The ability of 4-[(benzylamino)carbonyl]-1-methylpyridinium bromide salt to form hydrates was studied. Hirshfeld surfaces analysis was performed for identification of intermolecular interactions.
Keywords: 4-[(benzylamino)carbonyl]-1-methylpyridinium bromide, molecular structure, crystal structure, Hirshfeld surface analysis
Abstract
The hemihydrate of 4-[(benzylamino)carbonyl]-1-methylpyridinium bromide, C14H15N2O+·Br−·0.5H2O, was studied by single-crystal and powder X-ray diffraction methods. In the asymmetric unit, two organic cations of similar conformation, two bromide anions and one water molecule are present. In the crystal, N—H⋯Br hydrogen bonds link the cations and anions. The formation of a set of intermolecular C—H⋯Br and C—H⋯π interactions result in double chains extending parallel to [011]. A Hirshfeld surface analysis showed high contributions of H⋯H and C⋯H/H⋯C short contacts to the total Hirshfeld surfaces of the cations.
1. Chemical context
The 4-[(benzylamino)carbonyl]-1-methylpyridinium cation (Am+) has been shown to possess antiviral activity (Buhtiarova et al., 2003 ▸; Frolov et al., 2004 ▸; Boltz et al., 2018 ▸; te Velthuis et al., 2021 ▸). Being charged due to quartenization of the pyridine N atom, this type of cation is more stable than its protonated analogue formed by H-atom transition in the form of an acid–base pair. Halogenide anions can be used as simple counter-ions of the organic cation. In fact, the iodide salt of 4-[(benzylamino)carbonyl]-1-methylpyridinium (AmI) is known as a multimodal antiviral drug and has been studied by single-crystal X-ray diffraction, powder diffraction, IR spectroscopy, and DSC methods (Drebushchak et al., 2017 ▸). The search for polymorphic modifications, hydrates or solvates is of great importance for the pharmaceutical industry to improve the quality of a drug and to protect intellectual property. However, polymorphic screening performed for the AmI salt did not reveal any other crystalline form.
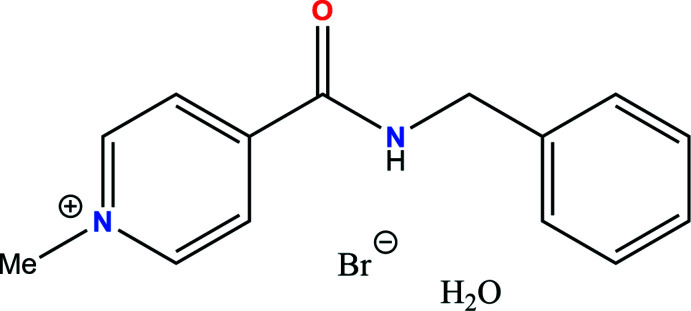
The 4-[(benzylamino)carbonyl]-1-methylpyridinium bromide (AmBr) salt is the closest analogue of AmI. Polymorphic screening for this salt resulted in the crystallization of a hemihydrate. In this communication we present the molecular and crystal structures of 4-[(benzylamino)carbonyl]-1-methylpyridinium bromide hemihydrate, (C14H15N2O)+Br−·0.5H2O.
2. Structural commentary
The asymmetric unit contains two molecules of the cation (denoted A and B), two bromide anions (A and B) and one water molecule (Fig. 1 ▸). The positive charge of the cation is located at the quaternized nitrogen atom of the pyridine ring. The carbamide group is slightly non-coplanar with the plane of the aromatic ring, as shown by the N2—C7—C4—C3 torsion angles given in Table 1 ▸. The non-planarity is caused by steric repulsion between the two constituents as revealed by the amideH2⋯H3pyridine and amideH2⋯C3pyridine short contacts (Table 1 ▸) as compared to the van der Waals radii sums (Zefirov, 1997 ▸) of 2.34 and 2.87 Å, respectively. The cations A and B have similar conformations of the benzyl substituent (Fig. 2 ▸). The phenyl fragment of the benzyl substituent is located in an −ac position in relation to the C7—N2 bond and is twisted in relation to the carbamide fragment in both cations A and B, as seen in the C7—N2—C8—C9 and N2—C8—C9—C10 torsion angles (Table 1 ▸).
Figure 1.
Molecular structure of the title compound, AmBr hemihydrate. Displacement ellipsoids are shown at the 50% probability level. C—H⋯Br and N—H⋯Br hydrogen bonds are indicated by dotted lines.
Table 1. Some geometrical characteristics (Å, °) of cations A and B in AmBr hemihydrate.
| Parameter | Cation A | Cation B |
|---|---|---|
| N1—C2 | 1.343 (6) | 1.323 (7) |
| N1—C6 | 1.330 (7) | 1.329 (7) |
| N2—C7—C4—C3 | 17.1 (7) | −1.4 (9) |
| C7—N2—C8—C9 | −102.6 (6) | −107.0 (6) |
| N2—C8—C9—C10 | −168.9 (5) | −167.4 (5) |
| H2⋯H3 | 2.11 | 2.04 |
| H2⋯C3 | 2.59 | 2.54 |
Figure 2.
Molecular overlay plot of cations A and B.
3. Supramolecular features
In the crystal, cations A and B interact with the bromide anions by N—H⋯Br hydrogen bonds. In addition, a set of C—H⋯Br and C—H⋯π interactions are found in the crystal structure (Table 2 ▸). The solvent water molecule forms one C—H⋯O hydrogen bond as a proton acceptor and O—H⋯Br and O—H⋯O hydrogen bonds as a proton donor (Table 2 ▸). All these hydrogen-bonding interactions result in the formation of double chains extending parallel to [011] (Fig. 3 ▸).
Table 2. Hydrogen-bond geometry (Å, °).
CgA and CgB are the centroids of the C9A–C14A and C9B–C14B rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2A—H2A⋯Br1A | 0.86 | 2.53 | 3.339 (5) | 158 |
| C3A—H3A⋯Br1A | 0.93 | 2.98 | 3.814 (5) | 150 |
| C2A—H2AA⋯Br1A i | 0.93 | 2.84 | 3.725 (6) | 159 |
| C1A—H1AA⋯Br1A i | 0.96 | 2.88 | 3.784 (6) | 157 |
| C6A—H6A⋯CgB ii | 0.93 | 2.65 | 3.510 (7) | 154 |
| N2B—H2B⋯Br1B | 0.86 | 2.60 | 3.419 (5) | 159 |
| C3B—H3B⋯Br1B | 0.93 | 2.83 | 3.753 (5) | 175 |
| C6B—H6B⋯CgA iii | 0.93 | 2.71 | 3.400 (7) | 132 |
| O1W—H1WA⋯Br1B iv | 0.85 | 3.03 | 3.473 (7) | 115 |
| C1A—H1AC⋯O1W v | 0.96 | 2.89 | 3.794 (10) | 157 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
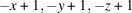 ; (v)
; (v)
 .
.
Figure 3.
Crystal packing of AmBr hemihydrate in a view along [100]. Hydrogen-bonding interactions are shown by dashed lines.
4. Hirshfeld surface analysis
Intermolecular interactions were analysed using Hirshfeld surface analysis and two-dimensional fingerprint plots by using CrystalExplorer17 (Turner et al., 2017 ▸). The Hirshfeld surfaces were calculated separately for cations A and B using a standard high surface resolution, mapped over d norm (Fig. 4 ▸). The red spots corresponding to contacts that are shorter than the van der Waals radii sum of the closest atoms are observed at the hydrogen atom of the amino group and at some phenyl and methyl hydrogen atoms. The two-dimensional fingerprint plots showed the absence of strong hydrogen bonds in the structure under study. To compare intermolecular interactions of different types in a more quantitative way, their contributions to the total Hirshfeld surfaces were analysed (Fig. 5 ▸). The main contribution is provided by H⋯H short contacts (Fig. 5 ▸ g,h). The contribution of C⋯H/H⋯C short contacts is also significant (Fig. 5 ▸ i,j). The Br⋯H/H⋯Br and O⋯H/H⋯O interactions contribute to the total Hirshfeld surface in the same way (Fig. 5 ▸ c,d and 5e,f).
Figure 4.
Hirshfeld surfaces mapped over d norm for cations A (left) and B (right) in the crystal structure of AmBr hemihydrate.
Figure 5.
Contributions of interactions of different types to the total Hirshfeld surface of cations A and B in the crystal structure of AmBr hemihydrate.
5. Database survey
A search of the Cambridge Structural Database (Version 5.42, update of November 2020; Groom et al., 2016 ▸) revealed the structure of the anhydrous AmI salt with an equimolar cation:iodine ratio (refcode BEBFIA; Drebushchak et al., 2017 ▸). A comparison of the molecular conformation of the cation showed its flexibility due to rotation about the N—Csp 3 and Csp 3—Caryl bonds.
6. Powder diffraction characterization
An X-ray powder diffraction pattern of the title compound was registered using a Siemens D500 powder diffractometer (Cu Kα radiation, Bragg–Brentano geometry, curved graphite monochromator on the counter arm, 4 < 2θ < 60°, D2θ = 0.02°). A Rietveld refinement (Fig. 6 ▸) on the basis of the obtained pattern was carried out with FullProf and WinPLOTR (Rodriguez-Carvajal & Roisnel, 1998 ▸) using data of an external standard (NIST SRM1976) for the calculation of the instrumental profile function and the single-crystal data as the structure model for refinement. The main results of the Rietveld refinement are shown in Table 3 ▸. On the basis of the Rietveld refinement, the experimental powder X-ray diffraction pattern coincides with the theoretical one calculated from the X-ray single crystal study.
Figure 6.
Final Rietveld plots for the title compound. Observed data points are indicated by red circles, the best-fit profile (black upper trace) and the difference pattern (blue lower trace) are shown as solid lines. The vertical green bars correspond to the Bragg reflections.
Table 3. Experimental data of the X-ray powder diffraction study performed at 293 K.
| Crystal system, space group | Triclinic, P

|
|---|---|
| a (Å) | 5.8858 (2) |
| b (Å) | 14.7604 (3) |
| c (Å) | 17.8118 (4) |
| α (°) | 65.819 (1) |
| β (°) | 85.321 (2) |
| γ (°) | 85.402 (1) |
| V (Å3) | 1405.09 (6) |
| D x (Mg m−3) | 1.499 |
| Refinement | |
| R p | 0.0359 |
| R wp | 0.0522 |
| R exp | 0.0120 |
| R B | 0.0371 |
| R F | 0.0171 |
7. Synthesis and crystallization
4-[(Benzylamino)carbonyl]-1-methylpyridinium iodide (57.7 g, 0.163 mol), silver bromide (33.77 g, 0.180 mol) and 700 ml of water were loaded into a glass flask. The mixture was stirred for 72 h, and the resulting precipitate was filtered off. The solvent was evaporated under reduced pressure. To the precipitate were added 300 ml of acetonitrile and refluxed for 2 h. The reaction then was spontaneously cooled to a temperature of 303 K and the precipitate filtered off and rinsed on the filter with 50 ml of cooled acetonitrile. The product was dried at 313 K for 12 h. Yield: 14 g of 4-[(benzylamino)carbonyl]-1-methylpyridinium bromide (28%); colourless crystals.
8. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 4 ▸. All of the hydrogen atoms were placed in calculated positions and treated as riding with C—H = 0.96 Å, U iso(H) = 1.5U eq for methyl groups and with Car—H = 0.93 Å, C sp 2—H = 0.97 Å, U iso(H) = 1.2U eq for all other hydrogen atoms.
Table 4. Experimental details.
| Crystal data | |
| Chemical formula | C14H15N2O+·Br−·0.5H2O |
| M r | 316.19 |
| Crystal system, space group | Triclinic, P

|
| Temperature (K) | 293 |
| a, b, c (Å) | 5.8891 (4), 14.7565 (10), 17.8090 (11) |
| α, β, γ (°) | 65.773 (6), 85.396 (6), 85.544 (6) |
| V (Å3) | 1405.08 (17) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 2.92 |
| Crystal size (mm) | 0.30 × 0.15 × 0.10 |
| Data collection | |
| Diffractometer | Xcalibur, Atlas |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2021 ▸) |
| T min, T max | 0.634, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 14465, 4925, 3547 |
| R int | 0.075 |
| (sin θ/λ)max (Å−1) | 0.595 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.060, 0.175, 1.06 |
| No. of reflections | 4925 |
| No. of parameters | 339 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 1.12, −0.45 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989022003784/wm5623sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022003784/wm5623Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989022003784/wm5623Isup3.cml
CCDC reference: 2164796
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors are grateful to Farmak JSC for support.
supplementary crystallographic information
Crystal data
| C14H15N2O+·Br−·0.5H2O | Z = 4 |
| Mr = 316.19 | F(000) = 644 |
| Triclinic, P1 | Dx = 1.495 Mg m−3 |
| a = 5.8891 (4) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 14.7565 (10) Å | Cell parameters from 4991 reflections |
| c = 17.8090 (11) Å | θ = 3.6–25.4° |
| α = 65.773 (6)° | µ = 2.92 mm−1 |
| β = 85.396 (6)° | T = 293 K |
| γ = 85.544 (6)° | Plate, colorless |
| V = 1405.08 (17) Å3 | 0.30 × 0.15 × 0.10 mm |
Data collection
| Xcalibur, Atlas diffractometer | 4925 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 3547 reflections with I > 2σ(I) |
| Detector resolution: 10.3779 pixels mm-1 | Rint = 0.075 |
| ω scans | θmax = 25.0°, θmin = 3.4° |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2021) | h = −6→6 |
| Tmin = 0.634, Tmax = 1.000 | k = −16→17 |
| 14465 measured reflections | l = −21→20 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.060 | H-atom parameters constrained |
| wR(F2) = 0.175 | w = 1/[σ2(Fo2) + (0.0802P)2 + 0.8154P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max = 0.001 |
| 4925 reflections | Δρmax = 1.12 e Å−3 |
| 339 parameters | Δρmin = −0.45 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1A | 0.68503 (10) | 1.06475 (4) | 0.84634 (4) | 0.0614 (2) | |
| Br1B | 0.37067 (11) | 0.44764 (5) | 0.67323 (4) | 0.0621 (2) | |
| O1A | 0.3136 (7) | 0.6956 (3) | 0.9887 (2) | 0.0611 (11) | |
| N1A | 0.9794 (7) | 0.7007 (3) | 1.1351 (2) | 0.0438 (10) | |
| N1B | 0.6285 (7) | 0.7967 (3) | 0.3685 (3) | 0.0496 (11) | |
| N2A | 0.3986 (7) | 0.8566 (3) | 0.9181 (3) | 0.0493 (11) | |
| H2A | 0.486736 | 0.902420 | 0.913909 | 0.059* | |
| O1B | −0.0865 (8) | 0.8195 (3) | 0.5244 (3) | 0.0766 (14) | |
| N2B | 0.0287 (8) | 0.6602 (4) | 0.5982 (3) | 0.0518 (11) | |
| H2B | 0.133971 | 0.614443 | 0.603968 | 0.062* | |
| C9B | −0.1252 (8) | 0.6264 (4) | 0.7400 (3) | 0.0413 (12) | |
| C9A | 0.2998 (8) | 0.8874 (4) | 0.7765 (3) | 0.0432 (12) | |
| C2A | 0.9195 (9) | 0.7968 (4) | 1.0910 (3) | 0.0473 (13) | |
| H2AA | 0.996826 | 0.846374 | 1.096125 | 0.057* | |
| C10B | −0.2902 (9) | 0.5848 (4) | 0.8037 (3) | 0.0505 (14) | |
| H10B | −0.423087 | 0.563252 | 0.792737 | 0.061* | |
| C4A | 0.6308 (8) | 0.7478 (4) | 1.0305 (3) | 0.0406 (12) | |
| C6A | 0.8719 (9) | 0.6286 (4) | 1.1285 (3) | 0.0440 (12) | |
| H6A | 0.915307 | 0.562491 | 1.159801 | 0.053* | |
| C7A | 0.4317 (8) | 0.7654 (4) | 0.9765 (3) | 0.0431 (12) | |
| C14A | 0.5157 (9) | 0.8564 (4) | 0.7586 (3) | 0.0484 (13) | |
| H14A | 0.618478 | 0.829285 | 0.800112 | 0.058* | |
| C3A | 0.7436 (9) | 0.8223 (4) | 1.0381 (3) | 0.0471 (13) | |
| H3A | 0.701404 | 0.888839 | 1.007835 | 0.057* | |
| C8A | 0.2168 (9) | 0.8807 (4) | 0.8609 (3) | 0.0516 (14) | |
| H8AA | 0.141562 | 0.943815 | 0.855334 | 0.062* | |
| H8AB | 0.104777 | 0.830227 | 0.883854 | 0.062* | |
| C4B | 0.2677 (9) | 0.7633 (4) | 0.4813 (3) | 0.0435 (12) | |
| C6B | 0.4700 (10) | 0.8688 (4) | 0.3605 (4) | 0.0576 (15) | |
| H6B | 0.481705 | 0.930081 | 0.315903 | 0.069* | |
| C13A | 0.5825 (10) | 0.8647 (4) | 0.6799 (4) | 0.0572 (15) | |
| H13A | 0.730428 | 0.845033 | 0.668565 | 0.069* | |
| C5A | 0.6988 (9) | 0.6501 (4) | 1.0763 (3) | 0.0456 (12) | |
| H5A | 0.626632 | 0.598873 | 1.071699 | 0.055* | |
| C8B | −0.1697 (9) | 0.6367 (4) | 0.6549 (3) | 0.0517 (14) | |
| H8BA | −0.226852 | 0.574898 | 0.658879 | 0.062* | |
| H8BB | −0.287947 | 0.688592 | 0.632632 | 0.062* | |
| C14B | 0.0695 (9) | 0.6561 (4) | 0.7590 (3) | 0.0495 (13) | |
| H14B | 0.182173 | 0.683315 | 0.717605 | 0.059* | |
| C10A | 0.1499 (9) | 0.9266 (4) | 0.7139 (3) | 0.0509 (14) | |
| H10A | 0.003602 | 0.948655 | 0.724536 | 0.061* | |
| C13B | 0.1036 (10) | 0.6469 (4) | 0.8380 (3) | 0.0560 (14) | |
| H13B | 0.236700 | 0.667865 | 0.849318 | 0.067* | |
| C11B | −0.2587 (10) | 0.5752 (5) | 0.8820 (4) | 0.0600 (16) | |
| H11B | −0.370718 | 0.547599 | 0.923613 | 0.072* | |
| C1A | 1.1677 (9) | 0.6740 (5) | 1.1928 (3) | 0.0537 (15) | |
| H1AA | 1.250770 | 0.732005 | 1.181669 | 0.080* | |
| H1AB | 1.268614 | 0.623796 | 1.185187 | 0.080* | |
| H1AC | 1.104851 | 0.649076 | 1.248552 | 0.080* | |
| C12B | −0.0633 (10) | 0.6060 (5) | 0.8998 (4) | 0.0591 (15) | |
| H12B | −0.043285 | 0.599418 | 0.953119 | 0.071* | |
| C7B | 0.0549 (9) | 0.7494 (4) | 0.5377 (3) | 0.0490 (13) | |
| C2B | 0.6158 (10) | 0.7092 (4) | 0.4317 (3) | 0.0575 (15) | |
| H2BA | 0.729127 | 0.659726 | 0.436849 | 0.069* | |
| C12A | 0.4310 (11) | 0.9020 (5) | 0.6185 (4) | 0.0653 (17) | |
| H12A | 0.473938 | 0.905876 | 0.565843 | 0.078* | |
| C3B | 0.4378 (10) | 0.6904 (4) | 0.4896 (3) | 0.0540 (14) | |
| H3B | 0.431425 | 0.628916 | 0.534200 | 0.065* | |
| C5B | 0.2912 (10) | 0.8543 (4) | 0.4166 (3) | 0.0560 (15) | |
| H5B | 0.184373 | 0.906179 | 0.411098 | 0.067* | |
| C11A | 0.2154 (11) | 0.9334 (5) | 0.6356 (4) | 0.0658 (17) | |
| H11A | 0.112389 | 0.959471 | 0.594095 | 0.079* | |
| C1B | 0.8149 (11) | 0.8123 (5) | 0.3041 (4) | 0.0697 (18) | |
| H1BA | 0.752677 | 0.819184 | 0.253735 | 0.105* | |
| H1BB | 0.922527 | 0.756296 | 0.321780 | 0.105* | |
| H1BC | 0.890242 | 0.871600 | 0.295088 | 0.105* | |
| O1W | 0.1099 (12) | 0.5752 (7) | 0.4262 (5) | 0.132 (3) | |
| H1WA | 0.222090 | 0.534776 | 0.446576 | 0.198* | |
| H1WB | 0.054401 | 0.561287 | 0.389761 | 0.198* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1A | 0.0648 (4) | 0.0476 (4) | 0.0676 (4) | −0.0142 (3) | −0.0086 (3) | −0.0165 (3) |
| Br1B | 0.0719 (4) | 0.0510 (4) | 0.0559 (4) | −0.0091 (3) | −0.0022 (3) | −0.0133 (3) |
| O1A | 0.068 (3) | 0.049 (2) | 0.061 (2) | −0.0199 (19) | −0.0167 (19) | −0.0110 (19) |
| N1A | 0.050 (2) | 0.047 (3) | 0.033 (2) | −0.0052 (19) | −0.0020 (17) | −0.015 (2) |
| N1B | 0.056 (3) | 0.052 (3) | 0.041 (2) | −0.016 (2) | 0.0023 (19) | −0.018 (2) |
| N2A | 0.054 (3) | 0.045 (3) | 0.045 (2) | −0.0116 (19) | −0.0074 (19) | −0.011 (2) |
| O1B | 0.069 (3) | 0.059 (3) | 0.075 (3) | 0.002 (2) | 0.018 (2) | −0.005 (2) |
| N2B | 0.058 (3) | 0.052 (3) | 0.040 (2) | −0.003 (2) | 0.0033 (19) | −0.015 (2) |
| C9B | 0.044 (3) | 0.038 (3) | 0.039 (3) | −0.007 (2) | −0.002 (2) | −0.011 (2) |
| C9A | 0.046 (3) | 0.033 (3) | 0.046 (3) | −0.007 (2) | −0.008 (2) | −0.009 (2) |
| C2A | 0.062 (3) | 0.036 (3) | 0.043 (3) | −0.008 (2) | −0.009 (2) | −0.012 (2) |
| C10B | 0.043 (3) | 0.054 (4) | 0.053 (3) | −0.015 (2) | 0.007 (2) | −0.020 (3) |
| C4A | 0.049 (3) | 0.041 (3) | 0.032 (3) | −0.007 (2) | 0.001 (2) | −0.015 (2) |
| C6A | 0.053 (3) | 0.033 (3) | 0.039 (3) | −0.003 (2) | −0.004 (2) | −0.008 (2) |
| C7A | 0.049 (3) | 0.045 (3) | 0.036 (3) | −0.009 (2) | 0.002 (2) | −0.016 (2) |
| C14A | 0.048 (3) | 0.041 (3) | 0.052 (3) | −0.002 (2) | −0.011 (2) | −0.013 (2) |
| C3A | 0.057 (3) | 0.034 (3) | 0.048 (3) | −0.006 (2) | −0.007 (2) | −0.013 (2) |
| C8A | 0.045 (3) | 0.051 (4) | 0.052 (3) | 0.001 (2) | −0.011 (2) | −0.013 (3) |
| C4B | 0.054 (3) | 0.043 (3) | 0.033 (3) | −0.005 (2) | −0.005 (2) | −0.015 (2) |
| C6B | 0.064 (4) | 0.040 (3) | 0.054 (3) | −0.007 (3) | 0.003 (3) | −0.005 (3) |
| C13A | 0.058 (3) | 0.051 (4) | 0.062 (4) | 0.000 (3) | −0.004 (3) | −0.023 (3) |
| C5A | 0.057 (3) | 0.037 (3) | 0.043 (3) | −0.012 (2) | 0.003 (2) | −0.016 (2) |
| C8B | 0.058 (3) | 0.049 (3) | 0.043 (3) | −0.009 (2) | 0.000 (2) | −0.012 (3) |
| C14B | 0.050 (3) | 0.043 (3) | 0.048 (3) | −0.010 (2) | 0.005 (2) | −0.012 (3) |
| C10A | 0.043 (3) | 0.050 (3) | 0.052 (3) | −0.001 (2) | −0.013 (2) | −0.011 (3) |
| C13B | 0.060 (3) | 0.058 (4) | 0.055 (4) | −0.009 (3) | −0.007 (3) | −0.025 (3) |
| C11B | 0.064 (4) | 0.063 (4) | 0.050 (3) | −0.019 (3) | 0.018 (3) | −0.021 (3) |
| C1A | 0.047 (3) | 0.060 (4) | 0.049 (3) | 0.000 (3) | −0.014 (2) | −0.016 (3) |
| C12B | 0.072 (4) | 0.062 (4) | 0.046 (3) | −0.004 (3) | −0.002 (3) | −0.025 (3) |
| C7B | 0.051 (3) | 0.051 (4) | 0.039 (3) | −0.001 (3) | −0.003 (2) | −0.013 (3) |
| C2B | 0.065 (4) | 0.045 (4) | 0.056 (4) | 0.000 (3) | 0.002 (3) | −0.015 (3) |
| C12A | 0.079 (4) | 0.061 (4) | 0.061 (4) | −0.012 (3) | 0.000 (3) | −0.029 (3) |
| C3B | 0.063 (3) | 0.045 (3) | 0.042 (3) | −0.003 (3) | 0.004 (2) | −0.007 (3) |
| C5B | 0.058 (3) | 0.045 (3) | 0.054 (3) | 0.001 (3) | 0.004 (3) | −0.011 (3) |
| C11A | 0.068 (4) | 0.063 (4) | 0.062 (4) | 0.002 (3) | −0.022 (3) | −0.019 (3) |
| C1B | 0.067 (4) | 0.072 (5) | 0.062 (4) | −0.012 (3) | 0.019 (3) | −0.021 (3) |
| O1W | 0.103 (5) | 0.189 (8) | 0.132 (6) | 0.013 (5) | −0.021 (4) | −0.095 (6) |
Geometric parameters (Å, º)
| O1A—C7A | 1.223 (6) | C4B—C5B | 1.373 (7) |
| N1A—C6A | 1.330 (7) | C4B—C3B | 1.382 (7) |
| N1A—C2A | 1.343 (6) | C4B—C7B | 1.514 (7) |
| N1A—C1A | 1.491 (7) | C6B—C5B | 1.357 (8) |
| N1B—C2B | 1.323 (7) | C6B—H6B | 0.9300 |
| N1B—C6B | 1.329 (7) | C13A—C12A | 1.372 (9) |
| N1B—C1B | 1.480 (7) | C13A—H13A | 0.9300 |
| N2A—C7A | 1.332 (6) | C5A—H5A | 0.9300 |
| N2A—C8A | 1.459 (7) | C8B—H8BA | 0.9700 |
| N2A—H2A | 0.8600 | C8B—H8BB | 0.9700 |
| O1B—C7B | 1.231 (6) | C14B—C13B | 1.386 (8) |
| N2B—C7B | 1.326 (7) | C14B—H14B | 0.9300 |
| N2B—C8B | 1.446 (7) | C10A—C11A | 1.382 (8) |
| N2B—H2B | 0.8600 | C10A—H10A | 0.9300 |
| C9B—C14B | 1.373 (7) | C13B—C12B | 1.384 (8) |
| C9B—C10B | 1.397 (7) | C13B—H13B | 0.9300 |
| C9B—C8B | 1.502 (7) | C11B—C12B | 1.375 (9) |
| C9A—C14A | 1.376 (7) | C11B—H11B | 0.9300 |
| C9A—C10A | 1.381 (7) | C1A—H1AA | 0.9600 |
| C9A—C8A | 1.507 (8) | C1A—H1AB | 0.9600 |
| C2A—C3A | 1.381 (8) | C1A—H1AC | 0.9600 |
| C2A—H2AA | 0.9300 | C12B—H12B | 0.9300 |
| C10B—C11B | 1.370 (8) | C2B—C3B | 1.370 (8) |
| C10B—H10B | 0.9300 | C2B—H2BA | 0.9300 |
| C4A—C5A | 1.379 (7) | C12A—C11A | 1.372 (9) |
| C4A—C3A | 1.385 (7) | C12A—H12A | 0.9300 |
| C4A—C7A | 1.515 (7) | C3B—H3B | 0.9300 |
| C6A—C5A | 1.365 (8) | C5B—H5B | 0.9300 |
| C6A—H6A | 0.9300 | C11A—H11A | 0.9300 |
| C14A—C13A | 1.383 (8) | C1B—H1BA | 0.9600 |
| C14A—H14A | 0.9300 | C1B—H1BB | 0.9600 |
| C3A—H3A | 0.9300 | C1B—H1BC | 0.9600 |
| C8A—H8AA | 0.9700 | O1W—H1WA | 0.8506 |
| C8A—H8AB | 0.9700 | O1W—H1WB | 0.8502 |
| C6A—N1A—C2A | 120.9 (4) | C6A—C5A—H5A | 120.0 |
| C6A—N1A—C1A | 119.3 (4) | C4A—C5A—H5A | 120.0 |
| C2A—N1A—C1A | 119.9 (5) | N2B—C8B—C9B | 114.0 (5) |
| C2B—N1B—C6B | 120.5 (5) | N2B—C8B—H8BA | 108.8 |
| C2B—N1B—C1B | 119.4 (5) | C9B—C8B—H8BA | 108.8 |
| C6B—N1B—C1B | 120.0 (5) | N2B—C8B—H8BB | 108.8 |
| C7A—N2A—C8A | 121.7 (5) | C9B—C8B—H8BB | 108.8 |
| C7A—N2A—H2A | 119.1 | H8BA—C8B—H8BB | 107.6 |
| C8A—N2A—H2A | 119.1 | C9B—C14B—C13B | 122.1 (5) |
| C7B—N2B—C8B | 122.7 (5) | C9B—C14B—H14B | 119.0 |
| C7B—N2B—H2B | 118.6 | C13B—C14B—H14B | 119.0 |
| C8B—N2B—H2B | 118.6 | C9A—C10A—C11A | 120.6 (5) |
| C14B—C9B—C10B | 117.7 (5) | C9A—C10A—H10A | 119.7 |
| C14B—C9B—C8B | 123.6 (4) | C11A—C10A—H10A | 119.7 |
| C10B—C9B—C8B | 118.7 (5) | C12B—C13B—C14B | 119.1 (6) |
| C14A—C9A—C10A | 118.3 (5) | C12B—C13B—H13B | 120.5 |
| C14A—C9A—C8A | 123.7 (4) | C14B—C13B—H13B | 120.5 |
| C10A—C9A—C8A | 118.0 (5) | C10B—C11B—C12B | 120.7 (5) |
| N1A—C2A—C3A | 120.4 (5) | C10B—C11B—H11B | 119.7 |
| N1A—C2A—H2AA | 119.8 | C12B—C11B—H11B | 119.7 |
| C3A—C2A—H2AA | 119.8 | N1A—C1A—H1AA | 109.5 |
| C11B—C10B—C9B | 120.8 (5) | N1A—C1A—H1AB | 109.5 |
| C11B—C10B—H10B | 119.6 | H1AA—C1A—H1AB | 109.5 |
| C9B—C10B—H10B | 119.6 | N1A—C1A—H1AC | 109.5 |
| C5A—C4A—C3A | 118.6 (5) | H1AA—C1A—H1AC | 109.5 |
| C5A—C4A—C7A | 116.8 (5) | H1AB—C1A—H1AC | 109.5 |
| C3A—C4A—C7A | 124.7 (5) | C11B—C12B—C13B | 119.7 (6) |
| N1A—C6A—C5A | 120.9 (5) | C11B—C12B—H12B | 120.2 |
| N1A—C6A—H6A | 119.5 | C13B—C12B—H12B | 120.2 |
| C5A—C6A—H6A | 119.5 | O1B—C7B—N2B | 123.4 (5) |
| O1A—C7A—N2A | 124.3 (5) | O1B—C7B—C4B | 119.2 (5) |
| O1A—C7A—C4A | 118.4 (5) | N2B—C7B—C4B | 117.4 (5) |
| N2A—C7A—C4A | 117.2 (5) | N1B—C2B—C3B | 120.8 (5) |
| C9A—C14A—C13A | 121.1 (5) | N1B—C2B—H2BA | 119.6 |
| C9A—C14A—H14A | 119.4 | C3B—C2B—H2BA | 119.6 |
| C13A—C14A—H14A | 119.4 | C13A—C12A—C11A | 119.2 (6) |
| C2A—C3A—C4A | 119.3 (5) | C13A—C12A—H12A | 120.4 |
| C2A—C3A—H3A | 120.4 | C11A—C12A—H12A | 120.4 |
| C4A—C3A—H3A | 120.4 | C2B—C3B—C4B | 119.6 (5) |
| N2A—C8A—C9A | 113.5 (4) | C2B—C3B—H3B | 120.2 |
| N2A—C8A—H8AA | 108.9 | C4B—C3B—H3B | 120.2 |
| C9A—C8A—H8AA | 108.9 | C6B—C5B—C4B | 120.2 (5) |
| N2A—C8A—H8AB | 108.9 | C6B—C5B—H5B | 119.9 |
| C9A—C8A—H8AB | 108.9 | C4B—C5B—H5B | 119.9 |
| H8AA—C8A—H8AB | 107.7 | C12A—C11A—C10A | 120.6 (5) |
| C5B—C4B—C3B | 117.8 (5) | C12A—C11A—H11A | 119.7 |
| C5B—C4B—C7B | 117.6 (5) | C10A—C11A—H11A | 119.7 |
| C3B—C4B—C7B | 124.6 (5) | N1B—C1B—H1BA | 109.5 |
| N1B—C6B—C5B | 121.0 (5) | N1B—C1B—H1BB | 109.5 |
| N1B—C6B—H6B | 119.5 | H1BA—C1B—H1BB | 109.5 |
| C5B—C6B—H6B | 119.5 | N1B—C1B—H1BC | 109.5 |
| C12A—C13A—C14A | 120.2 (6) | H1BA—C1B—H1BC | 109.5 |
| C12A—C13A—H13A | 119.9 | H1BB—C1B—H1BC | 109.5 |
| C14A—C13A—H13A | 119.9 | H1WA—O1W—H1WB | 109.4 |
| C6A—C5A—C4A | 120.0 (5) | ||
| C6A—N1A—C2A—C3A | 0.3 (8) | C14B—C9B—C8B—N2B | 12.8 (8) |
| C1A—N1A—C2A—C3A | −179.3 (5) | C10B—C9B—C8B—N2B | −167.4 (5) |
| C14B—C9B—C10B—C11B | 0.7 (8) | C10B—C9B—C14B—C13B | −0.7 (8) |
| C8B—C9B—C10B—C11B | −179.0 (6) | C8B—C9B—C14B—C13B | 179.0 (5) |
| C2A—N1A—C6A—C5A | 0.4 (8) | C14A—C9A—C10A—C11A | 0.6 (8) |
| C1A—N1A—C6A—C5A | −180.0 (5) | C8A—C9A—C10A—C11A | −179.1 (6) |
| C8A—N2A—C7A—O1A | −1.5 (8) | C9B—C14B—C13B—C12B | 0.3 (9) |
| C8A—N2A—C7A—C4A | 177.9 (5) | C9B—C10B—C11B—C12B | −0.3 (10) |
| C5A—C4A—C7A—O1A | 15.0 (7) | C10B—C11B—C12B—C13B | −0.1 (10) |
| C3A—C4A—C7A—O1A | −163.5 (5) | C14B—C13B—C12B—C11B | 0.1 (9) |
| C5A—C4A—C7A—N2A | −164.4 (5) | C8B—N2B—C7B—O1B | −0.2 (9) |
| C3A—C4A—C7A—N2A | 17.1 (7) | C8B—N2B—C7B—C4B | −178.4 (5) |
| C10A—C9A—C14A—C13A | 0.5 (8) | C5B—C4B—C7B—O1B | −0.9 (8) |
| C8A—C9A—C14A—C13A | −179.8 (6) | C3B—C4B—C7B—O1B | −179.7 (6) |
| N1A—C2A—C3A—C4A | −0.5 (8) | C5B—C4B—C7B—N2B | 177.4 (5) |
| C5A—C4A—C3A—C2A | −0.1 (8) | C3B—C4B—C7B—N2B | −1.4 (9) |
| C7A—C4A—C3A—C2A | 178.4 (5) | C6B—N1B—C2B—C3B | 0.9 (9) |
| C7A—N2A—C8A—C9A | −102.6 (6) | C1B—N1B—C2B—C3B | −176.0 (6) |
| C14A—C9A—C8A—N2A | 11.4 (8) | C14A—C13A—C12A—C11A | 1.9 (10) |
| C10A—C9A—C8A—N2A | −168.9 (5) | N1B—C2B—C3B—C4B | 0.9 (10) |
| C2B—N1B—C6B—C5B | −0.3 (9) | C5B—C4B—C3B—C2B | −3.1 (9) |
| C1B—N1B—C6B—C5B | 176.6 (6) | C7B—C4B—C3B—C2B | 175.6 (6) |
| C9A—C14A—C13A—C12A | −1.8 (9) | N1B—C6B—C5B—C4B | −2.0 (10) |
| N1A—C6A—C5A—C4A | −1.0 (8) | C3B—C4B—C5B—C6B | 3.7 (9) |
| C3A—C4A—C5A—C6A | 0.8 (8) | C7B—C4B—C5B—C6B | −175.1 (6) |
| C7A—C4A—C5A—C6A | −177.8 (5) | C13A—C12A—C11A—C10A | −0.8 (10) |
| C7B—N2B—C8B—C9B | −107.0 (6) | C9A—C10A—C11A—C12A | −0.5 (10) |
Hydrogen-bond geometry (Å, º)
CgA and CgB are the centroids of the C9A–C14A and C9B–C14B rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2A—H2A···Br1A | 0.86 | 2.53 | 3.339 (5) | 158 |
| C3A—H3A···Br1A | 0.93 | 2.98 | 3.814 (5) | 150 |
| C2A—H2AA···Br1Ai | 0.93 | 2.84 | 3.725 (6) | 159 |
| C1A—H1AA···Br1Ai | 0.96 | 2.88 | 3.784 (6) | 157 |
| C6A—H6A···CgBii | 0.93 | 2.65 | 3.510 (7) | 154 |
| N2B—H2B···Br1B | 0.86 | 2.60 | 3.419 (5) | 159 |
| C3B—H3B···Br1B | 0.93 | 2.83 | 3.753 (5) | 175 |
| C6B—H6B···CgAiii | 0.93 | 2.71 | 3.400 (7) | 132 |
| O1W—H1WA···Br1Biv | 0.85 | 3.03 | 3.473 (7) | 115 |
| C1A—H1AC···O1Wv | 0.96 | 2.89 | 3.794 (10) | 157 |
Symmetry codes: (i) −x+2, −y+2, −z+2; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+2, −z+1; (iv) −x+1, −y+1, −z+1; (v) x+1, y, z+1.
Funding Statement
Funding for this research was provided by: National Academy of Sciences of Ukraine (grant No. 0120U102660).
References
- Boltz, D., Peng, X., Muzzio, M., Dash, P., Thomas, P. G. & Margitich, V. (2018). Antivir. Chem. Chemother. 26, 1–9. [DOI] [PMC free article] [PubMed]
- Buhtiarova, T. A., Danilenko, V. P., Homenko, V. S., Shatyrkina, T. V. & Yadlovsky, O. E. (2003). Ukrainian Med. J. 33, 72–74.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Drebushchak, T. N., Kryukov, Y. A., Rogova, A. I. & Boldyreva, E. V. (2017). Acta Cryst. E73, 967–970. [DOI] [PMC free article] [PubMed]
- Frolov, A. F., Frolov, V. M., Buhtiarova, T. A. & Danilenko, V. P. (2004). Ukrainian Med. J. 39, 69–74.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst. 53, 226–235. [DOI] [PMC free article] [PubMed]
- Rigaku OD (2021). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, England.
- Rodriguez-Carvajal, J. & Roisnel, T. (1998). International Union of Crystallography Newsletter, 20, 35–36.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. A71, 3–8.
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17. University of Western Australia. http://Hirshfeldsurface.net.
- Velthuis, A. J. W., te Zubkova, T. G., Shaw, M., Mehle, A., Boltz, D., Gmeinwieser, N., Stammer, H., Milde, J., Müller, L. & Margitich, V. (2021). Antimicrob. Agents Chemother. 65, e02605–20. [DOI] [PMC free article] [PubMed]
- Zefirov, Yu. V. (1997). Kristallografiya, 42, 936–958.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989022003784/wm5623sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022003784/wm5623Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989022003784/wm5623Isup3.cml
CCDC reference: 2164796
Additional supporting information: crystallographic information; 3D view; checkCIF report








