Abstract
Analysis of 71 ciprofloxacin-resistant (MIC ≥ 4 μg/ml) Streptococcus pneumoniae clinical isolates revealed only 1 for which the quinolone resistance-determining regions of the parC, parE, and gyrB genes were genetically related to those of viridans group streptococci. Our findings support the occurrence of interspecies recombination of type II topoisomerase genes; however, its contribution to the emergence of quinolone resistance among pneumococci appears to have been minimal.
Fluoroquinolone resistance in Streptococcus pneumoniae and the viridans group streptococci (VGS) is increasing (1, 3, 7, 8, 11–14, 16, 17). Resistance to the fluoroquinolones has been associated with specific chromosomal mutations in the genes that encode the type II topoisomerase enzymes DNA gyrase and topoisomerase IV (12, 22, 25). In S. pneumoniae and VGS, these heterotetrameric enzymes are of the form GyrA2GyrB2 for DNA gyrase and ParC2ParE2 for topoisomerase IV (18, 24). Amino acid substitutions in the quinolone resistance-determining region (QRDR) of either the ParC subunit of topoisomerase IV or the GyrA subunit of DNA gyrase contribute to low-level resistance, depending on the quinolone used for selection (22, 23, 26). Higher levels of resistance are achieved with additional amino acid substitutions in either ParC or GyrA. Amino acid substitutions in the ParE and GyrB subunits have also been reported; however, their role in resistance is unclear (20, 25).
The exchange of chromosomal DNA between VGS and pneumococci has given rise to penicillin binding proteins (PBPs) with decreased affinity for penicillin and has contributed significantly to the spread and worldwide prevalence of penicillin resistance (9). However, it is only recently that fluoroquinolone-resistant pneumococcal isolates whose type II topoisomerase genes are genetically related to VGS have been reported (10)—this despite several in vitro studies which have reported the frequency of interspecies recombination of these genes to be 106 times that of spontaneous mutation (12, 15, 19, 27).
In this study, we found only 1 (isolate SPN1506) of 71 previously described (2, 3, 7) ciprofloxacin-resistant pneumococcal isolates whose QRDR nucleotide sequences had more than expected dissimilarity with pneumococcal sequences. Specifically, we show that the parC, parE, and gyrB sequences are genetically more closely related to the corresponding regions of selected VGS isolates than to S. pneumoniae.
The QRDR sequences of 71 pneumococcal isolates for which the ciprofloxacin MIC was ≥4 μg/ml (7) and 19 VGS blood isolates (6 S. sanguis, 8 S. mitis, and 5 S. oralis) for which the ciprofloxacin MIC was ≤2 μg/ml (8) were analyzed using the CLUSTAL multiple sequence alignment function of Vector NTI Suite 5.5 software (InforMax Inc., Bethesda, Md.). Phylogenetic analysis and tree construction were performed with the TREEVIEW software program using default parameters (21).
The QRDRs of the parCE and gyrAB genes were amplified from genomic DNA by PCR, as described previously (3). The following oligonucleotide primers, based on published sequences, were used for PCR and sequencing: M0363 and M4271 for parC (22); VGA3 and VGA4 for gyrA (22); H4025 and H4026 for gyrB (22); and SPPARE7 and SPPARE8 (25), as well as parE398 and parE483 (12), for parE. Due to the nucleotide sequence dissimilarities between the different species examined, not all VGS QRDRs could be amplified. In order to sequence the parC open reading frame of SPN1506, a 2.4-kb fragment was amplified by PCR using Taq polymerase and subsequently cloned into the pCR2.1-TOPO TA cloning vector (Invitrogen Corporation, Carlsbad, Calif.). The open reading frame was sequenced by “DNA walking” with a combination of primers specific for the vector and SPN1506.
All isolates were tested for active efflux of ciprofloxacin by the agar dilution-reserpine method, as previously described (3, 6). Only isolates for which the ciprofloxacin MIC decreased by fourfold or greater in the presence of reserpine were considered positive for reserpine-inhibited efflux.
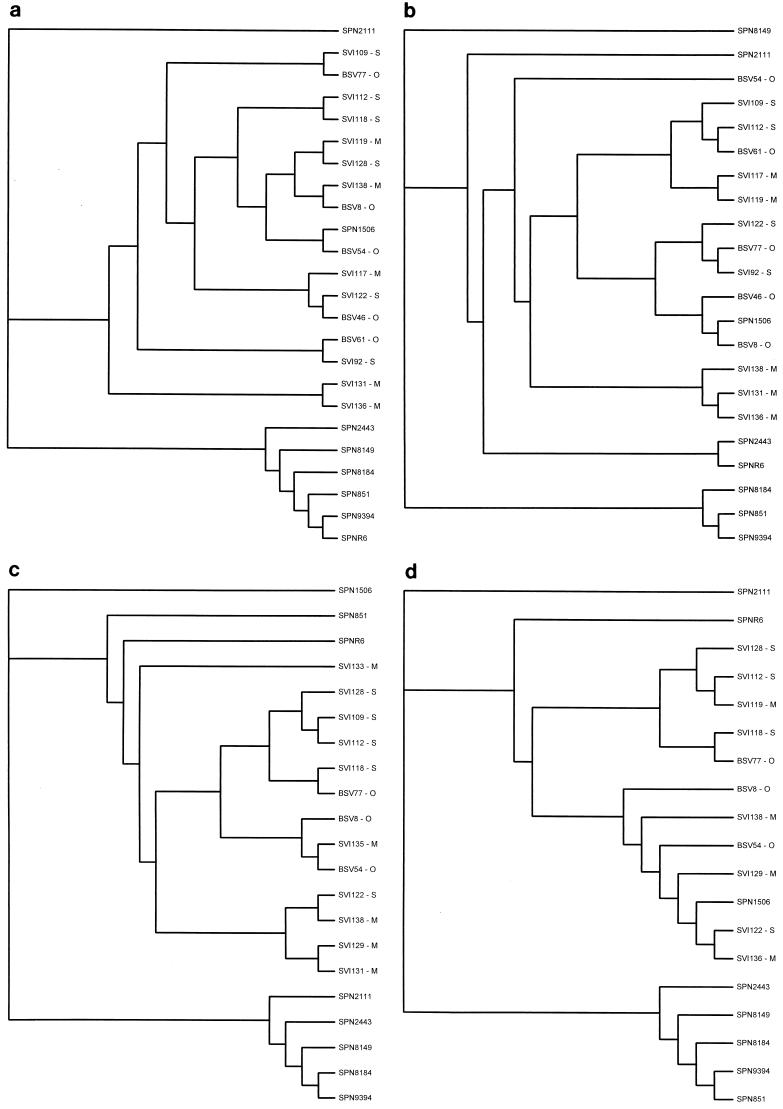
Of the 71 pneumococcal isolates examined in this study, only 1 (SPN1506) had QRDR nucleotide variations similar to VGS compared with SPNR6 (parC, 27 of 322 bp [8.4%]; parE, 15 of 185 bp [8.1%]; gyrB, 18 of 430 bp [4.2%]). This is in contrast to the relatively low nucleotide variation (≤1%) found in the QRDRs of the remaining 70 pneumococcal isolates and in the QRDR of gyrA from SPN1506. To determine whether the QRDR variations noted in SPN1506 were the result of recombinational events between VGS and S. pneumoniae, a comparison of the QRDR nucleotide sequences of selected pneumococcal and VGS isolates was performed (Fig. 1). We demonstrate that the parC, parE, and gyrB QRDR sequences of SPN1506 are more closely related to those of VGS, while the gyrA sequence is more closely related to that of S. pneumoniae. Incidentally, no amino acid substitutions known to confer quinolone resistance among pneumococci were identified in SPN1506. This, in conjunction with this isolate's negative-efflux phenotype, makes it difficult to speculate on the mechanism of resistance. Further work is required to determine whether any of the 27 amino acid substitutions within the ParC subunit contribute to the elevated MIC of ciprofloxacin for SPN1506 (Fig. 2).
FIG. 1.
Cladograms of pneumococcal and VGS parC (a), parE (b), gyrA (c), and gyrB (d) QRDR nucleotide sequences. QRDRs are as follows: parC, 322 bp encoding residues 49 to 155; parE, 185 bp encoding residues 429 to 489; gyrA, 318 bp encoding residues 55 to 160; gyrB, 430 bp encoding residues 375 to 517. VGS species are indicated as follows: O, S. oralis; S, S. sanguis; M, S. mitis.
FIG. 2.
Alignment of the predicted amino acid sequence of the parC open reading frame from SPN1506 with the published SPNR6 parC sequence (10). The QRDR used for analysis of genetic relatedness is underlined. Only the SPNR6 amino acids that differ from those of SPN1506 are shown; identical amino acids are indicated by dashes.
The frequency of horizontal gene transfer between S. pneumoniae and VGS in natural streptococcal populations is unknown. However, such DNA exchanges are known to perpetuate diversity and have contributed to the emergence and spread of penicillin resistance (9). In contrast, interspecies recombination has had little impact on the emergence of quinolone resistance among pneumococcal isolates despite high frequencies of recombination demonstrated in vitro (12, 15, 19, 27). Consistent with the reported low prevalence of quinolone-resistant pneumococcal isolates that likely evolved as a result of homologous recombination with variant alleles from VGS, we identified only 1 of 71 pneumococcal isolates with homology to VGS. The reason for this discrepancy between the expected in vitro frequency and the reported clinical frequency of recombination is unknown.
We speculate, however, that the difference between the prevalence of pneumococcal isolates that harbor PBPs of VGS origin and the prevalence of pneumococcal isolates that harbor topoisomerase genes of VGS origin is due in part to the multimeric nature of the DNA gyrase and topoisomerase IV enzymes. For example, only certain combinations of parC-parE and gyrA-gyrB alleles may encode functional tetrameric enzymes. Therefore, horizontal transfer of compatible alleles that results in viable isolates could be a rare occurrence. In contrast, the PBPs are encoded by a single gene and therefore do not require the association of multiple subunits to form a functional enzyme.
Interestingly, the sequence diversity of the QRDRs among the VGS isolates characterized in this study suggests that horizontal transfer may have played a significant role in their evolution. However, it is equally possible that the poor classification of VGS may have contributed to this observed sequence diversity among the type II topoisomerase genes. For these reasons, it is not possible to speculate on the origin of the variant parC, parE, and gyrB alleles identified in SPN1506. Nevertheless, it is apparent that the nucleotide variations identified in SPN1506 were likely the result of recombination with VGS DNA and not the result of spontaneous mutation. Furthermore, since the average length of DNA that is integrated in a single recombination event is often greater than 8 kb (4), the proximity of the parCE genes of SPN1506 to each other suggests that they were replaced in a single event (5). However, since over 40 kb separates parE from gyrB on the S. pneumoniae chromosome, it is likely that gyrB has recombined independently of parEC. We speculate that a third recombination event would have had to occur if gyrA had shown variations consistent with VGS, given that a distance of approximately 344 kb separates the gyrA gene from parC (The Institute for Genomic Research, personal communication).
In conclusion, our findings indicate that interspecies recombination of type II topoisomerase genes between VGS and S. pneumoniae has minimal impact on pneumococcal quinolone resistance. However, it is possible that as quinolone resistance continues to increase among these streptococcal species, so will the number of quinolone resistance-conferring alleles available for horizontal transfer. Hence, in due course we may see greater numbers of pneumococcal isolates that harbor topoisomerase genes of VGS origin. It is important therefore to provide sequence data on quinolone-resistant pneumococcal isolates so as to better monitor the role of interspecies transfer in the spread of resistance.
Nucleotide sequence accession number.
The open reading frame of the parC gene from isolate SPN1506 has been assigned GenBank accession no. AY035995.
Acknowledgments
This work was supported by grants from the Canadian Bacterial Diseases Network and the Bayer Corporation. D.J.B is a recipient of a Medical Research Council of Canada Postdoctoral Research Fellowship.
S. pneumoniae sequence data were obtained from the website of The Institute for Genomic Research (http://www.tigr.org). We offer special thanks to Kowthar Salim for help in the interpretation of nucleotide sequence relationships.
REFERENCES
- 1.Barry A L, Brown S D, Fuchs P C. Fluoroquinolone resistance among recent clinical isolates of Streptococcus pneumoniae. J Antimicrob Chemother. 1999;43:428–429. doi: 10.1093/jac/43.3.428. [DOI] [PubMed] [Google Scholar]
- 2.Bast D J, de Azavedo J C. Quinolone resistance: older concepts and newer developments. Curr Infect Dis Rep. 2001;3:20–28. doi: 10.1007/s11908-001-0055-y. [DOI] [PubMed] [Google Scholar]
- 3.Bast D J, Low D E, Duncan C L, Kilburn L, Mandell L A, Davidson R J, de Azavedo J C. Fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae: contributions of type II topoisomerase mutations and efflux to levels of resistance. Antimicrob Agents Chemother. 2000;44:3049–3054. doi: 10.1128/aac.44.11.3049-3054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas G D, Ravin A W. Heterospecific transformation of pneumococcus and streptococcus. IV. Variations in hybrid DNA produced by recombination. Mol Gen Genet. 1971;110:1–22. doi: 10.1007/BF00276040. [DOI] [PubMed] [Google Scholar]
- 5.Boor K J, Duncan M L, Price C W. Genetic and transcriptional organization of the region encoding the beta subunit of Bacillus subtilis RNA polymerase. J Biol Chem. 1995;270:20329–20336. doi: 10.1074/jbc.270.35.20329. [DOI] [PubMed] [Google Scholar]
- 6.Brenwald N P, Gill M J, Wise R. Prevalence of a putative efflux mechanism among fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2032–2035. doi: 10.1128/aac.42.8.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen D K, McGeer A, de Azavedo J C, Low D E. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. Canadian Bacterial Surveillance Network. N Engl J Med. 1999;341:233–239. doi: 10.1056/NEJM199907223410403. [DOI] [PubMed] [Google Scholar]
- 8.de Azavedo J C, Trpeski L, Pong-Porter S, Matsumura S, Low D E. In vitro activities of fluoroquinolones against antibiotic-resistant blood culture isolates of viridans group streptococci from across Canada. Antimicrob Agents Chemother. 1999;43:2299–2301. doi: 10.1128/aac.43.9.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowson C G, Coffey T J, Spratt B G. Origin and molecular epidemiology of penicillin-binding-protein-mediated resistance to beta-lactam antibiotics. Trends Microbiol. 1994;2:361–366. doi: 10.1016/0966-842x(94)90612-2. [DOI] [PubMed] [Google Scholar]
- 10.Ferrandiz M J, Fenoll A, Linares J, De La Campa A G. Horizontal transfer of parC and gyrA in fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 2000;44:840–847. doi: 10.1128/aac.44.4.840-847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrandiz M J, Oteo J, Aracil B, Gomez-Garces J L, De La Campa A G. Drug efflux and parC mutations are involved in fluoroquinolone resistance in viridans group streptococci. Antimicrob Agents Chemother. 1999;43:2520–2523. doi: 10.1128/aac.43.10.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez I, Georgiou M, Alcaide F, Balas D, Linares J, De La Campa A G. Fluoroquinolone resistance mutations in the parC, parE, and gyrA genes of clinical isolates of viridans group streptococci. Antimicrob Agents Chemother. 1998;42:2792–2798. doi: 10.1128/aac.42.11.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerin F, Varon E, Hoi A B, Gutmann L, Podglajen I. Fluoroquinolone resistance associated with target mutations and active efflux in oropharyngeal colonizing isolates of viridans group streptococci. Antimicrob Agents Chemother. 2000;44:2197–2200. doi: 10.1128/aac.44.8.2197-2200.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho P L, Que T L, Tsang D N, Ng T K, Chow K H, Seto W H. Emergence of fluoroquinolone resistance among multiple resistant strains of Streptococcus pneumoniae in Hong Kong. Antimicrob Agents Chemother. 1999;43:1310–1313. doi: 10.1128/aac.43.5.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janoir C, Podglajen I, Kitzis M D, Poyart C, Gutmann L. In vitro exchange of fluoroquinolone resistance determinants between Streptococcus pneumoniae and viridans streptococci and genomic organization of the parE-parC region in S. mitis. J Infect Dis. 1999;180:555–558. doi: 10.1086/314888. [DOI] [PubMed] [Google Scholar]
- 16.Jones M E, Sahm D F, Martin N, Scheuring S, Heisig P, Thornsberry C, Kohrer K, Schmitz F J. Prevalence of gyrA, gyrB, parC, and parE mutations in clinical isolates of Streptococcus pneumoniae with decreased susceptibilities to different fluoroquinolones and originating from worldwide surveillance studies during the 1997–1998 respiratory season. Antimicrob Agents Chemother. 2000;44:462–466. doi: 10.1128/aac.44.2.462-466.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jorgensen J H, Weigel L M, Ferraro M J, Swenson J M, Tenover F C. Activities of newer fluoroquinolones against Streptococcus pneumoniae clinical isolates including those with mutations in the gyrA, parC, and parE loci. Antimicrob Agents Chemother. 1999;43:329–334. doi: 10.1128/aac.43.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klevan L, Wang J C. Deoxyribonucleic acid gyrase-deoxyribonucleic acid complex containing 140 base pairs of deoxyribonucleic acid and an alpha 2 beta 2 protein core. Biochemistry. 1980;19:5229–5234. doi: 10.1021/bi00564a012. [DOI] [PubMed] [Google Scholar]
- 19.Munoz R, De La Campa A G. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagai K, Davies T A, Pankuch G A, Dewasse B E, Jacobs M R, Appelbaum P C. In vitro selection of resistance to clinafloxacin, ciprofloxacin, and trovafloxacin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2000;44:2740–2746. doi: 10.1128/aac.44.10.2740-2746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 22.Pan X S, Ambler J, Mehtar S, Fisher L M. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:2321–2326. doi: 10.1128/aac.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan X S, Fisher L M. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob Agents Chemother. 1997;41:471–474. doi: 10.1128/aac.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng H, Marians K J. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J Biol Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 25.Perichon B, Tankovic J, Courvalin P. Characterization of a mutation in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1166–1167. doi: 10.1128/aac.41.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pestova E, Millichap J J, Noskin G A, Peterson L R. Intracellular targets of moxifloxacin: a comparison with other fluoroquinolones. J Antimicrob Chemother. 2000;45:583–590. doi: 10.1093/jac/45.5.583. [DOI] [PubMed] [Google Scholar]
- 27.Tankovic J, Perichon B, Duval J, Courvalin P. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob Agents Chemother. 1996;40:2505–2510. doi: 10.1128/aac.40.11.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]