Abstract
Gout is a prevalent and painful inflammatory arthritis, and its global burden continues to rise. Intense pain induced by gout attacks is a major complication of gout. However, systematic studies of gout inflammation and pain are lacking. Using a monosodium urate (MSU) crystal-induced gout model, we performed genome-wide transcriptome analysis of the inflamed ankle joint, dorsal root ganglion (DRG), and spinal cord of gouty mice. Our results revealed important transcriptional changes, including highly elevated inflammation and broad activation of immune pathways in both the joint and the nervous system, in gouty mice. Integrated analysis showed that there was a remarkable overlap between our RNAseq and human genome-wide association study (GWAS) of gout; for example, the risk gene, stanniocalcin-1 (STC1) showed significant upregulation in all three tissues. Interestingly, when compared to the transcriptomes of human osteoarthritis (OA) and rheumatoid arthritis (RA) joint tissues, we identified significant upregulation of cAMP/cyclic nucleotide-mediated signaling shared between gouty mice and human OA with high knee pain, which may provide excellent drug targets to relieve gout pain. Furthermore, we investigated the common and distinct transcriptomic features of gouty, inflammatory pain, and neuropathic pain mouse models in their DRG and spinal cord tissues. Moreover, we discovered distinct sets of genes with significant differential alternative splicing or differential transcript usage in each tissue, which were largely not detected by conventional differential gene expression analysis approaches. Based on these results, our study provided a more accurate and comprehensive depiction of transcriptomic alterations related to gout inflammation and pain.
Keywords: Gout, inflammation, pain, RNAseq, arthritis, dorsal root ganglion, spinal cord
Introduction
Gout is the most common form of inflammatory arthritis with an approximately 2–4% prevalence worldwide, and it is characterized by the deposition of monosodium urate (MSU) crystals in and around joints. The natural history of gout is typically divided into four stages as follows: asymptomatic hyperuricemia (high uric acid levels in the blood), acute gout, interval period, and chronic tophaceous gout. 1 Gout flares can start suddenly and be extremely painful for days or weeks, followed by a long period of time with no symptoms, which is known as remission. Etiological studies of gout can be traced back to the eighteenth century. 2 It has been well established that hyperuricemia and other influencing factors, including reduced temperatures, lower pH, high concentration of sodium ions, and abnormal connective tissues, lead to crystals of uric acid to form and settle in the joints, eventually causing gouty arthritis.2–4
Half a century ago, multiple studies have shown that the injection of MSU crystals into the joints of normal men and gouty patients successfully induced acute inflammatory responses that closely resembled gouty attacks. 2 Since then, several animal models have been generated to mimic human gouty arthritis by direct injection of MSU-crystals into different anatomical structures, including ankles, knees, and paws.2,5–7
During the past few decades, gout-associated immune pathways and inflammatory responses have been gradually elucidated. A previous study has shown that MSU crystals are proinflammatory by activating the NALP3 inflammasome, resulting in the production of active IL-1β and IL-18. 4 Following the activation of the NLRP3 inflammasome and the release of cytokines (especially IL-1β) and chemokines, innate immune cells, such as monocytes and neutrophils, are recruited to the inflammatory sites. 8 Although MSU crystals activate NLRP3, which cleaves pro-IL-1β into mature IL-1β, it remained unclear how pro-IL-1β was induced in gout. Subsequent studies have shown that myeloid-related proteins S100A8 and S100A9, which are endogenous TLR4 ligands, secreted by MSU crystal-activated phagocytes, induced pro-IL-1β in a TLR4-dependent manner. 9 However, large deposits of tophi alone may not be sufficient to elicit gouty attacks. Dietary factors, such as high-purine foods and alcohol, can trigger gout flares. Joosten et al. reported that the interaction of free fatty acids with TLR2 synergized with MSU crystals to induce the release of a large amount of IL-1β. 10 Another study has suggested that through an NALP3 inflammasome independent pathway, the AIM2 inflammasome is involved in the full development of acute gouty inflammation. 5 Additionally, serum levels of IL-33 in gout patients are significantly higher than those in healthy controls. 11 In mouse gout model, IL-33 and its receptor, ST2, promote neutrophil-dependent ROS production and activate TRPA1 in the DRG. 7
To date, most studies have focused on the acute inflammatory responses induced by exogenous MSU crystals. However, little is known about the cellular and molecular mechanisms of gout pain. As an excruciatingly painful form of arthritis, gout attacks can prevent one from walking or going to work. If left untreated, gout can develop into chronic arthritis with chronic pain. Furthermore, previous transcriptomic analyses of gout animal models are limited to inflammatory tissues or joints.5,7 Therefore, it remains unclear how the DRG and spinal cord are affected to produce highly intensified and excruciating pain in gout. Taking all these factors into consideration, we decided to investigate gout from the perspectives of inflammation and pain. In the present work, we sought to systematically investigate the transcriptional changes by genome-wide expression profiling of the ankle joints, DRG, and spinal cord after MSU-induced acute inflammation in the ankle joints of mice. We then integrated our RNAseq data with large-scale GWAS of gout to identify the top candidate genes. To gain new insights into the molecular mechanisms of different types of arthritis, we performed comparative RNAseq analysis of joint tissue samples among the mouse gout model, human osteoarthritis (OA), and human rheumatoid arthritis (RA). Moreover, we integrated our RNAseq data of the DRG and spinal cord with public RNAseq datasets of complete Freund's adjuvant (CFA) and spared nerve injury (SNI) pain models of mice to discover common dysregulated genes and pathways in pain. In addition, by comparison with RNAseq of human OA with high knee pain, we aimed to identify genes for arthritis-related severe joint pain.
RNAseq based alternative splicing analysis provides robust detection of disease-associated splice events. Alternative splicing is known to be involved in regulating normal physiological functions as well as diseases. Aberrant alternative splicing events are associated with cancer, neurological and psychiatric diseases, inflammation, immune and metabolic disorders. Numerous alternatively spliced isoforms of ion channels and GPCRs in nociceptors have been recognized. 12 A recent study has shown that alternative splicing of the Nrcam gene in the DRG contributed to neuropathic pain genesis and targeting Nrcam alternative splicing effectively attenuated pain hypersensitivity. 13 We observed widespread and distinct alternative splicing events and differential transcript usage (DTU) in the ankle joints, DRG, and spinal cord, suggesting that tissue-specific alternative splicing may contribute to the tissue-specific manifestation of gout.
Materials and methods
Animals
C57BL/6 mice were purchased from the Laboratory Animal Center of Wenzhou Medical University. Mice were housed with a 12-h light/12-h dark cycle and have free access to rodent lab chow and water. Twelve-to sixteen-week-old mice were selected for the assay. The handling of mice and experimental procedures in this study were performed in accordance with the requirements of the Institutional Animal Care and Use Committee of Wenzhou Medical University.
MSU-induced gout mouse model
For the preparation of MSU crystals: 1.0 g uric acid (Sigma-Aldrich) was dissolved in 200 mL boiling distilled water containing 6.0 mL of 1 M NaOH. The pH of the solution was adjusted to 7.2 with HCl until the solution temperature dropped to room temperature. The solution was stirred on a magnetic stirrer for 24 h and then crystals formed. Crystals were then washed with ddH20, dry at 55°C for 24 h and autoclaved. Sterile MSU crystals were suspended in phosphate-buffered saline (PBS, 1x) at a concentration of 20 mg/mL. All reagents were prepared under pyrogen-free conditions.
In this model, the right ankle joint of mice in the MSU group received an intra-articular injection of 100 μL MSU suspension (2 mg MSU crystals in total). For the control group, 100 μL PBS solution was injected into the right ankle joint. Eight hours after injections, the degree of joint swelling was measured with an electronic vernier caliper and photographed. Mechanical hyperalgesia was evaluated using Von Frey filament tests.
RNAseq library preparation and RNAseq data analysis
Eight hours post-injections of MSU crystals or 1 x PBS, mice were anesthetized with isoflurane and decapitated. Ankle joints, DRG, and spinal cord were collected and stored in RNAlater (Thermo Fisher Scientific). Total RNAs were extracted using TRIzol reagent (Thermo Fisher Scientific). RNAseq libraries were prepared for sequencing using a VAHTS Universal V6 RNAseq Library Prep Kit (Vazyme) for Illumina. The library was sequenced on an Illumina HiSeq platform. Clean sequencing reads were aligned to the mouse genome (mm39) using the HISAT2 alignment program, and a >95% alignment rate was achieved for each sample. Differential expression analyses were performed with the DESeq2 R package. 14 For each type of tissue, genes with total counts less than 50 summarized for all 6 samples were filtered out first. For our own RNAseq and public RNAseq datasets, differential expression analysis was performed with the DESeq2 R package. 14 Genes were considered differentially expressed when they had a | log2(fold change) | > log21.5, and p-value < 0.05. Functional enrichment analysis was conducted using the clusterProfiler package. 15 Venn diagrams were generated by the VennDiagram package. 16
Gene regulatory network construction and visualization
Transcriptional regulatory interactions between transcription factors and their target genes were determined according to the TRRUST database. 17 The gene regulatory network was inferred using GENIE3 and visualized using Gephi (0.9.2). 18
Alternative splicing analysis
Differential alternative splicing and differential transcript usage (DTU) were analyzed by SUPPA2. 19 Thresholds of statistical significance for |ΔPSI| (PSI: percent spliced-in) and p-values were set at 0.1 and 0.05, respectively. Gene Ontology (GO) analysis for genes with significantly differential alternative splicing and DTU was performed using the clusterProfiler package. 15 Venn diagrams were generated by the VennDiagram package. 16
Statistical analysis
All data are expressed as the mean ± SEM. Data were analyzed by a two-tailed Student’s t-test between two groups (experimental and control). The criterion for statistical significance was p < 0.05.
Data availability statement
All data used to support the findings of this study are available within the paper and supplementary materials. Our own 18 RNAseq datasets have been deposited in the Gene Expression Omnibus (GEO) repository under access number GSE190138. RNAseq datasets for 18 normal and 20 OA human knee cartilage tissues were downloaded from the GSE114007 dataset. 20 RNAseq datasets of 5 OA patients with high knee pain and 5 OA patients with low knee pain were downloaded from the GSE99662 dataset. 21 RNAseq datasets of CFA and SNI pain mouse models were obtained from the GSE111216 dataset. 22 Human RA single-cell RNAseq data were downloaded from ImmPort (https://www.immport.org/shared/study/SDY998).
Results
Transcriptomics of the mouse gout model
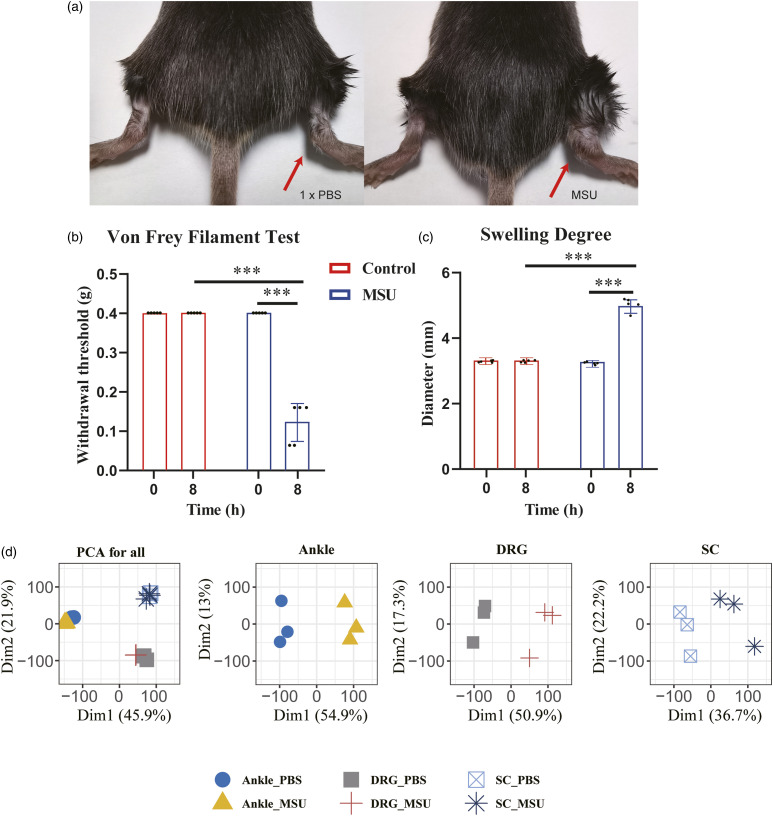
To study the pathophysiology of gouty inflammation and pain at the molecular level, we used an established mouse model of gout in which MSU crystals were injected into the ankle joints of C57BL/6 male mice as described. The control group received 100 μL of 1 x PBS injection. Eight hours after MSU injection, mice developed obvious joint edema and mechanical allodynia (Figure 1(a)-(c)). We next performed RNAseq from the MSU-induced acute inflammation model. For each injected mouse, total RNAs were extracted from three different types of tissues, namely, the ankle joint, DRG (L3-L5), and spinal cord. The quality of RNAseq was assessed by mapping rates on the mouse genome and unsupervised clustering of samples based on gene expression. On average, the alignment rate was >95% for each sample when the reads were mapped to the reference genome. Principal component analysis (PCA) effectively distinguished the samples from different tissues under control and gout conditions (Figure 1(d)). In particular, the three types of tissues presented clear separations with only two principal components. For each type of tissue, samples were well separated between the control and gout models (Figure 1(d)).
Figure 1.
MSU-induced mouse gout model and overview of transcriptomes of the injected ankle joint, DRG, and spinal cord. a. Representative photos showing mouse ankle joints injected with 1 x PBS or MSU. The pictures were taken 8 h after injections. b. Von Frey filament tests showing mechanic allodynia of MSU-injected mice (n = 6 mice/group). c. Swelling degree of MSU-injected mice as indicated by increased ankle diameter (n = 6 mice/group). d. Tissue-based principal component analyses (PCA) of mouse RNAseq under gout condition showed clustering and clear separation for three different types of tissues and between case and control samples of each tissue (n = 3 mice/group).
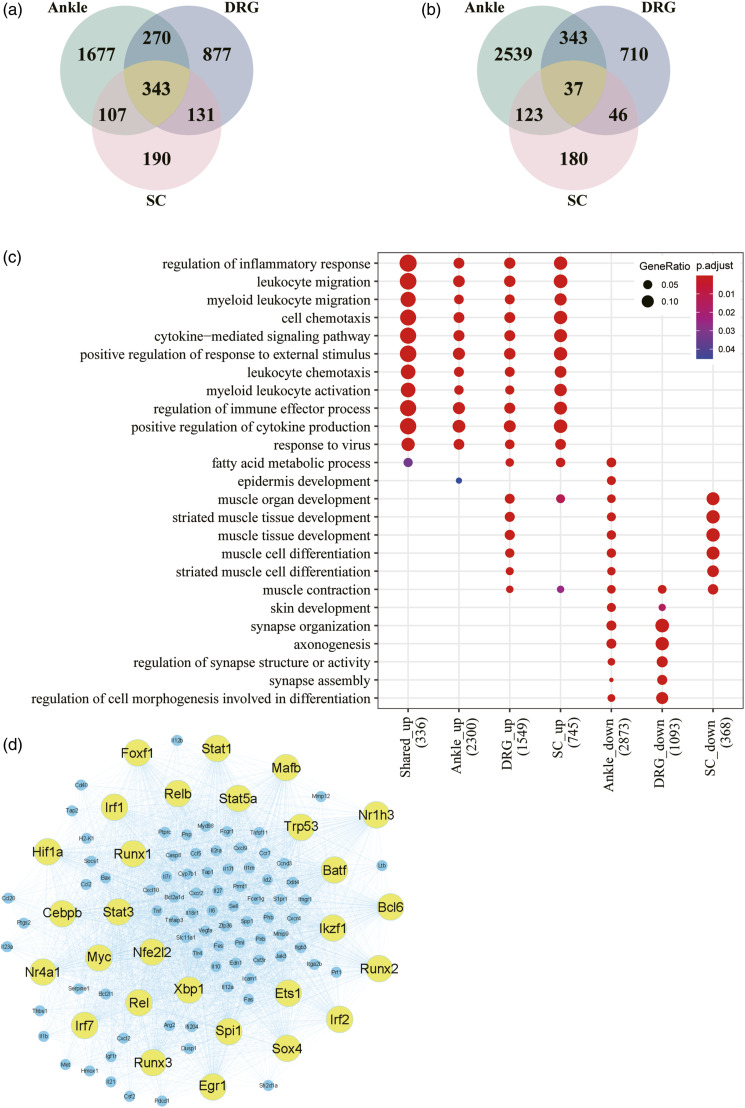
Differential gene expression was analyzed by DESeq2. Genes with a | log2(fold change) | > log21.5 and p-value < 0.05 were selected as DEGs. In total, we identified 5439 DEGs (2397 upregulated and 3042 downregulated) from the ankle joint, 2757 DEGs (1621 upregulated and 1136 downregulated) from the DRG, and 1157 DEGs (771 upregulated and 386 downregulated) from the spinal cord (Figure 2(a) and 2(b)). We then performed functional enrichment analysis for each tissue, which demonstrated that upregulated DEGs in all three tissues were mainly associated with the inflammatory response and immune pathways (Figure 2(c)). However, the top-ranked GO terms for downregulated DEGs were different (Figure 2(c)). Furthermore, the Venn diagrams in Figure 2(a) and 2(b) show the overlapping upregulated and downregulated DEGs from the ankle joint, DRG, and spinal cord. There were 343 shared upregulated DEGs and only 37 shared downregulated DEGs (Supplemental Table 1). Importantly, the 343 shared upregulated DEGs were also significantly enriched in the inflammatory response and immune pathways (Figure 2(c)), which suggested that severe inflammation was triggered in both the ankle joint and the nervous system.
Figure 2.
Transcriptomic analysis of the injected ankle joint, DRG, and spinal cord. a. Intersection of upregulated DEGs between the ankle joint, DRG, and spinal cord of gouty mice. b. Intersection of downregulated DEGs between the ankle joint, DRG, and spinal cord of gouty mice. c. GO functional enrichment analysis of up- and downregulated DEGs from the three tissues. d. Gene regulatory network for upregulated inflammatory and immune DEGs in the ankle joint.
To better understand the transcriptional regulation of gout inflammation, we further constructed a gene regulatory network for the upregulated DEGs of inflammatory and immune responses in the ankle joints. Consequently, 28 TFs were first extracted and their targeted genes were inferred based on the TRRUST database. 17 In total, 107 target genes were selected by intersecting inferred targeted genes with the upregulated DEGs related to inflammatory and immune responses in the ankle joints. We applied GENIE3, a top-ranked network inference algorithm, to build the gene regulatory network, and we visualized the network by Gephi (Figure 2(d)). The size of the circle represents the number of target genes. Many key inflammatory transcription factors, such as Irf1/2/7, Stat1/3/5a, Myc, and Nr4a1, are the most important regulators in the network that directly regulate a broad range of inflammatory mediators including cytokines, chemokines, and matrix metalloproteinases (MMPs) (Figure 2(d)).
Table 1 shows the top 25 DEGs for the ankle joint, DRG, and spinal cord in the mouse gout model based on log2FC. In addition to significantly upregulated chemokines and cytokines, serum amyloid A (SAA) family proteins, a major type of acute phase proteins and the most sensitive indicators in response to inflammation and tissue injury, were highly expressed in all three tissues. In the ankle joint, the expression of three SAA family proteins, SAA1/2/3 expression was increased approximately 500-fold. In the DRG, SAA1/2/3 were the most upregulated DEGs and their levels increased as much as in the ankle joint. Similarly, the gene expression levels of SAA1/2/3 significantly increased in the spinal cord. SAA proteins have strong proinflammatory properties and induce a wide range of cytokines and chemokines. 23 Such increases were not observed in the DRG and spinal cord of mouse CFA and SNI pain models, or in human OA and RA joint tissues analyzed in this study, indicating that there are important differences between gout and other related diseases.
Table 1.
List of top 25 DEGs for the ankle joint, DRG and spinal cord in the mouse gout model based on log2FC.
| Ankle joint | log2FC | DRG | log2FC | SC | log2FC |
|---|---|---|---|---|---|
| Csf3 | 11.5211 | Saa1 | 10.3678 | Csf3 | 9.6181 |
| Cxcl3 | 10.2888 | Saa2 | 9.0135 | Cxcl1 | 9.1630 |
| Il6 | 9.8986 | Saa3 | 8.4723 | Selp | 7.9485 |
| Saa3 | 9.8221 | Csf3 | 8.1977 | Saa3 | 7.7465 |
| Cxcl2 | 9.7280 | Odf3l2 | 8.1241 | Serpina3f | 7.0725 |
| Saa2 | 9.6969 | Tnfsf11 | 8.0714 | Mefv | 7.0175 |
| Saa1 | 8.7028 | Elobl | 7.1844 | Sele | 6.3845 |
| Acod1 | 8.3274 | Sele | 7.1192 | Il6 | 6.2670 |
| Ccl4 | 8.1948 | Fgb | 7.0158 | Slc10a6 | 6.2058 |
| Nos2 | 7.2523 | Cxcl1 | 6.8378 | Cxcl2 | 6.1489 |
| Cxcl1 | 7.2176 | Catsper4 | 6.6992 | Il1rn | 5.6162 |
| Klk1b1 | 7.1834 | Tmem52 | 6.3292 | Saa1 | 5.5035 |
| Ccl3 | 7.0601 | Myhas | 6.3137 | Plin4 | 5.3598 |
| Sult6b2 | 6.9770 | Prok2 | 6.2708 | F10 | 5.1351 |
| Ptx3 | 6.7073 | Dupd1 | 6.2434 | Ms4a8a | 5.0921 |
| Mbl1 | 6.6097 | Lrrc30 | 6.0803 | Hcar2 | 5.0827 |
| Cxcl5 | 6.5312 | Selp | 6.0472 | Lcn2 | 4.8363 |
| Slc22a6 | 6.3333 | Ptx3 | 5.9484 | Saa2 | 4.7556 |
| Il1b | 6.2989 | Myl1 | 5.7829 | Cd244a | 4.5848 |
| Mir7678 | 6.2929 | Plin4 | 5.6349 | Ms4a6d | 4.3855 |
| Slfn4 | 6.1824 | Ampd1 | 5.5826 | Plaur | 4.3416 |
| Ms4a20 | 6.0637 | Abra | 5.5299 | Ms4a4a | 4.0995 |
| Dmbt1 | 5.9288 | Cox6a2 | 5.4676 | Ms4a4c | 4.0984 |
| Ptgs2 | 5.8963 | Myf6 | 5.4250 | Ptgs2 | 4.0929 |
| Clec4e | 5.8540 | Mylpf | 5.3839 | Clec4d | 4.0369 |
Relevance of DEGs in the DRG and spinal cord of gouty mice to mouse pain models
Pain is first detected by DRG neurons and then transmitted to the dorsal horn of the spinal cord. The spinal dorsal horn is a major site of nociceptive signal processing and relay. From there, pain information is sent to the higher brain centers including the thalamus and cortex along the ascending pathway. Therefore, to better understand how excruciating pain is produced and transmitted with gout flares, it is crucial to investigate the transcriptional changes in the DRG and spinal cord in the gout model.
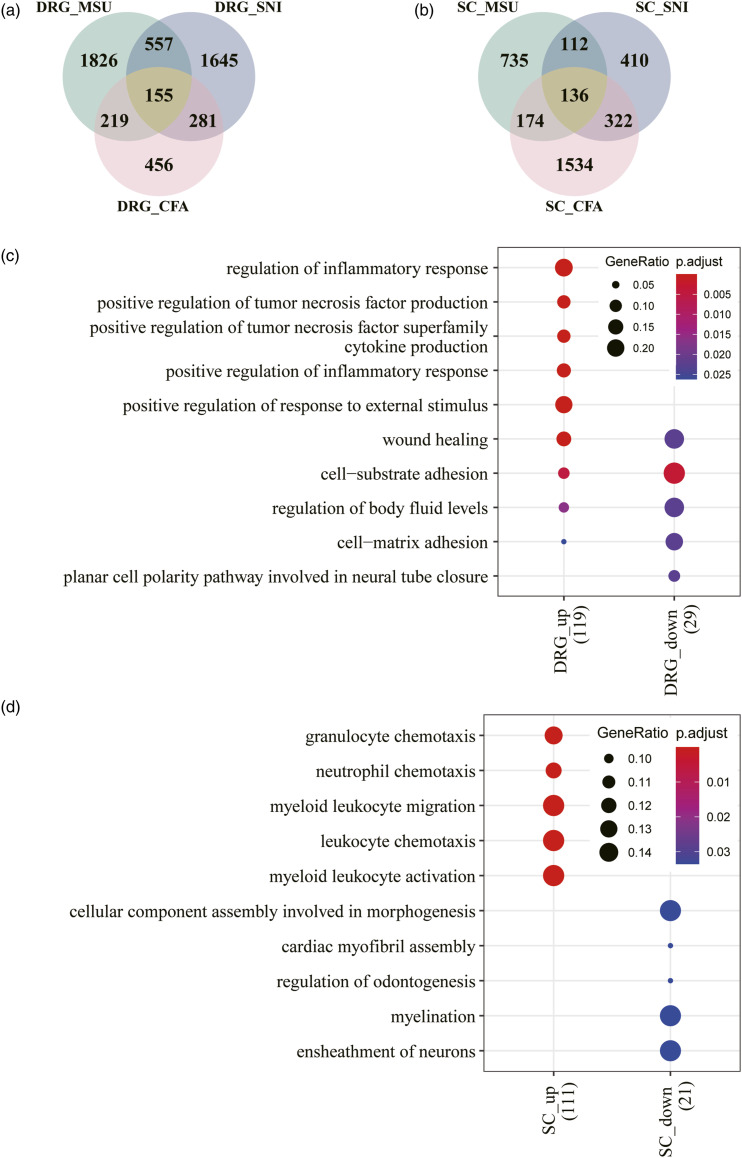
Complete Freund’s adjuvant (CFA)-induced inflammatory pain and spared nerve injury (SNI) induced neuropathic pain rodent models have been well established and widely used. Several transcriptomic studies have revealed transcriptional alterations in different pain models.22,24,25 Here, we conducted transcriptomic comparisons of the DRG and spinal cord among the gout, CFA and SNI models. Transcriptome profiles of the CFA and SNI models were retrieved from a previous article. 22 Overall, there were 155 overlapping DEGs in the DRG of the gout, CFA and SNI models (126 upregulated and 29 downregulated, Supplemental Table 2) (Figure 3(a)). The most enriched GO biological processes for the 129 upregulated DEGs included regulation of the inflammatory response, positive regulation of TNF and cytokine production (Figure 3(c)). For the spinal cord, 136 DEGs were shared among the three models (114 upregulated and 22 downregulated, Supplemental Table 3) (Figure 3(b)). The 111 upregulated DEGs were strongly associated with granulocyte, leukocyte, and neutrophil chemotaxis as well as, leukocyte migration and activation (Figure 3(d)). Our results indicated that upregulation of inflammatory and immune responses is a common feature in the DRG and spinal cord in the gout, CFA and SNI mouse models.
Figure 3.
Comparative transcriptomic analysis of the DRG and spinal cord between gout, CFA and SNI pain mouse models. a. Intersection of DEGs in the DRG between gout, CFA and SNI pain mouse models. b. Intersection of DEGs in the spinal cord between gout, CFA and SNI pain mouse models. c. GO enrichment analysis for shared DEGs of the DRG. d. GO enrichment analysis for shared DEGs of the spinal cord.
Integrative analysis of gouty mouse RNAseq and human gout GWAS
Genome-wide association studies (GWAS) have identified many susceptibility loci associated with serum urate concentrations and gout.26–28 In 2012, the Global Urate Genetics Consortium reported a large-scale GWAS from >140,000 individuals of European ancestry and identified 28 genome-wide significant loci. 27 Additionally, a GWAS conducted on a Japanese population has discovered 10 loci. 28 We compared our DEGs with the human GWAS top hits and revealed many overlaps, which mostly showed downregulation in our RNAseq data, including ATXN2, CNIH2, CUX2, HLF, KCNQ1, NIPAL1, NRXN2, PRKAG2, and SLC2A9. Several overlaps were upregulated in our study, including IGF1R, INHBB, STC1, UBE2Q2, and VEGFA. In total, among these candidate genes identified by GWAS, 14 of 37 (37.84%) were differentially expressed in at least one type of tissue of the mouse gout model. Notably, only stanniocalcin-1 (STC1) was significantly upregulated in all three types of tissues, namely, the ankle joint, DRG, and spinal cord (Table 2). STC1 is a secreted glycoprotein, implicated in several pathologies including cancer, retinal degeneration, and inflammation. It has been shown that tumor STC1 inhibits phagocytosis and promotes tumor immune evasion and resistance. 29 It will be interesting to determine its role during gout development.
Table 2.
Overlaps between the gouty mice RNAseq and human GWAS of gout, corresponding statistics of DEGs are bold, underlined and italic.
| Ankle joint | DRG | SC | ||||
|---|---|---|---|---|---|---|
| Symbol | log2FC | p-value | log2FC | p-value | log2FC | p-value |
| ATXN2 | −0.03389 | 0.81245 | −0.63983 | 0.00000 | −0.02006 | 0.82734 |
| CNIH2 | −0.96389 | 0.02244 | −0.21701 | 0.12839 | 0.13911 | 0.31001 |
| CUX2 | 0.01525 | 0.97320 | −0.70206 | 0.00000 | −0.08247 | 0.44834 |
| HLF | −1.86339 | 0.00000 | −0.01119 | 0.98194 | −0.08137 | 0.45980 |
| IGF1R | 0.77819 | 0.00000 | −0.07880 | 0.44310 | 0.12685 | 0.23950 |
| INHBB | 0.94277 | 0.00000 | −0.14309 | 0.36845 | −0.17877 | 0.28079 |
| KCNQ1 | −1.42708 | 0.00067 | −0.07852 | 0.80479 | −0.31812 | 0.37203 |
| NIPAL1 | −1.52108 | 0.00000 | 0.27202 | 0.35190 | −0.19287 | 0.65037 |
| NRXN2 | −1.71093 | 0.00002 | −1.07030 | 0.00000 | 0.16269 | 0.03693 |
| PRKAG2 | −0.58726 | 0.00001 | −0.29779 | 0.00042 | 0.08028 | 0.42038 |
| SLC2A9 | −1.05144 | 0.00012 | −0.56304 | 0.02822 | 0.06642 | 0.88348 |
| STC1 | 1.52280 | 0.00000 | 2.75347 | 0.00002 | 0.83847 | 0.00560 |
| UBE2Q2 | 1.04378 | 0.00000 | −0.10197 | 0.36463 | −0.05931 | 0.53433 |
| VEGFA | 0.85048 | 0.00000 | −0.06485 | 0.64870 | −0.56125 | 0.00000 |
The GWAS loci not shared with DEGs of the gouty mice RNAseq are listed as below.
A1CF, ABCG2, ACVR1BACVRL1, B3GNT4, BAZ1B, BCAS3, FAM35A, GCKR, HIST1H2BF-HIST1H4E, HNF4G, INHBC, MAF, NFAT5, PDZK1, RFX3, RREB1, SFMBT1, SLC16A9, SLC17A1, SLC22A11, SLC22A12, TMEM171, TRIM46.
The above comparisons between the DEGs identified in our study and human GWAS support the contention that some of the DEGs uncovered in the mouse gout model have biological relevance to human gout pathogenesis. The 14 genes simultaneously detected as DEGs and by GWAS have a higher probability of being involved in gout pathogenesis.
Comparative analysis between gouty mouse RNAseq and human OA transcriptomes
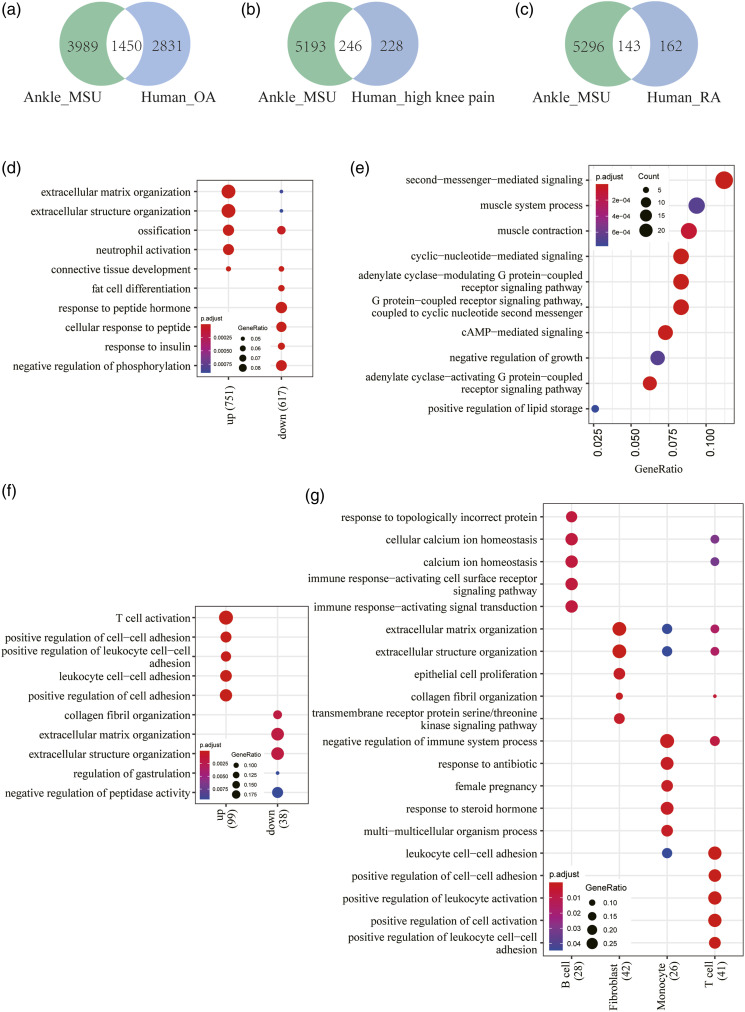
Osteoarthritis (OA) is the most common form of arthritis that can cause severe joint pain, swelling, and stiffness, affecting more than 10% of the population aged 60 years or older. 30 OA is a chronic “wear and tear” disease characterized by the deterioration of cartilage in joints. 31 A recent RNAseq study has been performed on 18 normal and 20 OA human articular cartilage samples in the knee joint to identify dysregulated genes and pathways. 20 We retrieved the RNAseq data from NCBI GEO (access no. GSE114007) and analyzed them with DESeq2. A Venn diagram showed 1450 overlapping DEGs between the ankle joint of the gout model and human OA (Figure 4(a)). Overrepresentation analysis was performed using GO databases for overlapping DEGs. The results showed that upregulated overlapping DEGs were mainly associated with extracellular matrix organization, ossification, and neutrophil activation (Figure 4(d)).
Figure 4.
Comparative transcriptomic analysis of the joint tissues between gouty mice, human OA and RA. a. Intersection of DEGs in the joint tissues between gouty mice and human OA. b. Intersection of DEGs in the joint tissues between gouty mice and human OA with high knee pain. c. Intersection of DEGs in the joint tissues between gouty mice and human RA. d. GO enrichment analysis for shared DEGs of the joint tissues between gouty mice and human OA. e. GO enrichment analysis for shared DEGs of the joint tissues between gouty mice and human OA with high knee pain. f. GO enrichment analysis for shared DEGs of the joint tissues between gouty mice and human RA. g. Cell-type specific GO enrichment analysis for shared DEGs of the joint tissues between gouty mice and human RA.
Severe joint pain is one of the leading causes of disability and morbidity in arthritis. The etiology of joint pain in OA and RA is still unclear. Transcriptomic comparison of OA patients with high knee pain and low knee pain has identified many candidate genes associated with joint pain in OA knee synovial tissues. 21 A total of 246 of these OA joint pain genes were overlapped with the ankle joint DEGs in the present study, and the overlapping DEGs are mostly upregulated (Figure 4(b)). Importantly, the upregulated DEGs are involved in cAMP/cyclic nucleotide-mediated signaling and GPCR signaling pathways (Figure 4(e) and Table 3). There is growing evidence indicating that pain processing is dependent upon activation of the cAMP signaling pathway in a dose-dependent manner.32–34 Inflammatory mediators reduce the threshold for nociceptor neurons to fire action potentials and lead to pain sensitivity. 35 Inflammatory pain behaviors were greatly reduced or abolished in adenylyl cyclase knock-out mice. 36 Therefore, cAMP-mediated signaling and GPCR signaling play a key role in nociceptor sensitization caused by inflammatory and immune mediators. The overlaps identified in the present study are promising targets of drug screening for effective pain relief (Table 3).
Table 3.
List of shared upregulated DEGs in the ankle joint between gouty mice and human OA with high knee pain, involved in cAMP/cyclic nucleotide-mediated signaling and GPCR signaling pathways.
| Symbol | Gene name | log2FC | p-value |
|---|---|---|---|
| GAL | Galanin and GMAP prepropeptide | 2.7442 | 0.0010 |
| MRAP | Melanocortin 2 receptor accessory protein | 1.9223 | 0.0177 |
| RASD1 | Ras related dexamethasone induced 1 | 1.5736 | 0.0000 |
| RAMP1 | Receptor activity modifying protein 1 | 1.4898 | 0.0315 |
| PRKAR2B | Protein kinase cAMP-dependent type II regulatory subunit beta | 1.3688 | 0.0382 |
| ATP1A2 | ATPase Na+/K+ transporting subunit alpha 2 | 1.2874 | 0.0036 |
| CNR1 | Cannabinoid receptor 1 | 1.2838 | 0.0103 |
| PTGER3 | Prostaglandin E receptor 3 | 1.2799 | 0.0121 |
| ADCY5 | Adenylate cyclase 5 | 1.1300 | 0.0055 |
| PDE4D | Phosphodiesterase 4D | 1.0775 | 0.0000 |
| PTGFR | Prostaglandin F receptor | 1.0622 | 0.0005 |
| CASQ2 | Calsequestrin 2 | 1.0067 | 0.0251 |
| NPR1 | Natriuretic peptide receptor 1 | 1.0042 | 0.0169 |
| PLN | Phospholamban | 0.9696 | 0.0056 |
| GNAZ | G protein subunit alpha z | 0.9567 | 0.0290 |
| ADRA1A | Adrenoceptor alpha 1A | 0.9532 | 0.0117 |
| DMD | Dystrophin | 0.9209 | 0.0000 |
| ADCY3 | Adenylate cyclase 3 | 0.8855 | 0.0000 |
| KSR1 | Kinase suppressor of ras 1 | 0.8558 | 0.0000 |
| S1PR3 | Sphingosine-1-phosphate receptor 3 | 0.7286 | 0.0309 |
| GNAL | G protein subunit alpha L | 0.7149 | 0.0002 |
| ADGRL3 | Adhesion G protein-coupled receptor L3 | 0.7018 | 0.0413 |
| PALM | Paralemmin | 0.6707 | 0.0123 |
| NFATC4 | Nuclear factor of activated T cells 4 | 0.6698 | 0.0019 |
| ACKR3 | Atypical chemokine receptor 3 | 0.6416 | 0.0150 |
| PTGIR | Prostaglandin I2 receptor | 0.6050 | 0.0071 |
Comparative analysis between gouty mouse RNAseq and human RA transcriptomes
Rheumatoid arthritis (RA) is a chronic autoimmune and inflammatory disease that causes swelling, stiffness, pain, and loss of function primarily in the joints. The prevalence of RA is relatively constant, accounting for approximately 1% of the world population. 37 RA is characterized by systemic inflammation and persistent synovitis, leading to serious complications in many other tissues as well. GWASs and transcriptomic studies have significantly improved the understanding of RA pathogenesis. Recently, synovial T cells, B cells, monocytes, and fibroblasts from 36 RA and 15 OA patients have been analyzed by utilizing an integrated approach with single-cell RNAseq, bulk RNAseq, flow cytometry, and mass cytometry. 38 Characterized by a high abundance of CD4+ T cells, CD8+ T cells, and B cells, leukocyte-rich RA had significantly higher Krenn inflammation scores than leukocyte-poor RA or OA samples, 38 and subsequently, single-cell RNAseq analysis defined 18 distinct cell subpopulations for the four types of cells. We selected enriched genes for leukocyte-rich RA samples, which had 143 overlapping genes with the ankle joint DEGs of gouty mice (Figure 4(c)). GO analysis of all 143 genes demonstrated that the top enriched biological processes included T-cell activation and positive regulation of cell-cell adhesion for upregulated genes; however, downregulated genes are significantly associated with extracellular matrix organization (Figure 4(f)). Cell type enrichment analysis demonstrated that distinct GO pathways were statistically overrepresented in each cell type (Figure 4(g)), as follow: the “immune response-activating cell surface receptor signaling pathway” and “immune response-activating signal transduction” GO terms were enriched in B cells; the “extracellular matrix organization” GO term was enriched in fibroblasts; the “negative regulation of immune system process” GO term was enriched in monocytes; and the “positive regulation of cell-cell adhesion” and “positive regulation of cell activation” GO terms were enriched in T cells. Together, the comparative studies between gouty mouse RNAseq and human RA single-cell RNAseq revealed shared inflammatory/immune response genes and pathways for specific cell types in the joint tissues.
Differential alternative splicing under gout conditions
Alternative splicing (AS) of pre-mRNA is an essential mechanism to generate genomic and proteomic diversity. In humans, more than 95% of multiexon genes undergo alternative splicing to encode two or more splice isoforms. 39 Compared to gene expression profiling alone, alternative splicing analysis of RNAseq provided robust detection of disease-associated splice events and allowed us to analyze genomic diversity in an unprecedented depth.
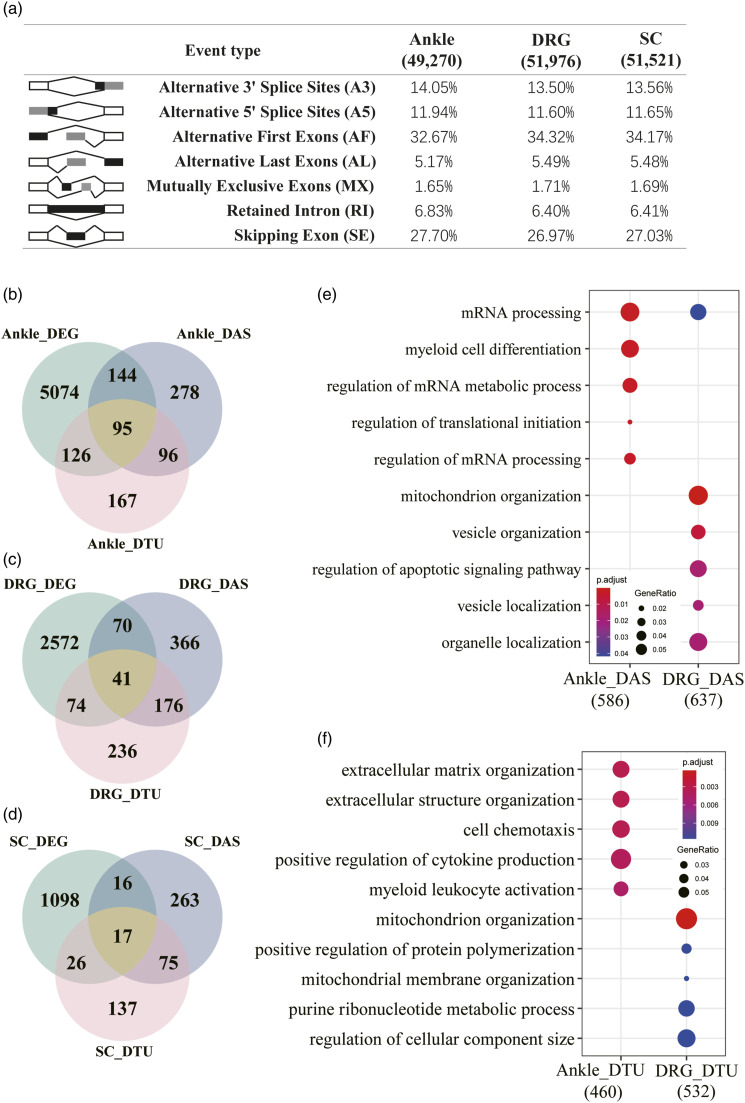
In the present study, we utilized our RNAseq data to incorporate splice variant analysis. In total, 49,270, 51,976, and 51,521 alternative splicing events were identified in the ankle joint, DRG, and spinal cord, respectively (Figure 5(a)). The distribution of seven different types of alternative splicing events in the three tissues was similar with alternative first exons (AF) at the highest frequency (>32%), followed by skipping exon (SE) (>26%), alternative 3′ splice sites (A3) (∼13%), and alternative 5′ splice site (A5) (∼11%) (Figure 5(a)). To define genes differentially alternatively spliced in response to gout conditions, differential alternative splicing events were detected by SUPPA2 using cutoffs of | ΔPSI | > 0.1 and p-value < 0.05. 19 We identified 613, 653, and 371 differentially alternatively spliced genes in the ankle joint, DRG, and spinal cord, respectively (Figure 5(b)-5(d) and Supplemental Table 6). We then examined the relationships between DEGs and differentially alternatively spliced genes. The number of DEGs was greater than that of differentially alternatively spliced genes. In the ankle joint, 239 out of 613 (38.99%) differentially alternatively spliced genes are also DEGs. However, for the DRG and spinal cord, only 111 out of 653 (17.00%) and 33 out of 371 (8.89%) differentially alternatively spliced genes are DEGs, respectively (Figure 5(b)-5(d)).
Figure 5.
Alternative splicing analysis for gouty mice. a. Overview of the seven different types of alternative splicing and their frequencies in the three types of tissues. b. Intersections between DEGs, differentially alternatively spliced (DAS) genes, and differential transcript usage (DTU) genes for the ankle joint of gouty mice. c. Intersections between DEGs, differentially alternatively spliced (DAS) genes, and differential transcript usage (DTU) genes for the DRG of gouty mice. d. Intersections between DEGs, differentially alternatively spliced (DAS) genes, and differential transcript usage (DTU) genes for the spinal cord of gouty mice. e. GO enrichment analysis for differentially alternatively spliced (DAS) genes in the ankle joint, DRG, and spinal cord of gouty mice. f. GO enrichment analysis for differential transcript usage (DTU) genes in the ankle joint, DRG, and spinal cord of gouty mice.
Functional enrichment analysis showed that the major overrepresented GO terms for differentially alternatively spliced genes in the ankle joint were “mRNA processing” and “myeloid cell differentiation”. The enriched GO biological processes for differentially alternatively spliced genes in the DRG included “mitochondrion organization”, “vesicle organization”, and “organelle localization”. However, no significant GO terms were identified for differentially alternatively spliced genes in the spinal cord (Figure 5(e)). Overall, these results showed that the ankle joint, DRG, and spinal cord underwent different alternative splicing changes under gout conditions, which may be related to different aspects of gout pathogenesis.
Differential transcript usage under gout conditions
Differential gene expression has been the primary focus of most transcriptomic studies. However, isoform diversity information is lost in conventional DEG analysis. Recent studies have demonstrated that specific transcript usage profiles are associated with cancer and neurological diseases.40,41 Differential transcript usage (DTU) measures and compares how much each transcript contributes to the total expression of the gene between conditions. In the present study, we performed a genome-wide analysis of DTU in gouty mice. A gene exhibiting at least one DTU event with statistical significance was defined as a DTU gene. As a result, 484, 527, and 255 DTU genes were detected in the ankle joint, DRG, and spinal cord, respectively. Interestingly, the majority of DTU genes were not differentially expressed, and DTU genes largely did not overlap with differentially alternatively spliced genes as well (Figure 5(b)-5(d)). Importantly, DTU genes from different tissues were enriched in different functional pathways. The most enriched biological processes for DTU genes from the ankle joint are “extracellular matrix organization”, “cell chemotaxis”, “positive regulation of cytokine production”, and “myeloid leukocyte activation”. In contrast, the most enriched GO terms for DTU genes from DRG are “mitochondrion organization”, “positive regulation of protein polymerization”, and “purine ribonucleotide metabolic process”. Moreover, no enriched GO terms were found for DTU genes from the spinal cord (Figure 5(f)). Thus, the DTU analysis provided additional insights into the transcriptomic landscape in gout.
Discussion
The present study is the first genome-wide transcriptomic analysis of the inflamed ankle joint, DRG, and spinal cord in a mouse model of gout. Sudden onset of severe inflammation and intense pain are characteristic features of gout arthritis. Although gout inflammation has been extensively studied, systematic analysis of transcriptomic alterations in gout is lacking and the mechanisms involved in gout pain are unclear.
According to The Global Burden of Disease Study 2017, there are more than 41 million adults with gout worldwide.42–44 Maintaining patient adherence to urate-lowering medications is an ongoing challenge.42,45 As a result, most gout patients continue to experience gout incidence and flares. In addition, increasing evidence supports that gout is strongly associated with metabolic disorders. The rise in obesity, hypertension, coronary artery disease, and diabetes is an important contributor to the marked increase in gout prevalence globally.43,46 Gout adds a considerably burden to the public healthcare costs. Currently, non-steroidal anti-inflammatory drugs (NSAIDs) and Colchicine are the most widely used pharmaceuticals for the treatment of gout inflammation and pain. However, accumulating clinical data suggested that benefits of these drugs were limited for gout patients and statistically significantly greater number of adverse events were reported for long term and/or high-dose usages.47,48 Management and outcomes in people with chronic gout are poor. Novel treatments with high efficacy and minimal side effects are urgently needed for gout patients. Therefore, a renewed interest and a deeper understanding of the pathogenesis of gout will help us develop novel therapies for better gout care.
Systematic transcriptional profiling of the inflamed ankle joint, DRG, and spinal cord in gouty mice
Even though it is well established that an excessively increased serum urate concentration is the major risk factor for the development of gout, the molecular mechanisms driving inflammatory manifestations and severe pain of gout are still poorly studied. Here, we performed subcutaneous injections of MSU crystals in the ankle joints of mice, a model of human gout, and then conducted systematic transcriptome analysis of the ankle joints, DRG and spinal cord, to investigate how intensive pain is triggered and transmitted under acute gouty inflammation. Our results demonstrated that the inflammatory responses and immune pathways were activated and significantly upregulated in all three tissues. For example, the transcriptional levels of the SAA family genes, SAA1/2/3, increased several hundred-fold in the gout model, which may be an indicator of the severity of acute gout inflammation and pain. In addition, such observation might be one of the unique features of gout that are not shared in other pain models or human arthritis. Furthermore, the gene regulatory network provided an overall picture of the interactions of upregulated inflammatory TFs and their target genes during MSU-induced acute inflammation.
Comparative studies of gouty mice and human GWAS of gouty
To assess whether the DEGs identified in our mouse study implicated similar genes as revealed in human GWAS data, we retrieved the genetic association summary statistics of two recent large-scale GWAS cohorts.27,28 Comparison of our DEGs with the human GWAS revealed 14 overlapping genes. Among these genes, SLC9A2 inhibition reduced serum urate, and activation of KCNQ1 relieved gout pain. 49 Overall, 12 out of the 14 genes were identified among the ankle joint DEGs. Two of the 14 genes, ATXN2 and CUX2 were only identified among the DRG DEGs. In particular, STC1, which is immune modulator and phagocytosis checkpoint that promotes tumor immunotherapy resistance, was the only molecule present in all three types of tissues . 29 Thus, the overlapping genes between our RNAseq DEGs and human GWAS may be excellent candidate genes worthy of further functional characterization.
Comparative gene expression analysis of gouty mice, human OA and human RA
OA and RA are the two most common forms of arthritis. OA is a degenerative disease of the joints due to the wear and tear of the cartilage. RA is an autoimmune disease in which the immune system attacks the joints. Gout is an inflammatory type of arthritis that mainly affects men over the age of 40. Multiple transcriptomic studies have examined the gene expression changes in OA and RA patients. However, comparative studies among OA, RA and gout are rare. We systematically compared our RNAseq data of the ankle joints from gouty mice with RNAseq data from human OA and RA knee joints. For human OA and our gout RNAseq, the upregulated overlapping genes are significantly associated with extracellular matrix organization, ECM-receptor interaction, ossification, and neutrophil activation. In contrast, when comparing our gout RNAseq data with human RA single-cell RNAseq data, enriched biological processes for the overlapping genes include immune response, T-cell activation, leukocyte activation, cell-cell adhesion, and extracellular matrix organization. The results highlight the key differences between human OA and RA, suggesting that gout arthritis may share distinctive features with human OA and RA.
Comparative gene expression analysis of gouty mice, CFA pain model and SNI pain model, and human OA with high knee pain
Sudden and intense pain is one of the characteristics of gout. To determine genes and pathways associated with high pain in gout attacks, we investigated the transcriptomes of DRG and the spinal cord of the gout mouse. For the first time, we revealed that cytokine, cell chemotaxis, leukocyte, and T cell activation related genes were all dramatically upregulated in both the DRG and spinal cord in the gouty mice. We further identified shared DEGs in the DRG and spinal cord among the gouty, CFA, and SNI mouse models. Our results showed that upregulation of cytokine and TNF production in the DRG and increasing cell chemotaxis and leukocyte activation in the spinal cord were shared by the three models. These results suggest that a higher level of inflammatory responses and broad immune activation in the DRG and spinal cord might be one of the underlying causes for severe pain with gout flares.
A previous transcriptomic study of OA patients with low and high pain has identified many DEGs in knee synovial tissues, which are mostly upregulated and likely associated with high pain. We further compared these DEGs with our RNAseq data and then performed functional enrichment analysis. We found that cyclic-nucleotide-mediated signaling, cAMP-mediated signaling, and GPCR signaling pathway coupled to cyclic nucleotide second messengers were significantly enriched for the overlapping upregulated DEGs. Inflammatory mediators abnormally elevate intracellular cAMP levels and activate cyclic nucleotide signaling which in turn causes either sensitization or increased expression of pain channels in sensory neurons.32,34,50 More importantly, the cAMP-dependent protein kinase (PKA) transduction cascade mediates pain sensation in a dose-dependent manner. 51 Together, our analysis suggests that markedly increased inflammatory responses, immune responses, cyclic nucleotide signaling (including cAMP and cGMP), and GPCR signaling pathways in the DRG and spinal cord may be the primary reasons for significant gout pain.
Alternative splicing analysis of gouty mice
RNAseq data have provided researchers unprecedented insight into the transcriptional landscape of cells. The current sequencing depth is sufficient to reveal the complexity of genomes and transcriptomes, such as gene fusions, mutations/indels, alternative splicing, posttranslational modification, and novel RNA species, in addition to differential gene expression. Alternative splicing is an important mechanism used to increase the diversity of transcripts and proteins in the genome. Alternative splicing is crucial in the regulation of normal cellular function. Aberrant splicing events are commonly reported in human diseases, particularly cancer and neurological diseases. Alternative splicing plays an important role in immune cell development and regulation of inflammatory responses. Targeting alternative splicing has led to the development of novel therapeutics. 52 In the present study, we discovered differential alternative splicing and differential transcript usage in the inflamed ankle joint, DRG and spinal cord of the mouse gout model, which would remain concealed in traditional transcriptomic studies. We found that distinct sets of genes were differentially spliced under gout conditions for different tissues, and these genes were enriched in different biological processes and may be relevant to gout pathogenesis.
Conclusions
In the present study, we systematically investigated the transcriptional profiles of the ankle joint, DRG, and spinal cord in gouty mice. The prominent result was the broad upregulation of immune and inflammatory responses in all three tissues. Integrative analysis with human GWAS data of gout showed that 14 of 37 known susceptibility loci were also DEGs. In particular, STC1 was the only locus upregulated in all three tissues. In addition, differentially alternatively spliced genes and genes with differential transcript usage were detected under gout conditions, which showed tissue specific patterns. Interestingly, for each tissue, DEGs, differentially spliced genes, and genes with differential transcript usage are largely not overlapped. Comparison with human OA and RA transcriptomic datasets indicated that significant upregulation of cAMP/cyclic nucleotide-mediated signaling was likely associated with high pain. Our findings provide key insight into how severe gouty inflammation and pain are triggered by whole-genome gene expression analysis, which not only provides candidate genes to further study gout pathogenesis but also aids in the development of novel therapies for better gout care.
Supplemental Material
Supplemental Material for Systematic analysis of inflammation and pain pathways in a mouse model of gout by Yuanlan Fan, Jiajun Yang, Song Xie, Jiangshuang He, Shenghui Huang, Jiawang Chen, Shangying Jiang, Lei Yu, Yujuan Zhou, Xuxia Cao, Xiang Ji and , Yi Zhang in Molecular Pain
Supplemental Material for Systematic analysis of inflammation and pain pathways in a mouse model of gout by Yuanlan Fan, Jiajun Yang, Song Xie, Jiangshuang He, Shenghui Huang, Jiawang Chen, Shangying Jiang, Lei Yu, Yujuan Zhou, Xuxia Cao, Xiang Ji and , Yi Zhang in Molecular Pain
Supplemental Material for Systematic analysis of inflammation and pain pathways in a mouse model of gout by Yuanlan Fan, Jiajun Yang, Song Xie, Jiangshuang He, Shenghui Huang, Jiawang Chen, Shangying Jiang, Lei Yu, Yujuan Zhou, Xuxia Cao, Xiang Ji and , Yi Zhang in Molecular Pain
Supplemental Material for Systematic analysis of inflammation and pain pathways in a mouse model of gout by Yuanlan Fan, Jiajun Yang, Song Xie, Jiangshuang He, Shenghui Huang, Jiawang Chen, Shangying Jiang, Lei Yu, Yujuan Zhou, Xuxia Cao, Xiang Ji and , Yi Zhang in Molecular Pain
Supplemental Material for Systematic analysis of inflammation and pain pathways in a mouse model of gout by Yuanlan Fan, Jiajun Yang, Song Xie, Jiangshuang He, Shenghui Huang, Jiawang Chen, Shangying Jiang, Lei Yu, Yujuan Zhou, Xuxia Cao, Xiang Ji and , Yi Zhang in Molecular Pain
Supplemental Material for Systematic analysis of inflammation and pain pathways in a mouse model of gout by Yuanlan Fan, Jiajun Yang, Song Xie, Jiangshuang He, Shenghui Huang, Jiawang Chen, Shangying Jiang, Lei Yu, Yujuan Zhou, Xuxia Cao, Xiang Ji and , Yi Zhang in Molecular Pain
Supplemental Material for Systematic analysis of inflammation and pain pathways in a mouse model of gout by Yuanlan Fan, Jiajun Yang, Song Xie, Jiangshuang He, Shenghui Huang, Jiawang Chen, Shangying Jiang, Lei Yu, Yujuan Zhou, Xuxia Cao, Xiang Ji and , Yi Zhang in Molecular Pain
Footnotes
Supplementary Material: Supplementary Material for this article is available online.
Author contributions
Conceptualization, Y.Z.; Methodology, Y.Z., Y.F., X.J.; Investigation, Y.F., J.Y., S.X., J.H., S.H., J.C., S.J., L.Y., Y.Z., X.C.; Resources, Y.Z.; Writing - Original Draft, Y.Z.; Writing – Review & Editing, Y.F., J.Y., S.H., X.C., X.J.; Supervision, Y.Z.; Funding Acquisition, Y.Z.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the National Natural Science Foundation of China (No. 31771165, No.81971064 to Y. Z.), and funding from Wenzhou Medical University.
References
- 1.Ragab G, Elshahaly M, Bardin T. Gout: An old disease in new perspective - A review. J Adv Res 2017; 8(5): 495–511, DOI: 10.1016/j.jare.2017.04.008, https://www.ncbi.nlm.nih.gov/pubmed/28748116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faires J, Jr, Mccarty D. Acute arthritis in man and dog after intrasynovial injection of sodium urate crystals. The Lancet 1962; 280(7258): 682–685. [Google Scholar]
- 3.Chhana A, Lee G, Dalbeth N. Factors influencing the crystallization of monosodium urate: a systematic literature review. BMC Musculoskelet Disord 2015; 16: 296, DOI: 10.1186/s12891-015-0762-4, https://www.ncbi.nlm.nih.gov/pubmed/26467213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 2006; 440(7081): 237–241, DOI: 10.1038/nature04516, https://www.ncbi.nlm.nih.gov/pubmed/16407889 [DOI] [PubMed] [Google Scholar]
- 5.Mariotte A, De Cauwer A, Po C, Abou-Faycal C, Pichot A, Paul N, Aouadi I, Carapito R, Frisch B, Macquin C, Chatelus E, Sibilia J, Armspach J-P, Bahram S, Georgel P. A mouse model of MSU-induced acute inflammation in vivo suggests imiquimod-dependent targeting of Il-1β as relevant therapy for gout patients. Theranostics 2020; 10(5): 2158–2171, DOI: 10.7150/thno.40650, https://www.ncbi.nlm.nih.gov/pubmed/32104502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pineda C, Fuentes-Gómez AJ, Hernández-Díaz C, Zamudio-Cuevas Y, Fernández-Torres J, López-Macay A, Alba-Sánchez I, Camacho-Galindo J, Ventura L, Gómez-Quiróz LE, Gutiérrez-Ruíz M, García-Vázquez F, Reginato AM, Gutiérrez M, López-Reyes A. Animal model of acute gout reproduces the inflammatory and ultrasonographic joint changes of human gout. Arthritis Res Ther 2015; 17: 37, DOI: 10.1186/s13075-015-0550-4, https://www.ncbi.nlm.nih.gov/pubmed/25889158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin C, Liu B, Li Y, Li X, Wang J, Chen R, Tai Y, Shou Q, Wang P, Shao X, Liang Y, Zhou H, Mi W, Fang J, Liu B. IL-33/ST2 induces neutrophil-dependent reactive oxygen species production and mediates gout pain. Theranostics 2020; 10(26): 12189–12203. DOI: 10.7150/thno.48028, https://www.ncbi.nlm.nih.gov/pubmed/33204337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalbeth N, Choi HK, Joosten LAB, Khanna PP, Matsuo H, Perez-Ruiz F, Stamp LK. Gout. Nat Rev Dis Primers 2019; 5(1): 69, DOI: 10.1038/s41572-019-0115-y, https://www.ncbi.nlm.nih.gov/pubmed/31558729 [DOI] [PubMed] [Google Scholar]
- 9.Holzinger D, Nippe N, Vogl T, Marketon K, Mysore V, Weinhage T, Dalbeth N, Pool B, Merriman T, Baeten D, Ives A, Busso N, Foell D, Bas S, Gabay C, Roth J. Myeloid-related proteins 8 and 14 contribute to monosodium urate monohydrate crystal-induced inflammation in gout. Arthritis Rheumatol 2014; 66(5): 1327–1339, DOI: 10.1002/art.38369, https://www.ncbi.nlm.nih.gov/pubmed/24470119 [DOI] [PubMed] [Google Scholar]
- 10.Joosten LAB, Netea MG, Mylona E, Koenders MI, Malireddi RKS, Oosting M, Stienstra R, van de Veerdonk FL, Stalenhoef AF, Giamarellos-Bourboulis EJ, Kanneganti T-D, van der Meer JWM. Engagement of fatty acids with Toll-like receptor 2 drives interleukin-1beta production via the ASC/caspase 1 pathway in monosodium urate monohydrate crystal-induced gouty arthritis. Arthritis Rheum 2010; 62(11): 3237–3248, DOI: 10.1002/art.27667, https://www.ncbi.nlm.nih.gov/pubmed/20662061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan L, Huang Y, Su Q, Lin Q, Liu W, Luo J, Yu B, He Y, Qian H, Liu Y, Chen J, Shi G. Potential of IL-33 for Preventing the Kidney Injury via Regulating the Lipid Metabolism in Gout Patients. J Diabetes Res 2016; 2016: 1–7, DOI: 10.1155/2016/1028401, https://www.ncbi.nlm.nih.gov/pubmed/27579324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donaldson LF, Beazley-Long N. Alternative RNA splicing: contribution to pain and potential therapeutic strategy. Drug Discov Today 2016; 21(11): 1787–1798, DOI: 10.1016/j.drudis.2016.06.017, https://www.ncbi.nlm.nih.gov/pubmed/27329269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang L, Wu S, Lin C, Chang Y-J, Tao Y-X. Alternative Splicing of Nrcam Gene in Dorsal Root Ganglion Contributes to Neuropathic Pain. J Pain 2020; 21(7–8): 892–904, DOI: 10.1016/j.jpain.2019.12.004, https://www.ncbi.nlm.nih.gov/pubmed/31917219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15(12): 550, DOI: 10.1186/s13059-014-0550-8, https://www.ncbi.nlm.nih.gov/pubmed/25516281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics-a J Integrative Biology 2012; 16(5): 284–287, DOI: 10.1089/omi.2011.0118, https://www.ncbi.nlm.nih.gov/pubmed/22455463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 2011; 12: 35, DOI: 10.1186/1471-2105-12-35, https://www.ncbi.nlm.nih.gov/pubmed/21269502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han H, Cho J-W, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, Lee S, Kang B, Jeong D, Kim Y, Jeon H-N, Jung H, Nam S, Chung M, Kim J-H, Lee I. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res 2018; 46(D1): D380–D386, DOI: 10.1093/nar/gkx1013, https://www.ncbi.nlm.nih.gov/pubmed/29087512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huynh-Thu VA, Irrthum A, Wehenkel L, Geurts P. Inferring regulatory networks from expression data using tree-based methods. PLoS One 2010; 5(9): e12776, DOI: 10.1371/journal.pone.0012776, https://www.ncbi.nlm.nih.gov/pubmed/20927193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trincado JL, Entizne JC, Hysenaj G, Singh B, Skalic M, Elliott DJ, Eyras E. SUPPA2: fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol 2018; 19(1): 40, DOI: 10.1186/s13059-018-1417-1, https://www.ncbi.nlm.nih.gov/pubmed/29571299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisch KM, Gamini R, Alvarez-Garcia O, Akagi R, Saito M, Muramatsu Y, Sasho T, Koziol JA, Su AI, Lotz MK. Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthritis Cartilage 2018; 26(11): 1531–1538, DOI: 10.1016/j.joca.2018.07.012, https://www.ncbi.nlm.nih.gov/pubmed/30081074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bratus-Neuenschwander A, Castro-Giner F, Frank-Bertoncelj M, Aluri S, Fucentese S, Schlapbach R, Sprott H. Pain-Associated transcriptome changes in synovium of knee osteoarthritis patients. Genes 2018; 9(7): 338, DOI: 10.3390/genes9070338, https://www.ncbi.nlm.nih.gov/pubmed/29973527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parisien M, Samoshkin A, Tansley SN, Piltonen MH, Martin LJ, El-Hachem N, Dagostino C, Allegri M, Mogil JS, Khoutorsky A, Diatchenko L. Genetic pathway analysis reveals a major role for extracellular matrix organization in inflammatory and neuropathic pain. Pain 2019; 160(4): 932–944, DOI: 10.1097/j.pain.0000000000001471, https://www.ncbi.nlm.nih.gov/pubmed/30763288 [DOI] [PubMed] [Google Scholar]
- 23.De Buck M, Gouwy M, Wang JM, Van Snick J, Proost P, Struyf S, Van Damme J. The cytokine-serum amyloid A-chemokine network. Cytokine Growth Factor Rev 2016; 30: 55–69, DOI: 10.1016/j.cytogfr.2015.12.010, https://www.ncbi.nlm.nih.gov/pubmed/26794452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bangash MA, Alles SRA, Santana-Varela S, Millet Q, Sikandar S, de Clauser L, ter Heegde F, Habib AM, Pereira V, Sexton JE, Emery EC, Li S, Luiz AP, Erdos J, Gossage SJ, Zhao J, Cox JJ, Wood JN. Distinct transcriptional responses of mouse sensory neurons in models of human chronic pain conditions. Wellcome Open Res 2018; 3: 78, DOI: 10.12688/wellcomeopenres.14641.1, https://www.ncbi.nlm.nih.gov/pubmed/30079380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berta T, Perrin FE, Pertin M, Tonello R, Liu Y-C, Chamessian A, Kato AC, Ji R-R, Decosterd I. Gene expression profiling of cutaneous injured and non-injured nociceptors in SNI Animal Model of Neuropathic Pain. Sci Rep 2017; 7(1): 9367, DOI: 10.1038/s41598-017-08865-3, https://www.ncbi.nlm.nih.gov/pubmed/28839165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C-J, Tseng C-C, Yen J-H, Chang J-G, Chou W-C, Chu H-W, Chang S-J, Liao W-T. ABCG2 contributes to the development of gout and hyperuricemia in a genome-wide association study. Sci Rep 2018; 8(1): 3137, DOI: 10.1038/s41598-018-21425-7, https://www.ncbi.nlm.nih.gov/pubmed/29453348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, Pistis G, Ruggiero D, O'Seaghdha CM, Haller T, Yang Q, Tanaka T, Johnson AD, Kutalik Z, Smith AV, Shi J, Struchalin M, Middelberg RP, Brown MJ, Gaffo AL, Pirastu N, Li G, Hayward C, Zemunik T, Huffman J, Yengo L, Zhao JH, Demirkan A, Feitosa MF, Liu X, Malerba G, Lopez LM, van der Harst P, Li X, Kleber ME, Hicks AA, Nolte IM, Johansson A, Murgia F, Wild SH, Bakker SJ, Peden JF, Dehghan A, Steri M, Tenesa A, Lagou V, Salo P, Mangino M, Rose LM, Lehtimäki T, Woodward OM, Okada Y, Tin A, Müller C, Oldmeadow C, Putku M, Czamara D, Kraft P, Frogheri L, Thun GA, Grotevendt A, Gislason GK, Harris TB, Launer LJ, McArdle P, Shuldiner AR, Boerwinkle E, Coresh J, Schmidt H, Schallert M, Martin NG, Montgomery GW, Kubo M, Nakamura Y, Tanaka T, Munroe PB, Samani NJ, Jacobs DR, Liu K, D’Adamo P, Ulivi S, Rotter JI, Psaty BM, Vollenweider P, Waeber G, Campbell S, Devuyst O, Navarro P, Kolcic I, Hastie N, Balkau B, Froguel P, Esko T, Salumets A, Khaw KT, Langenberg C, Wareham NJ, Isaacs A, Kraja A, Zhang Q, Wild PS, Scott RJ, Holliday EG, Org E, Viigimaa M, Bandinelli S, Metter JE, Lupo A, Trabetti E, Sorice R, Döring A, Lattka E, Strauch K, Theis F, Waldenberger M, Wichmann HE, Davies G, Gow AJ, Bruinenberg M, au fnm, Stolk RP, Kooner JS, Zhang W, Winkelmann BR, Boehm BO, Lucae S, Penninx BW, Smit JH, Curhan G, Mudgal P, Plenge RM, Portas L, Persico I, Kirin M, Wilson JF, Mateo Leach I, van Gilst WH, Goel A, Ongen H, Hofman A, Rivadeneira F, Uitterlinden AG, Imboden M, von Eckardstein A, Cucca F, Nagaraja R, Piras MG, Nauck M, Schurmann C, Budde K, Ernst F, Farrington SM, Theodoratou E, Prokopenko I, Stumvoll M, Jula A, Perola M, Salomaa V, Shin SY, Spector TD, Sala C, Ridker PM, Kähönen M, Viikari J, Hengstenberg C, Nelson CP, Meschia JF, Nalls MA, Sharma P, Singleton AB, Kamatani N, Zeller T, Burnier M, Attia J, Laan M, Klopp N, Hillege HL, Kloiber S, Choi H, Pirastu M, Tore S, Probst-Hensch NM, Völzke H, Gudnason V, Parsa A, Schmidt R, Whitfield JB, Fornage M, Gasparini P, Siscovick DS, Polašek O, Campbell H, Rudan I, Bouatia-Naji N, Metspalu A, Loos RJ, van Duijn CM, Borecki IB, Ferrucci L, Gambaro G, Deary IJ, Wolffenbuttel BH, Chambers JC, März W, Pramstaller PP, Snieder H, Gyllensten U, Wright AF, Navis G, Watkins H, Witteman JC, Sanna S, Schipf S, Dunlop MG, Tönjes A, Ripatti S, Soranzo N, Toniolo D, Chasman DI, Raitakari O, Kao WH, Ciullo M, Fox CS, Caulfield M, Bochud M, Gieger C. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet 2013; 45(2): 145–154, DOI: 10.1038/ng.2500, https://www.ncbi.nlm.nih.gov/pubmed/23263486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama A, Nakaoka H, Yamamoto K, Sakiyama M, Shaukat A, Toyoda Y, Okada Y, Kamatani Y, Nakamura T, Takada T, Inoue K, Yasujima T, Yuasa H, Shirahama Y, Nakashima H, Shimizu S, Higashino T, Kawamura Y, Ogata H, Kawaguchi M, Ohkawa Y, Danjoh I, Tokumasu A, Ooyama K, Ito T, Kondo T, Wakai K, Stiburkova B, Pavelka K, Stamp LK, Dalbeth N, Sakurai Y, Suzuki H, Hosoyamada M, Fujimori S, Yokoo T, Hosoya T, Inoue I, Takahashi A, Kubo M, Ooyama H, Shimizu T, Ichida K, Shinomiya N, Merriman TR, Matsuo H. GWAS of clinically defined gout and subtypes identifies multiple susceptibility loci that include urate transporter genes. Ann Rheum Dis 2017; 76(5): 869–877, DOI: 10.1136/annrheumdis-2016-209632, https://www.ncbi.nlm.nih.gov/pubmed/27899376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin H, Kryczek I, Li S, Green MD, Ali A, Hamasha R, Wei S, Vatan L, Szeliga W, Grove S, Li X, Li J, Wang W, Yan Y, Choi JE, Li G, Bian Y, Xu Y, Zhou J, Yu J, Xia H, Wang W, Alva A, Chinnaiyan AM, Cieslik M, Zou W. Stanniocalcin 1 is a phagocytosis checkpoint driving tumor immune resistance. Cancer Cell 2021; 39(4): 480–493, DOI: 10.1016/j.ccell.2020.12.023, https://www.ncbi.nlm.nih.gov/pubmed/33513345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Jordan JM. Epidemiology of osteoarthritis. Clin Geriatr Med 2010; 26(3): 355–369, DOI: 10.1016/j.cger.2010.03.001, https://www.ncbi.nlm.nih.gov/pubmed/20699159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berenbaum F. Osteoarthritis as an inflammatory disease (osteoarthritis is not osteoarthrosis!). Osteoarthritis Cartilage 2013; 21(1): 16–21, DOI: 10.1016/j.joca.2012.11.012, https://www.ncbi.nlm.nih.gov/pubmed/23194896 [DOI] [PubMed] [Google Scholar]
- 32.Malmberg AB, Brandon EP, Idzerda RL, Liu H, McKnight GS, Basbaum AI. Diminished inflammation and nociceptive pain with preservation of neuropathic pain in mice with a targeted mutation of the type I regulatory subunit of cAMP-dependent protein kinase. J Neurosci 1997; 17(19): 7462–7470, https://www.ncbi.nlm.nih.gov/pubmed/9295392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierre S, Eschenhagen T, Geisslinger G, Scholich K. Capturing adenylyl cyclases as potential drug targets. Nat Rev Drug Discov 2009; 8(4): 321–335, DOI: 10.1038/nrd2827, https://www.ncbi.nlm.nih.gov/pubmed/19337273 [DOI] [PubMed] [Google Scholar]
- 34.Sluka KA. Stimulation of deep somatic tissue with capsaicin produces long-lasting mechanical allodynia and heat hypoalgesia that depends on early activation of the cAMP pathway. J Neurosci 2002; 22(13): 568720026530–568720035693, https://www.ncbi.nlm.nih.gov/pubmed/12097520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinho-Ribeiro FA, Verri WA, Jr, Chiu IM. Nociceptor Sensory Neuron-Immune Interactions in Pain and Inflammation. Trends Immunol 2017; 38(1): 5–19, DOI: 10.1016/j.it.2016.10.001, https://www.ncbi.nlm.nih.gov/pubmed/27793571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei F, Qiu C-S, Kim SJ, Muglia L, Maas JW, Pineda VV, Xu H-M, Chen Z-F, Storm DR, Muglia LJ, Zhuo M. Genetic elimination of behavioral sensitization in mice lacking calmodulin-stimulated adenylyl cyclases. Neuron 2002; 36(4): 713–726, DOI: 10.1016/s0896-6273(02)01019-x, https://www.ncbi.nlm.nih.gov/pubmed/12441059 [DOI] [PubMed] [Google Scholar]
- 37.Xu Y, Wu Q. Prevalence trend and disparities in rheumatoid arthritis among us adults, 2005-2018. J Clin Med 2021; 10(15), DOI: 10.3390/jcm10153289, https://www.ncbi.nlm.nih.gov/pubmed/34362073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang F, Wei K, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, Goodman SM, Tabechian D, Hughes LB, Salomon-Escoto K, Watts GFM, Jonsson AH, Rangel-Moreno J, Meednu N, Rozo C, Apruzzese W, Eisenhaure TM, Lieb DJ, Boyle DL, Mandelin AM, Boyce BF, DiCarlo E, Gravallese EM, Gregersen PK, Moreland L, Firestein GS, Hacohen N, Nusbaum C, Lederer JA, Perlman H, Pitzalis C, Filer A, Holers VM, Bykerk VP, Donlin LT, Anolik JH, Brenner MB, Raychaudhuri S. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol 2019; 20(7): 928–942, DOI: 10.1038/s41590-019-0378-1, https://www.ncbi.nlm.nih.gov/pubmed/31061532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annual Rev Biochemistry 2015; 84: 291–323, DOI: 10.1146/annurev-biochem-060614-034316, https://www.ncbi.nlm.nih.gov/pubmed/25784052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dick F, Nido GS, Alves GW, Tysnes O-B, Nilsen GH, Dölle C, Tzoulis C. Differential transcript usage in the Parkinson’s disease brain. PLoS Genet 2020; 16(11): e1009182, DOI: 10.1371/journal.pgen.1009182, https://www.ncbi.nlm.nih.gov/pubmed/33137089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vitting-Seerup K, Sandelin A. The Landscape of Isoform Switches in Human Cancers. Mol Cancer Res 2017; 15(9): 1206–1220, DOI: 10.1158/1541-7786.MCR-16-0459, https://www.ncbi.nlm.nih.gov/pubmed/28584021 [DOI] [PubMed] [Google Scholar]
- 42.Danve A, Neogi T. Rising Global Burden of Gout: Time to Act. Arthritis Rheumatol 2020; 72(11): 1786–1788, DOI: 10.1002/art.41453, https://www.ncbi.nlm.nih.gov/pubmed/33150696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Safiri S, Kolahi AA, Cross M, Carson‐Chahhoud K, Hoy D, Almasi‐Hashiani A, Sepidarkish M, Ashrafi‐Asgarabad A, Moradi‐Lakeh M, Mansournia MA, Kaufman JS, Collins G, Woolf AD, March L, Smith E. Prevalence, incidence, and years lived with disability due to gout and its attributable risk factors for 195 Countries and Territories 1990-2017: a systematic analysis of the global burden of disease study 2017. Arthritis Rheumatol 2020; 72(11): 1916–1927, DOI: 10.1002/art.41404, https://www.ncbi.nlm.nih.gov/pubmed/32755051 [DOI] [PubMed] [Google Scholar]
- 44.Xia Y, Wu Q, Wang H, Zhang S, Jiang Y, Gong T, Xu X, Chang Q, Niu K, Zhao Y. Global, regional and national burden of gout, 1990-2017: a systematic analysis of the Global Burden of Disease Study. Rheumatology 2020; 59(7): 1529–1538, DOI: 10.1093/rheumatology/kez476, https://www.ncbi.nlm.nih.gov/pubmed/31624843 [DOI] [PubMed] [Google Scholar]
- 45.Keenan RT. Limitations of the current standards of care for treating gout and crystal deposition in the primary care setting: a review. Clin Ther 2017; 39(2): 430–441, DOI: 10.1016/j.clinthera.2016.12.011, https://www.ncbi.nlm.nih.gov/pubmed/28089200 [DOI] [PubMed] [Google Scholar]
- 46.Thottam GE, Krasnokutsky S, Pillinger MH. Gout and Metabolic Syndrome: a Tangled Web. Curr Rheumatol Rep 2017; 19(10): 60, DOI: 10.1007/s11926-017-0688-y, https://www.ncbi.nlm.nih.gov/pubmed/28844079 [DOI] [PubMed] [Google Scholar]
- 47.van Durme CM, Wechalekar MD, Buchbinder R, Schlesinger N, van der Heijde D, Landewé RB. Non-steroidal anti-inflammatory drugs for acute gout. Cochrane Database Syst Rev 2014; 9: CD010120, DOI: 10.1002/14651858.CD010120.pub2, https://www.ncbi.nlm.nih.gov/pubmed/25225849 [DOI] [PubMed] [Google Scholar]
- 48.van Echteld I, Wechalekar MD, Schlesinger N, Buchbinder R, Aletaha D. Colchicine for acute gout. Cochrane Database Syst Rev 2014; 8: CD006190, DOI: 10.1002/14651858.CD006190.pub2, https://www.ncbi.nlm.nih.gov/pubmed/25123076 [DOI] [PubMed] [Google Scholar]
- 49.Zheng Y, Xu H, Zhan L, Zhou X, Chen X, Gao Z. Activation of peripheral KCNQ channels relieves gout pain. Pain 2015; 156(6): 1025–1035, DOI: 10.1097/j.pain.0000000000000122. https://www.ncbi.nlm.nih.gov/pubmed/25735002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Linley JE, Rose K, Ooi L, Gamper N. Understanding inflammatory pain: ion channels contributing to acute and chronic nociception. Pflugers Arch 2010; 459(5): 657–669, DOI: 10.1007/s00424-010-0784-6, https://www.ncbi.nlm.nih.gov/pubmed/20162302 [DOI] [PubMed] [Google Scholar]
- 51.Sluka KA. Activation of the cAMP transduction cascade contributes to the mechanical hyperalgesia and allodynia induced by intradermal injection of capsaicin. Br J Pharmacol 1997; 122(6): 1165–1173, DOI: 10.1038/sj.bjp.0701486, https://www.ncbi.nlm.nih.gov/pubmed/9401782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet 2016; 17(1): 19–32, DOI: 10.1038/nrg.2015.3, https://www.ncbi.nlm.nih.gov/pubmed/26593421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material for Systematic analysis of inflammation and pain pathways in a mouse model of gout by Yuanlan Fan, Jiajun Yang, Song Xie, Jiangshuang He, Shenghui Huang, Jiawang Chen, Shangying Jiang, Lei Yu, Yujuan Zhou, Xuxia Cao, Xiang Ji and , Yi Zhang in Molecular Pain
Supplemental Material for Systematic analysis of inflammation and pain pathways in a mouse model of gout by Yuanlan Fan, Jiajun Yang, Song Xie, Jiangshuang He, Shenghui Huang, Jiawang Chen, Shangying Jiang, Lei Yu, Yujuan Zhou, Xuxia Cao, Xiang Ji and , Yi Zhang in Molecular Pain
Supplemental Material for Systematic analysis of inflammation and pain pathways in a mouse model of gout by Yuanlan Fan, Jiajun Yang, Song Xie, Jiangshuang He, Shenghui Huang, Jiawang Chen, Shangying Jiang, Lei Yu, Yujuan Zhou, Xuxia Cao, Xiang Ji and , Yi Zhang in Molecular Pain
Supplemental Material for Systematic analysis of inflammation and pain pathways in a mouse model of gout by Yuanlan Fan, Jiajun Yang, Song Xie, Jiangshuang He, Shenghui Huang, Jiawang Chen, Shangying Jiang, Lei Yu, Yujuan Zhou, Xuxia Cao, Xiang Ji and , Yi Zhang in Molecular Pain
Supplemental Material for Systematic analysis of inflammation and pain pathways in a mouse model of gout by Yuanlan Fan, Jiajun Yang, Song Xie, Jiangshuang He, Shenghui Huang, Jiawang Chen, Shangying Jiang, Lei Yu, Yujuan Zhou, Xuxia Cao, Xiang Ji and , Yi Zhang in Molecular Pain
Supplemental Material for Systematic analysis of inflammation and pain pathways in a mouse model of gout by Yuanlan Fan, Jiajun Yang, Song Xie, Jiangshuang He, Shenghui Huang, Jiawang Chen, Shangying Jiang, Lei Yu, Yujuan Zhou, Xuxia Cao, Xiang Ji and , Yi Zhang in Molecular Pain
Supplemental Material for Systematic analysis of inflammation and pain pathways in a mouse model of gout by Yuanlan Fan, Jiajun Yang, Song Xie, Jiangshuang He, Shenghui Huang, Jiawang Chen, Shangying Jiang, Lei Yu, Yujuan Zhou, Xuxia Cao, Xiang Ji and , Yi Zhang in Molecular Pain
Data Availability Statement
All data used to support the findings of this study are available within the paper and supplementary materials. Our own 18 RNAseq datasets have been deposited in the Gene Expression Omnibus (GEO) repository under access number GSE190138. RNAseq datasets for 18 normal and 20 OA human knee cartilage tissues were downloaded from the GSE114007 dataset. 20 RNAseq datasets of 5 OA patients with high knee pain and 5 OA patients with low knee pain were downloaded from the GSE99662 dataset. 21 RNAseq datasets of CFA and SNI pain mouse models were obtained from the GSE111216 dataset. 22 Human RA single-cell RNAseq data were downloaded from ImmPort (https://www.immport.org/shared/study/SDY998).