Abstract
Protein nanocages have been used as functional bio-templates for the synthesis or organization of nanomaterials. However, the stability of these protein nanocages is nonideal, which limits their applications. Herein, we characterized the high thermal stability of plant ferritin, soybean seed H-2 ferritin (SSFH-2), the melting point (Tm) of which is 106 °C. We demonstrated that the hyperthermostability of SSFH-2 is derived from extra peptides (EP) located on its outer surface. Indeed, removal of the EP domains resulted in a dramatic decrease in Tm to 88 °C. Similar to EP-deleted plant ferritin, human H-chain ferritin (HuHF) has a Tm of 82 °C. Excitingly, the graft of the EP domain on the exterior surface of HuHF pronouncedly improved its Tm to 103 °C, which represents a simple, efficient approach to the construction of protein architectures with high stability. The remarkable stability of protein nanocages will greatly facilitate their application as robust biotemplates in the field of nanoscience.
Ferritin nanocage exhibits hyperthermostability with EP domain located on its outer surface.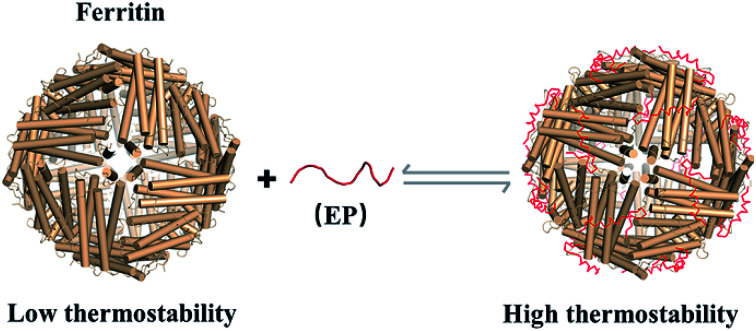
Introduction
Proteins that are highly stable to heat are particularly useful as biotemplates because they expand the range of processing conditions that may be used in the production of biomaterials. Protein nanocages have specific shell-like structures and are widely distributed in nature, representing a common architectural paradigm of the subcellular world. For example, clathrin cages are responsible for endocytosis, carboxysomes are associated with CO2 fixation, and viral capsids are involved in nucleic acid storage and transport.1–3 So far, nanotechnologists have subverted these natural functions and used self-assembled cage structures of nanometer dimensions for the preparation of nanomaterials and encapsulation of guest molecules with multiple potential applications because of their high symmetry, solubility and monodispersity.4–9
Of the protein nanocages, ferritins are a special class of diiron protein that play roles in both iron housekeeping and iron detoxification, which are universally distributed in animals, plants, and bacteria. All ferritins share the highly conserved architecture that comprises 24 similar or different subunits assembling into a quasi-spherical shell with an outer diameter of ∼12 nm and an inner diameter of ∼8 nm. In vertebrates, ferritins consist of two types of subunits, H and L, respectively, which share ∼55% sequence identity. The H-type subunit of vertebrate ferritins contains a ferroxidase center at which the rapid oxidation of Fe2+ to Fe3+ by dioxygen or hydrogen peroxide occurs. Unlike the H-subunit, the L-subunit lacks such ferroxidase center.10–12 Benefiting from the low immunogenicity and the high selectivity for cancer cells which overexpress transferrin receptor 1 (TfR1) for human H-chain ferritin (HuHF),13 ferritin especially human H-chain ferritin, has attracted attention in materials and medicinal chemistry with use of ferritin nanocages as a vehicle for tumor-imaging agents, anticancer drugs, and bioactive nutrients.14–19 The structure of plant ferritin is different from that of animal ferritin in that plant ferritin only consists of H-type subunits which usually contain an extra peptide (EP) (about 30 amino acid residues) at its N-terminal extremity, while animal ferritin is devoid of such EP domain.20 This EP domain exhibited the serine-like protease activity and contributed to the storage stability of ferritin.21
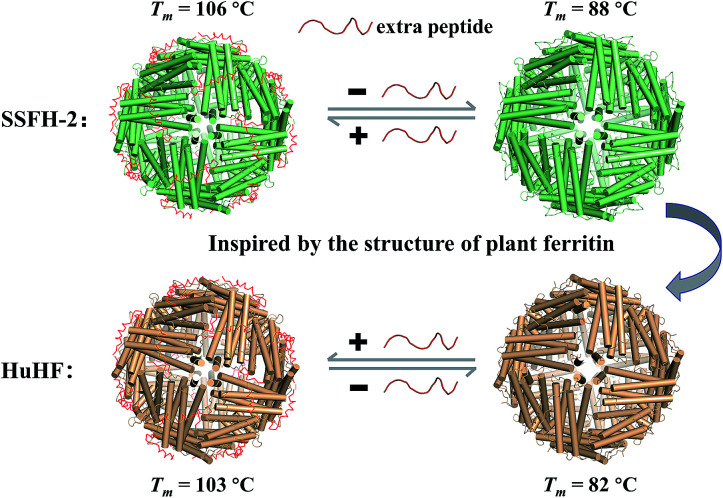
Although members of the ferritin family have been employed extensively in the formation and stabilization of inorganic nanomaterials, these applications are limited by the proteins' often nonideal thermostability. Therefore, it is of considerable importance to either find new thermostable protein architectures as biotemplates in nature or build an approach to engineer high stability into known protein architectures by genetic or chemical modification. Here, we found that soybean seed H-2 ferritin (SSFH-2) has an extremely high melting point (Tm) of 106 °C for the first time, a value being higher than the boiling point of water, and such high Tm of SSFH-2 is found to be derived from its EP domain. Bioinspired by high thermal stability of plant ferritin, the EP was grafted on the exterior surface of HuHF by genetic modification, causing a dramatic increase in its Tm from original 82 °C to 103 °C (Fig. 1). These properties greatly facilitate the utilization of HuHF as biotemplates for the preparation of nanomaterials.
Fig. 1. Schematic representation of reversible control of the hyperthermostability of ferritin nanocage by extra peptide (EP). With EP domain located on its outer surface, ferritin exhibits hyperthermostability (value of Tm above 100 °C).
Methodology
Cloning and purification of SSFH-2, SSFH-2-ΔEP, HuHF and HuHF-∇EP
The genes encoding SSFH-2-ΔEP and HuHF-∇EP were synthesized by Synbio Technologies, which have been inserted into the plasmid pET-3a. The plasmids corresponding to SSFH-2-ΔEP and HuHF-∇EP were transformed into E. coli strain BL21 (λ DE3) and verified by DNA sequencing.22 Native SSFH-2 and HuHF were purified as previously described.21 SSFH-2-ΔEP and HuHF-∇EP were purified as following. The E. coli strains BL21 (DE3) which contained SSFH-2-ΔEP or HuHF-∇EP expression plasmids were grown at 37 °C. Protein expression was likewise induced with 200 μM IPTG. After the cell density reached an absorbance as 0.6 at 600 nm, the cells were harvested by centrifugation (10 000 rpm) after 10 h of induction and the precipitate was re-suspended in 50 mM Tris–HCl (pH 8.0), followed by sonication, ammonium sulfate fractionation (40% saturated fraction), and dialysis to obtain crude protein. Then, the protein solution was applied to an ion-exchange column (Q-Sepharose Fast Flow, GE Healthcare), followed by gradient elution with 0–1.0 M NaCl. Finally, the protein solution was concentrated and purified on a gel filtration column (Superdex 200 pg 16/60, GE Healthcare), equilibrated with 50 mM Tris–HCl, 150 mM NaCl (pH 8.0). Finally, the protein solution was dialyzed against 50 mM MOPS (pH 7.9) at 4 °C, and protein concentrations were determined according to the Lowry method with bovine serum albumin (BSA) as standard.
Thermal treatment
After diluting SSFH-2 to the final concentration of 1.0 μM, protein solutions (1 mL) were placed in test plastic tubes (volume 2 mL) and then subjecting them to a heat treatment by placing the test tubes in a temperature-controlled water bath at different temperatures (60–100 °C) for different holding times (5–30 min), while 121 °C was carried out in an autoclave.23 Following thermal treatments, protein solutions were immediately cooled at 4 °C for further analysis.
Biophysical characterization of native protein and thermally treated samples
Polyacrylamide gel electrophoresis (PAGE)
The purity and molecular weight of protein samples was estimated by polyacrylamide gel electrophoresis. Native PAGE used a 4–20% polyacrylamide gradient gel running at 5 mA for 10 h at 4 °C, and employing Tris–HCl (25 mM, pH 8.3) as a running buffer. Then gels were stained with Coomassie brilliant blue R250. In addition, gel electrophoresis under denaturing conditions was carried out using a 15% polyacrylamide-SDS gel as reported by Laemmli.24
Circular dichroism (CD) spectrum analysis
CD spectra was performed on a Pistar π-180 spectrometer (Applied Photophysics, UK), using a quartz cuvette of 0.1 mm optical pathlength at 25 °C. The spectra were obtained as an average of three scans in the far UV range (190–260 nm) with the 50 mM MOPS background subtracted.
Fluorescence spectroscopy
Fluorescence measurements were run on a Cary Eclipse spectrofluorimeter (Varian). Excitation–emission spectra were measured at 25 °C and processed to obtain the emission spectra from 305 nm to 450 nm at excitation wavelength (295 nm). All measurements were performed in triplicate.
Transmission electron microscope (TEM)
Purified ferritin samples were diluted to 0.2 μM with 50 mM MOPS buffer (pH 7.9) prior to placing on carbon-coated copper grids and excess solution was removed with filter paper. Resulting samples were stained using 2% uranyl acetate for 5 min. Transmission electron micrographs were obtained at 80 kV through a Hitachi H-7650 transmission electron microscope.
Dynamic light scattering (DLS)
The DLS measurements were performed at 25 °C using a Viscotek model 802 dynamic light-scattering instrument (Viscotek Europe Ltd.) as previously described.25 OmniSIZE 2.0 software was used to calculate the size/hydrodynamic radius (RH) distribution of prepared solutions.
Differential scanning calorimetry (DSC)
The melting temperatures (Tm) of proteins are measured by using a differential scanning calorimeter (PerkinElmer DSC 8000).26 Protein samples were dialyzed against water for three times, deep frozen at −80 °C, and freeze dried by a freeze dryer. Then, a total of 3–5 mg of proteins is sealed inside an aluminum pan and tested from 30 to 150 °C at a temperature ramp rate of 10 °C min−1. An empty aluminum pan is used as reference to determine the difference in heat flow of the sample and the reference. The DSC experiments provided heat flow in melting processes and peak temperatures (Tm in °C) were taken from the obtained thermograms. Each test was conducted in triplicate.
Iron loading abilities
The fast kinetics experiments were conducted on a Varian Cary 50 spectrophotometer (Varian, USA) at 25 °C. Weakly acidic FeSO4 solution (5 mM, 100 μL) were quickly added to 1 mL buffered SSFH-2 or thermal treated SSFH-2 solutions (1.0 μM, 50 mM MOPS, pH 7.9) at 25 °C in the thermostated sample compartment with 1 cm optical path length. Iron loading experiments were performed by measuring the absorbance at 303 nm as previously reported.27 As for the iron loading ability of HuHF, the final concentration is 0.5 μM.
Crystal structure determination
Purified SSFH-2, TTH-2 and HuHF-∇EP were concentrated to 10 mg mL−1 in a buffer consisting of 20 mM MOPS at pH 7.9, and crystals were obtained using the hanging drop vapor diffusion method by mixing equal volumes of the protein sample and mother liquid. The mother liquid for each crystal was displayed in Table S1.† Diffraction data of the crystals of SSFH-2, TTH-2 and HuHF-∇EP were collected to resolutions of 2.1 Å, 2.6 Å and 4.0 Å, respectively.28 Data were processed, merged and scaled with the HKL-3000 (HKL Research). Data processing statistics are shown in Table S2.† The structure of native SSFH-2 was determined by molecular replacement using the Molrep program in CCP4 using the structure of soybean seed ferritin 4 (SFER4) (PDB code 3A68) as a search model. Structure refinement was conducted using the Refmac5 program and PHENIX software. The structure was rebuilt using COOT, which made the model manually adjusted. Figures of protein structures were prepared using the PyMOL program.
Results and discussion
Structural analyses of SSFH-2
To date, all known naturally occurring plant ferritins from various seeds such as soybean, pea, black bean, maize, and alfalfa consist of two subunits of H-1 and H-2, respectively, with high sequence identity (82% in soybean seed ferritin).20,29 Based on our previous study, SSFH-2 is more stable than its analogue, soybean seed H-1 ferritin (SSFH-1),21 so we chose SSFH-2 as a model to study its thermostability. SSFH-2 was prepared according to our reported method.21 SDS and native PAGE analyses showed that SSFH-2 was purified to homogeneity (Fig. S1a and b†). To obtain structural information on SSFH-2, we tried to crystallize it and eventually obtained large enough crystals suitable for X-ray diffraction. The snowflake-like crystals (Fig. S1c†) of SSFH-2 appeared after 21 days incubation. We solved the crystal structure at 2.1 Å (I4 space group, PDB ID: 6J4J) (Table S2†). Similar to the structures of ferritins from vertebrates and bacteria,10–12 the structure of SSFH-2 has a cage-like hollow shell composed of 24 subunits, which are related by 4-, 3-, and 2-fold symmetry (Fig. S1d–f†). The N-terminal EP from the N terminus to Thr33 cannot be observed because of no electron density, reflecting its high flexibility.
Characterization of the high thermostability of SSFH-2
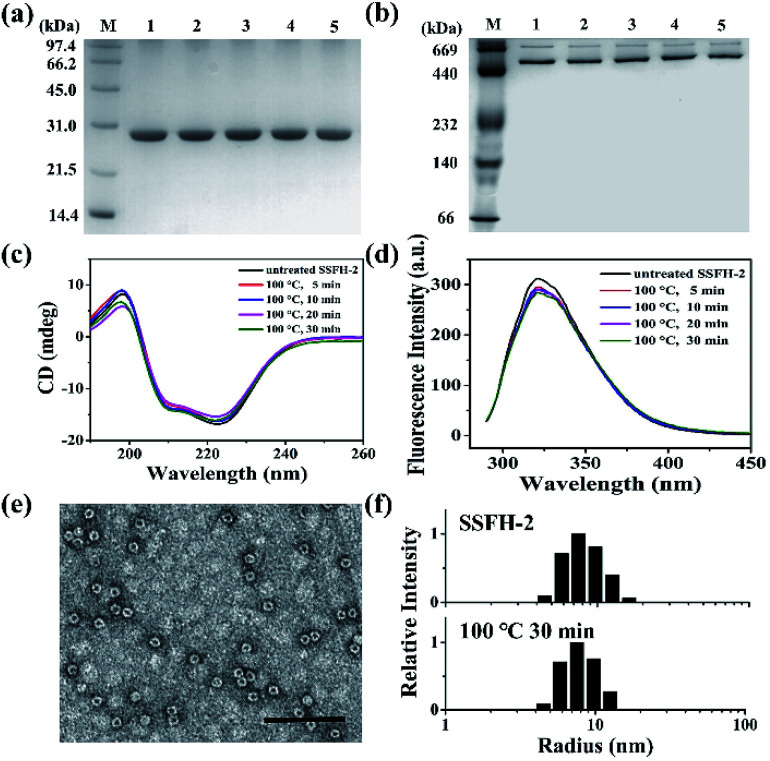
The intrinsic fluorescence emission from tryptophan residue (Trp) in proteins was sensitive to the microenvironment surrounding the fluorophore residue. By taking advantages of the specific location of sole Trp residue located at each E-helix of SSFH-2, the thermostability of SSFH-2 was studied by fluorescence titration at pH 7.9, 25 °C as a function of temperature (Fig. S2†). SSFH-2 exhibited a strong fluorescence peak at 324 nm when excited at 295 nm, indicating that most of the observed fluorescence was attributed to the Trp residue. Upon thermal treatment of SSFH-2 in the temperature range of 60 °C to 100 °C, the fluorescence intensity of SSFH-2 was just slightly decreased. These findings suggested that SSFH-2 is a hyperthermostable protein, which is able to resist high temperature such as 100 °C. To confirm this idea, we fixed treatment temperature at 100 °C and then heated protein for different times (5, 10, 20, and 30 min). SDS and native PAGE results showed that there is no change in molecular weight (MW) of subunit and the overall protein for all samples (Fig. 2a and b), indicating that protein primary structure is kept well. Consistent with this observation, CD and fluorescent analyses revealed that the secondary and tertiary structures kept almost unchanged upon thermal treatment of SSFH-2 at 100 °C for different times (Fig. 2c and d). Native ferritin usually consists of 24 subunits assembled to form a hollow spherical structure which can be observed by TEM. To probe the impact of thermal treatment on the morphology of SSFH-2, we visualized the morphology of SSFH-2 by TEM after protein was treated for 30 min at 100 °C. As revealed by TEM, SSFH-2 appeared as discrete protein cages with the exterior diameter of around 12 nm (Fig. 2e), which exhibited the same morphology as reported for other ferritin nanocage.14,18 And DLS analyses stated clearly that both untreated and thermal-treated SSFH-2 showed one population with the approximately equal hydrodynamic radius (RH) in solution (Fig. 2f). These findings approved the above view, demonstrating that SSFH-2 is a hyperthermostable protein nanocage for the first time. Support for this conclusion came from the observation that, similar to animal ferritin,14,18 SSFH-2 protein nanocage still possesses reversible disassembly and reassembly property controlled by pH upon its thermal treatment for 30 min at 100 °C. For example, TEM analyses showed that pH 2.0 induced thermal-treated SSFH-2 disassembly into subunits (Fig. S3a–c†), while these disassociated subunits reassembled into protein nanocage when pH was adjusted back to 7.9. These results were confirmed by DLS analyses (Fig. S3d–f†). Bear in mind that SSFH-2 is also an enzyme which is responsible for fast Fe2+ oxidation by O2.20,22 Enzymatic activity analyses showed that the initial rate of Fe2+ oxidation catalyzed by untreated SSFH-2 is nearly identical to that of thermal treated SSFH-2 (Fig. S4†), further demonstrating that SSFH-2 is hyperthermostable, the Tm of which is higher than 100 °C.
Fig. 2. Characterization of the structure of SSFH-2 after thermal treatment at 100 °C for different times. (a) SDS-PAGE and (b) native PAGE analyses of thermal-treated SSFH-2. Lane M, protein markers and their corresponding molecular masses; lanes 1, 2, 3, 4, and 5 correspond to SSFH-2 upon treated at 100 °C for 0, 5, 10, 20, and 30 min, respectively. (c) CD spectra and (d) Fluorescence spectra of SSFH-2 upon thermal treatment for 5, 10, 20, and 30 min. (e) TEM image of SSFH-2 at 100 °C for 30 min. Scale bar represent 100 nm. (f) Dynamic light scattering (DLS) analyses of untreated SSFH-2 and upon thermal treatment at 100 °C for 30 min. Conditions: 1.0 μM SSFH-2 in 50 mM MOPS, pH 7.9.
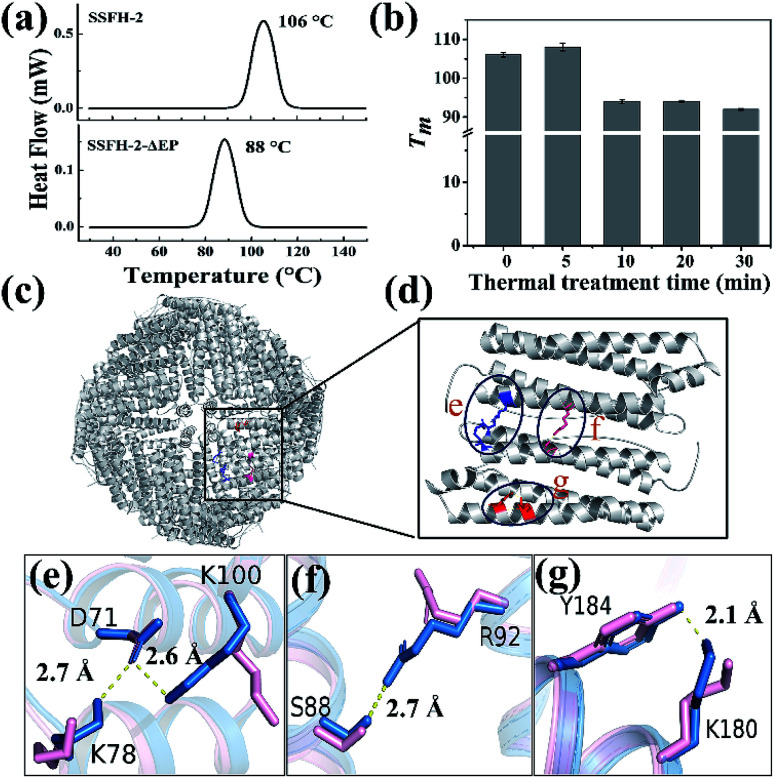
However, when SSFH-2 was heated at 121 °C (such high temperature has been used for sterilization during food processing) for 5 min, its fluorescence intensity decreased sharply, accompanied by a red shift of maximum emission wavelength, suggesting that protein nanocage was denatured (Fig. S5a†). Indeed, TEM analyses showed that most of inherent protein nanocages disappeared; instead, denatured protein filaments were formed (Fig. S5b†). Thus, it appears that the Tm of SSFH-2 should be between 100 and 121 °C. To corroborate this interpretation, differential scanning calorimeter (DSC) was used to directly measure the Tm of SSFH-2. As expected, we found that the Tm of SSFH-2 is around 106 °C (Fig. 3a).
Fig. 3. The structural basis of the hyperthermostability of SSFH-2. (a) DSC curves of SSFH-2 and its EP-deleted mutant SSFH-2-ΔEP, at scan rate of 10 °C min−1. (b) The thermal stability of SSFH-2 as a function of treatment time at fixed temperature 100 °C. (c) The crystal structure of SSFH-2 upon treated at 100 °C for 5 min, which is viewed down the 4-fold channels (PDB ID: 6J4M). (d) The position of newly formed noncovalent interactions upon thermal treatment. (e–g) Superposition of untreated (pink) and thermal-treated (blue) SSFH-2 subunit revealed the formation of several new types of new noncovalent interactions upon treated at 100 °C for 5 min.
Structural basis of the hyper-thermostable SSFH-2
Attempts to identify the factors responsible for the high thermostability of SSFH-2 have included structural comparisons of homologous proteins. The major difference in structure between plant ferritin and animal ferritin is that plant ferritin has the extra peptide at the N-terminal while animal ferritin lacks,20,29 so we envisioned that this peptide could play an important role in maintaining the high thermal stability of plant ferritin. To test this hypothesis, we made a mutant named SSFH-2-ΔEP where the extra peptide has been removed by genetic modification. Gene encoding the designed protein was constructed and cloned into an expression vector. After E. coli cells expressing the proteins were lysed, and resulting proteins were purified according to our reported method. SDS and native PAGE analyses revealed that this mutant has nearly the same electrophoretic behavior as SSFH-2, suggesting that it also has a similar 24-mer protein assembly to native SSFH-2 (Fig. S6a†). This conclusion was validated by TEM observation, showing that this mutant has the ability to form a shell-like protein nanocage (Fig. S6b†). Thus, deletion of the extra peptide domains seems to have no effect on the shell-like protein structure of plant ferritin. However, DSC analyses showed that the Tm of SSFH-2-ΔEP decreased to 88 °C (Fig. 3a). The difference in Tm between native SSFH-2 and its mutant confirmed the above view that the hyperthermostability of SSFH-2 originates from its extra peptide domains. This result is consistent with previous reports for an extra human glucagon at N terminus of ferritin.30
Surprisingly, we also found that the thermostability of SSFH-2 protein nanocage can be slightly enhanced by thermal treatment of protein for 5 min based on the fact that the Tm of SSFH-2 was increased from 106 to 108 °C, whereas prolonged thermal treatment of SSFH-2 (≥10 min) resulted in a decrease in its Tm as shown in Fig. 3b. To shed light on the reason why the stability of SSFH-2 can be increased with 5 min thermal treatment, we directly used the thermal-treated SSFH-2 (TTH-2) to screen for their suitable crystallization conditions by using commercially available protein crystallization screens, and eventually we found conditions which are suitable for the growth of crystals. We solved the crystal structure of TTH-2 at 2.6 Å (I23 space group, PDB ID: 6J4M) (Table S2†). Although the shell-like protein nanocage was kept intact in TTH-2 (Fig. 3c), the structure of its subunit is different from that of untreated ferritin in that several types of new noncovalent interactions were generated within subunit upon thermal treatment (Fig. 3d). For example, thermal treatment caused both Lys100 and Lys78 to move close to D71, consequently producing two salt bridges within subunit as shown in Fig. 3e. Additionally, both a new hydrogen bond between Ser88 and Arg92 (Fig. 3f) and polar π interaction between Tyr184 and Lys180 (Fig. 3g) were also formed within subunit due to thermal treatment. These newly formed noncovalent interactions gave a good explanation at atomic resolution for the increased Tm of TTH-2. Meanwhile, these results indicated that the subunit interactions are the most important ones in determining the stability of the assembled molecules.31
Enhanced thermostability of HuHF
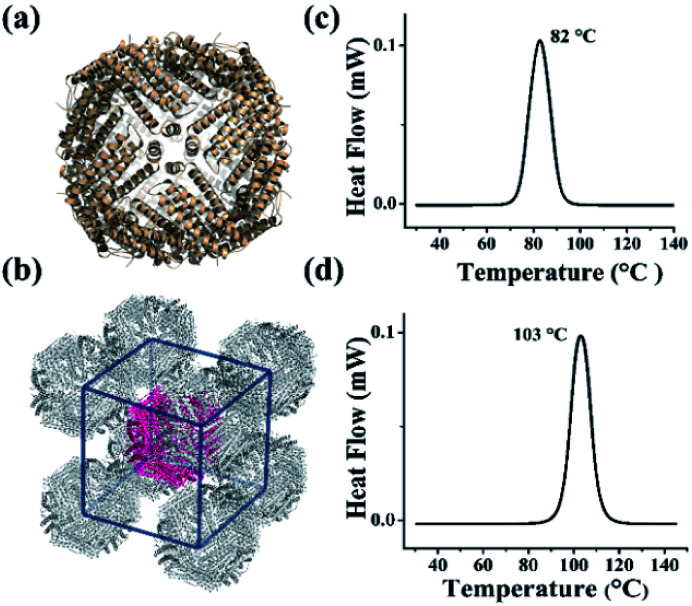
The above results demonstrated that the extra peptide in plant ferritin plays an important role in controlling protein thermostability. We wonder whether such extra peptide can also be utilized to improve the thermostability of animal ferritin such as HuHF because it has emerged as a popular biotemplate or vehicle in the field of nanotechnology and nanomedicine.14–19 To this end, we prepared a HuHF mutant termed HuHF-∇EP where EP was connected to the N-terminal of HuHF subunit by genetic modification. After E. coli cells expressing this mutant were lysed, we purified it to homogeneity as suggested by SDS and native PAGE (Fig. S7a and b†). The migration pattern for HuHF-∇EP on native PAGE was different from that of HuHF, indicating that both charge and mass were changed due to the insertion of EP peptide, which confirmed the successful preparation of HuHF-∇EP.32 Moreover, the iron loading ability of HuHF was improved by EP grafting (Fig. S7c†), which is consistent with our previous report that EP play a role in iron oxidation.33 To elucidate the effect of EP on the structure of HuHF, we found experimental conditions suitable for the growth of HuHF-∇EP crystals, and then solved its crystal structure (PDB IB: 6J4A). The inserted EP domain cannot be observed from the crystal structure, reflecting its high flexibility when it was fused to the N-terminal of ferritin subunit that is located on the exterior surface of protein nanocage closing to the 3-fold channels. However, the crystal structure revealed that the shell-like structure characteristic of ferritin nanocage keeps well (Fig. 4a), indicating that the EP domain hardly affects the structure of ferritin. Interestingly, the packing pattern of HuHF-∇EP in crystals is different from that of native HuHF, namely, native ferritin molecules tend to arrange in a face-centered-cubic (FCC) in crystals (Fig. S8†), while HuHF-∇EP can assemble into a body-centered-cubic (BCC) lattice (Fig. 4b). We believe that such difference in the packing pattern comes from the EP domain. Subsequently, we determined the thermostability of HuHF-∇EP by DSC with native HuHF as control under the same experimental conditions as plant ferritin. As expected, we found that the Tm of native HuHF was only 82 °C (Fig. 4c), a value being much lower than that of SSFH-2. Excitingly, the graft of the EP domain on HuHF protein nanocage markedly improved its Tm to 103 °C (Fig. 4d). Agreeing with this finding, native PAGE and TEM analyses revealed that the protein nanocage of native HuHF was damaged to a large extent upon thermal treatment at 100 °C for 30 min, whereas the nanocage structure of HuHF-∇EP remained unchanged under the same experimental conditions (Fig. S9†). The significant improvement of human ferritin nanocage in the thermal stability validated our design.
Fig. 4. The crystal structure and thermal stability of HuHF and EP-inserted mutant (HuHF-∇EP). (a) The crystal structure of HuHF-∇EP viewed down the 4-fold channels (PDB ID: 6J4A). (b) The body-centered-cubic (BCC) packing arrangement of HuHF-∇EP crystals. (c and d) DSC curves of native HuHF (c) and its mutant HuHF-∇EP (d), at scan rate of 10 °C min−1.
Conclusions
So far, most of known hyperthermostable proteins or enzymes have been isolated from hyperthermophilic organisms such as archaea and bacteria that thrives in extremely hot environments.34,35 In contrast, such hyperthermostable protein from animal and plant sources is rare. Herein, we found that SSFH-2 has high thermostability i.e. Tm of 106 °C, which may represent a new family of proteins with high thermostability. More importantly, we demonstrated that the hyperthermostability of SSFH-2 is originated from its EP domain, a specific peptide only occurring in mature plant ferritin nanocage. By taking advantage of the contribution of the EP domain to the stability of plant ferritin, we grafted the EP domain on the exterior surface of human ferritin nanocage, leading to a marked increase in its Tm, which represents a rational design for the creation of a high stable protein architecture. The development of protein nanocage templates that are stable under extreme conditions of heat will expand processing conditions and end-use applications for biomaterials that require exceptional stability and robustness.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31730069 and 31671805). The Shanghai Synchrotron Radiation Facility (SSRF) is especially acknowledged for beam time. We thank the staffs from BL17U1/BL18U1/19U1 beamline of National Center for Protein Sciences Shanghai (NCPSS) at Shanghai Synchrotron Radiation Facility for assistance during data collection, and the staffs in Tsinghua University Branch of China National Center for Protein Sciences Beijing for technical assist. We thank Masuda Taro for generously providing the SSFH-2 plasmids.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra04785a
Notes and references
- Royle S. J. Cell. Mol. Life Sci. 2006;63:1823–1832. doi: 10.1007/s00018-005-5587-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S. Kerfeld C. A. Sawaya M. R. Cai F. Heinhorst S. Cannon G. C. Yeates T. O. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- Canady M. A. Larson S. B. Day J. McPherson A. Nat. Struct. Mol. Biol. 1996;3:771–781. doi: 10.1038/nsb0996-771. [DOI] [PubMed] [Google Scholar]
- Rother M. Nussbaumer M. G. Renggli K. Bruns N. Chem. Soc. Rev. 2016;45:6213–6249. doi: 10.1039/C6CS00177G. [DOI] [PubMed] [Google Scholar]
- Douglas T. Young M. Science. 2006;312:873–875. doi: 10.1126/science.1123223. [DOI] [PubMed] [Google Scholar]
- Wörsdörfer B. Woycechowsky K. J. Hilvert D. Science. 2011;331:589–592. doi: 10.1126/science.1199081. [DOI] [PubMed] [Google Scholar]
- Wörsdörfer B. Pianowski Z. Hilvert D. J. Am. Chem. Soc. 2012;134:909–911. doi: 10.1021/ja211011k. [DOI] [PubMed] [Google Scholar]
- Dickerson M. B. Sandhage K. H. Naik R. R. Chem. Rev. 2008;108:4935–4978. doi: 10.1021/cr8002328. [DOI] [PubMed] [Google Scholar]
- Haskar S. Lim S. NPG Asia Mater. 2017;9:e371. doi: 10.1038/am.2016.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasteen N. D. Harrison P. M. J. Struct. Biol. 1999;126:182–194. doi: 10.1006/jsbi.1999.4118. [DOI] [PubMed] [Google Scholar]
- Bou-Abdallah F. Biochim. Biophys. Acta, Gen. Subj. 2010;1800:719–731. doi: 10.1016/j.bbagen.2010.03.021. [DOI] [PubMed] [Google Scholar]
- Arosio P. Ingrassia R. Cavadini P. Biochim. Biophys. Acta, Gen. Subj. 2009;1790:589–599. doi: 10.1016/j.bbagen.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Li L. Fang C. J. Ryan J. C. Niemi E. C. Lebrón J. A. Björkman P. J. Arase H. Torti F. M. Torti S. V. Nakamura M. C. Seaman W. E. Proc. Natl. Acad. Sci. U. S. A. 2010;107:3505–3510. doi: 10.1073/pnas.0913192107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida M. Kang S. Reichhardt C. Harlen K. Douglas T. Biochim. Biophys. Acta, Gen. Subj. 2010;1800:834–845. doi: 10.1016/j.bbagen.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M. Fan K. Zhou M. Duan D. Zheng J. Yang D. Feng J. Yan X. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14900–14905. doi: 10.1073/pnas.1407808111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Z. Tang W. Guo C. Chen H. Lin X. Liu G. Fei B. Chen X. Xu B. Xie J. ACS Nano. 2013;7:6988–6996. doi: 10.1021/nn402199g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P. Rong P. Jin A. Yan X. Zhang M. G. Lin J. Hu H. Wang Z. Yue X. Li W. Niu G. Zeng W. Wang W. Zhou K. Chen X. Adv. Mater. 2014;26:6401–6408. doi: 10.1002/adma.201400914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Bai G. Yang R. Zang J. Zhou T. Zhao G. Food Chem. 2014;149:307–312. doi: 10.1016/j.foodchem.2013.10.115. [DOI] [PubMed] [Google Scholar]
- Conti L. Lanzardo S. Ruiu R. Cadenazzi M. Cavallo F. Aime S. Crich S. G. Oncotarget. 2016;7:66713–66727. doi: 10.18632/oncotarget.10920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G. Biochim. Biophys. Acta, Gen. Subj. 2010;1800:815–823. doi: 10.1016/j.bbagen.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Fu X. Deng J. Yang H. Masuda T. Goto F. Yoshihara T. Zhao G. Biochem. J. 2010;427:313–321. doi: 10.1042/BJ20100015. [DOI] [PubMed] [Google Scholar]
- Masuda T. Goto F. Yoshihara T. Mikami B. Biochem. Biophys. Res. Commun. 2010;400:94–99. doi: 10.1016/j.bbrc.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Wang J. Xia N. Yang X. Yin S. Qi J. He X. Yuan D. Wang L. J. Agric. Food Chem. 2012;60:3302–3310. doi: 10.1021/jf205128v. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Yang H. Fu X. Li M. Leng X. Chen B. Zhao G. Plant Physiol. 2010;154:1481–1491. doi: 10.1104/pp.110.163063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. Wang C. Hong Y. Zhang M. Su M. Anal. Chem. 2010;82:1186–1190. doi: 10.1021/ac902724y. [DOI] [PubMed] [Google Scholar]
- Deng J. Liao X. Yang H. Zhang X. Hua Z. Masuda T. Goto F. Yoshihara T. Zhao G. J. Biol. Chem. 2010;285:32075–32086. doi: 10.1074/jbc.M110.130435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. Zhang K. Cui Y. Wang Z. Pan Q. Liu K. Sun B. Zhou H. Li M. Xu Q. Xu C. Yu F. He J. Nucl. Sci. Tech. 2018;29:3–9. doi: 10.1007/s41365-017-0339-z. [DOI] [Google Scholar]
- Liao X. Yun S. Zhao G. Crit. Rev. Food Sci. Nutr. 2014;54:1342–1352. doi: 10.1080/10408398.2011.635914. [DOI] [PubMed] [Google Scholar]
- Kim S. W. Kim Y. H. Lee J. Biochem. Biophys. Res. Commun. 2001;289:125–129. doi: 10.1006/bbrc.2001.5931. [DOI] [PubMed] [Google Scholar]
- Stefanini S. Cavallo S. Wang C. Q. Tataseo P. Vecchini P. Giartosio A. Chiancone E. Arch. Biochem. Biophys. 1996;325:58–64. doi: 10.1006/abbi.1996.0007. [DOI] [PubMed] [Google Scholar]
- Subhadarshanee B. Mohanty A. Jagdev M. K. Vasudevan D. Behera R. K. Biochim. Biophys. Acta, Proteins Proteomics. 2017;1865:1267–1273. doi: 10.1016/j.bbapap.2017.07.012. [DOI] [PubMed] [Google Scholar]
- Li C. Fu X. Qi X. Hu X. Chasteen N. D. Zhao G. J. Biol. Chem. 2009;284:16743–16751. doi: 10.1074/jbc.M109.011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild L. J. L Mancinelli R. Nature. 2001;409:1092–1101. doi: 10.1038/35059215. [DOI] [PubMed] [Google Scholar]
- Aguilar C. F. Sanderson I. Moracci M. Ciaramella M. Nucci R. Rossi M. Pearl L. H. J. Mol. Biol. 1997;271:789–802. doi: 10.1006/jmbi.1997.1215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.