Chronological age might be the most common health predictor, but are measures of aging built on molecular changes better predictors of health than chronological age alone? Epigenetics refers to the biological mechanisms which control gene transcription and cellular state without changing the underlying genetic code. Epigenetic aging biomarkers allow researchers to interrogate the aging process from a biological – as opposed to chronological - perspective and understand aging related health risks using epigenetic features such as DNA methylation. Epigenetic aging biomarkers are associated with a range of health outcomes including cancer, obesity, cardiovascular disease, and lung function.1, 2 One of the primary goals of research involving all aging biomarkers is to translate chronological age-related risks (which are unmodifiable) into biological aging-related risks that are grounded in biological processes which may be modifiable or even reversible. In this manuscript,3 the authors seek to translate the unmodifiable risks of chronological age for incident atrial fibrillation (AF) into modifiable risks using DNA methylation age, i.e. biological age as estimated by DNA methylation loci, which may be reversible under certain interventions,4 giving it significant clinical and public health benefits.
AF is the most common cardiac arrhythmia and the prevalence of AF is expected to rise to 12.1 million individuals in the United States 2030.5 AF is a risk factor for a variety of adverse health outcomes including stroke, myocardial infarction, heart failure, venous thromboembolism, and even dementia.6 One of the strongest and most consistent risk factors for incident AF is advancing age.7 While this knowledge is certainly useful in predicting the burden of AF in aging populations, it is limiting as chronological age is not a modifiable risk factor. Time stops for no one, as the saying goes, but modifiable biomarkers that capture the age-related risks of AF may better capture who is at most risk, environmental drivers of risks, and what interventions may alter biological aging-related AF risks. Telomere length, which measures biological age through the length of chromosomal “caps”, has been evaluated for associations with AF, but no associations have been found in the general population.8 Thus, the biological mechanisms underlying AF-chronological age associations have remained a mystery.
Roberts et al3 utilize four DNA methylation-based aging biomarkers to provide a (potentially) reversible, biological process-related basis for the known association between chronological age and incident AF. Though AF has often not shown associations with telomere length, DNA methylation-derived accelerated aging measures frequently have associations with health endpoints that are independent of telomere length.9, 10
The authors take a robust approach utilizing a meta-analysis of three population-based studies, combined with a nested confounder adjustment strategy. Evidence for causality is evaluated using Mendelian Randomization, and the authors evaluate several known AF risk factors for evidence of mediating the observed associations. A DNA methylation-based biomarker for plasma PAI-1 was also evaluated for associations with incident AF. It is worth noting at this point that GrimAge 11 differs slightly in its estimation from the other epigenetic age biomarkers (or clocks as they are commonly known due to their strong correlation with age across the lifespan). While the Horvath,12 Hannum,13 and PhenoAge14 epigenetic age biomarkers only require measures of DNA methylation for their estimation, GrimAge also requires the chronological age and sex of the sample. This is because GrimAge is specifically designed to estimate mortality risk, hence it’s often greater associations with mortality in as compared to the other biomarkers. Despite slight differences in its estimation, GrimAge is still measured in units of years, and has properties similar to the epigenetic age biomarkers which often warrant it being analyzed alongside the Horvath, Hannum, and PhenoAge biomarkers.
While aging biomarkers were associated with incident AF, chronological age remained a significant, and sometimes stronger predictor of incident AF according to the authors. However, the magnitude of the association between incident AF and chronological age was attenuated in models that also contained an epigenetic aging biomarker indicating that some of the association between chronological age and AF is captured by these biomarkers of biological aging. This highlights an important point to consider when evaluating any aging biomarker, which is that while the biomarkers capture biological processes that are altered as we age, no individual biomarker can capture the totality of the biological aging process which will be heterogeneous between individuals as well as within the cells and tissues of a single individual. Chronological age, on the other hand, is a proxy for many age-related biological changes. Thus, when examining any biological aging biomarker we should expect that associations between chronological age will persist unless age acceleration as captured by the aging biomarker(s) reflects most of the age-related risks of the outcome of interest. The authors stop short of including all aging biomarkers in a single model which could have provided additional information on whether the set of biomarkers further attenuated the chronological age associations.
Although chronological age may more completely capture age-related incident AF risks than any individual aging biomarker, the aging biomarkers do have the advantage of pointing towards more specific aging-related changes that can be explored further in future studies. All age acceleration measures, except for the one based on the Horvath DNA methylation age, were associated with incident AF after adjustment for chronological age, sex, race, smoking, and technical factors. After further adjustment for 7 traditional AF risk factors accelerated aging as determined by GrimAge and PhenoAge remained associated. PhenoAge is a weighted sum of 513 DNA methylation loci, each of which was selected based on associations with age or selected aging-related phenotypes.14 Thus, it may be possible to further examine the components of PhenoAge to better understand the drivers of its associations with incident AF. The authors did perform a mediation analysis for some clinical parameters, but these had limited overlap with the PhenoAge components leaving open the possibility of unexplored mediators.
The authors also evaluated evidence for causal associations with incidence AF among the biomarkers examined. The authors took the approach of evaluating the loci which compose each biomarker individually, as opposed to evaluating the biomarkers themselves – possibly due to a lack of well powered genome-wide associations studies for all the biomarkers. As the individual components certainly capture less of the aging-related signal than the biomarker as a whole, this may have contributed to the lack of evidence for any causal association. Of course, another explanation is simply that there is not a causal relationship between the biomarkers examined and incident AF risk, which is not wholly unexpected given the current understanding of DNA methylation-based aging biomarkers. Epigenetic aging biomarkers may be consequences of biological age as opposed to causal factors inducing these changes. Indeed, in an experiment where a human chromosome 21 was inserted into a mouse cell, the chromosome was seen to accumulate changes in DNA methylation much more rapidly than when in a human cell, in line with the shortened lifespan of mice as compared to humans.15 Additionally, Mendelian Randomization specifically evaluates causality from the lens of germline genetic variation. Only a small proportion of variation in the epigenetic biomarkers (< 5%) was captured by germline genetic variation and any causal relationships that would have been driven by environmental exposures (either in early life or adulthood) would not be captured by Mendelian Randomization analyses. In general, given the potential for DNA methylation biomarkers to be more consequence rather than cause of aging and the known relationship between environmental exposures and DNA methylation age,2 all causal analyses for these biomarkers should be carefully constructed and cautiously evaluated.
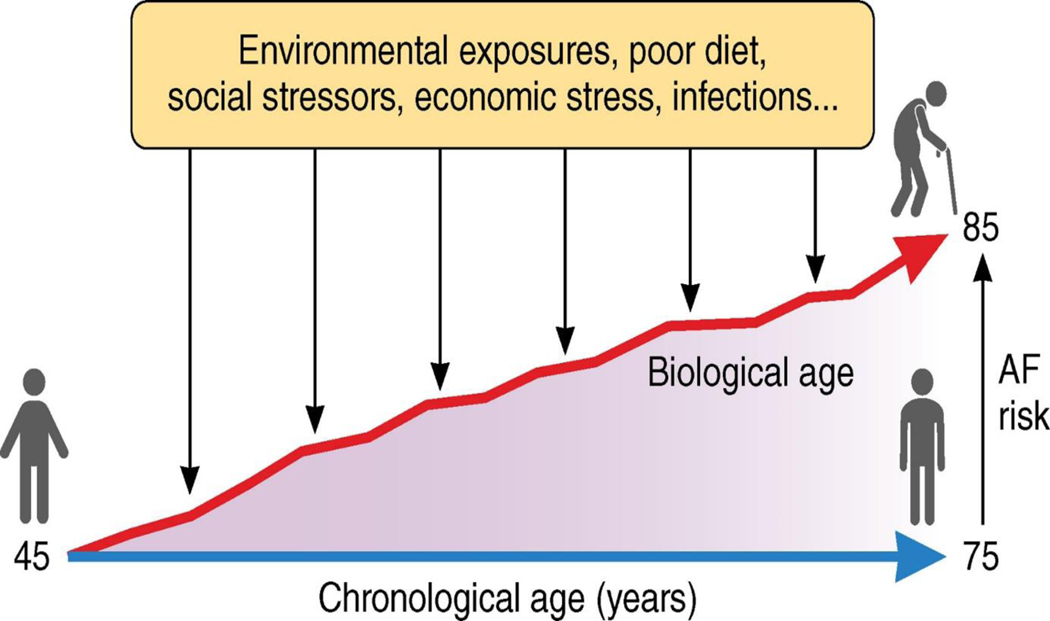
Overall, this manuscript highlights the potential contribution of biological age to estimating risks of incident AF (Figure 1). Although, chronological age remained a strong predictor in the models, epigenetic age was independently associated with incident AF. With emerging technologies for the consumer & clinical assessment of molecular profiles, it is likely that in the future epigenetic features will be utilized to better understand health status. Combined with the possibility for interventions that alter the biological aging process as measured using epigenetics, understanding changes in health risks as reflected by biological age may have important public and personalized health implications. This manuscript helps to lay the foundation for that understanding as it applies to incident AF, though more work on mediating pathways and causality is needed.
Figure 1.
While chronological age progresses at an immutable rate (blue line), biological aging (red line) – measured by epigenetic age in this manuscript – is modifiable by various exposures and life experiences. Thus, establishing a relationship between epigenetic age and atrial fibrillation risk gives a unique insight into modifiable ageing associated atrial fibrillation risks.
Acknowledgments
Dr. Cavin Ward-Caviness is a scientific advisor for the Clock Foundation. The Clock Foundation had no role in any aspect of this work. The views expressed here are those of the author alone and do not necessarily represent the views or policies of the US Environmental Protection Agency.
Footnotes
Conflict of Interest Disclosures:
References:
- 1.Ryan J, Wrigglesworth J, Loong J, Fransquet PD and Woods RL. A Systematic Review and Meta-analysis of Environmental, Lifestyle, and Health Factors Associated With DNA Methylation Age. The Journals of Gerontology: Series A. 2020;75:481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhingra R, Nwanaji-Enwerem JC, Samet M and Ward-Caviness CK. DNA Methylation Age-Environmental Influences, Health Impacts, and Its Role in Environmental Epidemiology. Curr Environ Health Rep. 2018;5:317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts JD, Vittinghoff E, Lu AT, Alonso A, Wang B, Sitlani CM, Mohammadi-Shemirani P, Fornage M, Kornej J, Brody JA, Arking DE, Lin H, Heckbert SR, Prokic I, Ghanbari M, Skanes AC, Bartz TM, Perez MV, Taylor KD, Lubitz SA, Ellinor PT, Lunetta KL, Pankow JS, Paré G, Sotoodehnia N, Benjamin EJ, Horvath S and Marcus GM. Epigenetic Age and the Risk of Incident Atrial Fibrillation. Circulation. 2021;0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fahy GM, Brooke RT, Watson JP, Good Z, Vasanawala SS, Maecker H, Leipold MD, Lin DTS, Kobor MS and Horvath S. Reversal of epigenetic aging and immunosenescent trends in humans. Aging Cell. 2019;18:e13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Virani SS, Alonso A, Aparicio HJ, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Cheng S, Delling FN, Elkind MSV, Evenson KR, Ferguson JF, Gupta DK, Khan SS, Kissela BM, Knutson KL, Lee CD, Lewis TT, Liu J, Loop MS, Lutsey PL, Ma J, Mackey J, Martin SS, Matchar DB, Mussolino ME, Navaneethan SD, Perak AM, Roth GA, Samad Z, Satou GM, Schroeder EB, Shah SH, Shay CM, Stokes A, VanWagner LB, Wang N-Y and Tsao CW. Heart Disease and Stroke Statistics - 2021 Update. Circulation. 2021;143:e254–e743. [DOI] [PubMed] [Google Scholar]
- 6.Staerk L, Sherer JA, Ko D, Benjamin EJ and Helm RH. Atrial Fibrillation: Epidemiology, Pathophysiology, and Clinical Outcomes. Circulation Research. 2017;120:1501–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schnabel RB, Yin X, Gona P, Larson MG, Beiser AS, McManus DD, Newton-Cheh C, Lubitz SA, Magnani JW, Ellinor PT, Seshadri S, Wolf PA, Vasan RS, Benjamin EJ and Levy D. 50 year trends in atrial fibrillation prevalence, incidence, risk factors, and mortality in the Framingham Heart Study: a cohort study. The Lancet. 2015;386:154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staerk L, Wang B, Lunetta KL, Helm RH, Ko D, Sherer JA, Ellinor PT, Lubitz SA, McManus DD, Vasan RS, Benjamin EJ and Trinquart L. Association Between Leukocyte Telomere Length and the Risk of Incident Atrial Fibrillation: The Framingham Heart Study. Journal of the American Heart Association. 2017;6:e006541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitling LP, Saum K-U, Perna L, Schöttker B, Holleczek B and Brenner H. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clinical Epigenetics. 2016;8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marioni RE, Harris SE, Shah S, McRae AF, von Zglinicki T, Martin-Ruiz C, Wray NR, Visscher PM and Deary IJ. The epigenetic clock and telomere length are independently associated with chronological age and mortality. International Journal of Epidemiology. 2016;45:424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, Hou L, Baccarelli AA, Li Y, Stewart JD, Whitsel EA, Assimes TL, Ferrucci L and Horvath S. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY). 2019;11:303–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvath S. DNA methylation age of human tissues and cell types. Genome Biology. 2013;14:3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan J-B, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T and Zhang K. Genome-wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates. Molecular Cell. 2013;49:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y, Whitsel EA, Wilson JG, Reiner AP, Aviv A, Lohman K, Liu Y, Ferrucci L and Horvath S. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10:573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowe R, Barton C, Jenkins CA, Ernst C, Forman O, Fernandez-Twinn DS, Bock C, Rossiter SJ, Faulkes CG, Ozanne SE, Walter L, Odom DT, Mellersh C and Rakyan VK. Ageing-associated DNA methylation dynamics are a molecular readout of lifespan variation among mammalian species. Genome Biology. 2018;19:22. [DOI] [PMC free article] [PubMed] [Google Scholar]