Abstract
Pseudomonas aeruginosa GW-1 was isolated in 2000 in South Africa from blood cultures of a 38-year-old female who developed nosocomial pneumonia. This isolate harbored a self-transferable ca. 100-kb plasmid that conferred an expanded-spectrum cephalosporin resistance profile associated with an intermediate susceptibility to imipenem. A β-lactamase gene, blaGES-2, was cloned from whole-cell DNA of P. aeruginosa GW-1 and expressed in Escherichia coli. GES-2, with a pI value of 5.8, hydrolyzed expanded-spectrum cephalosporins, and its substrate profile was extended to include imipenem compared to that of GES-1, identified previously in Klebsiella pneumoniae. GES-2 activity was less inhibited by clavulanic acid, tazobactam and imipenem than GES-1. The GES-2 amino acid sequence differs from that of GES-1 by a glycine-to-asparagine substitution in position 170 located in the omega loop of Ambler class A enzymes. This amino acid change may explain the extension of the substrate profile of the plasmid-encoded β-lactamase GES-2.
Clavulanic acid-inhibited extended-spectrum β-lactamases (ESBLs) conferring resistance to expanded-spectrum cephalosporins have been reported, first in Enterobacteriaceae and then in Pseudomonas aeruginosa (11, 23). Rare reports of TEM- and SHV-type ESBLs in P. aeruginosa are known (SHV-2a, TEM-4, TEM-24, and TEM-42 [15, 23, 31]), while they have been extensively described in Enterobacteriaceae (11). Three non-TEM-, non-SHV-type ESBLs have been reported in P. aeruginosa, i.e., PER-1, VEB-1, and OXA-18 β-lactamases (20, 25, 27, 38). The PER-1 β-lactamase gene is widespread in Turkey, although not reported as plasmid mediated in P. aeruginosa (39). VEB-1 β-lactamase, originally described in Escherichia coli and Klebsiella pneumoniae isolates in Vietnam, has been found in P. aeruginosa and enterobacterial isolates in Thailand (8, 29, 38).
We have recently identified another Ambler class A β-lactamase, GES-1, in a K. pneumoniae isolate in French Guiana (28). It was found to be remotely related to other ESBLs. This ESBL differs by two amino acid substitutions from IBC-1 β-lactamase recently found in an Enterobacter cloacae isolate in Greece (7). blaVEB-1, blaGES-1, and blaIBC-1 are plasmid located and are part of gene cassettes integrated into class 1 integrons (7, 28, 29).
Mobile cassettes contain genes most often mediating antibiotic resistance and a cassette recombination site, designated the 59-base element (59-be) (9, 10). The 59-be sites vary in length (57 to 141 bp) and structure, but they are all bounded by a core site (GTTRRRY) at the recombinant crossover point and an inverse core site (RYYYAAC) at the 3′ end of the inserted gene (4, 9). Integrons are genetic elements capable of integrating individual gene cassettes by a site-specific recombination mechanism that involves a DNA integrase, IntI; an integron-specific recombination site, attI; and 59-be (4, 9, 10). The 5′ conserved segment (5′-CS) of the integrons contains the integrase gene (intI) and the recombination site attI1. The 3′-CS of class 1 integrons carries the antisepsis resistance qacEΔ1 gene; the sul1 gene, which confers resistance to sulfonamides; and an open reading frame (ORF) of unknown function, ORF5 (26).
While analyzing carbapenem-resistant P. aeruginosa isolates from South Africa, we retained a P. aeruginosa isolate that was resistant to ceftazidime with an unusual substrate profile that included imipenem, according to preliminary analysis. Thus, the β-lactamase content of this strain was further characterized. A plasmid-mediated Ambler class A ESBL was identified with a substrate profile extended to imipenem but with hydrolysis rates lower than those of the chromosome-encoded carbapenem-hydrolyzing class A β-lactamases NmcA, SME-1/2, and IMI-1 identified in rare isolates of E. cloacae and Serratia marcescens (19, 21, 24, 33, 35) and of the recently reported plasmid-encoded β-lactamase KPC-1 from K. pneumoniae (40).
MATERIALS AND METHODS
Bacterial strains.
P. aeruginosa clinical isolate GW-1 was identified with the API-20 NE system (bioMérieux, Marcy l'Etoile, France). E. coli DH10B was the host for cloning experiments, and in vitro-obtained rifampin-resistant P. aeruginosa PU21 was used as a recipient strain for conjugative transfer (30).
Susceptibility testing.
Antibiotic-containing disks were used for routine antibiograms by the disk diffusion assay (Sanofi-Diagnostic Pasteur, Marnes-la-Coquette, France) as previously described (27). The double-disk synergy test was performed with disks containing ceftazidime and amoxicillin-clavulanic acid on Mueller-Hinton agar plates, and the results were interpreted as described previously (11). MICs were determined by an agar dilution technique with Mueller-Hinton agar (Sanofi-Diagnostic Pasteur) with an inoculum of 104 CFU, as described previously (27). All plates were incubated at 37°C for 18 h at ambient atmosphere. MICs of β-lactams were determined alone or in combination with a fixed concentration of clavulanic acid (2 μg/ml) and tazobactam (4 μg/ml). MIC results were interpreted according to the guidelines of the National Committee for Clinical Laboratory Standards (22).
PCR and hybridization experiments.
Whole-cell DNA of P. aeruginosa GW-1 was extracted as described previously (27). This DNA was used as a template in standard PCR conditions (36) with a series of primers designed for the detection of class A β-lactamase genes and their extended-spectrum derivatives found in enterobacterial and P. aeruginosa isolates: blaTEM, blaSHV, blaPER-1/−2, blaVEB-1, blaToho-1/−2, blaSFO-1, blaGES-1, and blaCTX-M-2 (8, 28). Similarly, a series of primers were designed for detection of genes coding for acquired carbapenem-hydrolyzing β-lactamases such as Van3 (5′-CCTGAGGGGATGACTAAA-3′) and Van6 (5′-GTTATGCACTAC GAAGGC-3′) for blaSME-1 (21); I4 (5′-CTAATGAAATAG GAGTAC-3′) and I5 (5′-AACAGATTTCAATGGCAGG-3′) for blaNMC-A (19), VIM-B and VIMF for blaVIM-1/VIM-2 (30) and Imp-1 (5′-CTACCGCAGCAGAGTCTTTGC-3′) and Imp-2 (5′-GAACAACCAGTTTTGCCTTAC C-3′) for blaIMP-1 (2). Since several β-lactamase genes are part of gene cassettes that are class 1 integron encoded, primers located in the 5′-CS (INT2F, 5′-TCTCGGGTAACATCAAGG-3′) and 3′-CS (5′-AAGCAGACTTGACCTGA-3′) regions were used for PCR amplifications (14). Southern hybridizations were performed as described by Sambrook et al. (36) using the ECL nonradioactive labeling and detection kit (Amersham Pharmacia Biotech, Orsay, France). Natural plasmid pGW-1 was hybridized with a PCR-generated probe consisting of the internal 860-bp PCR fragment for blaGES-1, as described previously (28).
Cloning experiments, recombinant plasmid analysis, and DNA sequencing.
The obtained PCR fragment (1.8 kb) with 5′-CS and 3′-CS integron primers was purified with a QIAquick column (Qiagen, Courtaboeuf, France) and cloned into the SrfI site of plasmid pPCR Cam SK(+) (Stratagene, Amsterdam, The Netherlands). Recombinant plasmids were selected onto Trypticase soy (TS) agar plates containing amoxicillin (100 μg/ml) and chloramphenicol (30 μg/ml). The cloned DNA fragment inserted into one of the recombinant plasmids (pLAP-1) was sequenced on both strands with an Applied Biosystems sequencer (ABI 377). The nucleotide and deduced amino acid sequences were analyzed and compared to sequences available over the Internet at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov).
Plasmid study.
Conjugation experiments were performed with P. aeruginosa GW-1 and in vitro-obtained rifampin-resistant P. aeruginosa strain PU21 in solid and liquid media at 37°C (27). Transconjugants were selected on TS agar plates containing 150 μg of rifampin per ml and 5 μg of ceftazidime per ml. Plasmid DNAs of P. aeruginosa GW-1 and transconjugant P. aeruginosa PU21 were extracted with the Qiagen plasmid DNA maxi kit and analyzed by electrophoresis with a 0.8% agarose gel (Gibco BRL-Life Technologies, Cergy-Pontoise, France), as previously described (36). Plasmid DNAs extracted from E. coli NCTC 50192 were used as size standards (6).
β-Lactamase purification and isoelectric focusing (IEF) analysis.
Cultures of E. coli DH10B(pLAP-1) were grown overnight at 37°C in 4 liters of TS broth containing amoxicillin (100 μg/ml). β-Lactamase was purified with exactly the same protocol as that described for GES-1 (28). Briefly, the β-lactamase extract was sonicated, cleared by ultracentrifugation, loaded on a Q-Sepharose column, and eluted with a linear NaCl gradient. The purity of the enzyme was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (36).
IEF analysis was performed with an ampholine polyacrylamide gel (pH 3.5 to 9.5), as described previously (27). Purified β-lactamase from a culture of E. coli DH10B(pLAP-1) and nonpurified extracts of 100-ml cultures of P. aeruginosa GW-1 and one of its P. aeruginosa PU21 transconjugants were submitted to IEF analysis. The focused β-lactamases were detected by overlaying the gel with 1 mM nitrocefin (Oxoid, Dardilly, France) in 100 mM phosphate buffer (pH 7.0). The pI values were determined and compared to those of known β-lactamases, including GES-1 β-lactamase.
Kinetic measurements.
Purified β-lactamase was used for kinetic measurements performed at 30°C with 100 mM sodium phosphate (pH 7.0) with an ULTROSPEC 2000 UV spectrophotometer (Amersham Pharmacia Biotech) as previously described for the biochemical analysis of GES-1 β-lactamase (28).
Fifty percent inhibitory concentrations (IC50s) were determined for clavulanic acid, tazobactam, sulbactam, and imipenem. Various concentrations of these inhibitors were preincubated with the purified enzyme for 3 min at 30°C to determine the concentrations that reduced the hydrolysis rate of 100 μM benzylpenicillin by 50%.
The specific activity of the purified β-lactamase from E. coli DH10B(pLAP-1) was obtained as described previously (29). One unit of enzyme activity was defined as the activity which hydrolyzed 1 μmol of benzylpenicillin per min per mg of protein. The total protein content was measured with the DC Protein assay kit (Bio-Rad, Ivry-sur-Seine, France). Specific activity was also determined with 100 μM ceftazidime as a substrate.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the GenBank nucleotide database under accession no. AF326355.
RESULTS
Properties of P. aeruginosa isolate GW-1.
GW-1 was isolated in May 2000 at the Pretoria Academic Hospital, Pretoria, Republic of South Africa, from a 38-year-old Zimbabwean refugee hospitalized for cerebral malaria. On the 12th day of her hospitalization in the intensive care unit, she developed nosocomial pneumonia, and blood cultures grew P. aeruginosa GW-1. Empirical treatment with a combination of imipenem and amikacin was unsuccessful. Imipenem was replaced after 3 days with aztreonam, and her clinical condition improved remarkably. She received no treatment prior to her hospitalization in South Africa.
P. aeruginosa GW-1 exhibited a broad spectrum of resistance to expanded-spectrum cephalosporins and an intermediate susceptibility to imipenem, according to antibiotic susceptibility testing by disk diffusion. Double-disk synergy testing remained negative with clavulanate- and ceftazidime-containing disks. P. aeruginosa GW-1 was also resistant to kanamycin, gentamicin, netilmicin, fluoroquinolones, sulfonamides, and tetracycline and susceptible to tobramycin. Preliminary experiments with crude extracts of a culture of P. aeruginosa GW-1 showed that imipenem hydrolysis was detectable (data not shown).
Cloning and sequencing of the β-lactamase gene.
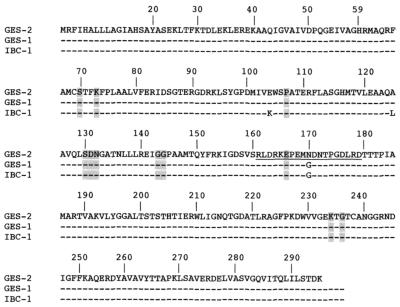
Preliminary PCR detection of most of the class A ESBLs and carbapenem-hydrolyzing β-lactamase genes failed (data not shown). However, PCR amplification was positive with primers for blaGES-1. With whole-cell DNA of P. aeruginosa GW-1 as a template and consensus primers for 5′-CS and 3′-CS ends of class 1 integrons, a 1.8-kb DNA fragment was obtained. It was cloned into the SrfI site of plasmid pPCR Cam SK(+), yielding recombinant plasmid pLAP-1. Sequence analysis of the cloned fragment revealed an 864-bp-long ORF encoding a 287-amino-acid preprotein. This protein was a β-lactamase with the STFK tetrad and structural elements characteristic of the active site of an Ambler class A β-lactamase (Fig. 1) (12). The β-lactamase, designated GES-2, had one amino acid change (glycine to asparagine in position 170) and three amino acid changes compared to GES-1 and IBC-1, respectively (Fig. 1). The G+C content of blaGES-2 was 51.5%, a value which is not within the range of G+C content of P. aeruginosa genes (60.1 to 69.5%).
FIG. 1.
Comparison of the amino acid sequence of GES-2 to those of GES-1 and IBC-1 β-lactamases (7, 28). The numbering is according to the Ambler designation (1). Highlighted amino acids are those strictly conserved in class A β-lactamases. The amino acids of the omega loop are underlined.
As found for GES-1 and IBC-1, GES-2 was distantly related to the class A β-lactamases (7, 28). The highest percentage of amino acid identity was 36%, found with either carbenicillinase GN79 from Proteus mirabilis, a constitutive penicillinase from Yersinia enterocolitica YENT, or the L-2 chromosomally encoded extended-spectrum β-lactamase from Stenotrophomonas maltophilia.
Transfer of β-lactam resistance.
Transconjugant P. aeruginosa PU21 strains were obtained with P. aeruginosa GW-1 as a donor. They showed a broad-spectrum β-lactam resistance phenotype, including an increased resistance against imipenem (Table 1). A slightly positive double-disk synergy test was done, with transconjugants indicating the presence of an ESBL (data not shown). Cotransferred antibiotic resistance markers were those carrying resistance for kanamycin, gentamicin, netilmicin, and sulfonamides. The analysis of plasmid content of P. aeruginosa GW-1 and its transconjugant revealed a ca. 100-kb plasmid, designated pGW-1 (data not shown). Hybridization using an internal probe for blaGES-1 confirmed the presence of a blaGES-1-like gene on plasmid pGW-1 found in P. aeruginosa GW-1 and its transconjugant (data not shown).
TABLE 1.
MICs of β-lactams
| β-Lactam(s)b | MIC (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|
| P. aeruginosa GW-1 | P. aeruginosa PU21(pGW-1) | P. aeruginosa PU21 | E. coli DH10B(pLAP-1) | E. coli DH10B(pC1) | E. coli DH10B | |
| Amoxicillin | >512 | >512 | >512 | >512 | >512 | 4 |
| Amoxicillin + CLA | >512 | >512 | >512 | 16 | >128 | 4 |
| Ticarcillin | >512 | >512 | 8 | 256 | >512 | 4 |
| Ticarcillin + CLA | >512 | 256 | 4 | 4 | 64 | 4 |
| Ticarcillin + TZB | >512 | 256 | 8 | 4 | 256 | 2 |
| Piperacillin | 128 | 64 | 8 | 8 | 64 | 1 |
| Piperacillin + CLA | 64 | 16 | 2 | 1 | 8 | 1 |
| Piperacillin + TZB | 128 | 16 | 16 | 8 | 8 | 1 |
| Cephalothin | >512 | >512 | >512 | 32 | 256 | 2 |
| Cefoxitin | >512 | >512 | >512 | 4 | 8 | 1 |
| Ceftazidime | 32 | 16 | 1 | 8 | 128 | 0.5 |
| Ceftazidime + CLA | 16 | 16 | 2 | 0.5 | 8 | 0.5 |
| Ceftazidime + TZB | 8 | 4 | 2 | 0.5 | 8 | 0.5 |
| Cefotaxime | 128 | 128 | 8 | 1 | 4 | 0.06 |
| Cefotaxime + CLA | 256 | 256 | 16 | 0.06 | 0.5 | 0.06 |
| Cefotaxime + TZB | 64 | 32 | 8 | 0.5 | 2 | 0.06 |
| Cefepime | 32 | 16 | 0.5 | 0.12 | 0.25 | 0.03 |
| Cefsulodin | >512 | 512 | 2 | 32 | 32 | 1 |
| Aztreonam | 16 | 8 | 4 | 0.5 | 1 | 0.12 |
| Imipenem | 16 | 16 | 2 | 0.25 | 0.06 | 0.06 |
| Meropenem | 16 | 2 | 0.5 | 0.06 | 0.06 | 0.06 |
| Moxalactam | 16 | 8 | 8 | 0.12 | 0.12 | 0.12 |
β-Lactamases: GES-2, AmpC type, pI 7.5, for P. aeruginosa GW-1; GES-2, AmpC type for P. aeruginosa PU21(pGW-1); AmpC type for P. aeruginosa PU21; GES-2 for E. coli DH10B(pLAP-1); GES-1 for E. coli DH10B(pC1).
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
β-Lactam susceptibility.
MICs of β-lactams for P. aeruginosa GW-1 mirrored those for its transconjugant (Table 1). The GES-2-producing P. aeruginosa strains were characterized by resistance to carbenicillin, ureidopenicillins, cefotaxime, and ceftazidime and by an intermediate susceptibility to aztreonam (Table 1). The MIC of imipenem was eightfold higher for P. aeruginosa PU21(pGW-1) than that for P. aeruginosa PU21 (Table 1). A similar trend of resistance was noted once blaGES-2 was expressed in E. coli DH10B. However, in this latter case, the recombinant strain was of intermediate susceptibility to extended-spectrum cephalosporins and susceptible to imipenem (Table 1). GES-2-producing E. coli DH10B(pLAP-1) was less resistant to cephalosporins than GES-1-producing E. coli DH10B(pC1) (Table 1). The imipenem MIC for E. coli DH10B(pLAP-1) was only slightly higher than that for E. coli DH10B (Table 1).
Clavulanate and tazobactam partially restored the β-lactam activities against the GES-2-producing E. coli strain as previously found for the GES-1-producing E. coli strain (Table 1). Clavulanate did not lower significantly the β-lactam MICs for the GES-2-producing P. aeruginosa strains (Table 1). This result was likely due to a concomitant induction of the chromosomal cephalosporinase of P. aeruginosa.
IEF analysis and kinetic parameters.
IEF analysis showed that P. aeruginosa GW-1, its transconjugant, and E. coli DH10B(pLAP-1) had β-lactamase activities with a pI value of 5.8, corresponding to that of GES-2 and identical to that of GES-1 (data not shown). β-Lactamase activities with a pI value of 8 to 8.5 were also detected for P. aeruginosa GW-1 and its transconjugant corresponding to the chromosomal cephalosporinase of P. aeruginosa. A nonidentified β-lactamase with a pI of 7.5 was found in GW-1 and in its transconjugant. Additional PCR experiments with whole-cell DNAs of P. aeruginosa GW-1 and its transconjugants as templates and primers for the detection of OXA-1, OXA-2, OXA-3, OXA-10, OXA-18, and OXA-20 genes failed (data not shown).
The specific activity of the purified β-lactamase GES-2 was 720 mU · mg of protein−1, determined with 100 μM benzylpenicillin as a substrate. Its overall recovery was 70% with a 20-fold purification. The purity of the enzyme was estimated to be 90% according to sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. Kinetic parameters of GES-2 showed its broad-spectrum activity against most β-lactams, except aztreonam and meropenem (Table 2). The hydrolysis rates of GES-2 for penicillins were similar to those of GES-1 because the Km values were 10-fold lower. However, the hydrolysis efficiency of GES-2 for extended-spectrum cephalosporins was slightly lower than that of GES-1 β-lactamase. Although ceftazidime was hydrolyzed, its kinetic parameters could not be determined precisely. This was due to a very high Km value (>3,000 μM), reflecting a very low affinity of GES-2 for ceftazidime. Nevertheless, its Vmax value was high (data not shown), and the specific activity of GES-2 was 57 mU · mg−1 with 100 μM ceftazidime as a substrate. As reported for GES-1 (28), high kcat/Km values were obtained for most cephalosporins for GES-2 when expressed in millimolar units and not, as usual for an ESBL activity of a class A enzyme, in micromolar units.
TABLE 2.
Steady-state kinetic parameters of GES-2a
| Substrate | GES-2
|
GES-1
|
||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (mM−1 · s−1) | kcat(s−1) | Km (μM) | kcat/Km (mM−1 · s−1) | |
| Benzylpenicillin | 0.4 | 4 | 96 | 2.8 | 40 | 70 |
| Amoxicillin | 0.7 | 25.8 | 26 | 13 | 200 | 65 |
| Ticarcillin | 0.06 | 13.3 | 4.5 | 0.3 | 400 | 0.7 |
| Piperacillin | 0.3 | 22.8 | 23 | 8 | 900 | 13 |
| Cephalothin | 0.3 | 3 | 112 | 179 | 3,400 | 52 |
| Cephaloridine | 0.5 | 7.7 | 65 | 53 | 2,000 | 26 |
| Cefoxitin | —b | — | — | 0.9 | 30 | 33 |
| Ceftazidime | NDc | >3,000 | ND | 380 | 2,000 | 188 |
| Cefepime | 1.1 | 1,900 | 0.6 | 2.8 | 1,800 | 1.6 |
| Cefotaxime | 2.2 | 890 | 2.5 | 68 | 4,600 | 15 |
| Imipenem | 0.004 | 0.45 | 9 | 0.003 | 45 | 0.07 |
| Meropenem | — | — | — | — | — | — |
| Aztreonam | — | — | — | — | — | — |
Parameters of GES-2 are compared to those of GES-1 β-lactamase, as previously published (28). Values are expressed as the mean of three independent measures (standard deviations of the values were within 15%).
—, not hydrolyzed (the initial rate of hydrolysis was lower than 0.001 μM−1 · s−1).
ND, not determinable due to very high Km values.
GES-2, unlike GES-1, measurably hydrolyzed imipenem (Table 2). Meropenem hydrolysis by GES-2 was not detected. The hydrolysis efficiency of GES-2 against imipenem was 100-fold higher than that of GES-1 due to a 100-fold-lower Km value (Table 2) but still remained marginal. GES-2 was probably saturated by imipenem, while GES-1 was not. Inhibition studies as measured by IC50s with benzylpenicillin as a substrate showed that GES-2 activity was inhibited by clavulanic acid and tazobactam more than GES-1 is, while GES-2 was less inhibited by imipenem (Table 3).
TABLE 3.
Inhibition profile of GES-2 compared with those of other β-lactamases
Genetic environment of blaGES-2.
Upstream of blaGES-2, two putative promoter sequences named P1 and P2 were located in the structural integrase gene (data not shown). Comparison of P1 with known promoters for which expression studies were performed (5, 13) identified P1 as a weak promoter. As described, the insertion of three guanosine molecules 119 bases downstream of the promoter P1 creates a secondary promoter (P2) for blaGES-1 expression. This triple nucleotide insertion brings the spacing between the −35 and −10 regions of P2 to 17 bp. Therefore, P2 expression may be responsible for 90% of blaGES-2 transcription, as shown for other genes (5, 13). The blaGES-2 gene cassette which was inserted at the attI recombination site has a core site (GTTAGAC) and an inverse core site (GTCTAAC). Downstream of this gene, the 59-be sequence was made up of 110 bp. It was exactly identical to that found downstream of blaIBC-1 and shared 70% nucleotide identity within the 19-bp-long and truncated 59-be of blaGES-1 (data not shown).
DISCUSSION
This report characterized another non-SHV-, non-TEM-type ESBL, showing that class A ESBLs are not limited to SHV and TEM derivatives. GES-2 β-lactamase is the fourth example of a non-TEM-, non-SHV-type ESBL in P. aeruginosa after PER-1, VEB-1, and OXA-18. A study of the GES-2-producing P. aeruginosa GW-1 isolate further indicated that ESBL genes in P. aeruginosa are difficult to detect by double-disk synergy tests (23) and may therefore be clinically underestimated.
GES-2 β-lactamase is also the third example of a GES-type ESBL, in addition to GES-1 from K. pneumoniae ORI-1 (28) and IBC-1 from E. cloacae HT9 (7). The natural producer of the GES-type enzymes remains to be determined. Indeed, isolation of a GES-2 gene in P. aeruginosa with a G+C content of non-P. aeruginosa origin indicated a horizontal transfer of blaGES genes in gram-negative species.
One interesting aspect of this study is the comparison of the substrate profiles of GES-1 and GES-2. A glycine-to-asparagine substitution in GES-2 extended its activity to imipenem. This substitution occurred in an amino acid position of the omega loop of class A β-lactamases that is of primary importance in the catalytic activity of these enzymes (3, 16). This substitution may enlarge the pocket that houses the hydroxyethyl moiety of imipenem on the alpha face of the acyl enzyme for GES-2 (17, 18).
GES-2, like GES-1 and IBC-1 β-lactamases and also like the class A enzymes with significant catalytic efficiency against imipenem (i.e., the chromosomally encoded β-lactamases SME-1/2, IMI-1, and Nmc-A and the plamid-encoded KPC-1 enzyme [40]), contains two cysteine residues in positions 69 and 238 that may form a disulfide bridge. The catalytic activity (kcal/Km, expressed in millimoles per second) of GES-2 versus that of imipenems (9 and 520, respectively) remained much lower than those of the carbapenem-hydrolyzing β-lactamases such as SME-1 (33). Thus, as reported (34), this cysteine bridge may enable the catalytic site to bind imipenem, but other amino acid residues are likely to be involved in the significant catalytic efficiency of the carbapenem-hydrolyzing enzymes against imipenem.
The IC50 of imipenem for GES-2 was similar to the values of other class A β-lactamases like TEM-1 (Table 3). Inhibitory activity profiles of GES-1, IBC-1, and GES-2 showed that clavulanic acid and tazobactam are similarly active against GES-2 and IBC-1 but less so against GES-1.
Comparison of the surrounding sequences of blaGES-2 to those of blaGES-1 and blaIBC-1 showed that blaGES genes are part of gene cassettes. Identification of an identical 59-be consisting of 110 bp for blaGES-2 and blaIBC-1 indicated the spread of an identical gene cassette among P. aeruginosa and enterobacterial isolates. Additionally, it may confirm the hypothesis that a deletion occurs in the 59-be of the blaGES-1 cassette (28).
A striking similarity may be drawn between VEB and GES β-lactamase genes. They encode class A ESBLs, are located on broad-host-range conjugative plasmids, are part of gene cassettes in class 1 integrons, and are found in gram-negative isolates, and their origin (natural producer) remains unknown. Additionally, they may have spread worldwide, since VEB β-lactamases have been isolated in southeastern Asia and recently in Kuwait (8, 20, 29, 32, 38); GES β-lactamases are found in French Guiana, Greece, and now South Africa (7, 28).
This work shows that plasmid- and integron-mediated genes encoding ESBLs with some carbapenem hydrolytic activity are not limited to the Ambler class B β-lactamase genes. GES-2 β-lactamase may contribute in part to the decreased susceptibility of P. aeruginosa to imipenem. However, once expressed from a multicopy vector in E. coli, blaGES-2 expression did not significantly increase the imipenem MIC, thus making its clinical detection in enterobacterial isolates by a simple susceptibility study unlikely.
Finally, this report indicates that a class A β-lactamase with a substrate profile extended to imipenem may be selected in vivo through a single amino acid substitution in an ESBL sequence.
ACKNOWLEDGMENTS
This work was financed by a grant from the Ministère de l'Education Nationale et de la Recherche (grant UPRES, JE-2227), Université Paris XI, Paris, France.
We thank M. J. Pitout, A. M. S. Van Straten, and A. Laubscher for their contributions.
REFERENCES
- 1.Ambler R P, Coulson A F, Frère J-M, Ghuysen J M, Joris B, Forsman M, Lévesque R C, Tiraby G, Waley S G. A standard numbering scheme for the class A β-lactamases. Biochem J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N, Otha M. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee S, Pieper U, Kapadia G, Pannell L K, Herzberg O. Role of the omega-loop in the activity, substrate specificity, and structure of class A β-lactamase. Biochemistry. 1998;37:3286–3296. doi: 10.1021/bi972127f. [DOI] [PubMed] [Google Scholar]
- 4.Collis C M, Hall R M. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol. 1992;174:1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collis C M, Hall R M. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–162. doi: 10.1128/aac.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danel F, Hall L M C, Gür D, Livermore D. OXA-14, another extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1881–1884. doi: 10.1128/aac.39.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giakkoupi P, Tzouvelekis L S, Tsakris A, Loukova V, Sofianou D, Tzelepi E. IBC-1, a novel integron-associated class A β-lactamase with extended-spectrum properties produced by an Enterobacter cloacae clinical strain. Antimicrob Agents Chemother. 2000;44:2247–2253. doi: 10.1128/aac.44.9.2247-2253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girlich D, Poirel L, Leelaporn A, Karim A, Tribuddharat C, Fennewald M, Nordmann P. Molecular epidemiology of the integron-located VEB-1 extended-spectrum β-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J Clin Microbiol. 2001;39:175–182. doi: 10.1128/JCM.39.1.175-182.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall R M, Brookes D E, Stokes H W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 10.Hall R M, Collis C M. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 11.Jarlier V, Nicolas M-H, Fournier G, Philippon A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 12.Joris B, Ledent P, Dideberg O, Fonze E, Lamotte-Brasseur J, Kelly J A, Ghuysen J M, Frère J-M. Comparison of the sequences of class A β-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob Agents Chemother. 1991;35:2294–2301. doi: 10.1128/aac.35.11.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lévesque C, Brassard S, Lapointe J, Roy P H. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 14.Lévesque C, Piché L, Larose C, Roy P H. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchandin H, Jean-Pierre H, De Champs C, Sirot D, Darbas H, Perigault P F, Carrière C. Production of a TEM-24 plasmid-mediated extended-spectrum β-lactamase by a clinical isolate of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2000;44:213–216. doi: 10.1128/aac.44.1.213-216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matagne A, Lamotte-Brasseur J, Frère J-M. Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem J. 1998;330:581–598. doi: 10.1042/bj3300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maveyraud L, Mourey L, Kotra L P, Pedelacq J-D, Guillet V, Mobashery S, Samama J-P. Structural basis for clinical longevity of carbapenem antibiotics in the face of challenge by the common class A β-lactamases from the antibiotic-resistant bacteria. J Am Chem Soc. 1998;120:9748–9752. [Google Scholar]
- 18.Mourey L, Miyashita K, Swaren P, Bulychev A, Samama J-P, Mobashery S. Inhibition of the NMC-A β-lactamase by a penicillanic acid derivative, and the structural bases for the increase in substrate profile of this antibiotic resistance enzyme. J Am Chem Soc. 1998;120:9343–9383. [Google Scholar]
- 19.Naas T, Nordmann P. Analysis of a carbapenem-hydrolyzing class A β-lactamase from Enterobacter cloacae and of its Lys-R type regulatory protein. Proc Natl Acad Sci USA. 1994;91:7693–7696. doi: 10.1073/pnas.91.16.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naas T, Poirel L, Karim A, Nordmann P. Molecular characterization of In50, a class 1 integron encoding the gene for the extended-spectrum β-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol Lett. 1999;176:411–419. doi: 10.1111/j.1574-6968.1999.tb13691.x. [DOI] [PubMed] [Google Scholar]
- 21.Naas T, Vandel L, Sougakoff W, Livermore D, Nordmann P. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A, β-lactamase, Sme-1 from Serratia marcescens S6. Antimicrob Agents Chemother. 1994;38:1262–1270. doi: 10.1128/aac.38.6.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 23.Nordmann P, Guibert M. Extended-spectrum β-lactamases in Pseudomonas aeruginosa. J Antimicrob Chemother. 1998;42:128–131. doi: 10.1093/jac/42.2.128. [DOI] [PubMed] [Google Scholar]
- 24.Nordmann P, Mariotte S, Naas T, Labia R, Nicolas M-H. Biochemical properties of a carbapenem-hydrolyzing β-lactamase from Enterobacter cloacae and cloning of the gene into Escherichia coli. Antimicrob Agents Chemother. 1993;37:939–946. doi: 10.1128/aac.37.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nordmann P, Naas T. Sequence analysis of PER-1 extended spectrum β-lactamase from Pseudomonas aeruginosa and comparison with class A β-lactamases. Antimicrob Agents Chemother. 1994;38:104–114. doi: 10.1128/aac.38.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulsen I T, Littlejohn T G, Radström P, Sundström L, Sköld O, Swedberg G, Skurray R A. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993;37:761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philippon L N, Naas T, Bouthors A-T, Barakett V, Nordmann P. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2188–2195. doi: 10.1128/aac.41.10.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44:622–632. doi: 10.1128/aac.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirel L, Naas T, Guibert M, Chaibi E B, Labia R, Nordmann P. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother. 1999;43:573–581. doi: 10.1128/aac.43.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo J-D, Nordmann P. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother. 2000;44:891–897. doi: 10.1128/aac.44.4.891-897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirel L, Ronco E, Naas T, Nordmann P. Extended-spectrum β-lactamase TEM-4 in Pseudomonas aeruginosa. Clin Microbiol Infect. 1999;5:651–652. doi: 10.1111/j.1469-0691.1999.tb00425.x. [DOI] [PubMed] [Google Scholar]
- 32.Poirel L, Rotimi V O, Mokaddas E M, Karim A, Nordmann P. VEB-1-like extended-spectrum β-lactamases in Pseudomonas aeruginosa, Kuwait. Emerg Infect Dis. 2001;7:468–470. doi: 10.3201/eid0703.010322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Queenan A M, Torres-Viera C, Gold H S, Carmeli Y, Eliopoulos G M, Moellering R C, Jr, Quinn J P, Hindler J, Medeiros A A, Bush K. SME-type carbapenem-hydrolyzing class A β-lactamases from geographically diverse Serratia marcescens strains. Antimicrob Agents Chemother. 2000;44:3035–3039. doi: 10.1128/aac.44.11.3035-3039.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raquet X, Lamotte-Brasseur J, Bouillenne F, Frère J-M. A disulfide bridge near the active site of carbapenem-hydrolyzing class A β-lactamases might explain their unusual substrate profile. Proteins. 1997;27:47–58. doi: 10.1002/(sici)1097-0134(199701)27:1<47::aid-prot6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 35.Rasmussen B A, Bush K, Keeney D, Yang Y, Hare R, O'Gara C, Medeiros A A. Characterization of IMI-1 β-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob Agents Chemother. 1996;40:2080–2086. doi: 10.1128/aac.40.9.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Taibi P, Mobashery S. Mechanism of turnover of imipenem by the TEM β-lactamase revisited. J Am Chem Soc. 1995;117:7600–7605. [Google Scholar]
- 38.Tribuddharat C, Fennewald M. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:960–962. doi: 10.1128/aac.43.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vahaboglu H, ÖOztürk R, Aygun G, Coskunkan F, Yaman A, Kaygusuz A, Leblebicioglu H, Balik I, Aydin K, Otkun M. Widespread detection of PER-1-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob Agents Chemother. 1997;41:2265–2269. doi: 10.1128/aac.41.10.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yigit H, Queenan A M, Anderson G J, Domenech-Sanchez A, Biddle J W, Steward C D, Alberti S, Bush K, Tenover F C. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]