) | 0.054 (3) | −0.009 (3) | 0.006 (2) | −0.023 (3) |
| C26 | 0.069 (3) | 0.068 (4) | 0.070 (3) | 0.003 (3) | −0.008 (3) | −0.015 (3) |
| C27 | 0.049 (3) | 0.070 (3) | 0.054 (3) | −0.004 (3) | −0.003 (2) | −0.012 (3) |
| C28 | 0.042 (3) | 0.063 (3) | 0.069 (3) | −0.006 (2) | −0.008 (2) | 0.001 (3) |
| C29 | 0.043 (2) | 0.045 (3) | 0.045 (3) | 0.005 (2) | 0.004 (2) | −0.003 (2) |
| C30 | 0.044 (3) | 0.063 (3) | 0.049 (3) | 0.006 (2) | 0.004 (2) | 0.007 (2) |
| C31 | 0.044 (3) | 0.071 (3) | 0.064 (3) | −0.005 (2) | −0.005 (2) | −0.001 (3) |
| C32 | 0.053 (3) | 0.064 (3) | 0.053 (3) | 0.012 (3) | −0.013 (3) | −0.008 (3) |
| C33 | 0.062 (3) | 0.067 (3) | 0.048 (3) | −0.001 (3) | −0.005 (2) | 0.005 (2) |
| C34 | 0.052 (3) | 0.055 (3) | 0.050 (3) | −0.004 (2) | −0.002 (2) | 0.006 (2) |
| C35 | 0.048 (3) | 0.040 (3) | 0.050 (3) | 0.009 (2) | −0.005 (2) | −0.002 (2) |
| C36 | 0.117 (5) | 0.125 (5) | 0.083 (4) | 0.008 (4) | −0.035 (3) | −0.004 (4) |
| N1 | 0.041 (2) | 0.055 (2) | 0.046 (2) | −0.0042 (18) | −0.0010 (17) | −0.0018 (17) |
| N2 | 0.055 (3) | 0.047 (2) | 0.070 (3) | −0.005 (2) | −0.0165 (19) | 0.004 (2) |
| N3 | 0.072 (3) | 0.081 (4) | 0.070 (3) | −0.002 (3) | −0.004 (2) | −0.001 (3) |
| N3' | 0.072 (3) | 0.081 (4) | 0.070 (3) | −0.002 (3) | −0.004 (2) | −0.001 (3) |
| N4 | 0.051 (2) | 0.072 (3) | 0.052 (2) | −0.005 (2) | 0.0048 (19) | −0.002 (2) |
| N5 | 0.059 (3) | 0.063 (3) | 0.061 (3) | −0.012 (2) | −0.009 (2) | 0.010 (2) |
| N6 | 0.048 (3) | 0.084 (3) | 0.064 (3) | −0.012 (3) | 0.003 (2) | 0.021 (2) |
| O1 | 0.075 (3) | 0.127 (7) | 0.092 (3) | −0.011 (5) | −0.020 (2) | −0.001 (6) |
| O1' | 0.075 (3) | 0.127 (7) | 0.092 (3) | −0.011 (5) | −0.020 (2) | −0.001 (6) |
| O2 | 0.097 (3) | 0.131 (6) | 0.062 (3) | −0.003 (5) | −0.015 (2) | 0.001 (5) |
| O2' | 0.097 (3) | 0.131 (6) | 0.062 (3) | −0.003 (5) | −0.015 (2) | 0.001 (5) |
| O3 | 0.073 (2) | 0.088 (3) | 0.084 (2) | −0.010 (2) | −0.0291 (18) | 0.0096 (18) |
| O4 | 0.106 (3) | 0.069 (2) | 0.060 (2) | 0.003 (2) | −0.0170 (18) | 0.0134 (18) |
| O5 | 0.082 (2) | 0.115 (3) | 0.070 (2) | −0.013 (2) | −0.0256 (19) | 0.0253 (19) |
| O6 | 0.072 (2) | 0.110 (3) | 0.066 (2) | −0.023 (2) | 0.0016 (18) | −0.002 (2) |
| O7 | 0.046 (2) | 0.128 (3) | 0.090 (2) | −0.001 (2) | 0.0000 (18) | 0.023 (2) |
| O8 | 0.0548 (18) | 0.061 (2) | 0.060 (2) | −0.0126 (17) | −0.0085 (14) | 0.0027 (15) |
| O9 | 0.0639 (19) | 0.066 (2) | 0.0473 (19) | 0.0027 (16) | −0.0024 (15) | 0.0080 (15) |
| O10 | 0.072 (2) | 0.102 (3) | 0.061 (2) | 0.0032 (19) | −0.0220 (18) | −0.0004 (19) |
| O11 | 0.063 (2) | 0.064 (2) | 0.069 (2) | 0.0012 (18) | 0.0089 (17) | −0.0012 (17) |
4-(4-Nitrophenyl)piperazin-1-ium 4-methoxybenzoate hemihydrate (V). Geometric parameters (Å, º)
| C1—N1 | 1.388 (4) | C21—H21 | 0.93 |
| C1—C6 | 1.391 (5) | C22—C23 | 1.373 (5) |
| C1—C2 | 1.398 (5) | C22—N6 | 1.448 (5) |
| C2—C3 | 1.375 (6) | C23—C24 | 1.363 (5) |
| C2—H2 | 0.93 | C23—H23 | 0.93 |
| C3—C4 | 1.359 (6) | C24—H24 | 0.93 |
| C3—H3 | 0.93 | C25—N4 | 1.459 (5) |
| C4—C5 | 1.354 (5) | C25—C26 | 1.509 (6) |
| C4—N3' | 1.471 (10) | C25—H25A | 0.97 |
| C4—N3 | 1.499 (9) | C25—H25B | 0.97 |
| C5—C6 | 1.375 (5) | C26—N5 | 1.474 (5) |
| C5—H5 | 0.93 | C26—H26A | 0.97 |
| C6—H6 | 0.93 | C26—H26B | 0.97 |
| C7—N1 | 1.453 (4) | C27—N5 | 1.476 (5) |
| C7—C8 | 1.499 (5) | C27—C28 | 1.499 (5) |
| C7—H7A | 0.97 | C27—H27A | 0.97 |
| C7—H7B | 0.97 | C27—H27B | 0.97 |
| C8—N2 | 1.469 (5) | C28—N4 | 1.455 (4) |
| C8—H8A | 0.97 | C28—H28A | 0.97 |
| C8—H8B | 0.97 | C28—H28B | 0.97 |
| C9—N2 | 1.470 (5) | C29—C30 | 1.379 (5) |
| C9—C10 | 1.499 (5) | C29—C34 | 1.383 (5) |
| C9—H9A | 0.97 | C29—C35 | 1.499 (5) |
| C9—H9B | 0.97 | C30—C31 | 1.374 (5) |
| C10—N1 | 1.457 (4) | C30—H30 | 0.93 |
| C10—H10A | 0.97 | C31—C32 | 1.379 (5) |
| C10—H10B | 0.97 | C31—H31 | 0.93 |
| C11—C12 | 1.372 (5) | C32—O10 | 1.366 (4) |
| C11—C16 | 1.387 (5) | C32—C33 | 1.380 (5) |
| C11—C17 | 1.506 (6) | C33—C34 | 1.377 (5) |
| C12—C13 | 1.388 (5) | C33—H33 | 0.93 |
| C12—H12 | 0.93 | C34—H34 | 0.93 |
| C13—C14 | 1.371 (5) | C35—O9 | 1.254 (4) |
| C13—H13 | 0.93 | C35—O8 | 1.258 (4) |
| C14—O5 | 1.363 (5) | C36—O10 | 1.421 (5) |
| C14—C15 | 1.375 (5) | C36—H36C | 0.96 |
| C15—C16 | 1.378 (5) | C36—H36B | 0.96 |
| C15—H15 | 0.93 | C36—H36A | 0.96 |
| C16—H16 | 0.93 | N2—H21N | 0.884 (18) |
| C17—O3 | 1.245 (5) | N2—H22N | 0.882 (18) |
| C17—O4 | 1.246 (5) | N3—O1 | 1.208 (9) |
| C18—O5 | 1.416 (5) | N3—O2 | 1.213 (9) |
| C18—H18C | 0.96 | N3'—O2' | 1.196 (9) |
| C18—H18B | 0.96 | N3'—O1' | 1.210 (10) |
| C18—H18A | 0.96 | N5—H51N | 0.872 (18) |
| C19—N4 | 1.374 (4) | N5—H52N | 0.910 (18) |
| C19—C20 | 1.399 (5) | N6—O6 | 1.228 (4) |
| C19—C24 | 1.410 (5) | N6—O7 | 1.232 (4) |
| C20—C21 | 1.354 (5) | O11—H11O | 0.836 (18) |
| C20—H20 | 0.93 | O11—H12O | 0.838 (18) |
| C21—C22 | 1.363 (5) | ||
| N1—C1—C6 | 121.1 (4) | C22—C23—H23 | 120.2 |
| N1—C1—C2 | 122.4 (4) | C23—C24—C19 | 121.9 (4) |
| C6—C1—C2 | 116.5 (4) | C23—C24—H24 | 119 |
| C3—C2—C1 | 121.6 (4) | C19—C24—H24 | 119 |
| C3—C2—H2 | 119.2 | N4—C25—C26 | 110.9 (3) |
| C1—C2—H2 | 119.2 | N4—C25—H25A | 109.5 |
| C4—C3—C2 | 119.6 (4) | C26—C25—H25A | 109.5 |
| C4—C3—H3 | 120.2 | N4—C25—H25B | 109.5 |
| C2—C3—H3 | 120.2 | C26—C25—H25B | 109.5 |
| C5—C4—C3 | 120.7 (4) | H25A—C25—H25B | 108 |
| C5—C4—N3' | 117.8 (6) | N5—C26—C25 | 108.9 (4) |
| C3—C4—N3' | 120.0 (6) | N5—C26—H26A | 109.9 |
| C5—C4—N3 | 120.2 (6) | C25—C26—H26A | 109.9 |
| C3—C4—N3 | 117.3 (6) | N5—C26—H26B | 109.9 |
| C4—C5—C6 | 120.2 (4) | C25—C26—H26B | 109.9 |
| C4—C5—H5 | 119.9 | H26A—C26—H26B | 108.3 |
| C6—C5—H5 | 119.9 | N5—C27—C28 | 109.5 (3) |
| C5—C6—C1 | 121.3 (4) | N5—C27—H27A | 109.8 |
| C5—C6—H6 | 119.3 | C28—C27—H27A | 109.8 |
| C1—C6—H6 | 119.3 | N5—C27—H27B | 109.8 |
| N1—C7—C8 | 112.5 (3) | C28—C27—H27B | 109.8 |
| N1—C7—H7A | 109.1 | H27A—C27—H27B | 108.2 |
| C8—C7—H7A | 109.1 | N4—C28—C27 | 110.5 (3) |
| N1—C7—H7B | 109.1 | N4—C28—H28A | 109.5 |
| C8—C7—H7B | 109.1 | C27—C28—H28A | 109.5 |
| H7A—C7—H7B | 107.8 | N4—C28—H28B | 109.5 |
| N2—C8—C7 | 111.5 (3) | C27—C28—H28B | 109.5 |
| N2—C8—H8A | 109.3 | H28A—C28—H28B | 108.1 |
| C7—C8—H8A | 109.3 | C30—C29—C34 | 117.5 (4) |
| N2—C8—H8B | 109.3 | C30—C29—C35 | 120.9 (4) |
| C7—C8—H8B | 109.3 | C34—C29—C35 | 121.6 (4) |
| H8A—C8—H8B | 108 | C31—C30—C29 | 121.5 (4) |
| N2—C9—C10 | 110.5 (3) | C31—C30—H30 | 119.3 |
| N2—C9—H9A | 109.5 | C29—C30—H30 | 119.3 |
| C10—C9—H9A | 109.5 | C30—C31—C32 | 120.3 (4) |
| N2—C9—H9B | 109.5 | C30—C31—H31 | 119.8 |
| C10—C9—H9B | 109.5 | C32—C31—H31 | 119.8 |
| H9A—C9—H9B | 108.1 | O10—C32—C31 | 115.4 (4) |
| N1—C10—C9 | 112.3 (3) | O10—C32—C33 | 125.3 (4) |
| N1—C10—H10A | 109.1 | C31—C32—C33 | 119.2 (4) |
| C9—C10—H10A | 109.1 | C34—C33—C32 | 119.6 (4) |
| N1—C10—H10B | 109.1 | C34—C33—H33 | 120.2 |
| C9—C10—H10B | 109.1 | C32—C33—H33 | 120.2 |
| H10A—C10—H10B | 107.9 | C33—C34—C29 | 121.8 (4) |
| C12—C11—C16 | 117.2 (4) | C33—C34—H34 | 119.1 |
| C12—C11—C17 | 120.2 (4) | C29—C34—H34 | 119.1 |
| C16—C11—C17 | 122.5 (4) | O9—C35—O8 | 123.4 (4) |
| C11—C12—C13 | 122.4 (4) | O9—C35—C29 | 118.2 (4) |
| C11—C12—H12 | 118.8 | O8—C35—C29 | 118.3 (4) |
| C13—C12—H12 | 118.8 | O10—C36—H36C | 109.5 |
| C14—C13—C12 | 119.3 (4) | O10—C36—H36B | 109.5 |
| C14—C13—H13 | 120.4 | H36C—C36—H36B | 109.5 |
| C12—C13—H13 | 120.4 | O10—C36—H36A | 109.5 |
| O5—C14—C13 | 124.7 (4) | H36C—C36—H36A | 109.5 |
| O5—C14—C15 | 116.0 (4) | H36B—C36—H36A | 109.5 |
| C13—C14—C15 | 119.3 (4) | C1—N1—C7 | 117.7 (3) |
| C14—C15—C16 | 120.7 (4) | C1—N1—C10 | 118.2 (3) |
| C14—C15—H15 | 119.6 | C7—N1—C10 | 111.6 (3) |
| C16—C15—H15 | 119.6 | C8—N2—C9 | 110.2 (3) |
| C15—C16—C11 | 121.0 (4) | C8—N2—H21N | 107 (3) |
| C15—C16—H16 | 119.5 | C9—N2—H21N | 112 (3) |
| C11—C16—H16 | 119.5 | C8—N2—H22N | 108 (3) |
| O3—C17—O4 | 124.6 (4) | C9—N2—H22N | 112 (3) |
| O3—C17—C11 | 117.5 (5) | H21N—N2—H22N | 107 (4) |
| O4—C17—C11 | 117.9 (5) | O1—N3—O2 | 121.2 (11) |
| O5—C18—H18C | 109.5 | O1—N3—C4 | 118.2 (10) |
| O5—C18—H18B | 109.5 | O2—N3—C4 | 120.6 (9) |
| H18C—C18—H18B | 109.5 | O2'—N3'—O1' | 122.0 (11) |
| O5—C18—H18A | 109.5 | O2'—N3'—C4 | 119.0 (9) |
| H18C—C18—H18A | 109.5 | O1'—N3'—C4 | 119.0 (10) |
| H18B—C18—H18A | 109.5 | C19—N4—C28 | 121.8 (3) |
| N4—C19—C20 | 121.8 (4) | C19—N4—C25 | 121.4 (4) |
| N4—C19—C24 | 122.2 (4) | C28—N4—C25 | 108.6 (3) |
| C20—C19—C24 | 116.0 (4) | C26—N5—C27 | 112.8 (3) |
| C21—C20—C19 | 121.5 (4) | C26—N5—H51N | 108 (3) |
| C21—C20—H20 | 119.3 | C27—N5—H51N | 114 (3) |
| C19—C20—H20 | 119.3 | C26—N5—H52N | 107 (3) |
| C20—C21—C22 | 121.0 (4) | C27—N5—H52N | 112 (3) |
| C20—C21—H21 | 119.5 | H51N—N5—H52N | 103 (4) |
| C22—C21—H21 | 119.5 | O6—N6—O7 | 122.1 (4) |
| C21—C22—C23 | 120.0 (4) | O6—N6—C22 | 118.5 (4) |
| C21—C22—N6 | 120.4 (4) | O7—N6—C22 | 119.3 (4) |
| C23—C22—N6 | 119.6 (4) | C14—O5—C18 | 118.7 (4) |
| C24—C23—C22 | 119.6 (4) | C32—O10—C36 | 116.7 (4) |
| C24—C23—H23 | 120.2 | H11O—O11—H12O | 108 (5) |
| N1—C1—C2—C3 | −176.3 (4) | O10—C32—C33—C34 | −177.5 (4) |
| C6—C1—C2—C3 | 1.0 (6) | C31—C32—C33—C34 | 1.3 (6) |
| C1—C2—C3—C4 | −0.3 (8) | C32—C33—C34—C29 | −1.7 (6) |
| C2—C3—C4—C5 | −0.7 (8) | C30—C29—C34—C33 | 0.7 (6) |
| C2—C3—C4—N3' | 164.9 (6) | C35—C29—C34—C33 | 179.9 (4) |
| C2—C3—C4—N3 | −165.5 (6) | C30—C29—C35—O9 | −4.2 (5) |
| C3—C4—C5—C6 | 0.9 (8) | C34—C29—C35—O9 | 176.6 (3) |
| N3'—C4—C5—C6 | −165.0 (6) | C30—C29—C35—O8 | 176.7 (3) |
| N3—C4—C5—C6 | 165.4 (6) | C34—C29—C35—O8 | −2.5 (5) |
| C4—C5—C6—C1 | −0.2 (7) | C6—C1—N1—C7 | 27.3 (5) |
| N1—C1—C6—C5 | 176.6 (4) | C2—C1—N1—C7 | −155.5 (4) |
| C2—C1—C6—C5 | −0.7 (6) | C6—C1—N1—C10 | 166.2 (4) |
| N1—C7—C8—N2 | −54.0 (5) | C2—C1—N1—C10 | −16.6 (6) |
| N2—C9—C10—N1 | 56.1 (5) | C8—C7—N1—C1 | −166.4 (3) |
| C16—C11—C12—C13 | 0.2 (6) | C8—C7—N1—C10 | 52.2 (4) |
| C17—C11—C12—C13 | 178.1 (4) | C9—C10—N1—C1 | 165.3 (3) |
| C11—C12—C13—C14 | −1.6 (7) | C9—C10—N1—C7 | −53.4 (5) |
| C12—C13—C14—O5 | −178.3 (4) | C7—C8—N2—C9 | 55.9 (5) |
| C12—C13—C14—C15 | 2.3 (7) | C10—C9—N2—C8 | −56.8 (4) |
| O5—C14—C15—C16 | 179.0 (4) | C5—C4—N3—O1 | 5.9 (11) |
| C13—C14—C15—C16 | −1.6 (7) | C3—C4—N3—O1 | 170.9 (8) |
| C14—C15—C16—C11 | 0.1 (7) | C5—C4—N3—O2 | −176.8 (7) |
| C12—C11—C16—C15 | 0.6 (6) | C3—C4—N3—O2 | −11.8 (11) |
| C17—C11—C16—C15 | −177.3 (4) | C5—C4—N3'—O2' | 166.2 (8) |
| C12—C11—C17—O3 | −177.7 (4) | C3—C4—N3'—O2' | 0.2 (12) |
| C16—C11—C17—O3 | 0.1 (6) | C5—C4—N3'—O1' | −15.5 (12) |
| C12—C11—C17—O4 | 2.6 (6) | C3—C4—N3'—O1' | 178.5 (9) |
| C16—C11—C17—O4 | −179.5 (4) | C20—C19—N4—C28 | −18.5 (6) |
| N4—C19—C20—C21 | 178.6 (4) | C24—C19—N4—C28 | 163.8 (4) |
| C24—C19—C20—C21 | −3.6 (6) | C20—C19—N4—C25 | −163.3 (4) |
| C19—C20—C21—C22 | 1.6 (6) | C24—C19—N4—C25 | 19.0 (6) |
| C20—C21—C22—C23 | 1.1 (6) | C27—C28—N4—C19 | −86.8 (5) |
| C20—C21—C22—N6 | −175.8 (4) | C27—C28—N4—C25 | 61.9 (4) |
| C21—C22—C23—C24 | −1.4 (6) | C26—C25—N4—C19 | 86.9 (4) |
| N6—C22—C23—C24 | 175.5 (4) | C26—C25—N4—C28 | −61.9 (4) |
| C22—C23—C24—C19 | −0.8 (6) | C25—C26—N5—C27 | −54.1 (5) |
| N4—C19—C24—C23 | −179.0 (4) | C28—C27—N5—C26 | 54.7 (4) |
| C20—C19—C24—C23 | 3.2 (6) | C21—C22—N6—O6 | 4.5 (6) |
| N4—C25—C26—N5 | 57.6 (5) | C23—C22—N6—O6 | −172.4 (4) |
| N5—C27—C28—N4 | −58.0 (4) | C21—C22—N6—O7 | −175.1 (4) |
| C34—C29—C30—C31 | 0.8 (6) | C23—C22—N6—O7 | 8.0 (6) |
| C35—C29—C30—C31 | −178.5 (4) | C13—C14—O5—C18 | 0.5 (7) |
| C29—C30—C31—C32 | −1.2 (6) | C15—C14—O5—C18 | 179.9 (4) |
| C30—C31—C32—O10 | 179.1 (4) | C31—C32—O10—C36 | 176.8 (4) |
| C30—C31—C32—C33 | 0.2 (6) | C33—C32—O10—C36 | −4.4 (6) |
4-(4-Nitrophenyl)piperazin-1-ium 4-methoxybenzoate hemihydrate (V). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C7—H7B···O7i | 0.97 | 2.54 | 3.451 (5) | 157 |
| C9—H9B···O4ii | 0.97 | 2.31 | 3.270 (5) | 169 |
| C20—H20···O9 | 0.93 | 2.53 | 3.461 (5) | 174 |
| C25—H25A···O2aiii | 0.97 | 2.5 | 3.206 (10) | 130 |
| C25—H25A···O2′biii | 0.97 | 2.49 | 3.212 (11) | 131 |
| C27—H27A···O7i | 0.97 | 2.58 | 3.548 (5) | 175 |
| C28—H28B···O9 | 0.97 | 2.55 | 3.489 (5) | 164 |
| C36—H36C···O1′bii | 0.96 | 2.49 | 3.395 (14) | 158 |
| N2—H21N···O8iv | 0.88 (2) | 1.83 (2) | 2.697 (4) | 166 (4) |
| N2—H21N···O9iv | 0.88 (2) | 2.57 (3) | 3.196 (4) | 129 (3) |
| N2—H22N···O11v | 0.88 (2) | 1.89 (2) | 2.758 (5) | 169 (4) |
| N5—H51N···O9vi | 0.87 (2) | 1.93 (2) | 2.778 (5) | 164 (4) |
| N5—H52N···O3vii | 0.91 (2) | 1.82 (2) | 2.724 (5) | 171 (4) |
| O11—H11O···O4 | 0.84 (2) | 1.83 (2) | 2.663 (4) | 176 (4) |
| O11—H12O···O8v | 0.84 (2) | 1.92 (2) | 2.754 (4) | 173 (4) |
Symmetry codes: (i) −x+1, −y+1, −z; (ii) −x+1, y+1/2, −z+1/2; (iii) x, −y+1/2, z−1/2; (iv) −x, −y+1, −z; (v) −x+1, y−1/2, −z+1/2; (vi) x, y−1, z; (vii) x−1, −y+1/2, z−1/2.
4-(4-Nitrophenyl)piperazin-1-ium 4-ethoxybenzoate (VI). Crystal data
| C10H14N3O2+·C9H9O3− | Z = 4 |
| Mr = 373.4 | F(000) = 792 |
| Triclinic, P1 | Dx = 1.324 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.874 (1) Å | Cell parameters from 4134 reflections |
| b = 9.263 (1) Å | θ = 2.4–28.0° |
| c = 27.996 (3) Å | µ = 0.10 mm−1 |
| α = 81.030 (6)° | T = 293 K |
| β = 85.675 (6)° | Plate, yellow |
| γ = 68.229 (5)° | 0.44 × 0.32 × 0.08 mm |
| V = 1872.8 (4) Å3 |
4-(4-Nitrophenyl)piperazin-1-ium 4-ethoxybenzoate (VI). Data collection
| Oxford Diffraction Xcalibur diffractometer | 3803 reflections with I > 2σ(I) |
| ω scans | Rint = 0.027 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | θmax = 25.4°, θmin = 2.4° |
| Tmin = 0.963, Tmax = 0.992 | h = −9→5 |
| 13344 measured reflections | k = −11→10 |
| 6868 independent reflections | l = −33→33 |
4-(4-Nitrophenyl)piperazin-1-ium 4-ethoxybenzoate (VI). Refinement
| Refinement on F2 | 0 constraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.061 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.137 | w = 1/[σ2(Fo2) + (0.0432P)2 + 0.7484P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max < 0.001 |
| 6858 reflections | Δρmax = 0.23 e Å−3 |
| 501 parameters | Δρmin = −0.22 e Å−3 |
| 16 restraints |
4-(4-Nitrophenyl)piperazin-1-ium 4-ethoxybenzoate (VI). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Nitrophenyl)piperazin-1-ium 4-ethoxybenzoate (VI). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | −0.3466 (3) | 0.6545 (4) | 0.40786 (9) | 0.1126 (10) | |
| O2 | −0.1630 (3) | 0.5811 (3) | 0.46683 (9) | 0.0901 (8) | |
| N3 | −0.1931 (4) | 0.6107 (3) | 0.42384 (10) | 0.0667 (7) | |
| N1 | 0.3914 (3) | 0.5404 (2) | 0.29014 (7) | 0.0412 (5) | |
| N2 | 0.6838 (3) | 0.4199 (3) | 0.22275 (9) | 0.0486 (6) | |
| C1 | 0.2485 (3) | 0.5547 (3) | 0.32330 (9) | 0.0364 (6) | |
| C2 | 0.0659 (4) | 0.6276 (3) | 0.30870 (10) | 0.0452 (7) | |
| H2 | 0.041054 | 0.663561 | 0.276117 | 0.054* | |
| C3 | −0.0759 (4) | 0.6467 (3) | 0.34138 (10) | 0.0487 (7) | |
| H3 | −0.195824 | 0.69631 | 0.330993 | 0.058* | |
| C4 | −0.0417 (4) | 0.5927 (3) | 0.38966 (10) | 0.0470 (7) | |
| C5 | 0.1352 (4) | 0.5197 (3) | 0.40558 (10) | 0.0460 (7) | |
| H5 | 0.157333 | 0.483265 | 0.438246 | 0.055* | |
| C6 | 0.2787 (4) | 0.5009 (3) | 0.37309 (10) | 0.0430 (7) | |
| H6 | 0.397837 | 0.451799 | 0.384052 | 0.052* | |
| C7 | 0.5772 (3) | 0.4477 (3) | 0.30663 (10) | 0.0473 (7) | |
| H7A | 0.588757 | 0.338899 | 0.315234 | 0.057* | |
| H7B | 0.597413 | 0.484321 | 0.33554 | 0.057* | |
| C8 | 0.7216 (4) | 0.4565 (4) | 0.26954 (10) | 0.0539 (8) | |
| H8A | 0.725864 | 0.561056 | 0.265102 | 0.065* | |
| H8B | 0.839991 | 0.382613 | 0.280859 | 0.065* | |
| C9 | 0.5080 (4) | 0.5395 (3) | 0.20545 (10) | 0.0528 (8) | |
| H9A | 0.482935 | 0.520499 | 0.174084 | 0.063* | |
| H9B | 0.516375 | 0.642512 | 0.201592 | 0.063* | |
| C10 | 0.3546 (4) | 0.5366 (4) | 0.23985 (9) | 0.0527 (8) | |
| H10A | 0.245506 | 0.626087 | 0.229683 | 0.063* | |
| H10B | 0.329888 | 0.442345 | 0.238274 | 0.063* | |
| O3 | −0.0728 (3) | 0.4331 (2) | 0.15512 (7) | 0.0578 (5) | |
| O4 | 0.1149 (2) | 0.2978 (2) | 0.21489 (7) | 0.0532 (5) | |
| O5 | 0.6778 (3) | 0.3001 (3) | 0.03965 (7) | 0.0734 (7) | |
| C11 | 0.2434 (4) | 0.3357 (3) | 0.13647 (9) | 0.0400 (6) | |
| C12 | 0.4193 (4) | 0.2483 (3) | 0.15096 (10) | 0.0518 (8) | |
| H12 | 0.438717 | 0.198421 | 0.182667 | 0.062* | |
| C13 | 0.5679 (4) | 0.2321 (4) | 0.11995 (10) | 0.0572 (8) | |
| H13 | 0.685415 | 0.171644 | 0.130663 | 0.069* | |
| C14 | 0.5409 (4) | 0.3059 (3) | 0.07314 (10) | 0.0524 (8) | |
| C15 | 0.3656 (4) | 0.3930 (4) | 0.05764 (10) | 0.0609 (9) | |
| H15 | 0.346406 | 0.442258 | 0.02587 | 0.073* | |
| C16 | 0.2192 (4) | 0.4072 (3) | 0.08898 (10) | 0.0520 (8) | |
| H16 | 0.101521 | 0.466057 | 0.078056 | 0.062* | |
| C17 | 0.0851 (4) | 0.3564 (3) | 0.17148 (10) | 0.0424 (7) | |
| C18 | 0.8615 (4) | 0.2090 (4) | 0.05351 (12) | 0.0820 (11) | |
| H18A | 0.874909 | 0.100553 | 0.064059 | 0.098* | |
| H18B | 0.894362 | 0.249403 | 0.079923 | 0.098* | |
| C19 | 0.9824 (5) | 0.2199 (5) | 0.01002 (14) | 0.1126 (16) | |
| H19A | 1.106148 | 0.152009 | 0.017216 | 0.169* | |
| H19B | 0.976652 | 0.32615 | 0.001769 | 0.169* | |
| H19C | 0.941525 | 0.188565 | −0.016723 | 0.169* | |
| O6 | 0.1516 (4) | 0.7336 (4) | 0.60499 (10) | 0.1157 (11) | |
| O7 | 0.1799 (4) | 0.9508 (4) | 0.61439 (10) | 0.1162 (11) | |
| N6 | 0.1845 (4) | 0.8525 (5) | 0.58929 (11) | 0.0830 (10) | |
| N4 | 0.2839 (3) | 0.9838 (3) | 0.38867 (8) | 0.0490 (6) | |
| N5 | 0.2937 (3) | 1.0699 (3) | 0.28629 (8) | 0.0462 (6) | |
| C20 | 0.2721 (4) | 0.9472 (3) | 0.43812 (10) | 0.0459 (7) | |
| C21 | 0.2534 (4) | 0.8084 (4) | 0.45928 (11) | 0.0653 (9) | |
| H21 | 0.259467 | 0.733784 | 0.439839 | 0.078* | |
| C22 | 0.2260 (5) | 0.7782 (4) | 0.50814 (12) | 0.0747 (10) | |
| H22 | 0.211482 | 0.684754 | 0.521174 | 0.09* | |
| C23 | 0.2199 (4) | 0.8824 (5) | 0.53759 (11) | 0.0636 (9) | |
| C24 | 0.2430 (5) | 1.0184 (5) | 0.51872 (13) | 0.0783 (11) | |
| H24 | 0.240692 | 1.089852 | 0.538938 | 0.094* | |
| C25 | 0.2700 (5) | 1.0504 (4) | 0.46951 (12) | 0.0711 (10) | |
| H25 | 0.287119 | 1.143254 | 0.457048 | 0.085* | |
| C26 | 0.3867 (4) | 1.0795 (3) | 0.36697 (10) | 0.0563 (8) | |
| H26A | 0.51213 | 1.012589 | 0.360971 | 0.068* | |
| H26B | 0.3881 | 1.149872 | 0.389107 | 0.068* | |
| C27 | 0.3030 (4) | 1.1732 (3) | 0.32039 (10) | 0.0539 (8) | |
| H27A | 0.180752 | 1.245608 | 0.326783 | 0.065* | |
| H27B | 0.375328 | 1.234293 | 0.305927 | 0.065* | |
| C28 | 0.1907 (4) | 0.9701 (3) | 0.30890 (10) | 0.0498 (7) | |
| H28A | 0.193308 | 0.897196 | 0.28723 | 0.06* | |
| H28B | 0.064102 | 1.035801 | 0.314055 | 0.06* | |
| C29 | 0.2714 (4) | 0.8803 (3) | 0.35626 (10) | 0.0529 (8) | |
| H29A | 0.196135 | 0.822751 | 0.371369 | 0.064* | |
| H29B | 0.392544 | 0.804857 | 0.35057 | 0.064* | |
| O8 | −0.3777 (3) | 0.8954 (2) | 0.25237 (8) | 0.0629 (6) | |
| O9 | −0.3341 (3) | 1.1185 (2) | 0.25354 (7) | 0.0569 (5) | |
| O10 | 0.2958 (3) | 0.8011 (3) | 0.10557 (8) | 0.0771 (7) | |
| C30 | −0.1331 (3) | 0.9365 (3) | 0.20503 (9) | 0.0408 (6) | |
| C31 | −0.1058 (4) | 0.8152 (3) | 0.17834 (10) | 0.0508 (7) | |
| H31 | −0.185001 | 0.760423 | 0.183038 | 0.061* | |
| C32 | 0.0357 (4) | 0.7741 (3) | 0.14506 (11) | 0.0555 (8) | |
| H32 | 0.049421 | 0.694181 | 0.127006 | 0.067* | |
| C33 | 0.1575 (4) | 0.8517 (3) | 0.13846 (10) | 0.0511 (7) | |
| C34 | 0.1341 (4) | 0.9716 (3) | 0.16508 (10) | 0.0512 (7) | |
| H34 | 0.215362 | 1.024354 | 0.161223 | 0.061* | |
| C35 | −0.0111 (4) | 1.0121 (3) | 0.19741 (10) | 0.0463 (7) | |
| H35 | −0.027019 | 1.09415 | 0.214785 | 0.056* | |
| C36 | −0.2919 (4) | 0.9871 (4) | 0.23973 (10) | 0.0478 (7) | |
| C37 | 0.4280 (5) | 0.8751 (5) | 0.09807 (14) | 0.0885 (12) | |
| H37A | 0.483742 | 0.868761 | 0.128435 | 0.106* | |
| H37B | 0.369923 | 0.984835 | 0.085011 | 0.106* | |
| C38 | 0.5698 (6) | 0.7907 (7) | 0.06316 (18) | 0.153 (2) | |
| H38A | 0.667443 | 0.830217 | 0.059976 | 0.229* | |
| H38B | 0.515701 | 0.807254 | 0.032204 | 0.229* | |
| H38C | 0.617246 | 0.680406 | 0.074893 | 0.229* | |
| H31N | 0.778 (5) | 0.425 (6) | 0.2007 (14) | 0.183* | |
| H32N | 0.677 (7) | 0.324 (3) | 0.2290 (18) | 0.183* | |
| H61N | 0.407 (4) | 1.009 (5) | 0.2766 (18) | 0.183* | |
| H62N | 0.236 (6) | 1.131 (5) | 0.2599 (12) | 0.183* |
4-(4-Nitrophenyl)piperazin-1-ium 4-ethoxybenzoate (VI). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0514 (15) | 0.173 (3) | 0.0801 (19) | −0.0138 (17) | 0.0103 (14) | 0.0040 (18) |
| O2 | 0.0855 (17) | 0.129 (2) | 0.0445 (15) | −0.0320 (16) | 0.0107 (13) | −0.0023 (14) |
| N3 | 0.0550 (18) | 0.076 (2) | 0.0573 (19) | −0.0143 (15) | 0.0078 (15) | −0.0036 (15) |
| N1 | 0.0399 (13) | 0.0448 (14) | 0.0380 (13) | −0.0148 (11) | −0.0030 (10) | −0.0033 (11) |
| N2 | 0.0450 (14) | 0.0536 (16) | 0.0466 (15) | −0.0175 (13) | 0.0038 (11) | −0.0089 (13) |
| C1 | 0.0440 (16) | 0.0302 (15) | 0.0369 (16) | −0.0153 (13) | −0.0019 (13) | −0.0053 (12) |
| C2 | 0.0486 (17) | 0.0463 (17) | 0.0378 (16) | −0.0152 (14) | −0.0064 (14) | −0.0001 (13) |
| C3 | 0.0392 (16) | 0.0525 (19) | 0.0513 (19) | −0.0132 (14) | −0.0007 (14) | −0.0071 (15) |
| C4 | 0.0488 (18) | 0.0463 (18) | 0.0441 (18) | −0.0167 (15) | 0.0066 (14) | −0.0065 (14) |
| C5 | 0.0584 (19) | 0.0406 (17) | 0.0386 (16) | −0.0186 (15) | −0.0016 (14) | −0.0020 (13) |
| C6 | 0.0433 (16) | 0.0381 (16) | 0.0457 (17) | −0.0127 (13) | −0.0066 (14) | −0.0027 (13) |
| C7 | 0.0407 (16) | 0.0543 (18) | 0.0470 (18) | −0.0162 (14) | −0.0043 (13) | −0.0088 (14) |
| C8 | 0.0440 (17) | 0.066 (2) | 0.057 (2) | −0.0243 (15) | 0.0013 (15) | −0.0150 (16) |
| C9 | 0.0544 (18) | 0.0547 (19) | 0.0451 (18) | −0.0186 (16) | 0.0029 (14) | 0.0005 (14) |
| C10 | 0.0452 (17) | 0.068 (2) | 0.0392 (17) | −0.0168 (15) | −0.0010 (13) | −0.0005 (15) |
| O3 | 0.0458 (12) | 0.0732 (15) | 0.0477 (12) | −0.0181 (11) | 0.0023 (10) | 0.0009 (11) |
| O4 | 0.0585 (12) | 0.0634 (13) | 0.0375 (12) | −0.0259 (11) | 0.0003 (9) | 0.0022 (10) |
| O5 | 0.0505 (13) | 0.0959 (17) | 0.0504 (13) | −0.0107 (12) | 0.0086 (11) | 0.0129 (12) |
| C11 | 0.0480 (17) | 0.0392 (16) | 0.0352 (15) | −0.0199 (14) | 0.0001 (13) | −0.0026 (13) |
| C12 | 0.0524 (18) | 0.064 (2) | 0.0362 (16) | −0.0234 (16) | −0.0022 (14) | 0.0074 (14) |
| C13 | 0.0453 (17) | 0.072 (2) | 0.0455 (19) | −0.0155 (16) | −0.0029 (15) | 0.0053 (16) |
| C14 | 0.0489 (18) | 0.060 (2) | 0.0424 (18) | −0.0173 (16) | 0.0060 (15) | −0.0001 (15) |
| C15 | 0.056 (2) | 0.075 (2) | 0.0327 (17) | −0.0089 (17) | 0.0009 (15) | 0.0090 (15) |
| C16 | 0.0458 (17) | 0.0560 (19) | 0.0431 (18) | −0.0091 (15) | −0.0006 (14) | 0.0016 (15) |
| C17 | 0.0498 (18) | 0.0416 (17) | 0.0418 (18) | −0.0237 (15) | 0.0000 (14) | −0.0054 (14) |
| C18 | 0.050 (2) | 0.106 (3) | 0.069 (2) | −0.014 (2) | 0.0095 (17) | 0.007 (2) |
| C19 | 0.058 (2) | 0.154 (4) | 0.088 (3) | −0.011 (2) | 0.022 (2) | 0.011 (3) |
| O6 | 0.125 (3) | 0.136 (3) | 0.0580 (18) | −0.028 (2) | 0.0049 (16) | 0.0186 (18) |
| O7 | 0.099 (2) | 0.208 (3) | 0.0619 (18) | −0.070 (2) | 0.0107 (15) | −0.049 (2) |
| N6 | 0.0584 (19) | 0.129 (3) | 0.049 (2) | −0.022 (2) | 0.0005 (15) | −0.010 (2) |
| N4 | 0.0744 (17) | 0.0449 (14) | 0.0381 (14) | −0.0343 (13) | −0.0005 (12) | −0.0046 (11) |
| N5 | 0.0553 (15) | 0.0425 (15) | 0.0435 (14) | −0.0231 (12) | 0.0049 (12) | −0.0033 (12) |
| C20 | 0.0459 (16) | 0.0463 (18) | 0.0443 (18) | −0.0147 (14) | 0.0001 (13) | −0.0083 (14) |
| C21 | 0.099 (3) | 0.054 (2) | 0.0440 (19) | −0.0301 (19) | 0.0055 (17) | −0.0063 (16) |
| C22 | 0.101 (3) | 0.070 (2) | 0.047 (2) | −0.031 (2) | 0.0048 (19) | 0.0032 (18) |
| C23 | 0.0510 (19) | 0.091 (3) | 0.0396 (19) | −0.0177 (19) | −0.0027 (15) | −0.0030 (19) |
| C24 | 0.087 (3) | 0.110 (3) | 0.051 (2) | −0.041 (2) | 0.0054 (19) | −0.038 (2) |
| C25 | 0.099 (3) | 0.074 (2) | 0.059 (2) | −0.049 (2) | 0.0046 (19) | −0.0196 (19) |
| C26 | 0.071 (2) | 0.058 (2) | 0.0524 (19) | −0.0381 (17) | 0.0000 (16) | −0.0085 (16) |
| C27 | 0.072 (2) | 0.0427 (17) | 0.0533 (19) | −0.0303 (16) | 0.0104 (16) | −0.0060 (15) |
| C28 | 0.0607 (19) | 0.0526 (18) | 0.0439 (18) | −0.0310 (16) | 0.0008 (14) | −0.0043 (14) |
| C29 | 0.077 (2) | 0.0475 (18) | 0.0436 (18) | −0.0344 (16) | −0.0019 (15) | −0.0032 (14) |
| O8 | 0.0597 (13) | 0.0531 (13) | 0.0781 (15) | −0.0273 (11) | 0.0142 (11) | −0.0062 (11) |
| O9 | 0.0627 (13) | 0.0524 (13) | 0.0573 (13) | −0.0226 (11) | 0.0088 (10) | −0.0129 (11) |
| O10 | 0.0744 (15) | 0.0862 (17) | 0.0732 (16) | −0.0304 (14) | 0.0256 (13) | −0.0281 (13) |
| C30 | 0.0447 (16) | 0.0366 (16) | 0.0382 (16) | −0.0133 (13) | −0.0061 (13) | 0.0021 (13) |
| C31 | 0.0524 (18) | 0.0469 (18) | 0.0559 (19) | −0.0229 (15) | −0.0043 (15) | −0.0020 (15) |
| C32 | 0.064 (2) | 0.0469 (18) | 0.056 (2) | −0.0192 (16) | −0.0013 (16) | −0.0139 (15) |
| C33 | 0.0507 (18) | 0.0488 (18) | 0.0463 (18) | −0.0113 (15) | 0.0020 (14) | −0.0032 (15) |
| C34 | 0.0482 (17) | 0.0498 (19) | 0.058 (2) | −0.0232 (15) | 0.0023 (15) | −0.0036 (16) |
| C35 | 0.0490 (17) | 0.0437 (17) | 0.0490 (18) | −0.0186 (14) | −0.0048 (14) | −0.0077 (14) |
| C36 | 0.0480 (18) | 0.0481 (19) | 0.0442 (18) | −0.0162 (15) | −0.0073 (14) | 0.0022 (15) |
| C37 | 0.075 (3) | 0.110 (3) | 0.082 (3) | −0.040 (2) | 0.025 (2) | −0.014 (2) |
| C38 | 0.122 (4) | 0.205 (6) | 0.147 (5) | −0.072 (4) | 0.083 (4) | −0.079 (4) |
4-(4-Nitrophenyl)piperazin-1-ium 4-ethoxybenzoate (VI). Geometric parameters (Å, º)
| O1—N3 | 1.220 (3) | O6—N6 | 1.231 (4) |
| O2—N3 | 1.213 (3) | O7—N6 | 1.223 (4) |
| N3—C4 | 1.447 (3) | N6—C23 | 1.456 (4) |
| N1—C1 | 1.384 (3) | N4—C20 | 1.378 (3) |
| N1—C7 | 1.462 (3) | N4—C26 | 1.449 (3) |
| N1—C10 | 1.467 (3) | N4—C29 | 1.452 (3) |
| N2—C9 | 1.475 (3) | N5—C27 | 1.476 (4) |
| N2—C8 | 1.481 (4) | N5—C28 | 1.486 (3) |
| N2—H31N | 0.937 (19) | N5—H61N | 0.910 (19) |
| N2—H32N | 0.895 (19) | N5—H62N | 0.896 (19) |
| C1—C2 | 1.405 (3) | C20—C21 | 1.384 (4) |
| C1—C6 | 1.412 (3) | C20—C25 | 1.391 (4) |
| C2—C3 | 1.366 (4) | C21—C22 | 1.370 (4) |
| C2—H2 | 0.93 | C21—H21 | 0.93 |
| C3—C4 | 1.376 (4) | C22—C23 | 1.349 (4) |
| C3—H3 | 0.93 | C22—H22 | 0.93 |
| C4—C5 | 1.377 (4) | C23—C24 | 1.359 (5) |
| C5—C6 | 1.372 (3) | C24—C25 | 1.381 (4) |
| C5—H5 | 0.93 | C24—H24 | 0.93 |
| C6—H6 | 0.93 | C25—H25 | 0.93 |
| C7—C8 | 1.498 (3) | C26—C27 | 1.497 (4) |
| C7—H7A | 0.97 | C26—H26A | 0.97 |
| C7—H7B | 0.97 | C26—H26B | 0.97 |
| C8—H8A | 0.97 | C27—H27A | 0.97 |
| C8—H8B | 0.97 | C27—H27B | 0.97 |
| C9—C10 | 1.493 (3) | C28—C29 | 1.498 (4) |
| C9—H9A | 0.97 | C28—H28A | 0.97 |
| C9—H9B | 0.97 | C28—H28B | 0.97 |
| C10—H10A | 0.97 | C29—H29A | 0.97 |
| C10—H10B | 0.97 | C29—H29B | 0.97 |
| O3—C17 | 1.261 (3) | O8—C36 | 1.264 (3) |
| O4—C17 | 1.253 (3) | O9—C36 | 1.252 (3) |
| O5—C14 | 1.365 (3) | O10—C33 | 1.363 (3) |
| O5—C18 | 1.426 (3) | O10—C37 | 1.431 (4) |
| C11—C12 | 1.375 (4) | C30—C35 | 1.372 (3) |
| C11—C16 | 1.383 (3) | C30—C31 | 1.387 (4) |
| C11—C17 | 1.499 (4) | C30—C36 | 1.500 (4) |
| C12—C13 | 1.379 (4) | C31—C32 | 1.376 (4) |
| C12—H12 | 0.93 | C31—H31 | 0.93 |
| C13—C14 | 1.373 (4) | C32—C33 | 1.383 (4) |
| C13—H13 | 0.93 | C32—H32 | 0.93 |
| C14—C15 | 1.378 (4) | C33—C34 | 1.381 (4) |
| C15—C16 | 1.374 (4) | C34—C35 | 1.378 (4) |
| C15—H15 | 0.93 | C34—H34 | 0.93 |
| C16—H16 | 0.93 | C35—H35 | 0.93 |
| C18—C19 | 1.502 (4) | C37—C38 | 1.496 (5) |
| C18—H18A | 0.97 | C37—H37A | 0.97 |
| C18—H18B | 0.97 | C37—H37B | 0.97 |
| C19—H19A | 0.96 | C38—H38A | 0.96 |
| C19—H19B | 0.96 | C38—H38B | 0.96 |
| C19—H19C | 0.96 | C38—H38C | 0.96 |
| O2—N3—O1 | 122.7 (3) | O7—N6—O6 | 123.5 (4) |
| O2—N3—C4 | 119.3 (3) | O7—N6—C23 | 118.2 (4) |
| O1—N3—C4 | 118.0 (3) | O6—N6—C23 | 118.2 (4) |
| C1—N1—C7 | 118.1 (2) | C20—N4—C26 | 121.3 (2) |
| C1—N1—C10 | 117.0 (2) | C20—N4—C29 | 121.3 (2) |
| C7—N1—C10 | 116.3 (2) | C26—N4—C29 | 111.9 (2) |
| C9—N2—C8 | 107.9 (2) | C27—N5—C28 | 110.0 (2) |
| C9—N2—H31N | 110 (3) | C27—N5—H61N | 112 (3) |
| C8—N2—H31N | 108 (3) | C28—N5—H61N | 110 (3) |
| C9—N2—H32N | 111 (3) | C27—N5—H62N | 108 (3) |
| C8—N2—H32N | 106 (3) | C28—N5—H62N | 110 (3) |
| H31N—N2—H32N | 113 (4) | H61N—N5—H62N | 107 (4) |
| N1—C1—C2 | 120.9 (2) | N4—C20—C21 | 121.8 (3) |
| N1—C1—C6 | 122.0 (2) | N4—C20—C25 | 122.0 (3) |
| C2—C1—C6 | 117.1 (2) | C21—C20—C25 | 116.2 (3) |
| C3—C2—C1 | 121.3 (3) | C22—C21—C20 | 121.6 (3) |
| C3—C2—H2 | 119.3 | C22—C21—H21 | 119.2 |
| C1—C2—H2 | 119.3 | C20—C21—H21 | 119.2 |
| C2—C3—C4 | 120.1 (3) | C23—C22—C21 | 120.9 (3) |
| C2—C3—H3 | 120 | C23—C22—H22 | 119.6 |
| C4—C3—H3 | 120 | C21—C22—H22 | 119.6 |
| C3—C4—C5 | 120.6 (3) | C22—C23—C24 | 119.7 (3) |
| C3—C4—N3 | 119.6 (3) | C22—C23—N6 | 120.8 (4) |
| C5—C4—N3 | 119.8 (3) | C24—C23—N6 | 119.5 (4) |
| C6—C5—C4 | 119.8 (3) | C23—C24—C25 | 119.9 (3) |
| C6—C5—H5 | 120.1 | C23—C24—H24 | 120 |
| C4—C5—H5 | 120.1 | C25—C24—H24 | 120 |
| C5—C6—C1 | 121.2 (3) | C24—C25—C20 | 121.6 (3) |
| C5—C6—H6 | 119.4 | C24—C25—H25 | 119.2 |
| C1—C6—H6 | 119.4 | C20—C25—H25 | 119.2 |
| N1—C7—C8 | 113.3 (2) | N4—C26—C27 | 110.5 (2) |
| N1—C7—H7A | 108.9 | N4—C26—H26A | 109.5 |
| C8—C7—H7A | 108.9 | C27—C26—H26A | 109.5 |
| N1—C7—H7B | 108.9 | N4—C26—H26B | 109.5 |
| C8—C7—H7B | 108.9 | C27—C26—H26B | 109.5 |
| H7A—C7—H7B | 107.7 | H26A—C26—H26B | 108.1 |
| N2—C8—C7 | 110.9 (2) | N5—C27—C26 | 111.0 (2) |
| N2—C8—H8A | 109.5 | N5—C27—H27A | 109.4 |
| C7—C8—H8A | 109.5 | C26—C27—H27A | 109.4 |
| N2—C8—H8B | 109.5 | N5—C27—H27B | 109.4 |
| C7—C8—H8B | 109.5 | C26—C27—H27B | 109.4 |
| H8A—C8—H8B | 108 | H27A—C27—H27B | 108 |
| N2—C9—C10 | 111.5 (2) | N5—C28—C29 | 111.1 (2) |
| N2—C9—H9A | 109.3 | N5—C28—H28A | 109.4 |
| C10—C9—H9A | 109.3 | C29—C28—H28A | 109.4 |
| N2—C9—H9B | 109.3 | N5—C28—H28B | 109.4 |
| C10—C9—H9B | 109.3 | C29—C28—H28B | 109.4 |
| H9A—C9—H9B | 108 | H28A—C28—H28B | 108 |
| N1—C10—C9 | 113.7 (2) | N4—C29—C28 | 111.5 (2) |
| N1—C10—H10A | 108.8 | N4—C29—H29A | 109.3 |
| C9—C10—H10A | 108.8 | C28—C29—H29A | 109.3 |
| N1—C10—H10B | 108.8 | N4—C29—H29B | 109.3 |
| C9—C10—H10B | 108.8 | C28—C29—H29B | 109.3 |
| H10A—C10—H10B | 107.7 | H29A—C29—H29B | 108 |
| C14—O5—C18 | 118.3 (2) | C33—O10—C37 | 118.5 (3) |
| C12—C11—C16 | 117.5 (2) | C35—C30—C31 | 117.1 (3) |
| C12—C11—C17 | 120.9 (2) | C35—C30—C36 | 120.9 (3) |
| C16—C11—C17 | 121.6 (3) | C31—C30—C36 | 122.0 (2) |
| C11—C12—C13 | 122.1 (3) | C32—C31—C30 | 121.5 (3) |
| C11—C12—H12 | 119 | C32—C31—H31 | 119.2 |
| C13—C12—H12 | 119 | C30—C31—H31 | 119.2 |
| C14—C13—C12 | 119.5 (3) | C31—C32—C33 | 120.0 (3) |
| C14—C13—H13 | 120.3 | C31—C32—H32 | 120 |
| C12—C13—H13 | 120.3 | C33—C32—H32 | 120 |
| O5—C14—C13 | 124.4 (3) | O10—C33—C34 | 124.5 (3) |
| O5—C14—C15 | 116.1 (3) | O10—C33—C32 | 116.1 (3) |
| C13—C14—C15 | 119.5 (3) | C34—C33—C32 | 119.4 (3) |
| C16—C15—C14 | 120.2 (3) | C35—C34—C33 | 119.2 (3) |
| C16—C15—H15 | 119.9 | C35—C34—H34 | 120.4 |
| C14—C15—H15 | 119.9 | C33—C34—H34 | 120.4 |
| C15—C16—C11 | 121.2 (3) | C30—C35—C34 | 122.7 (3) |
| C15—C16—H16 | 119.4 | C30—C35—H35 | 118.6 |
| C11—C16—H16 | 119.4 | C34—C35—H35 | 118.6 |
| O4—C17—O3 | 123.5 (2) | O9—C36—O8 | 124.0 (3) |
| O4—C17—C11 | 119.3 (3) | O9—C36—C30 | 118.3 (3) |
| O3—C17—C11 | 117.3 (2) | O8—C36—C30 | 117.7 (3) |
| O5—C18—C19 | 107.3 (3) | O10—C37—C38 | 107.4 (3) |
| O5—C18—H18A | 110.3 | O10—C37—H37A | 110.2 |
| C19—C18—H18A | 110.3 | C38—C37—H37A | 110.2 |
| O5—C18—H18B | 110.3 | O10—C37—H37B | 110.2 |
| C19—C18—H18B | 110.3 | C38—C37—H37B | 110.2 |
| H18A—C18—H18B | 108.5 | H37A—C37—H37B | 108.5 |
| C18—C19—H19A | 109.5 | C37—C38—H38A | 109.5 |
| C18—C19—H19B | 109.5 | C37—C38—H38B | 109.5 |
| H19A—C19—H19B | 109.5 | H38A—C38—H38B | 109.5 |
| C18—C19—H19C | 109.5 | C37—C38—H38C | 109.5 |
| H19A—C19—H19C | 109.5 | H38A—C38—H38C | 109.5 |
| H19B—C19—H19C | 109.5 | H38B—C38—H38C | 109.5 |
| C7—N1—C1—C2 | −173.1 (2) | C26—N4—C20—C21 | −149.7 (3) |
| C10—N1—C1—C2 | −26.5 (3) | C29—N4—C20—C21 | 2.0 (4) |
| C7—N1—C1—C6 | 8.5 (3) | C26—N4—C20—C25 | 33.3 (4) |
| C10—N1—C1—C6 | 155.0 (2) | C29—N4—C20—C25 | −175.1 (3) |
| N1—C1—C2—C3 | −177.9 (2) | N4—C20—C21—C22 | −174.3 (3) |
| C6—C1—C2—C3 | 0.6 (4) | C25—C20—C21—C22 | 2.8 (5) |
| C1—C2—C3—C4 | −0.7 (4) | C20—C21—C22—C23 | −1.3 (5) |
| C2—C3—C4—C5 | 0.3 (4) | C21—C22—C23—C24 | −0.7 (5) |
| C2—C3—C4—N3 | −178.7 (3) | C21—C22—C23—N6 | 177.8 (3) |
| O2—N3—C4—C3 | −170.8 (3) | O7—N6—C23—C22 | −179.8 (3) |
| O1—N3—C4—C3 | 9.6 (4) | O6—N6—C23—C22 | −3.4 (5) |
| O2—N3—C4—C5 | 10.2 (4) | O7—N6—C23—C24 | −1.3 (5) |
| O1—N3—C4—C5 | −169.4 (3) | O6—N6—C23—C24 | 175.1 (3) |
| C3—C4—C5—C6 | 0.2 (4) | C22—C23—C24—C25 | 0.9 (5) |
| N3—C4—C5—C6 | 179.2 (3) | N6—C23—C24—C25 | −177.6 (3) |
| C4—C5—C6—C1 | −0.2 (4) | C23—C24—C25—C20 | 0.8 (5) |
| N1—C1—C6—C5 | 178.3 (2) | N4—C20—C25—C24 | 174.6 (3) |
| C2—C1—C6—C5 | −0.2 (4) | C21—C20—C25—C24 | −2.6 (5) |
| C1—N1—C7—C8 | −173.0 (2) | C20—N4—C26—C27 | −148.9 (3) |
| C10—N1—C7—C8 | 40.3 (3) | C29—N4—C26—C27 | 57.0 (3) |
| C9—N2—C8—C7 | 62.2 (3) | C28—N5—C27—C26 | 56.4 (3) |
| N1—C7—C8—N2 | −51.8 (3) | N4—C26—C27—N5 | −57.5 (3) |
| C8—N2—C9—C10 | −61.5 (3) | C27—N5—C28—C29 | −54.8 (3) |
| C1—N1—C10—C9 | 173.4 (2) | C20—N4—C29—C28 | 150.1 (2) |
| C7—N1—C10—C9 | −39.5 (3) | C26—N4—C29—C28 | −55.8 (3) |
| N2—C9—C10—N1 | 50.3 (3) | N5—C28—C29—N4 | 54.6 (3) |
| C16—C11—C12—C13 | 0.5 (4) | C35—C30—C31—C32 | −1.0 (4) |
| C17—C11—C12—C13 | −177.6 (3) | C36—C30—C31—C32 | 177.1 (3) |
| C11—C12—C13—C14 | 0.4 (5) | C30—C31—C32—C33 | 1.6 (4) |
| C18—O5—C14—C13 | 1.3 (5) | C37—O10—C33—C34 | 0.7 (4) |
| C18—O5—C14—C15 | −178.7 (3) | C37—O10—C33—C32 | −178.7 (3) |
| C12—C13—C14—O5 | 178.9 (3) | C31—C32—C33—O10 | 178.6 (3) |
| C12—C13—C14—C15 | −1.0 (5) | C31—C32—C33—C34 | −0.8 (4) |
| O5—C14—C15—C16 | −179.2 (3) | O10—C33—C34—C35 | −179.8 (3) |
| C13—C14—C15—C16 | 0.7 (5) | C32—C33—C34—C35 | −0.5 (4) |
| C14—C15—C16—C11 | 0.2 (5) | C31—C30—C35—C34 | −0.3 (4) |
| C12—C11—C16—C15 | −0.8 (4) | C36—C30—C35—C34 | −178.5 (3) |
| C17—C11—C16—C15 | 177.3 (3) | C33—C34—C35—C30 | 1.1 (4) |
| C12—C11—C17—O4 | 2.7 (4) | C35—C30—C36—O9 | 15.2 (4) |
| C16—C11—C17—O4 | −175.3 (2) | C31—C30—C36—O9 | −162.9 (3) |
| C12—C11—C17—O3 | −177.3 (3) | C35—C30—C36—O8 | −166.4 (2) |
| C16—C11—C17—O3 | 4.6 (4) | C31—C30—C36—O8 | 15.5 (4) |
| C14—O5—C18—C19 | 178.7 (3) | C33—O10—C37—C38 | 176.2 (3) |
4-(4-Nitrophenyl)piperazin-1-ium 4-ethoxybenzoate (VI). Hydrogen-bond geometry (Å, º)
Cg2 and Cg6 are the centroids of the C1–C6 and C30–C35 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H31N···O3i | 0.94 (2) | 1.68 (2) | 2.613 (3) | 172 (5) |
| N2—H31N···O4i | 0.94 (2) | 2.51 (4) | 3.157 (3) | 127 (4) |
| N2—H32N···O9ii | 0.90 (2) | 1.96 (2) | 2.843 (3) | 171 (5) |
| N5—H61N···O8i | 0.91 (2) | 1.78 (2) | 2.686 (3) | 175 (5) |
| N5—H61N···O9i | 0.91 (2) | 2.59 (4) | 3.174 (3) | 122 (4) |
| N5—H62N···O4iii | 0.90 (2) | 1.83 (2) | 2.708 (3) | 165 (5) |
| C22—H22···O2iv | 0.93 | 2.6 | 3.502 (5) | 165 |
| C27—H27B···O9i | 0.97 | 2.59 | 3.215 (3) | 123 |
| C28—H28B···O7v | 0.97 | 2.65 | 3.410 (4) | 135 |
| C29—H29B···O1i | 0.97 | 2.53 | 3.249 (4) | 131 |
| C35—H35···O4iii | 0.93 | 2.52 | 3.263 (3) | 137 |
| C10—H10A···Cg6 | 0.97 | 2.82 | 3.746 (3) | 159 |
| C29—H29A···Cg2 | 0.97 | 2.76 | 3.556 (3) | 139 |
Symmetry codes: (i) x+1, y, z; (ii) x+1, y−1, z; (iii) x, y+1, z; (iv) −x, −y+1, −z+1; (v) −x, −y+2, −z+1.
Funding Statement
NM is grateful to the University of Mysore for research facilities. HSY thanks the UGC for a BSR Faculty fellowship for three years. SGG gratefully acknowledges financial support from the Spanish Ministerio de Ciencia e Innovación (PID2020–113558RB-C41) and Gobierno del Principado de Asturias (AYUD/2021/50997).
References
- Archana, S. D., Kumar, H. K., Yathirajan, H. S., Foro, S., Abdelbaky, M. S. M. & Garcia-Granda, S. (2021). Acta Cryst. E77, 1135–1139. [DOI] [PMC free article] [PubMed]
- O. Ayeni, A., M. Watkins, G. & C. Hosten, E. (2019). Bull. Chem. Soc. Eth. 33, 341.
- Berkheij, M., van der Sluis, L., Sewing, C., den Boer, D. J., Terpstra, J. W., Hiemstra, H., Iwema Bakker, W. I., van den Hoogenband, A. & van Maarseveen, J. H. (2005). Tetrahedron Lett. 46, 2369–2371.
- Bogatcheva, E., Hanrahan, C., Nikonenko, B., Samala, R., Chen, P., Gearhart, J., Barbosa, F., Einck, L., Nacy, C. A. & Protopopova, M. (2006). J. Med. Chem. 49, 3045–3048. [DOI] [PMC free article] [PubMed]
- Brockunier, L. L., He, J., Colwell, L. F. Jr, Habulihaz, B., He, H., Leiting, B., Lyons, K. A., Marsilio, F., Patel, R. A., Teffera, Y., Wu, J. K., Thornberry, N. A., Weber, A. E. & Parmee, E. R. (2004). Bioorg. Med. Chem. Lett. 14, 4763–4766. [DOI] [PubMed]
- Chaudhary, P., Kumar, R., Verma, K., Singh, D., Yadav, V., Chhillar, A. K., Sharma, G. L. & Chandra, R. (2006). Bioorg. Med. Chem. 14, 1819–1826. [DOI] [PubMed]
- Elliott, S. (2011). Drug Test. Anal. 3, 430–438. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Gupta, S., Pandey, D., Mandalapu, D., Bala, V., Sharma, V., Shukla, M., Yadav, S. K., Singh, N., Jaiswal, S., Maikhuri, J. P., Lal, J., Siddiqi, M. I., Gupta, G. & Sharma, V. L. (2016). Med. Chem. Commun. 7, 2111–2121.
- Harish Chinthal, C., Kavitha, C. N., Yathirajan, H. S., Foro, S., Rathore, R. S. & Glidewell, C. (2020). Acta Cryst. E76, 1779–1793. [DOI] [PMC free article] [PubMed]
- Kaya, B., Ozkay, Y., Temel, H. E. & &Kaplancikli, Z. A. (2016). J. Chem. Art. ID, pp. 5878410 http://dx.doi.org/10.1155/2016/5878410.
- Kharb, R., Bansal, K. & Sharma, A. K. (2012). Der Pharma Chem. 4, 2470–2488.
- Kiran Kumar, H., Yathirajan, H. S., Foro, S. & Glidewell, C. (2019). Acta Cryst. E75, 1494–1506. [DOI] [PMC free article] [PubMed]
- Kiran Kumar, H., Yathirajan, H. S., Harish Chinthal, C., Foro, S. & Glidewell, C. (2020). Acta Cryst. E75, 488–495. [DOI] [PMC free article] [PubMed]
- Lu, Y.-X. (2007). Acta Cryst. E63, o3611.
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst. 53, 226–235. [DOI] [PMC free article] [PubMed]
- Oxford Diffraction (2009). CrysAlis CCD and CrysAlis RED. Oxford Diffraction Ltd, Abingdon, England.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Upadhayaya, P. S., Sinha, N., Jain, S., Kishore, N., Chandra, R. & Arora, S. K. (2004). Bioorg. Med. Chem. 12, 2225–2238. [DOI] [PubMed]
- Wang, X.-Y., Wang, M.-Z., Guo, F.-J., Sun, J. & Qian, S.-S. (2014). Z. Kristallogr. New Cryst. Struct. 229, 97–98.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Wodtke, R., Steinberg, J., Köckerling, M., Löser, R. & Mamat, C. (2018). RSC Adv. 8, 40921–40933. [DOI] [PMC free article] [PubMed]
- Zhang, Y., Chao, J., Zhao, S., Xu, P., Wang, H., Guo, Z. & Liu, D. (2014). Spectrochim. Acta Part A, 132, 44–51. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I, II, III, IV, V, VI. DOI: 10.1107/S2056989022004157/ex2056sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022004157/ex2056Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989022004157/ex2056IIsup3.hkl
Structure factors: contains datablock(s) III. DOI: 10.1107/S2056989022004157/ex2056IIIsup4.hkl
Structure factors: contains datablock(s) IV. DOI: 10.1107/S2056989022004157/ex2056IVsup5.hkl
Structure factors: contains datablock(s) V. DOI: 10.1107/S2056989022004157/ex2056Vsup6.hkl
Structure factors: contains datablock(s) VI. DOI: 10.1107/S2056989022004157/ex2056VIsup7.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report