Abstract
The contribution of penicillin-binding protein 5 (PBP 5) to intrinsic and acquired β-lactam resistance was investigated by constructing isogenic strains of Enterococcus faecium producing different PBP 5. The pbp5 genes from three E. faecium clinical isolates (BM4107, D344, and H80721) were cloned into the shuttle vector pAT392 and introduced into E. faecium D344S, a spontaneous derivative of E. faecium D344 highly susceptible to ampicillin due to deletion of pbp5 (MIC, 0.03 μg/ml). Immunodetection of PBP5 indicated that cloning of the pbp5 genes into pAT392 resulted in moderate overproduction of PBP 5 in comparison to wild-type strains. This difference may be attributed to a difference in gene copy number. Expression of the pbp5 genes from BM4107 (MIC, 2 μg/ml), D344 (MIC, 24 μg/ml), and H80721 (MIC, 512 μg/ml) in D344S conferred relatively low levels of resistance to ampicillin (MICs, 6, 12, and 20 μg/ml, respectively). A methionine-to-alanine substitution was introduced at position 485 of the BM4107 PBP 5 by site-directed mutagenesis. In contrast to previous hypotheses based on comparison of nonisogenic strains, this substitution resulted in only a 2.5-fold increase in the ampicillin MIC. The reversed-phase high-performance liquid chromatography muropeptide profiles of D344 and D344S were similar, indicating that deletion of pbp5 was not associated with a detectable defect in cell wall synthesis. These results indicate that pbp5 is a nonessential gene responsible for intrinsic resistance to moderate levels of ampicillin and by itself cannot confer high-level resistance.
Enterococcus faecium is intrinsically resistant to moderate levels of ampicillin by production of the low-affinity penicillin-binding protein 5 (PBP 5). Bacterial growth occurs at β-lactam concentrations sufficient to inactivate all the other PBPs, suggesting that PBP5 is the only transpeptidase required for peptidoglycan synthesis under such conditions (4, 20). Acquired resistance to higher levels of ampicillin in clinical isolates of E. faecium has been associated with increased production of PBP 5 or decreased affinity for the β-lactam antibiotics (7, 8, 10, 13, 18, 20, 21). The latter mechanism was inferred from comparison of the pbp5 genes from clinical isolates in which amino acid substitutions at specific positions near or in the conserved motifs of the PBP module were associated with decreased interaction with β-lactams and expression of resistance (8, 13, 18, 21). However, the role of the PBP 5 in the level of resistance was not rigorously established since isogenic strains were not constructed. In the present study, pbp5 genes from E. faecium clinical isolates expressing various levels of ampicillin resistance were cloned into a shuttle vector and introduced into E. faecium D344S, a spontaneous mutant in which the chromosomal pbp5 locus is deleted. Expression of the different pbp5 genes in this host resulted in similar low levels of ampicillin resistance, indicating that alterations of PBP 5 alone do not account for acquired high-level β-lactam resistance in E. faecium.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are presented in Table 1. Strains were grown in brain heart infusion (BHI) broth or agar (Difco Laboratories, Detroit, Mich.) at 37°C. The MICs were determined three times on BHI agar using twofold dilutions of ampicillin and three additional intermediary concentrations within each twofold dilution (e.g., 16, 20, 24, 28, and 32 μg/ml) (20). An inoculum of approximately 104 CFU per spot was applied with a Steers replicator. Plates were incubated at 37°C for 18 h. The following antimicrobial agents were kindly provided: amoxicillin (Smith Kline Beecham Laboratories, Paris, France), and ceftriaxone (Roche S.A., Paris, France).
TABLE 1.
Properties of bacterial strains and plasmids used in this studya
| Strain or plasmid | Relevant property(ies) | Source or reference |
|---|---|---|
| Strains | ||
| E. faecium D344 | Clinical isolate | 20 |
| E. faecium BM4107 | Clinical isolate | 11 |
| E. faeciumH80721 | Clinical isolate | 18 |
| E. faecium D344S | Spontaneous mutant of D344 (Δpbp5) | 20 |
| Plasmids | ||
| pUC19 | Plasmid vector (Apr) | 15 |
| pAT392 | Shuttle expression vector (Gentr SpcroriRpUCoriRpAMβ1oriTRK2P2) | 1 |
| pSF1 | pbp5BM4107 cloned into pAT392 | This work |
| pSF2 | pbp5D344 cloned into pAT392 | This work |
| pSF3 | pbp5H80721 cloned into pAT392 | This work |
| pSF4 | pbp5BM4107M485A cloned into pAT392 | This work |
Abbreviations: Ap, ampicillin; Gent, gentamicin; oriRpUC, replication origin of pUC18; oriRpAMβ1, replication origin of pAMβ1 (active in E. faecium); oriTRK2, transfer origin of RK2; Spc, spectinomycin.
Molecular biology techniques.
Plasmid DNA isolation, digestion with restriction endonucleases (Roche Molecular Biochemicals, Paris, France), ligation of fragments with T4 DNA ligase (Roche Molecular Biochemicals), and transformation of Escherichia coli DH5a with recombinant plasmids were performed by standard methods (3). Genomic DNA of E. faecium strains was extracted and amplified (2) with oligodeoxyribonucleotide primers (Oligo Express, Paris, France) by using a PROGENE thermal cycler (Techne, Cambridge, United Kingdom).
Plasmid construction.
Plasmids pSF1, pSF2, and pSF3 were constructed by cloning the PCR-amplified pbp5 genes of E. faecium BM4107, D344, and H80721, respectively, into the shuttle vector pAT392. For these constructions, DNA fragments containing the pbp5 genes were amplified with the Pwo DNA polymerase (Roche Molecular Biochemicals) and primers ONJP16 (5′-CGGAATTCGTATTATGCAAGTATCA-3′) and ONJP23 (5′-CGCGGATCCTTATTATTGATAATTTTGGTT-3′). The amplified fragments were digested with EcoRI and BamHI (sites underlined), ligated with pUC19 DNA digested with the same enzymes, and introduced into E. coli DH5a. The inserts of the recombinant plasmids were sequenced on both DNA strands (Genome Express, Paris, France). The DNA fragments carrying the pbp5 genes were subcloned into pAT392 using EcoRI and BamHI.
Site-directed mutagenesis.
An ATG (Met)-to-GCG (Ala) mutation was introduced at codon 485 of the pbp5 gene of BM4107 as previously described (19). In brief, the 3′ terminus of the pbp5 gene of pSF1 was amplified using primers ONPJ42 (5′-CCGATAATATATATGCGGCACAAGAAACGTT-3′) and ONJP23 (above). The PCR fragment (595 bp) was used as a megaprimer with primer ONJP16 (above) to amplify the entire gene which was cloned into pUC19 and subcloned into pAT392 using EcoRI and BamHI, generating pSF4. The entire insert was sequenced to verify the presence of the expected mutation.
Strain construction.
Plasmids were introduced into E. faecium D344S by electrotransformation (1). Plasmid DNA from clones selected on gentamicin (100 μg/ml) were prepared and digested with EcoRI plus BamHI to screen for DNA rearrangements.
Immunodetection of PBP 5.
Strains were grown in BHI broth to an optical density of 0.5 at 600 nm. Membranes were prepared, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western blotting was performed with polyclonal antibodies raised to PBP 5, as previously described (18).
Analysis of peptidoglycan structure.
Muropeptides were prepared from cells grown at mid-exponential phase in BHI broth, separated by reversed-phase high-performance liquid chromatography, and analyzed by mass spectrometry (14). The respective abundance of the muropeptides were expressed as a percentage of the area under all the peaks using baselines connecting the lowest absorbance values at 210 nm.
RESULTS AND DISCUSSION
Complementation of the pbp5 deletion in E. faecium D344S.
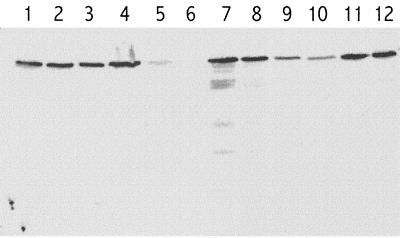
E. faecium D344 is a clinical isolate of E. faecium resistant to moderate levels of ampicillin (MIC, 24 μg/ml). E. faecium D344S is a spontaneous derivative of D344 lacking the entire pbp5 gene as shown by Southern blot hybridization and PCR analysis with various primers (data not shown). Deletion of pbp5 was associated with an 800-fold decrease in the MIC of ampicillin (from 24 to 0.03 μg/ml) (Table 2). A similar low MIC was previously reported for a mutant of Enterococcus hirae harboring a premature stop codon in pbp5 (12). For complementation analysis, the pbp5 gene of D344 (pbp5D344) was amplified by PCR and cloned into the shuttle vector pAT392, generating plasmid pSF2. The amplified fragment contained the promoter recently identified upstream from the pbp5 open reading frame by Northern blot and primer extension analyses (17). Transcription is not expected to originate from the vector since the P2 promoter of pAT392 was deleted upon cloning (see Materials and Methods) and transcription is barely detectable in its absence (1, 2). Introduction of pSF2(pbp5D344) into D344S resulted in a 400-fold increase in the level of ampicillin resistance (Table 2), confirming that pbp5 is the major determinant responsible for intrinsic resistance to ampicillin in D344. Western blot analysis showed that D344S/pSF2(pbp5D344) produced PBP 5 at a higher (approximately fourfold) level than the parental strain D344 (Fig. 1). This difference may be attributed to a gene dosage effect since the pbp5 gene was present at one copy per chromosome in D344 and on a low-copy-number vector containing the pAMβ1 origin of replication in D344S/pSF2(pbp5D344) (16). In spite of the high-level expression of pbp5 in D344S/pSF2(pbp5D344), the MIC of ampicillin was twofold lower in comparison to that for the parental strain D344.
TABLE 2.
MICs of ampicillin for various E. faecium strains
| E. faecium strain | MIC [μg/ml] of ampicillin (range for derivatives)a |
|---|---|
| BM4107 | 2 (ND) |
| D344 | 24 (ND) |
| H80721 | 512 (ND) |
| D344S | 0.03 (ND) |
| D344S/pAT392 | 0.03 (ND) |
| D344S/pSF1(pbp5BM4107) | 6 (8–12) |
| D344S/pSF2(pbp5D344) | 12 (20–56) |
| D344S/pSF3(pbp5H80721) | 20 (40–128) |
| D344S/pSF4(pbp5BM4107M485A) | 16 (40–56) |
MIC ranges were determined for derivatives only. Derivatives were selected on BHI agar containing ampicillin at a concentration two times the MIC. ND, not done.
FIG. 1.
Western blot detection of PBP 5 in membrane fractions from various E. faecium strains. Lanes: 1, derivative of D344S/pSF4(pbp5BM4107M485A) selected on ampicillin (MIC, 56 μg/ml); 2, D344S/pSF4(pbp5BM4107M485A); 3, derivative of D344S/pSF1(pb5pBM4107) (MIC, 12 μg/ml); 4, D344S/pSF1 (pb5pBM4107); 5, BM4107; 6, D344S; 7, derivative of D344S/pSF3(pbp5H80721) (MIC, 128 μg/ml); 8, D344S/pSF3(pbp5H80721); 9, H80721; 10, D344; 11, derivative of D344S/pSF2(pbp5D344) (MIC, 56 μg/ml); 12, D344S/pSF2(pbp5D344).
Comparison of wild-type pbp5 genes of BM4107, D344, and H80721.
The complementation test was used to compare the resistance phenotypes mediated by the PBP 5 from three clinical isolates with various levels of resistance to ampicillin (Table 2). The pbp5 genes of BM4107, D344, and H80721 were amplified, cloned into E. coli, and sequenced. The complete deduced amino acid sequence of the three PBP 5 differed in a total of 28 positions (Table 3). Twelve of the variable positions were located in the PBP module of the proteins, including position 485, which was reported to play a role in resistance (18, 21). As previously discussed, the presence of a Met, Ala, or Thr residue at this position was associated with low-level (BM4107), moderate-level (D344), or high-level (H80721) ampicillin resistance, respectively (18). The pbp5 genes were subcloned into pAT392, and the resulting plasmids were introduced into D344S, leading to moderate overproduction of the different PBP 5 (Fig. 1) and to 200- to 700-fold increases in the MIC of ampicillin (Table 2). The MIC was lowest (6 μg/ml) for D344S/pSF1, expressing the pbp5 gene of the low-level-resistant strain BM4107 (MIC, 2 μg/ml); intermediate (12 μg/ml) for D344S/pSF2, expressing the pbp5 gene of the moderately resistant strain D344 (MIC, 24 μg/ml); and highest (20 μg/ml) for D344S/pSF3, expressing the pbp5 gene of the highly resistant strain H80721 (MIC, 512 μg/ml) (Table 2). Qualitatively, these results indicate that structural differences in the PBP 5 of BM4107, D344, and H80721 contribute to the difference in the level of ampicillin resistance observed for these clinical isolates. However, structural differences in the PBP 5 appear to play only a marginal role, since the MIC ranged from 2 to 512 μg/ml for the clinical isolates and from 6 to 20 μg/ml for derivatives of D344S expressing the corresponding pbp5 genes. The pbp5 gene of HM80721 did not mediate high-level ampicillin resistance in D344S in spite of moderate overproduction of the protein (Fig. 1). Thus, neither the level of expression of pbp5HM80721 nor the structure of the gene product can fully account for high-level expression of ampicillin resistance in HM80721. It may be argued that some accessory factor required for PBP 5HM80721-mediated high-level ampicillin resistance was deleted with pbp5 in D344S. This does not appear to be the case, since introduction of pSF3(pbp5HM80721) into BM4107 also produced a moderate (fourfold) increase in the MIC of ampicillin (data not shown).
TABLE 3.
| Positionb | Amino acid
|
||
|---|---|---|---|
| BM4107 | D344 | H80721 | |
| Membrane anchor | |||
| 24 | V | A | A |
| 25 | T | A | A |
| 27 | S | G | G |
| 34 | R | Q | Q |
| 39 | S | T | T |
| N-terminal extension | |||
| 66 | G | E | E |
| 68 | A | T | T |
| 85 | E | D | D |
| 94 | A | T | A |
| 100 | E | Q | Q |
| 144 | K | Q | Q |
| N-terminal domain | |||
| 172 | T | A | A |
| 177 | L | I | I |
| 204 | D | G | G |
| 216 | A | S | S |
| 324 | T | A | A |
| C-terminal PBP domain | |||
| 404 | N | N | D |
| 466′ | None | None | S |
| 485 | Mc | T | A |
| 496 | N | K | K |
| 499 | A | T | T |
| 525 | E | D | D |
| 601 | N | Y | N |
| 626 | K | E | K |
| 629 | E | V | V |
| 654 | D | N | N |
| 663 | T | A | T |
| 667 | P | S | S |
Deduced amino acid sequence of pbp5 genes from E. faecium H80721 (GenBank accession no. X84862), D344 (GenBank accession no. AF362954), and BM4107 (GenBank accession no. AF364092).
Numbering of PBP 5 from E. faecium H80721. Positions were assigned to the different domains of the PBP as previously described (21).
A Met-to-Ala substitution was introduced by site-directed mutagenesis at this position (see pSF4 in Table 1).
Expression of the pbp5 gene of BM4107 conferred a higher level of resistance to ampicillin after cloning in D344s (MIC, 6 μg/ml) than in the parental strain BM4107 (2 μg/ml). Increased production of PBP5 (Fig. 1) may be responsible for this difference in accordance with previous studies which showed that increased amounts of PBP 5 was associated with increased β-lactam MICs in both E. hirae and E. faecium (6, 10, 12, 20).
Site-directed mutagenesis of the pbp5 gene of BM4107.
The presence of an alanine instead of a methionine at position 485 near the conserved SDN motif (positions 480 to 482) was proposed to play a critical role in PBP 5-mediated high-level ampicillin resistance (18, 21). The corresponding mutation was introduced into the pbp5 gene of BM4107 (Table 3). The MICs of ampicillin for derivatives of D344S harboring pSF1(pbp5BM4107) and pSF4(pbp5BM4107M485A) were 6 and 16 μg/ml, respectively (Table 2). Thus, the amino acid substitution led to a significant but limited increase in the level of resistance. These results indicate that the presence of an alanine residue at position 485 cannot account alone for high-level ampicillin resistance, at least in the context of the PBP5BM4107 sequence.
Selection of variants with increased resistance to ampicillin.
Derivatives of D344S harboring pSF1(pbp5BM4107), pSF2(pbp5D344), pSF3(pbp5H80721), and pSF4 (pbp5BM4107M485A) were selected on BHI agar containing ampicillin at concentrations twofold higher than the MICs (Table 2). For all four strains, variants were obtained at a high frequency (10−4 to 10−5), and the MICs of ampicillin were increased up to sixfold (Table 2). Western blot analysis performed for one highly resistant representative of each type of variant did not reveal any apparent increase in the level of PBP 5 production (Fig. 1). The highest MIC observed for derivatives of D344S/pSF3(pbp5H80721) (128 μg/ml) was considerably less than that for clinical isolate HM80721 (512 μg/ml). Thus, production of PBP5HM80721 in D344S did not restore the wild-type level of resistance, even after selection of variants with increased ampicillin resistance.
Structure of the peptidoglycan.
The overall structures of peptidoglycan from D344 and D344S were very similar, indicating that PBP 5 has no specific transpeptidase activity different from the other set of PBPs (Table 4). The only difference was a slight decrease of the proportion of monomers in D344S. The peptidoglycan of D344 was also analyzed after growth in the presence of ceftriaxone at 64 μg/ml. This concentration is 10 times higher than that required to saturate at 50% PBP 1, 2, 3, and 4 but significantly less than that required to inhibit growth or to saturate PBP 5 at 50% (>256 μg/ml) (reference 9 and data not shown). Comparison of the peptidoglycan isolated from D344 grown in the presence or absence of ceftriaxone did not reveal any significant difference, with the possible exception of a fourfold increase of the quantity of the monomer pentapeptide (0.6 versus 2.4%). Thus, PBP 5 was able to compensate for inhibition of PBP 1, 2, 3, and 4 by ceftriaxone. In Staphylococcus aureus, saturation of all PBPs except the low-affinity PBP 2A is associated with a virtual disappearance of oligomers (5), suggesting that PBP 2A, in contrast to PBP 5, is unable to assume all the transpeptidase functions involved in peptidoglycan synthesis.
TABLE 4.
Structures of muropeptides from E. faecium
| Peak no.b | Peptidoglycan structurec | Amt (% total)a
|
||
|---|---|---|---|---|
| D344 | D344 + CR064d | D344S | ||
| Monomers | 41.7 | 42.0 | 33.7 | |
| 1 | ds-di | 0.9 | 0.7 | 0.5 |
| 2 | ds-tri | 6.6 | 5.8 | 4.7 |
| 3 | ds-tetra | 2.5 | 3.2 | 3.4 |
| 4 | ds-Asp-tri | 4.1 | 2.8 | 1.5 |
| 5 | ds-Asn-tri | 16.3 | 16.2 | 13.0 |
| 6 | ds-Asp-tetra | 2.7 | 3.4 | 1.5 |
| 7 | ds-Asn-tetra | 3.2 | 5.5 | 4.4 |
| 8 | ds-Asn-penta | 0.6 | 2.4 | 1.8 |
| 9 | ds(Ac)-Asp-tri | 1.2 | 0.5 | 0.4 |
| 10 | ds(Ac)-Asn-tri | 3.6 | 1.5 | 2.5 |
| Dimers and trimers | 58.3 | 58.0 | 66.3 | |
| 11 | Bis-ds-tetra-Asn-tri | 4.5 | 6.3 | 3.2 |
| 12 | Bis-ds-Asp-tetra-Asn-trie | 4.3 | 3.6 | 5.2 |
| 13 | Bis-ds-Asn-tetra-Asn-tri | 17.1 | 18.2 | 19.6 |
| 14 | Bis-ds-Asp-tetra-Asn-tetrae | 1.4 | 1.6 | 1.3 |
| 15 | Bis-ds-(Ac)-tetra-Asn-tri | 1.1 | 1.7 | 1.1 |
| 16 | Bis-ds-Asn-tetra-Asn-tetra | 4.2 | 7.0 | 8.6 |
| 17 | Bis-ds-(Ac)-Asp-tetra-Asn-trie | 1.8 | 2.1 | 1.4 |
| 18 | Bis-ds-Asp-tetra-Asn-tetrae | 1.7 | 1.9 | 3.5 |
| 19 | Bis-ds-(Ac)-Asn-tetra-Asn-tri | 6.4 | 4.0 | 6.7 |
| 20 | Ter-ds-Asn-tetra-Asn-tetra-Asn-tri | 6.2 | 5.6 | 6.5 |
| 21 | Bis-ds-(AcX2)-Asp-tetra-Asn-trie | 2.7 | 2.5 | 4.0 |
| 22 | Bis-ds-(AcX2)-Asn-tetra-Asn-tri | 6.9 | 3.5 | 5.2 |
The values are presented as a percentage of the sum of all peaks presented in the table and are the means of two experiments.
Muropeptide peak designations as in reference 14.
Abbreviations: ds, disaccharide (N-acetylglucosamine-β-1.4-N-acetylmuramic acid); Bis, dimeric form; Ter, trimeric form; di, dipeptide (l-alanyl-d-isoglutamine); tri, tripeptide (l-alanyl-d-isoglutamyl-l-lysine); tetra, tetrapeptide (l-alanyl-d-isoglutamyl-l-lysyl-d-alanine); penta, pentapeptide (l-alanyl-d-isoglutamyl-l-lysyl-d-alanyl-d-alanine); Asn, d-asparagine; Asp, d-aspartate; Ac, O-acetylation located on N-acetylmuramic acid; ACx2, O-acetylation on both N-acetylmuramic acids of the dimer.
D344 was grown in presence of 64 μg of ceftriaxone per ml (CRO64).
Assignment of Asp and Asn residues to either stem peptide is arbitrary.
Conclusions.
Analysis of peptidoglycan indicated that the d,d-transpeptidase activity of the PBPs is largely redundant in E. faecium, since similar structures were obtained under conditions under which the full complement of the PBPs was active, PBP 5 was absent, or all PBPs except PBP 5 were inhibited by ceftriaxone. In contrast, PBP 5 strikingly differs from other PBPs by its low affinity for β-lactams (7, 18, 21). Accordingly, deletion of pbp5 was associated with an 800-fold reduction in the MIC of ampicillin. The deletion was complemented by a copy of pbp5 cloned on a plasmid, confirming that PBP 5 was responsible for intrinsic resistance. Use of this complementation test also confirmed that modification of the level of expression of pbp5 or alterations of the amino acid sequence of the protein near conserved motifs may lead to increased resistance to ampicillin. However, neither of these mechanisms could account alone or in association for the acquired high-level ampicillin resistance found in the clinical isolates (18). Thus, it is likely that other factors are necessary to obtain full expression of resistance.
ACKNOWLEDGMENT
This work was supported by a grant from INSERM (E0004).
REFERENCES
- 1.Arthur M, Depardieu F, Snaith H A, Reynolds P E, Courvalin P. Contribution of VanY d,d-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob Agents Chemother. 1994;38:1899–1903. doi: 10.1128/aac.38.9.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Molinas C, Courvalin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992;174:2582–2591. doi: 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, n.y.: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 4.Canepari P, Lleo M M, Cornaglia G, Fontana R, Satta G. In Streptococcus faecium penicillin-binding protein 5 alone is sufficient for cell growth at sub-maximal but not at maximal rate. J Gen Microbiol. 1986;132:625–631. doi: 10.1099/00221287-132-3-625. [DOI] [PubMed] [Google Scholar]
- 5.de Jonge B L M, Tomasz A. Abnormal peptidoglycan produced in a methicillin-resistant strain of Staphylococcus aureus grown in the presence of methicillin: functional role for penicillin-binding protein 2A in cell wall synthesis. Antimicrob Agents Chemother. 1993;37:342–346. doi: 10.1128/aac.37.2.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fontana R, Aldegheri M, Ligozzi M, Lopez H, Sucari A, Satta G. Overproduction of a low-affinity penicillin-binding protein and high-level ampicillin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1994;38:1980–1983. doi: 10.1128/aac.38.9.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontana R, Cerini R, Longoni P, Grossato A, Canepari P. Identification of a streptococcal penicillin-binding protein that reacts very slowly with penicillin. J Bacteriol. 1983;155:1343–1350. doi: 10.1128/jb.155.3.1343-1350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontana R, Ligozzi M, Pittaluga F, Satta G. Intrinsic penicillin resistance in enterococci. Microb Drug Resist. 1996;2:209–213. doi: 10.1089/mdr.1996.2.209. [DOI] [PubMed] [Google Scholar]
- 9.Gutmann L, Al-Obeid S, Billot-Klein D, Guerrier M L, Collatz E. Synergy and resistance to synergy between β-lactam antibiotics and glycopeptides against glycopeptide-resistant strains of Enterococcus faecium. Antimicrob Agents Chemother. 1994;38:824–829. doi: 10.1128/aac.38.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klare I, Rodloff A C, Wagner J, Witte W, Hakenbeck R. Overproduction of a penicillin-binding protein is not the only mechanism of penicillin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1992;36:783–787. doi: 10.1128/aac.36.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leclercq R, Derlot E, Weber M, Duval J, Courvalin P. Transferable vancomycin and teicoplanin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1989;33:10–15. doi: 10.1128/aac.33.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ligozzi M, Pittaluga F, Fontana R. Identification of a genetic element (psr) which negatively controls expression of Enterococcus hirae penicillin-binding protein 5. J Bacteriol. 1993;175:2046–2051. doi: 10.1128/jb.175.7.2046-2051.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ligozzi M, Pittaluga F, Fontana R. Modification of penicillin-binding protein 5 associated with high-level ampicillin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1996;40:354–357. doi: 10.1128/aac.40.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mainardi J L, Legrand R, Arthur M, Schoot B, van Heijenoort J, Gutmann L. Novel mechanism of β-lactam resistance due to by-pass of dd-transpeptidation in Enterococcus faecium. J Biol Chem. 2000;275:16490–16496. doi: 10.1074/jbc.M909877199. [DOI] [PubMed] [Google Scholar]
- 15.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 16.Poyart C, Trieu-Cuot P. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol Lett. 1997;156:193–198. doi: 10.1111/j.1574-6968.1997.tb12726.x. [DOI] [PubMed] [Google Scholar]
- 17.Rice L B, Carias L L, Hutton-Thomas R, Sifaoui F, Gutmann L, Rudin S D. Penicillin-binding protein 5 and expression of ampicillin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 2001;45:1480–1486. doi: 10.1128/AAC.45.5.1480-1486.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rybkine T, Mainardi J L, Sougakoff W, Collatz E, Gutmann L. Penicillin-binding protein 5 sequence alterations in clinical isolates of Enterococcus faecium with different levels of beta-lactam resistance. J Infect Dis. 1998;178:159–163. doi: 10.1086/515605. [DOI] [PubMed] [Google Scholar]
- 19.Smith A M, Klugman K P. “Megaprimer” method of PCR-based mutagenesis: the concentration of megaprimer is a critical factor. BioTechniques. 1997;22:438–442. doi: 10.2144/97223bm13. [DOI] [PubMed] [Google Scholar]
- 20.Williamson R, le Bouguenec C, Gutmann L, Horaud T. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to benzylpenicillin. J Gen Microbiol. 1985;131:1933–1940. doi: 10.1099/00221287-131-8-1933. [DOI] [PubMed] [Google Scholar]
- 21.Zorzi W, Zhou X Y, Dardenne O, Lamotte J, Raze D, Pierre J, Gutmann L, Coyette J. Structure of the low-affinity penicillin-binding protein 5 PBP5fm in wild-type and highly penicillin-resistant strains of Enterococcus faecium. J Bacteriol. 1996;178:4948–4957. doi: 10.1128/jb.178.16.4948-4957.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]