Correction to: Virology Journal (2022) 19:56 10.1186/s12985-022-01772-8
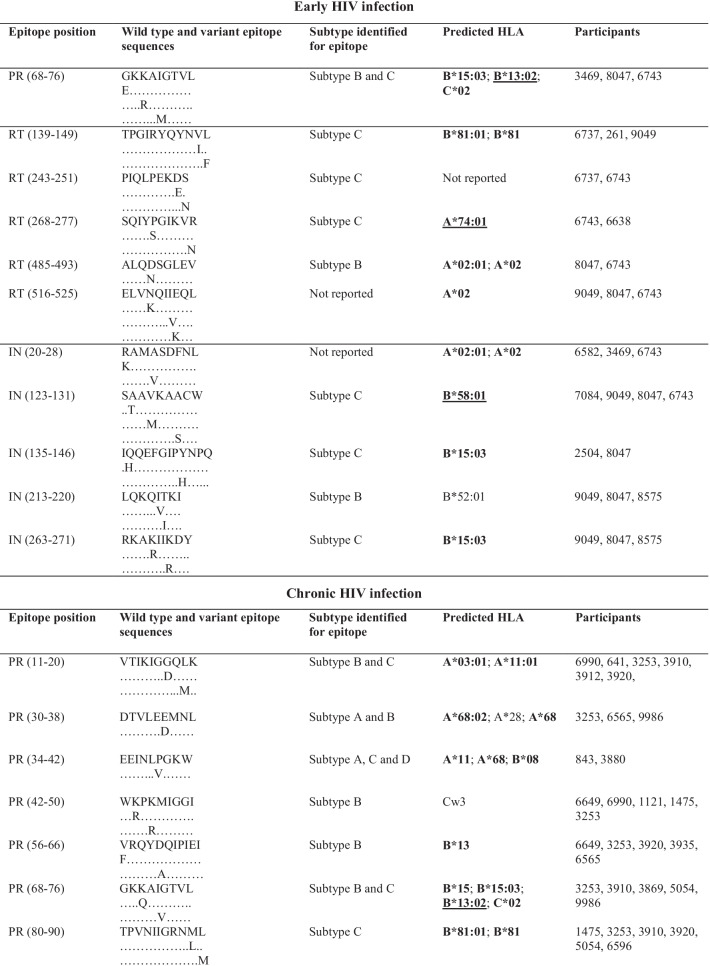
Following publication of the original article [1], the authors informed us that many epitopes from Table 3 have gross alignment issues, which were probably caused by formatting of this table before publication. The correct table is given below.
Table 3.
Frequently targeted Pol CTL epitopes and their predicted HLA alleles
HLA alleles in boldface are those that have been identified (reported) in South Africa or southern Africa [11,28–30]. Alleles underlined are those that were reported to be protective [11,28–29,49,50]. PR = protease; RT = reverse transcriptase; IN = integrase; HLA = human leukocyte antigen
The original article has been corrected.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reference
- 1.Nkone P, Loubser S, Quinn TC, et al. Deep sequencing of the HIV-1 polymerase gene for characterisation of cytotoxic T-lymphocyte epitopes during early and chronic disease stages. Virol J. 2022;19:56. doi: 10.1186/s12985-022-01772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]