Abstract
Background
Detecting SARS-CoV-2 using a simple real time molecular assay will be helpful for the mitigation efforts in low / middle income countries during the pandemic. We have developed and validated a rapid and simple real time loop mediated isothermal amplification assay (LAMP) for screening of SARS-CoV-2 infection in known infected and non-infected individuals.
Methods
Six sets of primers were designed targeting the N-gene of the SARS-CoV-2 (Accession ID MN994468). LAMP reactions were performed using Warm Start 2X Master Mix and real-time PCR machine at 65 °C for 60 cycles with 15 s for each cycle. Results were read by visualizing turbidity under ultraviolet light and real time fluorescence detection through FAM channel of the real time PCR machine. We tested a total of 320 including 240 SARS CoV-2 positive (Ct values <40) and 80 SARS CoV-2 negative samples as tested by a real time RT-PCR using the newly developed LAMP assay.
Results
A total of 206 out of 240 SARS CoV-2 positive samples were tested positive by the newly developed LAMP assay with a sensitivity of 86%. All 80 SARS CoV-2 negative samples were tested negative by the newly developed LAMP assay with a specificity of 100%.
Conclusion
The newly developed real time LAMP assay has a sensitivity of 86% and specificity of 100% compared to the real time RT-PCR for the detection of SARS CoV-2. The new assay will be useful to screen large number of samples if adopted to minimize the time and cost.
Keywords: SARS-CoV-2, Loop mediated amplification, rtRT-PCR, Sensitivity, Specificity
Abbreviations: SARS CoV-2, Severe acute respiratory syndrome coronavirus-2; rtRT-PCR, Real time reverse transcription polymerase chian reaction
1. Introduction
Covid-19 has posed unprecedented challenges to societies all over the world. It is a serious public health issue that requires simple diagnostic methods in detecting and preventing infections in different stages of the pandemic. The Covid-19 caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has been identified as the ‘20th century Spanish flu’ affecting the whole world. In mitigating the efforts to combat, developing a rapid and cost-effective diagnostic approach enabling the detection of the infection in the affected as well as asymptomatic individuals will be useful to assist preventive measures in resource limited settings. For the detection of SARS-CoV-2, the WHO and CDC recommend the use of nucleic acid based diagnostic methods, which are rapid and confirmatory [1].
Detecting the SARS-CoV-2 infection is important to assist preventive measures in affected areas. Numerous methods have been developed to detect SARS-CoV-2 infection and the commonly used method of laboratory diagnosis of Covid-19 is real time reverse transcription polymerase chain reaction (real time RT-PCR). Currently used real time RT-PCR assays have the advantage of accurate detection of the virus, however, the supply chain related issues and reporting time of more than 4 h might have a negative impact on the mitigating efforts. Real time RT-PCR assays are not cost effective for resource limited countries to continue to use these assays for screening large community samples [2]. Reverse transcription-loop mediated isothermal amplification (RT-LAMP) has been widely studied as an alternative. RT-LAMP of genomic sequence of interest, is a rapid and cost-effective method for the detection of pathogens and it has been used for the detection of dengue virus and pathogenic leptospirae in resource limited settings [3], [4], [5]. This method can amplify DNA or RNA in a single isothermal reaction from a few copies of DNA to 109 in less than an hour under isothermal conditions and is highly specific and sensitive with a constant temperature between 60 and 65 °C [6,7]. RT-LAMP assays use a “rolling hairpin” mechanism to allow amplification at a constant temperature using polymerase enzymes different from those used in PCR. This targeted isothermal nucleic acid amplification utilizes a combination of 4–6 primer sets and a DNA polymerase with high strand displacement activity to specifically replicate a region of DNA [8, 9].
Since the emergence of Covid-19, multiple primer sets for the detection of SARS-CoV-2 for the RT-LAMP have been published and some have received emergency use authorization by the Food and Drug Administration of the USA [10], [11], [12]. RT-LAMP workflow delivers results faster than real time RT-PCR. However, RT-LAMP reactions often result in non-specific amplification in the absence of the target, particularly at longer reaction times, limiting the sensitivity. This off-target amplification is problematic because LAMP reactions are commonly quantified using colorimetric dyes for a longer duration [13, 14]. According to a recent study, frequency of testing and speed of reporting are more important than the assay sensitivity for reducing the transmission of the virus [15].
In this study, we report areal time RT-LAMP for detecting SARS-CoV-2. We developed, validated and made a simple cost analysis to promote the use of this real time RT-LAMP for the detection of SARS-CoV-2 in resource limited settings. This assay can be used to screen symptomatic Covid-19 suspected as well as asymptomatic exposed individuals.
2. Materials and methods
2.1. Diagnostic samples
Ethical approval for use of archived samples was requested from the Ethics Review Committee of the Faculty of Medicine, University of Peradeniya, Sri Lanka. The committee exempted the study as only the de-identified samples were used for the optimization and validation of the assay (Exemption Protocol No: 2021/EC/46).
Our laboratory is one of the laboratories for testing for SARS-CoV-2 in collaboration with Ministry of Health (MoH), Sri Lanka. Samples sent for routine SARS-CoV-2 real time RT-PCR testing to our laboratory and the Diagnostic and Reference Virology Laboratory of National Hospital, Kandy were used for optimizing and validating the newly developed real time RT-LAMP assay. From November 2020 to March 2021, during the SARS-CoV-2 pandemic, a total of 320 RNA samples comprising 240 SARS-CoV-2 positive and 80 SARS-CoV-2 negative throat swab-based RNA extracts were tested by the newly developed real time RT-LAMP assay. A total of 210 out of 240 positives samples used for the validation of the assay had Ct value <25 and 30 positive samples had Ct values between 25 and 40.
To estimate the sample size required for testing by the real time RT-LAMP assay, the following formula was used - S = Z2 P(1-P)/d2. In here, ‘P’ is the percentage of the interested variable, and ‘d’ is the accepted level of error at 95% Confidence Interval [16]. The percentage of the interested variable for this study was considered as 24.5% based on the Covid-19 status in the country at the time of the study [17]. Taken these conditions, the calculated sample size was 284. Ten percent of the required sample was added for non-responses and thus final sample size was 312, however, we have tested 320 samples.
2.2. Viral RNA extraction and real time RT-lamp primers
Viral RNA was extracted using SpinStar™Viral Nucleic Acid Kit 1.0 (ADT Biotech sdnbhd, Malaysis) and BioFluxBiospin Virus DNA/RNA Extraction Kit (Hangzhou Bioer Technology Co., Ltd, China) at the Research and Diagnostic Virology Laboratory of the Department of Microbiology, Faculty of Medicine, University of Peradeniya, Sri Lanka and the Diagnostic and Reference Virology Laboratory of National Hospital, Kandy, Sri Lanka.
Six pairs of primers were designed to amplify the N gene of the virus with a loop formation as the read out. The homology of N gene was checked with other viruses like influenza virus, Bat-SL CoV, hCoV and SARS CoV using NCBI BLASTn. Mutations of primer regions were assessed by aligning all the available N gene sequences of the SARS-CoV-2 using ClustalWBio tool. The primer set consisted of one Forward Inner Primer (FIP) and Backward Inner Primer (BIP), a set of Forward and Backward outer primers (F3 and B3) and a set of Forward Loop (LF) and Backward Loop (LB) primers. Viral RNA was amplified using the real time LAMP with conditions following reverse transcription. The assay was performed in duplicates to optimize and to ensure reproducibility when necessary in the laboratory through different scientists / technologists. The primers (IDT, USA) corresponding to the N-gene of the SARS-CoV-2 are given in Table 1 .
Table 1.
Primers used to target the N gene of the SARS-CoV-2 in the new real time RT-LAMP.
| Target gene | Primers | Primer sequence |
|---|---|---|
| N-Gene | F3 | ACCGAAGAGCTACCAGACG |
| B3 | TGTAGCACGATTGCAGCATT | |
| FIP | TCTGGCCCAGTTCCTAGGTAGTAATTCGTGGTGGTGACGGTA | |
| BIP | AGACGGCATCATATGGGTTGCAGCGGGTGCCAATGTGATC | |
| LF | CCATCTTGGACTGAGATCTTTCAT | |
| LB | ACTGAGGGAGCCTTGAATACA |
2.3. Real time reverse transcription PCR and real time RT-lamp assays
All samples used had been tested by a real time RT-PCR kit (Real Star, Altona Diagnostics, Germany). This real time RT-PCR assay detects E and S genes and the assay was carried out according to the manufacturers’ instructions. Briefly, a 30 μL total reaction mixture containing 21.5μL detection mixture and 8.5μL target RNA template was mixed in a reaction tube. Then the assay was performed (45 °C for 10 min, 95 °C for 3 min, 95 °C for 15 s,58 °C for 30 s, 45 cycles) in CFX-96 real time PCR detection system (Bio-Rad, Mississauga, ON).
A single-target LAMP reaction was conducted using WarmStart LAMP kit (DNA & RNA), which has a combination of WarmStartRTx Reverse Transcriptase with Bst 2.0 WarmStart DNA Polymerase (New England Bio Labs). The assay was performed in a 25 μL LAMP reaction mixture containing 2.5μLof 10X LAMP primer mix (16 μM F1P and B1P, 4 μM LPF and LPB, 2 μMF3 and B3), 12.5 μL of Warm Start LAMP 2X Master mix, 1.5 μL of DMSO, 3 μL of nuclease free water and 0.5μL of 50X fluorescence dye supplied by the WarmStart LAMP kit and 5 μL of template. Amplification was measured through relative fluorescence units (RFU) per cycle using the same CFX-96 real time PCR machine at 65 °C for 60 cycles with 15 s for each cycle with an amplification time of 30 min.
2.4. Accuracy indices and cost analysis for the new real time RT-lamp assay
The newly developed real time RT-LAMP assay was validated using the real time RT-PCR as a comparator for the detection of SARS-CoV-2. The accuracy indices (sensitivity, specificity, positive predictive value (PPV) and the negative predictive value (NPV) were calculated (formulae given below) in comparison with the real time RT-PCR. Based on the results, a binary logistic regression model was developed to compare the real time RT-PCR Ct values (Predictor variable) and the LAMP results (Outcome). These estimates were compared with claims or internal criteria to judge the acceptability of the method. Receiver operating characteristic curve (ROC) was used to study the performance of the new real time RT-LAMP assay.
TP - True positives; TN -True negatives; FP - False positives; FN - False negatives
A cost analysis was performed for the newly developed real time RT-LAMP assay taking all costs including the equipment use, utilities, manpower and reagents.
3. Results
3.1. Detection of viral RNA by the new real time RT-lamp assay
In this study, we detected the amplification of viral RNA through RFU per cycle at 65 °C for 60 cycles with 15 s per cycle (∼ 30 min). Whenever real time amplification was noted under these conditions, the results were taken as SARS CoV-2 RNA detected by the new real time RT-LAMP assay. Whenever real time amplification was not noted under these conditions, the results were taken as SARS CoV-2 RNA not detected by the new assay. Additionally, results were read by visualizing turbidity under UV light and naked eye.
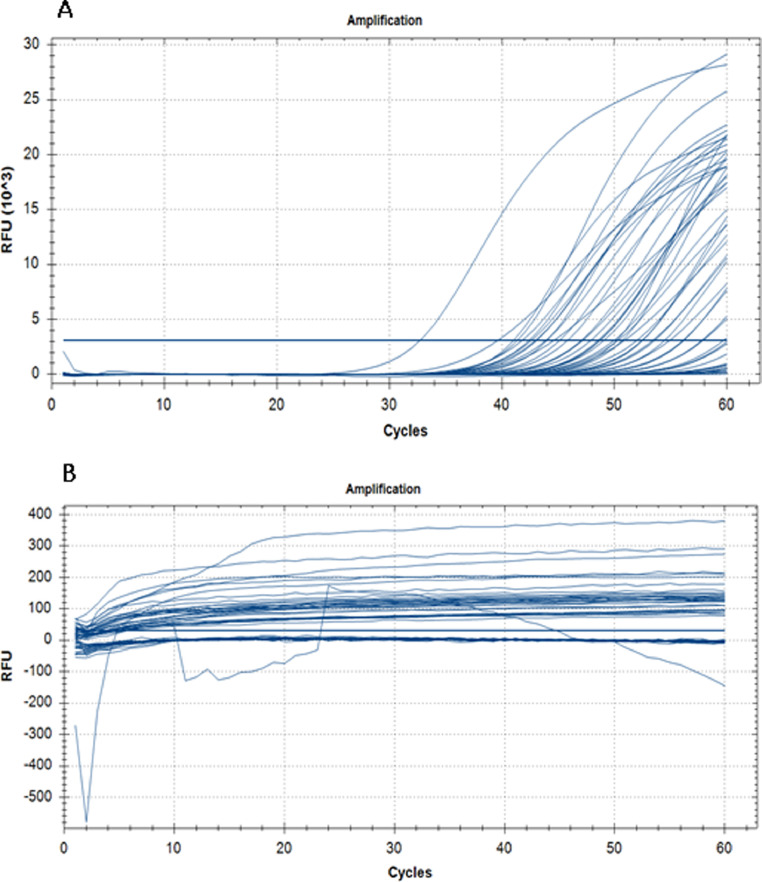
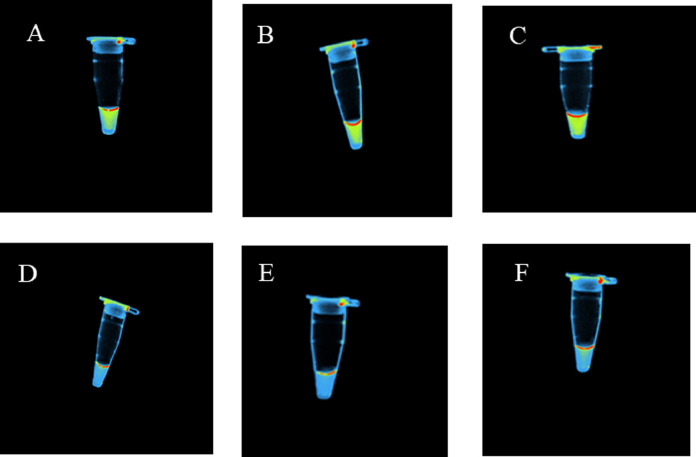
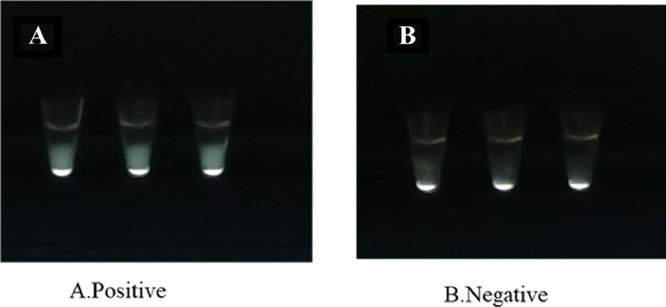
Presence of viral RNA was detected within 30 min compared to the real time RT-PCR, which took a reaction time of 2 h (Fig. 1 A). All negative samples detected by the real time RT-PCR were also negative by the newly developed real time RT-LAMP (Fig. 1B). Furthermore, visualization of these samples under UV light using a gel documentation system also gave similar results (Fig. 2 ). These samples were also visualized through naked eye for turbidity as instructed by the manufacturers (Fig. 3 ). All three read outs (real time RFU, UV light and naked eye turbidity) for the newly developed real time RT-LAMP agreed with each other.
Fig. 1.
Amplification of viral RNA detected through real time relative fluorescence units (RFU) per cycle. A - Amplification of the positive samples at 65 °C for 60 cycles with 15 s per cycle (30 min). B - Amplification of the negative samples under similar conditions.
Fig. 2.
Images of SARS CoV-2 positive and negative samples with multi-colour filter (UV light) in a gel documentation system (Applied Bio system). Positive sample showed a yellow colour (Tubes A, B & C) whereas the negative samples did not show yellow colour (Tubes D, E & F).
Fig. 3.
SARS-CoV-2 RNA detected by direct visualization for the presence and absence of turbidity. A - Positive samples showing turbidity. B -Negative samples not showing turbidity.
3.2. Sensitivity, specificity, ROC curve, PPV and NPV of the real time RT-LAMP
A total of 206 out of 240 SARS CoV-2 positive samples were tested positive by the newly developed real time RT-LAMP with a sensitivity of 85.8%. All 80 SARS CoV-2 negative samples were tested negative by the new LAMP with a specificity of 100%. A head-to-head comparison with the real time RT-PCR (Real Star, Altona Diagnostics, Germany) for the detection of SARS CoV-2 is summarized in Table 2 .
Table 2.
A comparison of real time RT-PCR and RT-LAMP for the detection of SARS CoV-2.
| Real time RT-PCR (Real Star, Altona Diagnostics, Germany) | ||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
|
Newly developed real time RT-LAMP assay |
Positive | 206 | – | 206 |
| Negative | 34 | 80 | 114 | |
| Total | 240 | 80 | 320 | |
Results obtained from the binary logistic regression model, which compared the real time RT-PCR Ct values and the RT-LAMP results for the detection of SARS-CoV-2 are summarized in Table 3 .
Table 3.
Results of the binary logistic regression model comparing the real time RT-PCR Ct values (Predictor variable) and the LAMP results (Outcome) for the detection of SARS CoV-2.
| Binary logistic regression model details | Values |
|---|---|
| Coefficient of RT-PCR Ct values | −0.242 |
| Standard error of coefficient RT-PCR Ct values | 0.035 |
| Odds ratio* | 0.78 |
| Constant | 7.589 |
| Standard error of constant | 0.918 |
| Chi-square p value | <0.05 |
| % Correct | 95.1 |
| Hosmer–Lemeshow p value | 0.454 |
| Sensitivity | 85.8% |
| Specificity | N/A |
| ROC area | 0.9 |
OR = Odds Ratio is coded as 1 for ‘yes’ and 0 for ‘no’.
The binary logistic regression model was significant (model X2, p<0.05) and fitted well (Hosmer & Lemeshow p value >0.05). A negative relationship was observed between the predictor and the real time RT-LAMP results indicating that the LAMP event becomes more likely as the predictor (RT-PCR Ct value) decreases. The newly developed real time RT-LAMP assay had a negative predictive value (NPV) of 0.701 (70%) and a positive predictive value (PPV) of 1(100%) for all samples tested.
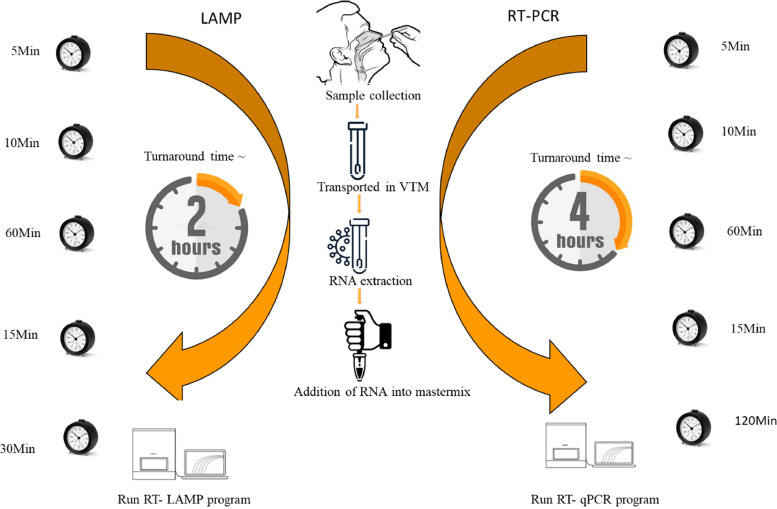
3.3. Rapid turnaround time
The workflow of the new real time RT-LAMP assay took 2 h from sample collection to reporting of the results compared to the real time RT-PCR, which took 4 h (Fig. 5).
Fig. 5.
Comparison of turnaround time for real time RT-LAMP and real time RT-PCR for the detection of SARS CoV-2.
3.4. Cost comparison between real time RT-lamp assay and real time RT-PCR
The newly developed real time RT-LAMP assay is a less expensive alternative molecular test to real time RT-PCR for detection of SARS-CoV-2 for large scale screening in resource limited settings (Table 4 ). Highly suspected real time RT-LAMP negative persons’ extracts can be subjected to real time RT-PCR for confirmation.
Table 4.
Cost comparison between the new real time RT-LAMP assay and the real time RT-PCR.
| Assay | No. of test per run | Cost per run for RNA extraction ($) | Cost per run for test reagents ($) | Cost per run consumables & primers ($) | Cost for labour per run ($) | Cost per test ($) |
|---|---|---|---|---|---|---|
| Real time RT-LAMP | 96 | 120 | 307.2 | 20 | 3 | 8.45 |
| Real time RT-PCR | 96 | 120 | 720 | 20 | 5 | 14.75 |
4. Discussion
Considering the infectious nature of SARS-CoV-2 and the crisis unfolding in the developing countries the utility of real time RT-PCR is costly and there is a need for a cost effective and a rapid assay, which can be performed with 1/2 of the cost of the real time RT-PCR to ease the screening pressure. A cost effective and a real time rapid RT-LAMP assay was developed, optimized and validated using a real time RT-PCR as a comparator for the detection of SARS-CoV-2 ensuring reproducibility in the laboratory.
The study describes the development of one-step, real-time RT-LAMP assay for rapid detection of the N gene of SARS-CoV-2. Just as in a one-step real time RT-PCR, cDNA synthesis and follow-up amplification are achieved in a single tube by incubating the viral RNA with a primer mixture containing 6 primers in the presence of WarmStartRTx Reverse Transcriptase with Bst 2.0 WarmStart DNA Polymerase simultaneously at a constant temperature of 65 °C for ~̴ 30 min in a real time thermal cycler. Amplification by the RT-LAMP assay is based on the auto-cycling strand displacement DNA synthesis. In the initial steps of the RT-LAMP reaction, all the primers are used, but later, during the cycling reaction, only the inner primers are used for strand displacement DNA synthesis. The current RT-LAMP assay used one of three methods for the detection of virus amplification including real-time detection of amplification by fluorescence detection through FAM channel or post-amplification detection of turbidity under naked eye or UV using a gel documentation system. We measured the amplification of SARS-CoV-2 RNA through increased RFU per cycle in the CFX-96 real time PCR detection system as previously described by Mohon et al. [18]. RFU values above 3 × 1 03 on CFX-96 instrument were considered as positive when associated with the typical amplification curve below 60 cycles. All our negative samples had a RFU value < 400.
The new real time LAMP assay has a sensitivity of 86% for all the samples tested up to Ct ≤40. The new real time RT-LAMP assay has a specificity of 100% for detecting the true SARS-CoV-2 negative persons without false-positive results with any of the real time RT-PCR tested negative persons’ samples. Apart from sensitivity and specificity, predictive values such as NPV and PPV were also calculated to understand how well the new test is useful to diagnoses the SARS CoV-2 infection based on the results of the comparator. The NPV for the assay was 0.71 (70%) (n = 240; Ct<40), hence, the non-infected individuals might be moderately predicted by the negative results. Whereas the PPV in this study was 1(100%), hence, there is a high probability that a person with the SARS CoV-2 infection being predicted when the person has a positive test result. As the PPV and the NPV of a new test depend on the prevalence of the disease in the population, if the prevalence of the disease is high in a given population, PPV increases and NPV decreases. Thus, the results of predictive values are not fixed characteristics of the test and cannot be generalized across populations with different prevalence rate of an evolving infection. Sri Lanka experienced a massive outbreak of Covid-19 from mid-April 2021 onwards and a rapid increase in cases was noted with the spread of delta variant fuelling the increase in cases and deaths in the country [19]. During the study period a seroprevalence study conducted in the western province showed a high seroprevalence rate of (24.5%) [17]. By the 25th of March 2022, a total of 663,091 infections and 16,500 Covid-19-related deaths have been reported in Sri Lanka since the beginning of the pandemic based on the Ministry of Health.
Our results coincide with the time when the samples were taken for the validation of the assay, a period of increasing Covid-19 prevalence (November 2020 to March 2021). Similar findings have been reported with respect to sensitivity, specificity, NPV and PPV by others too when evaluated these parameters during the increasing prevalence rates for SARS CoV-2 infection ([18, 20]; Bektaşet al., 2021).
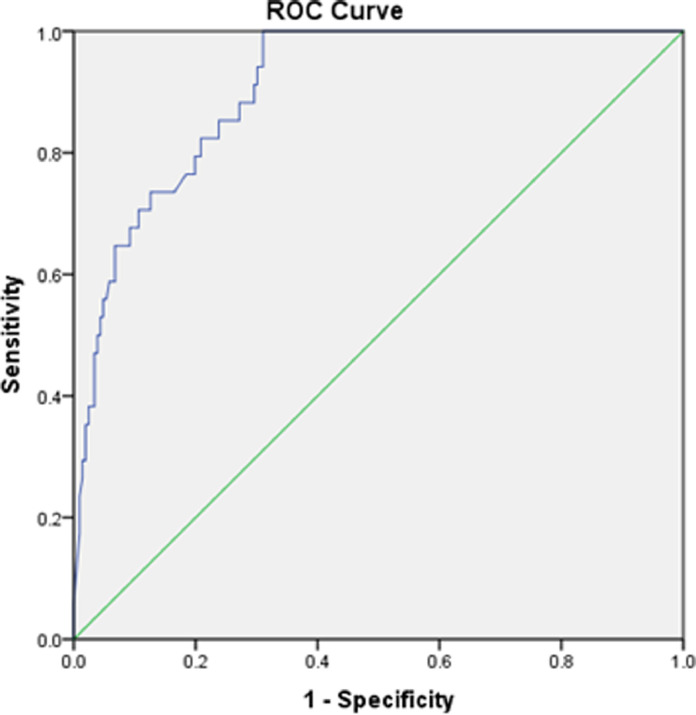
We then generated ROC curves for all samples tested (n = 240; Ct < 40) . The area under the curve (AUC) for the binary model was noted as 0.9. In general, an AUC of 0.5 suggests no discrimination, 0.7 to 0.8 is considered acceptable, 0.8 to 0.9 is considered excellent, and more than 0.9 is considered outstanding. Analysis by ROC curve confirmed a value of 0.9 for AUC for both models tested for the current results indicating the model is excellent (Table 3; Fig. 4 ).
Fig. 4.
Receiver operating characteristic curve (ROC curve) to study the performance of the newly developed LAMP assay for the detection of SARS CoV-2.
The current real time RT-LAMP has a turnaround time of 2 h from extraction to reporting and it is two times faster than the real time RT-PCR. The cost analysis provides evidence for the cost effectiveness of the real time RT-LAMP as a replacement for real time RT-PCR for SARS-CoV-2 detection. The average per test cost for the real time RT-LAMP was $8.45 and that of the real time RT-PCR was $14.75 when taking all related costs. Hence, the real time RT-LAMP is a less expensive alternative to real-time RT-PCR, suitable for use in resource limited countries. As most of the testing laboratories in resource limited countries including Sri Lanka have the real time reading systems established during the pandemic, the new assay can be used to screen samples in 2 h with more than 85% sensitivity and 100% specificity. Only the highly suspected selected negative samples can be subjected to real time RT-PCR saving the resources and time.
In this study, we noted 34 false negative tests for N gene target (n = 19 for Ct>25; n = 15 for Ct<25) and the majority of those were with higher Ct values. False negative tests for samples with Ct<25 (15/210) might be due to genetic polymorphism at key residues where the N gene target RT-LAMP primers bind. This issue could be overcome using two targets simultaneously as demonstrated by Mohon et al. [18]. False negative results of newly developed RT-LAMP for Ct >25 (19/30) might be due to the lower limit of detection (LOD) of the assay compared to the real-time RT-PCR used as the comparator. However, we could not enumerate the LOD for the RT-LAMP as our comparator was a semi-quantitative assay. Overall, getting false negative results in a screening assay for a highly infectious agent like SARS CoV-2 is a limitation of the assay. On the other hand, the real time RT-PCR used to validate the RT-LAMP targets E and S genes and the newly developed LAMP-based assay targets the N gene. This mismatch in the gene targets between the assays provides an indirect validation between the two technologies. However, it could also lead to the discrepancy with the sensitivity of the LAMP as we do not know if the standard RT-PCR assay would have had N-gene dropouts in LAMP negative samples. Ideally, the validation would have been done using the same target gene across both technologies. However, this mismatch was inevitable as this study was done with the public health workflow data and materials and the limited funding was not adequate to repeat assays with the same target.
LAMP assays have been reported to be highly susceptible to contamination [21, 22] and the recently reported SARS CoV-2 RT-LAMP assays also report such a problem. It is also difficult to avoid primer dimer formation and non-specific amplification when 4–6 primers are used. To overcome this, we have used 6% dimethyl sulfoxide (DMSO) in the final reaction mixture as recommended by previous LAMP based protocols [23]. DMSO in LAMP reaction mixtures increases amplification at low template concentration and inhibit Bst 2.0 WarmStart DNA polymerase activity at the latter part of the reaction. The latter decreases non-specific amplification and the current LAMP assay also used DMSO in the reaction mixture to minimize the non-specific amplification.
Improvements to the new developed real time RT-LAMP assay will enable robust, cost-effective and sensitive detection of SARS-CoV-2. A rapid extraction free colorimetric RT-LAMP may be used with our primers (Table 1) for a point of care analysis (POC) with minimal sophistication with a turnaround time of 40–60 min cutting off the RNA extraction process. Direct amplification from samples has also been reported for the detection of other RNA viruses, including African chikungunya virus [24], noroviruses [25] and bovine viral diarrhoea virus [26] using serum, throat swab and faecal samples. Though it is unlikely that DIRECT-PCR would completely replace conventional RNA extraction with real time RT-PCR in large diagnostic laboratories, ease of use and cost-effectiveness will be useful for patient screening and public health surveillance in resource limited countries [27, 28]. Extraction free colorimetric RT-LAMP for the prompt SARS-CoV-2 detection could address the challenges faced in resource limited countries in sustaining POC diagnostic tests. Moreover, POC diagnostics play an important role in the detection, diagnosis and control of the pandemic [29], [30], [31] as these provide a timely diagnosis in primary care facilities complementing the real-time RT-PCR.
In conclusion, the new real time RT-LAMP assay has a great potential to fill the technological gap, which hinders the large-scale testing as the assay enables nucleic acid detection with rapidity and cost-effectiveness. The new real time RT-LAMP would be an alternative test to screen large scale community samples in different parts of Sri Lanka and elsewhere.
Author contributions
Bushran N Iqbal: Performed bulk of the lab work, drafted & revised the manuscript
Shiyamalee Arunasalam: Sample scrutinization & performed lab work
Maduja V M Divarathna: Contributed to study design & performed lab work
AAOM Jabeer: Sample scrutinization & performed lab work
PDNN Sirisena: Contributed to study design & primer design
Thamarasi Senaratne: Contributed to study design & performed statistical analysis
Rohitha Muthugala: Contributed to study design & sample scrutinization
Faseeha Noordeen Designed the study, lead the project, acquired funding & critically revised the manuscript
Funding
This work was supported by National Research Council, Sri Lanka under Rapid response to COVID-19 (Grant No: NRC-RG-CVD-20–12).
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
We greatly appreciate the support from Dr. Jatin Shrinet from Florida state university USA for the help in designing the primers for the study. Support provided by Ms .Chalani Muthuwatta, Mr. AMSB Abeykoon and Mr.Sarath Ashoka from the Diagnostic and Research Virology Laboratory, Faculty of Medicine, University of Peradeniya are acknowledged.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcvp.2022.100081.
Appendix. Supplementary materials
References
- 1.Gandhi R.T., Lynch J.B., del Rio C. Mild or Moderate COVID-19. N. Engl. J. Med. 2020;383:1757–1766. doi: 10.1056/nejmcp2009249. [DOI] [PubMed] [Google Scholar]
- 2.Greene D.N. Selecting a SARS-CoV-2/COVID molecular testing method for your laboratory. J. Appl. Lab. Med. 2020;5:837–840. doi: 10.1093/jalm/jfaa076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen H.-.W., Weissenberger G., Atkins E., Chao C.-.C., Suputtamongkol Y., Ching .W..-M. Highly sensitive loop-mediated isothermal amplification for the detection of leptospira. Int. J. Bacteriol. 2015:1–6. doi: 10.1155/2015/147173. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau Y.L., Lai M.Y., Teoh B.T., Abd-Jamil J., Johari J., Sam S.S., Tan K.K., AbuBakar S. Colorimetric detection of dengue by single tube reverse-transcription-loop-mediated isothermal amplification. PLoS ONE. 2015;10:1–9. doi: 10.1371/journal.pone.0138694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suwancharoen D., Sittiwicheanwong B., Wiratsudakul A. Evaluation of loop-mediated isothermal amplification method (LAMP) for pathogenic Leptospira spp. detection with leptospires isolation and real-time PCR. J. Vet. Med. Sci. 2016;78:1299–1302. doi: 10.1292/jvms.15-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teoh B.T., Sam S.S., Tan K.K., Johari J., Danlami M.B., Hooi P.S., Md-Esa R., AbuBakar S. Detection of dengue viruses using reverse transcription-loop-mediated isothermal amplification. BMC Infect. Dis. 2013;13 doi: 10.1186/1471-2334-13-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson D., Lei Y. Mini review: recent progress in RT-LAMP enabled COVID-19 detection. Sensors and Actuators Reports. 2020;2 doi: 10.1016/j.snr.2020.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang, W.S., Lim, D.H., Yoon, J., Kim, A., Lim, M., Nam, J., Yanagihara, R., Ryu, S., Jung, B.K., Ryoo, H., Seung, C., Id, L., 2021. Development of a multiplex Loop-Mediated Isothermal Amplification (LAMP) assay for on- site diagnosis of SARS CoV-2 1–14. doi: 10.1371/journal.pone.0248042 [DOI] [PMC free article] [PubMed]
- 10.Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P.M., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Miranda I.B., Schnitzler P., Kräusslich H.G., Knop M., Anders S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12 doi: 10.1126/SCITRANSLMED.ABC7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang W.E., Lim B., Hsu C.C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kellner M.J., Ross J.J., Schnabl J., Dekens M.P.S., Heinen R., Tanner N.A., Fritsche-Polanz R., Traugott M., Seitz T., Zoufaly A., Foedinger M., Wenisch C., Zuber J., (VCDI), V.C.-19 D.I. Pauli A., Brennecke J. Scalable, rapid and highly sensitive isothermal detection of SARS-CoV-2 for laboratory and home testing. bioRxiv. 2020 2020.06.23.166397. [Google Scholar]
- 13.Meagher R.J., Priye A., Light Y.K., Huang C., Wang E. Impact of primer dimers and self-amplifying hairpins on reverse transcription loop-mediated isothermal amplification detection of viral RNA. Analyst. 2018;143:1924–1933. doi: 10.1039/c7an01897e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coelho B.D.O., Bruna H., Sanchuki S., Zanette D.L., Nardin J.M., Manuel H., Morales P., Fornazari B., Aoki M.N., Blanes L. Essential properties and pitfalls of colorimetric Reverse Transcription Loop ‑ mediated Isothermal Amplification as a point ‑ of ‑ care test for SARS ‑ CoV ‑ 2 diagnosis. Mol. Med. 2021 doi: 10.1186/s10020-021-00289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larremore, D.B., Wilder, B., Lester, E., Shehata, S., Burke, J.M., Hay, J.A., Tambe, M., Mina, M.J., Parker, R., 2021. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening 1–11. [DOI] [PMC free article] [PubMed]
- 16.Lwanga Stephen Kaggwa, Lemeshow Stanley, World Health Organization Sample size determination in health studies: a practical manual /S. K. Lwanga and S. Lemeshow. World Health Organization. 1991 https://apps.who.int/iris/handle/10665/40062 [Google Scholar]
- 17.Jeewandara C., Guruge D., Abyrathna I.S., Danasekara S., Gunasekera B., Pushpakumara P.D., Madhusanka D., Jayathilaka D., Ranasinghe T., Somathilake G., Tanussiya S., Jayadas T.T., Kuruppu H., Thashmi N., Harvie M., Wijayamuni R., Schimanski L., Tan T.K., Rijal P., Xiao J., Ogg G.S., Townsend A., Malavige G.N. Seroprevalence of SARS-CoV-2 infection in the colombo municipality region. Sri Lanka. Front. Public Heal. 2021;9:1–6. doi: 10.3389/fpubh.2021.724398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohon, A.N., Hundt, J., Marle, G.Van, Pabbaraju, K., Berenger, B., Griener, T., Lisboa, L., Church, D., Czub, M., Greninger, A., 2020. Development and validation of direct RT-LAMP for SARS-CoV-2 1. Department of microbiology, immunology, and infectious diseases, University of 2. Alberta Public Health Laboratory, Calgary, AB, Canada 3. Clinical Section of Microbiology, Alberta P 78, 1–26.
- 19.Gomes L., Jeewandara C., Jayadas T.P., Dissanayake O., Harvie M., Guruge D., Withanage V., Mahesh P.K.B., Rajapakse W., Ramachandran R., Dharmarajan V., Pathiraja I., Sanjeewani A., Bandara P., Nanayakkara G., Francis V.R., Kithsiri A., Edirisinghe D.A., De Silva K., Wijayamuni R., Ogg G.S., Waggoner J., Malavige G.N. Surveillance of SARS-CoV-2 variants of concern by identification of single nucleotide polymorphisms in the spike protein by a multiplex real-time PCR. J. Virol. Methods. 2022;300 doi: 10.1016/j.jviromet.2021.114374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, M., Pan, W., Arasthfer, A., Fang, W., Ling, L., Fang, H., 2020. Development and validation of a rapid, single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19 10, 1–6. doi: 10.3389/fcimb.2020.00331. [DOI] [PMC free article] [PubMed]
- 21.Jensen, M.A., Fukushima, M., Davis, R.W., 2010. DMSO and betaine greatly improve amplification of GC- rich constructs in de novo synthesis 5, 1–5. doi: 10.1371/journal.pone.0011024. [DOI] [PMC free article] [PubMed]
- 22.Hsieh K., Mage P.L., Csordas A.T., Eisenstein M., Soh H.T. Simultaneous elimination of carryover contamination and detection of DNA with uracil-DNA-glycosylase-supplemented loop-mediated isothermal amplification (UDG-LAMP) Chem. Commun. 2014;50:3747–3749. doi: 10.1039/c4cc00540f. [DOI] [PubMed] [Google Scholar]
- 23.Alekseenko A., Barrett D., Sanchez Y.P., Howard R.J., Strandback E., Korsah H.A., Rovšnik U., Veliz S.Z., Klenov A., Malloo J., Ye S., Liu X., Reinius B., Elsässer S.J., Nyman T., Sandh G., Yin X., Pelechano V. Direct detection of SARS ‑ CoV ‑ 2 using non ‑ commercial RT ‑ LAMP reagents on heat ‑ inactivated samples. Sci. Rep. 2021:1–10. doi: 10.1038/s41598-020-80352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pastorino B., Bessaud M., Grandadam M., Murri S., Tolou H.J., Peyrefitte C.N. Development of a TaqMan® RT-PCR assay without RNA extraction step for the detection and quantification of African Chikungunya viruses. J. Virol. Methods. 2005;124:65–71. doi: 10.1016/j.jviromet.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Nishimura N., Nakayama H., Yoshizumi S., Miyoshi M., Tonoike H., Shirasaki Y., Kojima K., Ishida S. Detection of noroviruses in fecal specimens by direct RT-PCR without RNA purification. J. Virol. Methods. 2010;163:282–286. doi: 10.1016/j.jviromet.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 26.Bachofen C., Willoughby K., Zadoks R., Burr P., Mellor D., Russell G.C. Direct RT-PCR from serum enables fast and cost-effective phylogenetic analysis of bovine viral diarrhoea virus. J. Virol. Methods. 2013;190:1–3. doi: 10.1016/j.jviromet.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 27.Ganguli A., Mostafa A., Berger J., Aydin M.Y., Sun F., Stewart de Ramirez S.A., Valera E., Cunningham B.T., King W.P., Bashir R. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wee, S.K., Sivalingam, S.P., Peng, E., Yap, H., 2020. Rapid direct nucleic acid amplification test without RNA extraction for SARS-CoV-2 Using a portable PCR thermocycler 2–6. [DOI] [PMC free article] [PubMed]
- 29.Mautner L., Baillie C.K., Herold H.M., Volkwein W., Guertler P., Eberle U., Ackermann N., Sing A., Pavlovic M., Goerlich O., Busch U., Wassill L., Huber I. Rapid point ‑ of ‑ care detection of SARS ‑ CoV ‑ 2 using reverse transcription loop ‑ mediated isothermal amplification (RT ‑ LAMP) Virol. J. 2020:1–14. doi: 10.1186/s12985-020-01435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moehling T.J., Choi G., Dugan L.C., Salit M., Robert J. Expert review of molecular diagnostics LAMP diagnostics at the point-of-care : emerging trends and perspectives for the developer community LAMP diagnostics at the point-of-care : emerging trends and perspectives for the developer community ABSTRACT. Expert Rev. Mol. Diagn. 2021;21:43–62. doi: 10.1080/14737159.2021.1873769. [DOI] [PubMed] [Google Scholar]
- 31.Dauner, A.L., Mitra, I., Gilliland, T., Seales, S., Pal, S., 2020. Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID- 19. The COVID-19 resource centre is hosted on Elsevier Connect, the company ’ s public news and information.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.